Abstract
Adiponectin mediates anti-diabetic effects via increasing hepatic insulin sensitivity and direct metabolic effects. In the present study, we conducted a comprehensive and unbiased metabolomic profiling of liver tissue from AdKO (adiponectin-knockout) mice, with and without adiponectin supplementation, fed on an HFD (high-fat diet) to derive insight into the mechanisms and consequences of insulin resistance. Hepatic lipid accumulation and insulin resistance induced by the HFD were reduced by adiponectin. The HFD significantly altered levels of 147 metabolites, and bioinformatic analysis indicated that one of the most striking changes was the profile of increased lysophospholipids. These changes were largely corrected by adiponectin, at least in part via direct regulation of PLA2 (phospholipase A2) as palmitate-induced PLA2 activation was attenuated by adiponectin in primary hepatocytes. Notable decreases in several glycerolipids after the HFD were reversed by adiponectin, which also corrected elevations in several diacyglycerol and ceramide species. Our data also indicate that stimulation of ω-oxidation of fatty acids by the HFD is enhanced by adiponectin. In conclusion, this metabolomic profiling approach in AdKO mice identified important targets of adiponectin action, including PLA2, to regulate lysophospholipid metabolism and ω-oxidation of fatty acids.
Keywords: adiponectin, high-fat diet, insulin, lipid, liver, phospholipase
INTRODUCTION
Adiponectin, now well established as a potent anti-diabetic hormone [1,2], is one of the most abundant circulating proteins in normal individuals, but levels are reduced in individuals with obesity and diabetes [3]. Under normal conditions, the 30 kDa adiponectin gene product undergoes post-translational modifications and oligomerization to form trimers, hexamers and higher-molecular-mass oligomers before secretion. The latter are thought to be the most physiologically active forms of adiponectin and mediate potent anti-diabetic effects via numerous mechanisms. These include direct regulation of substrate metabolism, improved insulin sensitivity and anti-inflammatory actions. Adiponectin mediates these effects via at least two AdipoR (adiponectin receptor) isoforms and subsequent signalling mediated via their direct binding partner APPL1 (adaptor protein, phosphotyrosine interaction, pleckstrin homology domain and leucine zipper-containing 1) and downstream targets including AMPK (AMP-activated protein kinase) [4]. The anti-diabetic effects of adiponectin also encompass actions on various target tissues, in particular liver [5–8] and skeletal muscle [9–16].
Although previous work has conclusively established that adiponectin exerts beneficial metabolic effects by enhancing hepatic insulin sensitivity, we still do not fully understand the precise biochemical changes that occur. We recently examined metabolomic profiles of skeletal muscle samples from AdKO (adiponectin-knockout) mice subjected to an HFD (high-fat diet) with or without adiponectin replenishment [9]. Hyperinsulinaemic–euglycaemic clamp studies confirmed exaggerated insulin resistance in AdKO mice fed on an HFD which was corrected by adiponectin administration. Metabolomics data established important changes in response to the HFD and we documented a signature of relatively normalized muscle metabolism across multiple metabolic pathways in response to adiponectin [9].
In the present study, we conducted a comprehensive analysis of metabolomic profiles in liver of AdKO mice after HFD feeding to discover the major lipid metabolic pathways targeted by adiponectin action. Our aims were to characterize changes in peripheral metabolism, hepatic steatosis and insulin sensitivity in these mice and use metabolomic profiling to both confirm studies using alternative approaches in the existing literature and extend them to uncover novel targets of adiponectin action.
EXPERIMENTAL
Animal models
Male AdKO mice, kindly provided by Dr Y. Matsuzawa [17], on a C57BL6 background and littermates were routinely bred and genotyped in-house before being allocated to experimental groups. Animal facilities met the guidelines of the Canadian Council on Animal Care, and all protocols used were approved by the Animal Care Committee, York University. Male AdKO mice at 6 weeks of age were fed either regular chow or on a 60 % HFD for up to 6 weeks. HFD mice were given either saline or full-length recombinant adiponectin at 3 μg per g of body weight, twice a day (at 09:00 and 18:00) via intraperitoneal injection, for an additional 2 weeks as described previously [9]. Before being killed, animals were deprived of food for 5–6 h and then subjected to hyperinsulinaemic–euglycaemic clamp. Insulin stimulation was administered as a 4 unit/kg of body weight bolus via tail vein injection, 15 min after which liver tissues were harvested and snap-frozen in liquid nitrogen. Serum samples were also collected and stored at − 80 °C.
In vitro studies in cultured cells
Primary mouse hepatocytes were isolated and cultured exactly as described previously [18]. HepG2 cells were generously provided by Dr K. Adeli (University of Toronto) and were maintained as described previously [19]. Once confluent, cells were seeded on to culture plates in 2 %(v/v) FBS-containing medium 24 h before treatment and treated as indicated in the Figure legends and by the additional methods described below.
Western blot analysis
Small amounts of snap-frozen liver tissue were cut, crushed using a mortar and pestle, collected in Eppendorf tubes and homogenized in lysis buffer containing 0.1 % NP-40, 30 mM Hepes (pH 7.4), 2.5 mM EGTA, 3 mM EDTA, 70 mM KCl, 20 mM β-glycerophosphate, 20 mM NaF, 1 mM sodium orthovanadate, 200 μM PMSF, 1 μM pepstatin A, 10 μM E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane] and 1 μM leupeptin. HepG2 cells were lysed, essentially as described previously [10], with buffer containing Tris/HCl (pH 6.8, 0.5 M), 10 % SDS, 15 % glycerol, 1 μM leupeptin, 200 μM PMSF, 10 μM E-64, 1 mM sodium orthovanadate and 100 μM EDTA. The protein concentration of all samples was determined using the BCA Protein Assay kit (Pierce/Thermo Scientific) as per the manufacturer’s instructions and 30 μg of total protein was resolved by SDS/PAGE and transferred on to PVDF membranes. Primary antibodies were diluted 1:1000 in a solution of one part 3 % BSA and two parts 1× Wash Buffer (6.067 g of Tris base and 8.766 g of NaCl in double-distilled water) with 1 %of Tween 20 and 1 %of NP-40. Immunodetection of proteins was visualized using an ECL kit (PerkinElmer) according to the manufacturer’s instructions.
Triacylglycerol quantification
Total tissue triacylglycerol content was analysed using a colorimetric Triglyceride Quantification Kit (Biovision) according to manufacturer’s instructions.
PCR analysis and array
Liver tissue samples were crushed to powder in liquid nitrogen, total RNA was extracted using TRIzol® reagent (Invitrogen Life Technologies) and cleaning was performed using an RNeasy Mini Kit (Qiagen). cDNAs were synthesized by RT (reverse transcription) with 1 μg of total RNA using RT2 First-Strand cDNA Synthesis Kit (Qiagen). cDNA and RT2 qPCR mastermix solutions were divided into aliquots in 96-well plates pre-coated with primer sequences encoding various genes involved in fatty acid metabolism (Qiagen). Genes were normalized to several housekeeping genes including GAPDH (glyceraldehyde-3-phosphate dehydrogenase), β-actin and HSP90α (heat-shock protein 90α). Cycling conditions consisted of an initial denaturation step at 95 °C for 3 min followed by 40 cycles of denaturation at 95 °C for 30 s, primer-specific annealing temperatures of 55–58 °C for 30 s and extension at 72 °C for 30 s, and the final extension was conducted at 72 °C for 5 min. Additional RT–PCR was performed using DyNAmo HS F-410L SYBR Green qPCR Kit (Thermo Scientific). cDNA was produced using 0.25 μl of oligo(dT) (Sigma), 0.12 μl of random primers, 1 μl of dNTP, 2 μl of 5× First Strand Buffer (Invitrogen), 1 μl of DTT (Invitrogen) and 5.33 μl of nuclease-free water and incubated at 42 °C for 2 min. Reverse transcriptase supermix (0.3 μl) (Invitrogen) was added to each PCR tube before incubation at 42 °C for 75 min and at 94 °C for 5 min. The resulting cDNA was diluted 10-fold in nuclease-free water before its use in the qPCR (quantitative real-time PCR) step. For qPCR, 10 μl of the F-410 (Thermo Scientific) mastermix, 25 pmol of forward and reverse primers and 4 μl of the diluted cDNA and 4 μl of double-distilled water were added into new PCR tubes. Initial denaturation was conducted at 95 °C for 15 min, further denaturation was conducted at 94 °C for 30 s, annealing at 57 °C for 30 s and extension at 72 °C for 30 s for 40 cycles. The final extension was performed at 72 °C for 5 min and a melting curve created from 56 °C to 95 °C, reading every 1 °C which was held for 1 s. The qPCR product was recorded and CT values were collected on the Opticon Monitor 3 Software. The following sequences were used in qPCR experiments: PLA1 (phospholipase A1) (captures all of the splice variants X1–X3), 5′-GGCTCATCCTAACCCACAGT-3′ (forward) and 5′-ACACGCAGGCTATTTTCAGG-3′ (reverse), and PLA2 [intracellular (cytosolic) group IVB], 5′-GAGGTTGCTGCTGGTGATG-3′ (forward) and 5′-AGTCT-GATGGGGTGTGTCG-3′ (reverse).
Phospholipase A2 activity assay
Primary rat hepatocytes were isolated from male Wistar rats (180–250 g) with collagenase perfusion as described above and previously [18], and seeded into six-well culture dishes. Cells were then grown in DMEM (Dulbecco’s modified Eagle’s medium) with 1 % serum treated without or with different concentrations of recombinant full-length adiponectin for 8 h, followed by treatment with palmitate (300 mM) or vehicle (lipid-free BSA) for 4 h. The cells were harvested and the intracellular activity of PLA2 was measured with EnzChek® Phospholipase A2 Assay Kit (Invitrogen).
Metabolomic and bioinformatic analysis
Global biochemical profiles were determined in liver as fee-for-service by Metabolon as previously described [9]. Scaled (to have a median of 1) metabolite expression values were used for further analysis. Two scenarios, comparing HFD with chow and HFD + adiponectin with HFD, were analysed to find pathways enriched with differentially expressed metabolites. Each sample was classified in terms of design type (i.e. HFD compared with chow or HFD + adiponectin compared with HFD). Metabolite expression samples in conjunction with the defined design type was then analysed using GlobalANCOVA [20], which constructs a general linear model consisting of expression data and group the sample belongs to (i.e. HFD or chow) compared with a model without the groups defined for each pathway to assess the significance of the grouping for the given set of metabolites. P-values were calculated using GlobalANCOVA’s permutation-based method with 1000 permutations, and genesets were only examined if there were at least three metabolites associated with it. Pathway sets were collated from two sources: pathways defined by Metabolon and supplied in the original dataset from Metabolon, and pathways from the MetaboAnalyst [21] website (http://www.msea.ca/MSEA/faces/Resources.jsp).
GlobalANCOVA was performed on each pathway set to assess its significance. P-values were corrected using Benjamini–Hochberg correction in R [22]. Resulting enrichments were formatted and visualized as a network in the Cytoscape network analysis and visualization software [23] using the Enrichment Map app [24] retaining only enrichments with P-value <0.005 and FDR (false discovery rate) <0.05. Nodes represent enriched pathways, and edges represent the metabolite overlap between two pathways. Only nodes that shared at least 30 % of their metabolites were connected (calculated using overlap coefficient >0.3).
Since the GlobalANCOVA test gives no indication which direction (i.e. up- or down-regulated) the enriched pathway is altered, to better aid visualization of the enrichments for each pathway, we calculated the ratio of significant up-regulated genes to significant down-regulated genes. In our data presentation, pathways with more up-regulated genes are coloured red in the enrichment map visualization, pathways with more down-regulated genes are blue and those that have equal amounts of up-and down-regulated genes (or have no significantly altered genes) are white.
Histological analysis
Liver tissues were fixed before embedding in paraffin and for cryosections, tissues were cryoprotected in 30 % sucrose for 24 h before embedding in OCT compound and freezing in liquid nitrogen. For haematoxylin and eosin staining (Sigma), 5 μm paraffin sections of liver tissues were used. Slides were placed in an oven (55 °C) for 10 min, deparaffinized in xylene twice for 15 min, rehydrated by placing in dilutions of ethanol ranging from 100 % to 70 %. Slides were then immersed in distilled water. Nuclei were stained by dipping slides in haematoxylin for 1 min. Slides were rinsed in water and excess haematoxylin was removed with 1 % hydrochloric acid in 70 % ethanol then transferred to water. Slides were then dipped several times in eosin, rinsed and dehydrated with ethanol (from 95 % to 100 %) and finally immersed in xylene. Stock solutions of Oil Red O (Fluka Chemica) were prepared using 0.5 g of Oil Red O and 100 ml of propan-2-ol. The working solutions contained 30 ml of stock solution diluted in 20 ml of distilled water. Slides were incubated in Coplin jars with Oil Red O working solution for 15 min then rinsed with propan-2-ol followed by haematoxylin staining five times and washing with distilled water.
Statistical analysis
All data were calculated as means±S.E.M. and analysed further using non-parametric one-way ANOVA or two-way ANOVA with Tukey’s post-hoc test. Differences were considered statistically significant at P < 0.05.
RESULTS
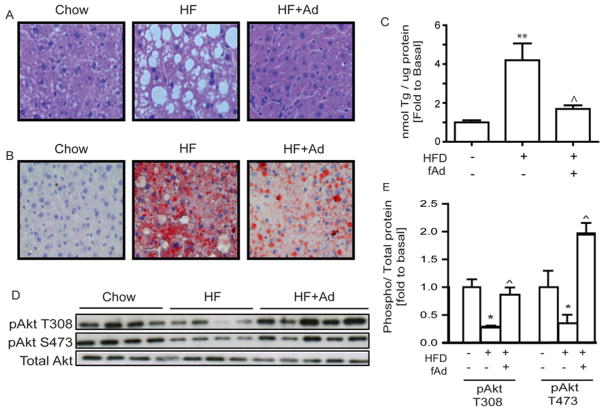
Haematoxylin and eosin staining to examine the morphology of liver tissue showed an increase in the number and size of vacuoles after the HFD (Figure 1A). These vacuoles were noticeably diminished in size and number in mice administered adiponectin (Figure 1A). Oil Red O staining of liver tissue sections demonstrated a clear increase in the number of lipid droplets in HFD-fed compared with chow-fed mice. Interestingly, the number of large lipid droplets was significantly reduced in mice administered adiponectin (Figure 1B). Quantitative analysis of triacylglycerols in these tissues confirmed the aforementioned observations as the liver of AdKO mice fed a HFD contained 4-fold more triacylglycerol than chow-fed mice (Figure 1C). HFD-fed mice treated for an additional 2 weeks with adiponectin demonstrated levels of triacylglycerol similar to that of chow-fed mice (Figure 1C). Compared with their chow-fed littermates, AdKO mice fed on the HFD demonstrated significantly reduced levels of insulin-stimulated phosphorylation of Akt on Thr308 and Ser473 (Figures 1D and 1E). In AdKO mice fed on the HFD that received adiponectin treatment, insulin-stimulated phosphorylation levels were similar to (Thr308) or exceeded (Ser473) those of mice fed on the chow diet (Figures 1D and 1E).
Figure 1. Lipid accumulation and insulin signalling in liver.
AdKO mice fed either regular chow or on a 60 % HFD for 6 weeks. HFD mice received an additional 2-week adiponectin or saline treatment at the dosage of 3 μg/g of body weight via intraperitoneal injection. Liver tissues were then collected 15 min after 4 units/kg insulin stimulation for later analysis. (A) Haematoxylin and eosin staining of paraffin-embedded cross-sections of liver tissues (×200). (B) Oil Red O staining of cryosections of liver tissues (×200). (C) Triacylglycerol content in liver tissues. (D and E) Representative Western blot and analysis of insulin-stimulated Akt at phosphorylation sites Ser473 and Thr308. Scale bars, 200 μm. Results are means±S.E.M. HF, HFD; Ad, adiponectin; fAd, full-length adiponectin; Tg, triacylglycerol. *Significant difference between chow and HFD group. ∧Significant difference between saline-treated and adiponectin-treated HFD groups. *∧P < 0.05, **∧∧P < 0.01.
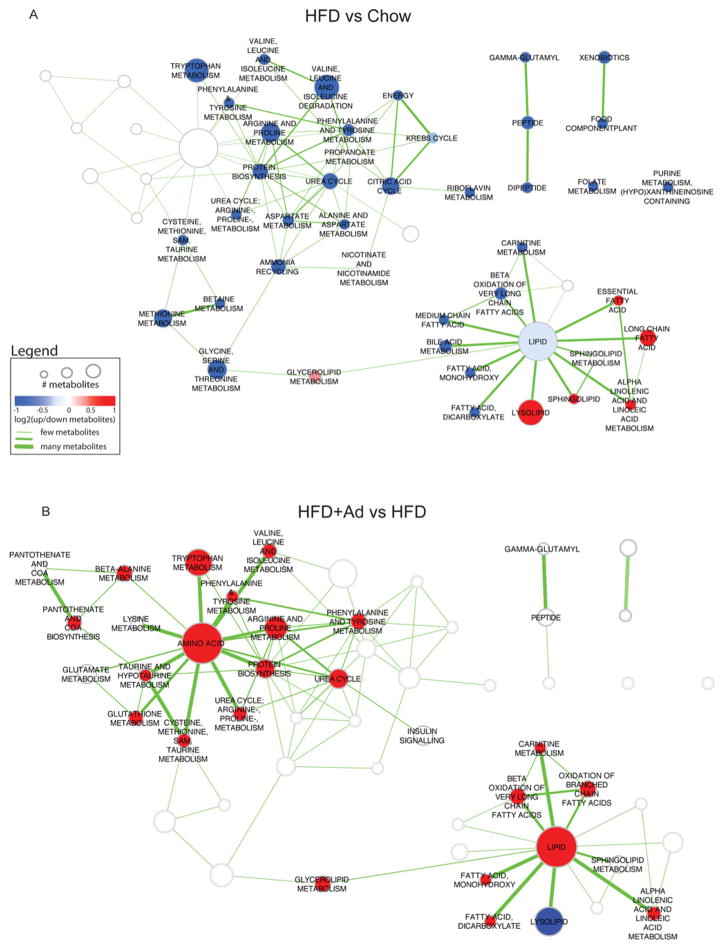
A global view of the metabolomic dataset (Table 1) indicated that the HFD significantly altered 147 metabolites in liver (48 elevated and 99 reduced). Superimposing adiponectin treatment on HFD-fed mice significantly altered the amount of 76 metabolites in liver (53 elevated and 23 reduced). To determine a global picture of how metabolite pathways are affected by diet and adiponectin, pathway enrichment analysis was performed on the set of differentially expressed metabolites for HFD compared with chow and for HFD + adiponectin compared with HFD. Results were visualized as an enrichment map network. To enable easier comparison between the two enrichment results, enriched pathways from both results are shown in each map, but pathways not enriched in a given comparison are represented as white nodes without labels (Figure 2). HFD compared with chow was enriched in 43 pathways whereas HFD + adiponectin compared with HFD was enriched in 32 pathways (P < 0.005 and FDR<0.05), of which 22 were common to both. More than half of the altered pathways were common to both comparisons, yet the direction of metabolite differential expression was opposite for all but three of the pathways with a predominant down-regulation of pathways in HFD compared with chow and an up-regulation of corresponding pathways in HFD + adiponectin compared with HFD. There was a larger subset of pathways (21 in HFD compared with chow in contrast with ten in HFD + adiponectin compared with HFD) unique to HFD compared with chow, thus indicating a set of pathways which were not influenced by the administration of adiponectin. We also performed analysis to highlight those pathways affected by adiponectin, or not (Figure 3).
Table 1. Global profiling of 341 metabolites in liver.
Welch’s two-sample t tests were used to determine whether the means of two groups are different where P ≤ 0.05 was taken as significant and a trend of 0.05<P < 0.10 identified biochemicals approaching significance.
| Welch’s two-sample t test | HFD compared with chow | HFD + adiponectin compared with chow | HFD + adiponectin compared with HFD |
|---|---|---|---|
| Total number of biochemicals with P ≤ 0.05 | 147 | 102 | 76 |
| Biochemicals (↑/↓) | 48/99 | 39/63 | 53/23 |
| Total number of biochemicals with 0.05<P < 0.10 | 23 | 27 | 35 |
| Biochemicals (↑/↓) | 8/15 | 13/14 | 26/9 |
Figure 2. Bioinformatic analysis of differentially expressed pathways.
Enriched pathways in HFD compared with chow (A) and HFD + adiponectin compared with HFD (B) data. Pathways are represented by nodes in the network. The size of the nodes directly correlates to the number of metabolites annotated to a pathway. The node colour indicates the direction of metabolite expression causing the significant enrichment, where red is up-regulation, blue is down-regulation and white indicates that there are equal numbers of up- and down-regulated metabolites (or no change). Edges (lines) represent known pathway cross-talk, defined by the number of metabolites shared between a pair of pathways (as calculated using overlap coefficient <0.3). To enable easier comparison between the two enrichment maps, all results were merged, but only the pathways relevant for a particular comparison are highlighted (colours and labels).
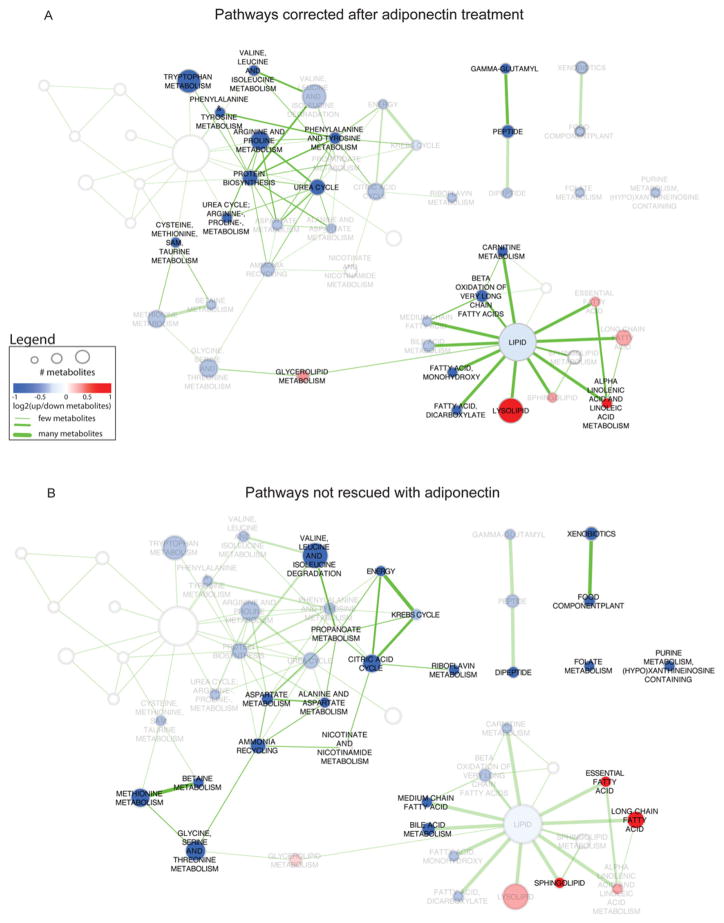
Figure 3. Bioinformatic analysis of pathways altered by adiponectin or not.
HFD compared with chow enriched pathway network highlighting pathways rescued (A) and not rescued (B) by adiponectin. Enriched pathways in HFD compared with chow are annotated with node labels and colours indicating the direction of metabolite expression, where red is up-regulated, blue is down-regulated and white indicates there was no change. The transparency of the nodes was increased in (A) if it was a pathway not seen in the HFD + adiponectin compared with HFD enriched pathway set to highlight pathways (common to both HFD compared with chow and HFD±adiponectin compared with HFD) that were rescued by adiponectin. The transparency of the nodes was increased in (B) if it was a pathway found in HFD + adiponectin compared with HFD enriched pathway set to highlight pathways unique to HFD compared with chow and not rescued by adiponectin.
Lipid analysis revealed that HFD-fed AdKO mice displayed a significant increase in C14:0, C20:0, C18:0, C18:1 and C16:0 ceramides in the liver (Supplementary Figure S1). Although there was an apparent trend towards a decrease in these levels with the administration of adiponectin, only C20:0 and C18:0 ceramides were significantly decreased. The total levels of ceramides remained unaltered between groups (Supplementary Figure S1). Levels of sphinganine and sphingosine, precursors of the ceramide/sphingosine 1-phosphate pathway, were significantly elevated in the HFD group compared with chow-fed mice and levels of these metabolites were significantly decreased in the HFD group treated with adiponectin. End-products of the palmitoyl/sphingomyelin pathway were significantly increased in the group treated with adiponectin compared with the group fed on the HFD alone (Supplementary Figure S1). In HFD-fed mice, the total levels of DAGs (diacylglycerols) were up-regulated. Specific types of DAGs, i.e. C16:0–C18:0, C18:0–C18:2, C18:1, C16:1 and C18:0–C20:4, were similarly elevated in the HFD group. Tissue from mice administered adiponectin displayed significantly attenuated levels of C18:0–C18:2 and C18:0–C20:4 DAGs (Supplementary Figure S1).
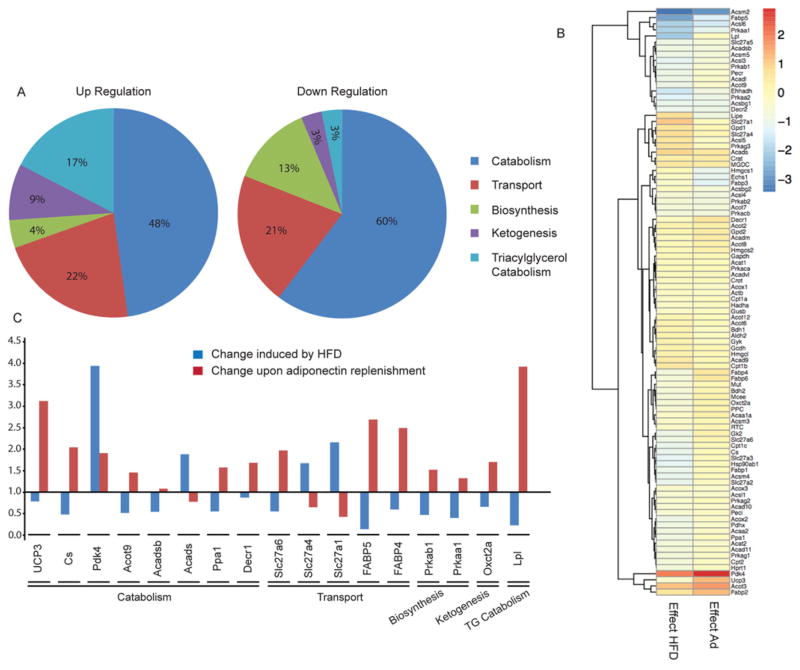
Analysis of metabolomic profiles using GlobalANCOVA indicated significant changes in fatty acid metabolism in response to HFD (Figure 2) and that adiponectin treatment had distinct effects on reversing some of these changes (Figure 3). Specifically, we found that the HFD elevated levels of numerous long-chain fatty acids; however, adiponectin did not have any significant impact on these changes (Supplementary Figure S2). Similarly, HFD-induced elevations in essential fatty acids EPA (eicosapentaenoic acid), n − 3 DPA (docosapentaenoic acid) and DHA (docosahexaenoic acid) were not altered by adiponectin (Supplementary Figure S2). The decreased deoxycarnitine and 3-dehydrocarnitine levels caused by the HFD were not altered by adiponectin (Supplementary Figure S1). However, adiponectin did counteract HFD-induced decreases in 13-HODE [(13S)-hydroxyoctadeca-(9Z,11E)-dienoic acid], 9-HODE [(9S)-hydroxyoctadeca-(10E,12Z)-dienoic acid], 16-hydroxypalmitate, hexadecanedioate and caproate (Supplementary Figure S2). Further analysis of changes related to fatty acid metabolism were performed using a PCR array of relevant enzymes. Figure 4(A) shows that the majority of changes in gene expression induced by the HFD occurred in those involved in fatty acid catabolism, and also in transport. A heat map showing changes in expression of all genes analysed is shown in Figure 4(B) with changes induced by the HFD in the left-hand column and the subsequent effect of adiponectin shown in the right-hand column. The principal changes are highlighted in Figure 4(C), and these included HFD-induced increases in Pdk4, Acads, Slc27a1 and Slc27a4 which were reduced by adiponectin. In contrast, adiponectin reversed HFD-induced decreases in UCP3, Cs, Acot9, Acadsb, Ppa1, Slc27a6, FABP5, FABP4, Prkab1, Prkaa1, Oxct2a and LPL gene expression (Figure 4C).
Figure 4. qPCR array analysis of genes involved in fatty acid metabolism.
AdKO mice fed either regular chow or on a 60 % HFD for 6 weeks. HFD mice received an additional 2 weeks’ adiponectin or saline treatment at the dosage of 3 μg/g of body weight via intraperitoneal injection. Liver tissues were then collected 5 min after 4 units/kg insulin stimulation for later analysis. (A) Effect of the HFD on genes involved in fatty acid metabolism. (B) Heat map revealed the effect of the HFD (left-hand column) and adiponectin (right-hand column) on genes involved in fatty acid metabolism. (C) List of genes involved in fatty acid metabolism that were significantly altered by the HFD and significantly corrected by adiponectin treatment. Results are fold changes. n ≥ 3.
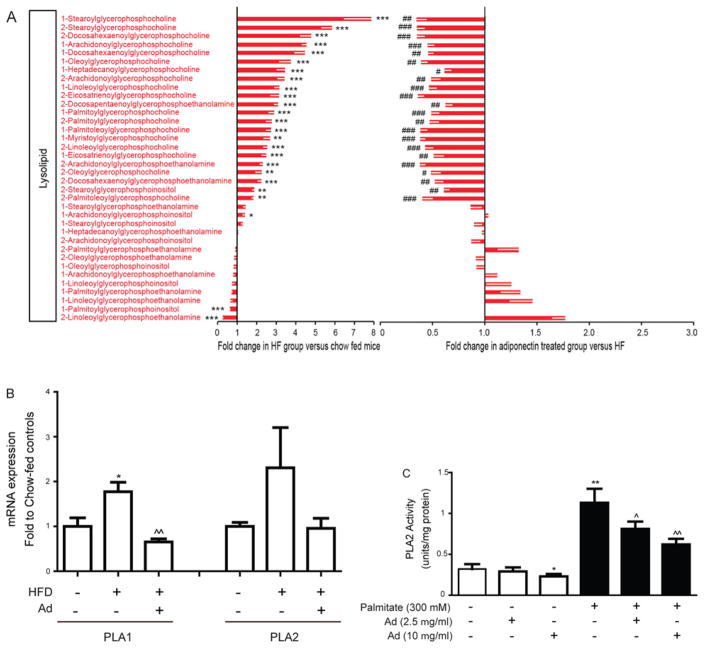
Bioinformatic analysis indicated that striking changes were observed in the category of lysolipids (Figure 2). In particular, those belonging to the group of lysophosphocholine and some lysophosphoethanolamines were significantly increased by the HFD and subsequently decreased in the adiponectin-treated groups (Figure 5A). Of note, significant changes in both 1- and 2-acylglycerophosphocholine/ethanolamine were observed. Glycerol 3-phosphate was decreased in the HFD groups compared with chow-fed. There were also no changes in the levels of glycerophosphocholine (Figure 5A). Precursors of phosphotidylethanolamine formation such as ethanolamine and phosphoethanolamine showed significant decreases in the HFD group and a subsequent increase in groups treated with adiponectin (Figure 5A).
Figure 5. Metabolomic analysis of lysolipid species profiles and analysis of phospholipase activity.
(A) Lysolipid metabolite changes in AdKO mice treated as described in Figure 1. Left panel: fold change between chow- and HFD-fed AdKO mice. Right panel: fold change between adiponectin-and saline-treated HFD-fed AdKO mice. Results are means±S.E.M. *Significant difference between chow and HFD group. #Significant difference between saline-treated and adiponectin-treated HFD group. *#P < 0.05, **##P < 0.01, ***###P < 0.001. (B) The hepatic mRNA expression of PLA1 and PLA2 was analysed in tissue samples from AdKO mouse groups matching data in (A). (C) Primary hepatocytes were isolated and treated without or with palmitate or without or with adiponectin as indicated, and the activity of PLA2 was determined. Results are means±S.E.M. *Significant difference between chow and HFD group or control cell group, #Significant difference between saline-treated and adiponectin-treated HFD groups. *#P < 0.05, **##P < 0.01, ***###P < 0.001.
Because lysophospholipid metabolism was a major target of the HFD and adiponectin, we next examined alterations in relevant phospholipase enzyme expression and activity. The relative expression levels of PLA1 and PLA2 were similar, based upon CT values. The HFD elicited a significant increase in hepatic expression of PLA1 and this was reversed in animals treated with adiponectin (Figure 5B), whereas PLA2 levels were apparently increased by the HFD and reduced by adiponectin (Figure 5B). To examine whether adiponectin regulated phospholipase enzyme activity directly, we used primary hepatocytes treated without or with palmitate (300 mM) and without or with adiponectin (2.5 μg/ml and 10 μg/ml). The activity of PLA2 was significantly elevated by palmitate, and adiponectin significantly reduced palmitate-induced PLA2 activity (Figure 5C).
Further metabolomic data analysis, focusing on metabolites in the superpathway of carbohydrate metabolism, showed a trend of suppression of various metabolites after HFD, with significant decreases in maltose, fucose, ribulose, 6-phosphogluconate, UDP-glucose, UDP-galactose, fructose/glucose 1,6-diphosphate and lactate (Supplementary Figure S3). There were fewer significant effects of adiponectin on metabolites in this class, although significant decreases in 2-phosphoglycerate and 3-phosphoglycerate were notable (Supplementary Figure S3). In addition, specific changes were observed in amino acids (Supplementary Figure S4) and other metabolites (Supplementary Figure S5).
DISCUSSION
Histological analysis as well as determination of total triacylglycerol levels clearly indicated that HFD induced hepatic steatosis in AdKO mice which, as expected, was associated with insulin resistance. These defects were corrected by replenishment of adiponectin, confirming the beneficial anti-steatotic and insulin-sensitizing actions of adiponectin in the liver. To probe the precise changes in metabolism occurring under these conditions, we used a metabolomic profiling approach to both confirm established targets and gain insight into new targets of adiponectin action in liver. Metabolomics is powerful in that it provides unbiased datasets which can be analysed to infer mechanistic information [25,26]. For example, metabolomic studies of liver from mice or rats subjected to an HFD have demonstrated significant diet induced changes in lysolipids, choline, fatty acids and associated intermediates and amino acids [27–29]. The global summary of the present analysis indicated that 48 of 341 metabolites were significantly elevated in liver of AdKO mice after 6 weeks of HFD, whereas 99 were reduced. The impact of replenishing adiponectin in these mice was to elevate 53 and reduce 23 metabolite levels. More informative data was then extracted by analysis of metabolomic alterations on the basis of super- and sub-pathway categorization.
The most striking changes were observed in the category of lysophospholipids with HFD-induced increases in many distinct species, including lysophosphatidylcholines and lysophosphatidylethanolamines. Our metabolomic analyses demonstrate that both isoforms of lysophospholipids (1- and 2-acylglycerophospholipid) are altered with the HFD and reversed with adiponectin treatment. Increased hepatic lysophospholipids have been reported previously in db/db mice and implicated as a cause of insulin resistance [30]. Interestingly, plasma lysophosphatidylcholine levels are reduced in obesity and Type 2 diabetes [31] and a recent study specifically identified circulating lysophosphatidylcholine levels as markers of metabolically benign non-alcoholic fatty liver [32]. Lysophospholipids are also of great interest as studies have demonstrated their functional roles beyond membrane structure and function, including impairment of insulin signalling independent of ceramide and DAG, increase in VLDL (very-low-density lipoprotein) assembly and secretion, fibrosis and inflammation [30,33,34]. Phospholipases and lysophosphatidylcholine acyltransferase, enzymes that co-ordinate the regulation of lysolipid levels, have been shown to moderate hepatic lipotoxicity, insulin resistance and fibrosis [30,35,36]. The altered lysophospholipid levels we observed in liver tissue are most likely to be a result of an imbalance between phospholipase and lysophosphatidylcholine acyltransferase activity [37]. Adiponectin is known to enhance phospholipase C signalling in skeletal muscle cells [38]; however, regulation of phospholipases A/B, lysophosphatidylcholine acyltransferase and lysolipid levels by adiponectin has not been shown previously. We focused on PLA2 given recent evidence implicating gain or loss of function in inducing [39] or preventing [40,41] hepatic insulin resistance respectively. We showed that adiponectin attenuated fatty- acid-induced PLA2 activity in primary hepatocytes, establishing PLA2 as a new target of adiponectin action.
We also presume that increased saturated lysophos-phatidylcholine would be accompanied by the release of arachidonate and modified eicosanoid synthesis. An interesting observation in our dataset was the HFD-induced increase in both 9-HODE and 13-HODE, which was reversed by adiponectin. These are hydroxy derivatives of unsaturated linoleic acid and major components of oxidized low-density lipoprotein [42]. Accordingly, they are considered good markers of lipid peroxidation [43], have potentially important roles in atherosclerosis [44] and have been shown to act as endogenous activators of PPARγ (peroxisome-proliferator-activated receptor γ) in various cell types [42,43,45], which is in keeping with our dataset indicating increased lipid uptake. These actions could also be considered beneficial in terms of insulin sensitivity and anti-inflammation.
Various lipids play an integral role in metabolic complications of hepatic steatosis [46] and we observed an expected overall increase in ceramides and DAGs in HFD-fed mouse liver [47]. Specifically, data showing increased levels of intermediates such as sphinganine and reduced amount of precursors including L-serine indicated that HFD induced de novo synthesis of ceramides (C14:0, C20:0, C18:0, C18:1 and C16:0) with less influence on the salvage pathway of ceramide synthesis inferred by no change in sphingomyelin [48]. Our data indicate that adiponectin selectively reduced the levels of specific ceramides (C20:0 and C18:0) and this appeared to be a result of influence on de novo and salvage pathways of ceramide synthesis [48]. In contrast, although effectively corrected here in liver, HFD-induced increases in ceramide levels were not significantly altered by adiponectin in skeletal muscle [9]. Adiponectin also selectively reduced specific DAGs (C18:0–C18:2 and C18:0–C20:4) in liver compared with our recent findings that in skeletal muscle, the HFD increased C16:0–C18:1, C18:1 and C18:0–C18:2 DAG species, all of which were reduced by adiponectin [9]. The differential effects of adiponectin supplementation in liver and skeletal muscle suggest that adiponectin regulates lipid metabolism in a tissue-specific manner.
The oxidation of long-chain fatty acids can occur via β-oxidation or the alternative pathway of ω-oxidation [49]. It has recently been suggested that ω-oxidation may act as a rescue pathway to reduce the accumulation of fatty acids and subsequent lipotoxic effects of their accumulation [49]. In this pathway, fatty acids, particularly very-long-chain fatty acids, are hydroxylated at the ω-carbon to produce ω-hydroxy-fatty acids and finally carboxylated to form ω-dicarboxylates in liver microsomes. Our metabolomics profile demonstrated increased levels of the intermediate, 16-hydroxypalmitate, and by-product, hexadecanedioate, of ω-oxidation [49]. Interestingly, adiponectin also increased both of these metabolites, suggesting that it may also up-regulate ω-oxidation and that, depending on the prevailing physiological and cellular milieu, this can be beneficial.
Having established the altered profile of fatty acid metabolites, we used a PCR array to detect changes in enzymes involved in fatty acid metabolism. First, we observed a decrease in pyrophosphatase, suggesting a decrease in energy demands [50]. This is congruent with the decrease in fatty acid oxidation after the HFD. Acyl-CoA thioesterase and acyl-CoA dehydrogenase, enzymes involved in fatty acid β-oxidation and in the inhibition of lipogenesis were reduced, which supports further an increase in lipogenesis, and a decrease in fatty acid β-oxidation in HFD-fed AdKO mice. Adiponectin significantly increased levels of enzymes involved in fatty acid uptake and oxidation, including uncoupling protein, citrate synthase, HSP90α B1, FATP (fatty acid transfer protein), 3-oxoacid CoA transferase 2A, FABP (fatty acid-binding protein) and 2,4-dienoyl reductase 1. Whereas an increase in fatty acid uptake with adiponectin treatment may seem to oppose the notion that adiponectin action is beneficial and reduces lipotoxicity, it is known that in the event of an increased systemic demand for fatty acid utilization, the liver takes up free fatty acids and increases oxidation while simultaneously breaking down stored triacylglycerols, packaging them as VLDLs and secreting them into the circulation [51]. Furthermore, this increased fatty acid uptake may be coupled to the increased ω-oxidation which we observed and described above.
Glucose metabolism across multiple metabolic routes was perturbed under HFD conditions. Known effects of HFD-induced insulin resistance on glucose metabolism in liver include elevated rates of gluconeogenesis and glycogen breakdown, even under hyperglycaemic conditions [52]. In the present study, HFD decreased levels of lactate and some amino acids which typically serve as precursors for gluconeogenesis, perhaps indicating elevated gluconeogenesis. Significantly lower levels for many intermediates of the TCA (tricarboxylic acid) cycle support the notion of poor oxidative energy metabolism with a HFD, consistent with glucose production by liver. Although adiponectin improves insulin sensitivity, the HFD-induced shifts in glycolytic/gluconeogenic pathway intermediates and changes in TCA cycle intermediates were not completely rectified by adiponectin supplementation. Our findings that treatment with full-length adiponectin for 2 weeks was unable to reverse all of the effects of the HFD in AdKO mice is consistent with our observation that glucose infusion rates in these mice were also only partially restored after full-length adiponectin treatment for 2 weeks. Thus we feel that it is important to point out that a longer duration of adiponectin treatment could conceivably elicit greater, or different, effects and this remains to be tested.
In addition to the consideration of individual metabolites discussed above, we determined major changes in groups of metabolites and applied an established pathway analysis pipeline, most often used with gene expression data, to further analyse our metabolomics data. Briefly, in liver from HFD-fed mice, predominant changes included decreased protein biosynthesis and fatty acid metabolism, but marked increases in lysolipids and long-chain fatty acids. This analysis confirmed that with the addition of adiponectin there was an increase in protein synthesis and fatty acid metabolism as well as a decrease in lysolipids. Although our pathway analysis provides a useful summary of our results at the systems level, some measured metabolites were not considered because they are not annotated to any pathway. A limitation of this approach is that a few metabolites were not annotated with Human Metabolite Database (HMDB) identifiers (~27 %) which eliminated a large proportion of our metabolites from subsequent pathway analysis. Another limitation was that the Metabolon database consisted of annotations that are metabolite-specific with only one annotation per metabolite. Metabolites belonging to multiple pathways are not specified, leading to a loss of information for metabolites that play a role in multiple pathways. Improved pathway annotations will improve metabolomics pathway analysis in the future. Nevertheless, this analysis rapidly identifies statistically significant changes in metabolic pathways and provides a useful comprehensive summary of our dataset.
In the present study, we have validated the HFD-induced development of insulin resistance and hepatic steatosis in AdKO mice and our metabolomics approach has proven useful in (i) confirming established literature regarding changes in lipid metabolites such as ceramides and DAGs in response to HFD and the ability of adiponectin to correct these, and (ii) uncovering novel targets of adiponectin action associated with lipid metabolism. The latter include the novel observation that up-regulation of lysophospholipids in response to HFD may have contributed to the phenotypes of HFD-induced insulin resistance, impaired glucose tolerance and hypertriglyceridaemia and that adiponectin-mediated reversal of these changes may occur partly via regulation of PLA2 activity. The stimulation of ω-oxidation of fatty acids by adiponectin and the ability of adiponectin to reverse changes in 9-HODE and 13-HODE may also have previously uncharacterized implications in systemic metabolic dysfunction and resulting complications.
Supplementary Material
Acknowledgments
We acknowledge the expert advice from Dr David Brindley (University of Alberta, Edmonton, Canada) and Dr Dominic Ng (University of Toronto) on lipid metabolism data and experimental planning.
FUNDING
This work was supported by an operating grant from the Canadian Institutes of Health Research to G.S., who is guarantor of this work. We also acknowledge HKU matching fund for State Key Laboratory of Pharmaceutical Biotechnology to A.X. and NRNB (U.S. National Institutes of Health, National Center for Research Resources) [grant number P41 GM103504 (to G.D.B.). S.W. received support from the Thailand Research Fund under the Royal Golden Jubilee Award PhD program and Mahidol University.
Abbreviations
- AdKO
adiponectin-knockout
- DAG
diacylglycerol
- FDR
false discovery rate
- HFD
high-fat diet
- 9-HODE
(9S)-hydroxyoctadeca-(10E,12Z)-dienoic acid
- 13-HODE
[(13S)-hydroxyoctadeca-(9Z,11E)-dienoic acid
- HSP90α
heat-shock protein 90α
- PLA
phospholipase A
- qPCR
quantitative real-time PCR
- RT
reverse transcription
- TCA
tricarboxylic acid
- VLDL
very-low-density lipoprotein
Footnotes
AUTHOR CONTRIBUTION
Experimental work was performed by Ying Liu, Sanjana Sen, Sivaporn Wannaiampikul, Rengasamy Palanivel and Ruby Hoo. Intellectual input and study design was by Ying Liu, Sanjana Sen, Aimin Xu and Gary Sweeney. Specific data analysis was by Ruth Isserlin and Gary Bader. Writing and pre-submission editing of the paper was by Ying Liu, Sanjana Sen, Rungsunn Tungtrongchitr, Ruth Isserlin, Gary Bader, Yves Deshaies, Aimin Xu and Gary Sweeney.
References
- 1.Turer AT, Scherer PE. Adiponectin: mechanistic insights and clinical implications. Diabetologia. 2012;55:2319–2326. doi: 10.1007/s00125-012-2598-x. [DOI] [PubMed] [Google Scholar]
- 2.Scheid MP, Sweeney G. The role of adiponectin signaling in metabolic syndrome and cancer. Rev Endocr Metab Disord. 2014;15:157–167. doi: 10.1007/s11154-013-9265-5. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Retnakaran R, Hanley A, Tungtrongchitr R, Shaw C, Sweeney G. Total and high molecular weight but not trimeric or hexameric forms of adiponectin correlate with markers of the metabolic syndrome and liver injury in Thai subjects. J Clin Endocrinol Metab. 2007;92:4313–4318. doi: 10.1210/jc.2007-0890. [DOI] [PubMed] [Google Scholar]
- 4.Deepa SS, Dong LQ. APPL1: role in adiponectin signaling and beyond. Am J Physiol Endocrinol Metab. 2009;296:E22–E36. doi: 10.1152/ajpendo.90731.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nawrocki AR, Rajala MW, Tomas E, Pajvani UB, Saha AK, Trumbauer ME, Pang Z, Chen AS, Ruderman NB, Chen H, et al. Mice lacking adiponectin show decreased hepatic insulin sensitivity and reduced responsiveness to peroxisome proliferator-activated receptor gamma agonists. J Biol Chem. 2006;281:2654–2660. doi: 10.1074/jbc.M505311200. [DOI] [PubMed] [Google Scholar]
- 6.Berg AH, Combs TP, Du X, Brownlee M, Scherer PE. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med. 2001;7:947–953. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- 7.Combs TP, Berg AH, Obici S, Scherer PE, Rossetti L. Endogenous glucose production is inhibited by the adipose-derived protein Acrp30. J Clin Invest. 2001;108:1875–1881. doi: 10.1172/JCI14120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001;7:941–946. doi: 10.1038/90984. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Turdi S, Park T, Morris NJ, Deshaies Y, Xu A, Sweeney G. Adiponectin corrects high-fat diet-induced disturbances in muscle metabolomic profile and whole-body glucose homeostasis. Diabetes. 2013;62:743–752. doi: 10.2337/db12-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Chewchuk S, Lavigne C, Brule S, Pilon G, Houde V, Xu A, Marette A, Sweeney G. Functional significance of skeletal muscle adiponectin production, changes in animal models of obesity and diabetes, and regulation by rosiglitazone treatment. Am J Physiol Endocr Metab. 2009;297:E657–E664. doi: 10.1152/ajpendo.00186.2009. [DOI] [PubMed] [Google Scholar]
- 11.Krause MP, Liu Y, Vu V, Chan L, Xu A, Riddell MC, Sweeney G, Hawke TJ. Adiponectin is expressed by skeletal muscle fibers and influences muscle phenotype and function. Am J Physiol Cell Physiol. 2008;295:C203–C212. doi: 10.1152/ajpcell.00030.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceddia RB, Somwar R, Maida A, Fang X, Bikopoulos G, Sweeney G. Globular adiponectin increases GLUT4 translocation and glucose uptake but reduces glycogen synthesis in rat skeletal muscle cells. Diabetologia. 2005;48:132–139. doi: 10.1007/s00125-004-1609-y. [DOI] [PubMed] [Google Scholar]
- 13.Vu V, Liu Y, Sen S, Xu A, Sweeney G. Delivery of adiponectin gene to skeletal muscle using ultrasound targeted microbubbles improves insulin sensitivity and whole body glucose homeostasis. Am J Physiol Endocrinol Metab. 2013;304:E168–E175. doi: 10.1152/ajpendo.00493.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vu V, Bui P, Eguchi M, Xu A, Sweeney G. Globular adiponectin induces LKB1/AMPK-dependent glucose uptake via actin cytoskeleton remodeling. J Mol Endocrinol. 2013;51:155–165. doi: 10.1530/JME-13-0059. [DOI] [PubMed] [Google Scholar]
- 15.Vu V, Kim W, Fang X, Liu YT, Xu A, Sweeney G. Coculture with primary visceral rat adipocytes from control but not streptozotocin-induced diabetic animals increases glucose uptake in rat skeletal muscle cells: role of adiponectin. Endocrinology. 2007;148:4411–4419. doi: 10.1210/en.2007-0020. [DOI] [PubMed] [Google Scholar]
- 16.Vu V, Dadson K, Odisho T, Kim W, Zhou X, Thong F, Sweeney G. Temporal analysis of mechanisms leading to stimulation of glucose uptake in skeletal muscle cells by an adipokine mixture derived from primary rat adipocytes. Int J Obes. 2011;35:355–363. doi: 10.1038/ijo.2010.160. [DOI] [PubMed] [Google Scholar]
- 17.Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y, et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002;8:731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- 18.Cheng KK, Iglesias MA, Lam KS, Wang Y, Sweeney G, Zhu W, Vanhoutte PM, Kraegen EW, Xu A. APPL1 potentiates insulin-mediated inhibition of hepatic glucose production and alleviates diabetes via Akt activation in mice. Cell Metab. 2009;9:417–427. doi: 10.1016/j.cmet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 19.Qiu W, Kohen-Avramoglu R, Rashid-Kolvear F, Au CS, Chong TM, Lewis GF, Trinh DK, Austin RC, Urade R, Adeli K. Overexpression of the endoplasmic reticulum 60 protein ER-60 downregulates apoB100 secretion by inducing its intracellular degradation via a nonproteasomal pathway: evidence for an ER-60-mediated and pCMB-sensitive intracellular degradative pathway. Biochemistry. 2004;43:4819–4831. doi: 10.1021/bi034862z. [DOI] [PubMed] [Google Scholar]
- 20.Hummel M, Meister R, Mansmann U. GlobalANCOVA: exploration and assessment of gene group effects. Bioinformatics. 2008;24:78–85. doi: 10.1093/bioinformatics/btm531. [DOI] [PubMed] [Google Scholar]
- 21.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 23.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS ONE. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patti GJ, Yanes O, Siuzdak G. Innovation. Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Metabolomics applied to diabetes research: moving from information to knowledge. Diabetes. 2009;58:2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HJ, Kim JH, Noh S, Hur HJ, Sung MJ, Hwang JT, Park JH, Yang HJ, Kim MS, Kwon DY, Yoon SH. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J Proteome Res. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 28.Bertram HC, Larsen LB, Chen X, Jeppesen PB. Impact of high-fat and high-carbohydrate diets on liver metabolism studied in a rat model with a systems biology approach. J Agric Food Chem. 2012;60:676–684. doi: 10.1021/jf203994k. [DOI] [PubMed] [Google Scholar]
- 29.Rubio-Aliaga I, Roos B, Sailer M, McLoughlin GA, Boekschoten MV, van Erk M, Bachmair EM, van Schothorst EM, Keijer J, Coort SL, et al. Alterations in hepatic one-carbon metabolism and related pathways following a high-fat dietary intervention. Physiol Genomics. 2011;43:408–416. doi: 10.1152/physiolgenomics.00179.2010. [DOI] [PubMed] [Google Scholar]
- 30.Han MS, Lim YM, Quan W, Kim JR, Chung KW, Kang M, Kim S, Park SY, Han JS, Park SY, et al. Lysophosphatidylcholine as an effector of fatty acid-induced insulin resistance. J Lipid Res. 2011;52:1234–1246. doi: 10.1194/jlr.M014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barber MN, Risis S, Yang C, Meikle PJ, Staples M, Febbraio MA, Bruce CR. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE. 2012;7:e41456. doi: 10.1371/journal.pone.0041456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann R, Franken H, Dammeier S, Rosenbaum L, Kantartzis K, Peter A, Zell A, Adam P, Li J, Xu G, et al. Circulating lysophosphatidylcholines are markers of a metabolically benign nonalcoholic fatty liver. Diabetes Care. 2013;36:2331–2338. doi: 10.2337/dc12-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pyne NJ, Dubois G, Pyne S. Role of sphingosine 1-phosphate and lysophosphatidic acid in fibrosis. Biochim Biophys Acta. 2013;1831:228–238. doi: 10.1016/j.bbalip.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Morris AJ, Panchatcharam M, Cheng HY, Federico L, Fulkerson Z, Selim S, Miriyala S, Escalante-Alcalde D, Smyth SS. Regulation of blood and vascular cell function by bioactive lysophospholipids. J Thromb Haemost. 2009;7(Suppl 1):38–43. doi: 10.1111/j.1538-7836.2009.03405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Ding T, Pan X, Li Y, Li R, Sanders PE, Kuo MS, Hussain MM, Cao G, Jiang XC. Lysophosphatidylcholine acyltransferase 3 knockdown-mediated liver lysophosphatidylcholine accumulation promotes very low density lipoprotein production by enhancing microsomal triglyceride transfer protein expression. J Biol Chem. 2012;287:20122–20131. doi: 10.1074/jbc.M111.334664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishihara K, Miyazaki A, Nabe T, Fushimi H, Iriyama N, Kanai S, Sato T, Uozumi N, Shimizu T, Akiba S. Group IVA phospholipase A2 participates in the progression of hepatic fibrosis. FASEB J. 2012;26:4111–4121. doi: 10.1096/fj.12-205625. [DOI] [PubMed] [Google Scholar]
- 37.Aloulou A, Ali YB, Bezzine S, Gargouri Y, Gelb MH. Phospholipases: an overview. Methods Mol Biol. 2012;861:63–85. doi: 10.1007/978-1-61779-600-5_4. [DOI] [PubMed] [Google Scholar]
- 38.Zhou L, Deepa SS, Etzler JC, Ryu J, Mao X, Fang Q, Liu DD, Torres JM, Jia W, Lechleiter JD, et al. Adiponectin activates AMP-activated protein kinase in muscle cells via APPL1/LKB1-dependent and phospholipase C/Ca2 + /Ca2 + /calmodulin-dependent protein kinase kinase-dependent pathways. J Biol Chem. 2009;284:22426–22435. doi: 10.1074/jbc.M109.028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hadad N, Burgazliev O, Elgazar-Carmon V, Solomonov Y, Wueest S, Item F, Konrad D, Rudich A, Levy R. Induction of cytosolic phospholipase A2α is required for adipose neutrophil infiltration and hepatic insulin resistance early in the course of high-fat feeding. Diabetes. 2013;62:3053–3063. doi: 10.2337/db12-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iyer A, Lim J, Poudyal H, Reid RC, Suen JY, Webster J, Prins JB, Whitehead JP, Fairlie DP, Brown L. An inhibitor of phospholipase A2 group IIA modulates adipocyte signaling and protects against diet-induced metabolic syndrome in rats. Diabetes. 2012;61:2320–2329. doi: 10.2337/db11-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song H, Wohltmann M, Bao S, Ladenson JH, Semenkovich CF, Turk J. Mice deficient in group VIB phospholipase A2 (iPLA2γ) exhibit relative resistance to obesity and metabolic abnormalities induced by a Western diet. Am J Physiol Endocrinol Metab. 2010;298:E1097–E1114. doi: 10.1152/ajpendo.00780.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 43.Negishi M, Shimizu H, Okada S, Kuwabara A, Okajima F, Mori M. 9HODE stimulates cell proliferation and extracellular matrix synthesis in human mesangial cells via PPARγ. Exp Biol Med. 2004;229:1053–1060. doi: 10.1177/153537020422901010. [DOI] [PubMed] [Google Scholar]
- 44.Vangaveti V, Baune BT, Kennedy RL. Hydroxyoctadecadienoic acids: novel regulators of macrophage differentiation and atherogenesis. Ther Adv Endocrinol Metab. 2010;1:51–60. doi: 10.1177/2042018810375656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itoh T, Fairall L, Amin K, Inaba Y, Szanto A, Balint BL, Nagy L, Yamamoto K, Schwabe JW. Structural basis for the activation of PPARγ by oxidized fatty acids. Nat Struct Mol Biol. 2008;15:924–931. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bikman BT, Summers SA. Sphingolipids and hepatic steatosis. Adv Exp Med Biol. 2011;721:87–97. doi: 10.1007/978-1-4614-0650-1_6. [DOI] [PubMed] [Google Scholar]
- 47.Turner N, Kowalski GM, Leslie SJ, Risis S, Yang C, Lee-Young RS, Babb JR, Meikle PJ, Lancaster GI, Henstridge DC, et al. Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia. 2013;56:1638–1648. doi: 10.1007/s00125-013-2913-1. [DOI] [PubMed] [Google Scholar]
- 48.Grosch S, Schiffmann S, Geisslinger G. Chain length-specific properties of ceramides. Prog Lipid Res. 2012;51:50–62. doi: 10.1016/j.plipres.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Wanders RJ, Komen J, Kemp S. Fatty acid ω-oxidation as a rescue pathway for fatty acid oxidation disorders in humans. FEBS J. 2011;278:182–194. doi: 10.1111/j.1742-4658.2010.07947.x. [DOI] [PubMed] [Google Scholar]
- 50.Kajander T, Kellosalo J, Goldman A. Inorganic pyrophosphatases: one substrate, three mechanisms. FEBS Lett. 2013;587:1863–1869. doi: 10.1016/j.febslet.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Liu M, Chung S, Shelness GS, Parks JS. Hepatic ABCA1 and VLDL triglyceride production. Biochim Biophys Acta. 2012;1821:770–777. doi: 10.1016/j.bbalip.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoo RL, Lee IP, Zhou M, Wong JY, Hui X, Xu A, Lam KS. Pharmacological inhibition of adipocyte fatty acid binding protein alleviates both acute liver injury and non-alcoholic steatohepatitis in mice. J Hepatol. 2013;58:358–364. doi: 10.1016/j.jhep.2012.10.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.