Abstract
Compared with normal cells, tumor cells have undergone an array of genetic and epigenetic alterations. Often, these changes underlie cancer development, progression, and drug resistance, so the utility of model systems rests on their ability to recapitulate the genomic aberrations observed in primary tumors. Tumor-derived cell lines have long been used to study the underlying biologic processes in cancer, as well as screening platforms for discovering and evaluating the efficacy of anticancer therapeutics. Multiple -omic measurements across more than a thousand cancer cell lines have been produced following advances in high-throughput technologies and multigroup collaborative projects. These data complement the large, international cancer genomic sequencing efforts to characterize patient tumors, such as The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC). Given the scope and scale of data that have been generated, researchers are now in a position to evaluate the similarities and differences that exist in genomic features between cell lines and patient samples. As pharmacogenomics models, cell lines offer the advantages of being easily grown, relatively inexpensive, and amenable to high-throughput testing of therapeutic agents. Data generated from cell lines can then be used to link cellular drug response to genomic features, where the ultimate goal is to build predictive signatures of patient outcome. This review highlights the recent work that has compared -omic profiles of cell lines with primary tumors, and discusses the advantages and disadvantages of cancer cell lines as pharmacogenomic models of anticancer therapies.
Introduction
Cell lines have a long history as models to study molecular mechanisms of disease. In some fields, such as cardiology and neuroscience, in vitro studies often use primary cultures with genetic perturbations or cells treated with an array of agents to induce a disease state. In cancer research, collections of tumor-derived cell lines are often used as models because they carry hundreds to thousands of aberrations that arose in the tumor from which they were derived. Cancer cell lines are used to study many biologic processes and have been widely used in pharmacogenomics studies. A recent review by Sharma and colleagues discussed the advantages and disadvantages of cell lines as a drug screening platform (1). Since this work, genomic measurements were made available for hundreds of cancer cell lines, and these data present new opportunities to link genomic profiles to therapeutic response.
The development and clinical implementation of Precision Medicine has become a national priority1. This will require the analysis of large-scale genomics data (2) from individuals and populations to identify features that predict individual cancer behavior, including probability of disease progression and response to treatment. Measurements relevant to Precision Medicine include, but are not limited to, gene expression, genome-wide RNAi screens, sequencing-based profiling, and measures of therapeutic response and patient outcome. These data are used to identify dysregulated genes and pathways with the goal of understanding the factors that drive tumor progression and underlie patient response to treatment. Given the ubiquity of these datasets in cancer, we are now in a position to study single cancer subtypes and to identify common and recurrent aberrations across cancers. This notion of “pan-cancer” analysis has sparked new interest in developing and repositioning anticancer drugs to target specific genetic aberrations or molecular subtypes, as opposed to the tumor tissue of origin (2).
Cell lines serve as models to study cancer biology, and connecting genomic alterations to drug response can aid in understanding cancer patient response to therapy. Accordingly, several large datasets have been generated to link genomic and pharmacologic profiles of cell lines. The first of these datasets was the NCI-60, a pharmacologic screen across 60 cancer cell lines (3). Later, genomic features of these cell lines were characterized and all NCI-60 related data were compiled in CellMiner (4). Targeted study of a panel of breast cancer cell lines have led to insights into the pathways and process directly affected by anticancer compounds (5, 6). Additional pharmacogenomics datasets such as the Connectivity Map (7), Genomics of Drug Sensitivity in Cancer (GDSC; ref. 8), the Cancer Cell Line Encyclopedia (CCLE; ref. 9), the Cancer Therapeutics Response Portal (CTRP; ref. 10), and the Cancer Target Discovery and Development Project2 have expanded the numbers of cell lines, drugs, and cancer types (Table 1). These studies have led to advances in our understanding of cellular response to drugs and have provided the necessary data to develop prediction algorithms that aim to match the response with genomic features.
Table 1.
Tissue representation of cell lines in large in vitro pharmacogenomics databases
| Malignancy | CCLE | GDSC | CTRP | NCI-60 | Unique |
|---|---|---|---|---|---|
| Bladder | 28 | 18 | 5 | 0 | 29 |
| Bone | 29 | 31 | 1 | 0 | 53 |
| Breast | 60 | 43 | 1 | 5 | 64 |
| Cervix | 0 | 12 | 0 | 0 | 12 |
| Colon | 63 | 35 | 37 | 7 | 70 |
| Endometrium | 28 | 10 | 11 | 0 | 29 |
| Head and neck | 35 | 23 | 2 | 0 | 44 |
| Hematopoietic and lymphoid | 181 | 113 | 24 | 6 | 227 |
| Kidney | 36 | 22 | 2 | 8 | 48 |
| Liver | 36 | 14 | 4 | 0 | 41 |
| Lung | 187 | 141 | 91 | 9 | 226 |
| Nervous system | 86 | 79 | 3 | 6 | 126 |
| Osephagus | 27 | 23 | 3 | 0 | 29 |
| Ovary | 52 | 20 | 26 | 7 | 57 |
| Pancreas | 46 | 17 | 10 | 0 | 47 |
| Prostate | 8 | 5 | 1 | 2 | 9 |
| Skin | 62 | 45 | 9 | 10 | 79 |
| Soft tissue | 21 | 17 | 3 | 0 | 28 |
| Stomach | 38 | 18 | 6 | 0 | 44 |
| Testis | 0 | 2 | 0 | 0 | 2 |
| Thyroid | 12 | 12 | 0 | 0 | 16 |
| Other | 11 | 7 | 3 | 0 | 20 |
| Total # of cell lines | 1,046 | 707 | 242 | 60 | 1300 |
| Total # of agents tested | 24 | 139 | 185 | −50,000 |
NOTE: The number of cell lines across all tissues is listed for each database along with the total number of agents tested. The final column identifies the number of unique cell lines across all four datasets for each cancer type.
Despite the ubiquitous use of cancer cell line models, we are still left with the same question that has been around since cell lines were first isolated in the 1950s: how well do in vitro cell line models recapitulate the biologic processes of in vivo disease and drug response? More specific to this review, are tumor-derived cell lines representative genomic models of in vivo disease and therapeutic response? Here, we summarize the work to date that has been aimed at addressing these questions.
Comparing -omic profiles of tumors and cell lines
The rich data resources mentioned above allow for a relatively complete assessment of how well, if at all, cell lines recapitulate the genomic aberrations observed in primary tumors. Several studies have shed light on how the genomic profiles, molecular subtypes, and heterogeneity of tumors compare with cell lines, and the result of this work can be used to identify the cell lines that best model the genomic features of particular diseases.
Cell lines model genomic features of tumors
One of the first efforts to systematically compare tumors with cell lines revealed global similarities in gene expression patterns between breast cancer cell lines and tumors (11, 12). Following this work, a comparison of copy number alterations (CNA) and transcriptional profiles demonstrated that breast cancer cell lines harbor the majority of functionally important alterations observed in breast cancer tumors. Of the genes that were significantly altered in both gene expression and CNA, there was 72% agreement between cell lines and tumors. However, while the global CNA profiles are remarkably similar between cell lines and breast tumors, in general, there are a greater number of CNAs in cell lines (13). These results underscore a common theme across tissue types: while cell lines mirror many genomic aspects of tumors, there are aberrations specific to cell lines that may play important roles in translational biology.
Panels of cell lines modeling ovarian cancer (14), head and neck cancer (15), and colorectal cancer (16) have also been comprehensively characterized in the context of solid tumors. Domcke and colleagues found that high-grade serous ovarian cancer (HGSOC) cell lines and tumors had similar recurrent mutations and alterations, but overall, the cell lines had a higher number of mutations and a wider distribution of CNAs (14). Similarly, in other tissue types, mutations are more abundant in cell lines as compared with primary tumors, including head and neck (15) and colorectal cancer (16) cell lines. Assessment in colorectal (16), melanoma (17), and non–small cell lung cancer (18) cell lines compared with their respective tumors revealed that CNAs were similar. However, vast differences between the CNAs found in head and neck cancer cell lines and tumors were observed (15).
The CCLE, representing approximately 1,000 cell lines across 36 cancer types, has characterized cell lines for gene expression, CNAs, and mutations in select genes using hybrid capture sequencing (9). A strong correlation was observed for each genomic measurement between cell lines and tumors, suggesting that the cell lines comprising this dataset contained most of the common genomic aberrations found in tumors. However, not all genomic measurements have equal similarity as there are notably fewer differences in transcriptional profiles (11–13) and CNAs (16, 18) when compared with the larger differences observed in mutational profiling (14, 15), although the differences in mutational profiling may be due to current technical difficulties in mutation calling from sequencing data. The effect of these genomic differences between cell lines and tumors in translating information from in vitro to in vivo is not fully understood. Comprehensive profiling of more cancer types, along with the associated analysis, is required for complete characterization of the impact of genomic aberrations that are different between cell lines and tumors.
Cell line DNA methylation show both similarities and differences to tumors
Epigenomic studies capture genome-wide, nongenetic modifications. While many forms of epigenetic alterations exist, DNA methylation is readily studied with high-throughput assays. An early study by Paz and colleagues showed that hypermethylation of the promoter region of 15 tumor suppressor genes across 12 cancer types and 70 cell lines was similar (19). More recently, a comparison of the complete methylomes of breast cancer cell lines and tumors revealed global concordance (20). While there are strong epigenetic similarities between cell lines and primary tumors, differences have been characterized and attributed to the process of cell culture itself. For example, Varley and colleagues identified DNA methylation differences between primary cultures and tissue, suggesting that the cell culturing process induces DNA methylome changes (21). Changes in DNA methylation will affect gene expression, thereby influencing cellular processes and potentially even response to therapy. The overall impact of differences in DNA methylation in cell lines is not clear, although the microenvironment along with external stimuli likely contributes and should be considered when interpreting cell line data.
Molecular subtypes of tumors are represented in cell lines
Cancer subtypes have traditionally been classified on histologic markers. Perou and colleagues (22), followed shortly by Golub and colleagues (23), presented a new way to molecularly subtype cancer through the clustering of gene expression patterns measured using microarrays. This approach advanced the field because molecular subtyping could be done in an unbiased manner based on gene expression patterns. Many studies have shown that therapeutic response to specific agents can be predicted on the basis of molecular subtype both in vitro (5, 6) and clinically (24–28). Because of the clinical impact, modeling and studying subtypes represented in cell culture has been the subject of many studies, and it is only recently that additional -omic profiles have been the basis for molecular subtyping of tumors.
As mentioned earlier, Domcke and colleagues focused their efforts on HGSOC, using gene expression, mutations, and CNAs to compare HGSOC cell lines and primary tumors (14). By designing a ranking system leveraging publicly available tumor (29) and cell line (9) data, the authors were able to identify the cell lines with the most similar genomic profiles of HGSOC. They found that the two most often used cell lines, accounting for 60% of HGSOC publications, showed poor genomic similarity to HGSOC tumor profiles, whereas the cell lines most similar to HGSOC account for less than 1% of publications studying this subtype. It is possible that translating in vitro results in HGSOC has been hindered because the cell lines most frequently studied are not genomically representative of the disease. Now that we have the ability to measure genome-wide molecular features, researchers can prioritize cell lines according to the target -omic profile for use in experimental studies.
Not all cancer subtypes are well represented by cell line models. The first study to demonstrate this was a gene expression clustering study of over 40 lung cancer cell lines and tumors that found only some of the cell lines clustered to the expected groups of tumors (30). Specifically, 50% of squamous cell carcinoma and over 80% of small-cell lung carcinoma cell lines clustered with their respective primary tumor types. However, none of the 11 adenocarcinoma cell lines clustered with primary adenocarcinomas. Therefore, either the adenocarcinoma subtype is lost in vitro as the authors suggest, or the adenocarcinoma lines were misclassified (30). In breast (31) and pancreatic ductal adenocarcinoma (32) cancer cell line panels, most of the subtypes common in these cancer were present, with the exception of one absent tumor subtype from each panel. The luminal A breast cancer subtype was not found in the breast cancer panel (31) and the exocrine-like subtype was not found in the panel of pancreatic ductal adenocarcinoma cell lines (32). While the reason for this is uncertain, it is clear that existing cell lines do not fully represent all tumor subtypes; thus, in vitro studies should be focused on the subtypes that are in common between cell lines and tumors.
Patterns of intertumor molecular subtypes in cell lines
Large, international cancer projects, such as The Cancer Genome Atlas (TCGA)3 and the International Cancer Genome Consortium (ICGC)4, have produced the data necessary to perform systematic analysis across many tumor types. This pan-cancer analysis has found both tissue-specific and recurrent patterns across many cancers. Data from the CCLE (9), GDSC (8), CTRP (10), and NCI-60 (3) provide the necessary material to mirror TCGA and ICGC pan-cancer analyses in cell lines. As displayed in Table 1, tumor types are well represented across these datasets.
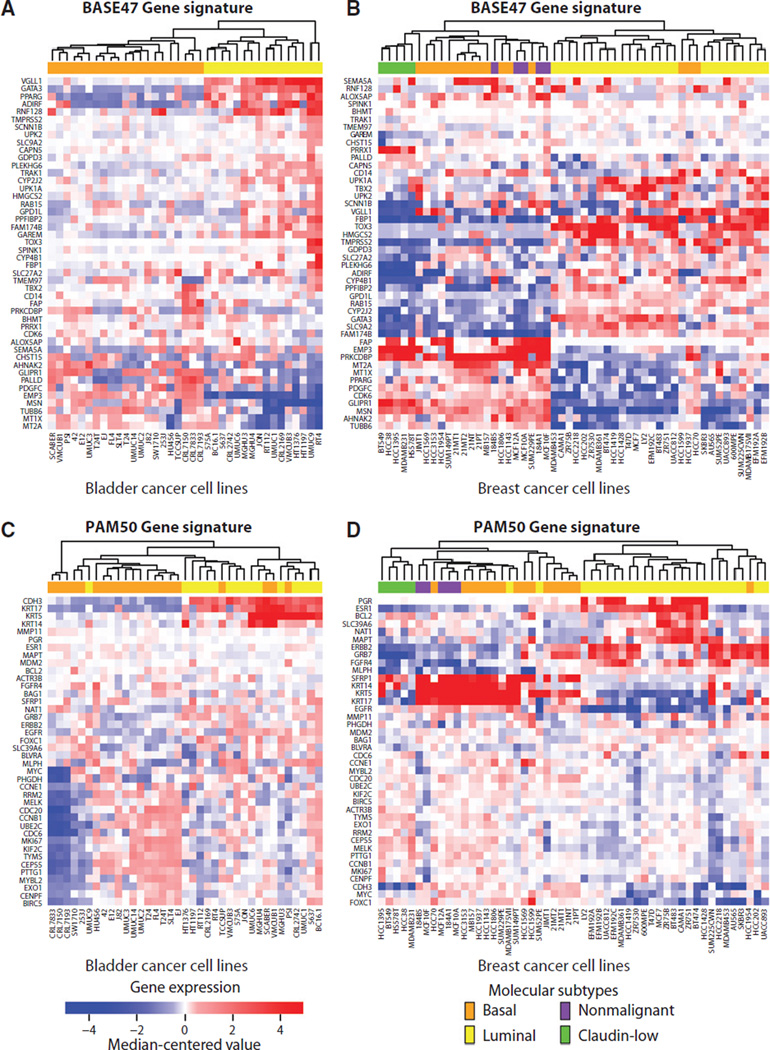
As a demonstration of intertumor molecular subtypes, two independent studies found a striking similarity in the gene expression profiles of basal and luminal lineages in breast tumors when compared with bladder tumors (25, 33). Similar to the PAM50 signature in breast cancer (34), Damrauer and colleagues developed the BASE47 signature to stratify basal and luminal subtypes in bladder cancer (33). The authors showed that the BASE47 correctly stratified breast cancer molecular subtypes, suggesting that similar transcriptional programs are operational across cancers of the breast and bladder. As an independent analysis, and shown in Fig. 1, we tested the ability of patient-derived BASE47 and PAM50 gene expression signatures to stratify bladder (35) and breast cancer (6) cell lines according to basal and luminal subtype gene expression patterns. When hierarchically clustered, the BASE47 and PAM50 signatures separate breast cancer cell lines into the same basal and luminal subtypes (Fig. 1B and D). Interestingly, when the gene signatures were applied to the bladder cancer cell lines, two main clusters were revealed, but the basal and luminal labels were not consistent. Putative basal lineage markers, KRT5, KRT14, and CDH3, are upregulated in both breast (36) and bladder (37) primary tissues; these markers are present in the PAM50 but not in the BASE47 signature. In Fig. 1A, we used the BASE47 signature to assign basal/ luminal subtypes to the bladder cancer cell lines. When we applied the PAM50 signature to the bladder cancer cell lines (Fig. 1C), KRT5, KRT14, and CDH3 were upregulated in the lines identified as luminal by the BASE47 signature, contrary to what was observed in both primary tissues as well as the breast cancer cell lines. In summary, we observed inconsistencies in the subtypes predicted by the PAM50 and BASE47 signatures in vitro, highlighting the need for a deeper study of the biologic basis of these signatures both across these tissue types as well as between cell lines and primary tumors.
Figure 1.
Breast and bladder cancer cell lines cluster into basal and luminal subtypes. Breast (6) or bladder (35) cancer gene expression data were log-transformed, median centered, then hierarchically clustered using Euclidean distance according to the genes present in the patient-derived (A and B) BASE47 bladder cancer signature or the (C and D) PAM50 breast cancer signature.
While molecular subtyping from genomic data has shown promise, contamination from surrounding stromal cells can influence the transcriptional profile of a sample. For example, Isella and colleagues showed that the stem/serrated/mesenchymal transcriptome subtype of colorectal cancer was predominantly produced by surrounding stromal cells rather than the colorectal cancer cells (38), highlighting the potential of stroma-contaminated samples contributing to the molecular characterization of tumors.
Patterns of intratumor heterogeneity are found in cell lines
Intratumor heterogeneity, the molecular differences between cells within an individual tumor, has clinical implications (39, 40). One model of drug resistance is that subpopulations in a heterogeneous tumor are selected during drug treatment (41). Heterogeneous cell line models may serve as an important tool for understanding and quantifying the biologic underpinnings and therapeutic implications of tumor heterogeneity.
Roschke and colleagues used spectral karyotyping to observe genomic variations within cell lines of the NCI-60 (3, 42), and identified heterogeneity in ploidy, rearrangements, and total number of chromosomes within particular cell lines (42). In a later targeted study, 12 of 30 lung cancer cell lines were found to have different EGFR mutations within the same cultures (43). These studies indicate that the biologic mechanisms underlying heterogeneity within primary tumors may be operational in cell line models, and that cell lines can be used to study heterogeneous responses to therapy.
Other studies have identified cancer stem cells as a specific form of heterogeneity in cell lines. Cancer stem cells are undifferentiated, can self-renew, and have the ability to initiate tumors. Subpopulations of cancer stem cells have also been identified in many cancer cell lines, including breast (44), glioma (45), and head and neck cancer (46). Evidence of cancer stem cells as a subpopulation in cell lines provides more support to the heterogeneity within cell lines. However, one study noted the loss of cancer stem cells during propagation of glioblastoma cell lines regardless of their presence in matching primary glioblastomas (47). This implies that the presence of cancer stem cells can be lost in vitro, and may also be cell line dependent.
Cell line genomics in translational medicine
Recent studies have begun to systematically incorporate larger amounts of genomics-level data into the interpretation of drug sensitivity data (3–5, 7–10). In this section, we highlight some of the studies that have relied heavily on in vitro pharmacogenomics as well as highlight some areas of need.
Large in vitro pharmacogenomics datasets
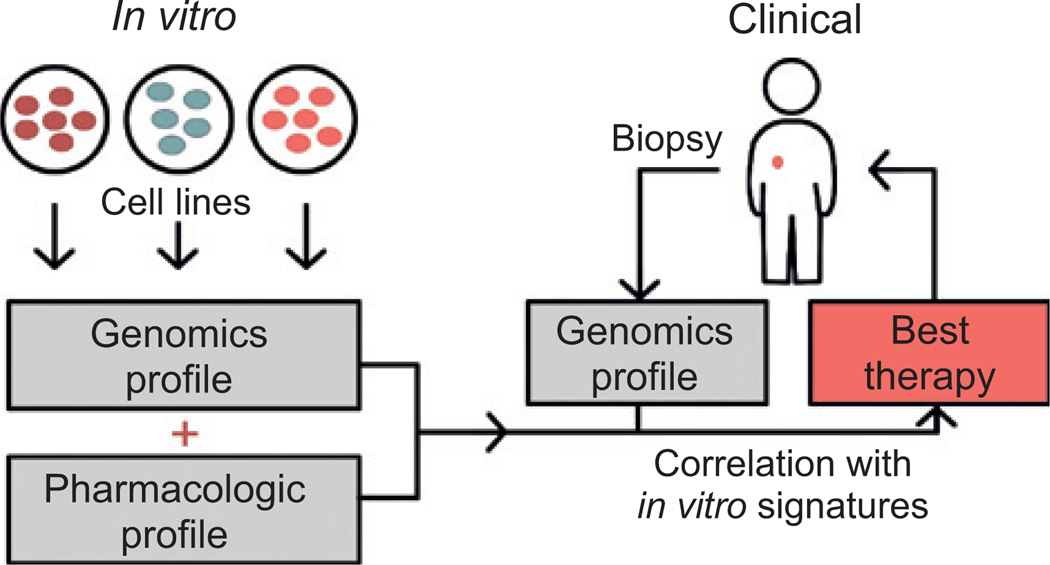
Translating cell line pharmacogenomics into in vivo therapeutics requires a range of tumor-derived cancer cell lines, genomics-level data, and therapeutic responses (Fig. 2). Accordingly, there are many large, public datasets that have characterized genomic profiles of cell lines and their susceptibility to compounds (3, 4, 8–10). It should be noted that the controlled and systematic study of cellular response after drug treatment is only scalable using cell line models. Table 1 outlines the available databases that link cell line genomics with pharmacologic data. The first of these databases was the NCI-60, which comprises 60 cell lines consisting of 9 cancer types (3, 4). Responses in this panel of cell lines were assessed for tens of thousands of compounds and drugs. One early algorithm developed for these data was COMPARE, which summarized multiple parameters of pharmacologic response into a single graph (48). They found that agents with similar mechanisms of action produced similar responses. COMPARE helped spur further algorithm advancement, which shifted the NCI-60 from a purely drug screening tool to one identifying molecular features predictive of drug response. Scherf and colleagues was the first study to link gene expression of the NCI-60 with pharmacologic profiles (49). This study inspired many of the studies discussed throughout this review.
Figure 2.
Process of using in vitro genomics for clinical treatment selection. Multiple prediction algorithms have been utilized to derive molecular features predictive of pharmacologic response of cells in vitro. After a tumor is biopsied and its genomic features are profiled, in vitro derived predictive signatures may be used to determine the best therapy for the individual’s tumor.
Other types of in vitro pharmacogenomics databases are unique in that they measure changes after cells are perturbed with various agents. The Connectivity Map (CMAP) is an effort to determine gene expression changes in response to over 6,000 drug candidates and genetic perturbations (7). The NIH Library of Integrated Network-based Cellular Signatures (LINCS) Program5 is actively pursuing similar efforts, and will assess molecular and cellular changes induced by small-molecule and genetic perturbations. Collectively, pharmacogenomics cell line datasets have been used in thousands of studies, and have led to novel drug predictions. For example, CMAP was used to predict that two HDAC inhibitors would be useful in treatment of childhood B-lymphoblastic leukemia, and this prediction was validated using primary patient samples (50).
Computational models for drug sensitivity prediction
The goal of Precision Medicine in cancer is to identify molecular features unique to a patient’s tumor that help to determine the best treatment option. Several examples of rational drug design have been based on specific genetic aberrations found within a cohort of patients, including the targeted therapies such as trastuzumab for HER2-overexpressing breast cancer (51, 52) and imatinib to inhibit BCR-ABL–activated leukemia (53). Large-scale genomics studies provide the data necessary to further develop Precision Medicine approaches.
Many research groups use genomic measurements to identify molecular signatures of tumors that classify patients based on their response to various treatments. Molecular features associated with tumor response can be discovered by comparing clinical responses to the -omic features of the responding tumors, or by comparing responses of cell lines treated with drugs to their -omic features (Fig. 2). Studies with patient tumors have been used to identify molecular subtypes associated with therapeutic response in colorectal (27), bladder (25), and breast cancers (26, 28). These studies demonstrate the promise of Precision Medicine and suggest possibilities for tailoring treatment to a patient’s tumor. However, associating clinical response to patient genomic features is limited by statistical power, availability of appropriate tissue samples, the time required to establish clinical outcomes, and the feasibility of testing the hundreds experimental compounds currently under development. Cell lines complement patient studies by serving as models to establish the causal role of identified genomic features on cellular response.
Preclinical studies of the responses of panels of well-characterized cancer cell lines have been used to build predictive computational models of drug response that can be tested clinically. This approach mitigates some of the logistical issues in clinical studies because cell lines can be readily manipulated, and also allow for identification of molecular events that are causally related to response. One of the first developments in this area was the coexpression extrapolation (COXEN) model that used gene expression from cell lines in the NCI-60 to build and test a chemosensitivity predictor for bladder cancer (35). COXEN was also retrospectively validated in vivo as it was able to predict response of breast cancer patients to common chemotherapeutics. More recently, Barretina and colleagues used information about gene expression, genome copy number, DNA sequence, and pharmacologic information to predict drug sensitivity in the CCLE (9). As an example, they demonstrated a link between SLFN11 expression and topoisomerase inhibitor sensitivity.
The CCLE (9) and GDSC (8) demonstrated the strength of an integrative genomics approach using measurements of multiple -omic endpoints to build predictive models. Molecular endpoints now being included in predictive model development include measurements of gene expression (microarray and sequencing), mutations, copy number alterations, epigenetic modifications, metabolomics, and proteomics. Despite the wide use of these datasets, important caveats have been reported. First, one should ensure the cell lines being studied are of the correct origin. Recent efforts led by Yu and colleagues have aimed to standardize a framework for cell line authentication, quality control, and annotation (54). This study also quantified similarities between cell lines and identified a total of 69 synonymous lines in the CCLE and 6 synonymous lines in the GDSC. Discrepancies in the pharmacologic profiles of cell lines and therapies tested in the CCLE and GDSC studies suggest that the details of the experimental approach for assessing drug sensitivity can have a major impact on the results (55, 56). However, in the context of targeted drugs and their target genes, both the CCLE and GDSC show strong consistency in drug response. As shown in Table 2, we compared the IC50s of cells aberrant for the gene of a targeted drug to cells that were wild type for the same gene. For the majority of drugs we analyzed, the groups of cell lines harboring an aberration in the target gene were significantly more sensitive to the targeted therapy. Even when the aberration did not predict drug response, the GDSC and CCLE results were concordant. Additional studies have shown that cell response to targeted therapies is associated with the presence of the therapeutic target. In two separate studies, sensitivity of non–small cell lung cancer cell lines to erlotinib was shown to correlate with EGFR mutations (18, 57); similar results were shown for vandetanib and lapatinib (18). PHA665752, a MET inhibitor, was also found to be more sensitive in gastric cancer cell lines with MET amplifications (57). These studies confirm that cell lines can model the effects of targeted agents and recapitulate the expected associations between mutations and drug sensitivity.
Table 2.
Sensitivity to targeted therapies correlates with the corresponding targeted alteration in vitro
| Alteration | Tissue | Drug | GDSC Wilcoxon | CCLE Wilcoxon |
|---|---|---|---|---|
| BCR-ABL | Blood | Imatinib | 0.0008 | NA |
| Blood | Nilotinib | 0.001 | NA | |
| Blood | Dasatinib | 0.001 | NA | |
| KIT CNV | All tissues | Imatinib | 0.051 | NA |
| FLT3 MUTATION | All tissues | Sorafenib | 0.16 | 0.0007 |
| All tissues | Sunitinib | 0.01 | NA | |
| ERBB2: MUT AMP | Breast | Lapatinib | 0.003 | 0.01 |
| Ovary | Lapatinib | NAb | 0.3 | |
| Lung | Lapatinib | NAb | 0.35 | |
| Stomach | Lapatinib | NAb | 0.047 | |
| All tissues | Lapatinib | 0.0002 | 0.0003 | |
| EGFR: MUT AMP | Lung | Erlotinib | 0.13 | 0.02 |
| CNSa | Erlotinib | 1 | 0.25 | |
| All tissues | Erlotinib | 0.04 | 1.40E-05 | |
| MET: MUT AMP | All tissues | PHA-665752 | 0.37 | 0.49 |
| All tissues | PF-2341066 | 0.07 | 0.15 | |
| ALK: MUT AMP | All tissues | PF-2341066 | 0.01 | 0.54 |
| BRAF: MUT AMP | All tissues | PLX4720 | <2.2e–16 | <2.2e–16 |
| Skin | PLX4720 | 0.003 | 0.027 |
NOTE: Cell lines across two large in vitro pharmacogenomics databases were separated based on being wild type or mutant for several alterations for which targeted therapy exists. Amplifications in each dataset (Affymetrix SNP 6.0 arrays) were defined according to the protocols in the GDSC (8) and CCLE (9). Data for this analysis were taken directly from the CCLE or GDSC web resources. The significance of mutant cell lines drug sensitivity compared with wild-type cell lines was determined using a Wilcoxon rank-sum test.
For the GDSC: Glioma, medulloblastoma, neuroblastoma.
IC50 not available in each group.
Molecular measurements assess different cellular processes, and integrative analyses seek to bring these data together for a more complete understanding of cell function and to build more robust predictive models (58, 59). PARADIGM is one analysis tool that integrates several types of genomics data to identify deregulated cellular pathways (59). One early study clustered glioblastoma patients using PARADIGM to reveal groups of patients with differential survival rates. Clustering with either gene expression by microarray or copy number alterations alone could not identify groups of patients with differential survival, suggesting an integrated, pathway-informed approach is more biologically relevant and informative. PARADIGM has also been used on cell line genomics to identify pathway differences between breast cancer subtypes (5).
Analytic procedures that accurately predict response to therapeutic inhibitors are critically needed for Precision Medicine. However, this field is still in its infancy. The 2012 NCI-DREAM Challenge supported international efforts to compare predictive analytic methodologies: measurements of a training set of responses of breast cancer cell lines to therapeutic inhibitors were made available to computational groups to predict responses of an undisclosed test dataset of cell lines (58). Forty-four teams from around the world submitted their solutions for evaluation. Of the 6 -omic measurements available, gene expression measured by microarrays was reported to be the most predictive single type of data. However, the top-performing methods integrated multiple -omic measurements to form response predictors. In addition, this work quantified the gain in performance when different data types were combined. For example, in a pairwise analysis, exome sequencing combined with gene expression data showed the biggest gain in performance for the top-performing Bayesian multitask multiple kernel learning method. Overall, the best performance was seen when all data types were integrated. Several other studies have suggested that integrative genomics surpasses the information obtained from any single genomics experiment (5, 6, 8, 9, 24, 60, 61). Integrative analyses will be required for a complete picture of therapeutic cellular responses, and the advent of multi-omic studies now makes this possible.
A new and challenging area of computational modeling is the prediction of drug synergies from genomics measurements and single drug profiles. The 2012 NCI-DREAM Challenge to predict drug synergies is the most comprehensive analysis to date of drug synergy methods (62). While the challenge identified methods that could predict synergism and antagonism, performances were not much better than random, indicating an ongoing need for improvement. Large, high-throughput combinatorial screening platforms (63) will offer the opportunity to systematically learn the genomic patterns that predict synergism, and measurements of cell and molecular responses of cell lines panels are ideal for this type of analytic development study.
Preclinical models to predict mechanisms of acquired therapeutic resistance
Unfortunately, even when patients are assigned to an effective therapy, they frequently become resistant to it over time, and this acquired resistance must be overcome to effectively eradicate or manage the disease. Cell lines are amenable to use as pharmacogenomic models of acquired resistance. For example, Engelman and colleagues treated a non–small cell lung cancer line with increasing concentrations of the EGFR inhibitor gefitinib to generate resistance clones (64). Genomic characterization of the derived resistant lines revealed that cells had amplified MET, which offered resistance to EGFR inhibition. Importantly, MET amplification was also found to offer acquired resistance to EGFR inhibition (64, 65). This example shows that mechanisms of in vitro resistance can recapitulate mechanisms of acquired resistance.
Several studies of acquired resistance to tamoxifen in breast cancer have identified IGF1R as an escape mechanism, an observation that has been made in both model systems as well as in primary patient samples. MCF7 breast cancer cell lines that have undergone selection to become resistant to tamoxifen may continue to proliferate because of changes in IGF1R signaling, in part by upregulating IGF1R (66). Similarly, tamoxifen-resistant MCF7 cell line xenograft models showed increased levels of phosphorylated IGF1R as compared with tamoxifen-sensitive models (67). However, protein levels of IGF1R were reduced in patient cohorts after the development of resistance to tamoxifen (68). This suggests that mechanisms of resistance in preclinical models may not directly replicate those found clinically. Nonetheless, cell line models provide a platform for gaining insights into this critical question in molecular cancer research.
Systematic, genome-scale functional knockdowns
A fundamental goal in cancer research is to identify the genetic factors that drive tumorigenesis. Project Achilles aims to identify genes required for cell viability in hundreds of cancer cell lines by systematic knockdown of genes through shRNAs (69). Identifying this set of genes provides a wealth of information to develop novel therapies. While single genes might not be essential on their own, the loss-of-function of two nonessential genes can have a synthetic lethal interaction resulting in cell death (70). Cancer is often defined by large numbers of genetic aberrations, so understanding how genes interact in the cell is critical for identifying effective therapeutic strategies in individual patients. Algorithms have been developed that use primary tumor data to predict synthetic lethal gene pairs (71), and a critical next step is to experimentally validate these predictions. High-throughput screens can be used to test gene-to-drug synthetic lethal interactions in cell lines. For instance, the drug nutlin-3, which activates p53 through inhibition of MDM2, was used along with a genome-wide shRNA library to determine genes that governed the cellular response of p53 activation. The authors found that ATM and MET were responsible for induction of cell arrest instead of apoptosis. Pharmacologic or genetic inhibition of ATM or MET induced apoptosis in tumor spheroids, highlighting the use of synthetic lethal screens in identifying therapeutic targets (72).
Results from functional knockdown screens performed in vitro have demonstrated clinical value. In contrast to BRAF (V600E) melanoma tumors, colon tumors with BRAF (V600E) mutations are resistant to vemurafenib. A synthetic lethal screen of BRAF-mutant colon cancer cell lines tested siRNA knockdown of kinome-related genes in the presence of vemurafenib, and identified EGFR as a resistance mechanism. The authors suggest that combinational therapy blocking BRAF (V600E) and EGFR may be effective in a subset of colon cancers with no targeted therapies (73). These observations indicate that functional screens in cancer cell lines can be used to identify therapeutic targets and cotreatment strategies that may be clinically impactful (72, 73).
Pharmacogenomics differences between in vitro methods
Monolayer culturing methods are limited in their capacity to accurately model the complexity of the in vivo tumor microenvironment (TME; ref. 1). Alternate approaches to modeling the TME have been developed, including three-dimensional (3D) cell cultures and coculturing systems (74, 75). The previously discussed large pharmacogenomics databases used cell lines grown in monolayer cultures on plastic, and ignore microenvironmental effects that likely influence how cells respond to therapy. Here, we describe these alterations to shed light on the pharmacogenomics differences across in vitro models.
Effect of the tumor microenvironment on pharmacogenomics studies
The TME is an important factor in vivo because cross-talk between the tumor and the microenvironment affects a number of processes. The TME includes cancer cells, fibroblasts, pericytes, immune cells, and stromal cells (76). Gene expression signatures of the TME has been shown to have prognostic value in breast cancer (77), which suggests that there may be a missing component in many in vitro studies.
Coculturing cancer cells with stromal cells, such as those that can be found in vivo, can affect gene expression. For example, coculturing breast cancer cell lines with primary osteoblasts leads to global changes in gene expression (78). In addition, genomic instability can be increased by elements of the TME (79). This suggests that observations from monolayer cell culture systems may be limited in their ability to translate to primary tumor samples. To study the effect of the TME in vitro, McMillin and colleagues cocultured stromal cells with tumor cells and showed that the TME can both enhance sensitivity and promote resistance depending on the cell, drug, and TME composition (80). These studies highlight the TME as a missing component in many in vitro studies, as well as the large-scale cell line pharmacogenomics studies (3, 4, 8–10). It is worth noting that these large databases were designed to study cell-intrinsic mechanisms of therapeutic response, and models such as those described above will be needed to consider the influence of cell-extrinsic factors on therapeutic response.
Pharmacogenomic differences between preclinical models
Several experimental model systems are available to study the molecular underpinnings of cancer, and each has strengths and weaknesses. Here, we describe observed pharmacogenomic differences between several preclinical models, such as xenografts, 3D culturing, and conventional monolayer culturing. Early studies showed differences in morphology and growth between monolayer culturing and xenograft models of cell lines (81), but the pharmacogenomics profiles between preclinical models have been compared only recently.
Alterations in cell lines that are grown in 3D culture (e.g., Matrigel) are similar to those found in cells grown in a monolayer. Kenny and colleagues grew matched cell lines in monolayer or 3D cultures and analyzed differences in gene expression using micro-arrays. Global changes were found to be minimal, as each cell line clustered with itself, regardless of culturing method. However, there were consistent changes in genes related to signal transduction between the two groups (82). Several other studies have indicated that cells grown in 3D versus monolayer culture have differences in global gene expression, notably increased extracellular matrix genes (83, 84). These differences are likely due to altered environmental signaling and mechanical composition compared with those in conventional monolayer culture.
The cell culture approach itself can also influence sensitivity to therapy. Alijitwi and colleagues showed leukemia cells were more resistant to chemotherapeutics when cocultured with human bone marrow mesenchymal stem cells in a 3D model versus monolayer (85). In addition, Chambers and colleagues showed that prostate cancer cell lines grown in 3D culture had different responses to docetaxel than those grown in monolayer (86). Other studies have observed pharmacologic differences in 3D culturing (87, 88). Altogether, these observations identify important differences between 3D and monolayer culture, indicating that response to therapy is driven by both the extracellular environment and intrinsic biology.
The genomic differences between xenograft and monolayer models have also been explored. Daniel and colleagues used primary xenografts of human small–cell lung cancer to derive cell lines and then used those cell lines in secondary xenograft experiments (89). The authors were able to identify gene expression differences between primary xenografts and monolayer culturing of the cell lines, suggesting genomic alterations between the two model systems. These genes remained differentially expressed in the secondary xenograft, suggesting stable changes had occurred in monolayer culture.
Ding and colleagues compared mutations between a matched primary breast tumor, a brain metastases, and a patient-derived xenograft model (PDX; ref. 90). While mutations were similar for each type, some mutations were enriched in the metastases and the PDX compared with the primary tumor. This study suggests that many of the genomic aberrations of the primary tumor are conserved during metastasis and at least early PDX models; however, selection that can have a strong impact on the model under study can occur. Pandita and colleagues used tumor glioblastoma multiforme (GBM) biopsies coupled with serial xenografts and the establishment of cell lines to find that EGFR amplifications are conserved in vivo but lost in vitro (91). Later studies in GBM confirmed that alterations of EGFR, PDGFRA, TP53, PTEN, and CDKN2A in patient tumors were conserved in flank and intracranial xenografts (92). These studies show that EGFR amplifications in GBM are selected against in monolayer culture, but are conserved across xenograft models.
PDX models and xenografts using human cell lines have some similar shortcomings. Immunooncology employs various ways to “educate” the immune system to better fight cancer. Because human cells cannot be xenografted into immunocompetent mice, immunooncology research most often is performed in mouse models lacking human cells, outlining one area of research where the use of human cancer cell lines is limited. However, the development of humanized mouse models has contributed to the study of immunooncology (reviewed by Lodhia and colleagues; ref. 93).
Another issue with these human xenograft models is that important components of the TME are lacking, including cells of the immune system. In addition, not all cancer cell lines are tumorigenic which limits the number of cell lines that are amenable for in vivo studies6. Xenograft models also display features of species incompatibility. For example, mouse growth factors do not activate human MET, which decreases the effectiveness of in vivo models in regards to MET-related studies (94). These disadvantages suggest to some that syngeneic or genetically engineered mouse models may be superior to human xenograft models because of species compatibility and intact immune systems. However, Voskoglou-Nomikos and colleagues have shown that the clinical translatability of murine allograft models were poorer compared to the advantages and disadvantages, and their vulnerabilities should be considered before their use (reviewed by Gould and colleagues; ref. 95) to models that contain human cells (96). The different in vivo models each offer an array of.
Overall, the studies we described outline the differences that occur in gene expression and pharmacologic profiling of mono-layer cultured cells versus other in vitro models. To a large degree, these differences are not surprising. The choice of experimental model must be dictated by the question being asked. Cell lines are not perfect models, but have strengths that include ease of use, low cost, and utility in diverse experimental studies. There is evidence in support of xenograft (96) and 3D models (75) more closely resembling tumor biology. However, like conventional cell line studies, these models fail to capture the diversity of microenvironments that tumor cells encounter in patients. In general, the limitations and differences associated with any model system should be considered when assessing the likelihood that observations made from it will be predictive of clinical behavior.
Discussion
Cell lines have long been used to study the mechanisms underlying biologic processes. Large collections of -omically functionally characterized cell lines are now available as a resource to the biomedical community. These data are now being used to develop analytic approaches to predict the behavior of complex adaptive systems, and to identify -omic features that provide insights into the mechanisms of therapeutic response. Central to these studies is the availability of well-characterized genomic measurements. Large, publicly available datasets, like the ones listed in Table 1, offer the level of genomic measurement needed to compare cell lines and tumors.
The comparisons of cell line and tumor genomics have offered insights into the relevance of in vitro models. In many instances, it is appropriate to use cell line genomics as a model for in vivo genomics; however, this is only true if due diligence is performed. Domcke and colleagues showed that the cell lines most commonly used to study HGSOC did not closely model the genomics of the disease, and that less frequently used models show stronger genomic similarities to primary disease (14). In addition, not all tumor subtypes are represented in current panels of cell lines (30– 32). This realization has spurred the generation of new cell lines and 3D culturing techniques that more closely reflect the features of the primary tumors from which they were derived.
In all cases, it is important to assess the extent to which cell lines accurately model the aspects of tumor biology being studied. With the advancement of high-throughput data generation techniques, we now have -omic profiles on over 1,000 human cancer cell lines to choose from, and the list is growing (97). These resources offer the data needed to systematically select the appropriate cell line genomic model for studying a primary tumor.
Omic studies have identified molecular features that are common to several different cancer types. These common features are likely related to the epithelial and mesenchymal origins for the tumors and persist in tumor-derived cancer cell lines. This has been demonstrated in breast and bladder cancers (25, 33). Molecular subtyping can inform patient treatment decisions, but caution must be used when applying molecular subtype signatures to cancer cell lines, as we demonstrated in Fig. 1.
Genomic measurements do not capture all aspects of cellular activity and integrative models provide greater predictive power than individual datasets. We expect that future pharmacogenomics studies will expand in the genomics space by leveraging next-generation sequencing to broaden the spectrum of molecules sampled, namely noncoding RNAs, alternative spliced transcripts, gene fusions, and allele-specific expression. Beyond genomics, we also expect to see an expansion into other -omic data types such as metabolomics and proteomics. From the analysis of data individually or from data integration approaches will come an increasing number of hypotheses to be tested. Cancer cell lines will remain a viable tool in this context and play a central role in cancer biology research into the future.
Acknowledgments
Grant Support
This study was financially supported by the Denver Chapter of the Golfers Against Cancer, NIH grants 5P30CA069533-16, U54 HG008100, U54 CA 112970, the Prospect Creek Foundation, and the Susan G. Komen Foundation SAC110012.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 2.The Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 4.Shankavaram UT, Varma S, Kane D, Sunshine M, Chary KK, Reinhold WC, et al. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics. 2009;10:277. doi: 10.1186/1471-2164-10-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heiser LM, Sadanandam A, Kuo W-L, Benz SC, Goldstein TC, Ng S, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daemen A, Griffith OL, Heiser LM, Wang NJ, Enache OM, Sanborn Z, et al. Modeling precision treatment of breast cancer. Genome Biol. 2013;14:R110. doi: 10.1186/gb-2013-14-10-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 8.Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–637. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ross DT, Perou CM. A comparison of gene expression signatures from breast tumors and breast tissue derived cell lines. Dis Markers. 2001;17:99–109. doi: 10.1155/2001/850531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 13.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Wawrose JS, Gooding WE, Garraway LA, Lui VWY, Peyser ND, et al. Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Mol Cancer Res. 2014;12:571–582. doi: 10.1158/1541-7786.MCR-13-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 17.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sos ML, Michel K, Zander T, Weiss J, Frommolt P, Peifer M, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, et al. A Systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- 20.Cope LM, Fackler MJ, Lopez-Bujanda Z, Wolff AC, Visvanathan K, Gray JW, et al. Do breast cancer cell lines provide a relevant model of the patient tumor methylome? PLoS ONE. 2014;9:e105545. doi: 10.1371/journal.pone.0105545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 24.Cho Y-J, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prat A, Bianchini G, Thomas M, Belousov A, Cheang MCU, Koehler A, et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH Study. Clin Cancer Res. 2014;20:511–521. doi: 10.1158/1078-0432.CCR-13-0239. [DOI] [PubMed] [Google Scholar]
- 27.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virtanen C, Ishikawa Y, Honjoh D, Kimura M, Shimane M, Miyoshi T, et al. Integrated classification of lung tumors and cell lines by expression profiling. Proc Natl Acad Sci. 2002;99:12357–12362. doi: 10.1073/pnas.192240599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kao J, Salari K, Bocanegra M, Choi Y-L, Girard L, Gandhi J, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS ONE. 2009;4:e6146. doi: 10.1371/journal.pone.0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci. 2014;111:3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JK, Havaleshko DM, Cho H, Weinstein JN, Kaldjian EP, Karpovich J, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proc Natl Acad Sci. 2007;104:13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 37.Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M, et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proc Natl Acad Sci. 2009;106:14016–14021. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312–319. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 39.Alsner J, Yilmaz M, Guldberg P, Hansen LL, Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Breast Cancer Res. 2000;2 doi: 10.1186/bcr109. P4.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maley CC, Galipeau PC, Finley JC, Wongsurawat VJ, Li X, Sanchez CA, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 41.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- 43.Nagai Y, Miyazawa H, Huqun, Tanaka T, Udagawa K, Kato M, et al. Genetic heterogeneity of the epidermal growth factor receptor in non–small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid-locked nucleic acid PCR clamp. Cancer Res. 2005;65:7276–7282. doi: 10.1158/0008-5472.CAN-05-0331. [DOI] [PubMed] [Google Scholar]
- 44.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci U S A. 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harper LJ, Piper K, Common J, Fortune F, Mackenzie IC. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med. 2007;36:594–603. doi: 10.1111/j.1600-0714.2007.00617.x. [DOI] [PubMed] [Google Scholar]
- 47.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 48.Paull KD, Shoemaker RH, Hodes L, Monks A, Scudiero DA, Rubinstein L, et al. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: development of mean graph and COMPARE algorithm. J Natl Cancer Inst. 1989;81:1088–1092. doi: 10.1093/jnci/81.14.1088. [DOI] [PubMed] [Google Scholar]
- 49.Scherf U, Ross DT, Waltham M, Smith LH, Lee JK, Tanabe L, et al. A gene expression database for the molecular pharmacology of cancer. Nat Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- 50.Bhatla T, Wang J, Morrison DJ, Raetz EA, Burke MJ, Brown P, et al. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood. 2012;119:5201–5210. doi: 10.1182/blood-2012-01-401687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz CC, Dantis L, et al. Phase II study of weekly intravenous recombinant humanized anti-p185HER2 monoclonal antibody in patients with HER2/neu-overexpressing metastatic breast cancer. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]
- 52.Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2639. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- 53.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 54.Yu M, Selvaraj SK, Liang-Chu MMY, Aghajani S, Busse M, Yuan J, et al. A resource for cell line authentication, annotation and quality control. Nature. 2015;520:307–311. doi: 10.1038/nature14397. [DOI] [PubMed] [Google Scholar]
- 55.Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJWL, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weinstein JN, Lorenzi PL. Cancer: discrepancies in drug sensitivity. Nature. 2013;504:381–383. doi: 10.1038/nature12839. [DOI] [PubMed] [Google Scholar]
- 57.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costello JC, Heiser LM, Georgii E, Go€nen M, Menden MP, Wang NJ, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol. 2014;32:1202–1212. doi: 10.1038/nbt.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:237–245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Consortium TEP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 61.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bansal M, Yang J, Karan C, Menden MP, Costello JC, Tang H, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol. 2014;32:1213–1222. doi: 10.1038/nbt.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Griner LAM, Guha R, Shinn P, Young RM, Keller JM, Liu D, et al. High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci. 2014;111:2349–2354. doi: 10.1073/pnas.1311846111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 65.Bean J, Brennan C, Shih J-Y, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parisot JP, Hu XF, DeLuise M, Zalcberg JR. Altered expression of the IGF-1 receptor in a tamoxifen-resistant human breast cancer cell line. Br J Cancer. 1999;79:693–700. doi: 10.1038/sj.bjc.6690112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Massarweh S, Osborne CK, Creighton CJ, Qin L, Tsimelzon A, Huang S, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function. Cancer Res. 2008;68:826–833. doi: 10.1158/0008-5472.CAN-07-2707. [DOI] [PubMed] [Google Scholar]
- 68.Drury SC, Detre S, Leary A, Salter J, Reis-Filho J, Barbashina V, et al. Changes in breast cancer biomarkers in the IGF1R/PI3K pathway in recurrent breast cancer after tamoxifen treatment. Endocr Relat Cancer. 2011;18:565–577. doi: 10.1530/ERC-10-0046. [DOI] [PubMed] [Google Scholar]
- 69.Cowley GS, Weir BA, Vazquez F, Tamayo P, Scott JA, Rusin S, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 71.Jerby-Arnon L, Pfetzer N, Waldman YY, McGarry L, James D, Shanks E, et al. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell. 2014;158:1199–1209. doi: 10.1016/j.cell.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 72.Sullivan KD, Padilla-Just N, Henry RE, Porter CC, Kim J, Tentler JJ, et al. ATM and MET kinases are synthetic lethal with non-genotoxic activation of p53. Nat Chem Biol. 2012;8:646–654. doi: 10.1038/nchembio.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 74.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 75.Weigelt B, Ghajar CM, Bissell MJ. The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv Drug Deliv Rev. 2014;69–70:42–51. doi: 10.1016/j.addr.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanahan D, Weinberg RA. Hallmarks of Cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 77.Winslow S, Leandersson K, Edsjo A, Larsson C. Prognostic stromal gene signatures in breast cancer. Breast Cancer Res. 2015;17:23. doi: 10.1186/s13058-015-0530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajski M, Vogel B, Baty F, Rochlitz C, Buess M. Global gene expression analysis of the interaction between cancer cells and osteoblasts to predict bone metastasis in breast cancer. PLoS ONE. 2012;7:e29743. doi: 10.1371/journal.pone.0029743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–5757. [PubMed] [Google Scholar]
- 80.McMillin DW, Delmore J, Weisberg E, Negri JM, Geer DC, Klippel S, et al. Tumor cell-specific bioluminescence platform to identify stroma-induced changes to anticancer drug activity. Nat Med. 2010;16:483–489. doi: 10.1038/nm.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tveit KM, Pihl A. Do cell lines in vitro reflect the properties of the tumours of origin? A study of lines derived from human melanoma xenografts. Br J Cancer. 1981;44:775–786. doi: 10.1038/bjc.1981.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hong H, Stegemann JP. 2D and 3D collagen and fibrin biopolymers promote specific ECM and integrin gene expression by vascular smooth muscle cells. J Biomater Sci Polym Ed. 2008;19:1279–1293. doi: 10.1163/156856208786052380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu H, Lin J, Roy K. Effect of 3D scaffold and dynamic culture condition on the global gene expression profile of mouse embryonic stem cells. Biomaterials. 2006;27:5978–5989. doi: 10.1016/j.biomaterials.2006.05.053. [DOI] [PubMed] [Google Scholar]
- 85.Aljitawi OS, Li D, Xiao Y, Zhang D, Ramachandran K, Stehno-Bittel L, et al. A novel three-dimensional stromal-based model for in vitro chemotherapy sensitivity testing of leukemia cells. Leuk Lymphoma. 2014;55:378–391. doi: 10.3109/10428194.2013.793323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chambers KF, Mosaad EMO, Russell PJ, Clements JA, Doran MR. 3D cultures of prostate cancer cells cultured in a novel high-throughput culture platform are more resistant to chemotherapeutics compared to cells cultured in monolayer. PLoS ONE. 2014;9:e111029. doi: 10.1371/journal.pone.0111029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ, et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855–860. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 88.Muranen T, Selfors LM, Worster DT, Iwanicki MP, Song L, Morales FC, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer Cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69:3364–3373. doi: 10.1158/0008-5472.CAN-08-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pandita A, Aldape KD, Zadeh G, Guha A, James CD. Contrasting in vivo and in vitro fates of glioblastoma cell subpopulations with amplified EGFR. Genes Chromosomes Cancer. 2004;39:29–36. doi: 10.1002/gcc.10300. [DOI] [PubMed] [Google Scholar]
- 92.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-Oncol. 2005;7:164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lodhia KA, Hadley AM, Haluska P, Scott CL. Prioritizing therapeutic targets using patient-derived xenograft models. Biochim Biophys Acta. 2015;1855:223–234. doi: 10.1016/j.bbcan.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gould SE, Junttila MR, de Sauvage FJ. Translational value of mouse models in oncology drug development. Nat Med. 2015;21:431–439. doi: 10.1038/nm.3853. [DOI] [PubMed] [Google Scholar]
- 96.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res. 2003;9:4227–4239. [PubMed] [Google Scholar]
- 97.Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2015;33:306–312. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]