Summary
A non-coding hexanucleotide repeat expansion in the C9ORF72 gene is the most common mutation associated with familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Patients harboring this expansion develop several unique histopathological hallmarks, including intranuclear foci composed of either sense or antisense RNA transcripts from the expanded repeats and dipeptide repeat proteins generated by non-canonical translation of the expanded RNA transcripts. To further investigate the pathological role of C9ORF72 in these diseases, we generated a line of mice carrying a bacterial artificial chromosome containing exons 1 to 6 of the human C9ORF72 gene with approximately 500 repeats of the GGGGCC motif. The mice showed no overt behavioral phenotype but recapitulated distinctive histopathological features that are the hallmark of C9ORF72 ALS/FTD, including sense and antisense intranuclear RNA foci and poly(glycine-proline) dipeptides repeat proteins. Finally, using a synthetic microRNA that targets human C9ORF72 in cultures of primary cortical neurons from the C9BAC mice, we have attenuated expression of the C9BAC transgene and the poly(GP) dipeptides. The C9ORF72 BAC transgenic mice will be a valuable tool in the study of ALS/FTD pathobiology and therapy.
Keywords: Amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FTD), C9ORF72, transgenic mice, RNA foci, RAN translation, repeat expansions, neurodegeneration
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that primarily affects motor neurons in the motor cortex, brainstem and spinal cord, whereas frontotemporal dementia (FTD) is caused by the degeneration of neurons in the fronto-temporal regions; intriguingly however, some individuals experience both disorders simultaneously. Our understanding of these diseases was transformed by the identification of an abnormally expanded GGGGCC repeat motif within the gene chromosome 9 open reading frame 72 (C9ORF72) [1-3]. Located in a non-coding region of C9ORF72 the mutation is now recognized as the most common cause of dominantly inherited ALS and FTD, referred to as c9ALS/FTD; in Caucasian populations it accounts for approximately 40% of familial ALS, 4-8% of sporadic ALS [1, 3, 4] and 20% of hereditary and 6% of sporadic FTD [5, 6]. In control individuals the GGGGCC repeat motif is believed to contain between 4 and 30 repeats [7]. Larger repeats are associated with pathology, although the precise threshold of GGGGCC repeats required for disease penetrance is unclear; c9ALS/FTD patients with greater than 2,000 have been reported [1, 3, 8]. There is no apparent association between expansion size and penetrance of ALS or FTD phenotypes [8].
The C9ORF72 gene encodes three transcript variants (Figure S1). Variants 2 and 3 (V2, V3) contain the intronic expansion and encode two distinct protein isoforms [1]. Presently the normal function of the C9ORF72 product is unknown, though structural homology with the rab-associated GDP/GTP exchange factor protein differentially expressed in normal and neoplastic cells (DENN) suggests a role in membrane trafficking [9, 10]. Decreased levels of C9ORF72 transcripts have been described in c9ALS/FTD patient tissues and derived cell lines [1, 2, 11, 12], suggesting that loss of C9ORF72 function via haploinsufficiency might contribute to disease. Indeed, deletion of the C9ORF72 homolog in C. elegans has been reported to result in axonopathy and motor deficits [13]. Reduced expression of C9ORF72 mRNA in c9ALS/FTD cases has been associated with hypermethylation of CpG islands upstream of the C9ORF72 promoter [14] and methylation of GGGGCC-binding histones [11]. Furthermore, the GGGGCC repeat forms highly stable DNA G-quadruplexes [15] that are reported to impair transcription [16].
It is also possible that the intronic C9ORF72 repeat expansion confers one or more toxic properties that compromise neuron viability. Bidirectional transcription of the mutated C9ORF72 generates GGGGCC sense and CCCCGG antisense repeat containing RNA transcripts that form intranuclear RNA foci [1, 17, 18]. These foci potentially act as new binding sites for specific RNA binding proteins, which may become abnormally sequestered, thereby impairing their function [19-22]. By the same token, it is conceivable that the expanded DNA tracts may also titrate out or otherwise sequester DNA-binding factors. Beyond that, it is also now evident that the expanded GGGGCC RNA transcripts undergo repeat-associated non-ATG (RAN) translation, a non-canonical mechanism of protein synthesis that yields dipeptide repeat (DPR) proteins (GP, GA, GR for the sense transcript, PA, PR and again GP for the anti-sense transcript) [20, 23-25]. These DPR proteins aggregate and form cytoplasmic inclusions in c9ALS/FTD and are toxic in varying degrees in different model systems; arginine-rich peptides are apparently the most toxic [26-30].
Model systems that recapitulate the pathobiology of the mutant C9ORF72 gene are particularly helpful in the quest to understand how such mutations cause neurodegeneration in vivo. Several invertebrate [13, 27, 29, 31], vertebrate [32] and patient derived cell models [12, 33, 34] have been reported. While each of these models will provide insight, it can be argued that a more informative model will permit studies of the C9ORF72 expanded GGGGCC repeat in the context of stable expression in an intact mammalian nervous system. For that reason, we have generated a novel transgenic mouse carrying an expanded form of the human C9ORF72 gene carrying approximately 500 GGGGCC motifs.
Results
BAC transgenic mice express the human mutant C9ORF72 gene
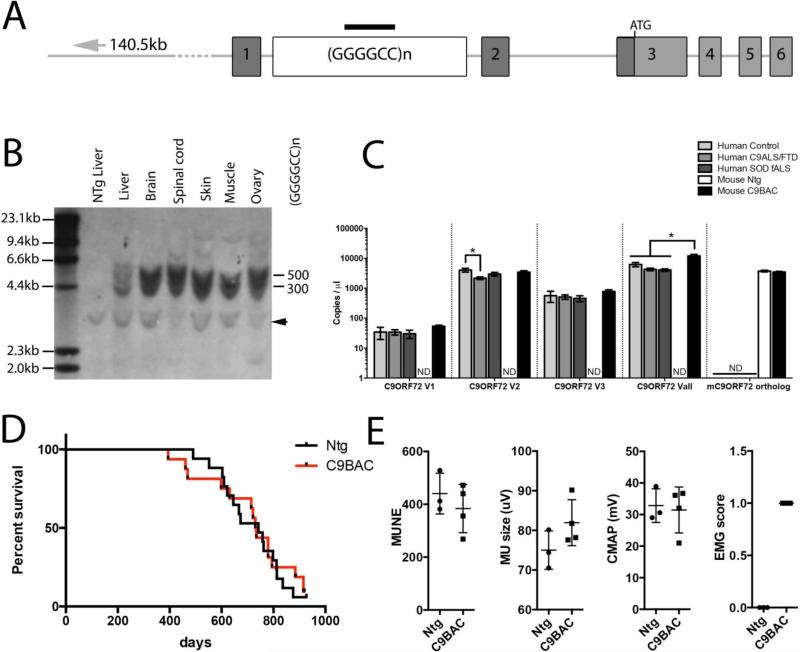
We generated a line of SJL/B6 transgenic mice using a 153.2kb bacterial artificial chromosome containing exons 1 through 6 of the human C9ORF72 with about 500 GGGGCC repeat motifs, including approximately 140.5kb of upstream sequence (Fig 1A, Fig S1A). Hemizygous C9BAC mice were viable, producing progeny at expected Mendelian frequencies (data not shown). Southern blot analysis of genomic DNA from these animals detected two distinct bands corresponding to about 500 and 300 GGGGCC repeat motifs (Fig 1B, Fig S1B). This profile showed intergenerational stability, with two consistently sized bands seen in distantly related F6 animals (Fig S1B). Similarly, no pronounced changes in the expansion size were evident in multiple tissues (brain, spinal cord, liver, skin, muscle, ovaries) (Fig 1B). Using a human specific probe for all variants (Vall) C9ORF72 mRNA transcripts were detected in the C9BAC mice in all tissues examined (Fig S1C). The truncated human C9ORF72 gene generates three transcript variants; V1 and V3 carry the repeat expansion (Fig S1A). Human specific probes targeting human C9ORF72 V1, V2 and V3 mRNAs detected all three transcripts in the C9BAC mice at expression levels relative to their abundances in human frontal cortex (Fig 1C). Moreover the total level of the transgenic human transcripts was roughly comparable to that of the endogenous mouse C9ORF72 ortholog transcripts, and to levels of expression of human control and c9ALS/FTD cases (Fig 1C).
Figure 1. Construct design, expansion size and expression profile for the C9ORF72 BAC transgene and phenotyping of C9BAC mice.
(A) Schematic of the bacterial artificial chromosome fragment used for generating the C9BAC mice. The 153.2kb construct contains (as shown from left to right) 140.5kb of human genomic DNA upstream of C9ORF72, the human C9ORF72 promoter region, and exons 1 to partial exon 6 with a (GGGGCC)500 repeat motif in C9ORF72 intron 1. The black bar indicates the target region for the Southern blotting probe used in B. (B) Southern blot of genomic DNA extracted from various tissues digested with HindIII, SacI, and TaqI and probed with a 5’-DIG-(GGGGCC)5-DIG-3’ DNA probe. Two bands of ~4.4kb and ~6.0kb, are stable in size after 6 generations representing expansions of 300 and 500 repeats (also Fig. S1B). The arrowhead indicates a non-specific band. (C) ddPCR analysis of expression of all human C9ORF72 variants (Vall), and variants V1, V2 and V3 and the mouse C9ORF72 ortholog in human frontal cortex tissues from non-neurological disease control, c9ALS/FTD and SOD fALS patients, and whole brain homogenate from C9BAC and Ntg mice (mean±SEM, n=4, Kruskal-Wallis test, Mann-Whitney U-test, * p<0.05, ND = not detectable). (D) Kaplan-Meier curve representing lifespan showing no significant difference in survival of C9BAC mice compared to Ntg littermates (Ntg n=17, C9BAC n=16, Mandel-Cox Log Rank, P=0.971). (E) Physiological studies reveal showed no significant change in motor unit number estimations (MUNE) motor units size or compound motor action potential, though a modest increase in electromyogram score was observed (mean±SEM, Ntg n=3, C9BAC n=4 unpaired t-test).
Survival, motor and cognitive systems are normal in mice bearing the mutant C9ORF72 gene
A cohort of F1 generation male C9BAC mice was aged for phenotypic characterization and analysis of survival. At all ages, the C9BAC and control mice remained healthy. The C9BAC mice trended towards a non-significantly elevated weight compared to Ntg littermates (Fig S1D, Ntg n=17, C9BAC n=15, 2-way ANOVA, Bonferroni's multiple comparison test) and lived beyond two years. Survival in the C9BAC and Ntg littermates was indistinguishable (Fig 1D, Ntg n=17, C9BAC n=16, Mantel-Cox Log-rank, P=0.971).
C9ORF72 mutations are associated with both ALS and FTD. We therefore tested for behavioral and histological abnormalities within the motor system of the C9BAC mouse. Overall, the C9BAC mice showed no significant motor deficits by rotarod performance and grip strength testing. (Fig S1E-F, Ntg n=17, C9BAC n=15, 2-way ANOVA, Bonferroni's multiple comparison test). Semi-thin sectioning of L5 ventral nerve motor axons from 24-month-old C9BAC and Ntg mice revealed an atypical morphology of thin myelination and some evidence of degenerative axons that were comparable in both aged groups (Fig S2A). Such changes were not observed in the sensory dorsal nerve roots of 24-month-old animals, or sciatic nerves of 10-month old animals (data not shown). Quantification of axon number and diameter revealed no significant differences in C9BAC mice (Fig S2A, Ntg n=3, C9BAC n=4, 2-way ANOVA, Bonferroni's multiple comparison). Immunostaining of the axon and post-synaptic receptors in neuromuscular junctions of the gastrocnemius muscle revealed only minimal denervation in 24-month C9BAC mice to a degree that matched that in controls (Fig S2B, Ntg n=4, 699 NMJs assessed, C9BAC n=4, 675 NMJs assessed; 2-way ANOVA). Motor unit number estimation (MUNE) recording in the hind limbs of 24-month C9BAC mice detected no changes in MUNE score, motor unit size or compound motor action potential, although there was a trend towards a modestly increased denervation score, as gauged by electromyography (Fig 1E, Ntg n=3, C9BAC n=4, unpaired t-tests).
We also assessed several pathological features in the motor cortex and spinal cord that are common features of ALS. No significant changes were seen in activation of microgliosis or astrogliosis (Fig S2C,E). Cytoplasmic mislocalization and aggregation of TDP-43, both identified in the majority of ALS patients [7, 35], were not detected in the motor cortex of our C9BAC mice (Fig S2D), nor did we see alteration in the levels of TDP-43 or the inclusion associated protein p62 (Fig S2E). To assess activation of programmed cell death within the spinal cord, we observed levels of cleaved capase-3 by Western blotting; levels in C9BAC and Ntg mice did not differ significantly (Fig S2F). Recent studies have demonstrated cortical hyperexcitability in patients with C9ORF72-ALS using transcranial magnetic stimulation [36]. Therefore, we next asked whether neurons from neonatal (P4-P5) C9BAC mice showed altered electrophysiological characteristics, targeting the two main excitatory projection neurons in layer 5 (L5) of motor cortex for analysis, pyramidal tract (PT)-type, which include corticospinal neurons, and intratelencephalically projecting (IT)-type neurons, which include corticocortical neurons [37]
To compare the intrinsic electrophysiological properties between non-transgenic control and C9BAC neurons, whole-cell patch clamp recordings were performed in acutely prepared brain slices (Fig. S2G-J). Their electrophysiological characteristics were tested in the presence of blockers of glutamatergic and GABAergic synaptic transmission (NBQX, 5 μM, AMPA receptor antagonist; CPP, 5 μM, NMDA receptor antagonist; and SR95531, 10 μM, GABAA receptor antagonist). We found no significant differences in the resting membrane potential, input resistance, sag amplitude, or action potential properties between control and C9BAC neurons for PT-type neurons (Fig. S2G, Table 1). The results were similar for IT-type neurons (Fig. S2I, Table 1). To compare the intrinsic excitability of neurons between genotypes, we evoked action potentials with a series of depolarizing current steps. We found that neither PT-type (Fig. S2H) nor IT-type neurons (Fig. S2J) showed a significant difference in the rheobase or the current-spike frequency relationship between control and mutant neurons. These results indicate that the electrophysiological properties of projection neurons in L5 of motor cortex are not altered in these neonatal C9BAC mice.
We next surveyed pathological findings that are common in FTD. No activation of gliosis was detected in the prefrontal cortex (PFC) or hippocampus (Fig S3A-B). Because changes in social interaction behavior have been described in rodent models of FTD [38, 39], we tested the response of C9BAC and Ntg mice to an intruder mouse. No deficits were detected in the duration of interaction between 18-22 month old C9BAC males and an unfamiliar juvenile male mouse inserted into the older mouse's home cage (Fig S3C, Ntg n=7, C9BAC n=9, unpaired t-test). Quantitative studies of Golgi preparations revealed no change in dendritic spine density of PFC layer 2-3 pyramidal neuron apical dendrites in aged C9BAC mice (Fig S3D).
The gene expression profile in C9BAC mice PFC is unchanged compared to non-transgenic littermates
It has been reported in other repeat disorders that the presence of multiple, small intranuclear RNA foci disturbs the function of some RNA binding proteins and the correct processing of their target RNA transcripts [40]. Altered gene expression has been shown in human fibroblast and induced pluripotent stem cells (iPSC)-derived motor neurons from C9ORF72 ALS patients [12, 33, 34, 41], with altered splicing and polyadenylation profiles demonstrated in cerebellum of c9ALS patients [42]. To determine whether a specific C9ORF72 RNA profile can be defined in our C9BAC transgenic mice, we sequenced total RNA extracted from the frontal cortex of five different six-month old C9BAC mice and three littermate controls. For each sample, at least 25 million reads mapped to a unique location of the mouse genome (version mm10). Hierarchical clustering of expression values of all genes failed to show a distinct RNA profile for the five C9BAC mice compared to their littermate controls. Statistical analysis of differentially expressed genes did not reveal any change between the two groups, including several genes identified as differentially expressed in iPSC-derived motor neurons ([12, 34], Figs S3E-H).
C9BAC mice recapitulate histopathological features of human C9ORF72 ALS-FTD
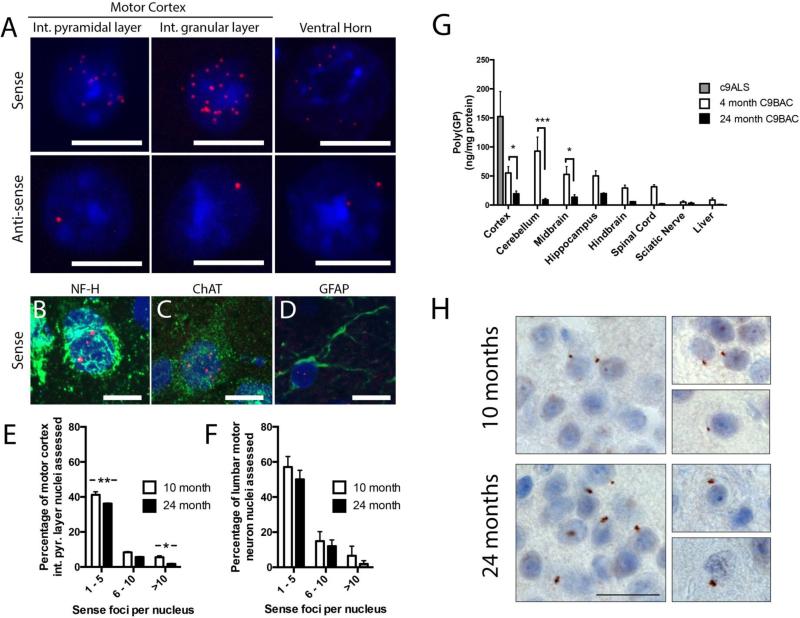
At autopsy, brains from c9ALS/FTD patients show several histopathological characteristics that are not detected in other forms of familial and sporadic ALS or FTD: (i) sense transcript nuclear and occasional cytoplasmic RNA foci, (ii) antisense transcript nuclear RNA foci and (iii) the generation of DPR proteins through RAN translation of the sense and antisense transcripts. We used fluorescence in situ hybridization (FISH) to detect C9ORF72 sense and antisense foci in the brain and spinal cords of C9BAC mice (Fig 2). Intranuclear, sense foci were detected at 3 months of age (data not shown) and by 10 and 24 months were abundant throughout the CNS (Fig 2A). Foci could be detected in c9ALS/FTD relevant regions, including internal and external pyramidal layer nuclei of the motor cortex and ventral horn motor neurons of the lumbar spinal cord. Sense foci were detected in neurofilament-H positive neurons throughout the brain (Fig 2B), cholinergic neurons of the cortex (Fig 2C) and activated astrocytes throughout the brain (Fig 2D). Antisense foci were also detected in 10- and 24-month old mice. However, by comparison with the sense foci, the anti-sense foci were more sparsely distributed throughout the brain (Fig 2A). Quantification of numbers of foci in nuclei of the external pyramidal layer (Fig 2E) and spinal motor neurons (Fig 2F) revealed that the majority of foci-positive nuclei contain one to five detectable sense foci, though in some cases greater than 20 foci were seen in individual nuclei. A trend towards a reduced number of nuclei containing foci was seen between 10 and 24-months, reaching statistical significance in the motor cortex external pyramidal layer (10-month, n=3, 821 nuclei assessed, 55.2%±2.3%; 24-month, n=3, 845 nuclei assessed, 43.7%±0.6; two-way ANOVA, Bonferroni's multiple comparison, p<0.05), but not spinal cord motor neurons (10-month, n=3, 124 nuclei assessed, 78.5%±4.7%; 24-month, n=3, 100 nuclei assessed 63.9%±9.8%, n=3; two-way ANOVA).
Figure 2. C9BAC mice recapitulate the histopathological hallmarks of C9ORF72 ALS and FTD patients.
(A) Fluorescence in situ hybridization (FISH) using Cy3-tagged DNA probes against sense and anti-sense abnormally expanded C9ORF72 transcripts (Red = probe, Blue = DAPI nuclear stain). The C9ORF72 repeat containing sense or anti-sense transcripts form intranuclear RNA inclusions, or foci, throughout the brain, including motor cortex internal pyramidal layer and internal granular layer, and lumbar motor neurons of 24-month old C9BAC mice. FISH coupled with cell type specific immunostaining revealed sense transcript foci in neurofilament heavy positive neurons (B), choline acetyl transferase (ChAT) positive pyramidal neurons (C) and GFAP positive astrocytes (D). The number of sense transcript foci per nucleus decreased between 10-month and 24-month old C9BAC mice in both motor cortex internal pyramidal layer (E) and lumbar motor neuron nuclei (F) (mean±SEM, 2-way ANOVA, Bonferroni multiple comparison * p<0.05, ** p<0.001). (G) Quantification of soluble poly(GP) proteins in various regions of the CNS and liver of 4- and 24-month old C9BAC mice and frontal cortex of C9ALS cases by immunoassay (mean±SEM, C9BAC groups n=3, c9ALS n=6, two-way ANOVA, Bonferroni's multiple comparison, * p<0.05, *** p<0.001). (H) Immunostaining for poly(GP) proteins in cortex of C9BAC mice. Staining revealed the presence of small perinuclear inclusion bodies throughout the brain at 10-months of age, with a pronounced elevation in the number observed in 24-month old mice. Such inclusions were not observed in age-matched Ntg mice. Scale bars A - D = 10 μm, H = 20 μm.
Because both sense and antisense C9ORF72 transcripts were present in the C9BAC mice, we next sought evidence of RAN translation products, using a sandwich immunoassay for the detection of poly(Glycine-Proline) (poly(GP)), which is synthesized from both sense and antisense transcripts. Tissues from 4- and 24-month old Ntg and C9BAC mice, and frontal cortex from six c9ALS case, were homogenized in buffer containing 2% SDS, with the detergent soluble fraction analyzed for poly(GP) content. At 4 months of age, poly(GP) was detected throughout the brain of C9BAC mice, with levels highest in the cerebellum, and also detected in the spinal cord, sciatic nerve and liver (Fig 2G). Though the mean concentration of poly(GP) levels in 4 month mice was lower than that observed in c9ALS frontal cortex (4-month C9BAC 55.2±10.9ng/mg protein, n=3; human c9ALS 152.4±43.1ng/mg protein, n=6), the 4-month C9BAC mean did fall within range of the lowest samples in the patient group (Fig 2G). All tissues from 24-month old C9BAC mice showed a significant reduction in SDS-soluble poly(GP) levels (4-month cortex 55.2±10.9ng/mg protein, n=3; 24-month cortex 19.4±4.7ng/mg protein, n=3; two-way ANOVA, Bonferroni's multiple comparison, p<0.05). One hypothesis for the observed decline in soluble poly(GP) is that the solubility of this DPR protein decreases with age. Immunostaining of brain tissues from 10- and 24-month old C9BAC mice revealed the presence of small perinuclear inclusion bodies stained for poly(GP) (Fig 2H). Such inclusions were detected throughout the brain, including cortex, cerebellum and striatum. The inclusions were observed to increase in frequency in affected brain regions in the 24-month mice, suggesting a heightened deposition of insoluble poly(GP) species.
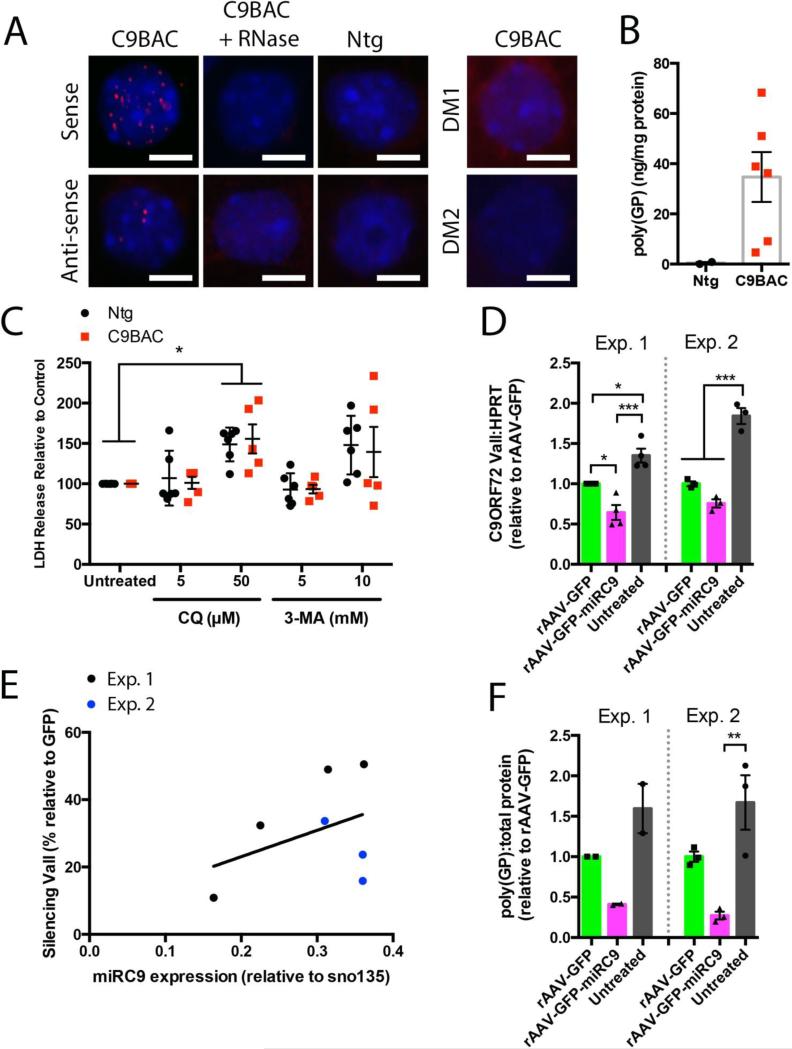
We next tested whether neurons from C9BAC mice are hypersensitive to cellular stress. In a previous study, neurons differentiated from induced pluripotent stem cells (iPSCs) derived from FTD patients showed an increased sensitivity to cell death induced by inhibition of autophagy in vitro [33]. We established primary cortical neuron cultures from C9BAC and non-transgenic littermate embryos. At 10 days in vitro (DIV) both nuclear foci containing sense or antisense C9ORF72 hexanucleotide expansion transcripts (Fig 3A) and poly(GP) RAN-translation products could be detected (Fig 3B). We treated 15 DIV primary cortical neuron cultures derived from individual embryos with two inhibitors of autophagy, chloroquine (Fig 3C) and 3-methyladenine. Both compounds increased cell death to the same degree in the C9BAC and Ntg neurons, as determined by an LDH assay (two independed experiments, individual embryos cultured; C9BAC n=6, Ntg n=5, 2-way ANOVA, Bonferroni's multiple comparison).
Figure 3. Silencing of C9ORF72 in C9BAC mice and primary cortical neurons.
(A) Sense and antisense foci are detected in cultured cortical neurons of C9BAC embryos at 10 days in vitro (DIV). These foci were not visible following RNase treatment and were not detected in Ntg neurons, or by probes targeting the myotonic dystrophy associated type-1 (CTG)n or type-2 (CCTG)n expansions (DM1 and DM2 respectively, scale bar = 5 μm). (B). Concentration of poly(GP) in cultured cortical neurons derived from individual C9BAC or Ntg littermate embryos measured by ELISA (mean±SEM, unpaired t-test, Ntg n=2, C9BAC n=6, unpaired t-test). (C) No significant differences were detected in the response of cultured cortical neurons derived from individual C9BAC and Ntg embryos to the autophagy inhibitors chloroquine or 3-methyladenine (two independed experiments, individual embryos cultured; C9BAC n=6, Ntg n=5, mean±SEM, 2-way ANOVA, Bonferroni's multiple comparison, * p<0.05). (D) ddPCR analysis of all transcripts of C9ORF72 (Vall) relative to HPRT in two independent experiments (Exp. 1. individual embryos n=4, mean±SEM, one-way ANOVA, Bonferroni's multiple comparison test, * p<0.05, *** p<0.0001; Exp. 2. mixed embryo culture, n=3, one-way ANOVA, Bonferroni's multiple comparison test *** p<0.001). (E) Levels of the mature microRNA product (normalized to snoRNA135) of rAAV-GFP-miRC9 correlated with percent silencing of C9ORF72 relative to rAAV-GFP from Exp. 1 (black) and Exp. 2 (blue). (F) Levels of poly(GP) quantified by ELISA from primary cortical neuron cultured derived from two independent experiments (Exp. 1. individual embryos n=2, mean±SEM, one-way ANOVA; Exp. 2. mixed embryo culture, n=3, one-way ANOVA, Bonferroni's multiple comparison test, ** p<0.001).
Silencing C9ORF72 transcripts and poly(DP) production in vitro
Though lacking cognitive or motor deficits, the C9BAC mouse robustly recapitulates the molecular pathology of c9ALS/FTD, with readily quantifiable expression of all three C9ORF72 transcripts, RNA foci and at least one of the poly(DP) RAN-translation products. Both of these read-outs rely on the expression of the C9ORF72 gene transcripts. We have therefore tested a gene therapy approach to silence expression of the C9ORF72 transgene in our C9BAC mice. We generated recombinant adeno-associated virus (rAAV) serotype 9 expressing an artificial microRNA (miR) targeting exon three of human C9ORF72 preceded by an eGFP reporter, under the transcriptional control of the CB-promoter and CMV-enhancer (rAAV-GFP-miRC9). For control groups, a vector expressing only the eGFP reporter was used (rAAV-GFP). We carried out two sets of experiments in DIV4 primary cortical neuron cultures (Fig. 3D, E, F). In the first (Exp 1), individual cultures were generated from separate C9BAC hemizygous embryos from the same litter of C9BAC mice; in the second (Exp 2), all transgenic embryos from a C9BAC litter were pooled to generate a mixed population of cortical neurons from multiple transgenic embryos. In both experiments, transgene transcripts and poly(GP) were reduced by expression of the GFP control vector alone in conjunction with some cell death; to account for this non-specific silencing, all silencing outcomes are normalized to this GFP control. In all experiments we consistently observed further depletion of C9ORF72 products in groups expressing the microRNA. Relative to the rAAV-GFP group, C9ORF72 mRNA transcript (Fig 3D) expression in individual embryo cultures (Exp 1) was decreased by 35% by rAAV-GFP-miRC9 (n=4, one-way ANOVA, Bonferroni's multiple comparison, p<0.05), and 24% in the pooled embryo cultures in Exp 2 (not statistically significant; n=3, one-way ANOVA, Bonferroni's multiple comparison). We detected mature miRNA only in the rAAV-GFP-miRC9 treated cultures, and found a correlation between its levels and percent silencing in the corresponding cultures (Fig. 3E). In two cultures derived from individual embryos (Exp 1), rAAV-GFP-miRC9 reduced levels of poly(GP) by ~60% (Fig. 3F). In mixed cultures (Exp 2), poly(GP) was reduced by 73% relative to GFP. Though not significant by one-way ANOVA (n=3, Bonferroni multiple comparison test), we note that in Exp 2 statistical testing of the rAAV-GFP vs rAAV-GFP-miRC9 (independent of the untreated group) detected a statistically significant decrease in poly(GP) levels (unpaired t-test, p=0.0008).
Discussion
The discovery of an expanded GGGGCC repeat motif in the non-coding region of the C9ORF72 gene in families in which ALS and FTD are co-inherited has defined a common molecular pathology that underlies both disorders. The mechanisms by which the mutated C9ORF72 gene is cytotoxic remain unclear but might include haploinsufficiency of the C9ORF72 gene, toxicities arising from the expanded tracts of RNA, and adverse effects of DPR proteins. One approach to understanding the pathological functions of GGGGCC-expanded C9ORF72 is to investigate this molecular defect in in vitro and in vivo model systems. Several such model systems have been reported, including iPSCs [12, 33, 34, 41], invertebrate models, such as the worm C. elegans and Drosophila [13, 27, 29, 31], zebrafish [32], mice lacking the C9ORF72 gene homolog [43] or expressing an exogenously delivered tract of GGGGCC repeats [44]. We report here a novel line of transgenic mice that harbor a portion of the human C9ORF72 gene with an expanded GGGGCC repeat motif. These repeats of approximately 500 and 300 motifs represent a stable insertion in this model; they are detected without changes in size across at least six generations and show no evidence for intergenerational anticipation.
Though the mice do not develop any overt motor phenotype, they recapitulate distinctive histopathological features seen in c9ALS/FTD patients, including both sense and antisense intranuclear RNA foci and the presence of RAN translated DPR proteins. These data indicate that the 300-500 GGGGCC repeat motifs are sufficient for the generation and deposition of abnormal tracts of RNA and DPR proteins but not to induce neurodegeneration. Why this is the case remains unclear. Possibly the pathology, seen quite clearly at the molecular level, is not sufficient to compromise motor neuron viability within the lifespan of the mice. A corollary is that higher levels of expression of the pathological RNA or DPR proteins may be required to induce neurodegenerative phenotypes. Supporting this view is a recent report that high levels of expression of the C9ORF72 transcript mediated by post-natal delivery of a (GGGGCC)60 repeat is associated with evidence motor neuron dysfunction (but not a paralytic phenotype) [44]. Alternatively, some other aspect of the model may be required. For example, if the pathology in humans entails sequestration of critical RNA binding proteins by intranuclear RNA aggregates, it is possible that these proteins are more abundant in the mouse, and thus less prone to be titrated to insufficiency in the RNA foci. Alternatively, it is possible that the DPR proteins are better tolerated in some manner in the mouse. For example, in humans with c9ALS/FTD one view is loss of function effects imparted by the hexanucleotide expansion within C9ORF72 might enhance the pathology caused by RNA foci and DPR proteins; in our C9BAC transgenic mice, the impact of loss of function from the transgene is mitigated by the fact that both endogenous mouse C9ORF72 ortholog alleles are intact. Finally, there may be some other species specific reasons why toxicity of the GGGGCC expansion is fundamentally less pronounced in mice as compared to humans [45].
Two aspects of the histological findings in these mice are intriguing. First, there were distinctly more cells containing sense foci than anti-sense RNA foci. This is broadly analogous to data from human cortex [18]. It is also conceivable that the relative paucity of anti-sense RNA foci reflects the structure of the human BAC in our mice, which contains the full 5’ expanse of the C9ORF72 locus but only extends through exons 1-6 and thus might lack 3’ regulatory regions necessary for generating anti-sense foci. Also of interest in the C9BAC mice were age-dependent changes in the abundance of RNA foci and the levels of soluble RAN-translated poly(GP) proteins. Our data indicate that the numbers of intranuclear foci composed of C9ORF72 RNA sense transcripts declined between 10 and 24 months. At 10 months, these foci were widespread in both neurons and glia throughout the central nervous system. The modest but statistically significant decline in RNA foci frequency between 10- and 24-months of age could reflect multiple factors, including an age-dependent increase in clearance of these foci, reduced expression of the offending transcripts with age, or loss of foci-containing cells.
There were also age-related shifts in the properties of poly(GP) DPR proteins. In 4-month C9BAC mice, SDS-soluble poly(GP) species were present at a concentration comparable to levels detected in frontal cortex of some c9ALS patients. Soluble poly(GP) levels in 24-month C9BAC mice were lower, a change that coincided with an increase in the number of poly(GP)-positive inclusions, suggesting conversion of soluble poly(GP) into insoluble species that aggregate. The reason for the age-related shift in poly(GP) from a soluble to insoluble fraction remains unclear but may reflect an accumulation of poly(GP) to levels that reach a critical threshold necessary for nucleation and aggregate formation.
Our RNAseq analyses did not define a transcriptome expression profile that distinguished the C9BAC from wild-type brains. This observation contrasts two reports of differential patterns of gene expression in iPSC-derived motor neurons with C9ORF72 hexanucleotide expansions [12, 34]. It is possible that these data sets differ because of a fundamental difference in the tissues analyzed: our study employed whole mouse frontal cortex at 6 months of age while the early studies were based on in vitro motor neurons that are developmentally equivalent to embryonic motor neurons. Alternatively, it may be that a detailed study of cell type specific transcriptomes though techniques such as translating ribosome affinity purification from the C9BAC mouse will disclose meaningful transcriptome profiles in the C9BAC motor neurons that are not evident in whole brain analyses.
Finally, we note that although the C9BAC mouse does not develop a discernible abnormal behavioral phenotype, the histopathological and transcript variant expression profile matches well with that of c9ALS/FTD patients. In successful in vitro experiments [12, 34, 41], the C9ORF72 expression has been reduced following treatment with antisense oligonucleotides (ASO). In the present study we have conducted a proof of principle experiment to test whether a microRNA (miR) targeting exon 3 of C9ORF72 delivered by recombinant adeno-associated virus (rAAV) might offer an alternative therapeutic strategy. With this intervention, primary cortical neuron cultures from the C9BAC transgenic mice demonstrated a consistent trend towards reduced expression of C9ORF72 transcripts and decreased production of the poly(GP) DPR. These studies suggest that this strategy (AAV-mediated delivery of a synthetic miR) merits further investigation and document that these C9 mice will be useful for assessing therapies that silence transcription or RAN translation across the expanded GGGGCC repeats, or otherwise inhibit the formation of RNA foci and DPR proteins. Combined with accurate analytical techniques to quantify levels of aberrant RNA transcripts, RNA foci and DPR proteins, the C9BAC mouse will be of considerable use for investigating the pathophysiology of C9ORF72-mediated neurodegeneration and much needed therapeutic approaches.
Experimental Procedures
Generation of C9ORF72 transgenic mice
A BAC library was generated from a familial FTD/ALS patient from a C9ORF72-linked ALS/FTD pedigree. Transgenic animals were produced using a BAC carrying a 153.2kb genomic DNA fragment including 140.5kb upstream of C9ORF72, exons 1 to exon 6 of the C9ORF72 gene, including part of the 3'UTR of the short isoform V1 and ~500 GGGGCC motifs in a non coding region between exons 1 and 2. Mice were maintained on a SJL/B6 background. Circular construct injections generated 3 out of 49 positive animals; one (C9BAC) resulted in germline transmission. Presence of the C9BAC transgene was detected using forward primers 5’ TTA ATT TCC TAC CCC TGC CC 3’ and reverse primers 5’ AGG CCT TGA CAA ATG TAG CC 3’, amplifying a 250bp fragment. F1 males used for longitudinal survival and behavior testing were housed individually. The University of Massachusetts Medical School Institutional Animal Care and Use Committee approved all experiments involving animals.
Electrophysiological recordings
MUNE electrophysiological recordings were as described [46]. Further details and description of whole-cell cortical neuron recordings are described in the Supplemental Experimental Procedures.
Droplet Digital PCR (ddPCR)
Frozen tissue samples were homogenized in a gentleMACS Dissociator (Miltenyi Biotec, USA) before total RNA extraction with Trizol (Life Technologies, USA). Reverse transcription to cDNA was performed using random hexamers and MultiScribe reverse transcriptase (High capacity RNA-to-cDNA Kit, Life Technologies). ddPCR was performed using ddPCR Supermix for Probes the QX100 Droplet Digital PCR system (Bio-Rad, USA).
RT-qPCR
Frozen tissue samples were homogenized in a gentleMACS Dissociator (Miltenyi Biotec, USA) before total RNA extraction with Trizol (Life Technologies). Reverse transcription to cDNA was performed using random hexamers and MultiScribe reverse transcriptase (High capacity RNA-to-cDNA Kit, Life Technologies).
Poly(GP) immunoassay and immunostaining
An immunoassay for poly(GP) dipeptides was performed as published [47]. (Details in Supplemental Experimental procedures). For immunostaining, eight micron-thick sagittal slices of mouse hemibrain were cut from formalin-fixed, paraffin-embedded blocks and mounted on glass slides. After drying, slides were deparaffinized and rehydrated in xylene and alcohol washes before being steamed for 30 min in 1X Tris–EDTA (pH 9) buffer solution for antigen retrieval. All slides were processed on a Dako Autostainer with the Dako EnVision™+ system and 3'3-diaminobenzidine chromogen. After staining with anti-GP serum (1:10,000) [23], slides were counterstained with Lerner hematoxylin and coverslipped with Cytoseal permanent mounting media.
Human tissues
Frozen frontal cortex tissues analyzed for for poly(GP) via ELISA assay were obtained from the brain bank for neurodegenerative disorders at Mayo Clinic in Jacksonville, Florida, which operates under protocols approved by the Mayo Clinic Institutional Review Board, in accordance with Health Insurance Portability and Accountability Act guidelines. Frozen frontal cortex tissues for ddPCR were obtained from the UCLA Human Brain and Spinal Fluid Resource center and University of Massachusetts and were collected under protocols approved by the UMMS Institutional Review Board.
RNA fluorescence in situ hybridization (FISH)
Frozen sections were hybridized overnight at 55°c with DNA probes against C9orf72 sense (GGCCCC) and anti-sense (GGGGCC) transcripts, with a Cy3 5’ end tag. Sense foci were quantified in the motor cortex external pyramidal layer and lumbar spinal cord motor neurons. For FISH coupled with fluorescent immunohistochemistry, sections were immunostained with primary antibodies followed by Alexa Fluor-488 conjugated secondary antibodies.
RNAseq
RNAseq was performed on frontal cortex tissue of six-month old C9BAC mice and their Ntg littermates (see Supplemental Experimental Procedures).
MicroRNA design and cloning
A 22-nucleotide artificial miRNA (targeting sequence AATGCAGAGAGTGGTGCTATA) in exon 3 of the C9ORF72 gene was designed and cloned into the miR-155 backbone. The artificial miR was cloned into the 3’UTR of a GFP encoding AAV proviral plasmid under the control of the chicken beta actin (CB) promoter with a CMV enhancer. The plasmid was used to package pseudotyped rAAV9 vectors. Plasmids and vectors can be requested for academic use at http://www.umassmed.edu/muellerlab
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the ALS Association, which funded the generation of these C9BAC mice as well as the ALS/FTD ALS genetics consortium (Lucie Bruijn, Bob Brown, Jonathan Haines, Chris Shaw, Teepu Siddique, Peggy Vance), which provided the C9ORF72 BAC used in this study. We are grateful to Pin-Tsun Lee for assistance with behavioral testing; the UMass Medical School Electron Microscopy Core Facility and the UMass Medical School Bioinformatics core for assistance; and Zuoshang Xu and Chunxing Yang for generously supplying antibodies to TDP-43 protein.
Funding sources
This work was supported by the National Institutes of Health / National Institute of Neurological Disorders and Stroke [R01NS088689 (RHB, LP, CM, OMP), R21NS089979 (TFG), RO1 NS057553 (F-BG), R21NS084528 (LP), R01NS063964 (LP); R01NS077402 (LP); P01NS084974 (LP), RO1FD004127 (RHB), RO1NS079836 (RHB), RO1NS065847(RHB), RO!NS073873(RHB)], National Institute of Environmental Health Services [R01ES20395 (LP)], Department of Defense [ALSRP AL130125 (LP)], Mayo Clinic Foundation (LP), Mayo Clinic Center for Individualized Medicine (LP), ALS Association (RB, LP, TFG), Robert Packard Center for ALS Research at Johns Hopkins (LP, F-BG), Target ALS (LP, JK, SPB), ALS Therapy Alliance (F-BG, RHB), Grant OD018259 (CM), the Angel Fund (RHB), Project ALS (RHB). HT is a Milton Safenowitz Postdoctoral Fellow funded by the ALS Association. OP is supported by the Michael J Fox Foundation. JK was supported by a National Research Foundation of Korea Fellowship (NRF-2011-357-E00005). SPB is supported by a Klingenstein-Simons Fellowship in the Neurosciences. PS is supported through the auspices of Dr. H. Robert Horvitz, an Investigator at the Howard Hughes Medical Institute.
Footnotes
Author contributions
Conceptualization: RHB, OMP, CM, GTC, HRH. Methodology: RHB, OMP, SPB, GTC, CM, CJ, PdJ, LP, TFG, HT, PCS. Investigation: OMP, GTC, HT, JEM, SPB, JK, ZK, JM, AW, LP, TFG, LD, Y-JZ, JS, NW. Resources: KB, DD. Writing – Original Draft: RHB, OMP. Writing – Review & Editing: GTC, CM, HRH, F-BG. Supervision: RHB, OMP, CM, PCS, TFG. Funding Acquisition: RHB, CM, LP, HRH.
References
- 1.Dejesus-Hernandez M, et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72(2):245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 3.Renton AE, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72(2):257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majounie E, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323–30. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci. 2014;17(1):17–23. doi: 10.1038/nn.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Damme P, Robberecht W. Clinical implications of recent breakthroughs in amyotrophic lateral sclerosis. Curr Opin Neurol. 2013;26(5):466–72. doi: 10.1097/WCO.0b013e328364c063. [DOI] [PubMed] [Google Scholar]
- 7.Rohrer JD, et al. C9orf72 expansions in frontotemporal dementia and amyotrophic lateral sclerosis. Lancet Neurol. 2015 doi: 10.1016/S1474-4422(14)70233-9. [DOI] [PubMed] [Google Scholar]
- 8.van Blitterswijk M, et al. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 2013;12(10):978–88. doi: 10.1016/S1474-4422(13)70210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farg MA, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23(13):3579–95. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine TP, et al. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29(4):499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belzil VV, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126(6):895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnelly CJ, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Therrien M, et al. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS One. 2013;8(12):e83450. doi: 10.1371/journal.pone.0083450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xi Z, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92(6):981–9. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fratta P, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gendron TF, et al. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127(3):359–76. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizielinska S, et al. C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 2013;126(6):845–57. doi: 10.1007/s00401-013-1200-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper-Knock J, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137(Pt 7):2040–51. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gendron TF, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126(6):829–44. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YB, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5(5):1178–86. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori K, et al. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126(6):881–93. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 23.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 25.Zu T, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci U S A. 2011;108(1):260–5. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.May S, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128(4):485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizielinska S, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345(6201):1192–4. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao Z, et al. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- 29.Wen X, et al. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014;84(6):1213–25. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YJ, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128(4):505–24. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110(19):7778–83. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciura S, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol. 2013 doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 33.Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126(3):385–99. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sareen D, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5(208):208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssens J, Van Broeckhoven C. Pathological mechanisms underlying TDP-43 driven neurodegeneration in FTLD-ALS spectrum disorders. Hum Mol Genet. 2013;22(R1):R77–87. doi: 10.1093/hmg/ddt349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geevasinga N, et al. Cortical Function in Asymptomatic Carriers and Patients With C9orf72 Amyotrophic Lateral Sclerosis. JAMA Neurol. 2015:1–7. doi: 10.1001/jamaneurol.2015.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris KD, Shepherd GM. The neocortical circuit: themes and variations. Nat Neurosci. 2015;18(2):170–81. doi: 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alfieri JA, Pino NS, Igaz LM. Reversible behavioral phenotypes in a conditional mouse model of TDP-43 proteinopathies. J Neurosci. 2014;34(46):15244–59. doi: 10.1523/JNEUROSCI.1918-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Filiano AJ, et al. Dissociation of frontotemporal dementia-related deficits and neuroinflammation in progranulin haploinsufficient mice. J Neurosci. 2013;33(12):5352–61. doi: 10.1523/JNEUROSCI.6103-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67(3):291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagier-Tourenne C, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013;110(47):E4530–9. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prudencio M, et al. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat Neurosci. 2015;18(8):1175–82. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki N, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci. 2013;16(12):1725–7. doi: 10.1038/nn.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chew J, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348(6239):1151–4. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105(19):6987–92. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia RH, Yosef N, Ubogu EE. Dorsal caudal tail and sciatic motor nerve conduction studies in adult mice: technical aspects and normative data. Muscle Nerve. 2010;41(6):850–6. doi: 10.1002/mus.21588. [DOI] [PubMed] [Google Scholar]
- 47.Su Z, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83(5):1043–50. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.