Abstract
Temporal and spatial control of gene expression can be achieved using an inducible system as a fundamental tool for regulated transcription in basic, applied and eventually in clinical research. We describe a novel “hit and run” inducible direct reprogramming approach. In a single step, 2 days post-transfection, transiently transfected Sox2FLAG under the Leu3p-αIPM inducible control (iSox2) triggers the activation of endogenous Sox2, redirecting primary astrocytes into abundant distinct nestin-positive radial glia cells. This technique introduces a unique novel tool for safe, rapid and efficient reprogramming amendable to regenerative medicine.
Keywords: FLP, tetracycline, doxycycline, CREERT2, embryonic stem cells, induced pluripotent stem cells, neural progenitor cells, tissue regeneration
Inducible gene expression systems
From the time of the first transcriptional regulatory systems (Gossen and Bujard, 1992), several inducible gene expression systems were developed (Clackson, 1997; Saez et al., 1997; Rossi and Blau, 1998) for applications in gene function analysis (Malleret et al., 2001), drug discovery (Aubel et al., 2001), gene therapy (Auricchio et al., 2002), engineering of desired phenotypes during development and in adult life (Niwa et al., 2000), trangenesis (Rossant and Nagy, 1995), stem cell programming and reprogramming (Brambrink et al., 2008; Hockemeyer et al., 2008; Maherali et al., 2008; Stadtfeld et al., 2008, 2010; Welstead et al., 2008; Wernig et al., 2008; Markoulaki et al., 2009; Carey et al., 2010). Regulatory systems developed to control the temporal and spatial gene expression use mostly fusion proteins as regulators and hormones or antibiotics as signals for gene expression. In mammals, the most commonly used inducible systems include the tetracycline system (Sprengel and Hasan, 2007), the systems of the recombination enzymes Cre/loxP (Sauer, 1998) and Flipase/FRT (Hummel and Klämbt, 2008), and the CRE-ERT2 system based on the function and modularity of the ligand-binding domain of the estrogen receptor (ER)(Chiba et al., 2000). The “OFF/ON” gene switches allow for the expression of dominant negative and cytotoxic proteins (Angrand et al., 1998), for the reversibility of the target gene expression (Kistner et al., 1996), for the study of “loss or gain” of function phenotypes (Caulin et al., 2007) and for the isolation of functional components (Meissner et al., 2003). The limitations of these systems lie within the fact that antibiotics are used as regulators of gene expression, resulting in cytotoxicity, interference with embryonic development, the high cost of the inducer, leakiness, chromosomal alterations, immune response and incompatibility in the integration into the regulatory and metabolic network of the target cell (Danielian et al., 1998; Gao et al., 1999; Loonstra et al., 2001; Wunderlich et al., 2001). Thus, tighter control of gene induction with no side effects is fundamental for gene function analysis, for the development of animal models and most importantly for the development of novel approaches in gene and stem cell therapy.
Leu3p-α-IPM inducible gene expression
We have previously reported a novel heterologous ligand-inducible regulatory “OFF-ON” genetic switch, based on the yeast transcription factor Leu3p (Leu3p-α-IPM; Poulou et al., 2010). This system is based on a transcription factor (Leu3p) involved in the regulation of the leucine pathway in yeast, whose function is controlled by α-IPM, a metabolite involved in leucine biosynthesis itself (Kohlhaw, 2003). Leu3p acts as an active repressor binding to its UASLEUDNA element, turning into an activator of the transcription in the presence α-IPM (Sze et al., 1992), an ideal inducer since it exhibits lipid solubility, metabolic stability, rapid “OFF-ON” kinetics with no apparent toxicity to mammalian cells, to fertilized mouse eggs cultured ex vivo and to animals alike (Poulou et al., 2010). Although the leucine biosynthetic pathway is found only in prokaryotes, fungi and plants, Leu3p has been shown to be fully functional In mammalian cells in culture (Sze and Kohlhaw, 1993; Remboutsika, 1994; Guo and Kohlhaw, 1996) and in primary mouse embryonic fibroblasts isolated from double transgenic mouse embryos bearing ubiquitously expressing Leu3p and a Leu3p regulated GFP reporter (Poulou et al., 2010). This system would be ideal for use in stem cell programming and reprogramming strategies transferable from the bench to the clinic.
Present study
Here, we show for the first time that the Leu3p-α-IPM inducible system can be used in direct reprogramming experiments.
Aim
To directly reprogram astrocytes to neural stem cells by “hit and run” Leu3p-α-IPM inducible Sox2 expression approach.
Methods
Experimental animals
Sox2COIN∕+ mice were bred with Tg(hGFAP:CRE) mice (Zhuo et al., 2001) to generate Sox2RG−INV∕+ mice harboring an ablation of Sox2 function in radial glia cells. All animals were handled in strict accordance with good animal practice as defined by the Animals Act 160/03.05.1991 applicable in Greece, revised according to the 86/609/EEC/24.11.1986 EU directive regarding the proper care and use of laboratory animals and in accordance to the Hellenic License for Animal Experimentation at the BSRC” Alexander Fleming” (Prot. No. 767/28.02.07) issued after protocol approval by the Animal Research Committee of the BSRC “Alexander Fleming” (Prot. No. 2762/03.08.05).
Primary astrocyte culture
Brains were carefully dissected from P3 mouse pups, the meninges were removed and the remaining brain tissue was digested with 0025% trypsin in CaMg-free HBSS. The tissue was pipetted up and down with a 10 ml pipette to generate a single cell suspension, shaken for 10 min at 37°C on a rotary shaker, in a waterbath (5 ml per brain). At the end of the incubation time, 5 ml of DMEM containing 10% FBS were added, the sample was pipetted a few times with a 10 ml pipette and passed through a 70 μm nylon mesh to remove the debris. After centrifugation at 200 g for 10 min, the cell pellet was washed once in complete media before the cells were re-suspended in 7 ml complete media, plated out in T 25 cm plates coated poly-L lysine (100 mg/ml). The astrocytes were allowed to grow to confluency (~10–12 days) with media changes every 3 to 5 days. P3 astrocytes were cultured at a density of 2 × 104 cells/cm2 in 4 well Sonic Shield culture dishes (Nunc-Thermo Fisher Scientific, Roskilde, Denmark). Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. α-IPM (20 mM final concentration) was added to the culture media 1 day post-transfection. Cells were incubated for another 24 h, rinsed with 1 × PBS, fixed in 4% ice-cold PFA in 0.12 M PB for 10 min on ice, rinsed again with 1 × PBS multiple times, before use for immunofluorescence or mounting in 50% glycerol before phase contrast photography on a Leica DMI3000 microscope.
RT-PCR
Total RNA was isolated using TriZol, according to the supplier's instructions, (Invitrogen) and RT-PCR was carried out using the Qiagen One-Step RT-PCR system. The following primers were used: for Gapdh (T = 57°C): 5′-CATCTCTGCCCC CTCTGCTG-3′ (forward) and 5′-CGACGCCTG CTTCACCACCT-3′ (reverse); for endoSox2 (T = 62°C): 5′- CCCCCGGCGGCA ATAGCA -3′ (forward) and 5′-TCGGCGCCG GGGAGATACAT-3′ (reverse); for Sox2FLAG (T = 60°C): 5′-CCCCCGGCGGCAATA GCA-3 (forward) and 5′-TCAAAG CTTGTCATCGTCGTCCTT-3′ (reverse); for wnt3a (T = 52°C) 5′- ATTGAATTTGGA GGAATGGT-3′ (forward) and 5′- CTTGAAGTA CGTGTAACGTG-3′ (reverse); for nestin (T = 60°C) 5′- CGCTGGAACAGAGAT TGGAAGG-3′ (forward) and 5′ –GTCTCAAGG GTATTAGGCAAG- 3′ (reverse).
Immunohistochemistry
Cells and cortical sections were fixed in 4% PFA in 0.12 M PB, pH 7.2 at 4°C for 5 min and incubated in Blocking buffer (BB; 0.12 M PB, pH 7.2, 0.15% glycine, 2 mg/ml BSA fragment V (Gibco-Invitrogen) and 0.1% Triton X-100) for 1 h on ice. Cells were incubated o/n at 4°C with primary antibodies in BB. After extensive washes with PBS at RT, cells were incubated with species specific secondary antibodies (Alexa 488-conjugated, 1:500; Invitrogen) for 1 h at RT. Samples were mounted in anti-fade DAPI mounting media (Invitrogen) and photographed on a Leica SP5 confocal microscope. Primary antibodies: anti-Nestin (mouse IgG 1:75, Developmental Studies Hybridoma Bank (DSHB), University of Iowa, http://dshb.biology.uiowa.edu), anti-RC2 (mouse IgM 1:100, DSHB), anti-GFAP, Cy3-linked (mouse IgG 1:100, SIGMA), anti-Sox2 (rabbit IgG 1:1000, Santa Cruz, INC).
Results and discussion
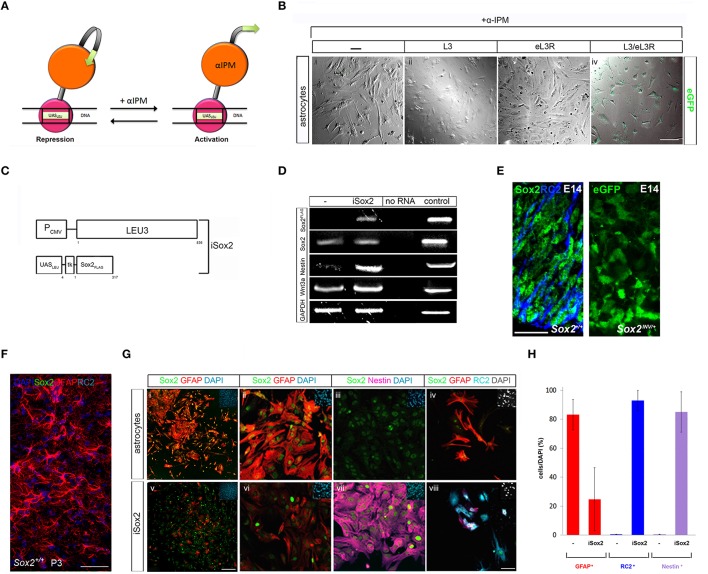
Nervous stem repair is an enormous challenge to regenerative medicine. Even though, neural stem/progenitor cells (NSCs) can be generated as an intermediate cell type from astrocytes on their way to generate neurons in vivo and in vitro, limitations including viral infection and integration, as well as uncontrolled reactivation of transgenes hamper their use in clinical applications (Tomanin and Scarpa, 2004; Huang and Tan, 2015). Here, we investigated the efficiency of the Leu3p-α-IPM as a “hit and run” gene switch to generate NSCs from astrocytes in vitro. We first demonstrated that α-IPM addition efficiently induced a Leu3p regulated eGFP reporter in transiently transfected primary astrocytes (Figures 1A,Bi–iv). NSCs develop and maintain their properties under the guidance of Sox2 (Ferri et al., 2004; Remboutsika et al., 2011; Mandalos et al., 2014; Figure 1E) and they can be generated by direct lineage reprogramming of astrocytes via in vivo overexpression of Sox2 (Niu et al., 2013). We then designed an iSox2 Leu3p inducible system to test the rapidness and efficiency in imposing Sox2 reprogramming ability to primary astrocytes ex vivo (Figure 1C). Although, in vivo at P3 astrocytes do not appear to express Sox2 (Niu et al., 2013; Figure 1F), in primary astrocyte cultures Sox2 (Figure 1D) and Sox2 protein (Figure 1Gi–iv) are expressed in low levels. Astrocytes do not express Nestin (0.6 ± 0.1%) or RC2 (0.5 ± 0.1%), markers of NSCs with a radial glia character (Figure 1D,Gi–iv). Two days post-transfection, high levels of Nestin (85 ± 13.89%) and RC2 (93 ± 6.97%) are expressed only in cells after Sox2 induction (iSox2) in astrocyte cultures (Figures 1D,Gv-viii,H). Concurrently, GFAP+ cells are reduced in iSox2 cultures (24.8 ± 21.73%; Figures 1Gv-viii,H), respectively, when these are compared to control astrocyte cultures (83.28 ± 10.43%; Figures 1Gi-viii,H). One would expect that only the cells with the highest expression of Sox2 could revert the astrocytic (GFAP+) to a NSC (Nestin+, RC2+) radial glia phenotype. However, we observed that the majority of cells have acquired a Nestin+ phenotype. This led us to believe that the iSox2 cells now express a signaling molecule that controls NSC identity. One of these could be wnt3a, known to be involved in the proliferation of NSCs (Lie et al., 2005), but also expressed in low levels in untransfected astrocytes cultures (Figure 1D). Indeed, wnt3a was induced in iSox2 NSCs, along with the induction of endogenous Sox2 and nestin (Figure 1D). Thus, iSox2 can generate NSCs from astrocytes rapidly and efficiently in vitro.
Figure 1.
Leu3p-α-IPM inducible fast - track direct reprogramming of astrocytes to neural stem cells. (A) Leu3p·α-IPM mode of action: Transcriptional repressor upon binding to the UASLEU DNA element and transcriptional activator upon α-IPM ligand binding. (B) Superimposed bright field and confocal images of transiently transfected primary P3 murine astrocytes (i-iv) with either Leu3p protein (L3) (ii) or UASLEU-eGFP reporter (eL3R) (iii) or both L3/eL3R (iv). eGFP is observed only in cells UASLEU with both L3/eL3R (iv). Scale bar 75 μm. (C) Generation iSox2 expression system under the control of Leu3p UASLEU elements. (D) iSox2 induces endogenous Sox2, nestin, and wnt3a 48 h post-transfection in P3 primary murine astrocytes. (E) Sox2+ and RC2+ neural progenitors in the proliferating zone of E14 cortex of wild type mouse embryos. eGFP expression detected in the proliferating zone of E14 cortex after Sox2 ablation in ragial glia cells. Scale bar 75 μm. (F) Sox2 is not expressed in GFAP+ cells in the proliferating zone of P3 mouse cortex. Scale bar 50 μm. (G) iSox2(v-viii) reduces the astrocytic marker GFAP (i,-ii, iv–vi) in P3 primary murine astrocytes and induces a Nestin+ (vii) radial glia (RC2+) (viii) NSC phenotype 48 h post-transfection. Dapi staining of nuclei is present in the upper right corner in all panels. Scale bars for panels i and v represent 250 μm and for panels ii-iv and vi-viii represent 75 μm. (H) Graphs depicting the GFAP+ and either nestin+ or RC2+ cells in untransfected astrocytes and in reprogrammed cultures.
To build an ideal regulatory system for clinical research three factors are important (a) activation by a highly specialized non-toxic bio-available exogenous ligand, (b) inactivation in the absence or removal of the ligand, and (c) no interference with endogenous mammalian gene expression and metabolic pathways. Evidently, Leu3p-α-IPM represents this novel unique tool for simple, safe, fast and efficient inducible programming and reprogramming in different cell types for gene and stem cell strategies directly amendable to the clinic.
Ethics statement
All animals were handled in strict accordance with good animal practice as defined by the Animals Act 160/03.05.1991 applicable in Greece, revised according to the 86/609/EEC/24.11.1986 EU directive regarding the proper care and use of laboratory animals and in accordance to the Hellenic License for Animal Experimentation at the BSRC“Alexander Fleming” (Prot. No. 767/28.02.07) issued after protocol approval by the Animal Research Committee of the BSRC “Alexander Fleming” (Prot. No. 2762/03.08.05).
Author contributions
MA, NM, TK, MS conducted research experiments; RM provided reagents; ER directed research and wrote the manuscript.
Funding
This research was financed by the Studentship Grant PENED 394, the European Union (ERDF) and Greek National funds through the Operational Program “Competitiveness and Entrepreneurship” of the NSRF (Cooperation Grant-09∑YN9D-12-966), the ADISC (THALIS) MIS380249 Grant funded by Framework Program “Education and Lifelong Learning,” co-financed European Social Fund and National funds (Ministry of Education) and a collaboration grant (NKUA-12439) from the Lieber Institute for Brain Development to E.R. (US).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Angrand P. O., Woodroofe C. P., Buchholz F., Stewart A. F. (1998). Inducible expression based on regulated recombination: a single vector strategy for stable expression in cultured cells. Nucleic Acid Res. 26, 3263–3269. 10.1093/nar/26.13.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubel D., Morris R., Lennon B., Rimann M., Kaufmann H., Folcher M., et al. (2001). Design of a novel mammalian screening system for the detection of bioavailable, non-cytotoxic streptogramin antibiotics. J. Antibiotics (Tokyo) 54, 44–55. 10.7164/antibiotics.54.44 [DOI] [PubMed] [Google Scholar]
- Auricchio A., Rivera V. M., Clackson T., O'Connor E. E., Maguire A. M., Tolentino M. J., et al. (2002). Pharmacological regulation of protein expression from adeno-associated viral vectors in the eye. Mol. Ther. 6, 238–242. 10.1006/mthe.2002.0660 [DOI] [PubMed] [Google Scholar]
- Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., et al. (2008). Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159. 10.1016/j.stem.2008.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B. W., Markoulaki S., Beard C., Hanna J., Jaenisch R. (2010). Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods 7, 56–59. 10.1038/nmeth.1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin C., Nguyen T., Lang G. A., Goepfert T. M., Brinkley B. R., Cai W. W., et al. (2007). An inducible mouse model for skin cancer reveals distinct roles for gain- and loss-of-function p53 mutations. J. Clin. Invest. 117, 1893–1901 10.1172/JCI31721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H., Chambon P., Metzger D. (2000). F9 embryonal carcinoma cells engineered for tamoxifen-dependent Cre-mediated site-directed mutagenesis and doxycycline-inducible gene expression. Exp. Cell Res. 260, 334–339. 10.1006/excr.2000.5022 [DOI] [PubMed] [Google Scholar]
- Clackson T. (1997). Controlling mammalian gene expression with small molecules. Curr. Opin. Chem. Biol. 1, 210–218. 10.1016/S1367-5931(97)80012-9 [DOI] [PubMed] [Google Scholar]
- Danielian P. S., Muccino D., Rowitch D. H., Michael S. K., McMahon A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326. [DOI] [PubMed] [Google Scholar]
- Ferri A. L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., et al. (2004). Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131, 3805–3819. 10.1242/dev.01204 [DOI] [PubMed] [Google Scholar]
- Gao X., Kemper A., Popko B. (1999). Advanced transgenic and gene-targeting approaches. Neurochem. Res. 24, 1181–1188. 10.1023/A:1020772706279 [DOI] [PubMed] [Google Scholar]
- Gossen M., Bujard H. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U.S.A. 89, 5547–5555. 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Kohlhaw G. B. (1996). Regulation of transcription in mammalian cells by yeast Leu3p and externally supplied inducer. FEBS Lett. 390, 191–195. 10.1016/0014-5793(96)00653-9 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Cook E. G., Gao Q., Mitalipova M., Jaenisch R. (2008). A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell 3, 346–353. 10.1016/j.stem.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Tan S. (2015). Direct lineage conversion of astrocytes to induced neural stem cells or neurons. Neurosci. Bull. 31, 357–367. 10.1007/s12264-014-1517-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T., Klämbt C. (2008). P-element mutagenesis. Methods Mol. Biol. 420, 97–117. 10.1007/978-1-59745-583-1_6 [DOI] [PubMed] [Google Scholar]
- Kistner A., Gossen M., Zimmermann F., Jerecic J., Ullmer C., Lübbert H., et al. (1996). Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 93, 10933–10938. 10.1073/pnas.93.20.10933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw G. B. (2003). Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67, 1–15, table of contents. 10.1128/MMBR.67.1.1-15.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie D. C., Colamarino S. A., Song H. J., Désiré L., Mira H., Consiglio A., et al. (2005). Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375. 10.1038/nature04108 [DOI] [PubMed] [Google Scholar]
- Loonstra A., Vooijs M., Beverloo H. B., Allak B. A., van Drunen E., Kanaar R., et al. (2001). Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 98, 9209–9214. 10.1073/pnas.161269798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Ahfeldt T., Rigamonti A., Utikal J., Cowan C., Hochedlinger K. (2008). A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 3, 340–345. 10.1016/j.stem.2008.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret G., Haditsch U., Genoux D., Jones M. W., Bliss T. V., Vanhoose A. M., et al. (2001). Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell 104, 675–686. 10.1016/S0092-8674(01)00264-1 [DOI] [PubMed] [Google Scholar]
- Mandalos N., Rhinn M., Granchi Z., Karampelas I., Mitsiadis T., Economides A. N., et al. (2014). Sox2 acts as a rheostat of epithelial to mesenchymal transition during neural crest development. Front. Physiol. 5:345. 10.3389/fphys.2014.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markoulaki S., Hanna J., Beard C., Carey B. W., Cheng A. W., Lengner C. J., et al. (2009). Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat. Biotechnol. 27, 169–171. 10.1038/nbt.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner U., Allabauer I., Repp R., Rascher W., Dötsch J. (2003). Inducible expression of hypoxia-inducible factor 1 (HIF-1) as a tool for studying HIF-1 -dependent gene regulation during Normoxia in vitro. Pharmacology 69, 74–78. 10.1159/000072359 [DOI] [PubMed] [Google Scholar]
- Niu W., Zang T., Zou Y., Fang S., Smith D. K., Bachoo R., et al. (2013). In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat. Cell Biol. 15, 1164–1175. 10.1038/ncb2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A. G. (2000). Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376. 10.1038/74199 [DOI] [PubMed] [Google Scholar]
- Poulou M., Bell D., Bozonelos K., Alexiou M., Gavalas A., Lovell-Badge R., et al. (2010). Development of a chromosomally integrated metabolite-inducible Leu3p-alpha-IPM “off-on” gene switch. PLoS ONE 5:e12488. 10.1371/journal.pone.0012488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remboutsika E. (1994). Transcriptional Regulator Leu3p of Yeast: Modular Architecture and Function. Thesis, Purdue University, U. S. A. [Google Scholar]
- Remboutsika E., Elkouris M., Iulianella A., Andoniadou C. L., Poulou M., Mitsiadis T. A., et al. (2011). Flexibility of neural stem cells. Front. Physiol. 2:16. 10.3389/fphys.2011.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J., Nagy A. (1995). Genome engineering: the new mouse genetics. Nat. Med. 1, 592–594. 10.1038/nm0695-592 [DOI] [PubMed] [Google Scholar]
- Rossi F. M., Blau H. M. (1998). Recent advances in inducible gene expression systems. Curr. Opin. Biotechnol. 9, 451–456. 10.1016/S0958-1669(98)80028-1 [DOI] [PubMed] [Google Scholar]
- Saez E., No D., West A., Evans R. M. (1997). Inducible gene expression in mammalian cells and transgenic mice. Curr. Opin. Biotechnol. 8, 608–616. 10.1016/S0958-1669(97)80037-7 [DOI] [PubMed] [Google Scholar]
- Sauer B. (1998). Inducible gene targeting in mice using the Cre/lox system. Methods 14, 381–392. 10.1006/meth.1998.0593 [DOI] [PubMed] [Google Scholar]
- Sprengel R., Hasan M. T. (2007). Tetracycline-controlled genetic switches. Handb. Exp. Pharmacol. 178, 49–72. 10.1007/978-3-540-35109-2_3 [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Borkent M., Hochedlinger K. (2010). A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Methods 7, 53–55. 10.1038/nmeth.1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K. (2008). Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240. 10.1016/j.stem.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze J. Y., Kohlhaw G. B. (1993). Purification and structural characterization of transcriptional regulator Leu3 of yeast. J. Biol. Chem. 268, 2505–2512. [PubMed] [Google Scholar]
- Sze J. Y., Woontner M., Jaehning J. A., Kohlhaw G. B. (1992). In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on alpha-isopropylmalate. Science 258, 1143–1145. 10.1126/science.1439822 [DOI] [PubMed] [Google Scholar]
- Tomanin R., Scarpa M. (2004). Why do we need new gene therapy viral vectors? Characteristics, limitations and future perspectives of viral vector transduction. Curr. Gene Ther. 4, 357–372. 10.2174/1566523043346011 [DOI] [PubMed] [Google Scholar]
- Welstead G. G., Brambrink T., Jaenisch R. (2008). Generating iPS cells from MEFS through forced expression of Sox-2, Oct-4, c-Myc, and Klf4. J. Vis. Exp. 734. 10.3791/734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Lengner C. J., Hanna J., Lodato M. A., Steine E., Foreman R., et al. (2008). A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 26, 916–924. 10.1038/nbt1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich F. T., Wildner H., Rajewsky K., Edenhofer F. (2001). New variants of inducible Cre recombinase: a novel mutant of Cre-PR fusion protein exhibits enhanced sensitivity and an expanded range of inducibility. Nucleic Acid Res. 29:E47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L., Theis M., Alvarez-Maya I., Brenner M., Willecke K., Messing A. (2001). hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis 31, 85–94. 10.1002/gene.10008 [DOI] [PubMed] [Google Scholar]