Abstract
Tissue and cell-type identity lie at the core of human physiology and disease. Understanding the genetic underpinnings of complex tissues and individual cell lineages is crucial for developing improved diagnostics and therapeutics. We present genome-wide functional interaction networks for 144 human tissues and cell types developed using a data-driven Bayesian methodology that integrates thousands of diverse experiments spanning tissue and disease states. Tissue-specific networks predict lineage-specific responses to perturbation, reveal genes’ changing functional roles across tissues, and illuminate disease-disease relationships. We introduce NetWAS, which combines genes with nominally significant GWAS p-values and tissue-specific networks to identify disease-gene associations more accurately than GWAS alone. Our webserver, GIANT, provides an interface to human tissue networks through multi-gene queries, network visualization, analysis tools including NetWAS, and downloadable networks. GIANT enables systematic exploration of the landscape of interacting genes that shape specialized cellular functions across more than one hundred human tissues and cell types.
Introduction
The precise actions of genes are frequently dependent on their tissue context, and human diseases result from the disordered interplay of tissue and cell-lineage-specific processes1–4. These factors combine to make the understanding of tissue-specific gene functions, disease pathophysiology, and gene-disease associations particularly challenging. Projects such as the Encyclopedia of DNA Elements (ENCODE)5 and The Cancer Genome Atlas (TCGA)6 provide comprehensive genomic profiles of cell lines and cancers, but the challenge of understanding human tissues and cell lineages in the multicellular context of a whole organism remains7. Integrative methods that infer functional gene interaction networks can capture the interplay of pathways, but existing networks lack tissue specificity8.
While direct assay of tissue-specific features remains infeasible in many normal human tissues, computational methods can infer them from large data compendia. We recently found that even samples measuring mixed cell lineages contain extractable information related to lineage-specific expression9. In addition to tissue-specificity, we10–13 and others14–17 have shown that heterogeneous genomic data contain functional information, e.g. of gene expression regulation by protein-DNA, protein-RNA, protein-protein and metabolite-protein interactions. Here we develop and evaluate methods that simultaneously extract functional and tissue/cell-type signals to construct accurate maps of both where and how proteins act.
We build genome-scale functional maps of human tissues by integrating a collection of datasets covering thousands of experiments contained in more than 14,000 distinct publications. To integrate these data, we automatically assess each dataset for its relevance to each of 144 tissue and cell-lineage-specific functional contexts. The resulting functional maps provide a detailed portrait of protein function and interactions in specific human tissues and cell lineages ranging from B-lymphocytes to the renal glomerulus to the whole brain. This allows us to profile the specialized function of genes in a high-throughput manner, even in tissues and cell lineages for which no or few tissue-specific data exist.
In contrast with tissue-naïve networks, which assume that the function of genes remains constant across tissues8, these maps can answer biological questions that are specific to a single gene in a single tissue. For example, we use these maps for the gene interleukin 1β (IL1B), in the blood vessel network, where it plays a key role in inflammation18, to predict lineage-specific responses to IL1B stimulation, which we experimentally confirmed. Examination of parallel networks shows changes in gene and pathway functions and interactions across tissues revealing tissue-specific rewiring. We demonstrate that several tissue-specific functions of the multifunctional gene lymphoid enhancer-binding factor 1 (LEF1) are evident from the way its connectivity changes in distinct tissues.
Tissue-specific networks provide a new means to generate hypotheses related to the molecular basis of human disease. We develop an approach, termed the network-wide association study (NetWAS). In NetWAS, statistical associations from a standard genome-wide association study (GWAS)19 guide the analysis of functional networks. This reprioritization method is discovery-driven and does not depend on prior disease knowledge. NetWAS, in conjunction with tissue-specific networks, effectively reprioritizes statistical associations from distinct GWAS to identify disease-associated genes, and tissue-specific NetWAS better identifies genes associated with hypertension than either GWAS or tissue-naïve NetWAS.
Our tissue-specific maps are available through the Genome-scale Integrated Analysis of Networks in Tissues (GIANT) interface, which provides interactive visualization and exploration of tissue-specific networks, including a comparative view that can highlight tissue-specific rewiring of genes and pathways. GIANT also provides NetWAS analysis for biomedical researchers to reprioritize their gene-based GWAS results in the context of our human tissue-specific networks.
Results
We integrated diverse genome-scale data in a tissue-specific manner to construct 144 human tissue and cell-lineage-specific networks and demonstrated their broad utility for generating specific, testable hypotheses, summarizing tissue-specific relationships between diseases, and reprioritizing results from genetic association studies (Fig. 1a). Our findings underscore the importance of considering tissue-specificity when integrating heterogeneous data to understand the pathophysiology of common human diseases.
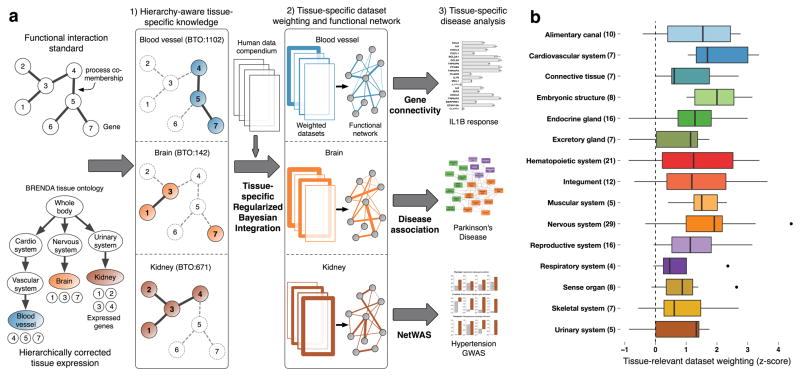
Figure 1. Tissue-ontology-aware regularized Bayesian integration.
(a) Our integration pipeline constructs tissue-specific functional interaction networks by (1) using tissue-specific knowledge to (2) identify and weight datasets by their tissue-relevant signal. We demonstrate the capabilities of the networks by (3, top panel) experimentally validating the gene connectivity scores, by (3, middle panel) demonstrating that they identify disease associations, and by (3, bottom panel) reprioritizing GWAS results. (b) Bayesian integration using tissue-specific knowledge automatically identifies and weighs tissue-relevant datasets. We validate our approach by evaluating the weights in a set of datasets with clear tissue-specificity. We calculate a z-score per tissue that measures how much the ‘relevant’ datasets are up-weighted relative to all datasets in the compendium for that tissue. Plotted here per organ system (y-axis) is the distribution of z-scores of tissues within that system in the form of a box-plot (x-axis). The thick line within each box indicates the median tissue z-score for that system; the lower and upper ends of the box indicate the first and third quartiles of the distribution; the extended lines on either side denote the limits of the distribution, with the outliers (dots) further away. Beyond automatically identifying relevant datasets, our method of automatic weighting constructed higher quality networks than an identical approach limited to only curation-identified tissue-relevant datasets (Supplementary Fig. 1).
Integrated tissue-specific functional interaction networks
We isolated tissue-relevant signals from data not resolved to the cell lineage or tissue using a Bayesian integration that incorporated the hierarchical relationships between tissues. We collected tissue-specific functional interactions for each tissue from known functional relationships and low-throughput tissue-gene expression data, and mapped tissue-gene annotations from the Human Protein Reference Database20 (HPRD) to the BRENDA Tissue Ontology21 (BTO). We leveraged this hierarchy to increase gene and tissue coverage, and to make the interactions consistent with tissue organization (see Methods). We used these known tissue-specific interactions to construct a Bayesian model of tissue-specific functional information from diverse experiments for each of 144 human tissues. Each tissue network represents the tissue-specific posterior probability of a functional relationship between each pair of genes from an ensemble of data covering more than 14,000 publications (Fig. 1a).
Our approach accurately identified tissue-relevant signals in the compendium (Fig. 1b; Supplementary Table 1), automatically up-weighting datasets from relevant tissues and prioritizing tissue-relevant signals over other data. Based on five-fold cross-validation, our method outperformed a Bayesian integration limited to only tissue-related datasets identified based on the experimental description (for 62 of 64 tissues; p = 3.2e–12; Supplementary Fig. 1). Our approach also substantially increased the number of tissues for which networks could be constructed. Only 64 tissues had sufficient labeled data to construct networks, but we were able to construct networks for 144 tissues by extracting tissue-specific information from hundreds of datasets. For example, our method constructed a network for the dentate gyrus (a tissue with limited data) by taking advantage of curated dentate gyrus-specific knowledge to extract relevant signals from other tissues and cell types in the nervous system. Networks for tissues with no or very limited amount of data had accuracies comparable to that of tissues with abundant tissue-specific data (Supplementary Fig. 1). Our approach generated diverse networks that reflected the tissue-specific connectivity of genes and pathways (Supplementary Table 2).
Tissue-specific networks predicted IL1B response
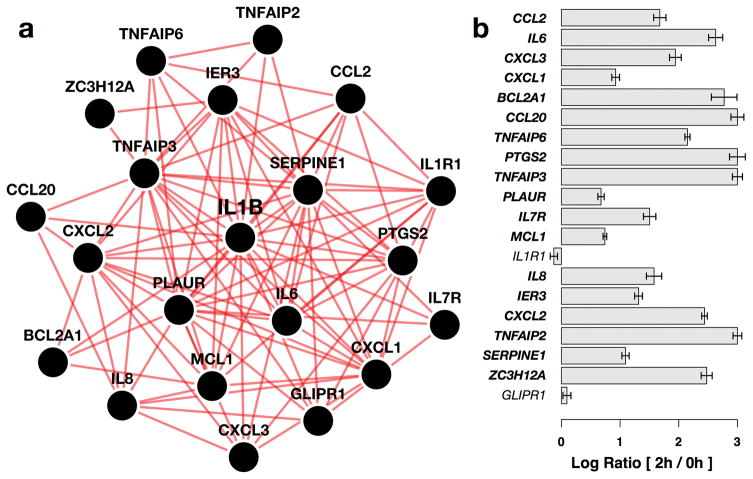
Our networks provided experimentally testable hypotheses about tissue-specific gene function and responses to pathway perturbations. We examined and experimentally verified the tissue-specific molecular response of blood vessel cells to stimulation by interleukin 1β (IL1B), a proinflammatory cytokine. We anticipated that the genes most tightly connected to IL1B in the blood vessel network would be among those responding to IL1B stimulation in blood vessel cells (Fig. 2a). We tested this hypothesis by profiling the gene-expression of human aortic smooth muscle cells (the predominant cell type in blood vessels) stimulated with IL1B. Examining the genes significantly up-regulated at 2h post-stimulation showed that 18 out of the 20 IL1B network neighbors were among the top 500 up-regulated genes in the experiment (p-value = 2.07e–23; Fig. 2b). The blood vessel network is the most accurate tissue in predicting this experimental outcome; none of the other 143 tissue-specific networks or the tissue-naïve network performs as well when evaluated by each network’s ability to predict the result of IL1B stimulation on these cells (Fig. 2a). Nine of IL1B’s top 20 neighbors in the blood vessel network are not top neighbors in the tissue-naïve network, and each has a key vasculature-specific role (Supplemental Table 3; Supplementary Note 1). Networks of other cardiovascular system tissues also captured IL1B response better than the tissue-naïve network, and this was consistent across a range of thresholds for top IL1B network neighbors as well as up-regulated genes in the experiment (Supplementary Fig. 2b & c).
Figure 2. Predicted IL1B functional interaction partners from the blood vessel network are significantly up-regulated after stimulation of blood vessel cells by IL1B.
(a) The 20 genes most tightly connected to IL1B in the blood vessel network are shown. These genes are predicted to respond to IL1B stimulation in blood vessel. (b) The barplot shows the differential expression levels of the 20 IL1B neighbors measured in a microarray experiment at 0h and 2h post IL1B stimulation in aortic smooth muscle cells which constitute most of the blood vessel. Each bar represents the gene’s log ratio of mean expression at 2h to that at 0h. Error bars represent regularized pooled standard errors estimated by LIMMA (n=4). 18 out of 20 IL1B network neighbors (labelled in bold) were found to be among the most significantly differentially expressed genes at 2h relative to 0h (p = 1.95e–23).
We also evaluated nine additional publicly available datasets that used modern genome-wide platforms to measure cellular response to IL1B stimulation in a diverse set of tissues. In all ten experiments, the appropriate tissue network identified a set of genes that responded significantly to IL1B, and randomly selected control sets of genes did not show a significant response to treatment (Supplementary Fig. 3).
Tissue-specific network rewiring of multifunctional genes
Complex multicellular organisms have multifunctional genes that participate in distinct cellular processes based on the developmental and anatomical context. For example, developmental programs are known to be controlled by broadly-expressed transcription factors that, in specific combinations, regulate cell-type-specific gene expression5,22,23 and have the potential to force cell-lineage conversions24,25. Multifunctional genes have also been implicated in pleiotropic disease phenotypes26,27. Such effects are likely to arise when a gene is ‘re-wired’ to associate with different functional partners in different tissues. Our genome-wide functional network maps of human tissues could potentially delineate tissue-specific wiring of multifunctional genes.
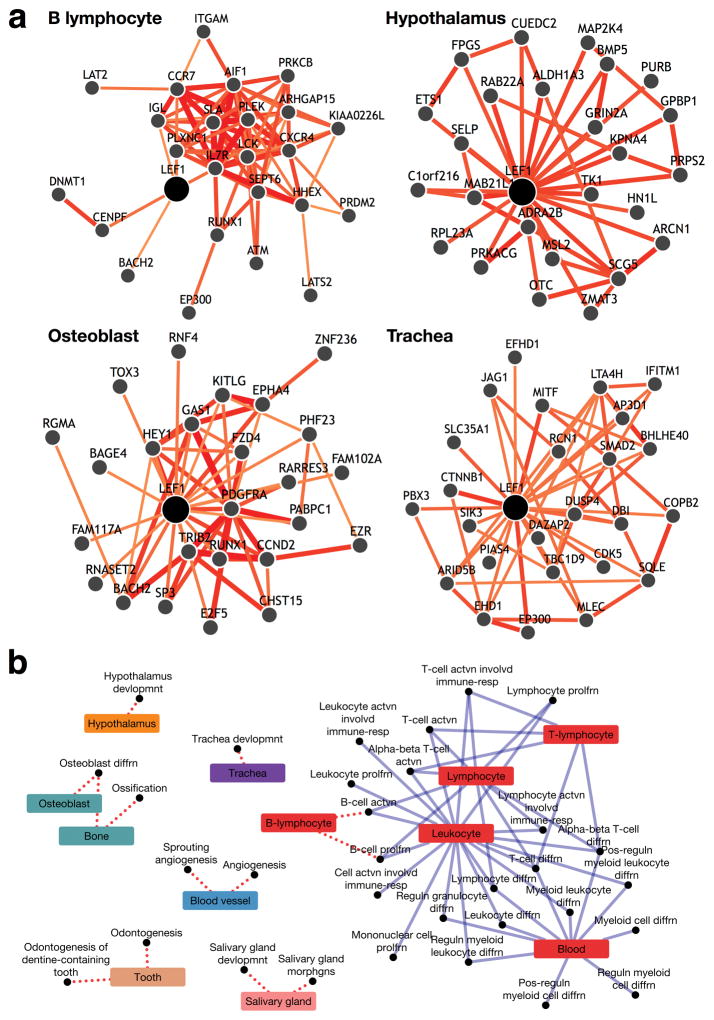
We focused on the transcription factor LEF1 that plays a key role in mediating the tissue-specific response to Wnt signaling28, a fundamental and highly conserved pathway known to elicit diverse cell-type-specific developmental responses29. Since very little is known about LEF1’s activity in human tissues, we probed its tissue-specific functional role by examining its neighbors across different networks (Fig. 3a shows LEF1’s neighbors in B-lymphocytes, hypothalamus, osteoblasts and trachea; Supplementary Fig. 4 shows a detailed view of LEF1 in B-lymphocytes). Analyzing LEF1 network partners (see Methods) revealed that, in twelve tissues, LEF1 was significantly associated with each tissue’s appropriate process (AUC 0.8; Fig. 3b), reflecting highly accurate representation of tissue-specific wiring of LEF1.
Figure 3. Tissue-networks capture tissue-specific functional rewiring.
(a) Multi-tissue view of LEF1 retrieved from GIANT, a web interface to our tissue-specific networks that facilitates user-directed analysis of human tissue-networks. Using the advanced functionality for comparing functional interactions across tissues, we queried GIANT with the multifunctional gene lymphoid enhancer-binding factor 1 (LEF1) in four tissues: B-lymphocyte, hypothalamus, osteoblast and trachea. The retrieved functional neighbors of LEF1 were indeed notably different across the four networks, leading us to the hypothesis that the tissue networks could recapitulate specific gene wiring. (b) The diverse tissue-specific functional rewiring of LEF1. This bipartite graph of tissues (colored rectangles) and processes (black circles) shows how LEF1 participates in different processes in distinct tissues. For example, in the blood vessel, LEF1 is most closely associated with angiogenesis, but in hypothalamus it is closely associated with hypothalamus development. In addition to prior knowledge about tissue-specific associations of LEF1 (solid blue edges), tissue networks also aid in the discovery of several novel tissue associations that have experimental support in model organisms (dotted red edges).
In addition to recapitulating current knowledge (solid blue edges in Fig. 3b), we identified several novel associations between LEF1 and tissue-specific processes in humans that have experimental support in model organisms (dotted red edges in Fig. 3b). Akin to the tissues in Figure 3a, we highlighted predicted functional associations of LEF1 in B-lymphocyte, hypothalamus, osteoblast and trachea (red, orange, green, and purple, respectively, in Fig. 3b). LEF1’s role in B cell activation in B-lymphocytes has already been characterized in mouse30,31, and further, LEF1 has been strongly linked with chronic lymphocytic leukemia (CLL)32–34. LEF1 mediated Wnt signaling has been shown to be critical for hypothalamic neurogenesis in zebrafish35,36. Numerous studies point to a pivotal role of LEF1 in osteoblast proliferation, maturation, function, and regeneration37–39, and potential involvement in bone disease40,41. Finally, several animal models support a clear association of LEF1 with development of submucosal glands42–44, which are epithelial secretory structures in the human tracheobronchial airways, involved in hypersecretory lung diseases such as asthma, chronic bronchitis, and cystic fibrosis45. Thus, tissue-specific networks can unravel the distinct functions of multifunctional genes such as LEF1 and provide opportunities to probe the tissue-specific pleiotropic effects of disease mutations.
Tissue networks can capture disease-disease associations
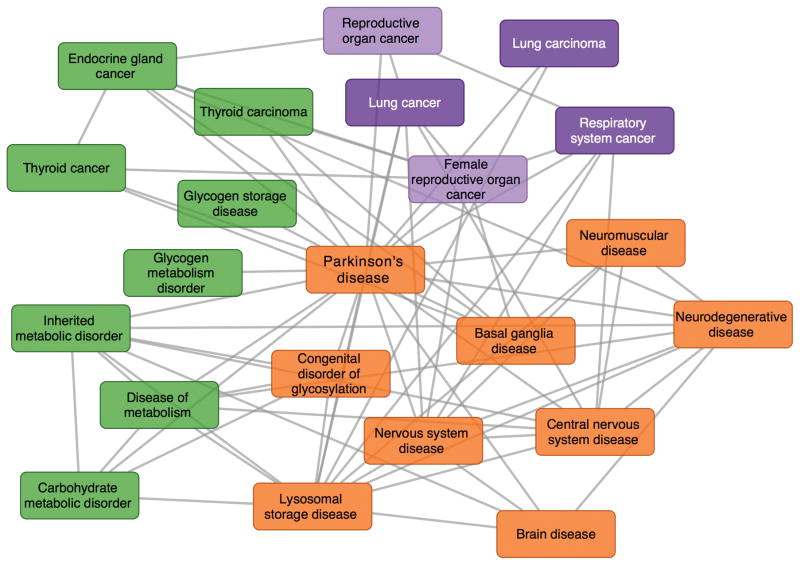
Most human diseases are syndromes with complex origins and manifestations in multiple tissues4,26,46. Diseases with common causative pathways or that are connected through crosstalk between pathways are expected to exhibit high functional associations in their relevant tissues26. We used tissue networks to quantify molecular interactions between diseases to derive a map of tissue-specific disease relationships. These were data-driven maps discovered from tissue-specific functional associations inferred from an integration of high-throughput data, making them relatively unbiased with respect to prior knowledge of disease associations. Here we focused on Parkinson’s disease (PD), a neurodegenerative disorder caused by progressive neuronal loss in the substantia nigra and subsequent reduction in dopamine production47. We created a functional disease map of PD based on the substantia nigra network (Fig. 4). Several documented disease associations were observed in the disease map: for example, PD is connected to ‘neurodegenerative disease’ and ‘basal ganglion disease’, classes of disease that include PD. The disease map also contained more subtle connections. For instance, PD is strongly connected to both lung and reproductive organ cancer, likely through the ubiquitin-protein ligase gene PARK2, which has been implicated in PD as well as brain, colorectal, lung and ovarian cancers48,49. We observed additional undocumented connections to thyroid cancer, driven by functional interactions involving the PD genes PARK2, PARK7 and HTRA2. A blinded literature evaluation showed that this disease map was significantly enriched (Fisher’s exact p= 0.001228) for associations strongly supported by the literature as compared to a control set of associations (Supplementary Fig. 5). Thus, modeling human complex diseases using tissue-specific networks provided several insights into disease genetics and crosstalk and highlighted avenues for discovery of novel molecular disease associations. We generated additional disease maps for Alzheimer’s disease, glomerulonephritis, and glycogen metabolism disorder (Supplementary Figure 6).
Figure 4. A disease map centered on Parkinson’s disease (PD) summarizing its molecular associations with other diseases in substantia nigra.
The disease map effectively identifies PD’s connection to both documented nervous system diseases as well as several cancers through the PARK gene. Parkinson’s disease is characterized by the death of dopaminergic neurons in substantia nigra. Associations between the genes associated with Parkinson’s disease and other diseases were tested by calculating the connectivity across the disease gene sets relative to their background connectivity in the substantia nigra network. All significant connections (edges) between diseases (nodes) are shown in this disease map.
Tissue networks are tools for data-driven analysis of GWAS
In the last decade, quantitative genetics – particularly GWAS – has emerged as a powerful approach to catalogue heritable and de novo sequence variation associated with a wide range of human traits and diseases50. However, due to the lack of statistical power to detect low-frequency mutations, small genetic effects, and epistatic interactions, GWAS findings usually only account for a small proportion of observed heritability51. Because most complex diseases have tissue-specific origins and manifestations, we hypothesized that tissue-specific networks could complement GWAS data in discovering disease-gene associations. The top GWAS hits, even those below a reasonable genome-wide significance cutoff, should be enriched with relevant (even ‘real’ causal) genes. Consequently, by identifying functional signatures associated with these top genes in the appropriate tissue-specific networks, we can further enrich for phenotype-associated genes in a genome-wide re-ranking of GWAS results. Thus, we developed a network-wide association study (NetWAS) approach consisting of a tissue-network classifier that learns network connectivity patterns associated with the phenotype of interest (using the top GWAS hits) and makes predictions for genes across the genome. The NetWAS approach is discovery-driven, as the genes used to identify connectivity patterns are derived from the GWAS itself rather than potentially biased/limited prior disease knowledge. This attribute allows NetWAS to be applied to any GWAS study, even those probing currently uncharacterized or minimally-characterized diseases and phenotypes as well as those for which no associations reached genome-wide significance.
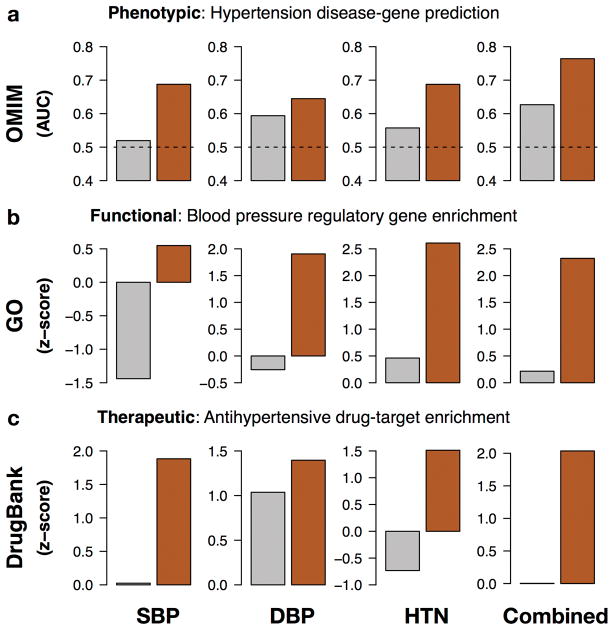
We applied the NetWAS strategy to a GWAS of hypertension52,53 (see Methods). Hypertension is a major cardiovascular risk factor and a complex trait involving a large number of genetic variants54. We converted SNP-level association statistics into gene-level statistics19 for each of three recorded phenotypes – diastolic blood pressure (DBP), systolic blood pressure (SBP) and hypertension (HTN). We applied support vector machines using genes with nominally significant p-values as positive examples and randomly selected genes as the negative examples. Using the tissue network for kidney, a tissue that plays a central role in blood pressure control55, this constructed a classifier that identifies tissue-specific network connectivity patterns associated with the phenotype of interest. Genes annotated to hypertension phenotypes in the Online Mendelian Inheritance in Man (OMIM) database were more highly ranked by this classifier than the initial GWAS (Fig. 5a). We hypothesized that the distinct endpoints might reveal different aspects of the disease. We evaluated whether or not combining the three phenotypes using a rank-sum approach resulted in predictions with a stronger association with known hypertension genetics than the individual phenotypes and found that it did. This performance is specific to the relevant tissues – when performing the same analysis on all tissue networks, we found that kidney, heart, and liver networks showed stronger performance than the tissue-naïve network (Supplementary Fig. 7a).
Figure 5. Network reprioritization of hypertension GWAS identifies hypertension-associated genes.
Genes ranked using GWAS (grey) and genes reprioritized using NetWAS (dark red) are assessed for correspondence to genes known to be associated with hypertension phenotypes, regulatory processes, and therapeutics. We compared individual (systolic blood pressure, SBP; diastolic blood pressure, DBP; hypertension, HTN) as well as combined hypertension end-points. (a) Gene rankings were compared to OMIM-annotated hypertension genes using area under the ROC curve (AUC). The AUC for the tissue-specific NetWAS is consistently higher than that for the original GWAS for all hypertension end-points. Merging the network-based predictions for the three hypertension-related endpoints into a combined phenotype results in the best performance (AUC = 0.77; original GWAS AUC = 0.62; dotted line at 0.5 denotes the AUC of a baseline random predictor). Gene rankings were also assessed for enrichment of genes involved in regulation of blood pressure (from GO) and targets of antihypertensive drugs (from DrugBank). The top NetWAS results were significantly enriched with genes involved in (b) blood pressure regulation as well as genes that are (c) targets of antihypertensive drugs. Enrichment was calculated as a z-score (see Methods), with higher scores indicating greater shift from expected ranking towards the top of the list. In nearly all cases, the NetWAS ranking was both significantly enriched with the respective gene sets and more enriched than in the original GWAS ranking.
In addition to well-documented hypertension genes like MTHFR and PPARG, NetWAS identified several additional candidates (Supplementary Table 4). Many lines of evidence in the literature link the top predicted genes to hypertension via mechanistic relationships to known disease genes and pathways or associations with hypertension risk factors. Several such examples are tabulated in Supplementary Table 5. Using functional annotations from the Gene Ontology (GO) to subsequently interpret NetWAS prioritized genes, we observed that NetWAS ranked genes annotated to the GO term ‘regulation of blood pressure’ significantly higher than GWAS (Fig. 5b). Since NetWAS provides a useful reprioritization of the genome in terms of phenotypic and functional association, we explored whether the re-ranking is also helpful in discovering appropriate therapeutics. Using drug-target data from DrugBank57, we found that targets of antihypertensive drugs were significantly enriched among the top-genes from NetWAS more than GWAS (Fig. 5c). We found similar results for targets from three other databases (PharmGKB58, TTD59, and CTD60; Supplementary Fig. 7b).
We evaluated NetWAS on four additional GWAS spanning diverse disease and tissue contexts and found that the approach consistently ranked documented disease genes higher than the GWAS (Supplemental Figure 8). Thus, NetWAS builds from nominally significant associations from GWAS to identify candidates by their connectivity in tissue-specific networks that are valuable for guiding research into disease mechanism and therapy.
A dynamic, interactive interface for biomedical researchers
To facilitate broad use of these networks by biomedical researchers, we have developed GIANT – a dynamic, interactive web interface. Researchers can query by individual genes or by gene sets of interest to analyze tissue-specific gene function and interactions. For example, GIANT can provide tissue-specific functional maps and predictions of tissue-specific gene function and disease association. Multi-tissue view allows for rapid examination of tissue-specific rewiring of functional connections across diverse tissues (Fig. 3a). Custom gene set functionality is implemented using the Tribe web service and is integrated into user analyses, such as biological process enrichment and querying by gene set. GIANT also provides a full NetWAS implementation, allowing users to upload gene-based association p-values to receive NetWAS association scores. Visualizations in the user-friendly dynamic web interface are implemented using the D3 library61, which enables use on any modern web browser without plugin installation. In addition to the interface, all of the underlying networks are provided for download, and the full list of input datasets and their sources is available through the webserver.
Discussion
Genes with tissue-specific expression and function play key roles in the physiological processes of complex organisms, and such genes are expected to underlie many human diseases62,63. Recent advances now allow for high-throughput discovery of genes expressed in specific lineages in solid tissues9,64. The next challenge is to understand the tissue-specific function of genes. This remains difficult because the precise functions of genes in multicellular organisms such as humans are defined by the context present in the cell lineage where they are expressed. Tissue-specific interactions are not well characterized because high-throughput interaction measurements are largely infeasible in solid tissues and their cell lineages. For direct studies of human genes, the available tools to assess tissue-specific function are generally confined to cell lines, many of which have diverged phenotypically from normal tissues. Moreover, many low-throughput experiments are highly skewed towards well-studied genes7,65.
We developed a data-driven approach that identifies tissue-specific interactions by integrating heterogeneous publicly available data using a tissue-specific regularized Bayesian framework. Our learned networks complement the tools of modern molecular genetics by allowing specific hypotheses about tissue-specific relationships to more precisely predict tissue-specific gene action. These lineage-specific networks also effectively connect genes’ roles in cell lineages to common diseases. We leverage this power in NetWAS, which uses genome-wide association studies as starting points for network analysis and provides a way to increase the value of existing GWAS. Other methods66 that reprioritize GWAS using networks are also expected to benefit substantially from tissue-specific networks. Analysis of genetic association data presents a key opportunity to apply tissue-specific networks to understand common human diseases. Because these networks accurately weigh and integrate diverse molecular data, they provide a more complete picture of the relationships between genes, phenotypes, and tissues and a clearer understanding of the etiology of complex disease. This is particularly important in the domain of phenome-wide association studies that rely on endpoints gleaned from electronic health records (EHRs)67. Tissue-specific networks can provide the necessary gene and tissue context to analyze such data and will help us scale methods to the repositories that we expect to see in the coming era of widespread EHR and genetic data.
Human health and disease states are the result of the interplay of genes within specific cell lineages and tissues, modulated by environmental exposures. Many of the key challenges in medicine involve tissue specificity. For example, identifying off-target effects of therapeutics requires an understanding of the therapeutics’ effect not just in the target tissue but also in all tissues. By disentangling the functions of genes in specific tissues, integrated tissue-specific networks learned from large data compendia present a means to address these challenges.
Methods
Data download/processing
We collected and integrated 987 genome-scale datasets encompassing approximately 38,000 conditions from an estimated 14,000 publications including both expression and interaction measurements. We downloaded interaction data from BioGRID1, IntAct2, MINT3, and MIPS4. BioGRID edges were discretized into five bins, labeled 0 to 4, where the bin number reflected the number of experiments supporting the interaction. For the remaining databases, edges were discretized into the presence or absence of an interaction.
Predicting transcriptional regulation based on DNA sequence is a major challenge to understanding transcription at a systems level. To estimate shared transcription factor (TF) regulation, binding motifs were downloaded from JASPAR5. Genes were scored for the presence of TF binding sites using the MEME software suite6. FIMO7 was used to scan for each TF profile within 1 kb upstream of each gene8. Motif matches were treated as binary scores (present if p < 0.001). The final score for each gene pair was obtained by calculating the Pearson correlation between the genes’ motif association vectors.
Chemical and genetic perturbation (c2:CGP) and miRNA target (c3:MIR) profiles were downloaded from the Molecular Signatures Database (MSigDB9). Each gene pair’s score was the sum of shared profiles weighted by the specificity of each profile (1/len(genes)). The resulting scores were converted to z-scores and discretized into bins ((-inf, −1.5), [−1.5, −0.5), [−0.5, 0.5), [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, 4.5), [4.5, inf)).
We downloaded all gene expression datasets from NCBI’s Gene Expression Omnibus10 (GEO) and collapsed duplicate samples. GEO contains 980 human datasets representing 20,868 conditions. Genes with more than 30% of values missing were removed, and remaining missing values were imputed using 10 neighbors11. Non-log transformed datasets were log transformed. Expression measurements were summarized to Entrez12 identifiers, and duplicate identifiers were merged. The Pearson correlation was calculated for each gene pair, normalized with Fisher’s z-transform, mean subtracted, and divided by the standard deviation. The resulting z-scores were discretized into bins ((-inf, −1.5), [−1.5, −0.5), [−0.5, 0.5), [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, inf)).
Knowledgebase construction and data integration
Hierarchically-aware Knowledgebase Construction via Ontological Pruning with Functional Knowledge Transfer
Functional Knowledge Extraction
We constructed a tissue-naïve functional relationship gold standard from a set of 564 expert-selected Gene Ontology (GO) biological process terms and experimentally derived gene annotations (GO evidence codes: EXP, IDA, IPI, IMP, IGI and IEP). Curators identified processes testable through specific molecular experiments (Supplementary Table 6). Pairs of genes that were co-annotated to expert-selected terms after propagation were treated as positive (i.e. functionally related) examples. Gene pairs not co-annotated to any of these terms were considered as negative examples, except in the following cases:
If two genes were annotated to two different GO terms with a significant number of shared genes (hypergeometric p-value < 0.05)
If two genes were co-annotated to a set of ‘negative’ GO terms that defined minimal relatedness13
Gene pairs that met either condition were excluded from the set of negative examples and treated as neither related nor unrelated.
Functional Knowledge Transfer
To increase the coverage of functional interactions, we transferred experimentally confirmed mouse GO annotations to human functional analogs identified by FKT14, a high-specificity annotation transfer method, for the 520 GO terms with mouse annotations. This resulted in a tissue-naïve gold standard of 604,038 functionally related gene pairs (positive examples) and 12,425,713 potentially un-related pairs (negative examples).
Ontology Pruning
Gene-to-tissue annotations were obtained from the Human Protein Reference Database (HPRD)15. HPRD tissues were mapped to the BRENDA Tissue Ontology16 (BTO) using direct matching where possible and manual curation where direct matches were unavailable (Supplementary Table 7). Tissues with fewer than ten directly annotated genes were pruned as non-informative from a molecular standpoint (e.g. BTO:0001493, trunk). Pruning resulted in an ontology containing functional, as opposed to structural, divisions of tissues and cell lineages (Supplementary Table 8). We defined ‘tissue categories’ from generic BRENDA terms, e.g. nervous system, to categorize tissues into organ systems for evaluation and analysis. For each tissue, we termed the set, T, as those genes directly annotated to that tissue or any of its descendants in the ontology. We used tissue categories to define unrelated tissues (those not associated with the same category as the tissue of interest). We defined T′ for each tissue as genes specifically annotated to unrelated tissues.
Annotation of Ubiquitously Expressed Genes
Genes ubiquitously expressed across tissues frequently carry out core biological processes and interact with tissue-specific genes to perform specialized functions17. We identified ubiquitous genes from a multi-tissue RNA-seq experiment18 and added ‘widely-expressed’ genes from a multi-cell-line mass spectroscopy experiment19, genes for proteins expressed in >75% of the tissues assayed in the human protein atlas,20 and curated ‘ubiquitous genes’ from HPRD15. These 8475 ubiquitous genes (U) were considered expressed in all tissues/cell-types, in addition to the curated tissue-specific genes (T). Sets T and U were made disjoint by retaining only genes in T genes that were not in U.
Integration of Tissue-specific and Functional Knowledge
We combined the curated gene-to-tissue annotations with the tissue-naïve functional gold standard to construct a hierarchical tissue-specific knowledgebase. We labeled each gene-pair (positive or negative) in the functional relationship standard as specifically co-expressed in a tissue if both genes were tissue-specific (T, T) or one was tissue-specific and the other ubiquitous (T, U). Interactions between ubiquitous gene-pairs were deemed non-tissue-specific and were ignored. After labeling specifically co-expressed gene-pairs/edges across all tissues, we considered four classes of edges – C1, C2, C3 and C4 – to constitute each tissue standard:
C1: positive functional edges between genes specifically co-expressed in the tissue [T–T and T–U]
C2: positive functional edges between a gene expressed in the tissue and another specifically expressed in an unrelated tissue [T–T′ and U–T′]
C3: negative functional edges between genes specifically co-expressed in the tissue [T–T and T–U]
C4: negative functional edges between one gene expressed in the tissue and another specifically expressed in an unrelated tissue [T–T′ and U–T′]
Among the four tissue classes, C1 represented tissue-specific functional relationships. To identify tissue-specific relationships, we constructed a specific gold standard for each tissue by labeling edges in C1 as positives and edges in the other classes as negatives. Because C3 is defined based on tissue-expressed genes and C2/4 on non-expressed genes, the number of edges in these classes varied across tissues based on how specific (cell-type/tissue/organ/system), well-studied (or easily-studied) and well-curated (literature bias) they are. To construct comparable networks across tissues, we used a negative set composed of equal proportions of edges from C2, C3 and C4. We limited all integrations to the set of 144 tissues (Supplementary Table 8) that contained at least 10 C1 edges between tissue-specific genes (T–T). This method incorporates the hierarchical relationships of tissues, allowing supervised methods to leverage these relationships.
Data integration
We constructed functional networks from genome-scale data by performing a tissue-specific Bayesian integration. We trained one naïve Bayesian classifier for each tissue using the tissue-specific standards described above. We also trained a classifier limited to only functional interactions to generate a tissue-naïve network. In each case, we constructed a class node, i.e. the presence or absence of a functional relationship between a pair of genes that is conditioned on nodes for each dataset. For large-scale genomics datasets, the assumption of conditional independence required for a naïve Bayes classifier is often not met, so we calculated and corrected for non-biological conditional dependency14.
Each tissue model trained on the hierarchy-aware tissue-specific knowledge was used to make genome-wide predictions by estimating the probability of tissue-specific functional interaction between all pairs of genes. We also estimated the probability of global functional interactions for the tissue-naïve network. We assigned a prior probability of a functional relationship of 0.01 for all models, allowing edge probabilities to be compared across tissues.
Code Availability
Integrations were performed with C++ naïve Bayesian learning implementations from the open source Sleipnir library for functional genomics21.
Evaluation of Tissue-specific Functional Relationships
We evaluated tissue-naïve and tissue-specific functional networks using five-fold cross-validation. The 6,062 genes represented in the tissue-specific knowledgebase were randomly partitioned into five sets. For each cross-validation run, gene pairs where neither gene was present in the holdout interval were used for training. Any gene pair where both genes were present in the holdout was used for evaluation of the area under the receiver operator characteristic curve (AUC). The estimated performance of each of the 144 functional networks was summarized as the median AUC of the five cross-validation runs (Supplementary Table 8).
Mapping datasets to tissues
We mapped datasets to tissues to compare with an integration of only tissue-specific data. Based on previous work22 that annotated samples from biological text, we extracted the title and description for each GDS dataset and annotated each using MetaMap23. This resulted in a mapping of GDS datasets to Unified Medical Language System (UMLS) terms. We applied the same process for the title and description of each BRENDA tissue and merged the two mappings by shared UMLS terms.
Characterization of tissue-specific molecular response
Network-based prediction
Top genes functionally connected to IL1B in a network (tissue-specific or naïve) were identified by ranking all genes based on the edge weight to IL1B normalized by their connection to all the genes. More precisely, in a network with V genes and interaction probabilities puv, the specific-connection (sv) of each gene v ∈ V in the network to a query gene u (IL1B, in this case) is:
This measure identified genes specifically connected to IL1B, which were compared to genes identified from the validation experiment described below.
Cell Culture
Human aortic vascular smooth muscle cells (HASMCs, Cambrex, Walkersville, MD) were maintained in smooth muscle cell growth medium with the manufacturer’s additives (SM-GM, Cambrex) and 10% Fetal Calf Serum (FCS) in 5% CO2 at 37 °C. Cells were expanded to sub-confluent cultures and split onto 100 mm culture dishes where they were grown to confluence. Subsequently, cells were rendered quiescent, by 24 hr incubation in serum-free medium, before stimulation with 10 ng/ml of IL1B (Sigma) for 2 hrs (n=4).
Gene Expression Analysis
Total RNA was isolated from HASMCs using Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA). Samples were prepared in one batch using the Nugen sample preparation protocol and hybridized to Affymetrix HG U-133A v2. CEL files were background corrected, normalized and summarized using RMA24 based on a custom CDF25. Differential expression analysis was carried out using LIMMA26, and genes induced at 2h post-stimulation compared to 0h were identified by ranking genes by their reported t-statistic. These data have been submitted to the GEO database (accession GSE59671).
Evaluation in Publicly Available Data
In addition to our validation experiment (GSE59671), we curated all series in GEO that included treatment of cells with IL1B and controls. This resulted in nine datasets: GSE13168 (airway smooth muscle), GSE26315 (amnion mesenchymal cells), GSE31679 (trophoblast cells), GSE40007 (endometrial stromal cells), GSE49604 osteoarthritis, GSE7216 (keratinocytes), GSE37624 (umbilical vein endothelial cells), GSE40560 (fibroblasts), and GSE40838 (peripheral blood mononuclear cells). Of these datasets, only GSE7216 was included in the data compendium used for integration. The rest were independent of the integration. To assess our networks’ ability to identify gene sets that would respond to IL1B treatment, we contrasted IL1B treatments with controls using GEO2R10. We queried the GIANT webserver for neighbors of IL1B in the tissue network that best corresponded to each dataset. In each tissue, genes were ranked based on the connectivity measure described above. We evaluated the mean fold change of the top twenty returned results. We evaluated randomly selected matched size sets of genes from each dataset as controls.
Evaluation of tissue-specific process, gene-level rewiring and disease-disease association
Mapping GO biological processes to tissues
To evaluate tissue-specific functional rewiring in our networks, we needed associations between tissues and tissue-specific processes. We used text matching followed by manual curation to map biological process (BP) terms in GO to tissue terms in the BRENDA Tissue ontology (Supplementary Table 9).
Network connectivity of tissue-specific processes
For each tissue, we constructed a tissue-minus-naïve network by subtracting edge probabilities of the naïve network from those of the tissue network. Negative weights were set to zero. In this subtracted network, positive scores corresponded to edges with a tissue-network interaction probability greater than the naïve-network probability. We expected relevant tissue-specific processes to be more connected in the tissue network than the naïve network and over processes that are not. For instance, for t-lymphocytes, ‘T cell receptor signaling pathway’ is a relevant process, while ‘neuron projection development’ is not. Within each subtracted tissue network, we ranked all tissue-specific processes by their edge density in the network and evaluated the extent to which relevant processes (positives) were ranked above processes specific to other tissues (negatives). Edge density for each process (with n genes) was calculated as the sum of weights divided by the total number of possible (n*(n-1)/2) edges between genes in that process. We measured the performance of the ranking using AUC and calculated a ‘best-AUC’ as the relative rank of the densest relevant process.
Gene-level rewiring across tissue-networks
Because tissue-specificity emanates from specialization of gene function, we identified genes with distinct functional neighborhoods in different tissue networks. We curated genes annotated to tissue-specific processes associated with at least two widely different tissues (descendants of different tissue categories). Using this gene-process-tissue mapping, we identified gene-tissue pairs, each with a set of relevant tissue-specific processes labeled positive and other processes annotated to the gene labeled negative. For example, the gene LEF1 is annotated to both tissues ‘blood vessel’ and ‘osteoblast’. In ‘blood vessel’, the term ‘angiogenesis’ would be a positive and ‘osteoblast differentiation’ would be a negative. Then, for a given gene-tissue pair (e.g. LEF1 and blood vessel), we calculated a z-score for the connectivity of a process (e.g. angiogenesis) using the formulation:
where μ and σ are the mean and standard deviation of the interaction probabilities of all genes to the query gene; n is the number of genes annotated to the process; m is the mean of the interaction probabilities of the process-genes to the query gene.
We ranked all processes by decreasing z-scores and quantified the separation between positively and negatively labeled processes using AUC. A high AUC for a gene across multiple associated networks showed that tissue networks reflected the gene’s annotation to multiple tissue-specific processes through preferential connectivity to the appropriate tissue-specific process in the matched tissue.
Disease-association map
We constructed a disease-association map, which represents a high-level view of functionally related diseases. As in Huttenhower et al.27, we calculated an association score between each disease pair using functional interactions between two diseases’ constituent genes. The score compared the means of two edge distributions: the edges between disease gene sets (between) and the edges that were incident to the disease gene sets genome-wide (background). We calculated a t-statistic as follows for disease pair i and j:
where Xw is the mean weight of edges between the two disease gene sets, Xb is the mean weight of all genome-wide edges incident to either gene set, and s and n are the respective standard deviations and sizes of the distributions.
We generated a bootstrapped null distribution for each disease pair by sampling 10,000 random gene set pairs of the same size and re-calculated the above t-statistic. With this null distribution, we calculated the final disease association score for each disease pair as follows:
where ti,j is the calculated t-statistic for a disease pair and μ and s are the mean and standard deviation of the null distribution. We applied a z-score cutoff of 2.5 to produce the Parkinson’s disease map in Figure 4.
Blinded Literature Evaluation of the Disease Association Map
To rigorously evaluate these maps, we constructed and shuffled a list of putative associations for Parkinson’s disease that combined associations from the disease map with ten randomly selected control associations. We provided this list to a researcher with no previous exposure to our manuscript or results. This researcher categorized disease associations from the literature as “strong” indicating there was clear evidence, “weak” indicating that there existed co-mentions but that the available evidence was limited, or “none” indicating that there were no publications with co-mentions.
Network Based Reprioritization of Genome-wide Association Study
We used tissue-specific networks to reprioritize gene candidates associated with hypertension endpoints in a GWAS. We hypothesized that disease-relevant genes would be enriched among the nominally significant genes, which would allow reprioritization through modern machine learning methods. We trained a support vector machine classifier using nominally significant (p < 0.01) genes as positive examples and 10,000 randomly selected non-significant (p >= 0.01) genes as negatives. The classifier was constructed using the tissue-network specific to kidney, a tissue associated with hypertension28, where the features of the classifier were the edge weights of the labeled examples to all the genes in the network. Genes were re-ranked using their distance from the hyperplane, which represented a network-based prioritization of a GWAS, termed NetWAS.
We applied NetWAS to a GWAS from the Women’s Genome Health Study to identify additional genes involved in hypertension.29 The study focused on three hypertension-related endpoints: systolic blood pressure, diastolic blood pressure, and hypertension diagnosis. To calculate per-gene p-values for each endpoint we used the versatile gene-based association study (VEGAS) system.30 To generate a combined list across phenotypes, we combined results from each hypertension-related endpoint using summed ranks. Performance was assessed by evaluating the ranking of genes annotated to ‘hypertension’ in OMIM. We performed functional evaluation by comparing NetWAS results to genes annotated to the term ‘regulation of blood pressure’ in GO. We performed an analogous calculation for therapeutics with targets of antihypertensive drugs from four different databases, DrugBank, TTD, PharmGKB, and CTD.
Evaluation of additional GWAS data
We performed NetWAS on four additional GWAS: C-reactive protein levels (lnCRP)29, type 2 diabetes (T2D)31, body mass index (BMI)32, and advanced age related macular degeneration (advanced AMD)33. Publicly available studies were obtained from their respective websites (BMI, advanced AMD) or dbGaP34 (T2D, phs000007-pha000418). NetWAS was applied as described for the hypertension NetWAS analysis. The relevant OMIM diseases were used to evaluate NetWAS results in relevant tissues.
Supplementary Material
Acknowledgments
The first three authors are co-first authors and are listed alphabetically.
We sincerely thank Young-suk Lee and Dima Gorenshteyn for help in curating disease-associations and Lars Bongo and Max Homilius for help in processing expression data. We are grateful to all members of the Troyanskaya Lab for help in curating specific GO biological processes and for valuable discussions.
This work was primarily supported by R01 GM071966, R01 HG005998 to OGT, and U54 HL117798 to GAF. CSG was supported in part by T32 CA009528 and P20 GM103534. AKW was supported in part by T32 HG003284. This work was supported in part by P50 GM071508 and by NIH contract number HHSN272201000054C. OGT is a senior fellow of the Genetic Networks program of CIFAR.
Footnotes
Author Contributions
C.S.G., A.K., A.K.W. and O.G.T., conceived and designed the research. C.S.G., A.K., and A.K.W. performed computational analyses with contributions from D.S.H and R.Z. and E.R. performed the molecular experiments. A.K.W., R.A.Z., and C.S.G. developed the web-interface. D.I.C., B.M.H, E.Z., S.C.S and K.D. provided data. C.S.G., A.K., A.K.W and O.G.T. wrote the manuscript with input from E.R., T.G., G.A.F., and K.D., and revisions from all co-authors.
Competing financial interests
The authors declare no competing financial interests.
URLs
GIANT, a web portal for tissue-specific functional networks: http://giant.princeton.edu Sleipnir, an open source library for functional genomics: http://libsleipnir.bitbucket.org. Tribe, a web service that provides cross-server analysis of gene sets: http://tribe.greenelab.com
Accession Codes
Gene expression measurements of HASMCs with and without IL1B stimulation are available in the GEO database under accession GSE59671.
References
- 1.D’Agati VD. The spectrum of focal segmental glomerulosclerosis: new insights. Curr Opin Nephrol Hypertens. 2008;17:271–81. doi: 10.1097/MNH.0b013e3282f94a96. [DOI] [PubMed] [Google Scholar]
- 2.Cai JJ, Petrov DA. Relaxed purifying selection and possibly high rate of adaptation in primate lineage-specific genes. Genome Biol Evol. 2010;2:393–409. doi: 10.1093/gbe/evq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lage K, et al. A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes. Proc Natl Acad Sci U S A. 2008;105:20870–5. doi: 10.1073/pnas.0810772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey AK, Lu L, Wang X, Homayouni R, Williams RW. Functionally enigmatic genes: a case study of the brain ignorome. PLoS One. 2014;9:e88889. doi: 10.1371/journal.pone.0088889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huttenhower C, et al. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ju W, et al. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013;23:1862–73. doi: 10.1101/gr.155697.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Troyanskaya OG, Dolinski K, Owen AB, Altman RB, Botstein D. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae) Proc Natl Acad Sci U S A. 2003;100:8348–53. doi: 10.1073/pnas.0832373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers CL, Troyanskaya OG. Context-sensitive data integration and prediction of biological networks. Bioinformatics. 2007;23:2322–30. doi: 10.1093/bioinformatics/btm332. [DOI] [PubMed] [Google Scholar]
- 12.Hibbs MA, et al. Directing experimental biology: a case study in mitochondrial biogenesis. PLoS Comput Biol. 2009;5:e1000322. doi: 10.1371/journal.pcbi.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park CY, et al. Functional knowledge transfer for high-accuracy prediction of understudied biological processes. PLoS Comput Biol. 2013;9:e1002957. doi: 10.1371/journal.pcbi.1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–53. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 15.Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science (80- ) 2004;306:1555–8. doi: 10.1126/science.1099511. [DOI] [PubMed] [Google Scholar]
- 16.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang S, Rhee SY, Marcotte EM, Lee I. Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network. Nat Protoc. 2011;6:1429–42. doi: 10.1038/nprot.2011.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kofler S, Nickel T, Weis M. Role of cytokines in cardiovascular diseases: a focus on endothelial responses to inflammation. Clin Sci. 2005;108:205–213. doi: 10.1042/CS20040174. [DOI] [PubMed] [Google Scholar]
- 19.Liu JZ, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–45. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gremse M, et al. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–13. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–57. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 23.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 24.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–94. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 25.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–63. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goh KI, et al. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunner HG, van Driel MA. From syndrome families to functional genomics. Nat Rev Genet. 2004;5:545–51. doi: 10.1038/nrg1383. [DOI] [PubMed] [Google Scholar]
- 28.Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492–504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 29.Van Amerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–14. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 30.Reya T, et al. Wnt signaling regulates B lymphocyte proliferation through a LEF-1 dependent mechanism. Immunity. 2000;13:15–24. doi: 10.1016/s1074-7613(00)00004-2. [DOI] [PubMed] [Google Scholar]
- 31.Park SK, Son Y, Kang CJ. A strong promoter activity of pre-B cell stage-specific Crlz1 gene is caused by one distal LEF-1 and multiple proximal Ets sites. Mol Cells. 2011;32:67–76. doi: 10.1007/s10059-011-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutierrez A, et al. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood. 2010;116:2975–83. doi: 10.1182/blood-2010-02-269878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erdfelder F, Hertweck M, Filipovich A, Uhrmacher S, Kreuzer KA. High lymphoid enhancer-binding factor-1 expression is associated with disease progression and poor prognosis in chronic lymphocytic leukemia. Hematol Rep. 2010;2:e3. doi: 10.4081/hr.2010.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandhirajan RK, et al. Small molecule inhibitors of Wnt/beta-catenin/lef-1 signaling induces apoptosis in chronic lymphocytic leukemia cells in vitro and in vivo. Neoplasia. 2010;12:326–35. doi: 10.1593/neo.91972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JE, Wu SF, Goering LM, Dorsky RI. Canonical Wnt signaling through Lef1 is required for hypothalamic neurogenesis. Development. 2006;133:4451–61. doi: 10.1242/dev.02613. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Lee JE, Dorsky RI. Identification of Wnt-responsive cells in the zebrafish hypothalamus. Zebrafish. 2009;6:49–58. doi: 10.1089/zeb.2008.0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahler RA, et al. Lymphocyte enhancer-binding factor 1 (Lef1) inhibits terminal differentiation of osteoblasts. J Cell Biochem. 2006;97:969–83. doi: 10.1002/jcb.20702. [DOI] [PubMed] [Google Scholar]
- 38.Hoeppner LH, et al. Runx2 and bone morphogenic protein 2 regulate the expression of an alternative Lef1 transcript during osteoblast maturation. J Cell Physiol. 2009;221:480–9. doi: 10.1002/jcp.21879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noh T, et al. Lef1 haploinsufficient mice display a low turnover and low bone mass phenotype in a gender- and age-specific manner. PLoS One. 2009;4:e5438. doi: 10.1371/journal.pone.0005438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 41.Cohen MM. Biology of RUNX2 and Cleidocranial Dysplasia. J Craniofac Surg. 2013;24:130–3. doi: 10.1097/SCS.0b013e3182636b7e. [DOI] [PubMed] [Google Scholar]
- 42.Duan D, et al. Submucosal gland development in the airway is controlled by lymphoid enhancer binding factor 1 (LEF1) Development. 1999;126:4441–53. doi: 10.1242/dev.126.20.4441. [DOI] [PubMed] [Google Scholar]
- 43.Driskell RR, et al. Wnt-responsive element controls Lef-1 promoter expression during submucosal gland morphogenesis. Am J Physiol Lung Cell Mol Physiol. 2004;287:L752–63. doi: 10.1152/ajplung.00026.2004. [DOI] [PubMed] [Google Scholar]
- 44.Driskell RR, et al. Wnt3a regulates Lef-1 expression during airway submucosal gland morphogenesis. Dev Biol. 2007;305:90–102. doi: 10.1016/j.ydbio.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verkman AS, Song Y, Thiagarajah JR. Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease. Am J Physiol Cell Physiol. 2003;284:C2–15. doi: 10.1152/ajpcell.00417.2002. [DOI] [PubMed] [Google Scholar]
- 46.Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forno LS. Neuropathology of Parkinson’s disease. J Neuropathol Exp Neurol. 1996;55:259–72. doi: 10.1097/00005072-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 48.Veeriah S, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denison SR, et al. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 2003;22:8370–8378. doi: 10.1038/sj.onc.1207072. [DOI] [PubMed] [Google Scholar]
- 50.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Seaghdha CM, Fox CS. Genome-wide association studies of chronic kidney disease: what have we learned? Nat Rev Nephrol. 2012;8:89–99. doi: 10.1038/nrneph.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ridker PM, et al. Rationale, design, and methodology of the Women’s Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem. 2008;54:249–55. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 53.Ho JE, et al. Discovery and replication of novel blood pressure genetic loci in the Women’s Genome Health Study. J Hypertens. 2011;29:62–69. doi: 10.1097/HJH.0b013e3283406927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oldham PD, Pickering G, Roberts JA, Sowry GS. The nature of essential hypertension. Lancet. 1960;1:1085–1093. doi: 10.1016/s0140-6736(60)90982-x. [DOI] [PubMed] [Google Scholar]
- 55.Guyton AC. Blood pressure control--special role of the kidneys and body fluids. Science (80- ) 1991;252:1813–1816. doi: 10.1126/science.2063193. [DOI] [PubMed] [Google Scholar]
- 56.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wishart DS, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorn CF, Klein TE, Altman RB. PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol Biol. 2013;1015:311–20. doi: 10.1007/978-1-62703-435-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qin C, et al. Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res. 2014;42:D1118–23. doi: 10.1093/nar/gkt1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davis AP, et al. The Comparative Toxicogenomics Database: update 2013. Nucleic Acids Res. 2013;41:D1104–14. doi: 10.1093/nar/gks994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bostock M, Ogievetsky V, Heer J. D3: Data-Driven Documents. IEEE Trans Vis Comput Graph. 2011;17:2301–9. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 62.Cai JJ, Petrov DA. Relaxed purifying selection and possibly high rate of adaptation in primate lineage-specific genes. Genome Biol Evol. 2010;2:393–409. doi: 10.1093/gbe/evq019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forrest ARR, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffmann R, Valencia A. Life cycles of successful genes. Trends Genet. 2003;19:79–81. doi: 10.1016/s0168-9525(02)00014-8. [DOI] [PubMed] [Google Scholar]
- 66.Kohler S, Bauer S, Horn D, Robinson PN. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008;82:949–58. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2012:gks1158. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kerrien S, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–6. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Licata L, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–61. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mewes HW, et al. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 1999;27:44–48. doi: 10.1093/nar/27.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Portales-Casamar E, et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–10. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–8. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber BR, Bulyk ML. Meta-analysis discovery of tissue-specific DNA sequence motifs from mammalian gene expression data. BMC Bioinformatics. 2006;7:229. doi: 10.1186/1471-2105-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrett T, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troyanskaya O, et al. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 12.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–7. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers CL, Barrett DR, Hibbs MA, Huttenhower C, Troyanskaya OG. Finding function: evaluation methods for functional genomic data. BMC Genomics. 2006;7:187. doi: 10.1186/1471-2164-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park CY, et al. Functional knowledge transfer for high-accuracy prediction of understudied biological processes. PLoS Comput Biol. 2013;9:e1002957. doi: 10.1371/journal.pcbi.1002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gremse M, et al. The BRENDA Tissue Ontology (BTO): the first all-integrating ontology of all organisms for enzyme sources. Nucleic Acids Res. 2011;39:D507–13. doi: 10.1093/nar/gkq968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bossi A, Lehner B. Tissue specificity and the human protein interaction network. Mol Syst Biol. 2009;5:260. doi: 10.1038/msb.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsköld D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burkard TR, et al. Initial characterization of the human central proteome. BMC Syst Biol. 2011;5:17. doi: 10.1186/1752-0509-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 21.Huttenhower C, Schroeder M, Chikina MD, Troyanskaya OG. The Sleipnir library for computational functional genomics. Bioinformatics. 2008;24:1559–61. doi: 10.1093/bioinformatics/btn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid PR, Palmer NP, Kohane IS, Berger B. Making sense out of massive data by going beyond differential expression. Proc Natl Acad Sci. 2012;109:5594–5599. doi: 10.1073/pnas.1118792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. D010001275 [pii] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 25.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 27.Huttenhower C, et al. Exploring the human genome with functional maps. Genome Res. 2009;19:1093–106. doi: 10.1101/gr.082214.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guyton AC. Blood pressure control--special role of the kidneys and body fluids. Science (80- ) 1991;252:1813–1816. doi: 10.1126/science.2063193. [DOI] [PubMed] [Google Scholar]
- 29.Ridker PM, et al. Rationale, design, and methodology of the Women’s Genome Health Study: a genome-wide association study of more than 25,000 initially healthy american women. Clin Chem. 2008;54:249–55. doi: 10.1373/clinchem.2007.099366. [DOI] [PubMed] [Google Scholar]
- 30.Liu JZ, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–45. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meigs JB, et al. Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S16. doi: 10.1186/1471-2350-8-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall JC, et al. Sex-stratified Genome-wide Association Studies Including 270,000 Individuals Show Sexual Dimorphism in Genetic Loci for Anthropometric Traits. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fritsche LG, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–9. 439e1–2. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mailman MD, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.