Abstract
We have fabricated protein polymer-gold nanoparticle (P-GNP) nanocomposites that exhibit enhanced binding and delivery properties of the small hydrophobic molecule drug, curcumin, to the model breast cancer cell line, MCF-7. These hybrid biomaterials are constructed via in situ GNP templated-synthesis with genetically engineered histidine tags. The P-GNP nanocomposites exhibit enhanced small molecule loading, sustained release and increased uptake by MCF-7 cells. When compared to the proteins polymers alone, the P-GNPs demonstrate a greater than 7-fold increase in curcumin binding, a nearly 50% slower release profile and more than 2-fold increase in cellular uptake of curcumin. These results suggest that P-GNP nanocomposites serve as promising candidates for drug delivery vehicles.
Keywords: Proteins, Gold nanoparticles, Drug delivery, Nanocomposites
Introduction
The fabrication of multifunctional, stimuli-responsive organic-inorganic hybrid materials that can self-assemble into defined structures bears tremendous potential in drug delivery and medicine [1–4]. The synthesis of hybrids combining stimuli responsive materials and gold nanoparticles (GNPs) has been explored in large part due to their unique properties [5–9]. For example, a composite hydrogel material comprised of temperature-sensitive copolymers, N-isopropylacrylamide and acrylamide, embedded with GNPs, bearing a gold sulfide nanoshell designed to absorb and convert near-IR light to heat has been developed [10]. Upon light triggered activation, the copolymer, when entrapped with a small molecule drug, undergoes a conformational change that in turn leads to drug release [10]. Another example of using gold nanoparticles for triggered drug release relies on liposomal nanoparticles composed 1,2-dipalmitoylsn-glycero-3-phosphocholine, 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[carboxy(polyethylene glycol)-2000] [11]. Such liposomes, when loaded with both GNPs and the hydrophilic drug calcein, when subjected to 532 nm laser treatments leads to light controlled calcein release due to microbubble cavitation of the liposome membrane[11]. While a wide range of synthetic materials have been developed and explored [10,12–14], proteins have attracted intense attention due to the fine molecular and conformational control of sequence and structure [15,16]. Recently, GNPs have been decorated with a library of cationic groups that complex non-covalently with green fluorescent protein (GFP) [17]. These GFP-GNP complexes have been employed in an array to chemically detect human serum proteins in complex serum. In this case, the strategy for construction of such protein-GNP hybrids rely on the covalent or non-covalent linkage of chemically pre-fabricated GNPs that have been synthesized under harsh organic solvents [18,19].
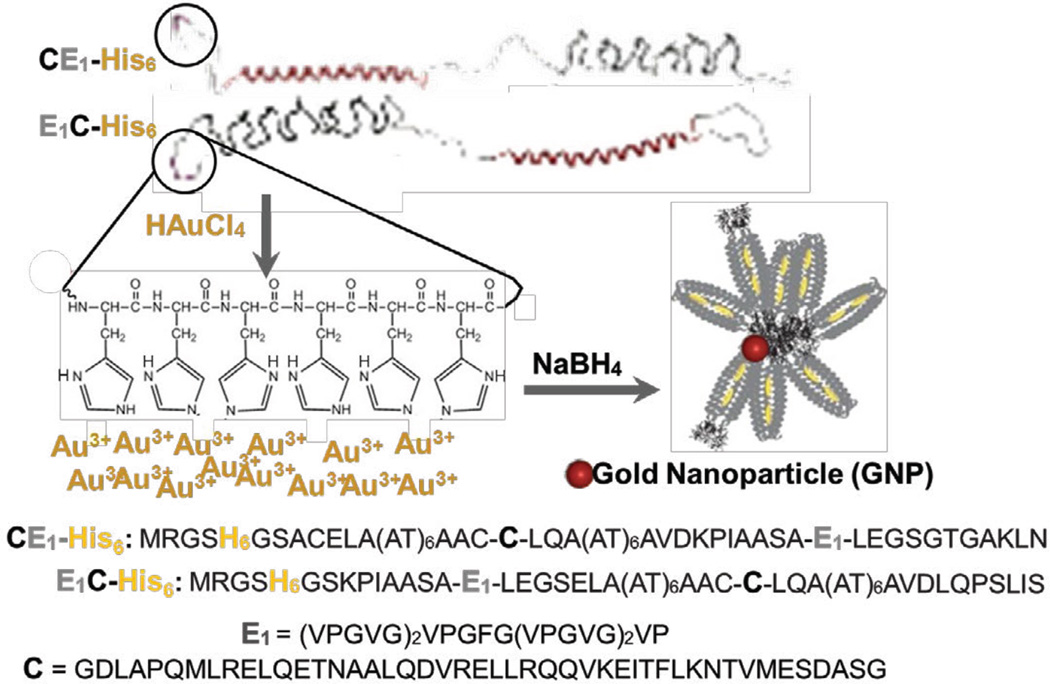
Specific chemical transformations are employed to prepare GNPs of discrete sizes and additional chemical steps are required to further decorate GNPs with key ligands as well as conjugate them with the macromolecule of interest [20]. Yet biological systems are able to fabricate GNPs under ambient conditions in situ through specific sequences [19,21–26]. We seek to generate multifunctional protein materials capable of: (i) templated-synthesis of inorganic nanoparticles in situ to fabricate organic-inorganic hybrids without the need for covalent bonding between each substituent part; (ii) encapsulating and stabilizing large payloads of small molecules; and (iii) modulating the delivery of small molecule chemotherapeutic drugs in clinically relevant cells (Figure 1).
Figure 1.
Protein polymer sequences of CE1-His6 and E1C -His6 and gold nanoparticle (GNP) templated-synthesis strategy.
Previously we have produced protein diblock copolymers comprised of two different self-assembling domains (SADs): 1) an elastin-like peptide (E); and 2) the coiled-coil region of Cartilage Oligomeric Matrix protein (C) [27,28]. While the diblocks, EC and CE, exhibit different temperature dependent conformations and self-assembly [27], they bind to curcumin [28], a naturally occurring anti-inflammatory agent bearing chemoprevention effects [29]. Curcumin has been chosen not only for its chemotherapeutic properties against the breast cancer cell line MCF-7 but also because it is insoluble and degrades rapidly under physiological conditions [29,30]. A drug delivery system that can solubilize and stabilize labile molecules such as curcumin would have beneficial therapeutic applications [29].
Here, we employ the two diblocks E1C-His6 and CE1-His6 each bearing an N-terminal hexahistidine tag for the templated-synthesis of gold nanoparticles (GNPs) in situ to yield the nanocomposites E1C-His6-GNP and CE1-His6-GNP, respectively (Figure 1). These protein polymers are selected due to their thermostability and superior small molecule binding abilities [31]. We hypothesize that such P-GNP nanocomposites will influence the thermoresponsiveness, drug binding capacity and release. Notably, E1C-His6-GNP and CE1-His6-GNP demonstrate elevated inverse temperature transitions, improved small molecule loading capacity, sustained release and enhanced uptake by cancer cells when compared to protein polymers alone.
Materials and Methods
General
Yeast extract and curcumin were obtained from Acros Organics (Geel, Belgium). Tryptic soy agar and gold(III) chloride trihydrate were acquired from MP Biomedicals (Santa Ana, CA). Ampicillin, isopropyl β-D-1-thiogalactopyranoside (IPTG), imidazole, sodium monobasic phosphate, sodium dibasic phosphate, sodium dodecyl sulfate, sodium hydroxide, sodium chloride, sucrose, tris-hydrochloride, tryptone, PFU high fidelity, DpnI, ACS grade methanol and urea were obtained from Fisher Scientific (Pittsburgh, PA). 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), magnesium sulfate, nickel chloride, sodium borohydride were purchased from Sigma Aldrich (St. Louis, MO). Tricine was purchased from Alfa Aesar (Ward Hill, MA). Glacial acetic acid and Factor Xa cleavage kit were purchased from EMD Millipore (Rockland, MA). Ethyl acetate was purchased from Pharmco-AAPER (Brookfield, CT). Ethylenediaminetetraacetic acid (EDTA) and hydrochloric acid were acquired from VWR (Radnor, PA). HPLC grade methanol was obtained from Ricca Chemical Company (Arlington, TX). Sephadex™ G-25 medium beads were purchased from Amersham Pharmacia Biotech AB (Piscataway, NJ). Columns were purchased from Bio-Rad (Hercules, CA).
Site-directed mutagenesis
pQE30/CE1 and pQE30/E1C were employed for production of CE1-His6 and E1C-His6 proteins in this study [31]. In order to generate proteins with Factor Xa IEGR cleavage site, site-directed mutagenesis was performed using the following primers: 5’-cgcagtagcagcgagctcgcgcccttctatgtgatggtgatggtg-3’ and 5’-cgcgctagccgcaatgcgcccttctatgtgatggtg atggtg-3’ and their reverse complements to generate PQE30/CE1-IGER and PQE30/E1C-IGER, respectively. Following the standard protocol for parent strand digestion using Dpn1, the resulting product was transformed into XL1-Blue cells for future use. Mutations were verified by DNA sequencing at Eurofins (Huntsville, AL).
Protein expression and purification
Biosynthesis and purification of CE1-His6, E1C-His6, CE1-IEGR and E1C-IEGR, was performed as previously described (Figure S1) [31]. Briefly, PQE30/CE1, PQE30/E1C, PQE30/CE1-IGER and PQE30/E1C-IGER were used to express the CE1-His6, E1C-His6, CE1-IEGR and E1C-IEGR proteins, respectively. All proteins were purified on a HiTrap IMAC FF column charged with nickel under denaturing conditions. For the negative control, CE1-IEGR and E1C-IEGR were dialyzed in 10 mM sodium phosphate buffer, pH 8.0, using SnakeSkin dialysis tubing (Thermo Scientific, 3.5 K MWCO). Factor Xa cleaves the protein after IEGR site, removing the His-tag. This reaction occurs in 1 µL of 0.5 unit/µL enzyme, 44 µL protein sample of 0.2 mg/mL concentration and 5 µL cleavage buffer (final cleavage buffer condition is 2 mM Tris-HCl, 50 mM NaCl, 0.5 mM CaCl2, pH 7.25). This ratio was scaled up to cleave 4 mL of the samples and cleavage reaction was allowed for 4 days at 4°C. This solution containing cleaved protein, His-tag and Factor Xa was transferred into Factor Xa capture resin and then passed through nickel beads to isolate the cleaved CE1 and E1C (Figure S2). After confirming the purity using sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE), CE1-His6, E1C-His6, CE1 and E1C were dialyzed into 10 mM sodium phosphate buffer, pH 8.0.
Gold nanoparticle templated-synthesis
A 0.1 M HAuCl4·3H2O solution (reactive gold solution) was prepared in dH2O. Approximately, 1.2 µL of the reactive gold solution was added into 300 µL of 10 µM protein sample, followed by gentle vortex for 10 minutes at room temperature. To the mixture, a 3.6 µL freshly prepared 0.1 M NaBH4 solution in dH2O, was added to reduce Au3+ to Au0. The mixture was then gently rotated to prevent aggregation or uneven templated-synthesis. The reaction was carried at room temperature for 1 hour. The molar ratio of Au3+ to protein was 40 to 1, while the NaBH4 to Au3+ ratio was 2.5 to 1. The resulting protein polymer-gold nanoparticle (P-GNP) nanocomposites were stored at room temperature for 1 hour before further characterization.
Absorbance spectroscopy
The absorbance spectrum from 200 nm – 1000 nm of each P-GNP nanocomposite was scanned using SpectraMax M2 (Molecular Devices) in UV-transparent 96 well microplate (Corning, half area flat bottom). As a control, buffer, CE1-His6 and E1C-His6, in addition to the cleaved CE1 and E1C proteins at pH 8 were scanned. All protein samples were prepared at 10 µM in 10 mM sodium phosphate buffer, pH 8.0.
Transmission electron microscopy
Transmission Electron Microscopy (TEM) was used to identify the nanometer-sized structures that resulted from self-assembly at room temperature. Samples were prepared in water at 10 µM concentrations in 10 mM sodium phosphate buffer pH 8.0. The samples were gently mixed and applied on a carbon coated 400 mesh Cu/Rh grids and negatively stained with 1% uranyl acetate as previously described [31]. The images of the samples were collected on a Phillips CM12 TEM instrument at 120 kV. The particle area and size were measured using Image J [32–34]. The protein particle sizes were determined from at least >130 particles, while sizes of the resulting GNPs were determined from at least >130 particles via Image J [32–34]. A histogram of the GNP sizes was generated to determine the average size distribution.
Circular dichroism (CD) spectroscopy
Wavelength-dependent Circular Dichroism (CD) spectra were collected on a Jasco J-815 CD Spectrometer equipped with a PTC-423S single position Peltier temperature control system and counter-cooled with an Isotemp 3016S (Fisher Scientific) water bath. Samples were loaded in a Hellma 218 quartz cuvette (500 µL, 1 mm path length). A far-UV temperature-dependent wavelength scan from 185–260 nm as a function of temperature was completed for CE1-His6 and E1C-His6 in the absence and presence of GNPs at 0.2 mg/mL in 10 mM sodium phosphate buffer pH 8.0 at scan rate of 50 nm/min for a range of temperatures (25–90°C) with 3 accumulation scans. At least two batches of separately purified proteins were measured. CD data was converted into mean residue molar ellipticity (MRW) via equation [θ]MRW = θ·MW/(10·n·C·l), where θ is in mdeg, MW is molecular weight, n is amino acid number in protein, C is concentration in mg/mL, l is path length in cm [35]. Fitting and calculation of protein secondary structure was processed with CDSSTR methods [36–38]. Parameters for the calculation using CDSSTR program were identical to our previously published work [31].
Turbidometry
The turbidometry, or inverse temperature transition (Tt), was determined via UV-Vis Spectrophotometer Cary-50 (Agilent Technology) equipped with TC 125 temperature controller (Quantum Northwest) in Type 21 quartz cuvette with 10 mm path length (Buck Science) by monitoring the change in turbidity at 800 nm from 25°C to 80°C at a rate of 1°C/min. Protein stock solutions for Tt measurement were prepared in 0.2 mg/mL (or 14.3 µM and 14.4 µM for CE1-His6 and E1C-His6, respectively) in 10 mM sodium phosphate buffer, pH 8.0. In order to bring Tt value of all the samples into instrument operation range, highly concentrated NaCl solution was added prior to Tt measurement (Table S1). Measurements were performed on at least two different protein sample preps to calculate the average Tt. The Tt was determined at the midpoint of the normalized turbidity [39].
Small molecule loading and release
Curcumin (6.5 nmol final concentration from 3 mM stock solution in HPLC grade methanol) was incubated with 1.3 nmol of CE1-His6, E1C-His6, CE1-His6-GNP and E1C-His6-GNP at room temperature for 2 hours and loaded onto Bio-Rad Spin6 columns packed with Sephadex G-25 medium beads 0.5 cm high. Bound protein polymer-curcumin complexes (in the presence or absence of GNP) were eluted by size, washed 3 times in 50 µL sodium phosphate buffer, followed by centrifugation for 5 min at 14000 rpm. The beads containing unbound curcumin were collected separately and resuspended back to buffer for solvent extraction. Both bound and unbound curcumin were extracted by adding 150 µL ethyl acetate and quantitatively determined by measuring absorbance at 416 nm. Absorbance was measured in a Hellma 105.201-QS type cuvette (10 mm light path, 100 µL sample) on SpectraMax M2. This binding study was performed on at least three different protein sample preparations to calculate the average loading capacities with errors represented as the standard deviation of the three trials.
Release of curcumin from CE1-His6, E1C-His6, CE1-His6-GNP and E1C-His6-GNP was then investigated. Curcumin (26 nmol) was added to 200 µL of 26 µM (5.2 nmol) protein sample. After 2 hours of incubation at room temperature, the solution was adjusted to contain a final concentration of 0.5 M NaCl. The protein polymer-curcumin complex (in the presence or absence of GNP) were incubated at 45°C (well above the Tt) for 30 min and centrifuged to separate protein polymer-curcumin complex from excess curcumin. The pellets were resuspended in 200 µL of 50 mM phosphate buffer, pH 7.4 and kept at room temperature in the dark for release. After 10 min, the suspensions were centrifuged and the supernatant was removed and used for extraction assessment of released curcumin. This resuspension-incubation-spin-release cycle was repeated for the next eight hours at the following time points: 10, 25, 55, 85, 135, 195, 255, 315 and 495 min. Release study was performed on two different protein sample preparations to obtain the averaged release profile. Error bars on the release data represented standard error of the two sample preparations.
Cell culture studies
MCF7 human breast cancer cells were obtained from ATCC and maintained at 37°C, 5% CO2 as monolayer cultures in Dulbecco’s modified Eagle’s medium (DMEM with high glucose containing phenol red) supplemented with 10% (v/v) fetal bovine serum (FBS), gentamicin (50 µg/mL), 100 U penicillin/ 100 µg/mL streptomycin. Because the loading capacities of curcumin for P-GNP nanocomposites are much larger than those of the proteins in the absence of GNP we prepared two corresponding curcumin controls that represented the bound curcumin levels in P-GNP and protein polymers alone (Table S2). To avoid any uptake of unbound curcumin by the cells directly, we limited the curcumin amount that is equivalent to the loading capacity of 26 µM of protein samples in 50 mM sodium phosphate buffer, pH 7.4 and allowed to bind for 2 hours at room temperature prior to cell culture studies.
Multiple sets of experiments were performed to record curcumin uptake by image acquisition using FITC filter (Em: 520 nm) under fluorescence microscopy and direct measurement of curcumin uptake in cell extractions. Cells were grown directly on 24-well culture plates (8 × 104 cells/well) for cell extraction or on cover slips for microscopy. After 24 hours of cell plating, cells were treated for 4 or 24 hours with different combination of proteins with or without GNP and/or curcumin. For all the treatments, the total volume of samples with DMEM in 24-well plates was kept constant at 300 µL, with proteins prepared at 10 µM concentrations. The ratio of sample amount to number of cells was also kept constant. The results are representative of two such independent sets of experiments.
For direct measurement of curcumin uptake, cells were washed with Dulbecco’s phosphate buffered saline and lysed with 200 µL RIPA/ well (25 mM TrisHCl pH7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) at room temperature for 20 min with gentle shaking. Lysed cells were then collected and vortexed. For curcumin extraction, 150 µL ethyl acetate was added the lysed cells. Thorough extraction was ensured by violently shaking the lysate-solvent mixture for 30 seconds. Curcumin containing solvent phase was then separated by centrifuging at 14,000 RPM for 2 minutes at room temperature. Absorbance of curcumin in ethyl acetate was measured using SpectraMax M2 (Molecular Devices) in Hellma 105.201-QS type quartz cuvette (100 µL volume, 10 mm light path) at 416 nm [40,41].
For fluorescent imaging of curcumin uptake, cells on coverslips were fixed with 300 µL 4% paraformaldehyde solution in DPBS for 20 minutes at room temperature on a plate rocker [42]. Following fixation, cell-containing coverslips were washed 3 × 300 µL DPBS and were mounted on glass slides using DAPI containing mounting medium (Southern Biotech Dapi Fluoromount-G). Coverslips were then sealed using clear nail polish for viewing under microscopy and long-term storage. Cells were viewed using fluorescence microscope IX71 (Olympus) using DAPI (for cell nuclei) and FITC (for curcumin uptake) at 60× magnification while keeping the exposure time for the FITC images constant at 200 milliseconds.
Cell viability measurements were carried out using a CellTiter 96® Aqueous One solution kit (Promega) in a 96-well plate, seeded 1 × 104 cells/well. After 24 hours, the cells were treated for 4 hours or 24 hours with protein polymers and P-GNP nanocomposites with and without curcumin along with control treatments of curcumin alone and media alone. After the treatment periods, 20 µL [3–(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) was added to each well, followed by incubation at 37°C for 3 hours. The plate was centrifuged for 3 minutes at 2500 rpm and then subjected to absorbance measurements at 490 nm (Tables S3 and S4).
Results
Fabrication of P-GNP nanocomposites
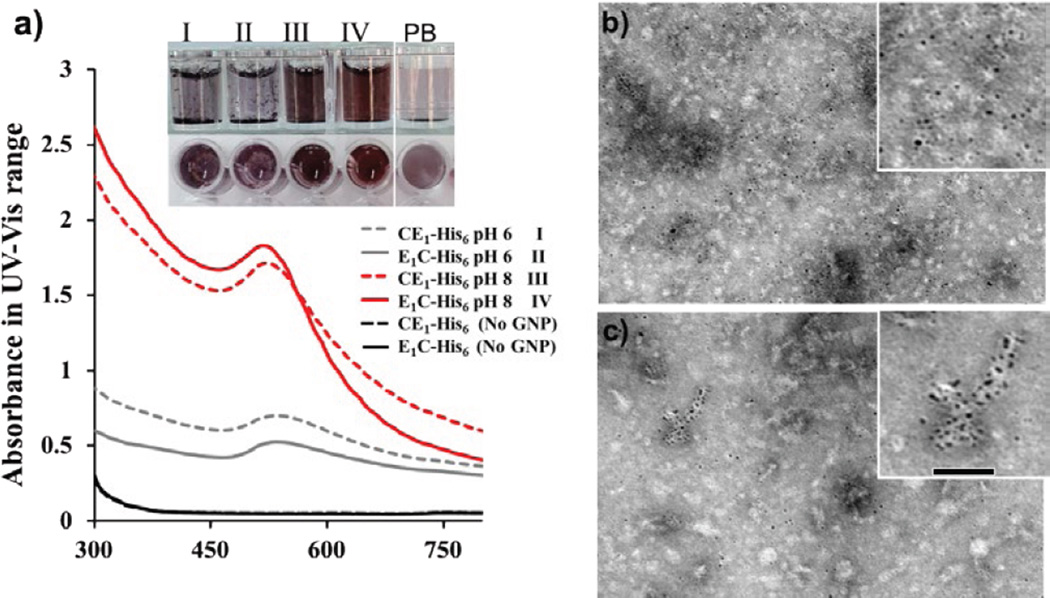
Both CE1-His6 and E1C-His6 were biosynthesized through recombinant bacterial expression and purified via nickel affinity resin. The protein diblock polymers were subject to GNP templated-synthesis without use of capping reagents. Gold salt (HAuCl4) solution was directly added to protein samples [24,25], followed by reduction with NaBH4 [43] under pH 6 and 8 (Figure 2a). Surprisingly, the P-GNP nanocomposites were stable at pH 8; within one week, absorbance spectra of complexes remained nearly the same with no observed precipitation even after one month when stored at room temperature (data not shown). The CE1-His6-GNP and E1C-His6-GNP exhibited successful templated-synthesis of GNPs with a distinct red-brown color change, confirmed by an observable peak at ~520 nm under pH 8 (Figure 2a). Since the lone pair electron on ε2N of histidine is protonated at pH ≤ 6, the protein polymer did not undergo GNP templated-synthesis very well under pH 6 conditions, consistent with literature [26]. Both CE1-His6 and E1C-His6 in the absence of gold salt did not lead to any detectable absorption peak at 520 nm (Figure S3); gold salt in the absence of protein did not produce signal indicating that the protein polymers were necessary for GNP templated-synthesis (Figure 2a). To affirm that the GNP templated-synthesis was due to the His6 tag, proteins lacking the N-terminal His6 sequence did not exhibit a strong signal at 520 nm (Figure S3).
Figure 2.
In situ gold nanoparticle (GNP) templated-synthesis by protein polymer sequences. (a) UV-Vis spectrum of protein polymer-GNP nanocomposites at pH6 and pH8 (inset shows the templated-synthesis products of CE1-His6-GNP pH6 (I), E1C-His6-GNP pH6 (II), CE1-His6-GNP pH8 (III), E1C-His6-GNP pH8 (IV) and phosphate buffer-GNP pH8 (V)). TEM of (b) CE1-His6-GNP and (c) E1C-His6-GNP at pH8.
Morphological characterization of P-GNP nanocomposites
To assess the morphology and sizes of the P-GNP nanocomposites, transmission electron microscopy (TEM) was performed (Figures 2b and 2c). As expected [31], the CE1-His6-GNP and E1C-His6-GNP assembled into nanoparticles with diameters of 23.8 ± 5.6 nm and 23.9 ± 5.2 nm, respectively (Table 1 and Figure S4). Average diameters of GNPs in both CE1-His6-GNP and E1C-His6-GNP were 3.4 ± 0.9 nm and 3.5 ± 0.9 nm, respectively (Table 1 and Figure S5). Consistent with published work, the observed absorption peak at 520 nm is due to the GNP diameters being within 2–10 nm range (Figure S6) [44].
Table 1.
Particle sizes, Tt and Loading capacities of proteins in the presence and absence of GNPs.
| Composite | Size of protein particles (nm) |
Size of GNPs (nm) |
Tt (°C) | CCM/Pc molar binding ratio |
|---|---|---|---|---|
| CE1-His6 | 26.0 ± 3.0a | N/A | 55.0 ± 0.8 | 0.40 ± 0.06 |
| E1C-His6 | 27.9 ± 3.7a | N/A | 33.8 ± 2.2 | 0.41 ± 0.10 |
| CE1-His6- GNP |
23.8 ± 5.6b | 3.4 ± 0.9 | 66.2 ± 0.8 | 3.16 ± 0.44 |
| E1C-His6- GNP |
23.9 ± 5.2b | 3.5 ± 0.9 | 42.1 ± 7.1 | 2.95 ± 0.42 |
Data from (ref 28).
Sizes were measured on P-GNP nanocomposites from > 130 particles.
Ratio of Curcumin to protein or P-GNP.
Secondary structure analysis of P-GNP nanocomposites
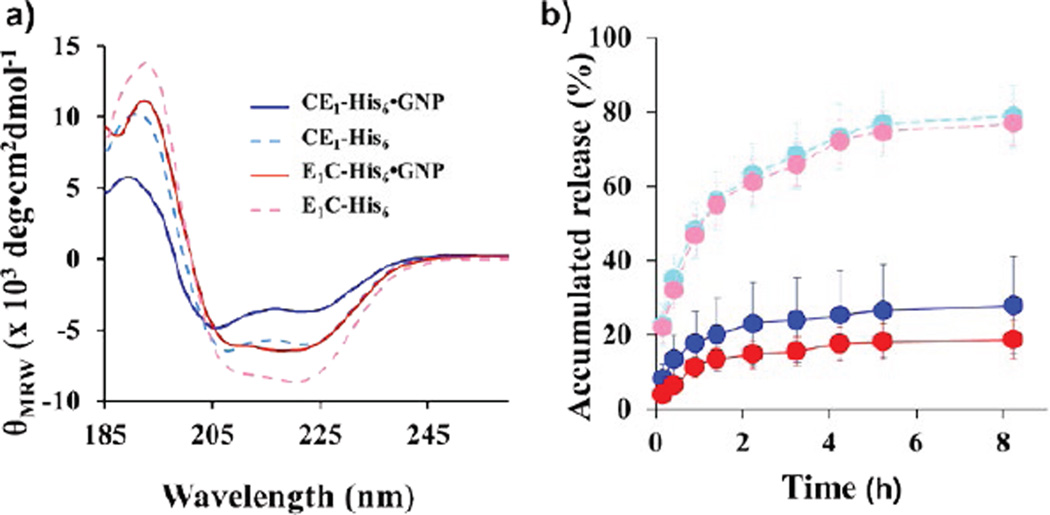
A comparison of the secondary structures in the presence and absence of GNP was performed via circular dichroism (CD) to determine whether GNP templated-synthesis affected the protein polymer conformations (Figure 3a). While the overall shape of the wavelength scans were maintained, a slight loss in structure was observed for CE1-His6-GNP and E1C-His6-GNP relative to CE1-His6 and E1C-His6, respectively (Figures 3a, S7 and S8). To assess the impact of GNP templated-synthesis on the inverse temperature transition (Tt), the UV/vis absorbance of CE1-His6-GNP and E1C-His6-GNP at 800 nm was monitored as a function of temperature (Table 1). Relative to the parent protein polymers, CE1-His6-GNP and E1C-His6-GNP revealed an increase in Tt by 11.2°C and 8.3°C, respectively.
Figure 3.
(a) CD wavelength scans of protein polymers in the absence and presence of GNP. (b) Accumulated release of CCM as a function of time.
Curcumin loading and release
To evaluate the loading capacity of the protein polymers in the absence and presence of GNP, curcumin was incubated with CE1-His6, E1C-His6, CE1-His6-GNP and E1C-His6-GNP for 2 hours. Unbound curcumin was then separated and quantified to determine the amount of curcumin bound to the protein polymer and P-GNP complexes (Table 1). Surprisingly, CE1-His6-GNP exhibited higher binding capacity than CE1-His6 by 8 fold, while E1C-His6-GNP demonstrated a 7.3 fold improvement over E1C-His6.
Release studies were performed by loading the protein polymers and P-GNPs with curcumin and assessing the amount of free curcumin over time. The protein polymers alone released >50% curcumin after 1.4 hours; both CE1-His6 and E1C-His6 showed rapid and nearly complete release of 77.0% and 78.8% free curcumin by 8.25 hours (Figure 3b). By contrast, CE1-His6-GNP and E1C-His6-GNP, revealed a slow and sustained release of 27.9% and 18.8% free curcumin by 8.25 hours (Figure 3b). Thus, the P-GNP nanocomposites not only increased the binding capacity for curcumin but also, slowed down its release.
Curcumin uptake by breast cancer cells
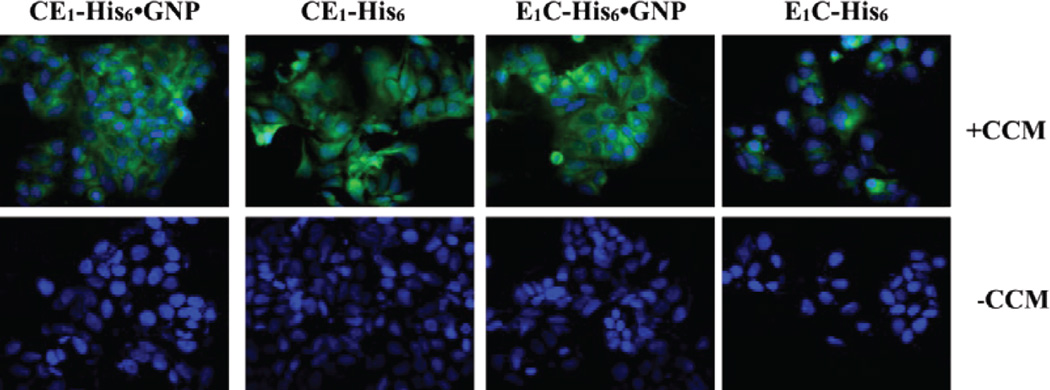
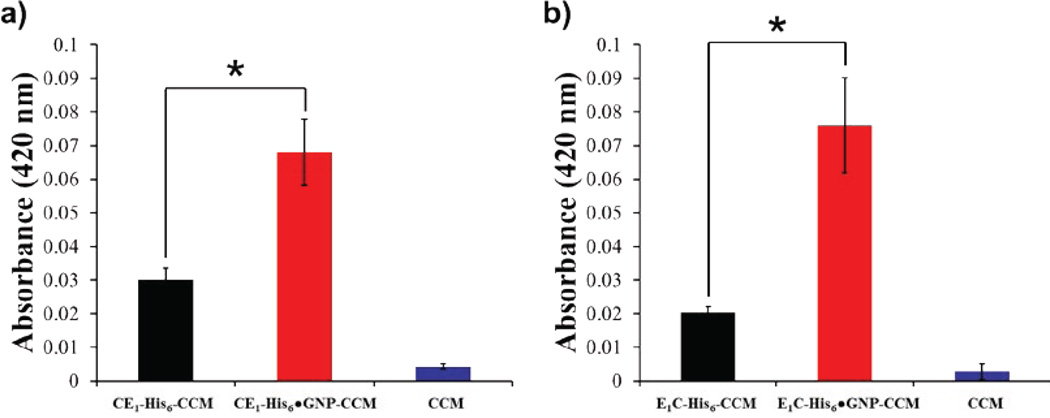
As curcumin is insoluble under aqueous conditions and does not effectively penetrate cancer cells alone [40], we investigated whether the P-GNP nanocomposites could enhance small molecule delivery and uptake by MCF7 breast cancer cells. Both CE1-His6-GNP and E1C-His6-GNP complexed with curcumin exhibited uptake as visualized by fluorescence (FITC channel); the curcumin appeared to be present in the cytoplasm as demonstrated by the overlay with DAPI stained cells (Figure 4). We also explored whether the protein polymers alone would deliver curcumin; both CE1-His6 and E1C-His6 revealed uptake albeit substantially less than the P-GNP nanocomposites (Figure 4). To assess whether CE1-His6-GNP, E1C-His6-GNP, CE1-His6 and E1C-His6 were themselves toxic to the cells, MTS assays were conducted; neither the protein polymer or P-GNP nanocomposites exhibited cytotoxicity (Table S3 and S4). Under identical conditions, the curcumin alone control did not show any uptake at the same concentrations of the protein polymers alone and the P-GNP nanocomposites. This was confirmed by quantifying curcumin extracted from the cells. Extraction of curcumin revealed 2.25-fold and 3.75-fold greater amount of available curcumin for CE1-His6-GNP and E1C-His6-GNP, respectively, relative to the protein polymers alone (Figure 5).
Figure 4.
Fluorescence microscopy images of MCF-7 cells treated with protein polymers alone or P-GNP nanocomposites in the absence and presence of curcumin (CCM).
Figure 5.
Quantification of uptake via extraction from cells. Absorption plots of (a) CE1-His6-CCM (black) CE1-His6-GNP-CCM (red), and CCM (blue) (p value < 0.05) and (b) E1C-His6 (black) E1C-His6-GNP-CCM (red) and CCM (blue) (p value < 0.05).
Discussion
Gold nanoparticle templated-synthesis influence on secondary structure and inverse temperature transition
We have produced stable P-GNP nanocomposites by GNP templated-synthesis through engineered N-terminal hexahistidine sequences within the protein diblocks CE1-His6 and E1C-His6. Either removing the hexahistidine sequence or decreasing the pH to alter the protonation state of the histidine residues does not lead to GNP production (Figure 2). After confirming GNP templated-synthesis to CE1-His6 and E1C-His6 spectroscopically, secondary structure analysis reveals that although a slight loss in alpha helicity is observed, the nanocomposites maintain overall conformation (Figure 3a). While in situ GNP templated-synthesis does not dramatically alter the protein polymer conformations, it does impact their thermoresponsive behavior. The marked effects on the thermoresponsiveness upon GNP templated-synthesis by the CE1-His6 and E1C-His6 protein polymers, regardless of the orientation of the domains can explain the improved loading capacity for curcumin. Upon GNP templated-synthesis, the P-GNP nanocomposites possessed elevated inverse temperature transitions (Table 1), indicative of heightened resistance to coacervative temperature-induced conformation changes. The enhanced stability against coacervation could impose greater mobility via increased hydration on the P-GNP nanocomposites thereby exposing more non-specific sites for curcumin binding leading to improved loading capacity.
Small molecule binding properties after gold nanoparticle templated-synthesis and in vitro delivery
The C domain present in both diblocks CE1-His6 and E1C-His6 is capable of binding small hydrophobic molecules such as curcumin [31]. This phytochemical possesses medically relevant pharmacological properties yet it fails to remain stable under physiological conditions [29]. Therefore, maximizing curcumin loading capacities and optimizing slower release profiles in carriers would be important for drug delivery. Upon GNP templated-synthesis of both protein diblocks with gold nanoparticles, there is a 7.3 and 8-fold increase in curcumin binding for CE1-His6-GNP and E1C-His6-GNP, respectively, when compared to the protein polymers alone (Table 1). Curcumin is interesting in that it only exhibits fluorescence upon binding to other molecules [40]. The curcumin bound P-GNP nanocomposites show quenching and a blue shift in the fluorescence spectra suggesting a proximity effect of the GNP on the fluorescence properties of curcumin (Figure S9). This further affirms that the P-GNP nanocomposites are binding to the curcumin.
The P-GNP nanocomposites demonstrate a prolonged release profile whereby nearly 70% of available curcumin was retained within both the P-GNP nanocomposites after 8.25 hours (Figure 3b). In contrast, the protein polymers alone released more than 50% of retained curcumin after 1.4 hours. The prolonged release profile could be due to: i) the binding of curcumin to GNPs and ii) the stabilization or increase in Tt observed upon GNP templation as mentioned. Previous work has demonstrated the ability of GNPs to bind small molecules directly [45], suggesting that the enhanced binding capacity of the P-GNP nanocomposites for curcumin could be attributed to the GNPs. The improved binding and stability provided by the GNP templation (Table 1), could cause to the prolonged release profile. These results translate to successful delivery into MCF-7 cells.
While it is unclear whether CE1-His6, E1C-His6, CE1-His6-GNP and E1C-His6-GNP get taken up by the cells, there is an improved delivery of curcumin by the P-GNP nanocomposites. Extraction of curcumin from treated MCF-7 cells reveals a greater than 2-fold increase in bioavailable phytochemical by both the nanocomposites relative to their protein polymers counterparts (Figures 5a and 5b). The high amount of curcumin recovered from the cells implies chemical protection and half-life extension of the labile, yet biologically active curcumin.
Conclusions
Remarkably, both CE1-His6-GNP and E1C-His6-GNP nanocomposites exhibit improved small molecule loading, slow and extended release as well as effective delivery when exposed to MCF-7 breast cancer cells. Further efforts are underway to elucidate the mechanisms by which P-GNP nanocomposites impact small molecule binding and releasing profile. These hybrid constructs can greatly broaden the biomaterials candidates for applications in targeted drug delivery. This can be achieved via the incorporation of tumor targeting domains in the solvent exposed residues of the protein polymer [46–48]. Furthermore, the drug loaded-nanocomposites, by way of templated-synthesis of GNP on the protein polymer, could be used for tandem chemotherapy and light-irradiated phototherapy [10,20,48].
Supplementary Material
Acknowledgments
We thank Youssef Wadghiri, Komal Jhaveri and Sungheon Kim for helpful discussions on cancer cell experiments. This work was supported by NSF DMR-1205384 (J.K.M.), Shiffrin Meyer Breast Cancer Discovery Fund (J.K.M.), and in part by the GK-12 Fellows Grant DGE-0741714 (J.A.F.), NIH CA175794 (A.S.) NSF MRSEC Program under Award Number DMR-1420073 as well as the NYU CTSA grant ULITR000038 from the National Center for Advancing Translational Sciences (NCATS), NIH.
Footnotes
Associated Content
Supporting information
SDS-PAGE gel images, circular dichroism spectra, secondary structure analysis via CDSSTR, Inverse transition temperature data, TEM images and analysis, curcumin fluorescence data, cell viability assay data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cole MA, Voelcker NH, Thissen H, Griesser HJ. Stimuli-responsive interfaces and systems for the control of protein-surface and cell-surface interactions. Biomaterials. 2009;30:1827–1850. doi: 10.1016/j.biomaterials.2008.12.026. [DOI] [PubMed] [Google Scholar]
- 2.Jeong B, Gutowska A. Lessons from nature: Stimuli-responsive polymers and their biomedical applications. Trends Biotechnol. 2002;20:305–311. doi: 10.1016/s0167-7799(02)01962-5. [DOI] [PubMed] [Google Scholar]
- 3.Hsieh DS, Langer R, Folkman J. Magnetic modulation of release of macromolecules from polymers. Proc Natl Acad Sci U S A. 1981;78:1863–1867. doi: 10.1073/pnas.78.3.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens MM, Allen S, Sakata JK, Davies MC, Roberts CJ, et al. pH-dependent behavior of surface-immobilized artificial leucine zipper proteins. Langmuir. 2004;20:7747–7752. doi: 10.1021/la030440e. [DOI] [PubMed] [Google Scholar]
- 5.Link S, El-Sayed MA. Size and temperature dependence of the plasmon absorption of colloidal gold nanoparticles. The Journal of Physical Chemistry B. 1999;103:4212–4217. [Google Scholar]
- 6.El-Sayed MA. Some interesting properties of metals confined in time and nanometer space of different shapes. Acc Chem Res. 2001;34:257–264. doi: 10.1021/ar960016n. [DOI] [PubMed] [Google Scholar]
- 7.Eustis S, el-Sayed MA. Why gold nanoparticles are more precious than pretty gold: noble metal surface plasmon resonance and its enhancement of the radiative and nonradiative properties of nanocrystals of different shapes. Chem Soc Rev. 2006;35:209–217. doi: 10.1039/b514191e. [DOI] [PubMed] [Google Scholar]
- 8.Zhu MQ, Wang LQ, Exarhos GJ, Li AD. Thermosensitive gold nanoparticles. J Am Chem Soc. 2004;126:2656–2657. doi: 10.1021/ja038544z. [DOI] [PubMed] [Google Scholar]
- 9.Surujpaul PP, Gutiérrez-Wing C, Ocampo-García B, Ramírez Fde M, Arteaga de Murphy C, et al. Gold nanoparticles conjugated to [Tyr3]octreotide peptide. Biophys Chem. 2008;138:83–90. doi: 10.1016/j.bpc.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Sershen SR, Westcott SL, Halas NJ, West JL. Temperature-sensitive polymer-nanoshell composites for photothermally modulated drug delivery. J Biomed Mater Res. 2000;51:293–298. doi: 10.1002/1097-4636(20000905)51:3<293::aid-jbm1>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 11.Mathiyazhakan M, Yang Y, Liu Y, Zhu C, Liu Q, et al. Non-invasive controlled release from gold nanoparticle integrated photo-responsive liposomes through pulse laser induced microbubble cavitation. Colloids Surf B Biointerfaces. 2015;126:569–574. doi: 10.1016/j.colsurfb.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg M, Langer R, Jia X. Nanostructured materials for applications in drug delivery and tissue engineering. J Biomater Sci Polym Ed. 2007;18:241–268. doi: 10.1163/156856207779996931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan R, Ringsdorf H, Satchi-Fainaro R. Polymer therapeutics--polymers as drugs, drug and protein conjugates and gene delivery systems: past, present and future opportunities. J Drug Target. 2006;14:337–341. doi: 10.1080/10611860600833856. [DOI] [PubMed] [Google Scholar]
- 14.Ma Y, Tang Y, Billingham NC, Armes SP, Lewis AL. Synthesis of biocompatible, stimuli-responsive, physical gels based on ABA triblock copolymers. Biomacromolecules. 2003;4:864–868. doi: 10.1021/bm034118u. [DOI] [PubMed] [Google Scholar]
- 15.Mart RJ, Osborne RD, Stevens MM, Ulijn RV. Peptide-based stimuli-responsive biomaterials. Soft Matter. 2006;2:822–835. doi: 10.1039/b607706d. [DOI] [PubMed] [Google Scholar]
- 16.More H, Yang CY, Montclare JK. Posttranslational Modification of Proteins Incorporating Nonnatural Amino Acids. Functional Polymers by Post-Polymerization Modification. 2012 [Google Scholar]
- 17.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, et al. Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein. Nat Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens MM, Flynn NT, Wang C, Tirrell DA, Langer R. Coiled-Coil Peptide-Based Assembly of Gold Nanoparticles. Advanced Materials. 2004;16:915–918. [Google Scholar]
- 19.Slocik JM, Moore JT, Wright DW. Monoclonal antibody recognition of histidine-rich peptide encapsulated nanoclusters. Nano Letters. 2002;2:169–173. [Google Scholar]
- 20.Stuchinskaya T, Moreno M, Cook MJ, Edwards DR, Russell DA. Targeted photodynamic therapy of breast cancer cells using antibody-phthalocyanine-gold nanoparticle conjugates. Photochemical & Photobiological Sciences. 2011;10:822–831. doi: 10.1039/c1pp05014a. [DOI] [PubMed] [Google Scholar]
- 21.Slocik JM, Wright DW. Biomimetic mineralization of noble metal nanoclusters. Biomacromolecules. 2003;4:1135–1141. doi: 10.1021/bm034003q. [DOI] [PubMed] [Google Scholar]
- 22.Djalali R, Chen YF, Matsui H. Au nanowire fabrication from sequenced histidine-rich peptide. J Am Chem Soc. 2002;124:13660–13661. doi: 10.1021/ja028261r. [DOI] [PubMed] [Google Scholar]
- 23.Hom N, Mehta KR, Chou T, Foraker AB, Brodsky FM, et al. Anisotropic nanocrystal arrays organized on protein lattices formed by recombinant clathrin fragments. J Mater Chem. 2012;22:23335–23339. doi: 10.1039/C2JM35019J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hainfeld JF, Liu W, Halsey CM, Freimuth P, Powell RD. Ni-NTA-gold clusters target His-tagged proteins. J Struct Biol. 1999;127:185–198. doi: 10.1006/jsbi.1999.4149. [DOI] [PubMed] [Google Scholar]
- 25.Minghui H, Luping Q, Raymond PB, Elena SL, James FH. Assembly of Nanoparticle–Protein Binding Complexes: From Monomers to Ordered Arrays. Angewandte Chemie International Edition. 2007;46:5111–5114. doi: 10.1002/anie.200701180. [DOI] [PubMed] [Google Scholar]
- 26.Djalali R, Chen YF, Matsui H. Au nanocrystal growth on nanotubes controlled by conformations and charges of sequenced peptide templates. J Am Chem Soc. 2003;125:5873–5879. doi: 10.1021/ja0299598. [DOI] [PubMed] [Google Scholar]
- 27.Haghpanah JSY, Civay DE, Barra H, Baker PJ, Khapli S, et al. Artificial Protein Block Copolymers Blocks Comprising Two Distinct Self-Assembling Domains. Chem Bio Chem. 2009;10:3. doi: 10.1002/cbic.200900539. [DOI] [PubMed] [Google Scholar]
- 28.Haghpanah JS, Yuvienco C, Roth EW, Liang A, Tu RS, et al. Supramolecular assembly and small molecule recognition by genetically engineered protein block polymers composed of two SADs. Mol Biosyst. 2010;6:1662–1667. doi: 10.1039/c002353a. [DOI] [PubMed] [Google Scholar]
- 29.Kawamori T, Lubet R, Steele VE, Kelloff GJ, Kaskey RB, et al. Chemopreventive effect of curcumin, a naturally occurring anti-inflammatory agent, during the promotion/progression stages of colon cancer. Cancer Res. 1999;59:597–601. [PubMed] [Google Scholar]
- 30.Syng-Ai C, Kumari AL, Khar A. Effect of curcumin on normal and tumor cells: role of glutathione and bcl-2. Mol Cancer Ther. 2004;3:1101–1108. [PubMed] [Google Scholar]
- 31.Dai M, Haghpanah J, Singh N, Roth EW, Liang A, et al. Artificial protein block polymer libraries bearing two SADs: effects of elastin domain repeats. Biomacromolecules. 2011;12:4240–4246. doi: 10.1021/bm201083d. [DOI] [PubMed] [Google Scholar]
- 32.Rasband WS. Image J. U. S. National Institutes of Health; 1997–2012. [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abrmoff MDM, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 35.Lee DL, Mant CT, Hodges RS. A novel method to measure self-association of small amphipathic molecules: temperature profiling in reversed-phase chromatography. J Biol Chem. 2003;278:22918–22927. doi: 10.1074/jbc.M301777200. [DOI] [PubMed] [Google Scholar]
- 36.Sreerama N, Venyaminov SY, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal Biochem. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 38.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 39.Urry DW. Molecular Machines: How Motion and Other Functions of Living Organisms Can Result from Reversible Chemical Changes. Angewandte Chemie International Edition in English. 1993;32:819–841. [Google Scholar]
- 40.Stankovic I. Curcumin: Chemical and Technical Assessment (CTA) Joint FAO/QHO Expert Committee on Food Additives. 2004;61:1–8. [Google Scholar]
- 41.Pan MH, Huang TM, Lin JK. Biotransformation of curcumin through reduction and glucuronidation in mice. Drug Metab Dispos. 1999;27:486–494. [PubMed] [Google Scholar]
- 42.Aubin-Tam ME, Hamad-Schifferli K. Gold nanoparticle-cytochrome C complexes: the effect of nanoparticle ligand charge on protein structure. Langmuir. 2005;21:12080–12084. doi: 10.1021/la052102e. [DOI] [PubMed] [Google Scholar]
- 43.Slocik JM, Kim SN, Whitehead TA, Clark DS, Naik RR. Biotemplated metal nanowires using hyperthermophilic protein filaments. Small. 2009;5:2038–2042. doi: 10.1002/smll.200900499. [DOI] [PubMed] [Google Scholar]
- 44.Daniel MC, Astruc D. Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem Rev. 2004;104:293–346. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 45.Zakaria HM, Shah A, Konieczny M, Hoffmann JA, Nijdam AJ, et al. Small molecule- and amino acid-induced aggregation of gold nanoparticles. Langmuir. 2013;29:7661–7673. doi: 10.1021/la400582v. [DOI] [PubMed] [Google Scholar]
- 46.Sudimack J, Lee RJ. Targeted drug delivery via the folate receptor. Adv Drug Deliv Rev. 2000;41:147–162. doi: 10.1016/s0169-409x(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Wang X, Zhang Y, Yang S, Wang J, et al. RGD-modified polymeric micelles as potential carriers for targeted delivery to integrinoverexpressing tumor vasculature and tumor cells. J Drug Target. 2009;17:459–467. doi: 10.1080/10611860902974085. [DOI] [PubMed] [Google Scholar]
- 48.El-Sayed IH, Huang X, El-Sayed MA. Surface plasmon resonance scattering and absorption of anti-EGFR antibody conjugated gold nanoparticles in cancer diagnostics: applications in oral cancer. Nano Lett. 2005;5:829–834. doi: 10.1021/nl050074e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.