Abstract
Vibrio are a genus of marine bacteria that have substantial environmental and human health importance, and there is evidence that their impact may be increasing as a consequence of changing environmental conditions. We investigated the abundance and composition of the Vibrio community within the Sydney Harbour estuary, one of the most densely populated coastal areas in Australia, and a region currently experiencing rapidly changing environmental conditions. Using quantitative PCR (qPCR) and Vibrio-specific 16S rRNA amplicon sequencing approaches we observed significant spatial and seasonal variation in the abundance and composition of the Vibrio community. Total Vibrio spp. abundance, derived from qPCR analysis, was higher during the late summer than winter and within locations with mid-range salinity (5–26 ppt). In addition we targeted three clinically important pathogens: Vibrio cholerae, V. Vulnificus, and V. parahaemolyticus. While toxigenic strains of V. cholerae were not detected in any samples, non-toxigenic strains were detected in 71% of samples, spanning a salinity range of 0–37 ppt and were observed during both late summer and winter. In contrast, pathogenic V. vulnificus was only detected in 14% of samples, with its occurrence restricted to the late summer and a salinity range of 5–26 ppt. V. parahaemolyticus was not observed at any site or time point. A Vibrio-specific 16S rRNA amplicon sequencing approach revealed clear shifts in Vibrio community composition across sites and between seasons, with several Vibrio operational taxonomic units (OTUs) displaying marked spatial patterns and seasonal trends. Shifts in the composition of the Vibrio community between seasons were primarily driven by changes in temperature, salinity and NO2, while a range of factors including pH, salinity, dissolved oxygen (DO) and NOx (Nitrogen Oxides) explained the observed spatial variation. Our evidence for the presence of a spatiotemporally dynamic Vibrio community within Sydney Harbour is notable given the high levels of human use of this waterway, and the significant increases in seawater temperature predicted for this region.
Keywords: Vibrio, seasonal variation, abundance and diversity, Vibrio cholerae, Vibrio vulnificus, Vibrio parahaemolyticus
Introduction
Marine microbes play essential ecological and biogeochemical roles in coastal and estuarine habitats (Ducklow and Carlson, 1992), but some species also pose a significant threat to ecosystem function and human health. A key group of marine pathogens responsible for diseases in a wide diversity of marine organisms, as well as illness in the human population, belong to the bacterial genus Vibrio. The Vibrio genus is comprised of a diverse group of gram-negative, largely marine and estuarine heterotrophic bacteria that frequently occur in close association with marine plants and animals, where they can act as both mutualistic symbionts (Lee and Ruby, 1994; Nyholm and Nishiguchi, 2008) or pathogens (Blackwell and Oliver, 2008; Vezzulli et al., 2012, 2015; Geng et al., 2014; Matteucci et al., 2015). As an important natural component of bacterioplankton communities they also contribute to biogeochemical cycling in aquatic habitats (Eiler et al., 2006; Thompson and Polz, 2006; Hasan et al., 2015). Due to their potentially significant impact on coastal ecosystems, marine animals, aquaculture (Higgins, 2000; Kushmaro et al., 2001; Ben-Haim and Rosenberg, 2002; Zorrilla et al., 2003; Austin and Zhang, 2006; Geng et al., 2014; Vezzulli et al., 2015) and human health (Daniels and Shafaie, 2000; Baker-Austin et al., 2013; Orata et al., 2014) an understanding of the spatiotemporal dynamics of Vibrios and their potential to bloom and cause disease outbreaks has become increasingly important (Lipp et al., 2002; Oberbeckmann et al., 2012; Takemura et al., 2014). This is particularly true in light of recent evidence that this group of organisms are increasing in abundance and impact in some regions as a consequence of environmental perturbations and climate change (Oberbeckmann et al., 2012; Froelich et al., 2013; Jacobs et al., 2014; Takemura et al., 2014).
Vibrios are copiotrophic bacteria that have the capacity to rapidly increase in abundance, shifting from a relatively rare component of coastal microbial communities to dominant members of the assemblage during “bloom” events (Thompson and Polz, 2006; Gilbert et al., 2011; Takemura et al., 2014). For example, during a 1 month period, the abundance of a single Vibrio sp. operational taxonomic unit (OTU), increased from <2 to 54% of the bacterial community inhabiting a coastal habitat, in apparent response to an increased abundance of a single diatom species (Gilbert et al., 2011).
As well as causing diseases in fish, corals, oysters, crustaceans, echinoderms, and other animals (Goarant et al., 2000; Kushmaro et al., 2001; Ben-Haim and Rosenberg, 2002; Becker et al., 2004; Frans et al., 2011; Geng et al., 2014; Vezzulli et al., 2015) multiple Vibrio spp. are also pathogenic to humans (Adebayo-Tayo et al., 2011; Baker-Austin et al., 2013; Geng et al., 2014; Orata et al., 2014; Kubota, 2015). Arguably the most clinically important pathogens for public surveillance matters are Vibrio cholerae, V. parahaemolyticus, and V. vulnificus (Daniels and Shafaie, 2000). In the USA, these three Vibrio spp. are responsible for an economic burden of $307 million USD per year (Ralston et al., 2011). Toxigenic V. cholerae is the causative agent of cholera, a disease for which around 2.8 million cases occur annually around the world, with an average of ∼93,500 deaths each year (Ali et al., 2012). Infections by V. parahaemolyticus and V. vulnificus are typically associated with either the consumption of undercooked seafood, or direct contact from swimming in coastal and estuarine waters (Daniels and Shafaie, 2000). In Japan, it is estimated that between 7,000 and 63,000 people experience foodborne illnesses caused by V. parahaemolyticus each year (Kubota, 2015). While in the USA, the estimated economic burden of medical costs associated with V. vulnificus infections represents approximately half of the total costs associated with food and water-borne marine pathogens and toxins (Ralston et al., 2011). Moreover, the capacity of V. vulnificus to cause wound infections that become septic is particularly notable, because these infections result in a ∼50% mortality rate (Daniels and Shafaie, 2000; Jones and Oliver, 2009; Horseman and Surani, 2011).
Understanding when and where outbreaks of pathogenic Vibrios will occur is essential for public health management (Hlady and Klontz, 1996; Ralston et al., 2011). The spatiotemporal dynamics of Vibrio populations and the occurrence of bloom events have been linked to several environmental drivers, including temperature, salinity, turbidity, dissolved oxygen (DO), pH, chlorophyll, and nutrients, as well as associations with potential host organisms (Takemura et al., 2014). In coastal environments, as well as estuaries and coastal rivers, elevated water temperatures and low salinity levels are also often significant explanatory factors for increases in Vibrio spp. abundance (Randa et al., 2004; Hsieh et al., 2008; Oberbeckmann et al., 2012; Froelich et al., 2013; Takemura et al., 2014), though there is evidence that the key environmental drivers can differ between species (Takemura et al., 2014). V. cholerae, V. vulnificus, and V. parahaemolyticus all generally prefer warm water temperatures (>20°C) and low (<10 ppt) salinity (Takemura et al., 2014), with infectious Vibrio outbreaks often tightly correlated with these parameters (Oberbeckmann et al., 2012; Jacobs et al., 2014; Takemura et al., 2014).
As a result of increasing sea surface temperatures driven by climate change, the Baltic Sea in Northern Europe has seen an increase in Vibrio-related wound infections with spikes in disease correlating with “heatwave” years (Baker-Austin et al., 2013). Similar correlations between Vibrio abundance (and infections) and temperature increases have been made in other parts of the world including Israel, the USA, Chile, Peru, and Spain (Centers for Disease Control and Prevention, 1998; González-Escalona et al., 2005; Paz et al., 2007; Martinez-Urtaza et al., 2008; Baker-Austin et al., 2010). It has been reported that an increase in sea surface water temperature of 3.7°C increases Vibrio-associated illness risk by 2–3 times (Sterk et al., 2015), while a 5°C temperature rise increases cholera risk by 3.3-fold (Huq et al., 2005; Colwell, 2009; Baker-Austin et al., 2013).
The links between pathogenic Vibrio spp. abundance and elevated seawater temperatures are particularly pertinent to south-eastern Australia, where seawater temperatures are currently rising more rapidly than in any other part of the southern hemisphere (Hobday and Lough, 2011). These increasing seawater temperatures are believed to be driven in part by the influence of the strengthening East Australian Current (EAC), a western boundary current that redistributes warm tropical waters into the temperate latitudes of the Australian east coast (Cai et al., 2005). The EAC is currently increasing in southerly extent as a result of climate change driven shifts in Pacific Ocean circulation patterns (Cai et al., 2005; Sun et al., 2012), and as a consequence southerly range expansions of tropical animal and plant species into the temperate latitudes of the Australian east coast have been observed (Poloczanska et al., 2007; Last et al., 2011). The EAC has also been implicated in the southward spread of populations of “tropical” microbial species into temperate latitudes of the Tasman Sea (Seymour et al., 2012).
It is predicted that by 2050 the average sea surface temperatures in south-eastern Australia will be 2°C higher than the 1990–2000 average, which will also result in higher temperature extremes during “heatwave” years (Hobday and Lough, 2011). Within the context of temperature-associated increases in the abundance of pathogenic Vibrios observed elsewhere (Huq et al., 2005; Oberbeckmann et al., 2012; Vezzulli et al., 2012; Baker-Austin et al., 2013; Jacobs et al., 2014; Takemura et al., 2014), current and predicted future increases in seawater temperatures raise the prospect for increased incidence of pathogenic Vibrios within Australian coastal waters. This is significant given that approximately 85% of the human population in this region reside within 50 km of the coast (Australian Bureau of Statistics, 2004). However, there is currently a severe lack of data regarding the natural occurrence and distributions of pathogenic Vibrios within the Australian marine environment or an understanding of the environmental processes underpinning them.
The Sydney Harbour estuary is one of the most densely populated coastal areas in Australia and the waters of this region are heavily used by commercial and recreational activities. This system has also been subject to significant contamination from heavy metals, organics, and a range of other pollutants (Birch, 1996; Thompson et al., 2011; Lee and Birch, 2014). The estuary hosts a diversity of habitats and high levels of biodiversity (Hutchings et al., 2013), but little is known about the microbiology of this environment. In this study, we investigated the dynamics of Vibrio spp., including human pathogen species, within Sydney Harbour.
Materials and Methods
Study Sites
This study was conducted within the Sydney Harbour estuary system, located on the central eastern coast of Australia (33°48′S 151°18′E). The Sydney Harbour estuary system spans an area of approximately 250 km2, and is completely encircled by the city of Sydney in all directions except to the east, where it meets the Tasman Sea. The estuary is exposed to high levels of anthropogenic impact (Lee and Birch, 2014) and is located in the temperate region of the east Australian coast – a region experiencing rapid increases in sea surface water temperatures (Hobday and Lough, 2011).
Sampling was performed at eight sites within the Sydney Harbour catchment and included four inner harbour and four outer harbour sites, spanning a salinity gradient from marine conditions near the mouth of the harbour at Chowder Bay (33°50′22″S 151°15′17″E) to the freshwater conditions in the Parramatta River to the west (33°48′36″S 150°59′46″E, Figure 1). Sites at Olympic Park (33°49′20″S 151°04′44″E) and Mcllwaine Park (Rhodes) (33°49′52″S 151°05′22″E) are situated furthest inland and near mangrove areas. The Hen and Chicken Bay (33°51′22″S 151°07′38″E) and Iron Cove (33°52′11″S 151°09′06″E) sites are directly adjacent to land areas that have been reclaimed using commercial and industrial waste (Suh et al., 2004). The sites at Rozelle Bay (33°52′20.0″S 151°10′32.6″E) and Iron Cove are adjacent to stormwater canals that are subject to regular discharges (Armitage and Rooseboom, 2000). The two most eastern sites at Mort Bay (33°51′13″S 151°11′05″E) and Chowder Bay are closest to the mouth of the Harbour. Sampling was conducted from jetties (piers) or pontoons at each site, whereby surface water samples were collected from points where the water depth was 2–5 m, with the exception of Hen and Chicken Bay where the water depth was only 1 m. Sampling was conducted on three occasions, corresponding to the late austral summer in March 2014, and the austral winter in June and August 2014.
FIGURE 1.
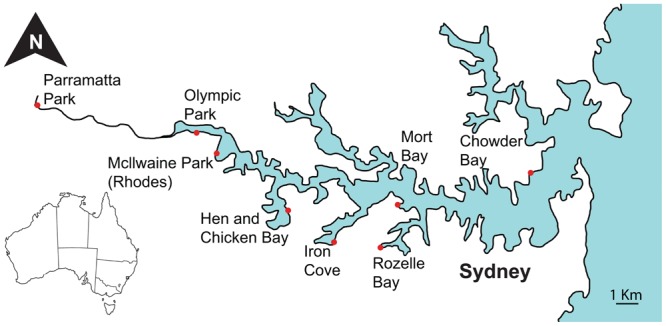
Map of sampling locations along the Parramatta River and within Sydney Harbour.
Sampling Protocols
At each of the sampling sites, approximately 16 L of water was collected from the surface using Nalgene bottles, and physicochemical parameters including temperature, DO, conductivity and pH were measured in situ using a WTW multiprobe meter (Multi 3430, Germany). Triplicate surface water samples (15 ml) were also collected at each site and filtered through 0.45 μm Sartorius Minisart filters (Satorius stedim Biotech) into 15 ml centrifuge tubes for nutrient analysis. All samples were immediately transported back to the laboratory on ice.
Nutrient Analysis
Concentrations of ammonium ion, phosphate, nitrite, and total nitrogen were determined using a colorimetric analysis with the Lachat Quikchem QC8500 Automated Ion Analyser (LACHAT Instruments, USA) in accordance with the manufacturer’s guidelines. The resulting data were interpreted using Omnion version 3 software (LACHAT Instruments, USA).
DNA Extraction
Microbial DNA was retrieved by filtering triplicate 2 L samples through 0.22 μm pore-size GP Sterivex membrane filters (Millipore) using a peristaltic pump. Filters were stored at -80°C until genomic DNA extraction was performed using the Powerwater DNA isolation Kit (MoBio Laboratoties, Inc.) in accordance with the manufacturer’s instructions, with the following minor modifications: (i) before extraction membrane filters were removed using sterilized scalpels and transferred into the bead tube and (ii) prior to the initial step of bead-beating and chemical lysis of cells, samples were heated to 60°C for 10 min to further aid cell lysis. DNA quantity and purity was evaluated using a Nanodrop-1000 Spectrophotometer.
Quantitative PCR (qPCR)
Quantitative PCR (qPCR) was used to track patterns in the abundance of the total Vibrio community as well as pathogenic V. cholerae, V. vulnificus, and V. parahaemolyticus. qPCR was performed using the StepOnePlusTM Real-Time PCR System (Applied Biosystems) and StepOne software version 2.3. All qPCR tests were run using three technical replicates, consisting of 20 μl reaction volumes containing 10 μl SYBR Select Master Mix, 3.4 μl nuclease free water, 0.4 μM for each forward and reverse primers and 5 μl of diluted (1:5) DNA template. Calibration curves were run with every plate and all extracted DNA was diluted fivefold to reduce pipetting errors.
To quantify patterns in the whole Vibrio community, the primer pair Vib1-f and Vib2-r (Thompson J.R. et al., 2004; Vezzulli et al., 2012) were used to amplify 16S rRNA genes specific to the Vibrio genus (Table 1). The qPCR cycling parameters involved: initial activation steps at 50°C for 120 s and 95°C for 120 s, followed by 40 cycles of a 2-step reaction involving denaturation at 95°C for 15 s and annealing/extension step at 60°C for 60 s. The amplification was concluded with a holding stage at 72°C for 120 s. To confirm that each primer pair produced only a single specific product, a melting curve was added to the end of every qPCR assay at every run (for all primers).
Table 1.
Primers for Q-PCR detection of Vibrio species and for next generation sequencing.
| Organism(s) | Target gene | Primers | Information on target gene | Reference |
|---|---|---|---|---|
| Vibrio genus | 16S rRNA | Vib1-f (5′-GGCGTAAAGCGCATGCAGGT-3′) Vib2-r (5′-GAAATTCTACCCCCCTCTACAG-3′) |
General Vibrio spp. (205-bp) | Thompson J.R. et al., 2004; Vezzulli et al., 2012 |
| Vibrio cholerae | ompW ctxA | OmpW-F (5′-AACATCCGTGGATTTGGCATCTG-3′) OmpW-R (5′-GCTGGTTCCTCAACGCTTCTG-3′) ctxA_F (5′-TTTGTTAGGCACGATGATGGAT-3′) ctxA-R (5′-ACCAGACAATATAGTTTGACCCACTAAG-3′) |
Species specific outer membrane protein (89-bp) Cholera toxin from toxigenic V.cholerae (84-bp) |
Gubala, 2006; Gubala and Proll, 2006; Blackstone et al., 2007 |
| Vibrio vulnificus | vvhA | Vul-F-1085 (5′-GGTTGCGGGTGGTTCGGT-3′) Vul-R-b1375 (5′-GATTTGCTTCATTTTCAGGGG-3′). |
Hemolysin A gene from pathogenic V. vulnificus (290-bp) | This study |
| Vibrio parahaemolyticus | tdhS | TDH-169 (5′-GTAAAGGTCTCTGACTTTTGGAC-3′) TDH-415 (5′-TGGAATATGAACCTTCATCTTCACC-3′) |
Thermostable direct haemolysin gene from pathogenic V. parahaemolyticus (270 bp) | Rizvi and Bej, 2010 |
| Vibrio Genus for MiSeq Illumina platform | 16S rRNA | Vib-169F (5′-GGATAACC/TATTGGAAACGATG-3′) Vib2-r (5′-GAAATTCTACCCCCCTCTACAG-3′) |
General Vibrio spp. (511-bp) | Thompson J.R. et al., 2004; Yong et al., 2006; Vezzulli et al., 2012 |
To quantify patterns in V. cholerae abundance we targeted the outer-membrane protein ompW gene (Gubala, 2006; Gubala and Proll, 2006; Table 1). qPCR cycling parameters were identical to the Vibrio 16S rRNA assay described above, with the exception of the annealing/extension step, which occurred at 61°C for 60 s. To test for the occurrence of toxigenic V. cholerae, we targeted the enterotoxin gene ctxA (Blackstone et al., 2007; Table 1), using the same cycling conditions employed for the ompW assay.
To quantify V. vulnificus we targeted the hemolysin A vvhA gene (Panicker et al., 2004; Panicker and Bej, 2005) using a new set of primers designed for this study (Table 1). The vvhA primers were designed using Primer3 software (Thornton and Basu, 2011) and assessed against available databases with cycling conditions identical to the general Vibrio 16S rRNA assay.
To specifically target V. parahaemolyticus, the thermostable direct haemolysin tdhS gene was targeted (Rizvi and Bej, 2010; Table 1). Cycling conditions for this assay involved initial activation steps at 50°C for 120 s and 95°C for 120 s, followed by 40 cycles of a 3-step reaction incorporating: denaturation at 95°C for 15 s, annealing at 56°C for 15 s and extension step at 72°C for 60 s. The amplification was concluded with a holding stage of 72°C for 120 s followed by a melting curve. Standard curves for each qPCR assay were prepared using cultures of V. cholerae (O1 El Tor N16961), V. parahaemolyticus (ATCC17802), and V. vulnificus (C71840), with V. parahaemolyticus (ATCC17802) genomic DNA used for the Vibrio community 16S rRNA assay. Each strain was inoculated into a 50 ml centrifuge tube containing 6 ml of 100% marine broth (Difco Marine Broth 2216) and incubated overnight at 37°C, while shaking at 150 rpm. Samples were homogenized by vortexing and each tube was sub-sampled (1 ml × 4) into four sterile 1.5 ml tubes, creating four replicates for each Vibrio spp. The media was washed three times with phosphate buffer solution (PBS × 3) and centrifuged at 5200 × g at 10°C for 10 min. One replicate (pellet + 1 ml PBS × 3) from each bacterial strain was serially diluted and enumerated by spread plating onto triplicate marine agar plates (Difco marine Broth 2216 and +1.5% Bacteriological agar) and incubating at 37°C overnight. DNA was extracted from the other three replicates for each Vibrio strain using the Powerwater DNA isolation Kit (MoBio Laboratories, Inc.) in accordance with the manufacturer’s instructions. The extracted DNA (n = 3) was pooled and used for the generation of standard curves.
Vibrio Diversity and Phylogenetic Analysis
To track shifts in the overall composition of the Vibrio community we combined the Vibrio-specific 16S rRNA primers Vib-169F (Yong et al., 2006) and Vib2-r (Thompson J.R. et al., 2004; Vezzulli et al., 2012; Table 1) to target the variable regions V2, V3, and part of V4. PCR reactions were performed in 50 μl volumes containing 25 μl of MangomixTM (Bioline), 0.4 μM of each of the forward and reverse primers and 1–5 μl of template DNA. PCR cycling conditions involved an initial activation step of 95°C for 120 s, followed by 30 cycles of: denaturation at 95°C for 15 s, annealing at 53°C for 30 s and extension at 72°C for 30 s, followed by a holding stage at 72°C for 10 min. After confirming positive amplification, the genomic DNA from the March and June samples was used to prepare DNA libraries with the Illumina TruSeq DNA library preparation protocol. Sequencing was performed on the Illumina MiSeq platform (at Molecular Research LP; Shallowater, TX, USA) following the manufacturer’s guidelines. Raw data files in FASTQ format were deposited in NCBI Sequence Read Archive (SRA) with the study accession number SRP069796 under Bioproject number PRJNA309925.
Vibrio 16S rRNA gene sequences were analyzed using the QIIME pipeline (Caporaso et al., 2010; Kuczynski et al., 2012). Briefly, paired-end DNA sequences were joined, de novo OTUs were defined at 97% sequence identity using UCLUST (Edgar, 2010) and taxonomy was assigned against the Greengenes database (version 13/8/2013) using BLAST (Altschul et al., 1990). Chimeric sequences were detected using ChimeraSlayer (Haas et al., 2011) and filtered from the dataset. Sequences were then rarefied to the same depth to remove the effect of sampling effort upon analysis.
Statistical Analysis
Seasonal differences in environmental parameters were tested with One-way ANOVA followed by Tukey HSD test, performed using STATISTICA version 10.0 (StatSoft, Tulsa, OK, USA). To compare total Vibrio spp. abundance, data was log| x + 1| transformed and a factorial ANOVA followed by Tukey HSD test was performed using STATISTICA version 10.0 (StatSoft, Tulsa, OK, USA). Data from the V. cholerae, V. parahaemolyticus, and V. vulnificus qPCR assays were analyzed using the Kruskal–Wallis (non-parametric) test, followed by a Multiple Comparisons Assessment (StatSoft, Tulsa, OK, USA). Correlations between qPCR results and environmental parameters were assessed using Minitab 17 (Minitab Inc.). For the Vibrio specific amplicon sequencing data, a non-metric MDS analysis was performed on the top 50 OTUs and normalized [(x-mean)/stdev] environmental parameters, with the Bray–Curtis similarity measurement using PAST (Hammer et al., 2001). In addition, PAST was also used for SIMPER analysis, with the Bray–Curtis similarity measurement used to identify the OTUs that contributed the most to differences between the winter and late summer samples. Alpha diversity parameters of the rarefied sequences and Jackknife Comparison of the weighted sequence data were calculated in QIIME (Caporaso et al., 2010; Kuczynski et al., 2012). Differences in the alpha diversity parameters were tested using the non-parametric Mann-Whitney U Test (StatSoft, Tulsa, OK, USA).
Results
Environmental Conditions
The environmental parameters measured during the study period are summarized in Supplementary Table S1. Water temperature, DO and pH varied significantly between seasons [ANOVA, F(4,16) = 115.3, p < 0.0001], with mean water temperatures decreasing by 8.8°C degrees (Tukey HSD, p < 0.0002), DO increasing by 1.6 mg L-1 (p < 0.0004) and pH increasing by 0.55 (Tukey HSD, p < 0.0021) from the summer to winter sampling periods. During summer, salinity decreased across the estuary from the two most eastern sites at Chowder Bay and Mort Bay, where salinities were near to marine conditions, to the western sites including Olympic Park, Rhodes, and Parramatta Park, where salinity dropped to 0–14.1 ppt. Olympic Park and Rhodes displayed the highest NH4+ levels during the winter (485.3 ± 105.3, 510.1 ± 192.6; average ± stdev μg L-1) while Olympic Park and Parramatta Park displayed the highest NH4+ levels during the summer (313 ± 23.8, 125.9 ± 13.1 μg L-1).
Total Vibrio spp. Abundance
Quantitative PCR analysis of the total Vibrio community (Figure 2) revealed significant differences between locations [ANOVA, F(4,48) = 19.4, p < 0.0001] and sampling periods [ANOVA, F(1,48) = 228, p < 0.0001], as well as a significant interactive effect between location and sampling periods [ANOVA, F(11,48) = 17.3, p < 0.0001]. Most locations (seven out of eight) exhibited higher Vibrio spp. abundances during the summer (March) than the winter (June and/or August) with five sites (Rhodes, Mort Bay, Rozelle Bay, and Chowder Bay) displaying significant differences between summer and winter (Tukey HSD, p < 0.05, asterisks in Figure 2). Mean abundances across the entire estuary were 2.6 × 105 cells ml-1 in the summer, compared to 4.5 × 104 cells ml-1 in the winter. The highest concentrations were measured in Rozelle Bay during the summer (1.6 × 106 cells ml-1) while the lowest concentration was recorded in Parramatta Park during the winter (6.5 × 102 cells ml-1).
FIGURE 2.
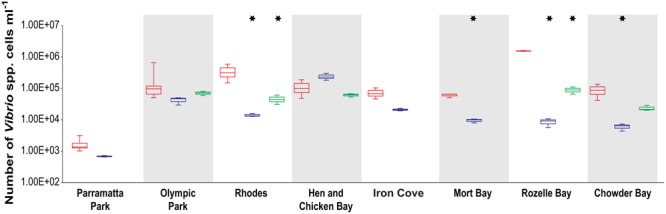
Total Vibrio abundance (mL-1) in Sydney Harbour determined by quantitative PCR. Red boxplots denote summer (March) samples, and blue and green boxplots denote winter [June and August (only at five location), respectively] samples. Asterisks indicate a significant difference between summer–winter samples.
Across the entire data set, total Vibrio spp. abundance demonstrated a positive correlation to temperature (r = 0.306, p = 0.011) and a negative correlation (r = -0.287, p = 0.017) to DO. Within sites, total Vibrio abundance was positively correlated to temperature (r ≥ 0.8, p < 0.05) and negatively correlated to pH (r ≤ -0.79, p ≤ 0.038) at Chowder Bay, Mort Bay, Rozelle Bay, Iron Cove and Rhodes, while it was negatively correlated to salinity (r ≥ 0.83, p < 0.05) at Iron Cove and Mort Bay.
Abundance of Pathogenic Vibrios
Vibrio cholerae was detected in the majority (71%) of samples when we targeted the species specific ompW gene, but levels of toxigenic V. cholerae (positive for ctxA) were below detection limit in all samples. In three out of the four sites with the highest V. cholerae abundance (Olympic Park, Rhodes, and Rozelle Bay), summer concentrations (72 ± 26, 98 ± 76, 372 ± 46; cells ml-1 average ± stdev) were higher than winter concentrations. Significant differences in V. cholerae abundance were observed between sites [Kruskal–Wallis test: H(7,69) = 15.5, p = 0.0297], with the highest numbers recorded at Rozelle Bay during the late summer, where concentrations reached 3.7 × 102 ± 46 (average ± stdev) cells ml-1 (Figure 3).
FIGURE 3.
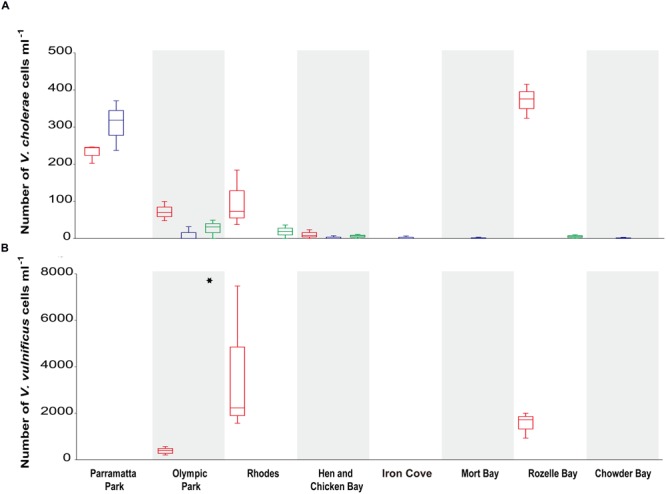
Vibrio cholerae and V. vulnificus abundance (mL-1) in Sydney Harbour determined by quantitative PCR. Specific qPCR assays were carried out on: (A) the V. cholerae outer-membrane protein gene ompW (Efficiency ≥ 93.3% and R2 ≥ 0.996; top graph) and for (B) the V. vulnificus hemolysin A vvhA gene (Efficiency = 92.8% and R2 = 0.995; bottom graph). Red boxplots denote summer (March) samples, and blue and green boxplots denote winter [June and August, respectively] samples. Asterisks indicate a significant difference in V. vulnificus between sample locations.
Across the entire data set, V. cholerae abundance was negatively correlated to DO (r < -0.428, p < 0.0001) and to pH [r < -0.328, p < 0.006]. The nature of correlations between V. cholerae abundance and environmental parameters observed over time varied significantly between locations, with three sites in particular characterized by significant correlations. Within Rozelle Bay, V. cholerae abundance was negatively correlated to DO, pH, and NH4+, but positively correlated to temperature (r ≥ 0.89, p < 0.005). Similarly, in Rhodes, V. cholerae abundance was negatively correlated to DO and pH, but positively correlated to temperature (r ≥ 0.75, p < 0.02). On the other hand in Parramatta Park, V. cholerae abundance was negatively correlated to DO, temperature and NO2, but positively correlated to pH and salinity (r ≥ 0.73, p < 0.05).
Vibrio vulnificus was detected in 14% of all samples (Figure 3), but was only observed during summer and at sites within the salinity range of 5–26 ppt (Supplementary Table S1). There were significant differences in V. vulnificus abundance between locations [Kruskal–Wallis test: H(2,12) = 8.6, p = 0.0134], with the highest concentration and variation (3.6 × 104 ± 3 × 104 cells ml-1; average ± stdev) recorded at Rhodes (Figure 3). V. parahaemolyticus levels were below detection limit in all samples.
Vibrio Community Diversity and Structure
Sequences were rarefied to 10,734 sequences per sample, to remove the effect of sampling effort upon analysis. Jackknife Comparison of the weighted 16S rRNA data sequences separated the sequence data into five different groups, (Figure 4A). In most (87%) locations and during most sampling times all three biological replicates grouped together, with a clear separation between summer and winter samples observed across the data set (Figure 4A). However, at two sites (Hen and Chicken Bay and Chowder Bay) the summer and winter samples grouped together (groups 3 and 4; Figure 4A). A non-metric MDS analysis of the top 50 OTUs, plotted together with normalized environmental parameters (representing 75.9% of the total rarefied sequence; Stress = 0.1237; Figure 4B) demonstrated clear divisions between summer and winter samples. However, it is notable that the three eastern most sampling sites (Chowder Bay, Rozelle Bay, and Mort Bay) displayed substantially more similarity across the seasons than the more western sampling sites (Figures 4B and 5), implying that seasonal shifts were more dramatic in the western region of the estuary. Temperature and salinity were the two most significant environmental drivers of differences in the Vibrio community between summer and winter (Supplementary Table S1, Figure 4B). The higher summer temperatures coincided with lower salinity, DO and pH levels (Supplementary Table S1, Figure 4B).
FIGURE 4.
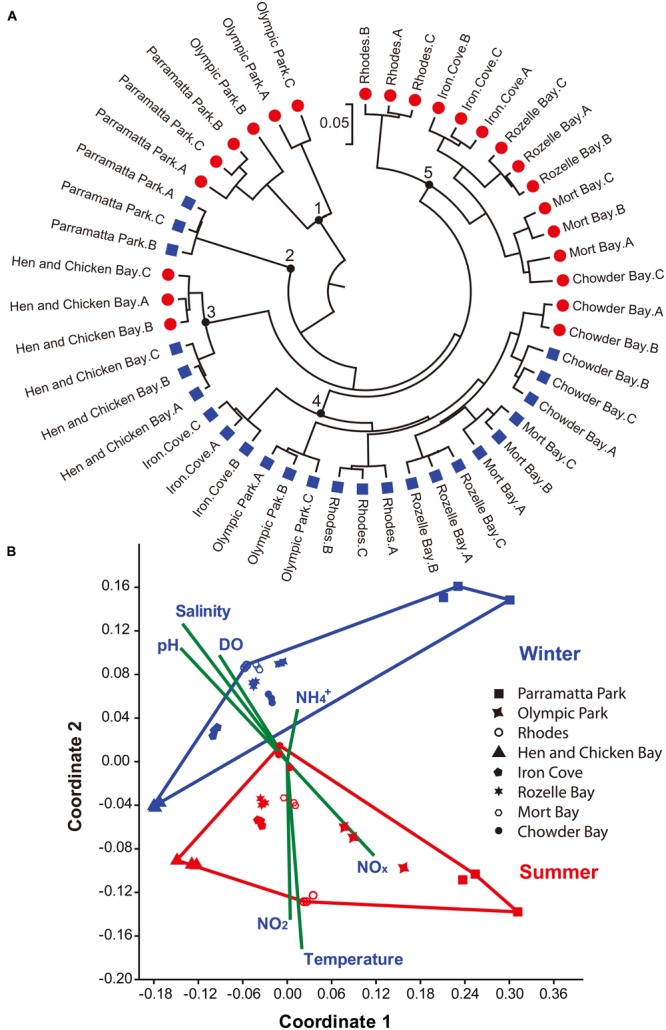
Sample assessment using 16S rRNA gene sequences and environmental parameters. (A) Jackknife Comparison on the weighted sequence data was carried out using QIIME (Caporaso et al., 2010) and constructed with MEGA 6.0 (Tamura et al., 2013). Red circles denote summer (March) and Blue squares denote winter (June) samples. The bar represents five substitutions per 100 nucleotide positions. Black circles numbered 1–5 highlight different groups of sequences. (B) Non-metric MDS plot of the top 50 OTUs. Red and Blue points represent summer and winter samples, respectively. A, B, and C represent the three biological replicates. Shepard plot stress value was 0.1237. Vectors indicate the contribution of each environmental parameter using PAST (Hammer et al., 2001).
FIGURE 5.
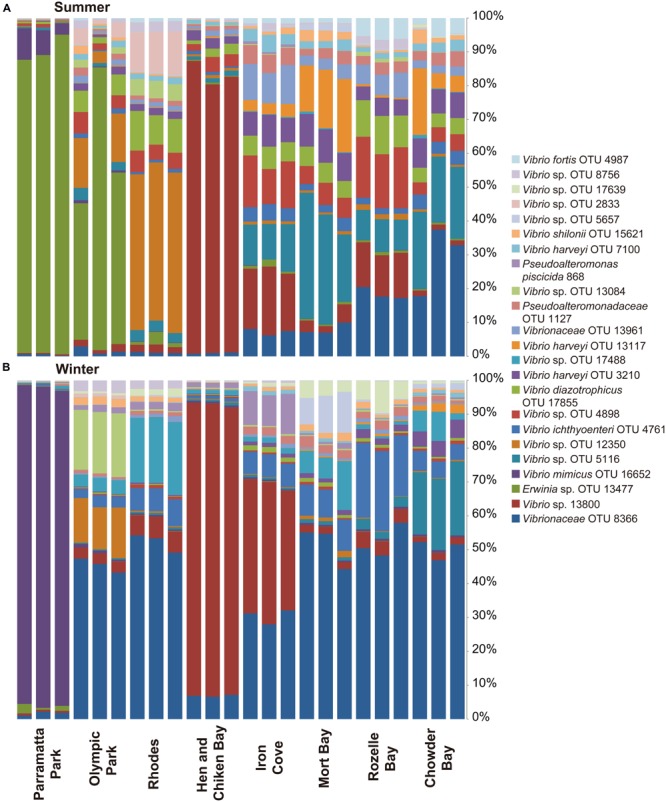
Most abundant Vibrio OTUs (representing 67.5% of the total normalized sequences during winter and summer. The figure displays the distribution of OTUs (for graphical reasons only OTUs that represent >0.5% of the total sequences were included) at the different sites, ranging from Parramatta Park in the west to Chowder Bay in the east: (A) summer sampling time (March), (B) winter (June).
Both spatial and temporal shifts in the composition of the Vibrio community were observed (Figures 4 and 5), with the majority (61%) of the most abundant OTUs (including all OTUs representing >0.5% of the sequences, which together comprised 67.5% of total sequences) observed during summer. However, it is notable that these dominant OTUs decreased in their relative importance during the winter (Figure 5). Moreover, Shannon Wiener index, observed species and species richness (Chao1), were all significantly higher in summer than winter (Mann–Whitney U Test, p ≤ 0.012). SIMPER analysis revealed that 4 OTUs, including OTU 8366 (16.5% contribution to the difference between winter and summer in Figure 4B), OTU 13800 (14.7%), OTU 13477 (12.4%), and OTU 16652 (8.8%), both represented the most abundant sequences in the data-set and together contributed 52.4% of the difference between winter and summer Vibrio communities. The Greengenes analysis assigned these sequences as follows: OTU 13800 – to the genus Vibrio, OTU 8366 – to the Vibrionaceae family, OTU 13477 – to the genus Erwinia and OTU 16652 – to V. mimicus.
Discussion
Our study revealed that Vibrios, including several human pathogens, are an abundant and dynamic component within the Sydney Harbour estuary, and that the community displays marked spatiotemporal heterogeneity that can often be clearly linked to environmental variables including temperature. Temperature significantly influenced both Vibrio abundance and community composition within Sydney Harbour, with highest concentrations observed during the warmer summer sampling period. This is consistent with evidence that many Vibrio species prefer warmer waters and is in-line with the seasonal dynamics observed in several other regions including the North Sea, Tyrrhenian coast brackish water, coastal regions of the Northern Baltic and along the east coast of the USA (Eiler et al., 2007; Turner et al., 2009; Caburlotto et al., 2012; Oberbeckmann et al., 2012; Vezzulli et al., 2012; Takemura et al., 2014; Matteucci et al., 2015). Indeed in other coastal regions it has been demonstrated that water temperature and salinity are the major drivers of patterns in Vibrio abundance (Oberbeckmann et al., 2012; Jacobs et al., 2014; Takemura et al., 2014).
While temperature was the most significant environmental determinant across the entire data-set, within sampling-periods and individual sites salinity was a key environmental determinant of patterns in Vibrio abundance and diversity. Negative correlations between Vibrio abundance and salinity were observed in two of the most easterly and ‘marine’ sites (Iron Cove and Mort Bay), while at the most western and fresh water site at Parramatta Park a positive correlation between salinity and V. cholerae was observed. Whereas, V. vulnificus occurred within the salinity range of 5–26 ppt (Olympic Park, Rhodes, and Rozelle Bay) supporting previous observations that salinity optimum varies between different Vibrio species (Kaspar and Tamplin, 1993; Randa et al., 2004; Hsieh et al., 2008; Takemura et al., 2014).
Presence of Pathogenic Vibrios in Sydney Harbour
We targeted three pathogenic Vibrio spp. known to have substantial relevance to human health. Several outbreaks of pathogenic Vibrios have previously been documented in Australia. In the 1970s, a small outbreak of cholera occurred in south-eastern Queensland (Rao and Stockwell, 1980), which was linked to local riverine toxigenic V. cholerae populations (Rao and Stockwell, 1980; Rogers et al., 1980), and the presence of toxigenic and non-toxigenic V. cholerae has subsequently been documented within several rivers and estuaries in eastern Australia (Desmarchelier and Reichelt, 1981; Bourke et al., 1986; Desmarchelier et al., 1988, 1995; Mallard and Desmarchelier, 1995; Islam et al., 2013). Other pathogenic Vibrios and Vibrio spp. related infections have been reported in Australia. Isolates of V. cholerae and V. parahaemolyticus have been extracted from Crassostrea commercialis (Sydney rock oysters) farmed at estuaries along the east coast of Australia (Eyles et al., 1985; Eyles and Davey, 1988) and several cases of V. vulnificus and V. parahaemolyticus infections of humans have been reported from estuaries in Northern Australia, following skin contact with river water and seawater (Ralph and Currie, 2007).
In our study, V. cholerae was detected at most sites within Sydney Harbour, with highest abundances observed during late summer, at sites with low to mid-range salinity levels (Parramatta Park, Olympic Park, Rhodes, and Rozelle Bay; 0–26 ppt). Similarly, temporal studies in another urban estuary along the Australian east coast identified the highest prevalence of V cholerae in water, sediment and in oysters during March–May (Eyles and Davey, 1988). It is possible that this high abundance of V cholerae in warm brackish waters followed an increase of dissolved organic matter following a phytoplankton bloom (Eiler et al., 2007). However, it is important to note that not all strains of V. cholerae are responsible for causing cholera, indeed only the serogroups O1/O139 are responsible. The ompW gene used here differentiates V. cholerae from other Vibrios, but does not necessarily identify disease causing strains of V. cholerae. Our subsequent analysis targeting the ctxA (cholera toxin) gene did not detect this gene in any samples, suggesting that during our study, toxigenic V. cholerae, which is the causative agent of cholera (Thompson F.L. et al., 2004; Nelson et al., 2009), was not present in the water of Sydney Harbour estuary. However, other, non-toxigenic strains of V. cholerae are also responsible for severe skin infections (Andersson and Ekdahl, 2006; Lukinmaa et al., 2006; Stypulkowska-Misiurewicz et al., 2006) and gastroenteritis (Dutta et al., 2013; Luo et al., 2013). Additionally, non-toxigenic V. cholerae strains from Sydney, that lack the ctxA gene, have been shown to be pathogenic through Rabbit Ileal Loop models, where pathogenesis (fluid accumulation) occurred due to the presence of other virulence factors (Islam et al., 2013). The presence of these alternate virulence factors in non-O1/non-O139 V. cholerae isolates has been observed in Iceland (Haley et al., 2012) and in the Botany Bay catchment (Islam et al., 2013), which is immediately adjacent to Sydney Harbour. Therefore, while not necessarily involved in cholera, the strains of V. cholerae identified here are still likely to be significant within the context of human health. Moreover, since Vibrios are particularly prone to lateral gene transfer, non-toxigenic strains of V. cholerae can acquire virulence through horizontal gene transfer especially following cellular interactions within localized microenvironments such as on zooplankton and within biofilms (Borgeaud et al., 2015; Metzger and Blokesch, 2016).
The links between V. cholerae abundance and season and temperature observed here are significant, because elsewhere warming patterns have coincided with outbreaks of Vibrio infections (including infections from non-toxigenic V. cholerae), with the distribution of infection cases closely corresponding to the temporal and spatial peaks in sea surface temperatures (Baker-Austin et al., 2013; Sterk et al., 2015). In regions where cholera occurs, there are also clear links between elevated temperatures and cholera outbreaks (Huq et al., 2005; Colwell, 2009).
The detection of the virulence gene vvhA by qPCR confirmed the presence of pathogenic V. vulnificus populations in three of the tested sites within the Sydney Harbour catchment during the late summer (March) sampling session. However, this organism was not detected at any sites during the winter sampling. This is consistent with patterns observed elsewhere, whereby the abundance of this species is generally correlated with elevated water temperatures (Jacobs et al., 2014), with the occurrence of V. vulnificus often limited to the summer months (Randa et al., 2004). As a consequence, highest frequencies of human infections typically occur during the warmer summer months (Strom and Paranjpye, 2000). The three sites where V. vulnificus was detected are located mid-estuary, where salinity levels were between 5 and 26 ppt. This also falls within the salinity range where V. vulnificus abundance has been shown to be highest elsewhere (Kaspar and Tamplin, 1993; Randa et al., 2004).
It has been suggested that increased V. vulnificus abundance within marine environments increases both the risk of infection from seafood consumption (Strom and Paranjpye, 2000; Chu et al., 2011) and direct exposure from swimming (Daniels and Shafaie, 2000). The highest concentration of V. vulnificus observed in this study was 7 × 104 cells ml-1. Whether these levels are sufficient to induce infection in humans is unclear since the infectious dose of this organism is currently unknown and probably dependent on host factors (Daniels and Shafaie, 2000). However, the high fatality rate associated with V. vulnificus infection (Daniels and Shafaie, 2000; Jones and Oliver, 2009; Horseman and Surani, 2011; Ralston et al., 2011) warrants a level of concern regarding the occurrence of this bacterium within the heavily used waters of Sydney Harbour.
We also expected to detect pathogenic V. parahaemolyticus within Sydney Harbour, at least during the summer, since this species is commonly found in estuarine waters above 20°C (Nigro et al., 2011; Matteucci et al., 2015) and has previously been identified in Sydney rock oysters (Eyles et al., 1985). However, in all samples, V. parahaemolyticus levels were below the detection limit of our qPCR assay. It is important to emphasize that we tested only the water while other studies detected these bacteria within oysters or associated with other marine animals (Eyles and Davey, 1984; Eyles et al., 1985). Moreover, given the high levels of spatiotemporal variability observed in our study, this finding does not eliminate the possibility that these bacteria occur in the Sydney Harbour catchment water at other times.
Patterns in Vibrio Community Diversity and Composition
In addition to the shifts in abundance described above, we observed substantial spatiotemporal changes in the composition of the Vibrio community within Sydney Harbour. Unfortunately, in some OTU’s the assigned taxonomy was above species and genus level due to the 16S rRNA gene’s limited resolution in the Vibrionaceae family (Sawabe et al., 2013). Vibrio diversity, as determined by our Vibrio-specific 16S rRNA gene sequencing, was higher in late summer than winter. Furthermore, Vibrio community composition was clearly separated into winter and summer groups within an MDS plot (Figure 4B). Analysis of the sequences without the inclusion of environmental parameters was not as conclusive (Figure 4A). While sequences from six of the locations separated between summer and winter groups, in two locations: Hen and Chicken Bay and Chowder Bay different patterns were observed. In Hen and Chicken Bay, the winter and summer samples grouped together, but separately from all other sites. However, in Chowder Bay individual replicates from the summer sampling period were divided between the winter and summer groups.
The eastern-most site, closest to the open ocean, at Chowder Bay, displayed the lowest seasonal variability in salinity, temperature and DO (Supplementary Table S1), which may explain the inter-seasonal consistency in Vibrio community composition at this site. The Vibrio community at Hen and Chicken Bay was universally separated from all other sites, which could be explained by the fact that this was the only sampling site where samples were obtained from relatively shallow water (∼1 m) leading to a potentially greater influence from resuspended sediments. We analyzed patterns in the Vibrio community collected on a 0.22 μm filter, meaning that both free-living and particle-attached populations were considered together. There is evidence that a particle attached lifestyle is common for many Vibrios (Takemura et al., 2014), and although data on suspended particle concentrations were not collected during this study it remains possible that some of the spatiotemporal patterns observed here were underpinned by variability in the concentrations of suspended particulate material in the estuary.
In accordance with the patterns in total Vibrio abundance, temperature was one of the most significant drivers of differences in the Vibrio community composition. This pattern supports previous observations that have indicated that elevated water temperature is a major factor explaining patterns in Vibrio community composition and abundance (Nishiguchi, 2000; Thompson J.R. et al., 2004; Tout et al., 2015).
Some of the differences in Vibrio abundance between the winter samples might be explained by differential influence of a rainfall event, where 58.3 mm of rain occurred during the week proceeding August samples collection (Observatory Hill station, Sydney, http://www.bom.gov.au/climate/data/index.shtml). Stormwater run-off associated with rainfall events influence salinity and nutrient levels within Sydney Harbour, which can drive heterogeneity in bacterioplankton community composition within the estuary (Jeffries et al., 2015). During each sampling period, and particularly during March (summer sample), spatial shifts in the composition of the Vibrio community were primarily governed by variability in salinity, which is consistent with patterns previously observed in other environments (Oberbeckmann et al., 2012; Jacobs et al., 2014; Takemura et al., 2014). While temperature and salinity were the most important drivers of shifts in overall Vibrio community structure, and total Vibrio abundance, not all Vibrio spp. displayed the same patterns. For instance, the relative proportion of the OTU (16652) with closest sequence matches to V. mimicus, and which comprised up to 10% of the community in three samples, increased significantly during winter in the Parramatta Park samples. This is consistent with observations in Bangladesh, where V. mimicus numbers spiked during lower temperature months (Chowdhury et al., 1989). This is particularly notable because V. mimicus has been implicated in outbreaks of vibriosis in freshwater catfish in China, leading to 80–100% mortality rates (Geng et al., 2014). V. mimicus is also pathogenic to humans, causing diarrheal disease (Hlady and Klontz, 1996; Adebayo-Tayo et al., 2011), with some isolates exhibiting the presence of the lysogenic filamentous bacteriophage that carries the cholera toxin genes in epidemic V. cholerae strains (Boyd et al., 2000). So the substantial proportion of Vibrio sequences matching this organism within regions of the Sydney Harbour estuary is also noteworthy.
Environmental and Human Health Implications
Previous studies that have examined Vibrio related infections occurring in coastal sites in the Baltic Sea, North Sea, Israel, and the Korean peninsula have revealed the link between high water temperatures and infection rates (Paz et al., 2007; Hashizume et al., 2008; Chu et al., 2011; Baker-Austin et al., 2013). Furthermore, recent models have revealed that elevated water temperature is a key factor explaining Vibrio abundance within aquatic samples (Oberbeckmann et al., 2012; Jacobs et al., 2014). These patterns and predictions are relevant within the context of the current, and predicted future, warming of seawater temperatures along the south-eastern Australian coast (Cai et al., 2005; Hobday and Lough, 2011; Last et al., 2011). We performed a simple analysis of surface seawater temperatures (SST) along the Sydney coastline (34°05′S 151°15′E) during the last 57 years (Supplementary Figure S1), which suggests that a rise in SST of 1–2°C will increase the number of days where SST ≥ 20°C, which is the preferred temperature regime of several pathogenic Vibrio species (Takemura et al., 2014), by up to 169%. This is particularly relevant within the context of recent evidence for temperature-induced shifts in bacterial community composition and function in this region (Seymour et al., 2012), and a precedent of previous Vibrio outbreaks within the waters of eastern Australia (Rao and Stockwell, 1980; Rogers et al., 1980; Desmarchelier et al., 1995).
Developing an understanding of the spatiotemporal dynamics of Vibrio populations and identifying key environmental drivers are essential for predicting future risks and hotspots for pathogen outbreaks within heavily used coastal ecosystems such as Sydney Harbour. Our observations suggest that several sites within Sydney Harbour, where significant abundances of V. cholerae and V. vulnificus already occur, are potentially at risk of pathogenic Vibrio outbreaks, particularly during warm summer months. Moreover, since many of the sites where pathogenic Vibrio were identified are situated near to river inputs or adjacent to stormwater canals, the combination of warm summer conditions and the typically high summer rainfall in Sydney (which will often reduce salinity levels and add nutrients) have the potential to provide a ‘perfect storm,’ within the context of conditions favoring pathogenic Vibrio outbreaks.
Author Contributions
NS and VB contributed equally to this research. NS, VB, JS, and ML conceived and designed the experiments. NS, VB, RC, and JS collected the samples. RC and VB performed the nutrient analysis. NS and VB performed the molecular analysis. NS analysed the data. NS, ML, and JS wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was funded by Australian Research Council grants DP120102764 and FT130100218 to JS.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00460
Distribution of the average seawater surface temperatures at Port-Hacking.
Sample location and environmental associated data.
References
- Adebayo-Tayo B., Okonko I., John M., Odu N., Nwanze J., Ezediokpu M. (2011). Occurrence of potentially pathogenic Vibrio species in Sea foods obtained from Oron Creek. Adv. Biol. Res. 5 356–365. [Google Scholar]
- Ali M., Lopez A. L., You Y., Kim Y. E., Sah B., Maskery B., et al. (2012). The global burden of cholera. Bull. World Health Organ. 90 209–218A. 10.2471/BLT.11.093427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andersson Y., Ekdahl K. (2006). Wound infections due to Vibrio cholerae in Sweden after swimming in the Baltic Sea, summer 2006. Euro Surveill. 11:E060803.2. [DOI] [PubMed] [Google Scholar]
- Armitage N., Rooseboom A. (2000). The removal of urban litter from stormwater conduits and streams: paper 1- The quantities involved and catchment litter management options. Water SA 26 181–188. [Google Scholar]
- Austin B., Zhang X. H. (2006). Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43 119–124. 10.1111/j.1472-765X.2006.01989.x [DOI] [PubMed] [Google Scholar]
- Australian Bureau of Statistics (2004). How Many People Live in Australia’s Coastal Areas? In Year Book Australia 2004 (Cat. No. 1301.0). Canberra, ACT: Commonwealth of Australia. [Google Scholar]
- Baker-Austin C., Stockley L., Rangdale R., Martinez-Urtaza J. (2010). Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: a European perspective. Environ. Microbiol. Rep. 2 7–18. 10.1111/j.1758-2229.2009.00096.x [DOI] [PubMed] [Google Scholar]
- Baker-Austin C., Trinanes J. A., Taylor N. G., Hartnell R., Siitonen A., Martinez-Urtaza J. (2013). Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Change 3 73–77. 10.1038/nclimate1628 [DOI] [Google Scholar]
- Becker P., Gillan D., Lanterbecq D., Jangoux M., Rasolofonirina R., Rakotovao J., et al. (2004). The skin ulceration disease in cultivated juveniles of Holothuria scabra (Holothuroidea, Echinodermata). Aquaculture 242 13–30. 10.1016/j.aquaculture.2003.11.018 [DOI] [Google Scholar]
- Ben-Haim Y., Rosenberg E. (2002). A novel Vibrio sp. pathogen of the coral Pocillopora damicornis. Mar. Biol. 141 47–55. 10.1007/s00227-002-0797-6 [DOI] [Google Scholar]
- Birch G. (1996). Sediment-bound metallic contaminants in Sydney’s estuaries and adjacent offshore, Australia. Estuar. Coast. Shelf Sci. 42 31–44. 10.1006/ecss.1996.0003 [DOI] [Google Scholar]
- Blackstone G. M., Nordstrom J. L., Bowen M. D., Meyer R. F., Imbro P., DePaola A. (2007). Use of a real time PCR assay for detection of the ctxA gene of Vibrio cholerae in an environmental survey of Mobile Bay. J. Microbiol. Methods 68 254–259. 10.1016/j.mimet.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Blackwell K. D., Oliver J. D. (2008). The ecology of Vibrio vulnificus, Vibrio cholerae, and Vibrio parahaemolyticus in North Carolina estuaries. J. Microbiol. 46 146–153. 10.1007/s12275-007-0216-2 [DOI] [PubMed] [Google Scholar]
- Borgeaud S., Metzger L. C., Scrignari T., Blokesch M. (2015). The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347 63–67. 10.1126/science.1260064 [DOI] [PubMed] [Google Scholar]
- Bourke A., Cossins Y. N., Gray B., Lunney T. J., Rostron N. A., Holmes R. V., et al. (1986). Investigation of cholera acquired from the riverine environment in Queensland. Med. J. Aust. 144 229–234. [DOI] [PubMed] [Google Scholar]
- Boyd E. F., Moyer K. E., Shi L., Waldor M. K. (2000). Infectious CTXΦ and the Vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect. Immun. 68 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburlotto G., Bianchi F., Gennari M., Ghidini V., Socal G., Aubry F. B., et al. (2012). Integrated evaluation of environmental parameters influencing Vibrio occurrence in the coastal Northern Adriatic Sea (Italy) facing the Venetian lagoon. Microb. Ecol. 63 20–31. 10.1007/s00248-011-9920-x [DOI] [PubMed] [Google Scholar]
- Cai W., Shi G., Cowan T., Bi D., Ribbe J. (2005). The response of the Southern Annular Mode, the East Australian Current, and the southern mid-latitude ocean circulation to global warming. Geophys. Res. Lett. 32 1–4. 10.1029/2005GL024701 [DOI] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (1998). Outbreak of Vibrio parahaemolyticus infections associated with eating raw oysters–Pacific Northwest, 1997. Morb. Mortal. Wkly. Rep. 47 457–462. [PubMed] [Google Scholar]
- Chowdhury M., Yamanaka H., Miyoshi S., Aziz K., Shinoda S. (1989). Ecology of Vibrio mimicus in aquatic environments. Appl. Environ. Microbiol. 55 2073–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C., Do Y., Kim Y., Saito Y., Lee S.-D., Park H., et al. (2011). Mathematical modeling of Vibrio vulnificus infection in Korea and the influence of global warming. Osong Public Health Res. Perspect. 2 51–58. 10.1016/j.phrp.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell R. R. (2009). “Viable but not cultivable bacteria,” in Uncultivated Microorganisms ed. Epstein S. S. (Heidelberg: Springer; ) 121–129. [Google Scholar]
- Daniels N. A., Shafaie A. (2000). A review of pathogenic Vibrio infections for clinicians. Infect. Med. 17 665–685. [Google Scholar]
- Desmarchelier P., Momen H., Salles C. (1988). A zymovar analysis of Vibrio cholerae isolated in Australia. Trans. R. Soc. Trop. Med. Hyg. 82 914–917. 10.1016/0035-9203(88)90041-7 [DOI] [PubMed] [Google Scholar]
- Desmarchelier P., Wong F., Mallard K. (1995). An epidemiological study of Vibrio cholerae O1 in the Australian environment based on rRNA gene polymorphisms. Epidemiol. Infect. 115 435–446. 10.1017/S0950268800058593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmarchelier P. M., Reichelt J. L. (1981). Phenotypic characterization of clinical and environmental isolates of Vibrio cholerae from Australia. Curr. Microbiol. 5 123–127. 10.1128/mBio.01824-14 [DOI] [Google Scholar]
- Ducklow H. W., Carlson C. A. (1992). “Oceanic bacterial production,” in Advances in Microbial Ecology ed. Marshall K. C. (New York, NY: Springer; ) 113–181. [Google Scholar]
- Dutta D., Chowdhury G., Pazhani G. P., Guin S., Dutta S., Ghosh S., et al. (2013). Vibrio cholerae non-O1 non-O139 serogroups and cholera-like diarrhea, Kolkata, India. Emerg. Infect. Dis. 19 464–467. 10.3201/eid1903.121156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Eiler A., Gonzalez-Rey C., Allen S., Bertilsson S. (2007). Growth response of Vibrio cholerae and other Vibrio spp. to cyanobacterial dissolved organic matter and temperature in brackish water. FEMS Microbiol. Ecol. 60 411–418. 10.1111/j.1574-6941.2007.00303.x [DOI] [PubMed] [Google Scholar]
- Eiler A., Johansson M., Bertilsson S. (2006). Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl. Environ. Microbiol. 72 6004–6011. 10.1128/AEM.00917-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyles M., Davey G., Arnold G. (1985). Behavior and incidence of Vibrio parahaemolyticus in Sydney rock oysters (Crassostrea commercialis). Int. J. Food Microbiol. 1 327–334. 10.1016/0168-1605(85)90004-2 [DOI] [Google Scholar]
- Eyles M. J., Davey G. R. (1984). Microbiology of commercial depuration of the Sydney rock oyster, Crassostrea commercialis. J. Food Prot. 47 703–712. [DOI] [PubMed] [Google Scholar]
- Eyles M. J., Davey G. R. (1988). Vibrio cholerae and enteric bacteria in oyster-producing areas of two urban estuaries in Australia. Int. J. Food Microbiol. 6 207–218. 10.1016/0168-1605(88)90013-X [DOI] [PubMed] [Google Scholar]
- Frans I., Michiels C. W., Bossier P., Willems K., Lievens B., Rediers H. (2011). Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. Fish Dis. 34 643–661. 10.1111/j.1365-2761.2011.01279.x [DOI] [PubMed] [Google Scholar]
- Froelich B., Bowen J., Gonzalez R., Snedeker A., Noble R. (2013). Mechanistic and statistical models of total Vibrio abundance in the Neuse River Estuary. Water Res. 47 5783–5793. 10.1016/j.watres.2013.06.050 [DOI] [PubMed] [Google Scholar]
- Geng Y., Liu D., Han S., Zhou Y., Wang K. Y., Huang X. L., et al. (2014). Outbreaks of vibriosis associated with Vibrio mimicus in freshwater catfish in China. Aquaculture 433 82–84. 10.1016/j.aquaculture.2014.05.053 [DOI] [Google Scholar]
- Gilbert J. A., Steele J. A., Caporaso J. G., Steinbrück L., Reeder J., Temperton B., et al. (2011). Defining seasonal marine microbial community dynamics. ISME J. 6 298–308. 10.1038/ismej.2011.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goarant C., Herlin J., Brizard R., Marteau A.-L., Martin C., Martin B. (2000). Toxic factors of Vibrio strains pathogenic to shrimp. Dis. Aquat. Organ. 40 101–107. 10.3354/dao040101 [DOI] [PubMed] [Google Scholar]
- González-Escalona N., Cachicas V., Acevedo C., Rioseco M. L., Vergara J. A., Cabello F., et al. (2005). Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg. Infect. Dis. 11 129–131. 10.3201/eid1101.040762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubala A. J. (2006). Multiplex real-time PCR detection of Vibrio cholerae. J. Microbiol. Methods 65 278–293. 10.1016/j.mimet.2005.07.017 [DOI] [PubMed] [Google Scholar]
- Gubala A. J., Proll D. F. (2006). Molecular-beacon multiplex real-time PCR assay for detection of Vibrio cholerae. Appl. Environ. Microbiol. 72 6424–6428. 10.1128/AEM.02597-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Gevers D., Earl A. M., Feldgarden M., Ward D. V., Giannoukos G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21 494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B. J., Chen A., Grim C. J., Clark P., Diaz C. M., Taviani E., et al. (2012). Vibrio cholerae in a historically cholera-free country. Environ. Microbiol. Rep. 4 381–389. 10.1111/j.1758-2229.2012.00332.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D. A. T., Ryan P. D. (2001). PAST-palaeontological statistics software package for education and data analysis. Palaeontol. Electronica 4 1–9. [Google Scholar]
- Hasan N. A., Grim C. J., Lipp E. K., Rivera I. N., Chun J., Haley B. J., et al. (2015). Deep-sea hydrothermal vent bacteria related to human pathogenic Vibrio species. Proc. Natl. Acad. Sci. U.S.A. 112 E2813–E2819. 10.1073/pnas.1503928112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume M., Armstrong B., Hajat S., Wagatsuma Y., Faruque A. S., Hayashi T., et al. (2008). The effect of rainfall on the incidence of cholera in Bangladesh. Epidemiology 19 103–110. 10.1097/EDE.0b013e31815c09ea [DOI] [PubMed] [Google Scholar]
- Higgins R. (2000). Bacteria and fungi of marine mammals: a review. Can. Vet. J. 41 105–116. [PMC free article] [PubMed] [Google Scholar]
- Hlady W. G., Klontz K. C. (1996). The epidemiology of Vibrio infections in Florida, 1981–1993. J. Infect. Dis. 173 1176–1183. 10.1093/infdis/173.5.1176 [DOI] [PubMed] [Google Scholar]
- Hobday A. J., Lough J. M. (2011). Projected climate change in Australian marine and freshwater environments. Mar. Freshw. Res. 62 1000–1014. 10.1016/j.marpolbul.2014.06.003 [DOI] [Google Scholar]
- Horseman M. A., Surani S. (2011). A comprehensive review of Vibrio vulnificus: an important cause of severe sepsis and skin and soft-tissue infection. Int. J. Infect. Dis. 15 e157–e166. 10.1016/j.ijid.2010.11.003 [DOI] [PubMed] [Google Scholar]
- Hsieh J. L., Fries J. S., Noble R. T. (2008). Dynamics and predictive modelling of Vibrio spp. in the Neuse River Estuary, North Carolina, USA. Environ. Microbiol. 10 57–64. 10.1111/j.1462-2920.2007.01429.x [DOI] [PubMed] [Google Scholar]
- Huq A., Sack R. B., Nizam A., Longini I. M., Nair G. B., Ali A., et al. (2005). Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl. Environ. Microbiol. 71 4645–4654. 10.1128/AEM.71.8.4645-4654.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings P. A., Ahyong S. T., Ashcroft M. B., McGrouther M. A., Reid A. L. (2013). Sydney Harbour: its diverse biodiversity. Aust. Zool. 36 255–320. 10.7882/AZ.2012.031 [DOI] [Google Scholar]
- Islam A., Labbate M., Djordjevic S. P., Alam M., Darling A., Melvold J., et al. (2013). Indigenous Vibrio cholerae strains from a non-endemic region are pathogenic. Open Biol. 3 120181 10.1098/rsob.120181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J. M., Rhodes M., Brown C. W., Hood R. R., Leight A., Long W., et al. (2014). Modeling and forecasting the distribution of Vibrio vulnificus in Chesapeake Bay. J. Appl. Microbiol. 117 1312–1327. 10.1111/jam.12624 [DOI] [PubMed] [Google Scholar]
- Jeffries T. C., Schmitz Fontes M. L., Harrison D. P., van Dongen-Vogels V., Eyre B. D., Ralph P. J., et al. (2015). Bacterioplankton dynamics within a large anthropogenically impacted urban estuary. Front. Microbiol. 6:1438 10.3389/fmicb.2015.01438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. K., Oliver J. D. (2009). Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar C., Tamplin M. (1993). Effects of temperature and salinity on the survival of Vibrio vulnificus in seawater and shellfish. Appl. Environ. Microbiol. 59 2425–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K. (2015). “Estimating the burden of foodborne illness for campylobacter, salmonella and Vibrio parahaemolyticus in Japan, 2006–2012,” in Proceedings of the Annual Meeting 2015 July 25–28, 2015 (Des Moines, IA: International Association for Food Protection; ). [Google Scholar]
- Kuczynski J., Stombaugh J., Walters W. A., González A., Caporaso J. G., Knight R. (2012). Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinformatics Chap. 10:Unit 10.7 10.1002/0471250953.bi1007s36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushmaro A., Banin E., Loya Y., Stackebrandt E., Rosenberg E. (2001). Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int. J. Syst. Evol. Microbiol. 51 1383–1388. 10.1099/00207713-51-4-1383 [DOI] [PubMed] [Google Scholar]
- Last P. R., White W. T., Gledhill D. C., Hobday A. J., Brown R., Edgar G. J., et al. (2011). Long-term shifts in abundance and distribution of a temperate fish fauna: a response to climate change and fishing practices. Glob. Ecol. Biogeogr. 20 58–72. 10.1111/j.1466-8238.2010.00575.x [DOI] [Google Scholar]
- Lee K.-H., Ruby E. G. (1994). Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl. Environ. Microbiol. 60 1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. B., Birch G. F. (2014). “Sydney estuary, australia: geology, anthropogenic development and hydrodynamic processes/attributes,” in Estuaries of Australia in 2050 and Beyond ed. Wolanski E. (Dordrecht: Springer; ) 17–30. [Google Scholar]
- Lipp E. K., Huq A., Colwell R. R. (2002). Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15 757–770. 10.1128/CMR.15.4.757-770.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinmaa S., Mattila K., Lehtinen V., Hakkinen M., Koskela M., Siitonen A. (2006). Territorial waters of the Baltic Sea as a source of infections caused by Vibrio cholerae non-O1, non-O139: report of 3 hospitalized cases. Diagn. Microbiol. Infect. Dis. 54 1–6. 10.1016/j.diagmicrobio.2005.06.020 [DOI] [PubMed] [Google Scholar]
- Luo Y., Ye J., Jin D., Ding G., Zhang Z., Mei L., et al. (2013). Molecular analysis of non-O1/non-O139 Vibrio cholerae isolated from hospitalised patients in China. BMC Microbiol. 13:52 10.1186/1471-2180-13-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard K. E., Desmarchelier P. M. (1995). Detection of heat-stable enterotoxin genes among Australian Vibrio cholerae O1 strains. FEMS Microbiol. Lett. 127 111–115. 10.1111/j.1574-6968.1995.tb07458.x [DOI] [PubMed] [Google Scholar]
- Martinez-Urtaza J., Huapaya B., Gavilan R. G., Blanco-Abad V., Ansede-Bermejo J., Cadarso-Suarez C., et al. (2008). Emergence of asiatic Vibrio diseases in South America in phase with El Niño. Epidemiology 19 829–837. 10.1097/EDE.0b013e3181883d43 [DOI] [PubMed] [Google Scholar]
- Matteucci G., Schippa S., Di Lallo G., Migliore L., Thaller M. C. (2015). Species diversity, spatial distribution, and virulence associated genes of culturable vibrios in a brackish coastal Mediterranean environment. Ann. Microbiol. 65 2311–2321. 10.1007/s13213-015-1073-6 [DOI] [Google Scholar]
- Metzger L. C., Blokesch M. (2016). Regulation of competence-mediated horizontal gene transfer in the natural habitat of Vibrio cholerae. Curr. Opin. Microbiol. 30 1–7. 10.1016/j.mib.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Nelson E. J., Harris J. B., Morris J. G., Calderwood S. B., Camilli A. (2009). Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7 693–702. 10.1038/nrmicro2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro O. D., Hou A., Vithanage G., Fujioka R. S., Steward G. F. (2011). Temporal and spatial variability in culturable pathogenic Vibrio spp. in lake Pontchartrain, Louisiana, following Hurricanes Katrina and Rita. Appl. Environ. Microbiol. 77 5384–5393. 10.1128/AEM.02509-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi M. K. (2000). Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl. Environ. Microbiol. 66 3550–3555. 10.1128/AEM.66.8.3550-3555.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm S., Nishiguchi M. (2008). The evolutionary ecology of a sepiolid squid-Vibrio association: from cell to environment. Vie Milieu Paris 58 175–184. [PMC free article] [PubMed] [Google Scholar]
- Oberbeckmann S., Fuchs B. M., Meiners M., Wichels A., Wiltshire K. H., Gerdts G. (2012). Seasonal dynamics and modeling of a Vibrio community in coastal waters of the North Sea. Microb. Ecol. 63 543–551. 10.1007/s00248-011-9990-9 [DOI] [PubMed] [Google Scholar]
- Orata F. D., Keim P. S., Boucher Y. (2014). The 2010 cholera outbreak in Haiti: how science solved a controversy. PLoS Pathog. 10:e1003967 10.1371/journal.ppat.1003967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker G., Bej A. K. (2005). Real-time PCR detection of Vibrio vulnificus in oysters: comparison of oligonucleotide primers and probes targeting vvhA. Appl. Environ. Microbiol. 71 5702–5709. 10.1128/AEM.71.10.5702-5709.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker G., Myers M. L., Bej A. K. (2004). Rapid detection of Vibrio vulnificus in shellfish and Gulf of Mexico water by real-time PCR. Appl. Environ. Microbiol. 70 498–507. 10.1128/AEM.70.1.498-507.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz S., Bisharat N., Paz E., Kidar O., Cohen D. (2007). Climate change and the emergence of Vibrio vulnificus disease in Israel. Environ. Res. 103 390–396. 10.1016/j.envres.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Poloczanska E., Babcock R., Butler A., Hobday A., Hoegh-Guldberg O., Kunz T., et al. (2007). Climate change and Australian marine life. Oceanogr. Mar. Biol. 45 407–478. [Google Scholar]
- Ralph A., Currie B. J. (2007). Vibrio vulnificus and V. parahaemolyticus necrotising fasciitis in fishermen visiting an estuarine tropical northern Australian location. J. Infect. 54 e111–e114. 10.1016/j.jinf.2006.06.015 [DOI] [PubMed] [Google Scholar]
- Ralston E. P., Kite-Powell H., Beet A. (2011). An estimate of the cost of acute health effects from food-and water-borne marine pathogens and toxins in the USA. J. Water Health 9 680–694. 10.2166/wh.2011.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randa M. A., Polz M. F., Lim E. (2004). Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70 5469–5476. 10.1128/AEM.70.9.5469-5476.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A., Stockwell B. (1980). The Queensland cholera incident of 1977: 1. The index case∗. Bull. World Health Organ. 58 663–664. [PMC free article] [PubMed] [Google Scholar]
- Rizvi A. V., Bej A. K. (2010). Multiplexed real-time PCR amplification of tlh, tdh and trh genes in Vibrio parahaemolyticus and its rapid detection in shellfish and Gulf of Mexico water. Antonie Van Leeuwenhoek 98 279–290. 10.1007/s10482-010-9436-2 [DOI] [PubMed] [Google Scholar]
- Rogers R., Cuffe R., Cossins Y., Murphy D., Bourke A. (1980). The Queensland cholera incident of 1977: 2. The epidemiological investigation∗. Bull. World Health Organ. 58 665–669. [PMC free article] [PubMed] [Google Scholar]
- Sawabe T., Ogura Y., Matsumura Y., Feng G., Amin A. K. M. R., Mino S., et al. (2013). Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp nov. Front. Microbiol. 4:414 10.3389/fmicb.2013.00414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour J. R., Doblin M. A., Jeffries T. C., Brown M. V., Newton K., Ralph P. J., et al. (2012). Contrasting microbial assemblages in adjacent water masses associated with the East Australian Current. Environ. Microbiol. Rep. 4 548–555. 10.1111/j.1758-2229.2012.00362.x [DOI] [PubMed] [Google Scholar]
- Sterk A., Schets F. M., Husman R., Maria A., Nijs T., Schijven J. F. (2015). Effect of climate change on the concentration and associated risks of Vibrio Spp. in Dutch recreational waters. Risk Anal. 35 1717–1729. 10.1111/risa.12365 [DOI] [PubMed] [Google Scholar]
- Strom M. S., Paranjpye R. N. (2000). Epidemiology and pathogenesis of Vibrio vulnificus. Microb. Infect. 2 177–188. 10.1016/S1286-4579(00)00270-7 [DOI] [PubMed] [Google Scholar]
- Stypulkowska-Misiurewicz H., Pancer K., Roszkowiak A. (2006). Two unrelated cases of septicaemia due to Vibrio cholerae non-O1, non-O139 in Poland, July and August 2006. Euro Surveill. 11:E061130.2. [DOI] [PubMed] [Google Scholar]
- Suh J.-Y., Birch G., Hughes K. (2004). Hydrochemistry in reclaimed lands of the 2000 Olympic games site, Sydney, Australia. J. Coast. Res. 20 709–721. 10.2112/1551-5036(2004)20[709:HIRLOT]2.0.CO;2 [DOI] [Google Scholar]
- Sun C., Feng M., Matear R. J., Chamberlain M. A., Craig P., Ridgway K. R., et al. (2012). Marine downscaling of a future climate scenario for Australian boundary currents. J. Climate 25 2947–2962. 10.1175/JCLI-D-11-00159.1 [DOI] [Google Scholar]
- Takemura A. F., Chien D. M., Polz M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5:38 10.3389/fmicb.2014.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson F. L., Iida T., Swings J. (2004). Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68 403–431. 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. R., Polz M. F. (2006). “Dynamics of Vibrio populations and their role in environmental nutrient cycling,” in The Biology of Vibrios eds Thompson F. L., Austin B., Swings J. (Washington, DC: ASM Press; ) 190–203. [Google Scholar]
- Thompson J. R., Randa M. A., Marcelino L. A., Tomita-Mitchell A., Lim E., Polz M. F. (2004). Diversity and dynamics of a North Atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70 4103–4110. 10.1128/AEM.70.7.4103-4110.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J., Roach A., Eaglesham G., Bartkow M. E., Edge K., Mueller J. F. (2011). Perfluorinated alkyl acids in water, sediment and wildlife from Sydney Harbour and surroundings. Mar. Pollut. Bull. 62 2869–2875. 10.1016/j.marpolbul.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Thornton B., Basu C. (2011). Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 39 145–154. 10.1002/bmb.20461 [DOI] [PubMed] [Google Scholar]
- Tout J., Siboni N., Messer L. F., Garren M., Stocker R., Webster N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432 10.3389/fmicb.2015.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. W., Good B., Cole D., Lipp E. K. (2009). Plankton composition and environmental factors contribute to Vibrio seasonality. ISME J. 3 1082–1092. 10.1038/ismej.2009.50 [DOI] [PubMed] [Google Scholar]
- Vezzulli L., Brettar I., Pezzati E., Reid P. C., Colwell R. R., Höfle M. G., et al. (2012). Long-term effects of ocean warming on the prokaryotic community: evidence from the vibrios. ISME J. 6 21–30. 10.1038/ismej.2011.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L., Pezzati E., Stauder M., Stagnaro L., Venier P., Pruzzo C. (2015). Aquatic ecology of the oyster pathogens Vibrio splendidus and Vibrio aestuarianus. Environ. Microbiol. 17 1065–1080. 10.1111/1462-2920.12484 [DOI] [PubMed] [Google Scholar]
- Yong L., Guanpin Y., Hualei W., Jixiang C., Xianming S., Guiwei Z., et al. (2006). Design of Vibrio 16S rRNA gene specific primers and their application in the analysis of seawaterVibrio community. J. Ocean Univ. China 5 157–164. 10.1007/BF02919216 [DOI] [Google Scholar]
- Zorrilla I., Chabrillón M., Arijo S., Dìaz-Rosales P., Martìnez-Manzanares E., Balebona M. C., et al. (2003). Bacteria recovered from diseased cultured gilthead sea bream (Sparus aurata L.) in southwestern Spain. Aquaculture 218 11–20. 10.1016/S0044-8486(02)00309-5 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of the average seawater surface temperatures at Port-Hacking.
Sample location and environmental associated data.


