Abstract
A faithful expression of the mitochondrial DNA is crucial for cell survival. Animal mitochondrial DNA (mtDNA) presents a highly compact gene organization. The typical 16.5 kbp animal mtDNA encodes 13 proteins, 2 rRNAs and 22 tRNAs. In the backyard pillbug Armadillidium vulgare, the rather small 13.9 kbp mtDNA encodes the same set of proteins and rRNAs as compared to animal kingdom mtDNA, but seems to harbor an incomplete set of tRNA genes. Here, we first confirm the expression of 13 tRNA genes in this mtDNA. Then we show the extensive repair of a truncated tRNA, the expression of tRNA involved in large gene overlaps and of tRNA genes partially or fully integrated within protein-coding genes in either direct or opposite orientation. Under selective pressure, overlaps between genes have been likely favored for strong genome size reduction. Our study underlines the existence of unknown biochemical mechanisms for the complete gene expression of A. vulgare mtDNA, and of co-evolutionary processes to keep overlapping genes functional in a compacted mitochondrial genome.
Keywords: coevolution, isopod, mtDNA, overlapping gene, RNA, transcription, tRNA repair
Introduction
Metazoan mitochondrial DNA (mtDNA) is highly compact. Its classical content is 37 genes comprising 2 rRNA (rRNA) genes, 13 genes coding for proteins of the respiratory chain, 22 tRNA (tRNA) genes and a D-loop regulatory region, the major noncoding part of the genome. The coding sequences of mitochondrial genes are usually separated by at most a few nucleotides.1 The genome is transcribed as 3 large polycistronic RNAs. Many mRNA (mRNA) coding sequences are immediately adjacent at their 3′ end to tRNA genes and according to the tRNA punctuation model, primary transcripts are processed by precise endonucleolytic cleavages by RNases P and Z before and after each tRNA to release mature rRNAs and mRNAs.2-4 In addition, several mRNAs excised from the precursor molecules end with U or UA at their 3′-termini, and require polyadenylation to create a functional stop codon.5 The 5′ extremity of mature mRNAs usually begins with the initiation codon and there is usually no 5′ UTR. However, even if the punctuation model is often used in the mt metazoan world, it is known since the sequence of the human mtDNA has been obtained that this mechanism cannot be applied in all cases (for a review, see ref. 6). For instance, in human mitochondria, 4 mRNA sites must be cleaved following tRNA-independent processes.4,7 Furthermore, in silico analyses allowed to speculate that overlapping sequences may code simultaneously for tRNAs and mRNAs in many metazoan species, but without any direct demonstration so far.8 A major reason why this question remains open is the difficulty to precisely annotate and localize expressed tRNA genes in animal mtDNA.
Indeed, while variations in protein-coding and rRNA genes number are rare, the number of identified tRNA genes varies in almost 16% of arthropod mitochondria (e.g. 9-11). Several reasons can explain this extensive variation: increase can be due to changes in the genetic code or a consequence of tRNA gene duplication, while decrease is generally due to the loss of mt tRNA genes during evolution, compensated by the import of nucleus-encoded tRNAs, a classical phenomenon in mitochondria.12,13 Alternatively, variation in tRNA gene number in annotated genomes may also be the consequence of defects of tRNA search algorithms that identify false positive tRNA or conversely fail to detect them. Metazoan mt tRNAs often appear degenerated, without the complete cloverleaf secondary structure: absence of the highly conserved D-loop and/or T-loop and even complete arms renders their detection problematic.14,15 As a matter of fact, in addition to the classical tool tRNAscan-SE16 that detects cloverleaf structures, other more specialized tools such as ARWEN17 and MiTFi14 were developed to significantly improve metazoan mt tRNA gene identification. Moreover, tRNA genes can also be missed because of extensive RNA editing that modifies the RNA sequence as compared to the genomic DNA sequence. In mitochondria, several editing processes have been observed: 3′- replacement editing (mainly due to polyadenylation) in various metazoans, 5′-replacement editing in protists and chytridomycete fungi and insertion editing in slime molds (e.g.,18,19). Finally, heteroplasmic mtDNA may allow for the presence of two tRNA genes with different anticodons at the same genome position, an example of “hidden” tRNA recently described in the mtDNA of Oniscidea.20-22 Unfortunately, in most cases, experimental data to validate or invalidate the expression of tRNA candidates are mostly unavailable yet.
A correct expression of the mtDNA is essential for life and its perturbation may cause multiple syndromes. The importance of RNA maturation in the regulation of metazoan mt gene expression is far from being fully understood (for a review, see refs. 6, 23). Here, we focused our work on the identification and expression of overlapping genes generated during the evolution of metazoan mitochondrial genomes. For that purpose we choose an appropriate model organism, the isopod crustacean Armadillidium vulgare. With a size of 13,939 bp, its mtDNA is among the smallest in arthropods,24 and the smallest described in crustacean malacostraca (average malacostraca mtDNA size of 15,747 bp, calculated from 136 species with complete sequences available on NCBI in April 2015). Although it codes for the usual set of 13 protein-coding genes and two rRNA subunits, only a partial set of mitochondrial tRNA genes was identified by computational means.20,25 This partial set of tRNA genes was however not sufficient to account for a size reduction of 1.8 kbp in comparison to average malacostraca mt genome. Additional size reduction is due to shortness of the control region,24 but also to putative gene overlaps, implicating tRNA genes. Based on these observations, we first reanalyzed the tRNA gene content and experimentally validated the tRNA candidates. Then we demonstrated that a tRNA expressed from a single truncated mt gene and overlapping with surrounding genes, is extensively repaired at its 3′ end. Likewise we showed that many expressed tRNA genes overlap with other genes. Extreme cases were encountered such as a large overlap between two tRNA genes transcribed in the same direction or tRNA genes either partially or fully inserted into a protein-coding gene in direct or opposite orientation. The data presented here support the existence of novel but still unknown biochemical mechanisms crucial for animal mt genes expression. From an evolutionary point of view, two remarkable consequences are worth to note. First, under selective pressure for genome size reduction, gene overlaps and dual-coding sequences may allow mtDNA genome compaction while avoiding the loss of tRNA genes. Second, this implies a strong co-evolution of both overlapping gene sequences and/or the development of repair system to keep the two genes functional. Finally, the existence of large gene overlaps may represent a powerful marker to perform phylogenetic studies on metazoan mtDNA.
Results
Mitochondrial tRNA gene expression in A. vulgare
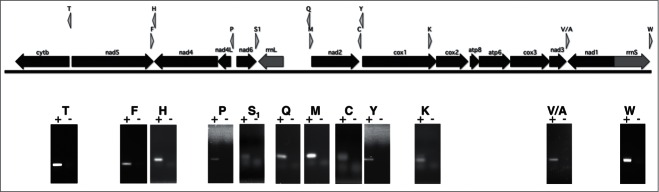
Using tRNAscan-SE26 and/or ARWEN17 software, Marcadé et al20 and Kilpert et al25 reported 9 and 13 tRNA genes from the A. vulgare mtDNA, respectively. Using the recently developed MiTFi pipeline under standard condition,14 10 tRNA genes (E-value cutoff of E <0.001) were recovered (updated NCBI A. vulgare genome EF643519.3). A common set of tRNA genes exists between the three annotations but also discrepancies and several tRNA genes are missing (Table S1). It was therefore difficult to assess through in silico analysis, the real set of tRNAs expressed from A. vulgare mt genes. To address this question, in the presence of a very limited amount of total RNA available, the expression of each putative tRNA gene was analyzed by RT-PCR (or circularized RT-PCR (cRT-PCR) when required); PCR products were cloned and sequenced. Only 13 mt tRNA genes were shown expressed (Fig. 1; Table S1). Among them, we confirmed the expression of both tRNAs valine and alanine from the previously identified valine/alanine alloacceptor tRNA gene,21 half of the clones analyzed being valine GTA anticodon and the others alanine GCA anticodon.
Figure 1.
Representation of A. vulgare mtDNA map with confirmed expressed genes. Sense of the arrows indicates sense of transcription. Genes coding for proteins of the respiratory chain, for rRNAs and for tRNAs are represented by black, dark gray and gray arrows respectively. Below the map, images of ethidium bromide stained gels of RT-PCR products demonstrating the expression of the tRNA genes are presented. Experiments were designed with pairs of primers specific for each tRNA (Supplementary Table 1). The presence (+) or absence (−) of reverse transcriptase (RT) during the cDNA synthesis is indicated. Amino acids specifying tRNAs are represented by the one-letter code.
Post-transcriptional maturation of tRNAHis sequence
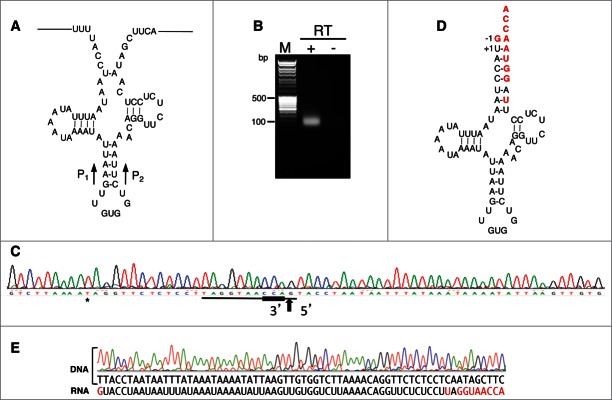
The identification of the tRNAHis gene was particularly difficult. First because its secondary structure (Fig. 2A) presents six mismatches in the acceptor stem, which is unexpected for a functional tRNA, and second because this gene was potentially overlapping with two other genes, tRNAPhe and nad4 (Fig. 3A). As the sequence of the specific reverse transcriptase-dependent product amplified by RT-PCR showed the expression of this gene (Fig. 1), we decided to determine the 5′- and 3′- extremities of the mature tRNAHis. For this purpose, total A. vulgare RNAs were circularized. A specific product was then amplified by RT-PCR, cloned, sequenced, and a cloverleaf structure of the mature tRNAHis deduced (Fig. 2D). In the mature tRNAHis, the base-pairing of the acceptor stem is fully restored thanks to nucleotide replacement at the 3′-end of the tRNA. In addition, the presence of a CCA triplet at the 3′ end and of an additional G-1 guanosine residue at the 5′ end, a nearly universal feature for aminoacylation by histidyl-tRNA synthetase27-29 confirms that A. vulgare mt tRNAHis encodes a functional tRNA. Out of 95 clones, 87 sequences correspond to the fully repaired tRNAHis while 8 represent intermediate products (Fig. S1). From this analysis, we propose the following steps of A. vulgare mt tRNAHis biogenesis (Fig. S2). The primary transcript is processed at the 5′- end by the mt RNase P, then a RNase Z activity may cleave unusually between the T stem and the acceptor stem. Although no processing intermediates have been found, we cannot exclude a classical cleavage of RNase Z followed by the involvement of a 3′- to 5′- exonuclease to remove the unpaired nucleotides. The 3′-end of the tRNA is then repaired. Two ways to restore base-pairing in the stem are possible. Either the truncated sequence is repaired by a trial and error process involving nucleotides elongation/degradation until the sequence is correct (a tRNA repair process demonstrated in metazoan mitochondria30), or the 5′-end of the acceptor stem is used as a template for synthesis of the 3′-end by a RNA-dependent RNA polymerase (RdRp) (as suggested in centipede mitochondria31). No tRNA processing intermediates showing mis-incorporated nucleotides were found, thus the action of the second repair process appears to be more likely. The two last nucleotides before the CCA-end are two As. The A72 pairs with the U+1 while the A73 is the discriminator nucleotide. To add these As, three enzymatic activities are possible, either a RdRp, a polyA polymerase, or the mt tRNA nucleotidyltransferase that classically adds the CCA-termini.30 Finally, the G-1 is incorporated thanks to a tRNAHis-dependent guanylyl transferase. Indeed, ESTs encoding a mt homolog of Thg1p, the cytosolic S. cerevisiae tRNAHis-dependent guanylyl transferase,32 were retrieved from A. vulgare EST libraries (Fig. S3). As in human and mouse, the A. vulgare enzyme possesses a putative N-terminal mt targeting sequence and this mt enzyme likely adds the essential G-1 residue.
Figure 2.
The biogenesis of A. vulgare mt tRNAHis requires repair at both the 5′- and 3′-ends. (A) Inferred tRNA secondary structure of the tRNAHis primary transcript deduced from the genomic sequence. The primers P1/P2 used for cRT-PCR analyses are depicted by black arrows, their precise positions are presented on Table S2. (B) Total nucleic acids were analyzed by cRT-PCR using primers P1 and P2. After circularization in the presence of T4 RNA ligase, cDNA was synthezized in the presence of primer P1. The image shows the ethidium bromide stained gel of the PCR product amplified using primers P1 and P2. The lane marked M shows the migration of the DNA ladder. (C) Sample sequence of one of the clones obtained from the cRT-PCR product shown in (B). The 3′- and 5′- extremities of the tRNA molecule are presented. The two extremities are separated by a vertical arrow. The CCA extremity is underlined in bold. Note that primers used for tRNA-His sequencing (Table S2) were designed from the online reference genome sequence (GenBank accession EF643519.3), which presents one nucleotide polymorphism in the variable loop (indicated by an asterisk). This did not affect the amplification of cDNA and the demonstration of post-transcriptional modification. (D) Inferred secondary structure of the mature tRNAHis deduced from the sequence of the cloned cRT-PCR product. Nucleotides differing between the genomic sequence and the RNA sequence are indicated in red. (E) Comparison between the sequence of the genomic DNA region coding for the A.vulgare mt tRNAHis and the sequence deduced from the corresponding RNA amplified by cRT-PCR.
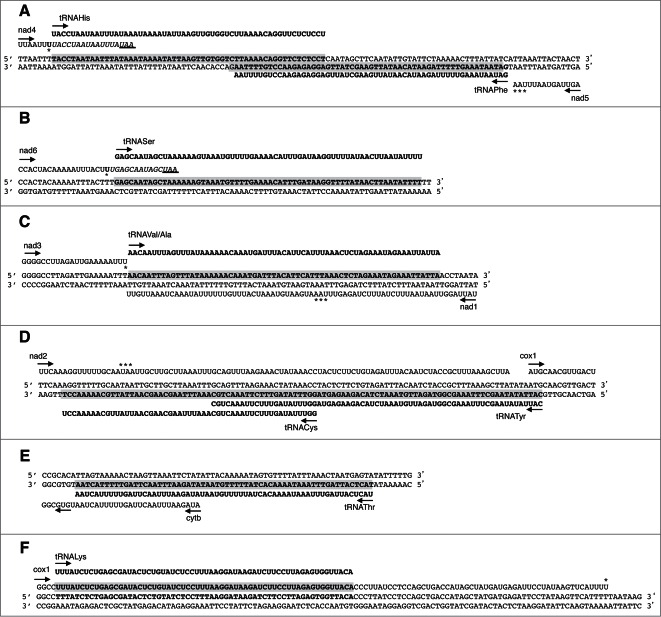
Figure 3.
Overlapping A. vulgare mt tRNA genes. Double stranded DNA regions coding for overlapping genes are presented. tRNA genes are under gray backgrounds. Horizontal arrows indicate the sense of transcription on RNA sequences. Stars indicate UAA stop codons or U residues requiring 2 As residues to generate them. For nad4 and nad6, RNA sequences not present in vivo are in italics and UAA stop codons not used in vivo are underlined. (A) overlaps nad4/tRNAHis and tRNAHis/tRNAPhe, (B) absence of overlap between nad6/tRNASer, (C) overlap tRNAVal/Ala/nad1, (D) overlaps tRNACys/tRNATyr and tRNATyr/cox1, (E) potential overlap cytb/tRNAThr, depending on the start codon used (AUA or GUG) (F) overlap cox1/tRNALys.
Multiple gene overlaps in the A. vulgare mt genome
Complete sequencing of A. vulgare mt genome revealed potential large overlaps between genes,20,25 with no functional demonstration. We further investigated the precise location of each expressed tRNA gene with regard to the other A. vulgare mt genes. To do so, we first determined the unknown extremities of the two rRNAs (Fig. S4) and the 3′ extremity of mRNAs (Fig. 3 and Fig. S5). It is to note that while the 3′ extremities of the two rRNAs are homogeneous in size and are polyadenylated in vivo, their 5′ extremities are rather heterogeneous. The 5′ extremity of the large rRNA ends within a stretch of 10 A residues (except for one sequence) while for the small rRNA, two types of 5′ extremity were found, one type about 30 nucleotides longer than the other. In both cases, whether this reflects in vivo situation, instability of the 5′ extremity or a cloning artifact is unknown.
With the exception of tRNAGln, tRNATrp and tRNASer, the other tRNA genes may overlap with other tRNA or mRNA genes. From the in silico analysis, the tRNAHis gene seems to overlap with two adjacent genes. On the opposite strand, tRNAHis overlaps with tRNAPhe over 19 nt (Fig. 3A). As mt tRNAHis and tRNAPhe are encoded on different strands, their processing from two independent primary transcripts is easily achieved. In the same orientation of transcription, tRNAHis shows an apparent minimum overlap with nad4 over 17 nt, while considering the UAA codon as a stop codon. However, after sequencing the exact 3′- end of nad4 mRNA (Fig. S5), we show that an alternative and truncated stop codon is polyadenylated (addition of two A residues to complete a UAA stop codon) (Fig. 3A), a common phenomenon in metazoan.6 This reduces the size of the gene and avoids overlap between nad4 and tRNAHis genes.
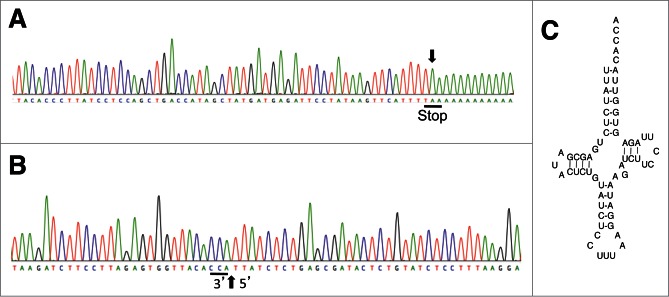
As shown for tRNAHis, in silico analysis suggested that tRNASer overlaps with nad6 (Fig. 3B). However, this was invalidated after the determination of the 3′- end of nad6 mRNA, showing a shortened version of the gene (with a stop codon generated by addition of a polyA tail), thus excluding any overlap. By contrast, the 65 nt alloacceptor tRNAVal/Ala gene fully overlaps with nad1 gene (in antisense orientation) while the overlap was expected to be of “only” 26 nt from in silico analysis when considering the UAA stop codon as the 3′ extremity of the mRNA (Fig. 3C). Even more surprising, tRNATyr overlaps with two genes, the tRNACys on the same strand, over 22 nt (Fig. 3D and Fig. S6), and cox1 on its opposite strand, over 3 nt. The tRNACys gene is completely included in the nad2 gene. The two genes are transcribed in opposite direction and the nad2 mRNA is 78 nt longer than predicted in silico. The tRNAThr and cytb genes, transcribed on the same strand (Fig. 3E), may share 26 nt if we consider codon AUA as a start codon. However, if the codon GUG is used to initiate translation, a situation already described for mt gene expression, (e.g.33,34) then there is no overlap and the processing of tRNAThr will release the 5′ extremity of the cytb mRNA. As we failed in determining the 5′ extremity of cytb mRNA, none of these two possibilities can be excluded. Finally the 60 nt long tRNALys is fully integrated into the coding sequence of cox1 (Fig. 3F). This latter observation was validated by the identification of the 3′ extremity of cox1 mRNA, where polyadenylation creates a stop codon (Fig. 4A). Moreover, the tRNALys gene is inserted into a conserved region of cox1 (Figure S7). Finally, the expression of a mature tRNALys was validated by cRT-PCR followed by cloning and sequencing of the PCR product (Figs. 4B and 4C).
Figure 4.
tRNALys gene lies within cox1 gene. (A) Sample sequence of one of the clones obtained from the analysis of the 3′ extremity of cox1 mRNA. The polyA tail found at the 3′ extremity is indicated by a vertical arrow and the generated stop codon underlined. This confirms the complete expression of cox1. (B) Sample sequence of one of the clones obtained from the cRT-PCR product amplified in the presence of primers specific of A. vulgare mitochondrial tRNALys. The 3′- and 5′- extremities of the tRNA molecule are separated by a vertical arrow. The post-transcriptionally added CCA extremity is underlined. (C) Deduced cloverleaf structure of the A. vulgare mitochondrial tRNALys.
Discussion
A. vulgare mt genome encodes a limited set of tRNA genes
A. vulgare mtDNA was shown to encode a limited set of 13 tRNA genes. Among them, degenerated tRNA genes were identified: several genes have reduced D- or T-arms, tRNAPhe, tRNAPro , tRNATrp and tRNAGln have no T-arm and tRNACys has no D-arm.20,25 This is in agreement with the multiple observations of tRNA genes with short D-and/or T-arms or even lacking at least one of them in metazoan mitochondria,14,35 one way toward genome size reduction. The extreme situation such as the existence of armless mt tRNA genes in nematodes Enoplea15,36 has not been observed here.
Although we cannot exclude that very short, post-transcriptionally modified and/or “bizarre” tRNA genes escaped our detection, it is unlikely that a full set of functional tRNAs is encoded by the mt genome of A. vulgare, where up to 9 mitochondria-encoded tRNAs are likely missing. Indeed, many mtDNAs have now lost at least few tRNA genes and the absence of an apparently full set of mt tRNA genes is rather frequent.10 For instance, in Cnidaria or Chaetognatha, nearly all mt tRNA genes have been lost (e.g.,14,37). In seven other isopod mt genomes, various numbers (from 1 to 8) of missing tRNA genes were observed.25 The missing genes are likely compensated by mitochondrial import of nucleus-encoded tRNAs, a widespread phenomenon in evolutionary divergent organisms.10 The loss of tRNA genes in A. vulgare mtDNA may have happened together with the reduction of the genome size and of the occurrence of multiple rearrangements within the mtDNA of isopod crustaceans.20,25
A. vulgare mt tRNAHis is extensively repaired
A second strategy to cope with genome size reduction and to maintain functional tRNA genes is to repair truncated tRNAs expressed from degenerated genes. A. vulgare mt tRNAHis is extensively repaired at its 3′ extremity in order to restore amino acid acceptor stem base-pairing, and classically a G-1 is added at the 5′ end to allow the recognition by the mt Histidyl-tRNA synthetase. This is an extreme case where both ends need to be edited. Integrity of the 3′- extremity of tRNAs is essential for their functionality and truncated mt metazoan tRNAs lacking up to 6 nt at their 3′- end were previously shown to be repaired.38 In most cases, repairs likely involve the tRNA nucleotidyltransferase and/or a polyA polymerase. However, a few observations are consistent with the existence of another type of polymerases able to add nucleotides to the 3′-end of tRNAs.30,39,40 The data presented here strongly suggests that a RdRp is involved in the synthesis of the 3′-end using the 5′-end of the tRNAHis acceptor stem as a template.
Overlapping genes in the A. vulgare mt genome
In highly compact metazoan mtDNAs, most mRNA coding sequences are immediately adjacent at their 3′ end to tRNA genes and according to the tRNA punctuation model (Fig. 5A), primary transcripts are processed by precise endonucleolytic cleavages by RNases P and Z before and after each tRNA to release mature mRNAs.2-4 These mRNAs possess no significant 5′UTR and several of them require polyadenylation to create a functional stop codon.5 As described above, the tRNA punctuation model can be applied to several A. vulgare genes (Fig. 5A; e.g. nad3 with the alloaceptor tRNAVal/Ala gene).
Figure 5.
RNA processing of overlapping mitochondrial genes. (A) In most cases of RNA processing of animal mtDNA, the DNA is transcribed into polycistronic RNAs. mRNAs are punctuated by tRNAs. The tRNAs are processed by RNases P and Z, thus excising mRNAs.2-4 When mRNAs are incomplete, polyadenylation is used to create a stop codon, and mRNA is translated.5 In A. vulgare, this tRNA punctuation model is valid for part of the RNAs, including the tRNAVal-Ala (V-A in the gray box), adjacent of the nad3 gene. In cases of gene overlaps, possible additional models are as follow. (B) When there is overlap between a tRNA and the 5′-end of the coding sequence of a mRNA, either cleavage occurs unusually releasing a full-length mRNA and a truncated tRNA that is further repaired or the tRNA is classically processed, thus excising an incomplete mRNA (ΔmRNA) that can be either translated thanks to an unusual initiation codon25 or degraded. This is the case of tRNAThr (T) which overlaps with the gene cytb. (C) When a tRNA is integrated within the coding sequence of a mRNA, alternative processing events are required to get both RNA molecules functional. If a tRNA cloverleaf structure is recognized by the tRNA processing machinery, a mature tRNA is generated and truncated mRNA enters the degradation pathway. If an alternative folding of the RNA molecule does not allow the cleavage by RNAses P and Z, the mRNA is engaged in the translation process. This is the case of tRNALys (K) embedded in the coding sequence of cox1. (D) When two tRNAs oriented in the same direction overlap, two alternative tRNA foldings may allow the processing of two tRNAs. The sequence common to both tRNAs is schematically presented in dark gray, and the sequences specific to tRNA1 and tRNA2 in black and pale gray respectively. This is the case for the overlapping tRNACys (C) and tRNATyr (Y). Stars represent initiation codon. To note, the other post-transcriptional modifications (CCA addition, nucleotides modification) required to get functional tRNAs are not presented here. Examples of A.vulgare genes where each potential model can be applied are presented under gray background.
The presence of overlapping genes represents another way to shorten a genome. Short overlaps of 1 to 3 nt have been identified, mainly in silico, in several metazoan mtDNAs.14 In A. vulgare mtDNA, potentially only three short overlaps exist. Cox1 and tRNATyr genes are in opposite direction and overlap over 3 nt. A 2 nt overlap between tRNAPro and nad4L and a 6 nt overlap between tRNAMet and nad2 may also exist (Fig. S8). Alternatively, if other downstream initiation codons are used, there will be no overlap. By contrast, the presence of potential larger overlaps between genes of various arthropods, including A. vulgare, has been postulated with no demonstration yet.25,41-44 Here we show that, indeed, the other overlaps are larger, from 19 to 65 nt, which is much more than usually described. When two overlapping genes are on different strands (e.g., the alloacceptor tRNAVal/Ala gene and nad1), cleaving the two independent primary transcripts will directly generate functional RNAs. By contrast, when the two overlapping genes are on the same strands (e.g., tRNACys and tRNATyr; tRNALys within cox1), the ways to produce functional RNAs are less obvious. Several non-exclusive processes are possible (Fig. 5). In the case of overlap between 2 tRNAs (e.g., tRNACys and tRNATyr), processing implicating alternative secondary structure of the primary transcript appears to be the most plausible explanation (Fig. 5D). Possible alternative processing has already been reported for the squid Loligo bleekeri tRNATyr and tRNACys but in that case it involves only a one nt overlap,40 while here the 22 nt overlap likely implies stronger constraints. If the overlap is between a tRNA and a mRNA (Fig. 5B), classical processing of the tRNA precursor will release a truncated mRNA and a non classical codon can be used to initiate translation, leading to a slightly shorter protein. The use of alternative start codons in isopod mt mRNAs has already been reported.25 Otherwise, if processing occurs at the expected initiation codon of the mRNA, a truncated tRNA is generated, and repair would imply the re-synthesis of more than one third of the complete tRNA, which is rather unlikely but cannot be excluded as observed for tRNAHis that requires extensive repair. Alternative processing might be possible for the production of either a complete mRNA or a complete tRNA (Fig. 5C), a mechanism that would need to be elucidated. This last explanation is the most likely for the tRNALys gene that is fully integrated into cox1 gene, as we confirmed the complete expression of both genes. In all potential alternative processing pathways proposed here, protein factors are likely implicated and need to be characterized.
From an evolutionary point of view, rather than to represent an exception, the situation found in A. vulgare mitochondria is likely more widespread than previously thought. Based on computational analysis, the presence of mitochondrial sequences that simultaneously code for tRNAs and proteins has already been envisaged in nematodes, arthropods or onychophora but was not demonstrated yet.8 Large overlaps between protein coding genes were also reported in cnidarians and vertebrates.7,45,46 When looking for bizarre tRNA genes expressed from mtDNA, the investigation is usually oriented toward intergenic regions. Here, we show the necessity to screen for tRNA genes all along the DNA sequence. This is particularly true for genomes where tRNA genes are apparently missing and we can speculate that the number of tRNA genes that do not follow the tRNA punctuation model will rapidly increase once we will start to look for them.
The use of overlapping genes for genome size reduction seems generally restricted to tRNA genes. Indeed, whether they are on the same or on the opposite strand, overlaps require a strong co-evolution between gene sequences. Such constraint is more likely to be too stringent for protein-coding genes overlaps. In contrast, tRNA gene sequences evolve rapidly and recent evidence showed that a simple secondary structure coupled with a specific anticodon may be sufficient to generate a functional tRNA,47,48 or that incomplete cloverleaf structure may be repaired post-transcriptionally (e.g., tRNAHis in this study).
Considering animal mt genome expression and regulation, the existence of a few animal mitochondrial mRNAs not delimited by tRNA genes and called tRNA-less RNA precursors, shows that their release cannot rely solely on the processing of tRNAs by RNase P and RNase Z. Indeed, the RNA binding protein GRSF1 (G-rich sequence factor 1) was recently shown to be involved in the processing of such primary transcripts.49 Here, the existence of tRNA genes partially or fully integrated within protein coding genes transcribed in the same direction represents another situation where the tRNA punctuation model of RNA processing described in metazoan mitochondria2,3 presents limitations. Furthermore, we also showed that A. vulgare nad1 mRNA does not end at the level of the stop codon but rather terminates at the level of the antisense sequence of the alloaceptor tRNAAla/Val gene sequence. This suggests that the antisense sequence of a true tRNA can be folded into a tRNA-like structure and can be recognized as a processing site to release the 3′ end of a mRNA. This intriguing but open question will need to be addressed in the future.
In addition to the tRNA punctuation model described as a major way for the release of functional RNAs, other mechanisms were early suspected to account for complete processing of animal mt primary transcripts. Our data strongly support the existence of still unknown alternative pathways to generate functional mRNAs and tRNAs in metazoan mitochondria. The importance and extent of alternative processing models such as the ones we propose here is likely more widespread than initially thought in evolutionary divergent metazoans. The pathways and protein factors involved in mitochondrial gene expression and regulation appear more complex than previously estimated and their characterization is a challenge for the future.
Materials and Methods
Total RNA extract
Five A. vulgare females from BF 2739 strain (BF) were used as starting material. The isofemale lines (i.e., a lineage of individuals all descended from a single female) are reared in the laboratory and are known to be asymbiotic for Wolbachia (i.e., A. vulgare endocellular symbiont) avoiding contamination. Total RNA was extracted from gonads, fat tissues, and nervous system using a Trizol protocol, as previously described.50 From five A. vulgare females, not more than 5 µg of total nucleic acid can be obtained.
Cloning of reverse transcription (RT)-PCR products
A. vulgare total nucleic acid was first treated with DNASE RQ1 according to manufacturer's intructions (Promega). The DNase-treated RNA sample was then used as substrate for RT-PCR amplification using relevant pairs of primers, cloned into pGEM-T Easy (Promega), as described in 51 and sequenced.
Cloning of circular RT-PCR products
Circular reverse transcription-PCR (cRT-PCR) was used to determine 5′- and 3′- termini of tRNAHis essentially as described in.51 Briefly, total RNA extracted from A. vulgare was incubated with 40U of T4 RNA ligase (New England Biolabs) in the supplied buffer supplemented with 2U of RNase inhibitor and in a total volume of 25 µl. Following circularization, all steps of RT-PCR were classically carried out using a relevant pair of primers. PCR products were cloned into pGEM-T Easy and sequenced.
Determination of 5′- and 3′-ends of rRNAs and mRNAs
Sequence determination was mostly performed as described in.36 Using T4 RNA ligase, total RNA was fused to a tagging DNA oligonucleotide that carried a ribonucleotide at the phosphorylated 5′-end and a 3′-end blocked by a 3′-3′- linked T. To determine RNA 3′-end, cDNA, synthesized by hybridizing RT primer to the tagging oligonucleotide, was then amplified in the presence of the RT primer and a second primer specific to the gene of interest. All steps of RT-PCR and cloning of PCR products were carried out as described above. To determine RNA 5′-end, the cDNA synthesized above was used in a tailing reaction with terminal deoxynucleotidyl-transferase and dCTP according to the manufacturer (Invitrogen). The reaction product was amplified by PCR using a C-tail primer and a second primer specific to the gene of interest. Cloning of PCR products were carried out as described above.
EST Library
EST data were extracted from the EST libraries previously performed in A. vulgare by SSH method.52
Miscellaneous
Oligonucleotides were synthesized by Eurofins. Oligonucleotide sequences used in this study are available as supplemental information (Tables S2 and S3).
Funding Statement
This work has been supported by Center National de la Recherche Scientifique (CNRS) in association with the University of Strasbourg. This work has been published under the framework of the LABEX (ANR-11-LABX-0057_MITOCROSS) and benefits from a funding from the state managed by the French National Research Agency as part of the Investiments for the future program. V.D. has been supported by the région Poitou-Charentes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Philippe Giegé, Anthony Gobert, Eric Westhof are acknowledged for critical reading of the manuscript and Myriam Badawi for informatic help. The authors wish to thank the membership of the ANR-2010-BLANC-170101 “ImmunSymbArt” for providing us the nuclear EST sequence.
Supplemental Material
Supplemental Material may be downloaded here: publisher's website
References
- 1.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res 1999; 27:1767-80; PMID:10101183; http://dx.doi.org/ 10.1093/nar/27.8.1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ojala D, Merkel C, Gelfand R, Attardi G. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell 1980; 22:393-403; PMID:7448867; http://dx.doi.org/ 10.1016/0092-8674(80)90350-5 [DOI] [PubMed] [Google Scholar]
- 3.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981; 290:470-4; PMID:7219536; http://dx.doi.org/ 10.1038/290470a0 [DOI] [PubMed] [Google Scholar]
- 4.Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al.. Sequence and organization of the human mitochondrial genome. Nature 1981; 290:457-65; PMID:7219534; http://dx.doi.org/ 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- 5.Schuster G, Stern D, RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol transl Sci; 2009; 85:393-422; PMID:19215778; http://dx.doi.org/ 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 6.Rorbach J, Minczuk M. The post-transcriptional life of mammalian mitochondrial RNA. Biochem J 2012; 444:357-73; PMID:22642575; http://dx.doi.org/ 10.1042/BJ20112208 [DOI] [PubMed] [Google Scholar]
- 7.Rackham O, Mercer TR, Filipovska A. The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. RNA 2012; 3:675-95; PMID:22777840 [DOI] [PubMed] [Google Scholar]
- 8.Morrison DA. How and where to look for tRNAs in Metazoan mitochondrial genomes, and what you might find when you get there. ; Internet. 2009. Available from: http://arxiv.org/abs/1001.3813. [Google Scholar]
- 9.Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 2008; 101:301-20; PMID:18612321; http://dx.doi.org/ 10.1038/hdy.2008.62 [DOI] [PubMed] [Google Scholar]
- 10.Huot JL, Enkler L, Megel C, Karim L, Laporte D, Becker HD, Duchêne AM, Sissler M, Maréchal-Drouard L. Idiosyncrasies in decoding mitochondrial genomes. Biochimie 2014; 100:95-106; PMID:24440477; http://dx.doi.org/ 10.1016/j.biochi.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 11.Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, Passamonti M. A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends in genetics : Trends in Genet 2014; 30:555-64; http://dx.doi.org/ 10.1016/j.tig.2014.09.002 [DOI] [PubMed] [Google Scholar]
- 12.Duchêne AM, Pujol C, Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet 2009; 55:1-18; PMID:Can't; http://dx.doi.org/ 10.1007/s00294-008-0223-9 [DOI] [PubMed] [Google Scholar]
- 13.Beagley CT, Wolstenholme DR. Characterization and localization of mitochondrial DNA-encoded tRNAs and nuclear DNA-encoded tRNAs in the sea anemone Metridium senile. Curr Genet 2013; 59:139-52; PMID:23801360; http://dx.doi.org/ 10.1007/s00294-013-0395-9 [DOI] [PubMed] [Google Scholar]
- 14.Jühling F, Pütz J, Bernt M, Donath A, Middendorf M, Florentz C, Stadler PF. Improved systematic tRNA gene annotation allows new insights into the evolution of mitochondrial tRNA structures and into the mechanisms of mitochondrial genome rearrangements. Nucleic Acids Res 2012; 40:2833-45; PMID:22139921; http://dx.doi.org/ 10.1093/nar/gkr1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jühling F, Putz J, Florentz C, Stadler PF. Armless mitochondrial tRNAs in Enoplea (Nematoda). RNA Biol 2012; 9:1161-6; PMID:23018779; http://dx.doi.org/ 10.4161/rna.21630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-64; PMID:9023104; http://dx.doi.org/ 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laslett D, Canback B. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 2008; 24:172-5; PMID:18033792; http://dx.doi.org/ 10.1093/bioinformatics/btm573 [DOI] [PubMed] [Google Scholar]
- 18.Gott JM, Somerlot BH, Gray MW. Two forms of RNA editing are required for tRNA maturation in Physarum mitochondria. RNA 2010; 16:482-8; PMID:20106952; http://dx.doi.org/ 10.1261/rna.1958810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betat H, Long Y, Jackman JE, Morl M. From end to end: tRNA editing at 5′- and 3′-terminal positions. Int J Mol Sci 2014; 15:23975-98; PMID:25535083; http://dx.doi.org/ 10.3390/ijms151223975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcadé I, Cordaux R, Doublet V, Debenest C, Bouchon D, Raimond R. Structure and evolution of the atypical mitochondrial genome of Armadillidium vulgare (Isopoda, Crustacea). J Mol Evol 2007; 65:651-9; PMID:Can't; http://dx.doi.org/ 10.1007/s00239-007-9037-5 [DOI] [PubMed] [Google Scholar]
- 21.Doublet V, Souty-Grosset C, Bouchon D, Cordaux R, Marcadé I. A thirty million year-old inherited heteroplasmy. PLoS One 2008; 3:e2938; PMID:18698356; http://dx.doi.org/ 10.1371/journal.pone.0002938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandler CH, Badawi M, Moumen B, Greve P, Cordaux R. Multiple Conserved Heteroplasmic Sites in tRNA Genes in the Mitochondrial Genomes of Terrestrial Isopods (Oniscidea). G3 2015; 5(7):1317; PMID:25911226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell CA, Nicholls TJ, Minczuk M. Nuclear-encoded factors involved in post-transcriptional processing and modification of mitochondrial tRNAs in human disease. Frontiers in Genet 2015; 6:79; http://dx.doi.org/ 10.3389/fgene.2015.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doublet V, Helleu Q, Raimond R, Souty-Grosset C, Marcadé I. Inverted repeats and genome architecture conversions of terrestrial isopods mitochondrial DNA. J Mol Evol 2013; 77:107-18; PMID:24068302; http://dx.doi.org/ 10.1007/s00239-013-9587-7 [DOI] [PubMed] [Google Scholar]
- 25.Kilpert F, Held C, Podsiadlowski L. Multiple rearrangements in mitochondrial genomes of Isopoda and phylogenetic implications. Mol Phyl Evol 2012; 64:106-17; http://dx.doi.org/ 10.1016/j.ympev.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 26.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 2005; 33:w686-9; PMID:15980563; http://dx.doi.org/ 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L'Abbé D, Lang BF, Desjardins P, Morais R. Histidine tRNA from chicken mitochondria has an uncoded 5′-terminal guanylate residue. J Biol Chem 1990; 265:2988-92; PMID:Can't [PubMed] [Google Scholar]
- 28.Yokobori S, Paabo S. Transfer RNA editing in land snail mitochondria. Proc Natl Acad Sci USA 1995; 92:10432-5; PMID:7479799; http://dx.doi.org/ 10.1073/pnas.92.22.10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Placido A, Sieber F, Gobert A, Galerani R, Giegé P, Maréchal-Drouard L. Plant mitochondria use two pathways for the biogenesis of tRNAHis. Nucleic Acids Res 2010; 38:7711-7; PMID:20660484; http://dx.doi.org/ 10.1093/nar/gkq646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reichert AS, Morl M. Repair of tRNAs in metazoan mitochondria. Nucleic Acids Res 2000; 28:2043-8; PMID:10773071; http://dx.doi.org/ 10.1093/nar/28.10.2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavrov DV, Brown WM, Boore JL. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc Natl Acad Sci USA 2000; 97:13738-42; PMID:11095730; http://dx.doi.org/ 10.1073/pnas.250402997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM. tRNAHis maturation: an essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev 2003; 17:2889-901; PMID:14633974; http://dx.doi.org/ 10.1101/gad.1148603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansen S, Guddal PH, Johansen T. Organization of the mitochondrial genome of Atlantic cod, Gadus morhua. Nucleic Acids Res 1990; 18:411-9; PMID:2308841; http://dx.doi.org/ 10.1093/nar/18.3.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desjardins P, Morais R. Nucleotide sequence and evolution of coding and noncoding regions of a quail mitochondrial genome. J Mol Evol 1991; 32:153-61; PMID:1706782; http://dx.doi.org/ 10.1007/BF02515387 [DOI] [PubMed] [Google Scholar]
- 35.He Y, Jones J, Armstrong M, Lamberti F, Moens M. The mitochondrial genome of Xiphinema americanum sensu stricto (Nematoda: Enoplea): considerable economization in the length and structural features of encoded genes. J Mol Evol 2005; 61:819-33; PMID:16315110; http://dx.doi.org/ 10.1007/s00239-005-0102-7 [DOI] [PubMed] [Google Scholar]
- 36.Wende S, Platzer EG, Jühling F, Putz J, Florentz C, Stadler PF, Mörl M. Biological evidence for the world's smallest tRNAs. Biochimie 2013; 100:151-8; PMID:23958440; http://dx.doi.org/ 10.1016/j.biochi.2013.07.034 [DOI] [PubMed] [Google Scholar]
- 37.Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem 2011; 80:14.1-.21; http://dx.doi.org/ 10.1146/annurev-biochem-060109-092838 [DOI] [PubMed] [Google Scholar]
- 38.Lavrov DV, Pett W, Voigt O, Worheide G, Forget L, Lang BF, Kayal E. Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code. Mol Biol Evol 2013; 30:865-80; PMID:23223758; http://dx.doi.org/ 10.1093/molbev/mss274 [DOI] [PubMed] [Google Scholar]
- 39.Yokobori SI, Paabo S. tRNA editing in metazoans. Nature 1995; 377:490; PMID:7566145; http://dx.doi.org/ 10.1038/377490a0 [DOI] [PubMed] [Google Scholar]
- 40.Tomita K, Ueda T, Watanabe K. RNA editing in the acceptor stem of squid mitochondrial tRNA(Tyr). Nucleic Acids Res 1996; 24:4987-91; PMID:9016670; http://dx.doi.org/ 10.1093/nar/24.24.4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satta Y, Ishiwa H, Chigusa SI. Analysis of nucleotide substitutions of mitochondrial DNAs in Drosophila melanogaster and its sibling species. Mol Biol Evol 1987; 4:638-50; PMID:2832697 [DOI] [PubMed] [Google Scholar]
- 42.Liu G, Jiang GF, Pang HC, Hong F. The mitochondrial genome of the Chinese special butterfly Luehdorfia chinensis Leech (Lepidoptera: Papilionidae). Mitochondrial DNA 2013; 24:211-3; PMID:23298143; http://dx.doi.org/ 10.3109/19401736.2012.748043 [DOI] [PubMed] [Google Scholar]
- 43.Xin T, Que S, Zou Z, Wang J, Li L, Xia B. Complete Mitochondrial Genome of Euseius nicholsi (Ehara et Lee) (Acari:Phytoseiidae). Mitochondrial DNA 2014; 538(1): 123- 137; http://dx.doi.org/ 10.1016/j.gene.2013.12.053 [DOI] [PubMed] [Google Scholar]
- 44.Easton EE, Darrow EM, Spears T, Thistle D. The mitochondrial genomes of Amphiascoides atopus and Schizopera knabeni (Harpacticoida: Miraciidae) reveal similarities between the copepod orders Harpacticoida and Poecilostomatoida. Gene 2014; 538:123-37; PMID:24389499; http://dx.doi.org/ 10.1016/j.gene.2013.12.053 [DOI] [PubMed] [Google Scholar]
- 45.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 1992; 141:173-216; PMID:1452431; http://dx.doi.org/ 10.1016/S0074-7696(08)62066-5 [DOI] [PubMed] [Google Scholar]
- 46.Capt C, Passamonti M, Breton S. The human mitochondrial genome may code for more than 13 proteins. Mitochondrial DNA 2015:1-4; PMID:25630734; http://dx.doi.org/ 10.3109/19401736.2014.1003924 [DOI] [PubMed] [Google Scholar]
- 47.Yona AH, Bloom-Ackermann Z, Frumkin I, Hanson-Smith V, Charpak-Amikam Y, Feng Q, Boeke JD, Dahan O, Pilpel Y. tRNA genes rapidly change in evolution to meet novel translational demands. eLife 2013; 2:e01339; PMID:24363105; http://dx.doi.org/ 10.7554/eLife.01339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, Xiao S, Li X, Li L, Shi W, Yu Z. Evolution of the tRNA gene family in mitochondrial genomes of five Meretrix clams (Bivalvia, Veneridae). Gene 2014; 533:439-46; PMID:24084366; http://dx.doi.org/ 10.1016/j.gene.2013.09.077 [DOI] [PubMed] [Google Scholar]
- 49.Jourdain AA, Koppen M, Wydro M, Rodley CD, Lightowlers RN, Chrzanowska-Lightowlers ZM, Martinou JC. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab 2013; 17:399-410; PMID:23473034; http://dx.doi.org/ 10.1016/j.cmet.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc 2006; 1:581-5; PMID:17406285; http://dx.doi.org/ 10.1038/nprot.2006.83 [DOI] [PubMed] [Google Scholar]
- 51.Placido A, Gagliardi D, Gallerani R, Grienenberger JM, Maréchal-Drouard L. Fate of a larch unedited tRNA precursor expressed in potato mitochondria. J Biol Chem 2005; 280:33573-9; PMID:16061472; http://dx.doi.org/ 10.1074/jbc.M505269200 [DOI] [PubMed] [Google Scholar]
- 52.Chevalier F, Herbiniere-Gaboreau J, Charif D, Mitta G, Gavory F, Wincker P, Grève P, Braquart-Varnier C, Bouchon D. Feminizing Wolbachia: a transcriptomics approach with insights on the immune response genes in Armadillidium vulgare. BMC Microbiol 2012; 12 Suppl 1:S1; PMID:22375708; http://dx.doi.org/ 10.1186/1471-2180-12-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.