ABSTRACT
RNA sequencing has revolutionized genome-wide transcriptome analyses, and the identification of non-coding regulatory RNAs in bacteria has thus increased concurrently. Here we reveal the transcriptome map of the lactic acid bacterial paradigm Lactococcus lactis MG1363 by employing differential RNA sequencing (dRNA-seq) and a combination of manual and automated transcriptome mining. This resulted in a high-resolution genome annotation of L. lactis and the identification of 60 cis-encoded antisense RNAs (asRNAs), 186 trans-encoded putative regulatory RNAs (sRNAs) and 134 novel small ORFs. Based on the putative targets of asRNAs, a novel classification is proposed. Several transcription factor DNA binding motifs were identified in the promoter sequences of (a)sRNAs, providing insight in the interplay between lactococcal regulatory RNAs and transcription factors. The presence and lengths of 14 putative sRNAs were experimentally confirmed by differential Northern hybridization, including the abundant RNA 6S that is differentially expressed depending on the available carbon source. For another sRNA, LLMGnc_147, functional analysis revealed that it is involved in carbon uptake and metabolism. L. lactis contains 13% leaderless mRNAs (lmRNAs) that, from an analysis of overrepresentation in GO classes, seem predominantly involved in nucleotide metabolism and DNA/RNA binding. Moreover, an A-rich sequence motif immediately following the start codon was uncovered, which could provide novel insight in the translation of lmRNAs. Altogether, this first experimental genome-wide assessment of the transcriptome landscape of L. lactis and subsequent sRNA studies provide an extensive basis for the investigation of regulatory RNAs in L. lactis and related lactococcal species.
KEYWORDS: dRNA-seq, lactic acid bacteria, small regulatory RNA, transcriptome, 6S
Introduction
Genome-wide transcriptome analyses using RNA sequencing (RNA-seq) has allowed adding to previously annotated genomes numerous novel elements, such as non-coding regulatory RNAs (sRNAs), antisense RNAs (asRNAs), small open reading frames (sORFs) and riboswitches. In addition, RNA-seq provides excellent opportunities to correct errors in annotated ORFs, to determine operon structures and to identify alternative internal transcription start sites (TSS) within coding genes.
An important next step in these studies is the validation of these novel RNAs and to unravel their functions in the cell. Functional studies of regulatory RNAs from a variety of bacterial genomes now reveal an ever-increasing number of new regulatory mechanisms. A lot of this research has been devoted to the sRNAs, which have been shown to post-transcriptionally control numerous cellular processes. They act mostly by base pairing with their target mRNAs, thereby influencing transcription termination, mRNA stability and/or mRNA translation.1-3 In addition, some sRNAs, such as the 6S sRNA and members from the CsrB family, have been reported to bind to and thereby influence the functionality of RNA polymerase and CsrA, respectively.4 Regulatory RNAs can function as signaling regulators responding to a changing environment and preparing the cell for altered conditions, as seen e.g. in pathogenic bacteria.5,6 Most regulatory RNAs are not translated into proteins and are therefore called non-coding regulatory RNAs although there are a number of exceptions of so-called dual-function RNA regulators; well-studied sRNAs such as RNAIII7,8 and SgrS9 have been reported to act as RNA regulators but also code for (small) proteins. The dual-function sRNAs can provide valuable insights into the evolutionary development of these RNAs by studying the physiological roles of the encoded peptide and the non-coding regulatory part.10
Different types of non-coding regulatory RNAs can be distinguished. For example, regulatory RNAs that derive from intergenic regions (IGRs) are generally named sRNAs. They are trans-encoded and affect one or more mRNA targets via imperfect base pairing. The RNA chaperone protein Hfq is often required to enable the interaction between the sRNA and its target mRNA.11-13 Hfq seems to be mainly present in GC-rich bacteria,14 and an Hfq homolog is absent in lactococcal genomes. Hfq has been used to purify and identify novel bacterial regulatory RNAs through Hfq-RNA immunoprecipitation and subsequent RNA-seq.15,16 By studying Hfq-bound transcripts, it has recently been shown that the 3′-untranslated regions (3′-UTRs) of mRNAs can harbor functional regulatory RNAs that act in trans. Such an sRNA can be derived by cleavage of the 3′-UTR of the original mRNA molecule. Alternatively, a separate promoter in the 3′-end of the gene can lead to an sRNA that overlaps the 3′-UTR of the mRNA.17
The second class of regulatory RNAs comprises the cis-encoded RNAs or antisense RNAs (asRNAs). An asRNA derives from the non-coding strand of a gene and its sequence is, thus, complementary to (part of) the gene’s mRNA. Although most regulatory RNAs are small and range in size from 50 to 350 nucleotides, antisense transcription can cover whole operons.18 The functions of many asRNAs still remain to be elucidated. The fraction of asRNAs in the total RNA pool of a bacterium is significant albeit variable between bacterial species.19 Even within species the total amount of asRNAs can greatly vary, which was very recently illustrated in an E. coli study that devoted special attention to asRNAs.20 Base pairing between an sRNA or asRNA and its partner mRNA usually involves the repression or activation of translation of the mRNA. The binding via 16S rRNA of the small 30S ribosomal subunit can be negatively affected by blocking of the ribosomal binding site (RBS) by the regulatory RNA.21 Activation can occur through the unfolding of a secondary structure in the mRNA via interaction with the regulatory RNA and the consequent liberation of the RBS.22 Moreover, base pairing of the 2 RNAs can lead to degradation of both by the endoribonuclease RNase E.23 Cleavage can occur near the RBS in the 5′ leader, in the coding region,24 or it can even take place downstream of the region where sRNA and mRNA interact.25 Translation-independent stabilization of mRNAs has also been reported in which the sRNA-mRNA hybrid interferes with RNase E-mediated degradation.26,27 Another ribonuclease that is important in mRNA regulation by sRNAs and asRNAs is RNase III, an enzyme that cleaves double stranded structures such as (a)sRNA-mRNA hybrids.
Another class of cis-encoded regulatory RNAs are sequences at the 5′ end of mRNAs that are able to change their conformation in response to an environmental cue. So-called thermometers react to changes in temperature,28 whereas a variety of riboswitches operate as intracellular sensors by binding to small metabolites or ions. Binding of the effector molecule influences the secondary structure of the riboswitch part of the mRNA, which affects the fate of transcription and/or determines whether the coding part of the mRNA is actually translated. Riboswitches can also influence mRNA stability.29,30 Two SAM riboswitches involved in the regulation of methionine and cysteine biosynthesis in L. monocytogenes were reported to act in-trans.31 Another surprising form of RNA regulation was reported in S. aureus, in which the 5′-UTR of the icaR mRNA interacts with the 3′-UTR of the same mRNA. This may either occur in cis within one mRNA molecule or in trans, involving 2 copies of the icaR transcript.32
RNA-seq and, to a lesser extent, tiling arrays have recently greatly increased the number of sRNAs in various microorganisms such as E. coli,33 B. subtilis,34 H. pylori35 and P. aeruginosa.36 The techniques also allowed, by exact determination of transcription start sites, the description of novel sORFs,37 operon structures and have in certain cases led to re-annotation of known ORFs.
Lactococcus lactis is an AT-rich, Gram-positive, mesophilic lactic acid bacterium with a relatively small genome size of 2.53 Mbp.38 It is widely applied in the dairy industry where its main function is to convert lactose into lactic acid and to provide texture, flavors and aromas. Previous studies using DNA microarray and proteomics technologies have identified genes and proteins involved in various (environmental) stress responses in L. lactis.39,40 The functioning in L. lactis of global regulators such as CcpA41 and CodY42 in carbon and nitrogen metabolism, as well as quite a number of other protein regulators has been described in considerable detail.43 Notwithstanding this, the presence and roles of regulatory RNAs L. lactis has not yet been reported, while it is becoming increasingly clear that these molecules play pivotal roles in gene regulation in many microorganisms, especially also in coping with stressful conditions. A better understanding of whether and how regulatory RNAs are involved in the regulation of stress responses and metabolic processes in L. lactis could lead to an improvement of the gene regulatory model of this organism44 and may have practical (industrial) implications. Using differential RNA sequencing (dRNA-seq), we uncovered 375 novel RNAs including sRNAs, asRNAs, long 5′-UTRs, putative regulatory 3′-UTRs, novel (small) ORFs, internal promoters, transcription start sites and operon structures.
Results and discussion
Determination of the primary transcriptome of L. lactis
In order to obtain deep insight in novel RNA elements in Lactococcus lactis, the organism was grown in GM17 and the cultures were harvested at 6 time-points during growth, 3 each in the exponential- and stationary phases, and mixed in equal OD equivalents prior to total RNA isolation and subsequent cDNA library preparation. Selective enrichment of primary transcripts was achieved by a Terminator 5′-phosphate-dependent exonuclease (TEX) treatment that specifically degrades processed 5′-monophosphate (5′P) RNA molecules.35 In addition to primary transcript enrichment, TEX treatment also results in enriched 5′-ends of mRNAs and ncRNAs in the RNA pool. In total, 10.5 million reads were generated, of which 7.2 million reads with a PHRED score > 28 were mapped onto the genome of L. lactis MG1363.38 Both in silico methods and visual inspection of the data were used to classify the L. lactis transcripts.
Identification of L. lactis sRNAs from intergenic regions and 3′-UTRs
The TEX-treated RNA was mapped on the genome of L. lactis MG1363 together with the in silico regulatory RNA prediction output from SIPHT45 to aid in the mining for potential regulatory RNAs. The genome-wide map was then visually inspected for sRNAs, asRNAs, long 5′-UTRs and to review and correct open reading frame (ORF) boundaries. See Fig. 1A for an overview of the results of the transcription start site (TSS) typing. The RNA-seq data and mining results have been integrated in a webpage using the JBrowse viewer, and can be assessed by http://jbrowse.molgenrug.nl/.
Figure 1.

TSS mining, 5′-UTR distribution and promoter analysis. (A) Different types of transcription start sites identified in the L. lactis MG1363 genome from mapped reads of the TEX-treated RNA-seq data set (gray arrows: annotated ORFs, blue: Reads from the + strand, green: Reads from the - minus strand, red blocks: Positions of putative regulatory RNAs predicted by SIPHT. (B) Length distribution of 5′-UTRs. 5′-UTRs up to a length of 100 nt are plotted in stepwise increments of 10 nt in gray, those larger than 100 nt are shown with increments of 25 nt (separated by the dotted line). Color code is given in the inset. The RBS consensus sequence and the consensus sequence in the first 7 nt of the 111 leaderless mRNAs were determined by MEME. (C) Top: Analysis using MEME of motifs in the 50 nt upstream of all 1819 TSSs predicted by TSSer. Curved dotted line: periodic AT stretches. Bottom: Reconstruction of the L. lactis promoter consensus using MAST. In both: −35 and −10 sequences are indicated.
RNAs in IGRs were annotated as sRNAs (denoted: LLMGnc_001–186, Table S1). Ten of these putatively trans-acting sRNAs overlap the 3′-UTR of mRNAs. They were identified on the basis of a high number of reads within the 3′-UTR, suggesting that a promoter exists for these sRNAs, although the possibility that they derive from processing of the overlapping longer mRNA cannot be excluded. Three sRNAs (LLMGnc_012/013/014) are located within a region of only 6 genes and show exceptionally high sequence similarity, suggesting they might have a common function. As a means to verify that the 186 sRNAs are genuine and to assess their conservation, a blast search was performed on 10 related L. lactis genomes. These genomes cover 5 strains each of the L. lactis subspecies lactis and cremoris. Most of the identified sRNA sequences are conserved in the subsp. cremoris strains, while this is to a lesser extent so for the 5 strains of the subspecies lactis (Table S1).
To examine the consensus of sRNA promoters, we evaluated the region from −100 to −1 upstream of the sRNA TSSs using MEME.46 We found no significant difference between the sRNA promoter consensus and the canonical L. lactis promoter. Subsequently, we screened the sRNA-promoter regions for the presence of known L. lactis transcription factor binding sequences (TFBSs). TFBSs for CcpA (carbon catabolite repression),41 CodY (nitrogen metabolism),42 ArgR (arginine metabolism)47 or FlpAB (metal ion homeostasis and oxidative stress)48 were identified upstream of 16 sRNA genes (Table S1 and S2). These putative regulation sites provide a link between transcription factors and sRNAs49 and underpin the versatility of gene regulation in this bacterium. To assess the relation between the predicted TFBSs in sRNA promoters and the function of the sRNA itself, 2 sRNA candidates (LLMGnc_147 and S6), which are predicted to be controlled by CcpA, were studied more in detail (see below).
To evaluate the presence in L. lactis of RNAs homologous to regulatory RNAs from other prokaryotes, we used the Bacterial Small Regulatory RNA Database (BSRD).50 A total of 37 of such homologous regulatory RNAs could thus be identified in the genome of L. lactis MG136 (Table S3). Although most of these correspond to regulatory elements located in 5′-UTRs, such as riboswitches, 5 RNAs from the BSRD matched to sRNAs identified in this study. These include the high-abundant housekeeping RNA 6S51 (LLMGnc_004), the noncoding catalytic subunit of RNase P52 (LLMGnc_059) and the tmRNA or SsrA RNA53 (LLMGnc_074), which had not been annotated previously in L. lactis.
L. lactis antisense RNAs and functional classification
RNAs that overlap in an antisense fashion with transcripts (including their 3′- or 5′-UTRs) were annotated as antisense RNAs (asRNAs) (Table S4). In total, 60 of such asRNAs were identified in the RNA-seq dataset derived from the TEX-treated RNA sample. The asRNAs were classified as being located at 5′-, internal or 3′- positions relative to the gene on the opposite strand. Although many single or low-abundant antisense reads were specified throughout the genome, we only took those reads into consideration when a TSS was present immediately downstream of the conserved promoter motifs −10 (TATAAT) and/or −35 (TTGACA), allowing 2 mismatches. In comparison with the reads from sRNAs located in IGRs, antisense transcripts were generally less abundant. This may be explained by assuming that the perfect match between asRNAs and their target mRNAs is more stable and therefore makes them better substrates for degradation by RNases. In an E. coli study, the abundancy of functional asRNAs that form a duplex with other RNAs appears to be very low due to cleavage by RNase III.54 Further studies on the stability of the L. lactis asRNAs are needed to draw reliable conclusions on this matter. That asRNAs appear to be (relatively) more abundant in other organisms,19 might be species-specific and/or may have a technical origin in the different automated or manual annotation approaches used in the various studies. On the basis of what will follow, we propose to distinguish 3 functional classes of asRNAs; in addition to the “regulatory” asRNAs, which have a role comparable to that of trans-encoded sRNAs, asRNAs can have a “protective” or “meta-regulatory” function.
We identified a relatively large number of novel asRNAs and sRNAs in regions in the genome of L. lactis that carry (remnants of) pro-phages. More than a third of all asRNAs are specified in these areas, while 5.5% of the genome of L. lactis MG1363 are bacteriophage-derived sequences.38 These asRNAs target the 5′- or 3′-UTR or the coding parts of phage transcripts. The asRNAs may be native to the phage genomes but could also have evolved after integration of the phage in L. lactis MG1363, via mutations in AT-rich regions leading to novel promoters driving asRNA synthesis. Gene silencing by antisense transcription might suppress any harmful phage induction by targeting essential transcripts necessary for the phage to enter the lytic phase. Interestingly, all 6 (defective) pro-phages contain asRNAs against their respective integrase genes. L. lactis MG1363 is known to lack active pro-phages, although 2 of the 6 phage genomes appear to be complete.55 In addition to the 19 asRNAs, 28 sRNAs were detected in the IGRs in the genomes of these (defective) pro-phages. Although the function of these sRNAs is still unknown, they could operate such that they create a lysogen that serves as an optimal host for silent phage propagation throughout all cells in the culture. These asRNAs could serve a protective role.
Ten asRNAs were detected in loci coding for transcriptional regulators. In these cases, antisense transcription could function as a rapid off-switch under those conditions where the regulators are no longer required, and this would represent asRNAs with a meta-regulatory function.
Finally, 2 pairs of mRNAs (llmg_0538 and llmg_0539 (fabI), both involved in fatty acid biosynthesis, and llmg_0529 and llmg_0530 (gapA)) overlap in an antisense fashion at their 5′-UTRs, with respectively 31 and 37 complementary nucleotides. Transcription of these mRNA pairs might be influenced due to variable promoter strengths.56 Also, the consequence for translation of the mRNA-pairs is unclear. Even though the overlapping sequences do not include the RBS regions, secondary structures might affect RBS-accessibility upon interaction of the 2 transcripts of a pair. In addition, transcript stabilization by the base pairing may occur, as well as degradation after processing by e.g., RNaseIII. With only these 2 examples, overlapping 5′-UTRs are a relatively rare phenomenon in the transcriptome of L. lactis MG1363. Other types of overlapping transcripts have been identified in L. monocytogenes,57 where both 3′-UTR and 5′-UTR overlapping transcripts occur. Overlapping transcripts of operons have also been observed in S. aureus.18
Long 5′-UTRs and conserved riboswitches
The RNA-seq data set from the TEX-treated sample was also used to evaluate the 5′-untranslated regions in a genome-wide manner in order to identify putative cis-encoded intracellular sensors such as riboswitches, which can affect the expression of downstream gene(s). To detect 5′-UTRs carrying potential regulatory elements, those containing ≥100 nucleotides were examined, resulting in the identification of 129 leader sequences (LLMG_R001–129, Table S5). As mentioned above, most of the 36 regulatory RNA homologs in the BSRD database specified by the genome of L. lactis MG1363 are present among this selection of 129 leader sequences (Table S3). For example, several T-box sequences were identified (LLMG_R001/041/071/113/124) that use tRNA molecules to regulate the expression of aminoacyl-tRNA synthetase genes and genes involved in amino acid uptake and biosynthesis.58 Leaders were observed with putative riboswitches for flavin mononucleotide (FMN), fluoride, lysine, purine, thiamine pyrophosphate (TPP) and pre-queuosine 1 (preQ1). Four mRNAs with leaders from the pyrimindine biosynthesis pathway (LLMG_R046/054/055/081 or pyrR, carB, pyrK and pyrE) contain binding domains for the pyrimidine biosynthesis regulator protein PyrR. These leaders can form structures that result in anti-terminators in the absence of UMP-bound PyrR, after which transcription can proceed.59 When pyrimidines and UMPs are abundant in the cell, PyrR can form a stabilizing anti-antiterminator structure, preventing the RNA polymerase from further transcribing the downstream genes involved in pyrimidine biosynthesis.
Only a limited number of riboswitches have been reported to date and this figure seems an underestimate considering the enormous potential contained within riboswitch RNA-ligand interactions. Clearly, current bioinformatics and genetic approaches need to be adapted to uncover novel RNA sensing structures.30 The challenge is to be able to predict binding ligand-RNA interactions, which when successful would allow devising synthetic riboswitches for novel applications in e.g. medicine or biotechnology.29 Reporting experimentally validated leader sequences could serve as guideline for further experimental research in L. lactis.
RNA-seq reveals 134 new (s)ORFs in L. lactis MG1363
Small proteins of 50 amino acid residues or less are normally not automatically annotated in bacterial genomes while biochemical detection is challenging.60 The rapid increase in the amount of high-resolution transcriptome data and the use of novel techniques such as ribosomal profiling61 have led to the identification in bacterial genomes of ever more potentially novel genes for small proteins (≤50 amino acid residues). We scanned the leader sequences, internal promoters, sRNAs and asRNAs recognized here for the presence of small open reading frames (sORFs). To this end, the regions comprising the 250 nucleotides downstream of the TSS were analyzed using the following criteria: occurrence of a minimal RBS sequence (NNGGN5–14(A/T/G)TG), the presence of an ORF of ≥20 codons using any of the 3 START codons AUG, UUG, GUG and a STOP codon within a 250-nt distance from the TSS. Leaderless transcripts were also mined for ORFs ≥20 codons. A total of 134 novel ORFs were identified in this way, ranging in size from 21 to 61 codons (Table S6/S7). The putative gene products were then examined for the presence of conserved protein domains using InterProScan 5.62 None of the 134 deduced proteins, however, contained a known protein domain. This is most likely due to the small size of the proteins as well as to the fact that only a limited number of such small proteins have been characterized in other organisms. Notwithstanding this, some of the ORFs identified here might not be actually protein encoding.
A total of 23 in-sense promoters, internal to known ORFs, were recognized on the basis of transcript abundance relative to surrounding reads and the presence of a promoter upstream of the TSS. Fifteen of these lead to transcripts that are predicted to encode a shorter version of the full-length protein. Interestingly, in 4 cases the shorter transcript carries an ORF in another reading frame, which would lead to a new protein. The remaining transcripts contain an ORF but lack a minimal RBS (Table S7).
The transcriptome map was also inspected for differences with the currently available genome annotation of L. lactis MG1363,38 genbank accession number NC_009004 (Table S8). The translation start site of 17 published ORFs is corrected here, since their start codons were either up- or downstream of the TSS determined in this study, leading to a shorter or longer ORF. For three genes with unknown functions (llmg_0305, llmg_2022 and llmg_2202), the transcriptome map suggests that they are non-existing for the following reasons. Firstly, no sequence reads were observed for these genes. Secondly, their predicted translation products do not contain any conserved protein domain, and thirdly the TSS of the gene up- or downstream starts within the ORF itself (Fig 1A). Therefore, we propose to remove these locus tags from the genome of L. lactis MG1363. A corrected annotation file was created containing the information on the non-coding RNAs and antisense RNAs discussed above and used to update the genbank file with accession number NC_009004. The corrected genome has also been integrated in a JBrowse webpage (http://jbrowse.molgenrug.nl/).
Leaderless mRNAs are abundant in L. lactis and carry a distinct 5′-motif
To enable fast genome-wide TSS prediction, a dataset of a non-TEX-treated, strand-specific RNA-seq experiment was combined with that of the TEX-treated RNA-seq sample. This method, also referred to as differential RNA-seq (dRNA-seq), was applied in combination with the TSS prediction program TSSer.63 TSSer predicted 1819 TSSs, of which 884 are primary, 744 are orphan, 66 are internal and 125 are antisense (Table S9). A significantly higher number of orphan TSSs and antisense TSSs were found with TSSer than by manual mining. Most likely, the latter is more restrictive because a TSS together with the probable RNA species was only annotated when a (variation of a) promoter sequence was also present at a correct distance from 5′-enriched sequence reads. Also, the data set used for manual mining consisted only of reads derived from primary transcripts, excluding processed RNAs.
TSSs resulting in coding transcripts were used to assess the 5′-UTR length distribution in L. lactis (Fig. 1B). Leader sequences larger than 10 nt contain a Shine-Delgarno sequence with the consensus aaGGAg. The most remarkable observation from the length distribution of 5′-UTRs is the relatively high number of mRNAs in L. lactis that do not contain a leader sequence (13% of the transcripts derived from primary TSSs), although even much higher percentages of leaderless mRNAs (lmRNAs) have been reported in other microbes such as the desert bacterium Deinococcus deserti.64 The gene products of lmRNAs of Campylobacter jejuni have been implicated in stress-responses such as DNA repair.65 Genome2D [http://genome2D.molgenrug.nl] predicts the 111 L. lactis lmRNAs to be predominantly involved in the Gene Ontology (GO) classes “Nucleotide Metabolism” and “DNA/RNA binding” (Table S10). A motif search on these lmRNAs using MEME46 uncovered an AUGaaaa motif overlapping the AUG start codon (Fig. 1B). This A-rich sequence might play a role in binding of ribosomes to the lmRNA, in addition to the conserved AUG reported earlier to be necessary for ribosomal binding.66
L. lactis contains only one sigma factor, RpoD (σ,70 Llmg_0521). A motif search in the genome regions 50 nt upstream of all 1819 TSSs predicted by TSSer using MEME revealed the known extended tgnTAtAAT consensus for the −10 or Pribnow box while a MAST search67 with this promoter consensus pinpointed the standard L. lactis constitutive promoter sequences −35 (TTGACA) and −10 (TATAAT) (Fig. 1C). As is clear from the sequence in Fig. 1C, a number of AT-rich stretches can be observed in the promoter region. In contrast to the promoter consensus in the Gram-negative bacteria C. jejuni and H. pylori, a clear −35 box is present overlapping one of these AT-stretches.35,68
Recently, more insight in the complexity of bacterial operon structures is rapidly gained from genome-wide transcription studies employing DNA microarray technology and, lately, RNA-seq. Transcripts of operons can vary in length as a consequence of transcriptional read-through or the presence of secondary promoters, while internal promoters can lead to additional “suboperons.” We used Rockhopper69 on both RNA-seq datasets and identified 1288 monocistrons and 432 polycistronic operons containing 2 or more genes in L. lactis (Table S11 and S12). The average gene number for polycistronic operons was 2.7, identical to that established in Neisseria gonorrhoeae.70 The actual number of operons is expected to be significantly higher as a result of the variations mentioned above.
Experimental validation of 12 novel RNA candidates
Northern hybridization was performed on a selection of 12 of the newly identified RNAs to verify the RNA-seq results and to determine the lengths of these RNAs. The expression of 7 sRNAs from IGRs, 2 sRNAs from 3′-UTRs and 3 asRNAs was examined in different phases of growth of L. lactis in GM17 medium and under various stress conditions. All 12 RNAs are present under at least one of the conditions employed (Fig. 2).
Figure 2.

Experimental validation of novel RNAs. Detection of 9 sRNAs and 3 asRNAs by Northern hybridization in total RNA isolated from L. lactis MG1363 grown in GM17 under various conditions (Ex: exponential phase, St: stationary phase, pH: 10 min acid (pH 4.5) stress, Sa: 10 min salt (2.5% w/v extra NaCl) stress, St*: 10 min starvation in PBS). The positions of the sRNAs and asRNAs are indicated with asterisks (*). As a control, all blots were probed with an oligonucleotide targeting 5S RNA. Visualization of the relevant chromosomal locus and the expression levels of the genes (as derived from the TEX-treated RNA-seq dataset) are given below each blot, // signifies that not the entire gene is shown. Probes used to identify the various RNAs are given in Table S13.
Of the 7 sRNAs from IGRs, LLMGnc_064 (∼160 nt) and LLMGnc_138 are constitutively and most highly expressed. According to the Northern blot LLMGnc_138 comprises ∼75 nt, albeit that the LLMGnc_138 sequence reads and the presence of a terminator structure suggest that it has a size of ∼100 nt. Possibly this 100-nt RNA molecule is processed into its mature form of ∼75 nt. LLMGnc_064 contains an ORF starting 61 nt from its 5′-end that theoretically encodes a small protein of 24 amino acid residues. LLMGnc_010 (∼110 nt) is mainly expressed during exponential phase and at low pH. LLMGnc_072 (∼290 nt) is most abundant under a high salt condition, suggesting that it is involved in regulating processes related to osmolarity. Two sRNA candidates located in 3′-UTRs, LLMGnc_172 within argR and LLMGnc_177 within zitRS, are each represented with 2 major RNA species on the Northern blots. The upper band identified with the LLMGnc_172 probe represents the entire argR-LLMGnc_172 transcript of ∼575 nt as it also hybridized to a probe for the argR gene (data not shown), while the smallest represents LLMGnc_172 (∼65 nt). In the blot probed for LLMGnc_177 (∼68 nt), the upper band is likely zitRS, which is ∼1400 nt in length. The middle band of a transcript of around 130 nt may be derived from zitRS transcript processing while the lowest band corresponds in size to LLMGnc_177.
LLMGnc_087 (∼200 nt) is expressed solely during the high-salt or low-pH conditions employed here and could, thus, play a role in L. lactis MG1363 coping with sudden changes in osmolarity and pH. The possibility that this RNA encodes a small protein of 31 AA residues is currently under investigation. The protein does not contain a conserved domain and, thus, no function could be predicted.
Two distinct bands are also visible on the Northern blot when probing for the LLMGnc_094 transcript. The upper one corresponds to a size of ∼180 nt. Manual inspection of the RNA-seq reads of LLMGnc_094 shows that transcription termination and/or processing occurs after ∼72 nt for a small fraction of the RNA. This 72-nt-long molecule is more abundant during the exponential phase and might therefore function as a small regulatory RNA during this phase of growth. Probing for LLMGnc_184 revealed several bands, the upper one of which (∼200 nt) is constitutively expressed and seems to correspond with the sequence reads and a predicted terminator structure. The origin of the RNA in the double bands at around 65 nt is unknown but both seem to be more abundant during the stationary phase of growth and in starvation.
The putative as-ps118 is specified by the incomplete L. lactis MG1363 prophage MG-1.55 It starts 80 nt upstream of the stop codon of ps118, directing toward the 3′-end of ps118, a gene encoding a predicted transcriptional regulator. Based on the Northern analysis, as-ps118 is ∼70 nt. The RNA-seq data shows that the genes ps119 and ps118 surrounding as-ps118 are silent under the conditions examined here. It is therefore not possible to predict whether or not as-ps118 overlaps with the 3′-UTR of ps118, or even with the 5′-UTR of ps119, although this might be a very likely scenario.
The gene for the antisense RNA as-ps108 is also located on the defective prophage MG-1. The transcript overlaps the 5′-end of ps108. Since the ps108 gene is part of a putative large operon starting with ps115, as-ps108 might affect the stability of the entire transcript by an antisense mode of action. The RNA as-llmg_1727 is a long antisense RNA that is constitutively expressed under the various conditions tested here. It has an estimated size of ∼2 kb and could cover both the llmg_1726 and llmg_1727 transcripts encoding a galactose-1-phosphate uridylyltransferase and a putative ABC transporter permease, respectively.38
The expression of L. lactis 6S RNA is carbon source dependent
The widely conserved global regulator 6S (LLMGnc_004) is located in the L. lactis chromosome downstream of the mtl operon and upstream of the genome of prophage MG-1. A catabolite-responsive element (cre) sequence was predicted immediately upstream of the putative −35 box of the promoter of 6S (Fig. 3A and Table S2). This suggests that expression of the 6S RNA is under the control of the carbon catabolite repression protein CcpA. Transcriptome analysis using RNA-seq indeed showed that 6S is upregulated ∼3-fold after deletion of the ccpA gene (unpublished data). Earlier work in E. coli has shown that 4 transcriptional regulators (FIS, H-NS, LRP and StpA) can affect the expression of 6S.71 Northern hybridization using RNA from cells collected at 6 different points in time during growth on 3 alternative carbon sources revealed that L. lactis S6 is not only abundant during the stationary phase, as is the case in many other organisms, but also highly expressed in the exponential phase when galactose or cellobiose (but not fructose) is provided as the sole carbon source (Fig. 3C). This further confirms involvement of CcpA, since CcpA repression in L. lactis is relieved by galactose and cellobiose, but not by fructose.72 As observed in for example S. pneumoniae,73 2 bands of 6S could represent processed forms of the RNA since the predicted length of 6S is 202 nt (Fig. 3B), which is slightly longer than calculated from the Northern analysis (Fig. 3C). This might also explain why the secondary structure prediction of 6S does not contain a typical central region.74 The longest fragment changes under some of the conditions employed while the short form does not, except when the cells are growing with galactose.
Figure 3.

Analysis of the L. lactis non-coding 6S RNA. (A) Genomic region of the 6S (LLMGnc_004) gene of L. lactis MG1363. Open reading frames are depicted as grey arrows, 6S is shown in blue. Solid black arrows: promoters. The nucleotide sequence of the 6S promoter (P6S) is given in capitals, including a predicted cre-site (small letter type) upstream of the −35 box. (B) Structure of 6S RNA predicted by Mfold.83 (C) Detection of 6S RNA by Northern hybridization in samples of L. lactis grown in GM17 until the indicated ODs at 600 nm, or until OD600 = 0.6 in M17 with 1% of the indicated sugars. 5S RNA served as an RNA concentration control. In addition, one lane of the 8% polyacrylamide gel was stained after electrophoresis with ethidium bromide to visualize the relative amounts of 5S and 6S RNAs. O.N.: overnight culture, OD 2h 2.0: cells taken from a culture maintained at OD600 = 2.0 for 2 hours. Primers used for these experiments are given in Table S13. (D) Volcano plot showing the differentially (p-value <0.01 and ≥2 -fold) expressed genes upon overexpression of 6S RNA in comparison with the control, using a short (10-min) pulse of nisin addition to a culture of L. lactis SVDM2001 in GM17 and at an OD600 = 0.45. Indicated in yellow: differentially expressed genes, gray circles: measure of expression level.
Overexpression of 6S for 10 min affected only a limited number a transcripts, as determined by RNA-seq (Fig. 3D). Besides three tRNA species and an sRNA with unknown function, 2 genes were significantly affected ≥2 fold. The gene for peptide deformylase (llmg_0532) was downregulated and that of a hypothetical protein (llmg_1640) transcribed as part of an operon together with an ABC transporter gene (llmg_1639) was upregulated. The relatively short pulse of overexpression of 6S could explain these subtle changes in the L. lactis transcriptome. Also, S6 could play a role in fine-tuning of the CcpA regulon, acting when CcpA repression is relieved during stationary phase and/or growth on alternative carbon sources.
The sRNA LLMGnc_147 is involved in carbon uptake and metabolism
As a further demonstration of the validity of the reported sRNAs and to initiate the functional analysis of these putative regulator molecules in L. lactis, we characterized one of them in more detail. LLMGnc_147 is a 102-nt-long sRNA of which the gene is located between llmg_1960 and the transcriptional activator gene tenA (Fig. 4B). Its promoter carries a possible cre site overlapping the −35 box, suggesting that LLMGnc_147 is under control of CcpA (Fig. 4A) and related to carbon utilization. Northern analysis shows that LLMGnc_147 is highly expressed in cells growing on cellobiose (Fig. 4C). A transcriptional fusion of the promoter of LLMGnc_147 to gfp confirmed that it was most active in the presence of cellobiose and to a lesser extent with galactose (Fig. 4D).
Figure 4.
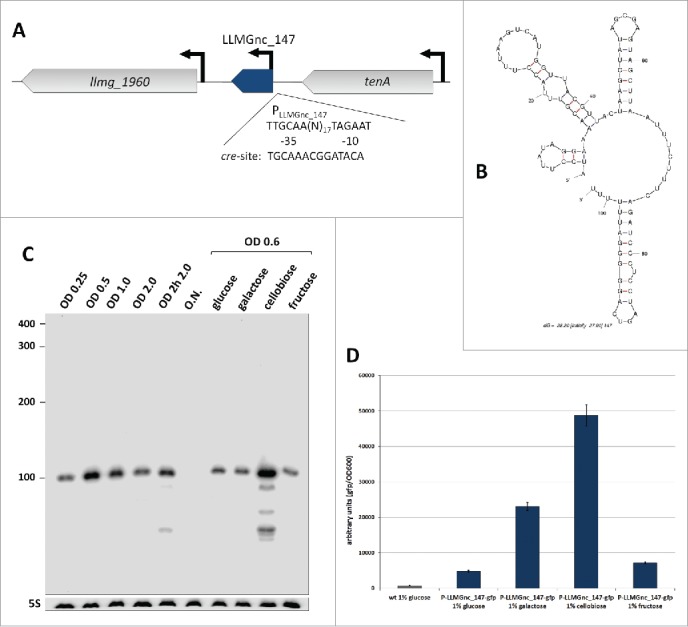
The sRNA LLMGnc_147 is involved in carbon metabolism. (A) Genomic region of LLMGnc_147. Open reading frames are depicted as gray arrows, LLMGnc_147 in shown in blue. Solid black arrows: promoters. The nucleotide sequence of the LLMGnc_147 promoter (PLLMGnc_147) is given, including a predicted cre-site that overlaps the −35 box. (B) Structure of LLMGnc_147 using Mfold.83 (C) Detection of LLMGnc_147 by Northern hybridization in samples of L. lactis grown in GM17 until the indicated ODs at 600 nm, or until OD600 = 0.6 in M17 with 1% of the indicated sugars. 5S RNA serves as a loading control. Labeled primers used for identification of the RNAs are given in Table S13. (D) PLLMGnc_147::gfp activity in L. lactis MG1363 (wt) and SVDM2003 cells grown in M17 containing 1% (w/v) of the indicated carbon source. Fluorescence and optical density were measured 5 hours after re-inoculation from an overnight culture growing in GM17. The experiment was repeated 3 times and error bars are indicated
To gain insight in potential mRNA targets, LLMGnc_147 was pulse-expressed for 10 min after which total RNA was isolated from the cells and subjected to RNA-seq. Clearly, one operon was highly upregulated (23 to 60-fold) as a consequence of the pulse of LLMGnc_147 RNA (Fig. 5A). The six genes of this operon specify the following predicted functions: a PTS transporter (llmg_0963), 2 β-glucosidases (llmg_0959/0960), a ribulose-phosphate 3-epimerase (llmg_0957), a ribose-5-phosphate isomerase B (llmg_0962) and an AraC transcriptional regulator (llmg_0962). Table 1 provides a complete list of differentially expressed genes upon pulse-expression of LLMGnc_147. Since a specific substrate has not yet been identified for this putative carbon utilization operon, we determined the effect of LLMGnc_147 overexpression on the ability of L. lactis to switch from glucose to another carbon source. Of the various sugars tested (Fig. 5C) only galactose seemed to have a beneficial effect on growth, without the lag-phase seen for the control strain, upon overexpression of LLMGnc_147 (Fig. 5B). As the operon llmg_0957-llmg_0963 specifies a putative ribulose epimerase and a ribose isomerase, a pentose sugar rather than galactose, a C-4 epimer of glucose, was expected to be its substrate. Possibly, galactose is imported via the PTS IIC component specified by llmg_0963. On the other hand, overexpression of LLMGnc_147 led to a slower growth phenotype on glucose, mannose, fructose and the di-saccharide cellobiose. Only the latter sugar induced the LLMGnc_147 (Fig. 4C/D). We assume that slower growth on these sugars after LLMGnc_147 overexpression is caused by a titration effect of the PTS IIC component on cytoplasmic components of specific phosphotransferase systems, as has been reported previously for CelB/PtcAB.75,76
Figure 5.

LLMGnc_147 is involved in the utilization of galactose. (A) Volcano plot of genes that are differentially expressed (p-value <0.001 and ≥2 -fold change) after pulse-expression of LLMGnc_147 via a 10-min addition of nisin to a culture at an OD600 = 0.45. For clarification of symbols, see the legend to Fig. 3. (B) Nisin-induced overexpression for 20 min of LLMGnc_147 (red triangles) in L. lactis SVDM2002 in comparison with the empty vector control (blue squares), after which the strain was re-inoculated 1:20 in fresh M17 medium containing 1% galactose. (C) Identical experimental set-up as described in (B) for growth after re-inoculation in M17 with 1% (w/v) of the indicated carbon sources. The experiments were repeated twice and the lines represent averages of 4 microtiter plate measurements.
Table 1.
Differentially expressed genes upon pulse-expression of LLMGnc_147.
| GeneID | logFC | logCPM | LR | pvalue | adj_pvalue | Fold | minFDR |
|---|---|---|---|---|---|---|---|
| LLMGnc_147 | 7.72 | 8.18 | 440 | 1.20E-97 | 3.30E-94 | 211 | 310.55 |
| llmg_0960 | 6.07 | 3.66 | 151 | 9.10E-35 | 8.60E-32 | 67.1 | 103.2 |
| llmg_0959 | 5.87 | 3.56 | 111 | 7.20E-26 | 4.10E-23 | 58.5 | 74.38 |
| llmg_0957 | 5.8 | 3.61 | 193 | 7.20E-44 | 1.00E-40 | 55.6 | 132.84 |
| llmg_0958 | 5.42 | 3.97 | 123 | 1.40E-28 | 9.80E-26 | 42.8 | 83.07 |
| llmg_0962 | 4.69 | 0.89 | 30.3 | 3.70E-08 | 1.30E-05 | 25.9 | 16.21 |
| llmg_0963 | 4.54 | −0.26 | 17.5 | 2.90E-05 | 4.30E-03 | 23.3 | 7.85 |
| llmg_2339 | 1.64 | 3.48 | 25.8 | 3.80E-07 | 1.10E-04 | 3.1 | 13.17 |
| llmg_2150 | 1.29 | 3.88 | 17.1 | 3.60E-05 | 5.10E-03 | 2.4 | 7.61 |
| llmg_2432 | 1.22 | 6.38 | 18.4 | 1.80E-05 | 3.10E-03 | 2.3 | 8.33 |
| llmg_0629 | 1.1 | 6.31 | 13.2 | 2.80E-04 | 2.90E-02 | 2.1 | 5.09 |
| llmg_0294 | −1.14 | 8.29 | 13.3 | 2.60E-04 | 2.90E-02 | −2.2 | 5.11 |
| llmg_0253 | −1.15 | 8.67 | 16.8 | 4.10E-05 | 5.50E-03 | −2.2 | 7.5 |
| llmg_1424 | −1.21 | 9 | 17.6 | 2.80E-05 | 4.30E-03 | −2.3 | 7.85 |
| llmg_1775 | −1.3 | 4.61 | 19.7 | 9.20E-06 | 1.70E-03 | −2.5 | 9.17 |
| LLMGnc_103 | −1.31 | 6.01 | 13.8 | 2.00E-04 | 2.30E-02 | −2.5 | 5.41 |
| llmg_0931 | −1.42 | 3.92 | 24.2 | 8.60E-07 | 2.20E-04 | −2.7 | 12.14 |
| llmg_tRNA_33 | −1.48 | 11.2 | 20.7 | 5.40E-06 | 1.20E-03 | −2.8 | 9.71 |
| as-llmg_1269 | −1.48 | 3.69 | 17.7 | 2.60E-05 | 4.30E-03 | −2.8 | 7.86 |
| llmg_0091 | −1.68 | 2.13 | 13.6 | 2.30E-04 | 2.60E-02 | −3.2 | 5.28 |
| llmg_1776 | −1.99 | 3.93 | 42.7 | 6.30E-11 | 3.00E-08 | −4 | 24.99 |
| llmg_1570 | −3.2 | 3.88 | 12.3 | 4.60E-04 | 4.30E-02 | −9.2 | 4.53 |
| LLMGnc_179 | −6.2 | −0.09 | 21.8 | 3.00E-06 | 7.20E-04 | −73.3 | 10.44 |
| LLMGnc_128 | −7.77 | −1.96 | 12.6 | 3.80E-04 | 3.70E-02 | −218.6 | 4.76 |
| llmg_0797 | −8.6 | −2.27 | 14.2 | 1.70E-04 | 2.10E-02 | −387.6 | 5.6 |
| LLMGnc_121 | −8.72 | −2.1 | 15.5 | 8.20E-05 | 1.10E-02 | −423 | 6.56 |
| llmg_0981 | −9.24 | −1.45 | 20.3 | 6.60E-06 | 1.30E-03 | −605.8 | 9.54 |
| llmg_0132 | −10.3 | −1.05 | 28.1 | 1.10E-07 | 3.60E-05 | −1225 | 14.78 |
| LLMGnc_137 | −10.4 | −0.9 | 30.7 | 3.00E-08 | 1.20E-05 | −1329 | 16.31 |
Altogether, we show that the expression of LLMGnc_147 is controlled by galactose and cellobiose, the latter potentially via a cellobiose-specific transcriptional activator such as the AraC transcriptional regulator (Llmg_0962) from the operon controlled by LLMGnc_147. This might be a remnant of hemicellulose utilization, as L. lactis has a plant origin.38 CcpA might be negatively involved in the regulation of LLMGnc_147 and llmg_0957-llmg_0963, since a cre site is located in their promoter regions. We hypothesize that LLMGnc_147 is necessary for the llmg_0957-llmg_0963 operon in order to stabilize its long transcript.
Conclusions
Differential RNA sequencing uncovered hundreds of novel RNAs in the genome of the lactic acid bacterium L. lactis, for which no sRNAs have been described so far. These may shed light on the interpretation of specific (point) mutations obtained e.g., after evolutionary studies and genome re-sequencing. We have confirmed the expression of 14 of the RNAs by Northern hybridization, and show that the abundant non-coding RNA 6S is expressed in dependency of the available carbon source. Functional analysis on sRNA LLMGnc_147 shows that it is involved in carbon uptake and metabolism. The output of this study provides an excellent basis for further investigations on the molecular biology of L. lactis and a starting point for the characterization of the putative regulatory RNAs in this organism.
Materials and methods
Bacterial strains, growth conditions, cloning and nisin induction
For an overview of strains and plasmids used in this study, see Table 2. L. lactis was routinely grown as standing cultures at 30°C in M17 broth (Difco, Becton Dickinson, Le Pont de Claix, France) containing 0,5% (w/v) glucose (GM17). The restriction and ligation independent USER fusion cloning strategy77 was employed for vector constructions. In short, vector backbone and insert fragments were amplified with PfuX7 polymerase (see Table S13 for oligonucleotides used in this study). PCR fragments were then purified using a PCR clean-up kit (Macherey-Nagel GmbH, Germany) and inspected for correct size by agarose gel electrophoresis. PCR backbone and sRNA gene or promoter inserts were mixed at a 1:3 molar ratio, treated with the USER enzyme mix (New England Biolabs, Hitchin, UK) and directly introduced into competent cells of L. lactis NZ9000 by electroporation (at 2.5 kV, 25 uF, 200 Ohm) or by heat-shock (45 sec at 42°C) into chemically competent E. coli Dh5α cells. Colony PCR and subsequent sequencing (Macrogen, Amsterdam, The Netherlands) was used to verify construct correctness.
Table 2.
Strains and plasmids used in this study.
| Strain or plasmid | Relevant phenotype/genotype | Source |
|---|---|---|
| Strains | ||
| L. lactis NCDO712 | L. lactis subsp. cremoris | Gasson et al., 1983 |
| L. lactis MG1363 | Plasmid-free derivative of NCDO712 | Gasson et al., 1983 |
| L. lactis NZ9000 | MG1363 pepN::nisRK | Kuipers et al., 1998 |
| SVDM2001 | Cmr, NZ9000 with 6S gene in pNZ8048 | This work |
| SVDM2002 | Cmr, NZ9000 with LLMGnc_147 gene in pNZ8048 | This work |
| SVDM2003 | Emr, MG1363 with pSVDM5003 integrated in pseudo_10 locus | This work |
| Plasmids | ||
| pNZ8048 | Cmr, nisin-inducible expression vector | de Ruyter et al., 1996 |
| pSVDM5001 | Cmr, pNZ8048 with 6S gene downstream of PnisA | This work |
| pSVDM5002 | Cmr, pNZ8048 with LLMGnc_147 gene downstream of PnisA | This work |
| pSEUDO-GFP | Emr, pCS1966 derivative, genomic integration plasmid | Pinto et al., 2011 |
| pSVDM5003 | Emr, pSEUDO-GFP in which PLLMGnc_147 drives GFP expression | This work |
Cmr, chloramphenicol resistance marker
Emr, erythromycin resistance marker.
The promoter of LLMGnc_147, PLLMGnc_147, was PCR-amplified and cloned upstream of the GFP gene in plasmid pSEUDO_10, which was subsequently integrated in the chromosome of L. lactis MG1363 via single cross-over recombination.78 To stably maintain the integrated plasmid, cells were selected and continuously grown in the presence of 3 µg/ml erythromycin.
L. lactis NZ9000 was used for overexpression of sRNAs from the high-copy number plasmid pNZ8048. A single colony from a GM17 agar plate containing 5 µg/ml chloramphenicol was used to inoculate 10 ml of fresh GM17 medium. After overnight growth, the culture was diluted 1:100 and incubated until an OD600 between 0.4–0.5 was reached. Subsequently, the cells were induced for 10 min with 7.5 ng/ml nisin (Sigma-Aldrich, Munich, Germany), and harvested as described above.
Optical density and fluorescence were measured in a Tecan F200 (Tecan Group, Männedorf, Switzerland).
RNA isolation
For dRNA-seq, single colonies of Lactococcus lactis MG1363 or L. lactis NCDO712, grown on GM17 (1.5%)agar plates, were used to inoculate 10 ml fresh GM17 media for overnight growth at 30°C. The overnight cultures were each diluted 1:100 in 500 ml GM17. L. lactis MG1363 was sampled at 3 points in time in the exponential phase (OD600 of 0.9, 1.3 and 1.7) and at 3 time points in the stationary phase (at 30, 60 and 90 min after an OD600 of >2.5 was reached). To compensate for cell density, equivalent OD units were harvested by centrifugation at 10.000 rpm for 1 min; the cell pellets were immediately frozen in liquid nitrogen. Cells from L. lactis NCDO712 were harvested at the mid-exponential growth phase (OD600 of 1.0).
For RNA isolation, cell pellets were re-suspended in 400 μl TE-buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.4), after which 50 μl 10% Sodium dodecyl sulfate (SDS), 500 μl phenol/chloroform and 0.5 g glass beads (75–150 μm in diameter) were added. The mixture was cooled on ice and the cells were subsequently disrupted by 2 consecutive rounds of shaking for 45 sec in a Mini-BeadBeater (Biospec Products, Bartlesville, OK, USA) at 4°C, with intervening cooling on ice. After centrifugation (14.000 rpm for 10 min), the supernatants were treated with 500 μl chloroform, centrifuged as above, and the water phase was collected. Total RNA from the water phase was incubated for 30 min at 37°C with RNase-free DNase I supplemented with RiboLock RNase inhibitor (Fermentas/Thermo Scientific, Vilnius, Lithuania). The RNA was subsequently purified using standard phenol/chloroform extraction followed by sodium acetate/ethanol precipitation. RNA pellets were dissolved in TE-buffer. All solutions were treated with DEPC and autoclaved.
RNA treatment, library preparation and RNA deep sequencing
RNA concentrations were measured using a Nanodrop ND-1000 (Thermo Fischer Scientific, Rockford, IL, USA), after which the integrity of the 16S/23S rRNA and DNA contamination were assessed using an Agilent 2100 Bioanalyser (Agilent Technologies, Waldbronn, Germany). The RNA sample from L. lactis MG1363 was TEX-treated at Eurofins MWG GmbH (Ebersberg, Germany). rRNA depletion was done with the Terminator System kit (Epicentre, Madison, WI, USA), preparation of a 5′-fragment cDNA library was performed as previously described.35 After PCR amplification and library purification, the library was sequenced on an Illumina HiSeq2000 v3 platform (Illumina, San Diego, CA, USA), with a paired-end protocol and read length of 101 nt, resulting in a total output of 10.5 million (M) reads. The RNA sample for the non-TEX treated library (RNA sample from L. lactis NCDO712) was sequenced at Otogenetics Corporation (Norcross, GA, USA) on an Illumina HiSeq2000, with a ScriptSeq™ Complete Kit (Bacteria) (Epicentre, Madison, WI, USA) including Ribo-Zero™ rRNA removal and ScriptSeq v2 library preparation for directional RNA-Seq, resulting in a total of 15.7 M reads. RNA samples from the pulse-expression of LLMGnc_147, after Ribo-Zero™ rRNA removal and library preparation using the AmpliSeq™ kit (ThermoFischer Scientific), were sequenced at the PrimBio Research Institute (Exton, PA, USA) on a Ion Proton sequencer. This resulted in 13–23 M reads per sample.
TSS calling and data analysis
RNA-seq data of TEX-treated and untreated samples was used for automated TSS calling by TSSer,63 using default parameters. Predicted TSSs were used to perform a MEME search to identify promoter motifs and Shine-Delgarno sequences in the regions −50 to −1 upstream of all TSSs using a zero or one occurrence model. From the 111 leaderless mRNAs predicted by TSSer, a MEME motif search was performed in the region +1 to +15 downstream of the TSS, based on a one occurrence per sequence model, using an E-value threshold of 0.001. Operons in L. lactis were predicted by Rockhopper,69 using default settings.
For manual qualitative mining, raw data reads of 101 nt of the TEX-treated sample were quality trimmed with a PHRED score >28 and subsequently aligned to the genome of L. lactis MG136338 using Bowtie2.79 The resulting reads were visualized with Genome2D80 displaying known ORFs and putative regulatory RNA elements predicted in silico for L. lactis MG1363 using SIPHT.45 This data was manually inspected for novel RNA elements and re-annotation purposes. To prevent false positive calling of transcription start sites (TSS), especially with respect to lowly expressed RNAs, TSSs were inspected manually for promoter motifs (−10 and −35 boxes), after which transcription start sites were extracted using Tablet.81 Promoters of sRNAs and asRNAs were assessed for the presence of transcription factor binding sites (TFBS) by performing TFBS searches on http://genome2d.molgenrug.nl/, using −100 to −1 upstream of the TSS.
Comparative transcriptome data was quality checked as above and trimmed with a cut-off >30 nt, after which analyses were performed using the Transcriptome analysis webserver for RNA-seq expression data (T-REx).82
Northern hybridization
Total RNA (10 μg) was separated on an 8% denaturing polyacrylamide-(7 M)urea gel in Tris-acetate-EDTA buffer (TAE). RNAs were transferred to positively charged Zeta-Probe nylon membranes (Bio-Rad Laboratories BV, Veenendaal, The Netherlands) using a semi-dry electroblotting apparatus (Bio-Rad Laboratories BV). RNAs were covalently cross-linked to the membranes at 1200 mJ using a UVC-508 Ultraviolet Crosslinker (Ultra-Lum Inc.., Carson, CA, USA), after which the blots were hybridized overnight at 42°C in PerfectHyb Plus Hybridization buffer (Sigma-Aldrich Chemie Gmbh, Munich, Germany), using appropriate 32P-labeled DNA oligonucleotides (See Table S13). DNA probes were labeled with 32P-γATP using Polynucleotide kinase (Fermentas/Thermo Scientific), according to the manufacturer. Nylon membranes were washed twice in 2x saline sodium citrate (SSC) buffer with 0.1% SDS, exposed to a Phosphor Screen and imaged using a Cyclone Plus Phosphor Imager and OptiQuant software (PerkinElmer, Groningen, NL).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We kindly thank Akos T. Kovacs and Herwig Bachmann for helpful discussions and Mikkel Jørgensen for his help with the Northern analyses. We also would like to thank Ana Solopova for her insights in L. lactis carbon metabolism
References
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell 2009; 136:615-28; http://dx.doi.org/ 10.1016/j.cell.2009.01.043; PMID:1923988420220305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caron MP, Lafontaine DA, Masse E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol 2010; 7:140-4; PMID:20220305; http://dx.doi.org/ 10.4161/rna.7.2.11056 [DOI] [PubMed] [Google Scholar]
- 3.Desnoyers G, Bouchard M, Massé E. New insights into small RNA-dependent translational regulation in prokaryotes. TRENDS Genet 2013; 29:92-8; PMID:23141721; http://dx.doi.org/20980440 10.1016/j.tig.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 4.Gottesman S, Storz G. Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol 2011; 3:pii: a003798; PMID:20980440; http://dx.doi.org/ 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, Johansson J. RNAs: Regulators of bacterial virulence. Nat Rev Microbiol 2010; 8:857-66; PMID:21079634; http://dx.doi.org/7691599 10.1038/nrmicro2457 [DOI] [PubMed] [Google Scholar]
- 6.Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell host & microbe 2010; 8:116-27; PMID:20638647; http://dx.doi.org/7691599 10.1016/j.chom.2010.06.008 [DOI] [PubMed] [Google Scholar]
- 7.Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 1993; 12:3967-75; PMID:7691599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J 1995; 14:4569-77; PMID:7556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 2007; 104:20454-9; PMID:18042713; http://dx.doi.org/ 10.1073/pnas.0708102104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanderpool CK, Balasubramanian D, Lloyd CR. Dual-function RNA regulators in bacteria. Biochimie 2011; 93:1943-9; PMID:21816203; http://dx.doi.org/22123973 10.1016/j.biochi.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol 2007; 10:125-33; PMID:17395525; http://dx.doi.org/22123973 10.1016/j.mib.2007.03.015 [DOI] [PubMed] [Google Scholar]
- 12.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nature Reviews Microbiology 2011; 9:578-89; PMID:21760622; http://dx.doi.org/22123973 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aiba H. Mechanism of RNA silencing by hfq-binding small RNAs. Curr Opin Microbiol 2007; 10:134-9; PMID:17383928; http://dx.doi.org/22123973 10.1016/j.mib.2007.03.010 [DOI] [PubMed] [Google Scholar]
- 14.Jousselin A, Metzinger L, Felden B. On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol 2009; 17:399-405; PMID:19733080; http://dx.doi.org/22123973 10.1016/j.tim.2009.06.003 [DOI] [PubMed] [Google Scholar]
- 15.Dambach M, Irnov I, Winkler WC. Association of RNAs with Bacillus subtilis Hfq. PloS one 2013; 8:e55156; PMID:23457461; http://dx.doi.org/22123973 10.1371/journal.pone.0055156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS genetics 2008; 4:e1000163; PMID:18725932; http://dx.doi.org/22123973 10.1371/journal.pgen.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of hfq‐bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 2012; 31:4005-19; PMID:22922465; http://dx.doi.org/22123973 10.1038/emboj.2012.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penades JR, Valle J, et al.. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A 2011; 108:20172-7; PMID:22123973; http://dx.doi.org/ 10.1073/pnas.1113521108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georg J, Hess WR. Cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 2011; 75:286-300; PMID:21646430; http://dx.doi.org/ 10.1128/MMBR.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomason MK, Bischler T, Eisenbart SK, Forstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 2015; 197:18-28; PMID:25266388; http://dx.doi.org/ 10.1128/JB.02096-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottesman S. Micros for microbes: Non-coding regulatory RNAs in bacteria. TRENDS in Genetics 2005; 21:399-404; PMID:15913835; http://dx.doi.org/21325130 10.1016/j.tig.2005.05.008 [DOI] [PubMed] [Google Scholar]
- 22.Prévost K, Salvail H, Desnoyers G, Jacques J, Phaneuf É, Massé E. The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol Microbiol 2007; 64:1260-73; PMID:17542919; http://dx.doi.org/21325130 10.1111/j.1365-2958.2007.05733.x [DOI] [PubMed] [Google Scholar]
- 23.Morita T, Aiba H. RNase E action at a distance: Degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev 2011; 25:294-8; PMID:21325130; http://dx.doi.org/ 10.1101/gad.2030311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandyra KJ, Said N, Pfeiffer V, Górna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 2012; 47:943-53; PMID:22902561; http://dx.doi.org/21289064 10.1016/j.molcel.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prevost K, Desnoyers G, Jacques JF, Lavoie F, Masse E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 2011; 25:385-96; PMID:21289064; http://dx.doi.org/ 10.1101/gad.2001711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fröhlich KS, Papenfort K, Fekete A, Vogel J. A small RNA activates CFA synthase by isoform‐specific mRNA stabilization. EMBO J 2013; 32:2963-79; PMID:24141880; http://dx.doi.org/21665928 10.1038/emboj.2013.222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 2013; 153:426-37; PMID:23582330; http://dx.doi.org/21665928 10.1016/j.cell.2013.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narberhaus F, Waldminghaus T, Chowdhury S. RNA thermometers. FEMS Microbiol Rev 2006; 30:3-16; PMID:16438677; http://dx.doi.org/21665928 10.1111/j.1574-6976.2005.004.x [DOI] [PubMed] [Google Scholar]
- 29.Serganov A, Nudler E. A decade of riboswitches. Cell 2013; 152:17-24; PMID:23332744; http://dx.doi.org/21665928 10.1016/j.cell.2012.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell 2011; 43:867-79; PMID:21925376; http://dx.doi.org/21665928 10.1016/j.molcel.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 2009; 139:770-9; PMID:19914169; http://dx.doi.org/21665928 10.1016/j.cell.2009.08.046 [DOI] [PubMed] [Google Scholar]
- 32.de los Mozos, Igor Ruiz, Vergara-Irigaray M, Segura V, Villanueva M, Bitarte N, Saramago M, Domingues S, Arraiano CM, Fechter P, et al.. Base pairing interaction between 5′-and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS genetics 2013; 9:e1004001; PMID:24367275; http://dx.doi.org/21665928 10.1371/journal.pgen.1004001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res 2011; 21:1487-97; PMID:21665928; http://dx.doi.org/ 10.1101/gr.119370.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, et al.. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 2012; 335:1103-6; PMID:22383849; http://dx.doi.org/ 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- 35.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiß S, Sittka A, Chabas S, Reiche K, Hackermüller J, Reinhardt R. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010; 464:250-5; PMID:20164839; http://dx.doi.org/17307855 10.1038/nature08756 [DOI] [PubMed] [Google Scholar]
- 36.Gómez‐Lozano M, Marvig RL, Molin S, Long KS. Genome‐wide identification of novel small RNAs in Pseudomonas aeruginosa. Environ Microbiol 2012; 14:2006-16; PMID:22533370; http://dx.doi.org/17307855 10.1111/j.1462-2920.2012.02759.x [DOI] [PubMed] [Google Scholar]
- 37.Hobbs EC, Fontaine F, Yin X, Storz G. An expanding universe of small proteins. Curr Opin Microbiol 2011; 14:167-73; PMID:21342783; http://dx.doi.org/17307855 10.1016/j.mib.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, et al.. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 2007; 189:3256-70; PMID:17307855; http://dx.doi.org/ 10.1128/JB.01768-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders JW, Venema G, Kok J. Environmental stress responses in Lactococcus lactis. FEMS Microbiol Rev 1999; 23:483-501; PMID:7665513; http://dx.doi.org/17028270 10.1111/j.1574-6976.1999.tb00409.x [DOI] [Google Scholar]
- 40.Smith WM, Dykes GA, Soomro AH, Turner MS. Molecular mechanisms of stress resistance in Lactococcus lactis. Current research, technology and education topics in applied microbiology and microbial biotechnology 2010; 2:1106-18. [Google Scholar]
- 41.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 2007; 189:1366-81; PMID:17028270; http://dx.doi.org/ 10.1128/JB.01013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.den Hengst CD, van Hijum SA, Geurts JM, Nauta A, Kok J, Kuipers OP. The Lactococcus lactis CodY regulon: Identification of a conserved cis-regulatory element. J Biol Chem 2005; 280:34332-42; PMID:16040604; http://dx.doi.org/ 10.1074/jbc.M502349200 [DOI] [PubMed] [Google Scholar]
- 43.van Hijum SA, Medema MH, Kuipers OP. Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiol Mol Biol Rev 2009; 73:481,509; PMID:19721087; http://dx.doi.org/ 10.1128/MMBR.00037-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Jong A, Hansen ME, Kuipers OP, Kilstrup M, Kok J. The transcriptional and gene regulatory network of Lactococcus lactis MG1363 during growth in milk. PloS one 2013; 8:e53085; PMID:23349698; http://dx.doi.org/19458158 10.1371/journal.pone.0053085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livny J, Teonadi H, Livny M, Waldor MK. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PloS one 2008; 3:e3197; PMID:18787707; http://dx.doi.org/19458158 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 2009; 37:W202-8; PMID:19458158; http://dx.doi.org/ 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Larsen R, van Hijum SA, Martinussen J, Kuipers OP, Kok J. Transcriptome analysis of the Lactococcus lactis ArgR and AhrC regulons. Appl Environ Microbiol 2008; 74:4768-71; PMID:18539789; http://dx.doi.org/ 10.1128/AEM.00117-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott C, Rawsthorne H, Upadhyay M, Shearman CA, Gasson MJ, Guest JR, Green J. Zinc uptake, oxidative stress and the FNR‐like proteins of Lactococcus lactis. FEMS Microbiol Lett 2000; 192:85-9; PMID:11040433; http://dx.doi.org/23203879 10.1111/j.1574-6968.2000.tb09363.x [DOI] [PubMed] [Google Scholar]
- 49.Mandin P, Guillier M. Expanding control in bacteria: Interplay between small RNAs and transcriptional regulators to control gene expression. Curr Opin Microbiol 2013; 16:125-32; PMID:23415757; http://dx.doi.org/23203879 10.1016/j.mib.2012.12.005 [DOI] [PubMed] [Google Scholar]
- 50.Li L, Huang D, Cheung MK, Nong W, Huang Q, Kwan HS. BSRD: A repository for bacterial small regulatory RNA. Nucleic Acids Res 2013; 41:D233-8; PMID:23203879; http://dx.doi.org/ 10.1093/nar/gks1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell 2000; 101:613-23; PMID:10892648; http://dx.doi.org/24453212 10.1016/S0092-8674(00)80873-9 [DOI] [PubMed] [Google Scholar]
- 52.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983; 35:849-57; PMID:6197186; http://dx.doi.org/24453212 10.1016/0092-8674(83)90117-4 [DOI] [PubMed] [Google Scholar]
- 53.Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA‐binding protein essential for the peptide‐tagging activity of SsrA (tmRNA). EMBO J 1999; 18:3793-9; PMID:10393194; http://dx.doi.org/24453212 10.1093/emboj/18.13.3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lybecker M, Zimmermann B, Bilusic I, Tukhtubaeva N, Schroeder R. The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci U S A 2014; 111:3134-9; PMID:24453212; http://dx.doi.org/ 10.1073/pnas.1315974111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ventura M, Zomer A, Canchaya C, O'Connell-Motherway M, Kuipers O, Turroni F, Ribbera A, Foroni E, Buist G, Wegmann U, et al.. Comparative analyses of prophage-like elements present in two Lactococcus lactis strains. Appl Environ Microbiol 2007; 73:7771-80; PMID:17933937; http://dx.doi.org/ 10.1128/AEM.01273-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bendtsen KM, Erdossy J, Csiszovszki Z, Svenningsen SL, Sneppen K, Krishna S, Semsey S. Direct and indirect effects in the regulation of overlapping promoters. Nucleic Acids Res 2011; 39:6879-85; PMID:21609952; http://dx.doi.org/ 10.1093/nar/gkr390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K. The listeria transcriptional landscape from saprophytism to virulence. Nature 2009; 459:950-6; PMID:19448609; http://dx.doi.org/19213877 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- 58.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 2010; 584:318-24; PMID:19932103; http://dx.doi.org/19213877 10.1016/j.febslet.2009.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kilstrup M, Hammer K, Ruhdal Jensen P, Martinussen J. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol Rev 2005; 29:555-90; PMID:15935511; http://dx.doi.org/19213877 10.1016/j.fmrre.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 60.Storz G, Wolf YI, Ramamurthi KS. Small proteins can no longer be ignored. Annu Rev Biochem 2014; 83:753-77; PMID:24606146; http://dx.doi.org/19213877 10.1146/annurev-biochem-070611-102400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009; 324:218-23; PMID:19213877; http://dx.doi.org/ 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al.. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014; 30:1236-40; PMID:24451626; http://dx.doi.org/ 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jorjani H, Zavolan M. TSSer: An automated method to identify transcription start sites in prokaryotic genomes from differential RNA sequencing data. Bioinformatics 2014; 30:971-4; PMID:24371151; http://dx.doi.org/ 10.1093/bioinformatics/btt752 [DOI] [PubMed] [Google Scholar]
- 64.de Groot A, Roche D, Fernandez B, Ludanyi M, Cruveiller S, Pignol D, Vallenet D, Armengaud J, Blanchard L. RNA sequencing and proteogenomics reveal the importance of leaderless mRNAs in the radiation-tolerant bacterium Deinococcus deserti. Genome Biol Evol 2014; 6:932-48; PMID:24723731; http://dx.doi.org/ 10.1093/gbe/evu069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Porcelli I, Reuter M, Pearson BM, Wilhelm T, Van Vliet AH. Parallel evolution of genome structure and transcriptional landscape in the epsilonproteobacteria. BMC Genomics 2013; 14:616; PMID:24028687; http://dx.doi.org/18755843 10.1186/1471-2164-14-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brock JE, Pourshahian S, Giliberti J, Limbach PA, Janssen GR. Ribosomes bind leaderless mRNA in escherichia coli through recognition of their 5′-terminal AUG. RNA 2008; 14:2159-69; PMID:18755843; http://dx.doi.org/ 10.1261/rna.1089208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey TL, Gribskov M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 1998; 14:48-54; PMID:9520501; http://dx.doi.org/ 10.1093/bioinformatics/14.1.48 [DOI] [PubMed] [Google Scholar]
- 68.Petersen L, Larsen TS, Ussery DW, On SL, Krogh A. rpo D promoters in Campylobacter jejuni exhibit a strong periodic signal instead of a− 35 box. J Mol Biol 2003; 326:1361-72; PMID:12595250; http://dx.doi.org/23716638 10.1016/S0022-2836(03)00034-2 [DOI] [PubMed] [Google Scholar]
- 69.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-seq data. Nucleic Acids Res 2013; 41:e140; PMID:23716638; http://dx.doi.org/ 10.1093/nar/gkt444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Remmele CW, Xian Y, Albrecht M, Faulstich M, Fraunholz M, Heinrichs E, Dittrich MT, Muller T, Reinhardt R, Rudel T. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res 2014; 42:10579-95; PMID:25143534; http://dx.doi.org/ 10.1093/nar/gku762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neusser T, Gildehaus N, Wurm R, Wagner R. Studies on the expression of 6S RNA from E. coli: Involvement of regulators important for stress and growth adaptation. Biol Chem 2008; 389:285-97; PMID:18177266; http://dx.doi.org/17028270 10.1515/BC.2008.023 [DOI] [PubMed] [Google Scholar]
- 72.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 2007; 189:1366-81; PMID:17028270; http://dx.doi.org/ 10.1128/JB.01013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Acebo P, Martin-Galiano AJ, Navarro S, Zaballos A, Amblar M. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA 2012; 18:530-46; PMID:22274957; http://dx.doi.org/ 10.1261/rna.027359.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cavanagh AT, Wassarman KM. 6S RNA, a global regulator of transcription in Escherichia coli, Bacillus subtilis, and beyond. Annu Rev Microbiol 2014; 68:45-60; PMID:24742053; http://dx.doi.org/22660716 10.1146/annurev-micro-092611-150135 [DOI] [PubMed] [Google Scholar]
- 75.Aleksandrzak-Piekarczyk T, Polak J, Jezierska B, Renault P, Bardowski J. Genetic characterization of the CcpA-dependent, cellobiose-specific PTS system comprising CelB, PtcB and PtcA that transports lactose in Lactococcus lactis IL1403. Int J Food Microbiol 2011; 145:186-94; PMID:21262549; http://dx.doi.org/22660716 10.1016/j.ijfoodmicro.2010.12.011 [DOI] [PubMed] [Google Scholar]
- 76.Solopova A, Bachmann H, Teusink B, Kok J, Neves AR, Kuipers OP. A specific mutation in the promoter region of the silent cel cluster accounts for the appearance of lactose-utilizing Lactococcus lactis MG1363. Appl Environ Microbiol 2012; 78:5612-21; PMID:22660716; http://dx.doi.org/ 10.1128/AEM.00455-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA. USER fusion: A rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res 2007; 35:e55; PMID:17389646; http://dx.doi.org/ 10.1093/nar/gkm106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pinto JP, Zeyniyev A, Karsens H, Trip H, Lolkema JS, Kuipers OP, Kok J. pSEUDO, a genetic integration standard for Lactococcus lactis. Appl Environ Microbiol 2011; 77:6687-90; PMID:21764949; http://dx.doi.org/ 10.1128/AEM.05196-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nature methods 2012; 9:357-9; PMID:22388286; http://dx.doi.org/15128451 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baerends RJ, Smits WK, de Jong A, Hamoen LW, Kok J, Kuipers OP. Genome2D: A visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol 2004; 5:R37; PMID:15128451; http://dx.doi.org/ 10.1186/gb-2004-5-5-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform 2013; 14:193-202; PMID:22445902; http://dx.doi.org/ 10.1093/bib/bbs012 [DOI] [PubMed] [Google Scholar]
- 82.de Jong A, van der Meulen S, Kuipers OP, Kok J. T-REx: Transcriptome analysis webserver for RNA-seq expression data. BMC Genomics 2015; 16:663; PMID:26335208; http://dx.doi.org/12824337 10.1186/s12864-015-1834-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


