Abstract
Rsp5 ubiquitin ligase is required for ubiquitination of a wide variety of proteins involved in essential processes. Rsp5 was shown to be involved in regulation of lipid biosynthesis, intracellular trafficking of proteins, response to various stresses, and many other processes. In this article, we provide a comprehensive review of the nuclear and cytoplasmic functions of Rsp5 with a focus on biogenesis of different RNAs. We also briefly describe the participation of Rsp5 in the regulation of the RNA polymerase II complex, and its potential role in the regulation of other RNA polymerases. Moreover, we emphasize the function of Rsp5 in the coordination of the different steps of rRNA, mRNA and tRNA metabolism in the context of protein biosynthesis. Finally, we highlight the involvement of Rsp5 in controlling diverse cellular mechanisms at multiple levels and in adaptation of the cell to changing growth conditions.
Keywords: mRNA, RNA nuclear/cytoplasmic transport, RNA processing, rRNA, Rsp5 ubiquitin ligase, tRNA, ubiquitination
Abbreviations
- Ub
ubiquitin
- DUBs
deubiquitinating enzymes
- HECT
homologous to E6AP C-terminus
- ARTs
arrestin-related trafficking adaptors
- pre-rRNA
precursor rRNA
- MVB
multivesicular body
- Nups
nucleoporins
- RNAPII
RNA polymerase II
- TC-NER
transcription-coupled nucleotide excision repair
- CTD
C-terminal domain
- rRNA
ribosomal RNA
- NPCs
nuclear pore complexes
- pre-mRNAs
mRNA precursors
- mRNPs
messenger ribonucleoprotein particles
- TREX
transcription/export complex
- HSE
heat shock elements
- STRE
stress response element
- UBA
ubiquitin - associated domain
- tRNAs
transfer RNAs
- pre-tRNA
precursor tRNA
- tDNAs
tRNA genes.
Introduction
Ubiquitination is the process of conjugating the small peptide ubiquitin (Ub) to lysine residue of the substrate protein. It can be reversed by the removal of Ub by deubiquitinating enzymes (DUBs) which are ubiquitin-specific proteases. Ubiquitination can regulate diverse properties of a protein such as its half-life, activity, subcellular localization, or ability to form complexes. A protein can be modified by a single Ub (monoubiquitination), a chain of Ubs formed by attachment of consecutive Ub molecules to lysines (K6, K11, K27, K29, K33, K48, or K63) present in Ub itself (polyubiquitination), and by attachement of several Ub molecules individually to several lysine residues of the substrate protein (multiubiquitination). Ub chains linked via K48 or K63 are the most common. Mono- and multiubiquitination can act as signals for endocytosis, histone regulation, DNA repair, or nuclear export. However, substrates modified by K48-linked polyubiquitin chains composed of at least 4 Ub moieties are recognized by the proteasome and directed for proteasomal degradation.1 Attachment of Ub chains linked through lysines other than K48 can regulate cellular processes in both proteasomal-dependent2,3 as well as an independent manner, e.g., endocytosis.4
Ubiquitination is accomplished by a cascade of enzymatic reactions consisting of 3 steps. First, Ub is activated by the E1 enzyme and then, accepted by the conjugating enzyme E2. In the third step, Ub is transferred from E2 to the substrate protein in the presence of ubiquitin ligase E3 (reviewed in ref.5). One of the major subfamilies of E3 ligases is the homologous to E6AP C-terminus (HECT) domain-containing family of proteins.6 The HECT ligases actively participate in the ubiquitination by forming a thioester intermediate with Ub prior to attaching it to the substrate. The best studied subgroup of HECT E3s is the Nedd4-like family of ubiquitin ligases which exhibits a characteristic C2-WW-HECT modular structure (reviewed in ref.7).
The Saccharomyces cerevisiae Nedd4 homolog, Rsp5, is an essential yeast ubiquitin ligase. It is composed of an N-terminal C2 domain, 3 WW domains, and a C-terminal catalytic HECT domain (Fig. 1). The C2 domain is responsible for binding lipids8 and proteins. WW domains are involved in diverse protein-protein interactions (to date, 124 such interactions have been reported) and recognize proline-rich sequences called PY motifs9 in Rsp5 substrates or Rsp5 adaptor proteins, such as the arrestin-related trafficking adaptors (ARTs) that mediate Rsp5 - substrate binding10 (reviewed in ref.11). Therefore, Rsp5 recognizes a wide variety of substrates and is a key protein implicated in various signaling pathways. Consequently, regulation of Rsp5 will affect numerous cellular processes. Thus, Rsp5 can coordinate the processes inside the cell with changing conditions and provide response of the cell at multiple levels.12 The HECT domain is essential for Rsp5 activity in ubiquitination13 and its structural flexibility underlies the ability to adapt to different substrates and modify them by attachment of ubiquitin moieties.14 Rsp5 preferentially forms K63-linked Ub chains.15,16
Figure 1.
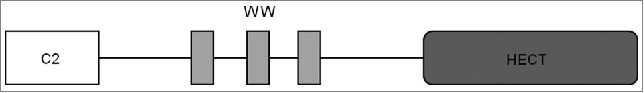
Domain structure of Rsp5 ubiquitin ligase. C2 domain at the N-terminal region binds lipids and proteins; WW domains are responsible for protein-protein interactions; HECT is a catalytic domain.
Rsp5 is localized to the cytoplasm, in cortical patches, perivacuolar endosomal structures, and has also been reported to have a nuclear pool. Cholbinski et al.17 showed that Rsp5 contains one nuclear export signal (NES) and 2 nuclear localization signals (NLS), both located in the HECT domain and therefore, the protein is able to shuttle between the nucleus and cytoplasm. Rsp5 nuclear export is Crm1-dependent17 and intriguingly, Crm1 is mainly involved in the export of precursor rRNA (pre-rRNA).18 The many processes in which Rsp5 plays a key role are highly diverse, including biosynthesis of unsaturated fatty acids and other lipids (reviewed in ref.19), endocytosis, multivesicular body (MVB) sorting, lysosomal degradation of plasma membrane proteins (reviewed in ref.20), and actin cytoskeleton organization and function.21,22 Recently, Lu et al.23 have shown that Rsp5 is also involved in degradation of protein aggregates in the autophagy pathway.
Rsp5 can affect both cytoplasmic and nuclear processes and coordinate many of them with each other to ensure efficient response of the cell to changing conditions. This review will focus on the function of Rsp5 ubiquitin ligase in RNA biology. We present Rsp5 involvement in regulation of Rpb1, the largest subunit of RNA polymerase II (RNAPII) and the novel insights gained from the in vivo, in vitro and microarray studies on the role of Rsp5 in rRNA, mRNA and tRNA biogenesis. The article describes Rsp5 requirement in transcription, processing and transport of different RNA species, placing this enzyme as an important regulator of RNA biology.
Rsp5 is involved in regulation of RNAPII
Transcription elongation by RNAPII is not a continuous process and the polymerase is often stalled leading to transcriptional arrest (reviewed in ref.24). RNAPII stalling can be caused by numerous factors, e.g., difficult to transcribe sequences,25 chromatin structure26 or DNA adducts27 (reviewed in ref.28). Upon DNA damage the RNAPII complex does not dissociate from the site of damage and remains attached to the template, blocking access of repair factors to the lesion. On the other hand, the high stability of the complex is crucial for transcription fidelity.29 When the transcription-coupled nucleotide excision repair (TC-NER) pathway is unable to remove stalled RNAPII, its largest subunit Rpb1 is polyubiquitinated and degraded.30,31 Interestingly, this event is induced not only by DNA damage, but it also occurs in different situations leading to transcriptional stalling,32,33 e.g., a mutation of the gene encoding transcript cleavage factor TFIIS.34
RNAPII ubiquitination and degradation is a regulated multi-step process. In the first step, Rpb1 is ubiquitinated by Rsp530,31 (Table 1), which associates with the C-terminal domain (CTD) of Rpb1 via its WW domain.35 Although Rpb1 is not ubiquitinated and degraded upon Rsp5 inactivation,31,36 Rsp5 catalyzes only monoubiquitination or K63-linked polyubiquitination of Rbp1,37 which usually do not direct proteins for proteasomal degradation. In the case of polyubiquitinated Rpb1 with Ubs linked via K63, the Rsp5-associated DUB, Ubp2 trims the Ub chains and leaves a single ubiquitin moiety.15 Rpb1 is then polyubiquitinated with K48-linked Ubs by the Elc1-Ela1-Cul3-Rbx1 complex.38,39 Intriguingly, only the pre-monoubiquitinated Rpb1, but not the unmodified protein, can be polyubiquitinated.36 Moreover, Ubp3 DUB is able to completely remove K48-linked polyubiquitin chains and rescue Rpb1 from degradation. Thus, Ubp3 provides a proof-reading role and can prevent unnecessary degradation.40 Finally, the ATPase Cdc48-Ubx5 and 26S proteasome are recruited to the polyubiquitinated RNAPII. The polymerase complex disassembles from DNA, ubiquitinated Rpb1 dissociates from RNAPII and is degraded in the proteasome while other subunits are released.41 Taken together, Rsp5 plays an important role in the RNAPII transcription by removing stalled RNAPII complexes and allowing repair of lesions. Moreover, some subunits of RNA polymerase I (RNAPI) or RNA polymerase III (RNAPIII) have also been reported to be Rsp5 substrates, e.g., Rrn subunits (Rrn3, Rrn5, Rrn6, Rrn9, Rrn10 and Rrn11) of RNAPI42 or different RNAPIII subunits42,43 (Table 1). Therefore, future research is required to provide information whether Rsp5 regulates transcription by affecting all 3 RNA polymerases.
Table 1.
Polymerase I, II and III subunits and nucleoporins ubiquitinated by Rsp5
| Protein | Process | Method | Reference |
|---|---|---|---|
| Pol I complex | |||
| Rrn3, Rrn5, Rrn6, Rrn7, Rrn9, Rrn10, Rrn11 | transcription | in vitro | 42 |
| Pol II complex | |||
| Rpb1 | transcription | in vitro, in vivo | 30,31 |
| Rpb2, Rpb3, Rpb5, Rpb7, Rpb8, Rpb9, Rpb10, Rpb11 | in vitro | 42 | |
| Pol III complex | |||
| Bdp1, Brf1, Maf1, Pzf1, Ret1, Rpc11, Rpc17, Rpc34, Rpc37, Rpc82, Sfp1, Tfc1, Tfc3, Tfc4, Tfc6, Tfc7, Tfc8 | transcription | in vitro | 42 |
| Rpc25 | in vitro, microarray | 42,43 | |
| Nucleoporins | |||
| Nup1, Nup2, Nup42, Nup49, Nup57, Nup82, Nup84, Nup85, Nup116, Nup120,Nup133,Nup145, Nup157, Nup159, Nup188 | mRNA export | in vitro | 42 |
| Nup53 | in vitro, microarray | 42,43 | |
| Nup60 | nucleocytoplasmic transport | in vitro | 42 |
| Nup192 | nuclear pore organization | ||
Rsp5 affects rRNA processing and transport
Synthesis of eukaryotic ribosomes is a complex but crucial process for growing cells. Ribosomes are composed of a 60S large subunit containing 3 ribosomal RNA (rRNA) species: 25S, 5.8S, and 5S rRNAs, and a 40S small subunit with 18S rRNA. A large polycistronic transcript containing 18S, 5.8S and 25S rRNAs, generated by RNAPI, undergoes endo- and exonucleolytic processing to yield the mature rRNAs (reviewed in ref.44,45). However, 5S rRNA is transcribed independently by RNAPIII.46
Export of precursor 40S (pre-40S) and pre-60S ribosomal subunits through nuclear pore complexes (NPCs) in yeast and higher eukaryotes requires Crm1 exportin (also known as Xpo1), which is a member of the β-importin family.18 The only known adaptor between Crm1 and pre-60S is Nmd3 protein,47 while the Crm1 adaptor for pre-40S subunit has not been identified yet.48 Mex67/Mtr2 (mRNA heterodimeric export receptor) and Arx1 proteins also participate in the export of pre-60S subunit.49 Furthermore, other proteins have been suggested to function in the export of pre-60S subunits, e.g., the mRNA export factor Npl350 and Exp-5, the vertebrate ortholog of yeast Msn5 protein that is involved in tRNA transport.51 Thus, ribosome nuclear export is likely connected to the export of mRNAs (Fig. 2) and tRNAs.
Figure 2.
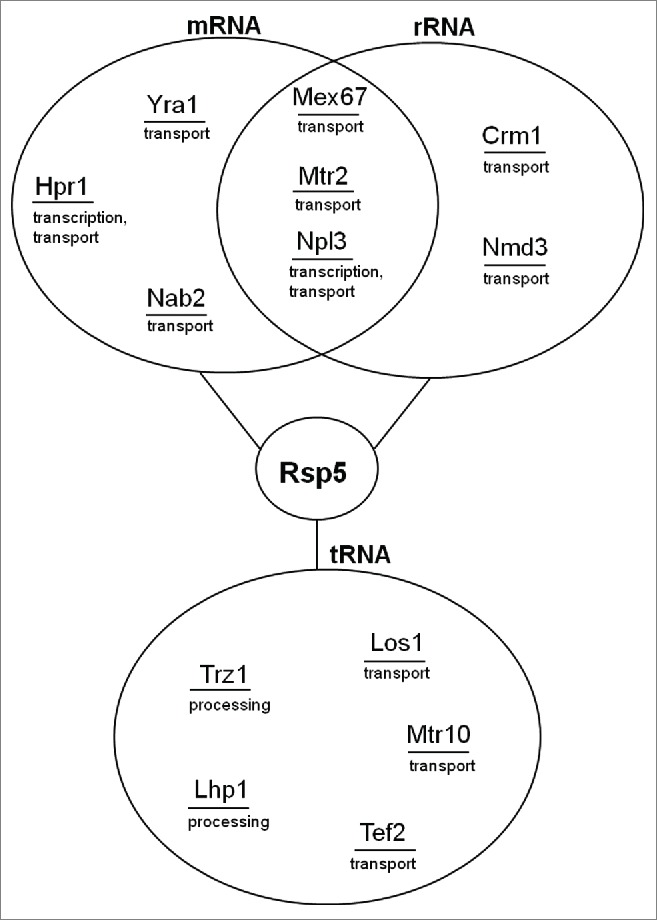
Proteins identified as Rsp5 substrates and involved in biogenesis of rRNA, mRNA and tRNA. The same proteins can participate in biology of various RNAs thus providing a link between different processes.
Several links between Rsp5 and the protein biosynthesis machinery have been reported. Neumann et al.52 showed that Rsp5 influences pre-rRNA maturation and its transport from the nucleus to the cytoplasm. Nuclear export of the pre-60S subunit was inhibited in rsp5-3 mutant at the restrictive temperature52 (Table 2 and Table 3). Thus, Rsp5 is involved in nuclear export of pre-rRNA. Moreover, pre-rRNA analysis showed that 35S pre-rRNA accumulated in the rsp5-3 mutant and the products of 35S processing (20S and 27S pre-rRNAs) were strongly depleted at the elevated temperature (Table 2). These results show that the early pre-rRNA cleavages at some sites can be inhibited in the rsp5-3 strain. Consistent with the pre-rRNA processing defects, the level of mature 18S, 5.8S and 25S rRNAs was reduced in rsp5-3 cells52 (Table 2).
Table 2.
Rsp5 alleles and their phenotypes
| Phenotype |
||||
|---|---|---|---|---|
| Mutant | rRNA | mRNA | tRNA | Reference |
| rsp5-1 | degradation of 25S rRNA | 53 | ||
| accumulation of poly(A)+ RNA in the nucleus | 71 | |||
| rsp5-19 | degradation of 18S and 25S rRNA (Northern and FISH analysis) | 53 | ||
| decreased total rate of ribosomes | 53 | |||
| reduced level of polysomes and 80S ribosomes | 53 | |||
| accumulation of poly(A)+ RNA in the nucleus | 90 | |||
| accumulation of tRNALeu precursors (Northern analysis) | 53 | |||
| accumulation of tRNATyr and tRNAMet in the nucleus (FISH analysis) | 90 | |||
| rsp5-3 | inhibition of nuclear export of the pre-60S subunit | 52 | ||
| accumulation of 35S pre-rRNA | 52 | |||
| decreased level of 20S and 27S pre-rRNAs | 52 | |||
| reduced level of mature 18S, 5.8S and 25S rRNAs | 52 | |||
| degradation of 18S and 25S rRNA | 53 | |||
| accumulation of poly(A)+ RNA in the nucleus | 52 | |||
| accumulation of tRNALeu and tRNATyr in the nucleus (FISH analysis) | 52 | |||
| accumulation of primary transcripts for tRNALeu and tRNAArg-Asp (Northern analysis) | 52 | |||
| rsp5-ww2 | accumulation of poly(A)+ RNA in the nucleus | 71 | ||
| rsp5-ww3 | accumulation of poly(A)+ RNA in the nucleus | 71 | ||
| rsp5-A401E | accumulation of HSF1 and MSN2/4 mRNAs in the nucleus | 72 | ||
| rsp5Δ | accumulation of poly(A)+ RNA in the nucleus | 71 | ||
| accumulation of mRNAs encoding Pgk1, Ssa1, Ssa2 and Ssa3 in the nucleus | 71 | |||
Table 3.
RSP5 mutants
| Mutant | Description of mutation | Reference |
|---|---|---|
| rsp5-1 | L733S substitution in HECT domain | 13 |
| rsp5-19 | P418L substitution in WW3 domain | 90 |
| rsp5-3 | T104A substitution between C2 and WW1 domain, E673G and Q716P substitutions in HECT domain | 52 |
| rsp5-ww2 | W359F and P362A substitutions in WW2 domain | 71 |
| rsp5-ww3 | W415F and P418A substitutions in WW3 domain | 71 |
| rsp5-A401E | A401E substitution in WW3 domain | 72 |
| rsp5Δ | deletion of RSP5 gene | 71 |
A significant depletion of 18S and 25S rRNA was also reported for cells harboring the rsp5-19 mutation (Table 3) after a shift to the restrictive temperature53 (Table 2). Furthermore, a smear of rRNA-derived decay products was clearly visible. This rRNA destabilization was not an effect of the elevated temperature itself because the rRNA level remained unchanged in other temperature-sensitive strains tested.53 It was reported that other mutations in RSP5, namely rsp5-1 and rsp5-3, also led to rRNA degradation53 (Table 2 and Table 3). Moreover, ribosomes were not degraded in rsp5-19 cells transformed with plasmid encoding RSP5. Based on these results, Shcherbik and Pestov53 concluded that Rsp5 is important for maintaining rRNA stability in yeast. Noteworthy, Rsp5 affects multiple RNAs,52 but mature rRNAs are apparently more vulnerable to degradation than other stable RNA species. Consistent with the decrease in the level of mature rRNA observed in rsp5-19 cells, also the total rate of ribosomes was diminished and the levels of polysomes and 80S ribosomes were strongly reduced in this mutant53 (Table 2) which is in agreement with the studies showing that in yeast rRNA degradation is complex and regulated process.54
To verify whether rsp5 mutations could enhance general autophagy, which directs cytoplasmic ribosomes to destruction in the vacuole,55 or induce ribophagy,56 a ribosome-specific autophagy pathway, deletions of key autophagy genes were introduced into the rsp5-19 strain. Those mutations, however, did not suppress the rRNA degradation caused by the rsp5 defect. Thus, the enhanced degradation of ribosomes caused by a lack of functional Rsp5 in rich medium utilizes a mechanism different rather than ribophagy.53 The importance of Rsp5 for ribosome stability is well proven since rRNA degradation was observed in different rsp5 temperature sensitive mutants and could be prevented by expression of wild-type RSP5.
In conclusion, there are considerable data supporting the role of Rsp5 in ribosome assembly, nuclear export and cytoplasmic stability. Moreover, some proteins involved in pre-rRNA nuclear export, e.g., exportin Crm1 and its adaptor, Nmd3, have been shown in vitro to be Rsp5 substrates42 (Table 4 and Fig. 2). Thus, Rsp5 could influence rRNA transport via ubiquitination of export proteins. Intriguingly, Bai et al.57 demonstrated that rRNA export affects rRNA synthesis. In HeLa cells nucleolar localization of Crm1 and Nmd3 was increased by inhibition of RNAPI. In addition, inhibition of Crm1 and depletion of Nmd3 reduced the level of 47S rRNA precursor, and inhibition of Crm1 compromised the processing of 28S rRNA. The data indicate that Crm1 and Nmd3 function in pathways that couple rRNA synthesis, processing and nuclear transport, and that RNA export machinery modulates rRNA synthesis probably through some yet unidentified signaling pathways. Altogether, a lack of ubiquitination of these proteins could lead to defects in rRNA processing and transport in rsp5 mutants. Therefore, in vivo studies on ubiquitination of Crm1 and Nmd3 proteins by Rsp5 could facilitate our understanding of the role of this modification in rRNA biology. Finally, it is also possible that changes in pre-rRNA processing, transport and the level of ribosomes in rsp5 cells could be caused by the action of signaling pathways which could be affected in these mutants.
Table 4.
Proteins involved in RNA biogenesis and ubiquitinated by Rsp5
| Protein | Process | Method | Reference |
|---|---|---|---|
| rRNA biogenesis | |||
| Crm1 | export | in vitro | 42 |
| Mex67 | |||
| Mtr2 | |||
| Nmd3 | |||
| Npl3 | in vitro; microarray | 42,43,9 | |
| mRNA biogenesis | |||
| Hpr1 | transcription, export | in vivo, in vitro | 73 |
| Mex67 | export | in vitro | 42 |
| Mtr2 | |||
| Nab2 | |||
| Yra1 | |||
| Npl3 | transcription, export | in vitro; microarray | 9,42,43 |
| tRNA biogenesis | |||
| Lhp1 | processing | microarray | 43,9 |
| Los1 | transport | in vitro | 42 |
| Mtr10 | |||
| Tef2 | transport | in vitro, microarray | 42,43 |
| Trz1 | processing | microarray | 43 |
Rsp5 regulates mRNA nuclear-cytoplasmic trafficking
It is well established that mRNA synthesis and processing are coupled to its nuclear export, which requires that the newly synthesized mRNA precursor (pre-mRNA) undergoes several processing steps, including 5′ capping, splicing, 3′ end cleavage and polyadenylation. During transcription nascent mRNA interacts with mRNA-binding proteins and noncoding RNAs leading to the formation of messenger ribonucleoprotein particles (mRNPs) that are transported to the cytoplasm (reviewed in ref.58). Various steps leading to mRNP formation are linked and mediated by interactions with the RNAPII elongation complex and the processing machinery. The coupling of different steps of mRNA biogenesis could be a part of quality control mechanisms ensuring that only mature and export-competent transcripts are transported to the cytoplasm.
Nuclear export of mRNA requires the heterodimeric export receptor Mex67/Mtr2, a transporter that is distinct from β-importins.59 Mex67 interacts with mRNPs while Mtr2 promotes Mex67 interaction with NPCs.60 As Mex67 exhibits low affinity for RNA, its interaction with mRNA is mediated by adaptor proteins, for example Yra1.61 The recruitment of Yra1 and its partner Sub2 occurs co-transcriptionally and is splicing-independent.62,63 Yra1 and Sub2 proteins are associated with the THO complex, implicated in transcription elongation and mRNA export. In yeast, the THO complex and the mRNA export factors Sub2 and Yra1 are components of the multi-subunit transcription/export (TREX) complex engaged in coupling transcription to mRNA export.64,65 TREX is recruited to an active gene during transcription elongation. It has been proposed that the binding of Mex67 to Yra1 at a later step displaces Sub2 from Yra1 before mRNA export.63 Furthermore, additional adaptors are implicated in mRNA export since Yra1 does not associate with all yeast transcripts and is dispensable for mRNA export in Caenorhabditis elegans and Drosophila melanogaster.66-68 Interestingly, 2 identified adaptors of Mex67, Npl3 and Nab2 are regulated by ubiquitination (reviewed in ref.69).
The involvement of Rsp5 and the ubiquitin/proteasome system in mRNA export is well characterized. Initial evidence for the role of the ubiquitin pathway in mRNA nuclear export was provided by studies performed using a Schizosaccharomyces pombe temperature-sensitive ubiquitin-activating E1 enzyme (Ptr3) mutant, in which the mRNA export was defective.70 To determine the role of the ubiquitin pathway in mRNA export, the function of each ubiquitin-protein ligase from the HECT family was analyzed in S. cerevisiae. The temperature sensitive rsp5-1 mutant accumulated poly(A)+ RNA in the nucleus only at the restrictive temperature, whereas rsp5Δ71 cells (Table 3) also at normal temperature, even when the essential role of Rsp5 in unsaturated fatty acid biosynthesis was bypassed by adding oleic acid to the medium71 (Table 2). Thus, the nuclear mRNA accumulation caused by RSP5 mutations is not due to a shortage of unsaturated fatty acids. It was indicated using rsp5-1, rsp5-ww2 and rsp5-ww3 mutants (Table 3) that 3 domains of Rsp5 are implicated in the export of mRNA from the nucleus to the cytoplasm: WW2, WW3 and HECT.71 Strong nuclear accumulation of poly(A)+ RNA was also observed in the rsp5-3 strain52 (Table 2).
The role of Rsp5 in the export of specific mRNAs has also been investigated. For example, Rodriguez et al.71 demonstrated that a lack of functional Rsp5 leads to defect in the nuclear export of mRNA encoding the glycolytic enzyme Pgk1 as well as mRNAs encoding Ssa heat shock proteins (Ssa1, Ssa2 and Ssa3) (Table 2). In addition, other authors have shown that in response to environmental stresses, Rsp5 regulates at the post-transcriptional level the expression of genes for 2 major transcription factors, Hsf1 and Msn2/4 which are responsible for stress-induced gene expression. Hsf1 binds to heat shock element (HSE) whereas Msn2/4 binds to the stress response element (STRE) in the promoters of many stress protein genes.72 In the rsp5-A401E mutant (Table 3) that is hypersensitive to various stresses, including high temperature, transcription of stress protein genes is defective. Although mRNA levels of HSF1 and MSN2/4 in rsp5-A401E are only slightly lower than in wild type cells, the respective protein levels are remarkably decreased in this mutant because HSF1 and MSN2/4 mRNAs accumulate in the nucleus of rsp5-A401E cells (Table 2). Thus, by modulating nuclear mRNA export Rsp5 regulates the expression of transcription factors required for stress responses.72
Rsp5 ubiquitin ligase activity is required for mRNA export, suggesting that Rsp5 substrates might include mRNA nuclear export factors. Indeed, Rsp5-dependent regulation of a component of THO complex, Hpr1, was shown.73 Degradation of Hpr1 is enhanced at high temperature and is linked to on-going RNAPII-mediated transcription while the stability of other components of THO complex is not affected under these conditions suggesting that Hpr1 turnover could control the formation of the TREX complex and mRNA export. Thus, Hpr1 appears to be a key factor of the THO complex and its stability controls the activity of the whole complex. Hpr1 is polyubiquitinated by Rsp5 in conjugation with E1 and Ubc4 as an E2 and degraded in the proteasome both in vivo and in vitro73 (Fig. 2). Although the data demonstrate that interactions within the enzyme-substrate complex do not require any adaptor in vitro, it cannot be excluded that ubiquitination of Hpr1 can be regulated by an additional ubiquitin ligase or other proteins in vivo, as it is for Rpb1 protein. It is known that ubiquitination of proteins with K63-linked Ub chains usually does not lead to their degradation in the proteasome. Nevertheless, it is possible that K63-linked Ub chains could serve as a proteasome targeting signal in vivo.3 Therefore, further analysis of Hpr1 ubiquitination in vivo is required to decipher fully its degradation mechanism. The ubiquitin - associated (UBA) domain of Mex67 binds the Hpr1-attached polyubiquitin chain and transiently protects the protein from degradation.74 Probably, upon mRNA 3′ end formation, Mex67-Mtr2 is transferred onto nascent mRNA exposing the ubiquitinated Hpr1 to the proteasome. Degradation of Hpr1 releases mRNA and causes disassembly of the THO complex which can then participate in a next transcription event (reviewed in ref.69). The Mex67-Hpr1 interaction coordinates transcription with recruitment of the mRNA export machinery.74 Noteworthy, it has been shown that the human ortholog of Hpr1, Thoc1, is polyubiquitinated by Nedd4 and then, degraded by the proteasome, suggesting that regulation of THO activity by HECT ligases and the ubiquitin-proteasome pathway is evolutionarily conserved between yeast and mammals.75
The export receptor Mex67-Mtr2 utilizes 2 adaptor proteins regulated by the ubiquitin pathway. One of them, Npl3, a shuttling protein implicated in coordinating transcription and mRNA export,76 has been proposed to be an Rsp5 substrate9,42,43 (Table 4 and Fig. 2). In addition, Npl3 can be phosphorylated and dephosphorylated which influences its interaction with Mex67 and has an impact on mRNA export. It is not clear how Rsp5 affects Npl3 and what the biological function of this interaction is; so further studies are required. However, the data provide evidence that post-translational modifications are important to coordinate recruitment of Mex67 with transcription and export. Also another Mex67 adaptor, a shuttling mRNA binding protein Nab2,77 which stabilizes the adaptor-receptor interaction by directly binding Mex67 and Yra1, was shown in vitro to be an Rsp5 substrate42 (Table 4 and Fig. 2).
The above examples exemplify Rsp5 contribution to mRNA transport. In addition to Hpr1 or Npl3, other proteins involved in mRNA export have been shown to be Rsp5 substrates. In vitro studies have indicated that Mex67, Mtr2 and Yra1 can be ubiquitinated by Rsp542 (Table 4 and Fig. 2). The question that remains to be resolved concerns how their modifications could affect mRNA biology. Noteworthy, no defects have been observed in the splicing of the ACT1 mRNA or pre-mRNA-like intron-containing U3 precursor. According to Neumann et al.52 Rsp5 seems not to be involved in mRNA processing. However, little is known about the involvement of Rsp5 in mRNA maturation and further studies are necessary to fully understand its role in this process.
Rsp5 modulates tRNA processing and transport
Transfer RNAs (tRNAs) are crucial in protein synthesis and take part in other processes. They deliver amino acids to the ribosome for incorporation into a polypeptide chain and participate in signaling pathways. tRNA biogenesis in yeast involves the synthesis of initial transcript by RNAPIII followed by post-transcriptional alterations to generate mature tRNA. The post-transcriptional steps comprise the removal of the 5′ leader, trimming the 3′ trailer, addition of CCA to the 3′ terminus, removal of introns (from tRNAs encoded by intron-containing genes), modification of multiple nucleoside residues, and export to the cytoplasm where the tRNA functions in translation (reviewed in ref.78). In fact, the subcellular traffic of tRNA is bidirectional and occurs in 3 steps. First, the newly synthesized and end-processed tRNAs are exported from the nucleus to the cytoplasm in the primary tRNA export process. Then, spliced tRNAs are imported back to the nucleus via retrograde tRNA nuclear import, likely to be repaired or degraded.79-83 Finally, mature tRNAs that were imported to the nucleus are re-exported to the cytoplasm.84
Twenty percent of yeast tRNAs are transcribed with an intron. In yeast, splicing of tRNA introns occurs on the cytoplasmic surface of mitochondria where the tRNA splicing endonuclease complex resides,85-87 so end-processed intron-containing tRNAs have to be exported from the nucleus to undergo splicing in the cytoplasm. The primary export of intron-containing precursor tRNA (pre-tRNA), involves the β-importin family member, Los1.79,88,89 Because LOS1 is non-essential gene in yeast, there is at least one additional unidentified nuclear exporter for intron-containing pre-tRNAs. Mtr10, a member of the yeast β-importin family, is implicated in the retrograde nuclear import but whether Mtr10 directly interacts with tRNA remains unknown (reviewed in ref.78). tRNA re-export is mediated in yeast by both Los1 and another β-importin family member, Msn5.79 Again, at least one unidentified transporter involved in the re-export process must exist because msn5Δ los1Δ double mutants are viable.79,83 Thus, tRNA biogenesis is highly complex and only partially understood.
Rsp5 is involved in tRNA biology. Neumann et al.52 reported nuclear accumulation of tRNALeu and tRNATyr, encoded by intron-containing genes, as well as tRNAGlu and tRNAGly, encoded by intron-less genes, in rsp5-3 cells at the nonpermissive temperature (Table 2). To determine whether the nuclear accumulation of tRNA in the mutant was a consequence of defects in RNA maturation, tRNA processing was investigated. The analyses revealed that the primary transcripts for tRNALeu and tRNAArg-Asp accumulated in the rsp5-3 mutant at the elevated temperature52 (Table 2). In addition, Shcherbik and Pestov53 showed that tRNALeu precursors accumulated also in rsp5-19 mutant (Table 2). Moreover, nuclear accumulation of tRNATyr, encoded by intron-containing genes, and tRNAMet, encoded by intron-lacking genes, was observed in this mutant after a temperature up-shift90 (Table 2). Thus, defects in pre-tRNA processing in rsp5 cells might be the cause of tRNA nuclear accumulation.
Other authors showed that the rsp5-19 mutation alters cell sensitivity to antibiotics that affect protein synthesis and increases the fidelity of translation.90 The mutant deficient in Rsp5 ubiquitin protein ligase activity was hypersensitive to anisomycin and cycloheximide that inhibit translation and resistant to paromomycin, which causes ribosomal misreading. The altered sensitivity of the rsp5-19 strain to some translational antibiotics is in agreement with other data showing that Rsp5 affects translation.91 The rsp5-19 strain exhibits about a 70% decrease in the rate of total protein synthesis that could be caused by defects in translation or nuclear accumulation of tRNA.90 Intriguingly, an additional copy of TEF2 (Table 4), one of 2 genes encoding elongation factor eEF-1A which delivers tRNAs to the ribosome, suppressed the defect in the subcellular distribution of tRNA as well as the growth defects in the presence and in the absence of antibiotics caused by rsp5-19 mutation.90 This is consistent with earlier studies showing that eEF-1A and tRNA aminoacylation are required for efficient export of mature tRNAs and suggests a coordination between translation and tRNA transport. Inhibition of tRNA aminoacylation by amino acid starvation, lack of CCA addition at the 3′ end of tRNAs or specific inhibition of aminoacyl - tRNA synthetase caused nuclear accumulation of tRNA.92 Finally, tRNAs that fail to be exported, e.g., because they are not aminoacylated, accumulate in the nucleolus where rRNA synthesis and some processing steps occur. Because the transcription and processing machineries of tRNA and rRNA share several components with each other,93 the presence of tRNA in the nucleolus could affect rRNA biogenesis.92 Thus, tRNA accumulation could act as a signal for RNAPI leading to cessation of its transcriptional activity and in this way negatively affect rRNA biogenesis. Therefore, cellular distribution of tRNA is important for the communication and signaling between the cytoplasm and the nucleus and modifications of proteins involved in tRNA dynamics could affect various processes.
Diverse approaches have revealed that some proteins involved in tRNA biogenesis are Rsp5 substrates, including those engaged in tRNA 3′ end processing (Trz1 and Lhp1) and tRNA transport (Mtr10)44 (Table 4 and Fig. 2). There is no clear evidence for Ub modification of Los1 protein. Huang and Hopper94 did not find by mass spectrometry Ub modifications for Los1 either in fed or amino acid starved cells – however, the protein coverage was not complete (∼60%). On the other hand, Lu et al.44 identified in vitro Los1 as Rsp5 substrate (Table 4 and Fig. 2). Taken together, Rsp5 could be involved in both tRNA processing and nuclear transport and studies of these processes will result in understanding how they can be coordinated by this ligase.
Concluding remarks
Different classes of RNA (rRNA, mRNA and tRNA) accumulate in the nucleus in temperature-sensitive rsp5 cells at the restrictive temperature. Several mechanisms of Rsp5 involvement in RNA biogenesis are possible. Rsp5 may have multiple targets which participate in the processing and/or export of different RNAs. Indeed, as the studies described above have shown, many proteins involved in the biogenesis of RNA can be Rsp5 substrates, e.g., Crm1, Nmd3, Hpr1, Npl3, Mex67, Mtr10 (Table 4 and Fig. 2). Rsp5 could also act at multiple levels and indirectly influence RNA biology, e.g., by modulating nutrient transport and cellular signaling. The defective export of various RNAs in rsp5 mutants could also, in principle, be due to Rsp5 influence on the composition of lipids in the nuclear membrane. However, the nuclear membrane organization is apparently unaltered in rsp5 cells52 and thus, the role of Rsp5 in the export of various RNAs through its impact on the nuclear membrane can be rather excluded.
Alternatively, Rsp5 could affect nuclear RNA export globally by ubiquitination of the NPC component(s) engaged in all 3 nuclear export systems. Indeed, a systematic analysis showed that more than 50% of nucleoporins (Nups) are ubiquitinated.95 Prevalence of monoubiquitination and involvement of distinct components of the ubiquitin conjugation machinery suggest that ubiquitination of the Nups is not only connected to proteasome-dependent protein degradation but rather associated with regulation of the NPC function.95 Some nucleoporins have been postulated to be ubiquitinated by Rsp5 in vitro, e.g., Nup2, Nup60, Nup116, Nup192, while the proposed ubiquitination of others was not confirmed in vivo, e.g., Nup1, Nup42, Nup49 (Table 1). For Nup159, postulated to be ubiquitinated by Rsp5 in vitro, another ligase was identified in vivo (Table 1). However, this does not exclude possibility that investigated Nups are Rsp5 substrates in different conditions. Interestingly, it has been revealed recently that similarly to mRNA, tRNA transcription can be coordinated with its export at NPCs in yeast. Mutants in which tRNA genes (tDNAs) do not establish a contact with NPCs are still viable and therefore the transcription at the NPCs is not essential but rather might facilitate the export of pre-tRNAs and its coordination with transcription.96 Intriguingly, it has been suggested that tRNA processing might occur at the NPCs as well. Studies in Caenorhabditis elegans have shown that nucleoporins of the NPC can associate with tDNAs to regulate 3′ end processing of nascent tRNA.97
Thus, the different steps of tRNA biogenesis could be coordinated with each other and modification of one protein could influence numerous processes. Also, the different steps of biogenesis of mRNA or rRNA known to be coupled with each other could be coordinated at the NPC by modification of a single protein. It will be interesting to decipher the mechanism by which Rsp5 controls the biology of RNA classes at multiple levels. Although the ubiquitination by Rsp5 of many individual components implicated in RNA biogenesis has been suggested or proven, there are still many unknowns. For instance: How does Rsp5 coordinate different steps of biogenesis of an individual RNA? How is the biogenesis of one RNA coordinated with that of others? What environmental signals are important for Rsp5 function? Despite the progress that has been made in the last few years, still much remains to be clarified about the exact role of Rsp5 in RNA biogenesis and further studies are required to determine the full potential of this ligase in RNA biology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Anita Hopper, Joanna Kufel and Teresa Zoladek for critical reading of the manuscript.
Funding
This work was supported by Foundation for Polish Science within International PhD Project “Studies of nucleic acids and proteins - from basic to applied research” (MPD/2009-3/2).
References
- 1.Finley D, Ulrich HD, Sommer T, Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 2012; 192:319-60; PMID:23028185; http://dx.doi.org/ 10.1534/genetics.112.140467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chastagner P, Israel A, Brou C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep 2006; 7:1147-53; PMID:17028573; http://dx.doi.org/ 10.1038/sj.embor.7400822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, Tanaka K. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J 2009; 28:359-71; PMID:19153599; http://dx.doi.org/ 10.1038/emboj.2008.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauwers E, Jacob C, Andre B. K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J Cell Biol 2009; 185:493-502; PMID:19398763; http://dx.doi.org/ 10.1083/jcb.200810114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreiber A, Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochim Biophys Acta 2014; 1843:163-81; PMID:23545414; http://dx.doi.org/ 10.1016/j.bbamcr.2013.03.019 [DOI] [PubMed] [Google Scholar]
- 6.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A 1995; 92:5249; PMID:7761480; http://dx.doi.org/ 10.1073/pnas.92.11.5249-a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 2009; 10:398-409; PMID:19436320; http://dx.doi.org/ 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- 8.Dunn R, Klos DA, Adler AS, Hicke L. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J Cell Biol 2004; 165:135-44; PMID:15078904; http://dx.doi.org/ 10.1083/jcb.200309026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S. Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol 2006; 7:R30; PMID:16606443; http://dx.doi.org/ 10.1186/gb-2006-7-4-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nikko E, Pelham HR. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 2009; 10:1856-67; PMID:19912579; http://dx.doi.org/ 10.1111/j.1600-0854.2009.00990.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leon S, Haguenauer-Tsapis R. Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp Cell Res 2009; 315:1574-83; PMID:19070615; http://dx.doi.org/ 10.1016/j.yexcr.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 12.Iesmantavicius V, Weinert BT, Choudhary C. Convergence of Ubiquitylation and Phosphorylation Signaling in Rapamycin-treated Yeast Cells. Mol Cell Proteomics 2014; 13:1979-92; PMID:24961812; http://dx.doi.org/ 10.1074/mcp.O113.035683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol 1999; 19:342-52; PMID:9858558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raimondo D, Giorgetti A, Bernassola F, Melino G, Tramontano A. Modelling and molecular dynamics of the interaction between the E3 ubiquitin ligase Itch and the E2 UbcH7. Biochem Pharmacol 2008; 76:1620-7; PMID:18805400; http://dx.doi.org/ 10.1016/j.bcp.2008.08.026 [DOI] [PubMed] [Google Scholar]
- 15.Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J 2005; 24:2414-24; PMID:15933713; http://dx.doi.org/ 10.1038/sj.emboj.7600710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kee Y, Munoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem 2006; 281:36724-31; PMID:17028178; http://dx.doi.org/ 10.1074/jbc.M608756200 [DOI] [PubMed] [Google Scholar]
- 17.Cholbinski P, Jastrzebska Z, Wysocka-Kapcinska M, Plochocka D, Gornicka A, Hopper AK, Zoladek T. Yeast ubiquitin ligase Rsp5 contains nuclear localization and export signals. Eur J Cell Biol 2011; 90:834-43; PMID:21868125; http://dx.doi.org/ 10.1016/j.ejcb.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 2003; 116:2409-19; PMID:12724356; http://dx.doi.org/ 10.1242/jcs.00464 [DOI] [PubMed] [Google Scholar]
- 19.Kaliszewski P, Zoladek T. The role of Rsp5 ubiquitin ligase in regulation of diverse processes in yeast cells. Acta Biochim Pol 2008; 55:649-62; PMID:19039336 [PubMed] [Google Scholar]
- 20.Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, Andre B. The ubiquitin code of yeast permease trafficking. Trends Cell Biol 2010; 20:196-204; PMID:20138522; http://dx.doi.org/ 10.1016/j.tcb.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 21.Kaminska J, Spiess M, Stawiecka-Mirota M, Monkaityte R, Haguenauer-Tsapis R, Urban-Grimal D, Winsor B, Zoladek T. Yeast Rsp5 ubiquitin ligase affects the actin cytoskeleton in vivo and in vitro. Eur J Cell Biol 2011; 90:1016-28; PMID:22000681; http://dx.doi.org/ 10.1016/j.ejcb.2011.08.002 [DOI] [PubMed] [Google Scholar]
- 22.Jarmoszewicz K, Lukasiak K, Riezman H, Kaminska J. Rsp5 ubiquitin ligase is required for protein trafficking in Saccharomyces cerevisiae COPI mutants. PloS one 2012; 7:e39582; PMID:22761830; http://dx.doi.org/ 10.1371/journal.pone.0039582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu K, Psakhye I, Jentsch S. Autophagic Clearance of PolyQ Proteins Mediated by Ubiquitin-Atg8 Adaptors of the Conserved CUET Protein Family. Cell 2014; 158:549-63; PMID:25042851; http://dx.doi.org/ 10.1016/j.cell.2014.05.048 [DOI] [PubMed] [Google Scholar]
- 24.Wilson MD, Harreman M, Svejstrup JQ. Ubiquitylation and degradation of elongating RNA polymerase II: the last resort. Biochim Biophys Acta 2013; 1829:151-7; PMID:22960598; http://dx.doi.org/ 10.1016/j.bbagrm.2012.08.002 [DOI] [PubMed] [Google Scholar]
- 25.Hawley DK, Wiest DK, Holtz MS, Wang D. Transcriptional pausing, arrest, and readthrough at the adenovirus major late attenuation site. Cell Mol Biol Res 1993; 39:339-48; PMID:8312969 [PubMed] [Google Scholar]
- 26.Kireeva ML, Hancock B, Cremona GH, Walter W, Studitsky VM, Kashlev M. Nature of the nucleosomal barrier to RNA polymerase II. Mol Cell 2005; 18:97-108; PMID:15808512; http://dx.doi.org/ 10.1016/j.molcel.2005.02.027 [DOI] [PubMed] [Google Scholar]
- 27.Svejstrup JQ. Transcription repair coupling factor: a very pushy enzyme. Mol Cell 2002; 9:1151-2; PMID:12086609; http://dx.doi.org/ 10.1016/S1097-2765(02)00553-1 [DOI] [PubMed] [Google Scholar]
- 28.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol 2008; 9:958-70; PMID:19023283; http://dx.doi.org/ 10.1038/nrm2549 [DOI] [PubMed] [Google Scholar]
- 29.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc Natl Acad Sci U S A 1994; 91:8502-6; PMID:8078911; http://dx.doi.org/ 10.1073/pnas.91.18.8502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci U S A 1997; 94:3656-61; PMID:9108033; http://dx.doi.org/ 10.1073/pnas.94.8.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beaudenon SL, Huacani MR, Wang G, McDonnell DP, Huibregtse JM. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol Cell Biol 1999; 19:6972-9; PMID:10490634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell 2007; 28:386-97; PMID:17996703; http://dx.doi.org/ 10.1016/j.molcel.2007.10.008 [DOI] [PubMed] [Google Scholar]
- 33.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell 2010; 38:202-10; PMID:20417599; http://dx.doi.org/ 10.1016/j.molcel.2010.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 2005; 121:913-23; PMID:15960978; http://dx.doi.org/ 10.1016/j.cell.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 35.Chang A, Cheang S, Espanel X, Sudol M. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. J Biol Chem 2000; 275:20562-71; PMID:10781604; http://dx.doi.org/ 10.1074/jbc.M002479200 [DOI] [PubMed] [Google Scholar]
- 36.Harreman M, Taschner M, Sigurdsson S, Anindya R, Reid J, Somesh B, Kong SE, Banks CA, Conaway RC, Conaway JW, et al.. Distinct ubiquitin ligases act sequentially for RNA polymerase II polyubiquitylation. Proc Natl Acad Sci U S A 2009; 106:20705-10; PMID:19920177; http://dx.doi.org/ 10.1073/pnas.0907052106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol 2010; 11:479-89; PMID:20551964; http://dx.doi.org/ 10.1038/nrm2921 [DOI] [PubMed] [Google Scholar]
- 38.Ribar B, Prakash L, Prakash S. Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae. Mol Cell Biol 2006; 26:3999-4005; PMID:16705154; http://dx.doi.org/ 10.1128/MCB.00293-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribar B, Prakash L, Prakash S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol Cell Biol 2007; 27:3211-6; PMID:17296727; http://dx.doi.org/ 10.1128/MCB.00091-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kvint K, Uhler JP, Taschner MJ, Sigurdsson S, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol Cell 2008; 30:498-506; PMID:18498751; http://dx.doi.org/ 10.1016/j.molcel.2008.04.018 [DOI] [PubMed] [Google Scholar]
- 41.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell 2011; 41:82-92; PMID:21211725; http://dx.doi.org/ 10.1016/j.molcel.2010.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu JY, Lin YY, Qian J, Tao SC, Zhu J, Pickart C, Zhu H. Functional dissection of a HECT ubiquitin E3 ligase. Mol Cell Proteomics 2008; 7:35-45; PMID:17951556; http://dx.doi.org/ 10.1074/mcp.M700353-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta R, Kus B, Fladd C, Wasmuth J, Tonikian R, Sidhu S, Krogan NJ, Parkinson J, Rotin D. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol 2007; 3:116; PMID:17551511; http://dx.doi.org/ 10.1038/msb4100159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turowski TW, Tollervey D. Cotranscriptional events in eukaryotic ribosome synthesis. Wiley Interdiscip Rev RNA 2014; PMID:25176256 [DOI] [PubMed] [Google Scholar]
- 45.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al.. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999; 285:901-6; PMID:10436161; http://dx.doi.org/ 10.1126/science.285.5429.901 [DOI] [PubMed] [Google Scholar]
- 46.Thomson E, Tollervey D. The final step in 5.8S rRNA processing is cytoplasmic in Saccharomyces cerevisiae. Mol Cell Biol 2010; 30:976-84; PMID:20008552; http://dx.doi.org/ 10.1128/MCB.01359-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 2001; 21:3405-15; PMID:11313466; http://dx.doi.org/ 10.1128/MCB.21.10.3405-3415.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altvater M, Schutz S, Chang Y, Panse VG. Dissecting ribosome assembly and transport in budding yeast. Methods Cell Biol 2014; 122:437-61; PMID:24857742; http://dx.doi.org/ 10.1016/B978-0-12-417160-2.00020-5 [DOI] [PubMed] [Google Scholar]
- 49.Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgärtel V, Boese G, Bassler J, Wild K, et al.. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell 2007; 27:767-79; PMID:17803941; http://dx.doi.org/ 10.1016/j.molcel.2007.06.034 [DOI] [PubMed] [Google Scholar]
- 50.Hackmann A, Gross T, Baierlein C, Krebber H. The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO Rep 2011; 12:1024-31; PMID:21852791; http://dx.doi.org/ 10.1038/embor.2011.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol 2010; 8:e1000522; PMID:21048991; http://dx.doi.org/ 10.1371/journal.pbio.1000522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neumann S, Petfalski E, Brugger B, Grosshans H, Wieland F, Tollervey D, Hurt E. Formation and nuclear export of tRNA, rRNA and mRNA is regulated by the ubiquitin ligase Rsp5p. EMBO Rep 2003; 4:1156-62; PMID:14608372; http://dx.doi.org/ 10.1038/sj.embor.7400026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shcherbik N, Pestov DG. The ubiquitin ligase Rsp5 is required for ribosome stability in Saccharomyces cerevisiae. RNA 2011; 17:1422-8; PMID:21665996; http://dx.doi.org/ 10.1261/rna.2615311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mroczek S, Kufel J. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic acids research 2008; 36:2874-88; PMID:18385160; http://dx.doi.org/ 10.1093/nar/gkm1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 1992; 119:301-11; PMID:1400575; http://dx.doi.org/ 10.1083/jcb.119.2.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol 2008; 10:602-10; PMID:18391941; http://dx.doi.org/ 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- 57.Bai B, Moore HM, Laiho M. CRM1 and its ribosome export adaptor NMD3 localize to the nucleolus and affect rRNA synthesis. Nucleus 2013; 4:315-25; PMID:23782956; http://dx.doi.org/ 10.4161/nucl.25342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell 2014; 54:547-58; PMID:24856220; http://dx.doi.org/ 10.1016/j.molcel.2014.04.033 [DOI] [PubMed] [Google Scholar]
- 59.Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J 1997; 16:3256-71; PMID:9214641; http://dx.doi.org/ 10.1093/emboj/16.11.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol 2000; 150:695-706; PMID:10952996; http://dx.doi.org/ 10.1083/jcb.150.4.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J 2000; 19:410-20; PMID:10722314; http://dx.doi.org/ 10.1093/emboj/19.3.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen TH, Boulay J, Rosbash M, Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol 2001; 11:1711-5; PMID:11696331; http://dx.doi.org/ 10.1016/S0960-9822(01)00529-2 [DOI] [PubMed] [Google Scholar]
- 63.Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 2001; 413:648-52; PMID:11675790; http://dx.doi.org/ 10.1038/35098113 [DOI] [PubMed] [Google Scholar]
- 64.Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondón AG, Aguilera A, Struhl K, Reed R, et al.. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002; 417:304-8; PMID:11979277; http://dx.doi.org/ 10.1038/nature746 [DOI] [PubMed] [Google Scholar]
- 65.Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 2002; 21:3526-35; PMID:12093753; http://dx.doi.org/ 10.1093/emboj/cdf335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gatfield D, Izaurralde E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 2002; 159:579-88; PMID:12438415; http://dx.doi.org/ 10.1083/jcb.200207128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet 2003; 33:155-61; PMID:12524544; http://dx.doi.org/ 10.1038/ng1080 [DOI] [PubMed] [Google Scholar]
- 68.Longman D, Johnstone IL, Caceres JF. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 2003; 9:881-91; PMID:12810921; http://dx.doi.org/ 10.1261/rna.5420503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iglesias N, Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS Lett 2008; 582:1987-96; PMID:18394429; http://dx.doi.org/ 10.1016/j.febslet.2008.03.038 [DOI] [PubMed] [Google Scholar]
- 70.Azad AK, Tani T, Shiki N, Tsuneyoshi S, Urushiyama S, Ohshima Y. Isolation and molecular characterization of mRNA transport mutants in Schizosaccharomyces pombe. Mol Biol Cell 1997; 8:825-41; PMID:9168469; http://dx.doi.org/ 10.1091/mbc.8.5.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez MS, Gwizdek C, Haguenauer-Tsapis R, Dargemont C. The HECT ubiquitin ligase Rsp5p is required for proper nuclear export of mRNA in Saccharomyces cerevisiae. Traffic 2003; 4:566-75; PMID:12839499; http://dx.doi.org/ 10.1034/j.1600-0854.2003.00115.x [DOI] [PubMed] [Google Scholar]
- 72.Haitani Y, Takagi H. Rsp5 is required for the nuclear export of mRNA of HSF1 and MSN2/4 under stress conditions in Saccharomyces cerevisiae. Genes Cells 2008; 13:105-16; PMID:18233954; http://dx.doi.org/ 10.1111/j.1365-2443.2007.01154.x [DOI] [PubMed] [Google Scholar]
- 73.Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, Rodriguez MS. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J Biol Chem 2005; 280:13401-5; PMID:15713680; http://dx.doi.org/ 10.1074/jbc.C500040200 [DOI] [PubMed] [Google Scholar]
- 74.Gwizdek C, Iglesias N, Rodriguez MS, Ossareh-Nazari B, Hobeika M, Divita G, Stutz F, Dargemont C. Ubiquitin-associated domain of Mex67 synchronizes recruitment of the mRNA export machinery with transcription. Proc Natl Acad Sci U S A 2006; 103:16376-81; PMID:17056718; http://dx.doi.org/ 10.1073/pnas.0607941103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song F, Fan C, Wang X, Goodrich DW. The Thoc1 encoded ribonucleoprotein is a substrate for the NEDD4-1 E3 ubiquitin protein ligase. PloS one 2013; 8:e57995; PMID:23460917; http://dx.doi.org/ 10.1371/journal.pone.0057995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell 2004; 13:201-12; PMID:14759366; http://dx.doi.org/ 10.1016/S1097-2765(04)00030-9 [DOI] [PubMed] [Google Scholar]
- 77.Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Von Dach E, Corbett AH, Dargemont C, Stutz F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev 2010; 24:1927-38; PMID:20810649; http://dx.doi.org/ 10.1101/gad.583310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 2013; 194:43-67; PMID:23633143; http://dx.doi.org/ 10.1534/genetics.112.147470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell 2010; 21:639-49; PMID:20032305; http://dx.doi.org/ 10.1091/mbc.E09-07-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2005; 102:11290-5; PMID:16040803; http://dx.doi.org/ 10.1073/pnas.0503836102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci U S A 2007; 104:8845-50; PMID:17502605; http://dx.doi.org/ 10.1073/pnas.0700765104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zaitseva L, Myers R, Fassati A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol 2006; 4:e332; PMID:17020411; http://dx.doi.org/ 10.1371/journal.pbio.0040332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 2005; 309:140-2; PMID:15905365; http://dx.doi.org/ 10.1126/science.1113346 [DOI] [PubMed] [Google Scholar]
- 84.Whitney ML, Hurto RL, Shaheen HH, Hopper AK. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 2007; 18:2678-86; PMID:17475781; http://dx.doi.org/ 10.1091/mbc.E07-01-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dhungel N, Hopper AK. Beyond tRNA cleavage: novel essential function for yeast tRNA splicing endonuclease unrelated to tRNA processing. Genes Dev 2012; 26:503-14; PMID:22391451; http://dx.doi.org/ 10.1101/gad.183004.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 2003; 14:3266-79; PMID:12925762; http://dx.doi.org/ 10.1091/mbc.E02-11-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 2007; 12:285-97; PMID:17352735; http://dx.doi.org/ 10.1111/j.1365-2443.2007.01056.x [DOI] [PubMed] [Google Scholar]
- 88.Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 1998; 18:6374-86; PMID:9774653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sarkar S, Hopper AK. tRNA nuclear export in saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 1998; 9:3041-55; PMID:9802895; http://dx.doi.org/ 10.1091/mbc.9.11.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kwapisz M, Cholbinski P, Hopper AK, Rousset JP, Zoladek T. Rsp5 ubiquitin ligase modulates translation accuracy in yeast Saccharomyces cerevisiae. RNA 2005; 11:1710-8; PMID:16177134; http://dx.doi.org/ 10.1261/rna.2131605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krsmanovic T, Kolling R. The HECT E3 ubiquitin ligase Rsp5 is important for ubiquitin homeostasis in yeast. FEBS Lett 2004; 577:215-9; PMID:15527788; http://dx.doi.org/ 10.1016/j.febslet.2004.10.006 [DOI] [PubMed] [Google Scholar]
- 92.Grosshans H, Hurt E, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev 2000; 14:830-40; PMID:10766739 [PMC free article] [PubMed] [Google Scholar]
- 93.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev 1998; 12:1678-90; PMID:9620854; http://dx.doi.org/ 10.1101/gad.12.11.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang HY, Hopper AK. Separate responses of karyopherins to glucose and amino acid availability regulate nucleocytoplasmic transport. Mol Biol Cell 2014; 25:2840-52; PMID:25057022; http://dx.doi.org/ 10.1091/mbc.E14-04-0948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hayakawa A, Babour A, Sengmanivong L, Dargemont C. Ubiquitylation of the nuclear pore complex controls nuclear migration during mitosis in S. cerevisiae. J Cell Biol 2012; 196:19-27; PMID:22213798; http://dx.doi.org/ 10.1083/jcb.201108124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen M, Gartenberg MR. Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev 2014; 28:959-70; PMID:24788517; http://dx.doi.org/ 10.1101/gad.236729.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ikegami K, Lieb JD. Integral nuclear pore proteins bind to Pol III-transcribed genes and are required for Pol III transcript processing in C. elegans. Mol Cell 2013; 51:840-9; PMID:24011592; http://dx.doi.org/ 10.1016/j.molcel.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]


