ABSTRACT
Streptococcus pyogenes is a human pathogen responsible for a wide spectrum of diseases ranging from mild to life-threatening infections. During the infectious process, the temporal and spatial expression of pathogenicity factors is tightly controlled by a complex network of protein and RNA regulators acting in response to various environmental signals. Here, we focus on the class of small RNA regulators (sRNAs) and present the first complete analysis of sRNA sequencing data in S. pyogenes. In the SF370 clinical isolate (M1 serotype), we identified 197 and 428 putative regulatory RNAs by visual inspection and bioinformatics screening of the sequencing data, respectively. Only 35 from the 197 candidates identified by visual screening were assigned a predicted function (T-boxes, ribosomal protein leaders, characterized riboswitches or sRNAs), indicating how little is known about sRNA regulation in S. pyogenes. By comparing our list of predicted sRNAs with previous S. pyogenes sRNA screens using bioinformatics or microarrays, 92 novel sRNAs were revealed, including antisense RNAs that are for the first time shown to be expressed in this pathogen. We experimentally validated the expression of 30 novel sRNAs and antisense RNAs. We show that the expression profile of 9 sRNAs including 2 predicted regulatory elements is affected by the endoribonucleases RNase III and/or RNase Y, highlighting the critical role of these enzymes in sRNA regulation.
Keywords: Antisense RNAs, gene expression regulation, leader RNAs, RNA sequencing, riboswitches, Streptococcus pyogenes, small RNAs, T-boxes
Introduction
Regulatory RNAs are present in all kingdoms of life. In bacteria, the family of small RNAs (sRNAs) is typically composed of 50 to 300 nucleotide (nt) long transcripts. Bacterial sRNAs can modulate gene expression through a diversity of mechanisms; for example they can control adaptive responses and play critical roles in metabolic and virulence pathways.1-6 Although mostly characterized in Gram-negative bacteria, novel mechanisms of sRNA action were recently discovered in Gram-positive bacteria.7 sRNA-driven regulations include mechanisms at the transcriptional, post-transcriptional, translational and post-translational levels. While some regulatory RNAs bind target proteins, most of the investigated sRNAs have been shown to act directly on RNA targets. RNA-targeting sRNAs can act in cis at the level of their own locus.8,9 For example, riboswitches respond to intracellular concentrations of metabolites, and antisense RNAs (asRNAs) are encoded on the opposite strand of their RNA targets sharing long, 100% complementary stretches.10 RNA-targeting sRNAs can also act in trans at the post-transcriptional level usually by base pairing with mRNA targets encoded from other loci through complementarity over a small region of 6 to 8 nt.8 Another family of sRNAs consists of the CRISPR RNAs (crRNAs) involved in bacterial and archaeal adaptive immunity against mobile genetic elements. In the interference stage of the immunity, mature short crRNAs guide single or complexed CRISPR-associated (Cas) protein(s) to target invading DNA or RNA.11
The genome of the M1 clinical isolate SF370 of the human pathogen S. pyogenes is 1852441 bp in size, of which 15% consist of intergenic regions (IGRs).12 Computational predictions of sRNAs in this pathogen were done previously using sRNApredict,13,14 SIPHT,15 sRNA scanner in SF370,16 MOSES in M49 GAS17 and sRNAPredict, eQRNA and RNAz in MGAS315.18 Three experimental searches for sRNAs in M1T1GAS (MGAS2221)19 and M49GAS20 using microarrays, in SF370 using the 454 RNA sequencing technology21 and in M1T1GAS (MGAS2221) using Illumina RNA sequencing22 were also performed. The microarray screens19,20 and computational predictions13–16,18 of sRNAs were further analyzed whereas the RNA sequencing analyses were only presented as raw data.21,22 A large number of novel putative sRNAs in S. pyogenes are now available for analysis in phenotypic and mechanistic studies (for details on sRNAs with known regulatory functions in S. pyogenes, see ref.23). Remarkably, none of the published screens of predicted or validated sRNAs in S. pyogenes have reported asRNA expression in this pathogen, most probably because the performed microarray analyses could reveal only sRNAs encoded within IGRs.24
Until now, only a few sRNAs have been studied in detail in S. pyogenes. Pel/sagA RNA (pleiotropic effect locus/streptolysin-associated gene A) and RivX RNA play critical roles in the expression of virulence factors, however their direct targets still remain unknown.25,26 Pel/sagA RNA encodes the virulence factor SLS responsible for the β-hemolytic phenotype of S. pyogenes. Pel RNA also acts as a positive regulator of virulence gene expression and the regulation was described to be strain-specific.25,27 RivX is an sRNA that is encoded immediately downstream of the gene encoding the response regulator RivR (RALP4, a RofA-like protein family of transcriptional regulators) in an M6 serotype, and data suggest that RivX corresponds to a processed form of the rivRX transcript.26 RivR and RivX both activate the Mga (multiple gene activator) regulon that positively controls the expression of a number of virulence-associated regulators and factors.26 The mechanism of FasX (fibronectin/fibrinogen-binding/hemolytic-activity/streptokinase-regulator-X) sRNA is well described.27–29 FasX is part of an operon comprises genes encoding 2 putative histidine kinases (FasBC) and one putative response regulator (FasA).28 FasX RNA affects virulence factor expression using 3 mechanisms. (I) FasX enhances streptokinase virulence factor expression by stabilizing the mRNA of the ska gene encoding streptokinase29; (II) FasX regulates pilus expression and adherence by base pairing to the mRNA of the pilus biosynthesis operon resulting in the destabilization of the mRNA27 and (III) FasX inhibits the translation of cpa mRNA encoding a pilin protein.27,30 It was recently demonstrated that regulation by FasX sRNA is serotype-specific and depends on the presence of genes encoding FasX targets in clinical isolates.30 In addition to these S. pyogenes specific sRNAs, the family of cia-dependent sRNAs (csRNAs), described first in Streptococcus pneumoniae, is present in all streptococcal genomes (with 2 to 6 csRNAs encoded per genome).31,32 In S. pneumoniae, the expression of the csRNAs is regulated by the 2-component system CiaRH (Cia, Competence induction and altered cefotaxime susceptibility) and the csRNAs inter alia target the comC mRNA encoding the precursor of the competence stimulating peptide (CSP).33
The first member of a novel CRISPR-associated sRNA family, trans-activating crRNA (tracrRNA), was initially identified in S. pyogenes and found associated to the type II CRISPR-Cas systems present in hundreds of bacterial species.21,34-36 In S. pyogenes, tracrRNA is involved in the virulence fitness of the pathogen by interfering with the horizontal transfer of temperate phages encoding pathogenicity factors. tracrRNA is critical in both the RNA maturation step of the immune pathway and the interference with invading target phage DNA. tracrRNA forms duplex RNAs with the precursor and processed forms of crRNA. Processed tracrRNA duplexed with mature crRNA forms the dual-tracrRNA:crRNA that guides the Cas9 endonuclease to cleave site-specifically the target DNA.37 The dual-tracrRNA:crRNA was simplified into a single-guide RNA to form with Cas9 the CRISPR-Cas9 complex that has now been widely harnessed for genome editing purposes in a large variety of cells and organisms.21,34,37–41
In Gram-negative bacteria, sRNA/mRNA interactions are often stabilized by the RNA chaperone Hfq.42 As many other low-GC Gram-positive bacteria, S. pyogenes does not encode an Hfq homolog.43 However, streptococcal sRNAs may be assisted by proteins substituting for Hfq, like RNA-binding proteins with chaperone, helicase or other RNA-related activities. The activity of regulatory RNAs in bacteria can also be controlled by ribonucleases (RNases) such as RNase III, RNase E or RNase Y. RNases have critical roles in the stabilization, degradation or maturation of target mRNAs of sRNAs (reviewed in refs.44,45). The highly conserved RNase III is an endoribonuclease capable of cleaving double stranded RNA (dsRNA). One of its main functions is processing of the precursor rRNA (pre-rRNA).46 RNase III also cleaves sRNA/mRNA and asRNA/mRNA duplexes.47,48 In Gram-negative bacteria, RNase E is involved in the turnover of sRNA/mRNA duplexes in complex with Hfq.49 Although streptococci do not encode RNase E, they encode an endoribonuclease having similar cleavage specificity, RNase Y. RNase Y is involved in mRNA and sRNA decay, and riboswitch turnover.50,51 Both RNase III and RNase Y were shown to affect virulence gene expression in bacteria.21,52-54
In this study, we present an analysis of high-throughput sRNA sequencing in the S. pyogenes SF370 clinical isolate. We identified novel putative regulatory sRNAs and demonstrate for the first time expression of asRNAs in this pathogen. The expression of selected asRNAs and novel sRNA candidates was experimentally validated by Northern blot analysis, revealing growth-phase dependent expression of some of the sRNAs investigated. The processing patterns of the asRNAs and sRNAs in RNase III and RNase Y deletion mutants were also investigated.
Results
sRNA sequencing reveals asRNAs and sRNAs in S. pyogenes
To identify novel sRNAs expressed in S. pyogenes SF370, we constructed cDNA libraries from total RNA (depleted from rRNAs) prepared from mid logarithmic phase cultures using the ScriptMiner™ Small RNA-Seq kit. The RNAs were treated with Tobacco Acid Pyrophosphatase (TAP) to incorporate both processed and primary transcripts in the cDNA libraries. The libraries were subjected to Illumina 100 bp single end sequencing. This method is designed for sequencing of small transcripts and strand-specific mapping. Since long transcripts are not sequenced using this protocol, the retrieved reads in this study can represent either single sRNAs or processed parts of longer RNAs.
A total of 302 600 reads were mapped to the reference genome and the number of reads starting (5' start) and ending (3' end) at each single nucleotide position were calculated. The maximal length of the mapped reads was 85 nt with the 3' ends of longer RNAs that could not be identified with certainty. We performed both a visual and a bioinformatics screening for novel sRNAs. For the bioinformatics screening, the reads were analyzed to identify potential 5' ends of RNAs. By screening for 5' ends with at least 10 reads located at a specific position (see Materials and Methods for more details), we identified 560 potential sRNA starts on the chromosome. sRNAs were annotated either as located in coding sequences (CDSs), antisense to CDSs, located in IGRs, as part of 5' untranslated regions (UTRs) (upstream) if they were located less than 50 nt upstream of an annotated gene, or as part of 3' UTRs (downstream) if they were located less than 50 nt downstream of an annotated gene. 406 start sites were located in intergenic regions (137), 5' UTRs (220) and 3' UTRs (49), which is 4.9 times more than expected if the locations were randomly distributed over the chromosome. In contrast, 129 locations were located in CDSs, which is 1.8 times less than expected if the locations were randomly distributed over the chromosome. We also identified 22 putative sRNA starts located antisense to CDSs.
We annotated putative promoters and Rho-independent terminators for the sRNAs identified by bioinformatic screening (Table S1). 76.8% of the start locations of the identified intergenic RNAs were associated with a predicted promoter element located just upstream of the sRNA. This frequency is statistically significantly higher (p-value <2.2 e-16, Kolmogorov-Smirnov test) than the frequency of predicted promoter elements found when the location is randomly chosen on the chromosome (0.5%). 86.8% of predicted sRNAs part of 5' UTRs were associated with a predicted promoter element. Furthermore, 21.1% of all sRNAs had a predicted terminator ≤300 nt downstream of their start location, which is significantly more (p-value <2.2 e-16, Kolmogorov-Smirnov test) than the 4.8% of predicted transcriptional terminators found when the location is randomly chosen on the chromosome. sRNAs found in the 3' UTRs had the highest percentage (44.9%) of predicted terminators (Fig. S1).
In the visual inspection screen for sRNAs, we selected transcripts encoded within IGRs and antisense to annotated transcripts. We identified a total of 197 putative regulatory RNAs including 28 sRNAs predicted to be encoded directly opposite to a transcript, possibly corresponding to asRNAs. 141 sRNAs were found in both visual and bioinformatics screenings of the sRNA sequencing data, of which 14 were annotated as asRNAs. Table S2 gives a summary of the various characteristics of the reads for the visually screened putative sRNAs including size, coordinates, coverage, direction of transcription, predicted location and the identity of genes located upstream and downstream of the sRNA loci. 97 sRNAs are predicted to be part of 5' or 3' UTRs. These sRNAs could be cis-acting RNAs regulating the expression of the associated genes. 58 sRNAs were encoded within IGRs and could function as trans-acting sRNAs. We compared our list of predicted sRNAs identified by visual screening with the lists of previously published sRNAs identified in high-throughput in silico and microarray analyses of S. pyogenes clinical isolates (Table S2). 105 sRNAs were previously identified, from which only 16 were confirmed experimentally by Northern blot analysis14-16,18-21 and 92 are novel sRNAs. In the visual screening, we detected a total of 21 sRNAs encoded within the 4 different prophages regions (370.1 – 370.4) present in strain SF370 (Table S2). These sRNAs could have regulatory functions in the biology of the prophages or in the adaptation of S. pyogenes to its environment.
sRNAs with predicted functions
To validate the quality of our screening methods, we used the Rfam database to retrieve S. pyogenes sRNAs homologous to known sRNAs.55 We annotated 35 sRNAs with predicted functions including known functional sRNAs, ribosomal protein leaders, T-boxes and putative riboswitches (Table S2).
The expression of the 4.5S RNA signal recognition particle (SRP) (Spy_sRNA190336), tmRNA (Spy_sRNA1065030), RNA component of RNase P (Spy_sRNA1365555) and 6S RNA (Spy_sRNA1663522) were validated by Northern blot analysis from S. pyogenes cultures grown at different growth phases (Fig. S2). These sRNAs appeared to be constitutively expressed during bacterial growth with a slight decrease of tmRNA and RNase P RNA expression at the late stationary phase of growth. As previously described, 6S RNA accumulates during the stationary phase.56 Ribosomal protein leader L19, L20, L21, L10 and L13 homologues and thrS, pheST, alaS, valS, glyQS, serS and trsA T-boxes were also identified (Table S2). Northern blot analysis shows that expression of the tsrA T-box is considerably reduced from late logarithmic through stationary phase of growth (Fig. S2). We also performed Northern blot analysis of the purine and glycine riboswitches, the yybP-ykoY element, the metK2 riboswitch, PyrR1 and PyrR2, the asd element and the TPP and FMN riboswitches (Table S1 and Fig. S2). We show that the expression of these sRNAs undergoes reduction starting early stationary phase of growth (Fig. S2).
Regulatory elements regulated by RNases
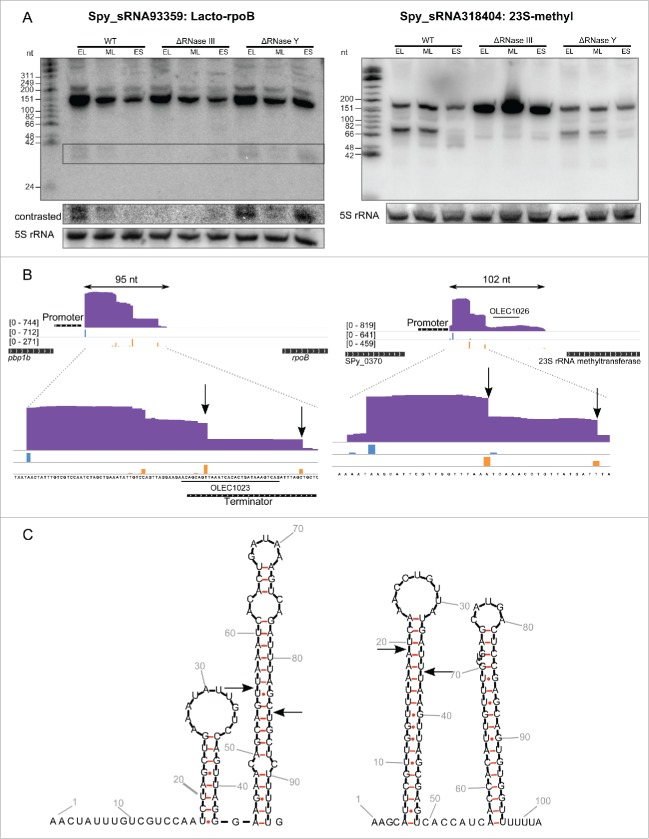
Regulatory elements including Lacto-rpoB (Spy_sRNA93359) and 23S-methyl (Spy_sRNA318404) were detected in our screen (Table S2), and we propose below that the expression of these elements is regulated by RNase III in S. pyogenes.
The Lacto-rpoB motif was first identified by bioinformatics analysis in Lactobacillales, always in the 5' UTR of the rpoB encoding an RNA polymerase subunit, and was suggested to participate in the regulation of rpoB expression.57 In strain SF370, Lacto-rpoB is located between, and in the same orientation as, pbp1b and rpoB and we predicted a putative promoter and a terminator for this element (Fig. 1). We observed several transcript forms of Lacto-rpoB by Northern blot analysis. We visualize 2 long transcripts, and an additional small transcript (<42 nt) present in the wild type and Δrny (RNase Y deletion) strains but absent in the Δrnc (RNase III deletion) mutant (Fig. 1). The small processed transcript (<42 nt) could correspond to the RNase IIIcleavage product of the long primary transcript of the Lacto-rpoB element. We predicted the structure of the Lacto-rpoB motif using RNAfold webserver58,59 and observed that this RNA forms a well-structured RNA with dsRNA regions (Fig. 1). Internal 5' and 3' ends obtained in sRNA sequencing data were positioned on the structure and we observed that these ends could correspond to the 2 nt 3' overhang characteristic of RNase III cleavage (Fig. 1). RNase III cleavage of Lacto-rpoB, based on structure prediction and sRNA sequencing data, would lead to the formation of a 29 nt transcript, corresponding to the <42 nt transcript detected by Northern blot analysis.
Figure 1.
For figure legend see page .Figure 1. (see previous page) Lacto-rpoB and 23S methyl RNA elements are regulated by RNase III in S. pyogenes. A. Northern blot analysis (polyacrylamide gel electrophoresis) of Lacto-rpoB and 23S-methyl RNA expression in WT (SF370), ΔRNase III (Δrnc) and ΔRNase Y (Δrny) strains grown to early logarithmic (EL), mid logarithmic (ML) and early stationary (ES) phases. 5S rRNA is used as loading control. B. Expression profiles of Lacto-rpoB and the 23S-methyl motif with surrounding genes captured using the Integrative Genomics Viewer (IGV) software. The sequence coverage was calculated using BEDTools-Version-2.15.0 and the scale is given in number of reads per million. The distribution of reads starting (5') and ending (3') at each nucleotide position is represented in blue and orange, respectively. The position of the oligonucleotide probes (OLEC) used in Northern blot analysis is indicated. C. Prediction of RNA secondary structure using RNAfold (rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The arrows represent putative cleavages by RNase III (nucleotides determined by analyzing the 5' and 3' ends of the sRNAs in sRNA sequencing data).
Similarly to the Lacto-rpoB element, the 23S-methyl motif is found upstream of the 23S rRNA methyltransferase in Lactobacillales and might regulate its expression.60 This RNA interacting enzyme could use the 23S-methyl motif to autoregulate its expression, comparable to the autoregulation of ribosomal protein genes.60 We observed 2 transcripts of <200 nt and ∼70 nt by Northern blot. In the Δrnc mutant strain, the <200 nt transcript is accumulating while the ∼70 nt of the transcript is absent (Fig. 1). We propose that the <200 nt (102 nt in sRNA sequencing) corresponds to a primary transcript while the ∼70 nt form corresponds to a processed transcript that could be generated by RNase III-mediated cleavage. The structure of the 23S-methyl motif was predicted using the RNAfold webserver58,59 and showed that this RNA contains 2 long hairpins as previously described.60 The ending of the second hairpin with a stretch of uridines may indicate a Rho-independent transcription terminator that could be involved in a transcription regulation mechanism.60 The internal 5' and 3' ends obtained in sRNA sequencing data are located within the first hairpin with a 2 nt 3' overhang characteristic of RNase III. A processed transcript of 66 nt results from this cleavage, which is in accordance with the ∼70 nt form observed by Northern blot analysis.
CiaR regulated sRNAs (csRNAs)
We detected 2 sRNAs, Spy_sRNA195261 and Spy_sRNA1721621, corresponding to cia-dependent sRNAs (csRNAs), csRNA15 and csRNA25 from the S. pyogenes clinical isolate MGAS31532 (Fig. S3). These sRNAs contain the TTTAAG-N5-TTTAAG consensus binding sequence of CiaR in their promoter region (note that for Spy_sRNA1721621, the first T is mutated and replaced by a C). The two sRNAs also contain the putative anti-RBS and anti-start codon sequences, previously described. We performed in vitro electrophoretic mobility shift assays of the Spy_sRNA195261 and Spy_sRNA1721621 promoter regions with purified CiaR (using a DNA template without the consensus binding sequence as a negative control). Other potential binding sequences containing various degrees of degenerated consensus sequences recognized by CiaR were selected as controls for the binding specificity (Fig. S3). We show that CiaR binds to all promoters albeit with different affinities. Higher binding affinity was observed for promoters with a sequence having a higher degree of similarity to the consensus sequence, and mutations within the consensus motif decreased the affinity of CiaR to the DNA. We created an in-frame ciaR deletion mutant and show in vivo by Northern blot analysis (Fig. S3) that the expression of the 2 csRNAs, Spy_sRNA195261 and Spy_sRNA1721621, is down-regulated in the absence of CiaR in S. pyogenes, as described in S. pneumoniae.32 A csRNA Spy_sRNA195261-deficient mutant was constructed in S. pyogenes SF370. A complete proteomic analysis of the mutant did not show any variation of protein expression after false positive reduction compared to the wild type strain in the culture conditions tested (data not shown).
Novel sRNAs in S. pyogenes
We validated the expression of 15 novel sRNA candidates by Northern blot analysis, from 31 sRNAs that were analyzed (Table 1, Table S2, Fig. S4). Only three sRNAs showed a constant expression during growth and 6 sRNAs had their expression reduced during early stationary growth phase compared to early- and mid-logarithmic growth phases. Some of the sRNAs were detected as several transcript forms of different sizes, which suggests, as mentioned previously, that a large number of identified sRNAs could be processed forms originating from longer transcripts.
Table 1.
List of selected sRNAs. The sRNA identifier, location in prophages, strand, surrounding genes (immediately located upstream and downstream of the sRNAs) and coverage (number of cDNA reads calculated using SAMtools) are indicated.
| Name | Phage | Strand | Upstream | Downstream | Coverage | ||
|---|---|---|---|---|---|---|---|
| Spy_sRNA542268 | 370.1 | − | SPy_0671 | hypothetical protein | SPy_0672 | hypothetical protein | 8 |
| Spy_sRNA544833 | 370.1 | + | SPy_0677 | phage associated protein | SPy_0678 | phage associated protein | 33 |
| Spy_sRNA694150 | + | SPy_0841 | RNA binding protein | SPy_0843 | hypothetical protein | 23 | |
| Spy_sRNA779816 | 370.3 | + | SPy_0937 | int3: Integrase | SPy_0938 | hypothetical protein | 405 |
| Spy_sRNA1110925 | − | SPy_1340 | major facilitator superfamily permease | SPy_1343 | hypothetical protein | 82 | |
| Spy_sRNA1186876 | − | SPy_1432 | pyrD: dihydroorotate dehydrogenase 1A | SPy_1434 | heavy metal-transporting ATPase | 23 | |
| Spy_sRNA1212757 | 370.2 | + | SPy_1469 | hypothetical protein | SPy_1470 | hypothetical protein | 37 |
| Spy_sRNA1222613 | 370.2 | − | SPy_1488 | int2: integrase | SPy_1489 | hlpA: histone-like DNA binding protein | 116 |
| Spy_sRNA1260092 | − | SPy_1531 | dpr: Dps-like peroxide resistance protein | SPy_1532 | leader peptidase family protein | 219 | |
| Spy_sRNA1300419 | + | SPy_1577 | aroB: 3-dehydroquinate synthase | SPy_1580 | acetate kinase | 90 | |
| Spy_sRNA1571135 | − | SPy_1894 | pyrG: CTP synthetase | SPy_1895 | rpoE: DNA-directed RNA polymerase subunit delta | 26 | |
| Spy_sRNA1755445 | − | SPy_2099 | trehalose operon transcriptional repressor | SPy_2102 | transcriptional regulator MarR family | 2176 | |
| Spy_sRNA1774740 | 370.4 | + | SPy_2122 | int4: integrase | SPy_2125 | repressor protein | 46 |
| Spy_sRNA1775122 | 370.4 | − | SPy_2122 | int4: integrase | SPy_2125 | repressor protein | 102 |
| Spy_sRNA1786666 | 370.4 | + | SPy_2147 | hypothetical protein | SPy_2148 | mutS: DNA mismatch repair protein MutS | 206 |
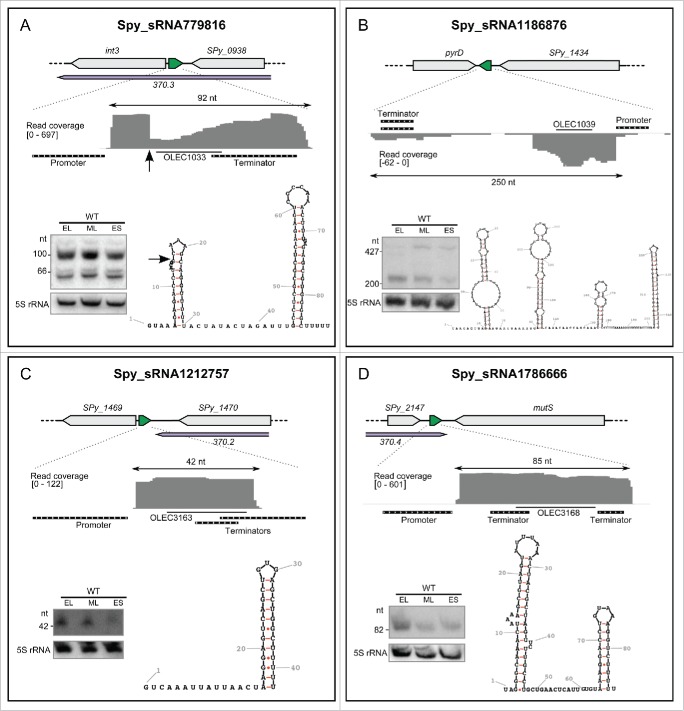
For a detailed analysis, we selected 4 distinct sRNAs as examples, Spy_sRNA779816, Spy_sRNA1186876, Spy_sRNA1212757 and Spy_sRNA1786666, that have both a predicted promoter element and a predicted terminator, and represent different lengths and regions (i.e both prophage and non-prophage regions) (Fig. 2, Fig. S5). Their expression was confirmed by Northern blot analysis using total RNA extracted from S. pyogenes SF370 cultured at different growth phases (early logarithmic, mid logarithmic and early stationary phases), and their secondary structures were predicted with RNAfold58,59 (Fig. 2, Fig. S5).
Figure 2.
Expression profiles of selected sRNAs. For each sRNA, the locus is depicted with the sRNA in green and the surrounding genes in gray. The prophage regions are indicated in purple. The RNA sequencing expression profiles are captured using the Integrative Genomics Viewer (IGV) software. The sequence coverage was calculated using BEDTools-Version-2.15.0 and the scale is given in number of reads per million. The putative promoters and terminators are indicated in black. Northern blot analysis (PAGE) were performed in WT (SF370) strains grown to early logarithmic (EL), mid logarithmic (ML) and early stationary (ES) phases. The position of the oligonucleotide probes (OLEC) used in Northern blot analysis is indicated. The 5S rRNA is used as a loading control. Folding for the sRNAs was predicted using RNAfold (rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The arrow in (A) represent a putative cleavage site in the RNA. See also Figures S4 and S5 for additional information regarding sRNA sequencing (expression profiles by Northern blot and RNA sequencing analyses, sequence conservation).
Spy_sRNA779816 is encoded between the int3 and Spy_0938 genes of prophage 370.3. Two main forms of the sRNA, with sizes approximately corresponding to those inferred from RNAseq data, were detected by Northern blot. We propose that the longer form (89 nt in sRNA sequencing data) is a primary transcript, which is processed into the smaller form at a position identified in the sRNA sequencing data (Fig. 2A). This sRNA is also present in Manfredo and NZ131 S. pyogenes strains and Streptococcus parauberis KCTC 11537.
Spy_sRNA1186876 is encoded between SPy_1432 (pyrD dihydroorotate dehydrogenase 1A) and SPy_1434 (heavy metal-transporting ATPase). We detected 2 forms of this sRNA of ∼500 nt and >200 nt in size by Northern blot analysis. We could not identify any reads or predominant 5' ends in our sequencing data that would correspond to the longer detected transcript, and thus we are unable to comment on its origin. Since the size of the shorter transcript perfectly corresponds to the size of transcript starting from the predicted promoter to the predicted terminator of the sRNA (143 nt), we hypothesize that the shorter RNA is a primary transcript and does not originate from the longer RNA. This sRNA is also present in Manfredo and NZ131 S. pyogenes strains and in Streptococcus dysgalactiae subsp. equisimilis GGS_124.
Spy_sRNA1212757 is encoded between 2 hypothetical genes in the vicinity of prophage 370.2 and we observed by Northern blot analysis a small transcript of 42 nt corresponding to the size visualized in the sRNA sequencing data (Fig. 2, Fig. S4, Fig. S5). This sRNA is unusually short, but a defined Northern blot signal, convincing promoter and terminator predictions, together with the unstructured 5' end that should be prone to degradation indicates that this transcript is functional. This RNA is also present with 100% conservation in the S. pyogenes clinical isolates, Manfredo, A20, and HSC5.
The last example, Spy_sRNA1786666, is encoded at one extremity of the 370.4 prophage region between SPy_2147 and SPy_2148 (mutS: DNA mismatch repair protein MutS). In accordance with the sRNA sequencing data, we observed a single transcript of >82 nt by Northern blot analysis (Fig. 2, Fig. S4, Fig. S5). This RNA is also present with 100% conservation in Manfredo and MGAS10270 S. pyogenes strains and in S. dysgalactiae subsp. equisimilis 167.
Novel putative sRNAs regulated by RNases
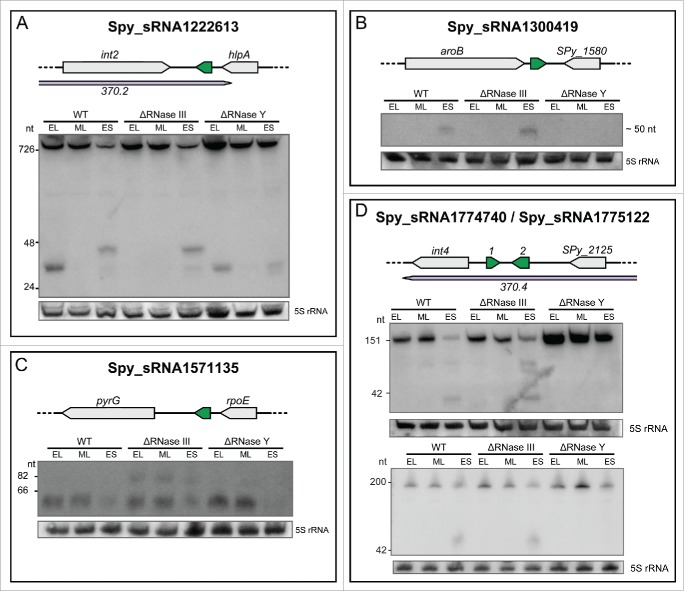
Northern blot analysis shows that the expression of 2 novel sRNAs is regulated by RNase III (Spy_sRNA1222613 and Spy_sRNA1571135) and the expression of 4 novel sRNAs is regulated by RNase Y (Spy_sRNA1222613, Spy_sRNA1300419, Spy_sRNA1774740 and Spy_sRNA1775122) (Fig. 3 and Fig. S4).
Figure 3.
Expression profiles of sRNAs regulated by RNases. Northern blot analysis of selected sRNAs showing a variation in expression or processing between WT (SF370), ΔRNase III (Δrnc) and ΔRNase Y (Δrny) strains grown to early logarithmic (EL), mid logarithmic (ML) and early stationary (ES) phases. 5S rRNA is used as a loading control. For each sRNA, the locus is depicted with the sRNA in green and the surrounding genes in gray. The prophage regions are indicated in purple. For a detailed set of Northern blots and sRNA sequencing expression patterns, refer to Figure S4.
Spy_sRNA1222613 is encoded within the prophage 370.2 between the int2 and hlpA genes and overlaps with the 3' UTR of hlpA (histone-like DNA-binding protein). HlpA is described to bind to lipoteichoic acid and epithelial cells and was shown to play a role in the virulence of S. pyogenes.61 In the wild type strain, Northern blot analysis shows that Spy_sRNA1222613 is part of a long transcript (>800 nt) expressed at all phases of growth. In addition, 2 smaller transcripts (between 24 and 48 nt) are visible, one small transcript (∼30 nt) during the early logarithmic growth phase and the other transcript (∼48 nt) during the early stationary growth phase. However, no transcript was detected during the mid logarithmic phase of growth (Fig. 3A and Fig. S4). The smallest transcript (∼30 nt) is absent in the Δrnc strain, suggesting that this short form originates from the 3' UTR hlpA mRNA by processing by RNase III. The ∼48 nt form is absent in the Δrny strain, probably resulting from RNase Y processing of the 3' UTR hlpA mRNA (Fig. 3A). The 3' UTR of the hlpA mRNA seems to be tightly regulated during growth in an RNase III and RNase Y dependent manner. It is still to be determined whether the 2 small transcripts are products originating through processing via an interaction with a target or whether they originate from hlpA growth dependent autoregulation.
Spy_sRNA1300419 is encoded within the 3' UTR of aroB (3-dehydroquinate synthase) and its expression was detected by Northern blot only during early stationary phase of growth (Fig. 3B). In the Δrny mutant, this ∼50 nt transcript is absent, indicating that Spy_sRNA1300419 probably corresponds to a processed form of 3' UTR aroB mRNA generated by this endoribonuclease (Fig. 3B).
Spy_sRNA1571135 is encoded in the 3' UTR of rpoE (DNA-directed RNA polymerase subunit delta). Its expression is comparable between wild type, Δrnc and Δrny strains, however in the Δrnc mutant, a longer transcript is also present (Fig. 3C). We hypothesize that expression of this transcript is regulated by RNase III in the wild type and Δrny strains (Fig. 3C).
Spy_sRNA1774740 and Spy_sRNA1775122 are encoded next to each other on opposite strands between int4 (integrase) and SPy_2125 (a repressor protein) in the genome of prophage 370.4. Northern blot analysis shows 3 different transcripts for Spy_sRNA1774740 and 2 forms for Spy_sRNA1775122 (Fig. 3D). The expression of the longest transcript form (∼150 nt) of Spy_sRNA1774740 is upregulated in the absence of RNase Y. The two smaller transcripts are only visible during the early stationary phase of growth in the wild type and Δrnc but are absent in the Δrny mutant (Fig. 3D). The smallest transcript form of Spy_sRNA1775122 is also present during the early stationary phase of growth and is absent in the Δrny mutant. The small transcript forms of both sRNAs probably result from RNase Y cleavage.
Antisense RNAs
Remarkably, our analysis of the sRNA sequencing data allowed identification of novel sRNAs (Table S2, Table 2) located antisense to annotated transcripts, thus being putative asRNAs. We divided the asRNAs into 3 categories based on their location (Table 2), namely directly antisense to the CDS, or antisense to the 5' or 3' UTRs of transcripts. A total of 36 putative asRNAs were retrieved, 28 by visual screening (Table S2) and 22 by the bioinformatics screening (Table S1), with 14 found in both screenings. We selected 21 transcripts from the visual screening for further analysis (Table S2). The expression of 10 RNAs antisense to UTRs and 5 RNAs antisense to CDSs was validated at different growth phases (early logarithmic, mid logarithmic and early stationary phases) in the wild type, Δrnc and Δrny strains by Northern blot analysis (Table 2, Fig. S6).
Table 2.
List of selected asRNAs. The asRNA identifier, location in prophages, strand, putative target genes and coverage (number of cDNA reads calculated using SAMtools) are indicated. Classification: asRNAs from 3 different families were retrieved from the sRNA sequencing data. asRNAs are depicted in red and putative target mRNAs are indicated in purple. 5' UTR and 3' UTR correspond to asRNAs that are complementary to the 5' and 3' UTRs of their putative target mRNAs, respectively. CDS corresponds to asRNAs that are complementary to the coding sequences of their putative target mRNAs. UTR: untranslated region; CDS: coding sequence.
| Classification | Name | Phage | Strand | mRNA putative target gene | Coverage |
|---|---|---|---|---|---|
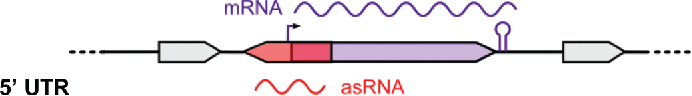 |
Spy_sRNA392987 | + | SPy_0481: transposase | 220 | |
| Spy_sRNA480642 | + | SPy_0598: phosphoglycerate mutase | 108 | ||
| Spy_sRNA480696 | − | SPy_0596: hypothetical protein | 71 | ||
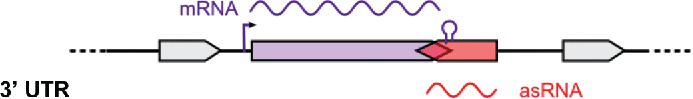 |
Spy_sRNA477741 | − | SPy_0593: hypothetical protein | 100 | |
| Spy_sRNA531081 | 370.1 | + | SPy_0656: hypothetical protein | 140 | |
| Spy_sRNA532788 | 370.1 | − | SPy_0658: hypothetical protein | 68 | |
| Spy_sRNA800369 | 370.3 | − | SPy_0978: hypothetical protein | 878 | |
| Spy_sRNA1221370 | 370.2 | − | SPy_1487: hypothetical protein | 136 | |
| Spy_sRNA1718031 | − | SPy_2063: hypothetical protein | 46 | ||
| Spy_sRNA1778807 | − | SPy_2129: hypothetical protein | 334 | ||
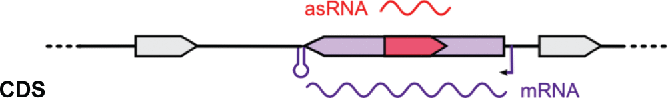 |
Spy_sRNA728180 | + | SPy_0880: mvaS.1, 3-hydroxy-3-0methylglutaryl-coenzyme A | 21 | |
| Spy_sRNA924338 | − | SPy_1128: eutD phosphotransacetylase | 29 | ||
| Spy_sRNA1032107 | + | SPy_1251: tRNA pseudouridine synthase B | 37 | ||
| Spy_sRNA1063616 | − | SPy_1287: ABC transporter permease | 155 | ||
| Spy_sRNA1167444 | + | SPy_1408: comEC, competence protein | 68 |
Most asRNAs seem to be constitutively expressed during growth, with 3 asRNAs that were downregulated. Several transcript forms were visualized for 7 asRNAs. The asRNA sizes inferred from the Northern blot data are sometimes higher than the ones retrieved from RNA sequencing, which is an artifact of our sequencing method directed toward detection of short transcripts. For most of the asRNA candidates, it is likely that they are products of processing or degradation of longer RNAs. Interestingly, 9 asRNAs were expressed as short, defined RNA species of up to 150 nt in length.
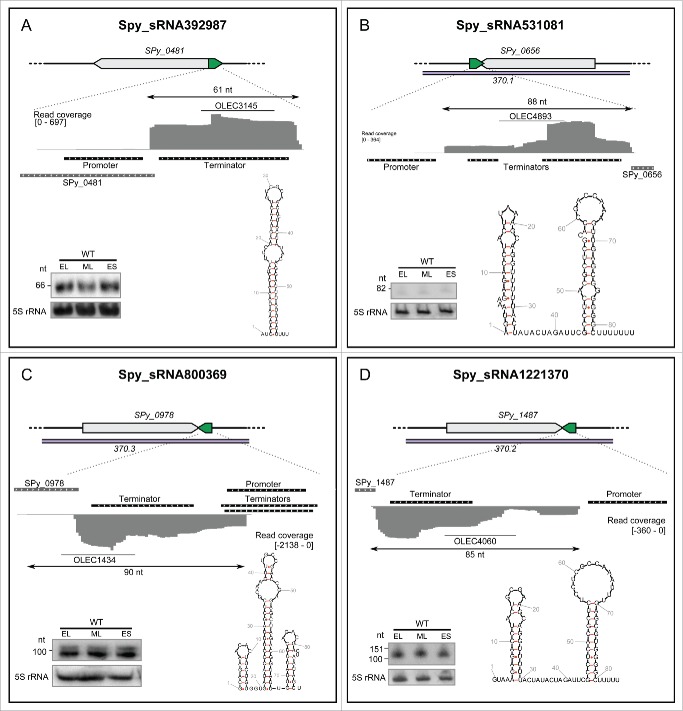
Four asRNAs harboring both a promoter and terminator, with a defined size smaller than 150 nt as observed by Northern blot analysis were selected as examples (Fig. 4, Fig. S7). We hypothesize that these 4 asRNAs are primary transcripts. The secondary structures of the selected asRNAs was predicted using RNAfold58,59 (Fig. 4). Spy_sRNA392987 is located antisense to the 5' UTR of the pseudogene SPy_0481 (transposase). The sizes of this sRNA retrieved by sRNA sequencing and Northern blot analysis are identical (61 nt) (Fig. 4). Spy_sRNA392987 is present at one or 2 loci in numerous strains of S. pyogenes, and in Streptococcus agalactiae and S. dysgalactiae (Fig. S8). Spy_sRNA531081, Spy_sRNA800369 and Spy_sRNA1221370 are encoded antisense to the 3' UTRs of phage hypothetical genes (respectively SPy_0656, SPy_0978 and SPy_1487). Spy_sRNA531081 is conserved in various Streptococcus. pyogenes strains, and in S. agalactiae and S. equi. Spy_sRNA800369 is only found in S. pyogenes Alab49. Spy_sRNA1221370 is present in various S. pyogenes strains and in S. equi and S. agalactiae (Fig. S8).
Figure 4.
Expression profiles of selected asRNAs. For each asRNA, the locus is depicted with the asRNA in green and the antisense genes in gray. The prophage regions are indicated in purple. The RNA sequencing expression profiles are captured using the Integrative Genomics Viewer (IGV) software. The sequence coverage was calculated using BEDTools-Version-2.15.0 and the scale is given in number of reads per million. The putative promoters and terminators are indicated in black. Northern blot analysis (polyacrylamide gel electrophoresis) was performed in WT (SF370) strains grown to early logarithmic (EL), mid logarithmic (ML) and early stationary (ES) phases. The position of the oligonucleotide probes (OLEC) used in Northern blot analysis is indicated below the sequence coverage. The 5S rRNA is used as a loading control. Folding for the sRNAs was predicted using RNAfold (rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). See also Figures S6, S7 and S8 for additional information regarding sRNAs.
asRNAs regulated by RNases
We observed by Northern blot analysis that 2 asRNAs, Spy_sRNA477741 and Spy_sRNA480696, are differently regulated in the RNase deletion mutants (Fig. 5).
Figure 5.
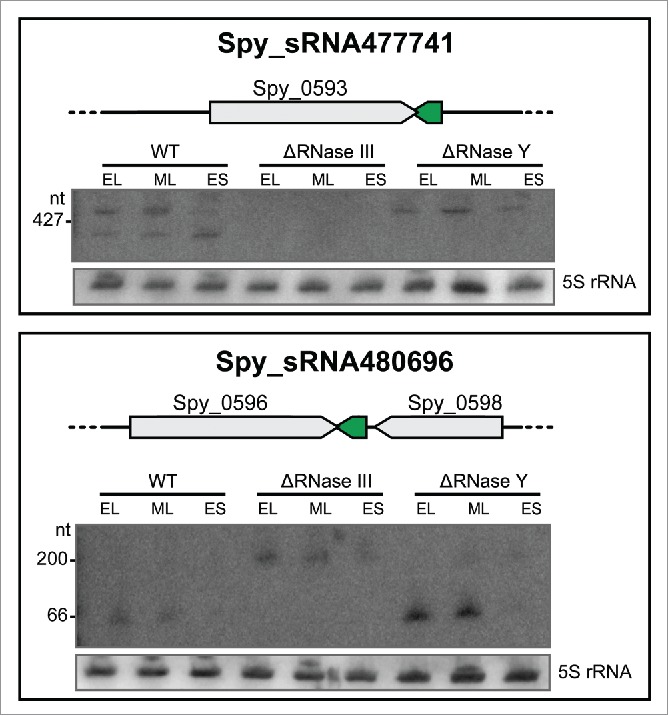
Expression profiles of asRNAs regulated by RNases. Northern blot analysis of selected asRNAs showing a variation in expression or processing between WT (SF370), ΔRNase III (Δrnc) and ΔRNase Y (Δrny) strains grown to early logarithmic (EL), mid logarithmic (ML) and early stationary (ES) phases. 5S rRNA is used as a loading control. For a detailed set of Northern blots and sRNA sequencing expression patterns, refer to Figure S6.
Spy_sRNA477741 is located antisense to the 3' UTR of SPy_0593 hypothetical protein. Two long transcripts of >400 nt are detected in the wild type strain (Fig. 5). Both transcripts are absent in the Δrnc strain, and only the longer transcript is present in the Δrny strain (Fig. 5), suggesting that the longest Spy_sRNA477741 transcript originates from RNase III cleavage and the smallest transcript is a product of RNase Y cleavage.
Spy_sRNA480696 is encoded between SPy_0596 (hypothetical protein) and SPy_0598 (phosphoglycerate mutase). The 3' UTRs of both SPy_0596 and SPy_0598 are overlapping. Spy_sRNA480696 is antisense to SPy_0596 and Northern blot analysis shows a ∼66 nt transcript in the wild type and Δrny strains. In the Δrnc deletion mutant, the transcript is longer, suggesting that the ∼66 nt form may be produced by RNase III cleavage (Fig. 5).
The defined sizes and convincing promoter and terminator predictions of these described asRNAs indicate that at least a subset of the new identified RNAs constitute products of specific and not pervasive transcription. The differential expression patterns and detected processings by RNases further suggest that these asRNAs could be functional, regulatory sRNA species. The biological function and exact nature of longer asRNA transcripts remain to be determined.
Discussion
In this study, we aimed to identify novel sRNAs expressed in S. pyogenes. An analysis of sRNA sequencing in strain SF370 cultured to mid logarithmic phase revealed a total of 197 putative sRNAs from which 92 are new, located in 5' UTRs, 3' UTRs, IGRs or antisense to transcripts.
The sRNA screening was validated by the retrieving of a few regulatory RNAs that were already identified and studied in S. pyogenes, namely FasX RNA (Spy_sRNA213738), Pel RNA (Spy_sRNA598538) and tracrRNA (Spy_sRNA854545). However, we could not detect the expression of RivX RNA in the sRNA sequencing data of SF370. RivX could either be specific to strain JRS4 (M6 serotype) or expressed only in specific conditions in strain SF370.21,37 The quality of our screening method was also validated by the retrieval of homologues of known functional sRNAs such as the 4.5S RNA, tmRNA, the RNA from RNase P, the 6S RNA and the 2 pRNAs (RNA products) that are transcribed from both strands of the 6S RNA during the sequestration of the RNA polymerase.62 Ribosomal protein leaders, T-boxes and known predicted riboswitches (purine, glycine, yybP-ykoY element, metK2, Pyr1 and Pyr2, asd, TPP and FMN) were also retrieved in our data.
We also found CiaR regulated csRNAs, as previously described in ref.32. CiaR is the response regulator of the 2-component system CiaRH. CiaR binds to the DNA sequence TTTAAG-N5-TTTAAG in promoter regions and regulates the expression of the associated downstream gene.32 CiaRH has regulatory functions in various phenotypes like ß-lactam resistance, autolysis, virulence and competence development, however the molecular mechanisms involved are still unknown.32 In S. pneumoniae, CiaR can bind to the promoters of 5 small non-coding csRNAs.31 In this bacterial species, the expression of 6 genes is under the control of csRNA-mediated regulation, including the comC mRNA, which encodes the precursor of CSP, defining a link between CiaR and competence control.33 It has also been described that all streptococcal genomes contain between 2 and 6 csRNA genes.32 All csRNAs are highly similar to one another as they all contain a UCCUCC sequence (putative anti-RBS) and a CCUC(N6)CAU sequence (putative anti-start codon). In this study, we observe that both Spy_sRNA195261 and Spy_sRNA1721621 show several transcript forms that are expressed from the lag to the early stationary phase of growth (Fig. S3). The Spy_sRNA1721621 expression also decreases during the transition between logarithmic and stationary phases of growth. The expression of both sRNA transcripts is reduced in a ciaR deficient mutant (Fig. S3). The study of the proteome of the Spy_sRNA195261 deletion mutant did not provide any indication with regard to a putative function for this RNA. Unlike S. pneumoniae, S. pyogenes is not naturally transformable and the function of the CiaRH homolog in this species remains still unclear.
For the first time, we show expression of asRNAs in S. pyogenes. asRNAs are encoded on the DNA strand opposite to their putative target, therefore being perfectly complementary. Antisense transcripts have been described opposite to transcripts encoding transposases, potentially toxic proteins, transcription regulators, metabolic enzymes, or transcription factors. asRNAs can overlap either with the 5' UTRs, 3' UTRs or CDSs of their target and interfere with mRNA transcription, stability or translation at the condition that both RNAs are expressed at the same time in the same cell. Although the exact function of most asRNAs still remain to be investigated,10 they act mostly by reducing the expression of the sense gene.63 Their function is often associated with RNase activity.64 For example, in Staphylococcus aureus, sense/antisense overlapping transcripts are processed by RNase III, generating a collection of short RNA fragments (20 nt on average).65,66 More than 300 asRNAs, forming a >40 nt complementary duplex with their target that is recognized by RNase III, were also identified in E. coli.48 asRNAs can also form RNA duplexes with target mRNAs, resulting in inhibition of target decay.64,67 Surprisingly, none of the published screens of S. pyogenes sRNAs have reported the expression of asRNAs, probably because of differences in the methods used (microarray analysis and computational predictions).24 In general, technical difficulties to distinguish from which strand the RNAs are expressed delayed the discovery of bacterial asRNAs compared to other trans-acting small RNAs. However, insilico searches led to the identification of 63 asRNAs in S. agalactiae, from which 3 were validated by Northern blotting analysis.68 Two sRNAs and one sRNA were shown to regulate their target expression negatively and positively, respectively. In the present study, we identified 28 putative asRNAs encoded antisense to 5' UTRs, 3' UTRs or CDSs by visual screening, from which 15 were validated by Northern blot analysis. Although some asRNAs have their own promoter and terminator, we show that most of these small transcripts could originate from longer transcripts (not detected in the sRNA sequencing data) (Fig. S6). A class of asRNAs that overlap with the 5' UTRs of transposase genes was described in various bacteria and archaea. In Methanosarcina mazei, these asRNAs are differentially expressed in response to nitrogen, and therefore transposon mobility could be regulated at the post-transcriptional level in a nitrogen-dependent manner.69 In E. coli, asRNAs were shown to regulate the translation of transposases in Tn10 and Tn30 transposons70,71 and similar asRNAs were also found in Sulfolobus solfataricus.72 We speculate that asRNA Spy_sRNA392987, which is also located in the 5' UTR of the transposase mRNA, could have a similar role.
To characterize sRNAs further, we studied the expression of selected sRNAs in strains lacking the genes encoding RNase III or RNase Y. These two endoribonucleases are not essential in S. pyogenes, and they have been shown to have critical roles in the regulation of sRNA expression in other bacterial species. RNase III plays a role in the degradation of mRNAs during translational silencing by sRNAs.46 RNase Y acts in the degradation and processing of mRNAs and sRNAs, as well as in the initiation of riboswitch turnover.51,52,73 We found that expression of the Lacto-rpoB and the 23S methyl putative regulatory elements is regulated by RNase III. The expression of 2 additional sRNAs (Spy_sRNA1222613 and Spy_sRNA1571135) is also regulated by RNase III, and the expression of 4 sRNAs (Spy_sRNA1222613, Spy_sRNA1300419, Spy_sRNA1774740 and Spy_sRNA1775122) is regulated by RNase Y. We also show that RNase III regulates the expression of 2 asRNAs, Spy_sRNA477741 and Spy_sRNA480696, and that RNase Y regulates the expression of one asRNA, Spy_sRNA477741. These regulations may suggest the involvement of these sRNAs or asRNAs in the direct regulation of target mRNA expression. In addition, the possible processing and degradation of sRNAs from all classes (cis-acting, putative trans-acting regulatory RNAs and asRNAs) by either of these enzymes highlight the role of sRNA regulation by processing in S. pyogenes.
Conclusion
To conclude, we identified asRNA candidates and new putative regulatory RNAs in the human pathogen S. pyogenes during growth in axenic cultures using sRNA sequencing. Our analysis increases the inventory of putative sRNAs with regulatory functions in this pathogen. We established a link to the mechanism of action of selected sRNAs and asRNAs by studying their transcription in two RNase deletion mutants (RNase III and RNase Y). Our observations that expression of these RNAs is tightly regulated, by either growth phase or RNases may predict that they have regulatory roles in S. pyogenes. Future studies should aim to sRNA target identification and elucidation of specific mechanisms of action in this pathogen. Despite the constant improvement of target prediction methods, the challenge remains to demonstrate the regulatory roles and molecular mechanisms of action of this increasing collection of sRNAs and asRNAs.
Materials and methods
Bacterial strains and growth conditions
Table S3 describes the bacterial strains used in this study. S. pyogenes M1 GAS SF370 (wild type, ATCC 700294) and derivative deletion mutants were cultured at 37°C without agitation in a 5% CO2 atmosphere. Todd Hewitt broth supplemented with 0.2% yeast extract and plates containing tryptic soy agar supplemented with 3% sheep blood were used as liquid and solid media, respectively. When required, antibiotics were added to the medium at the following final concentrations: erythromycin, 3 μg/ml; kanamycin, 300 μg/ml. Bacterial growth was monitored at regular time intervals at OD620nm with a microplate reader using 200 μl of culture (Eon™, biotek®).
Bacterial transformation
When not stated otherwise, electrocompetent S. pyogenes cells were prepared as ref.74 For the chromosomal deletion of rny (encoding RNase Y), bacterial transformation was performed as follows. Cells were grown overnight in 5 ml of THY medium containing 0.5 M sucrose and 80 mM L-Threonine. The overnight culture was diluted 1/100 in 500 ml of the same media and grown until mid logarithmic phase. The culture was centrifuged and the pellets were washed 3 times in 50 ml of ice cold 0.5 M sucrose and resuspended in 2 ml of ice cold 0.5 M sucrose. 100 μl of GAS electro-competent cells were transformed with 1 μg DNA under electroporation conditions of 1.8 kV, 25 μF and 400 Ω. The bacteria were grown in 6 ml of THY immediately after transformation and incubated for 2 hours at 37°C, 5% CO2 before being spread on the appropriate selection plates.
Chromosomal deletion of RNase III and RNase Y
The rnc (encoding RNase III) deletion mutant used in this study was previously generated in S. pyogenes SF370.21 The in-frame deletion of rny (encoding RNase Y) in strain SF370 was performed as follows. Amplification of DNA sequences located upstream and downstream of the rny coding sequence was performed using Phusion® polymerase (Thermo Scientific) and primer pairs OLEC1998/OLEC1999 and OLEC2000/OLEC2501, respectively. An erythromycin resistance cassette flanked by the lox71 and lox66 sites75 was amplified using OLEC1943 and OLEC1932. The three fragments (upstream of the coding sequence, erythromycin resistance cassette and downstream of the coding sequence) were ligated together by PCR-mediated ligation using OLEC1998 and OLEC2501. The linear fragment obtained was purified and used in transformation experiments using electrocompetent S. pyogenes SF370 as described above. Following homologous recombination at the level of upstream and downstream sequences of the rny gene, erythromycin resistant cells were selected and the gene replacement event was verified by PCR analysis. Clones harboring the rny gene replaced with the erythromycin resistance cassette were transformed with pEC455, a plasmid encoding kanamycin resistance and the Cre recombinase under the control of a constitutive promoter to promote excision of the erythromycin cassette through resolution at the lox sites. Clones resistant to kanamycin and sensitive to erythromycin were selected and cultured in THY without antibiotic to eliminate pEC455. Clones sensitive to erythromycin and kanamycin were further selected, and the replacement of the rny gene with the resolved lox72 site was verified by PCR and sequencing analyses.
Chromosomal deletion of ciaR
The in-frame deletion of the ciaR gene in S. pyogenes SF370 was constructed as follows. Amplification of the upstream and downstream sequences of the ciaR coding sequence was performed using Phusion® polymerase (Thermo Scientific) and primer pairs OLEC1350/OLEC1351 and OLEC1352/OLEC1353, respectively. The two fragments were ligated by PCR using OLEC1350 and OLEC1353. The PCR product was digested by BamHI and EcoRI and cloned into pEC214 to generate pEC277. The ciaR coding sequence overlaps the ciaH coding sequence from the same operon. To avoid a mis-translation of ciaH, a silent mutation of the stop codon of ciaR (within the ciaH coding sequence) was created without modifying the ciaH coding sequence. pEC277 was purified and introduced into S. pyogenes SF370. Kanamycin-resistant and blue clones were selected on TSA blood plates containing X-gal at 28°C. A series of temperature shifts at 37°C (integration of pEC277), 28°C (excision of the plasmid) and 37°C (elimination of the plasmid) were performed. Kanamycin sensitive, white clones with excised pEC277 were selected and the correct deletion of ciaR in the mutant strains was verified by PCR analysis. The deletion event was further confirmed by Southern blot, Northern blot, sequencing and RT-PCR analyses.
RNA extraction
Total RNA from S. pyogenes SF370 wild type, deletion mutants and complemented strains was prepared using TRIzol (Sigma-Aldrich)/chloroform extraction and isopropanol precipitation from samples collected at several phases of growth (early logarithmic phase, EL; mid logarithmic phase, ML, and early stationary phrase, ES). RNA concentration and integrity were determined using nanodrop and agarose gel electrophoresis.
Polyacrylamide northern blot analysis
Northern blot analysis was carried out as described in.36 Briefly, total RNA was separated on 10% polyacrylamide gels (8 M urea) and transferred onto nylon membranes (Hybond™ N+, GE healthcare; Trans-Blot® SD semi-dry transfer apparatus, Biorad; 1X TBE, 45 min, 20 V). The crosslinking was performed using EDC (1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride)76 and prehybridization was done using Rapid-hyb buffer (GE healthcare; 1 h at 42°C). The T4-polynucleotide kinase (Fermentas) was used to label 40 pmol of oligonucleotide probes with 32P (0.75 MBq) according to the manufacturer's protocol. The probes were purified with G-25 columns (GE Healthcare). Visualization of the radioactive signal was done using FLA-9000 (Fujifilm), including 5S rRNA as loading control. The approximate sizes of RNA fragments were estimated according to the ΦX174 DNA/HinfI Marker (Fermentas).
Identification of putative CiaR binding sites
Putative CiaR binding sites in the S. pyogenes SF370 genome were determined by search for the consensus binding motif TTTAAG-N5-TTTAAG31 using the VectorNTI software. Up to 2 mismatches to the consensus sequence were allowed and only sequences located within the putative promoters of target genes and sRNAs were selected for further analysis.
CiaR purification
The ciaR gene was amplified from S. pyogenes SF370 chromosomal DNA with Phusion polymerase (Finnzymes) using primers OLEC1471 and OLEC1472 and cloned into the pET21 (C-terminal His-Tag) overexpression vector using NdeI and XhoI. Following verification by sequencing analyses, the recombinant plasmid was transformed into E. coli BL21 overexpression strain using a heat-shock transformation protocol. Bacteria were grown at 37°C with shaking in LB medium containing 100 μg/ml ampicillin to an OD600nm of 0.8. After induction with 0.5 mM IPTG, bacteria were further grown for 4 hours at 37°C with shaking. Cells were collected by centrifugation at 4000 rpm for 10 min at 4°C and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl pH 8.0) supplemented with EDTA-free protease inhibitor cocktail (Roche). After 30 min incubation on ice, cells were lysed by sonication for 2×2 min. The lysate was cleaned from cell debris by a 10 min centrifugation at 4°C and 15000 × g. The supernatant was incubated for an hour at 4°C with Ni-NTA agarose beads (Qiagen) previously washed with lysis buffer. The resin was loaded on the gravity column and washed with lysis buffer supplemented with increasing amounts of imidazole (twice 20 mM and once 40 mM). The protein was eluted with lysis buffer containing 250 mM imidazole and checked for purity by SDS-PAGE analysis. The protein concentration was determined using the Bradford reagent (BioRad).
CiaR electrophoretic mobility shift assays
Putative promoter fragments (∼125 bp) encompassing the determined CiaR binding motif (see above) and the coding sequence of Spy_sRNA195261 (negative control) were amplified from S. pyogenes SF370 chromosomal DNA by PCR using primers listed in Table S3. Phusion polymerase (Finnzymes) was used and the amplified fragments were purified using a PCR purification kit (Qiagen). 40 ng (0.5 pmol) of DNA fragments were incubated in 15 μl reactions in band shift buffer (20 mM HEPES, 10 mM (NH4)2SO4, 1 mM DTT, 30 mM KCl, 10 mM MgCl2, pH 7.9) as previously described31 with varying amounts of purified His-tagged CiaR. Reactions were incubated at room temperature for 30 min and loaded on native 8% polyacrylamide gel in 1× TGE (50 mM Tris, 400 mM glycine, 1.73 mM EDTA). Bands were visualized with a 3 min EtBr staining. PCR fragment without the CiaR binding motif served as a DNA negative control.
cDNA library preparation, sequencing analysis and criteria for putative sRNA selection
cDNA library preparation and analysis were carried out essentially as described previously.34 Briefly, S. pyogenes SF370 was cultured until mid logarithmic (ML) growth phase. Total RNA was extracted with TRIzol (Sigma-Aldrich)/chloroform and treated with TURBO™ DNase (Ambion) and Ribo-Zero™ rRNA Removal Kit® for Gram-positive bacteria (Epicentre). ScriptMiner™ Small RNA-Seq Library Preparation Kit (Multiplex, Illumina® compatible, Epicentre) was used to prepare the library using Tobacco Acid Pyrophosphatase (TAP) (Epicentre). cDNAs were sequenced at the Next Generation Sequencing (CSF NGS Unit; http://csf.ac.at/)) facility of the Vienna Biocenter, Vienna, Austria (Illumina single end sequencing). Cutadapt (version 1.0)77 was used to remove adapter sequences. For reads where an adapter sequence was identified, only reads longer than 15 nt were kept for further analysis. For reads were no adapter sequence was identified, the last 15 nt were removed to increase the overall quality of the reads. The remaining reads were aligned to the S. pyogenes genome (GenBank: NC_002737) using Bowtie (version 1.1.0) with 2 mismatches allowed (-v 2). An in-house script was used to count the total coverage, the 5' and 3' ends of all mapped reads at each nucleotide position, for each DNA strand.
The reads were visualized using the Integrative Genomics Viewer (IGV).78 For the analysis of sRNA sequencing data using bioinformatics, putative sRNA candidates were selected based on the following criteria: location in the IGRs, total read counts above 10 and a distinct 5' end. A distinct 5' end was defined by 2 criteria: 50% of all the reads at a defined position should start at that position and 50% of all reads that start within a window of 11 nt (5 nt upstream and downstream of the sRNA 5' end) should start at that position. Predicted Rho-independent terminators by TransTermHP v2.07 (downloaded from http://transterm.cbcb.umd.edu/tt/Streptococcus_pyogenes_M1_GAS.tt) were analyzed.
In silico analysis of sRNAs
All sRNAs predicted by visual inspection of the sRNA sequencing data were compared to the Rfam database, using the RFAM web service79 to identify RNAs with known functions. For five sRNAs with defined 5' and 3' ends, the structure of the sRNA was predicted with RNAfold (v. Two.1.1)64 using settings (-p -d2 –noLP). Regions of 100 nt upstream of the visually identified sRNAs were extracted and analyzed using MEME (v 4.9.1) to identify putative promoter motifs. We used FIMO (v 4.9.1) to search for these motifs upstream of the sRNAs identified by bioinformatics analysis of the sRNA sequencing data. Homologues in other species were identified using BLAST (v.2.28+)80,81 against all whole RefSeq streptococcal genomes available. The sequences were downloaded from NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). sRNA homologues in other species were aligned using MAFFT (mafft-ginsi) (v7.220).82
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Acknowledgments
We are grateful to Aman Zare and Ido Tamir (Vienna BioCenter Campus Science Support Facilities GmbH) for their support with sRNA sequencing data processing. We thank Nikola Zlatkov Kolev for his help with the rny deletion mutant. We thank Elitza Deltcheva, Maria Eckert and Karine Gonzales for the high-throughput in silico predictions of sRNAs performed earlier in the laboratory and for the Northern blot analysis of housekeeping and putative riboswitch sRNAs presented in the supplementary information. We thank EC lab members for critical reading of the manuscript. This study was supported by the Wallenberg Advanced Bioinformatics Infrastructure (WABI), the Alexander von Humboldt (AvH) Foundation (AvH Professorship), the German Federal Ministry for Education and Research (BMBF), “the Helmholtz Association, the Goran Gustafsson”, the Swedish Research Council and Umeå University. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project b20133265.
References
- 1. Romby P, Vandenesch F, Wagner EGH. The role of RNAs in the regulation of virulence-gene expression. Curr Opin Microbiol 2006; 9:229-36; PMID:16529986; http://dx.doi.org/ 10.1016/j.mib.2006.02.005 [DOI] [PubMed] [Google Scholar]
- 2. Toledo-Arana A, Repoila F, Cossart P. Small noncoding RNAs controlling pathogenesis. Curr Opin Microbiol 2007; 10:182-8; PMID:17383223; http://dx.doi.org/ 10.1016/j.mib.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 3. Waters LS, Storz G. Regulatory RNAs in Bacteria. Cell 2009; 136:615–28; PMID:19239884; http://dx.doi.org/ 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet TIG 2005; 21:399–404; PMID:15913835; http://dx.doi.org/ 10.1016/j.tig.2005.05.008 [DOI] [PubMed] [Google Scholar]
- 5. Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem 2005; 74:199–217; PMID:15952886; http://dx.doi.org/ 10.1146/annurev.biochem.74.082803.133136 [DOI] [PubMed] [Google Scholar]
- 6. Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 2011; 43:880–91; PMID:21925377; http://dx.doi.org/ 10.1016/j.molcel.2011.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mellin JR, Cossart P. The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol 2012; 9:372–8; PMID:22336762; http://dx.doi.org/ 10.4161/rna.19235 [DOI] [PubMed] [Google Scholar]
- 8. Brantl S, Brückner R. Small regulatory RNAs from low-GC Gram-positive bacteria. RNA Biol 2014; 11:443–56; PMID:24576839; http://dx.doi.org/ 10.4161/rna.28036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol 2007; 10:102–9; PMID:17387036; http://dx.doi.org/ 10.1016/j.mib.2007.03.012 [DOI] [PubMed] [Google Scholar]
- 10. Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu Rev Genet 2010; 44:167–88; PMID:20707673; http://dx.doi.org/ 10.1146/annurev-genet-102209-163523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sorek R, Kunin V, Hugenholtz P. CRISPR -a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol 2008; 6:181–6; PMID:18157154; http://dx.doi.org/ 10.1038/nrmicro1793 [DOI] [PubMed] [Google Scholar]
- 12. Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A 2001; 98:4658–63; PMID:11296296; http://dx.doi.org/ 10.1073/pnas.071559398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Livny J, Fogel MA, Davis BM, Waldor MK. sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes. Nucleic Acids Res 2005; 33:4096–105; PMID:16049021; http://dx.doi.org/ 10.1093/nar/gki715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res 2006; 34:3484–93; PMID:16870723; http://dx.doi.org/ 10.1093/nar/gkl453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Livny J, Teonadi H, Livny M, Waldor MK. High-throughput, kingdom-wide prediction and annotation of bacterial non-coding RNAs. PLoS ONE 2008; 3:e3197; PMID:18787707; http://dx.doi.org/ 10.1371/journal.pone.0003197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sridhar J, Narmada SR, Sabarinathan R, Ou H-Y, Deng Z, Sekar K, Rafi ZA, Rajakumar K. sRNAscanner: a computational tool for intergenic small RNA detection in bacterial genomes. PLoS ONE 2010; 5:e11970; PMID:20700540; http://dx.doi.org/ 10.1371/journal.pone.0011970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Raasch P, Schmitz U, Patenge N, Vera J, Kreikemeyer B, Wolkenhauer O. Non-coding RNA detection methods combined to improve usability, reproducibility and precision. BMC Bioinformatics 2010; 11:491; PMID:20920260; http://dx.doi.org/ 10.1186/1471-2105-11-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tesorero RA, Yu N, Wright JO, Svencionis JP, Cheng Q, Kim J-H, Cho KH. Novel Regulatory Small RNAs in Streptococcus pyogenes. PLoS ONE 2013; 8:e64021; PMID:23762235; http://dx.doi.org/ 10.1371/journal.pone.0064021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Perez N, Treviño J, Liu Z, Ho SCM, Babitzke P, Sumby P. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS ONE 2009; 4:e7668; PMID:19888332; http://dx.doi.org/ 10.1371/journal.pone.0007668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patenge N, Billion A, Raasch P, Normann J, Wisniewska-Kucper A, Retey J, Boisguérin V, Hartsch T, Hain T, Kreikemeyer B. Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC Genomics 2012; 13:550; PMID:23062031; http://dx.doi.org/ 10.1186/1471-2164-13-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011; 471:602–7; PMID:21455174; http://dx.doi.org/ 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 2013; 41:e140; PMID:23716638; http://dx.doi.org/ 10.1093/nar/gkt444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Le Rhun A, Charpentier E. Small RNAs in streptococci. RNA Biol 2012; 9:414–26; PMID:22546939; http://dx.doi.org/ 10.4161/rna.20104 [DOI] [PubMed] [Google Scholar]
- 24. Cho KH, Kim J-H. Cis-encoded non-coding antisense RNAs in streptococci and other low GC Gram (+) bacterial pathogens. Front Genet 2015; 6:110; PMID:25859258; http://dx.doi.org/ 10.3389/fgene.2015.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mangold M, Siller M, Roppenser B, Vlaminckx BJM, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol Microbiol 2004; 53:1515–27; PMID:15387826; http://dx.doi.org/ 10.1111/j.1365-2958.2004.04222.x [DOI] [PubMed] [Google Scholar]
- 26. Roberts SA, Scott JR. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol Microbiol 2007; 66:1506–22; PMID:18005100 [DOI] [PubMed] [Google Scholar]
- 27. Liu Z, Treviño J, Ramirez-Peña E, Sumby P. The small regulatory RNA FasX controls pilus expression and adherence in the human bacterial pathogen group A Streptococcus. Mol Microbiol 2012; 86:140–54; PMID:22882718; http://dx.doi.org/ 10.1111/j.1365-2958.2012.08178.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol 2001; 39:392–406; PMID:11136460; http://dx.doi.org/ 10.1046/j.1365-2958.2001.02226.x [DOI] [PubMed] [Google Scholar]
- 29. Ramirez-Peña E, Treviño J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol 2010; 78:1332–47; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07427.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Danger JL, Cao TN, Cao TH, Sarkar P, Treviño J, Pflughoeft KJ, Sumby P. The small regulatory RNA FasX enhances group A Streptococcus virulence and inhibits pilus expression via serotype-specific targets. Mol Microbiol 2015; 96:249–62; PMID:25586884; http://dx.doi.org/ 10.1111/mmi.12935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Halfmann A, Kovács M, Hakenbeck R, Brückner R. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol 2007; 66:110–26; PMID:17725562; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05900.x [DOI] [PubMed] [Google Scholar]
- 32. Marx P, Nuhn M, Kovács M, Hakenbeck R, Brückner R. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics 2010; 11:661; PMID:21106082; http://dx.doi.org/ 10.1186/1471-2164-11-661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schnorpfeil A, Kranz M, Kovács M, Kirsch C, Gartmann J, Brunner I, Bittmann S, Brückner R. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol 2013; 89:334–49; PMID:23710838; http://dx.doi.org/ 10.1111/mmi.12277 [DOI] [PubMed] [Google Scholar]
- 34. Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol 2013; 10:726–37; PMID:23563642; http://dx.doi.org/ 10.4161/rna.24321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 2014; 42:6091–105; PMID:24728998; http://dx.doi.org/ 10.1093/nar/gku241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fonfara I, Le Rhun A, Chylinski K, Makarova KS, Lécrivain A-L, Bzdrenga J, Koonin EV, Charpentier E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res 2014; 42:2577–90; PMID:24270795; http://dx.doi.org/ 10.1093/nar/gkt1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816–21; PMID:22745249; http://dx.doi.org/ 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods 2013; 10:957–63; PMID:24076990; http://dx.doi.org/ 10.1038/nmeth.2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charpentier E, Doudna JA. Biotechnology: Rewriting a genome. Nature 2013; 495:50–1; PMID:23467164; http://dx.doi.org/ 10.1038/495050a [DOI] [PubMed] [Google Scholar]
- 40. Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346:1258096; PMID:25430774; http://dx.doi.org/ 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- 41. Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014; 157:1262–78; PMID:24906146; http://dx.doi.org/ 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol 2011; 9:578–89; PMID:21760622; http://dx.doi.org/ 10.1038/nrmicro2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res 2002; 30:3662–71; PMID:12202750; http://dx.doi.org/ 10.1093/nar/gkf508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev 2010; 34:883–923; PMID:20659169; http://dx.doi.org/ 10.1111/j.1574-6976.2010.00242.x [DOI] [PubMed] [Google Scholar]
- 45. Caron M-P, Lafontaine DA, Massé E. Small RNA-mediated regulation at the level of transcript stability. RNA Biol 7:140–4; PMID:20220305; http://dx.doi.org/ 10.4161/rna.7.2.11056 [DOI] [PubMed] [Google Scholar]
- 46. Court DL, Gan J, Liang Y-H, Shaw GX, Tropea JE, Costantino N, Waugh DS, Ji X. RNase III: genetics and function; structure and mechanism. Annu Rev Genet 2013; 47:405–31; PMID:24274754; http://dx.doi.org/ 10.1146/annurev-genet-110711-155618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Viegas SC, Arraiano CM. Regulating the regulators: How ribonucleases dictate the rules in the control of small non-coding RNAs. RNA Biol 2008; 5:230–43; PMID:18981732; http://dx.doi.org/ 10.4161/rna.6915 [DOI] [PubMed] [Google Scholar]
- 48. Lybecker M, Zimmermann B, Bilusic I, Tukhtubaeva N, Schroeder R. The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci 2014:201315974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bandyra KJ, Said N, Pfeiffer V, Gorna MW, Vogel J, Luisi BF. The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol Cell 2012; 47:943–53; PMID:22902561; http://dx.doi.org/ 10.1016/j.molcel.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marincola G, Schäfer T, Behler J, Bernhardt J, Ohlsen K, Goerke C, Wolz C. RNase Y of Staphylococcus aureus and its role in the activation of virulence genes. Mol Microbiol 2012; 85:817–32; PMID:22780584; http://dx.doi.org/ 10.1111/j.1365-2958.2012.08144.x [DOI] [PubMed] [Google Scholar]
- 51. Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J 2009; 28:3523–33; PMID:19779461; http://dx.doi.org/ 10.1038/emboj.2009.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen Z, Itzek A, Malke H, Ferretti JJ, Kreth J. Multiple roles of RNase Y in Streptococcus pyogenes mRNA processing and degradation. J Bacteriol 2013; 195:2585–94; PMID:23543715; http://dx.doi.org/ 10.1128/JB.00097-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lehnik-Habrink M, Schaffer M, Mader U, Diethmaier C, Herzberg C, Stulke J. RNA processing in Bacillus subtilis: identification of targets of the essential RNase Y. Mol Microbiol 2011; 81:1459–73; PMID:21815947; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07777.x [DOI] [PubMed] [Google Scholar]
- 54. Durand S, Gilet L, Condon C. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet 2012; 8:e1003181; PMID:23300471; http://dx.doi.org/ 10.1371/journal.pgen.1003181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 2005; 33:D121–4; PMID:15608160; http://dx.doi.org/ 10.1093/nar/gki081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wassarman KM. 6S RNA: a regulator of transcription. Mol Microbiol 2007; 65:1425–31; PMID:17714443; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05894.x [DOI] [PubMed] [Google Scholar]
- 57. Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol 2010; 11:R31; PMID:20230605; http://dx.doi.org/ 10.1186/gb-2010-11-3-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA Websuite. Nucleic Acids Res 2008; 36:W70–4; PMID:18424795; http://dx.doi.org/ 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorithms Mol Biol AMB 2011; 6:26; PMID:22115189; http://dx.doi.org/ 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res 2007; 35:4809–19; PMID:17621584; http://dx.doi.org/ 10.1093/nar/gkm487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stinson MW, McLaughlin R, Choi SH, Juarez ZE, Barnard J. Streptococcal histone-like protein: primary structure of hlpA and protein binding to lipoteichoic acid and epithelial cells. Infect Immun 1998; 66:259–65; PMID:9423866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wassarman KM, Saecker RM. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science 2006; 314:1601–3; PMID:17158328; http://dx.doi.org/ 10.1126/science.1134830 [DOI] [PubMed] [Google Scholar]
- 63. Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol 2013; 11:75–82; PMID:23268228; http://dx.doi.org/ 10.1038/nrmicro2934 [DOI] [PubMed] [Google Scholar]
- 64. Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 2011; 75:286–300; PMID:21646430; http://dx.doi.org/ 10.1128/MMBR.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A 2011; 108:20172–7; PMID:22123973; http://dx.doi.org/ 10.1073/pnas.1113521108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lasa I, Toledo-Arana A, Gingeras TR. An effort to make sense of antisense transcription in bacteria. RNA Biol 2012; 9:1039–44; PMID:22858676; http://dx.doi.org/ 10.4161/rna.21167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brantl S. Acting antisense: plasmid- and chromosome-encoded sRNAs from Gram-positive bacteria. Future Microbiol 2012; 7:853–71; PMID:22827307; http://dx.doi.org/ 10.2217/fmb.12.59 [DOI] [PubMed] [Google Scholar]
- 68. Pichon C, du Merle L, Caliot ME, Trieu-Cuot P, Le Bouguénec C. An in silico model for identification of small RNAs in whole bacterial genomes: characterization of antisense RNAs in pathogenic Escherichia coli and Streptococcus agalactiae strains. Nucleic Acids Res 2012; 40:2846–61; PMID:22139924; http://dx.doi.org/ 10.1093/nar/gkr1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jäger D, Sharma CM, Thomsen J, Ehlers C, Vogel J, Schmitz RA. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci U S A 2009; 106:21878–82; http://dx.doi.org/ 10.1073/pnas.0909051106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Arini A, Keller MP, Arber W. An antisense RNA in IS30 regulates the translational expression of the transposase. Biol Chem 1997; 378:1421–31; PMID:9461691097 1341; http://dx.doi.org/ 10.1515/bchm.1997.378.12.1421 [DOI] [PubMed] [Google Scholar]
- 71. Ma C, Simons RW. The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. EMBO J 1990; 9:1267–74; PMID:1691097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tang T-H, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Hüttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol 2005; 55:469–81; PMID:15659164; http://dx.doi.org/23326572 10.1111/j.1365-2958.2004.04428.x [DOI] [PubMed] [Google Scholar]
- 73. Laalami S, Bessieres P, Rocca A, Zig L, Nicolas P, Putzer H. Bacillus subtilis RNase Y activity in vivo analysed by tiling microarrays. PLoS One 2013; 8:e54062; PMID:23326572; http://dx.doi.org/ 10.1371/journal.pone.0054062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Caparon MG, Scott JR. Genetic manipulation of pathogenic streptococci. Methods Enzymol 1991; 204:556–86; PMID:1658571; http://dx.doi.org/ 10.1016/0076-6879(91)04028-M [DOI] [PubMed] [Google Scholar]
- 75. Lambert JM, Bongers RS, Kleerebezem M. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol 2007; 73:1126–35; PMID:17142375; http://dx.doi.org/ 10.1128/AEM.01473-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat Protoc 2008; 3:1077–84; PMID:18536652; http://dx.doi.org/ 10.1038/nprot.2008.67 [DOI] [PubMed] [Google Scholar]
- 77. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011; 17:10–2; http://dx.doi.org/ 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 78. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative Genomics Viewer. Nat Biotechnol 2011; 29:24–6; PMID:21221095; http://dx.doi.org/ 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res 2003; 31:439–41; PMID:12520045; http://dx.doi.org/ 10.1093/nar/gkg006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403–10; PMID:2231712; http://dx.doi.org/ 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 81. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics 2009; 10:421; PMID:20003500; http://dx.doi.org/ 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772–80; PMID:23329690; http://dx.doi.org/ 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol 2007; 8:R22; PMID:17313685; http://dx.doi.org/ 10.1186/gb-2007-8-2-r22 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.