ABSTRACT
Eggplant latent viroid (ELVd), like other members of family Avsunviroidae, replicates in plastids through a symmetric rolling-circle mechanism in which elongation of RNA strands is most likely catalyzed by a nuclear-encoded polymerase (NEP) translocated to plastids. Here we have addressed where NEP initiates transcription of viroid strands. Because this step is presumably directed by sequence/structural motifs, we have previously determined the conformation of the monomeric linear (+) and (−) RNAs of ELVd resulting from hammerhead-mediated self-cleavage. In silico predictions with 3 softwares led to similar bifurcated conformations for both ELVd strands. In vitro examination by non-denaturing PAGE showed that they migrate as prominent single bands, with the ELVd (+) RNA displaying a more compact conformation as revealed by its faster electrophoretic mobility. In vitro SHAPE analysis corroborated the ELVd conformations derived from thermodynamics-based predictions in silico. Moreover, sequence analysis of 94 full-length natural ELVd variants disclosed co-variations, and mutations converting canonical into wobble pairs or vice versa, which confirmed in vivo most of the stems predicted in silico and in vitro, and additionally helped to introduce minor structural refinements. Therefore, results from the 3 experimental approaches were essentially consistent among themselves. Application to RNA preparations from ELVd-infected tissue of RNA ligase-mediated rapid amplification of cDNA ends, combined with pretreatments to modify the 5′ ends of viroid strands, mapped the transcription initiation sites of ELVd (+) and (−) strands in vivo at different sequence/structural motifs, in contrast with the situation previously observed in 2 other members of the family Avsunviroidae.
KEYWORDS: Catalytic RNAs, hammerhead ribozymes, non-coding RNAs, RNA secondary structure, SHAPE
Abbreviations
- ASBVd
avocado sunblotch viroid
- PLMVd
peach latent mosaic viroid
- CChMVd
chrysanthemum chlorotic mottle viroid
- ELVd
eggplant latent viroid
- PAGE
polyacrylamide gel electrophoresis
- SHAPE
selective 2'-hydroxyl acylation analyzed by primer extension
Introduction
Having just a circular RNA genome of minimal size (ca. 250–400 nt) without protein coding ability, viroids rely exclusively on sequence and structural motifs for being recognized, replicated and spread in their host plants, as well as for circumventing the defensive response they mount.1-6 These sequence and structural motifs are embedded in compact secondary structures —adopted by viroid genomes as a consequence of their extensive self-complementarity— in which double-stranded segments are flanked by seemingly disordered loops.7,8 Therefore, determining the structure of viroid genomic RNAs is important for getting insights into how they may determine function. This aim was pursued since the very beginning with potato spindle viroid (PSTVd), the first viroid discovered9 and sequenced (by direct RNA sequencing).10 Searching the conformation with maximal base-pairing, and in vitro probing with RNases and bisulphite resulted in a rod-like secondary structure for PSTVd RNA,10,11 in consonance with thermodynamics-based predictions12,13 and electron microscopy observations.12,14 The rod-like secondary structure proposed for PSTVd appears characteristic, although with some exception,15,16 of members of the family Pospiviroidae, which as additional features show replication in the nucleus through an asymmetric rolling-circle mechanism without hammerhead ribozymes.17 However, the rod-like structure is the exception rather than the rule in the family Avsunviroidae, which clusters viroids replicating in plastids through a symmetric rolling-circle mechanism mediated by hammerhead ribozymes.18 Particularly notorious are peach latent mosaic viroid (PLMVd)19 and chrysanthemum chlorotic mottle viroid (CChMVd),20 for which multibranched most-stable conformations were predicted, and eggplant latent viroid (ELVd), with a predicted secondary structure containing 2 bifurcations.21
Examination of viroid RNA structure can be tackled essentially with 3 approaches. First, in silico, with algorithms searching for the secondary structures of minimal free energy,22-25 a relatively simple methodology that provides a first candidate structure(s). Second, in vitro, by probing in solution with RNases and bisulphite10,26 and, more recently, by selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), which interrogates local backbone RNA flexibility at single-nucleotide resolution27-29 and facilitates coupling the data to computer-assisted prediction.30 And, third, in vivo, by looking for sequence heterogeneity in natural variants of the same or homologous RNAs, particularly for co-variations that preserve double-stranded stems,21,31-34 or substitutions that do not alter the shape of loops according to isostericity matrices that predict recurrent 3-dimensional motifs more conserved in structure (the spatial arrangement their nucleotides adopt) than in sequence.35,36 When sufficient natural variability is available —as it happens in the family Avsunviroidae due to an extremely high mutation rate—37 the in vivo approaches are amenable and serve to validate or refine candidate structures obtained by the other 2 approaches, or even to propose new ones. For instance, PLMVd variability supports the existence of 2 alternative structures involving nucleotides from the so-called hammerhead arm of the (+) strand, one extended and the other cruciform;33,38 and subsequent SHAPE probing is consistent with these structures.39 On the other hand, co-variations between sequence variants of the same viroid40 and between different viroids support the conservation of the metastable motifs hairpin I within the family Pospiviroidae,41,42 and hairpin II within the genus Pospiviroid.43 These 2 motifs, initially described in thermal denaturation analyses,13 have been associated with important functional roles.43,44-46
One aspect on which viroid structure most likely plays a critical role is in transcription. This first replication step is catalyzed in the family Avsunviroidae by a nuclear-encoded RNA polymerase (NEP) translocated into chloroplasts.47,48 There is evidence for 2 members of this family indicating that NEP starts transcription at specific sites located in: i) (A+U)-rich terminal loops in the rod-like secondary structure adopted by both polarity strands of avocado sunblotch viroid (ASBVd),49 and ii) short double-stranded RNA stems in both polarity strands of PLMVd that also include the hammerhead self-cleavage sites.50,51 These results exemplify that 2 distinct sequence/structural motifs may have similar functional roles, as well as that one single motif may act in more than one function (initiation and self-cleavage of PLMVd RNAs). Moreover, because RNA folding occurs during transcription,52 the initiation sites of nascent viroid strands may affect the adoption of transient, albeit functionally relevant metastable structures, like the hammerhead structures,50 hairpins I and II13 and tertiary loop-loop interactions.26,53 There is evidence indicating that, at least in vitro, this is indeed the case with some of these metastable structures.54
Here we have determined the initiation sites of the ELVd (+) RNA (by convention the strand accumulating to higher levels in infected tissue) and of its complementary (−) counterpart. Examination of where these sites map in the secondary structure of ELVd (+) and (−) RNAs obtained by a combination of in silico, in vitro and in vivo approaches, has revealed that they are neither within nor flanked by conserved sequence/structural motifs, thus showing the versatility of eggplant NEP (or some associated transcription factor) in template binding and subsequent transcription initiation.
Results
In silico predictions lead predominantly to similar bifurcated conformations for ELVd (+) and (−) strands
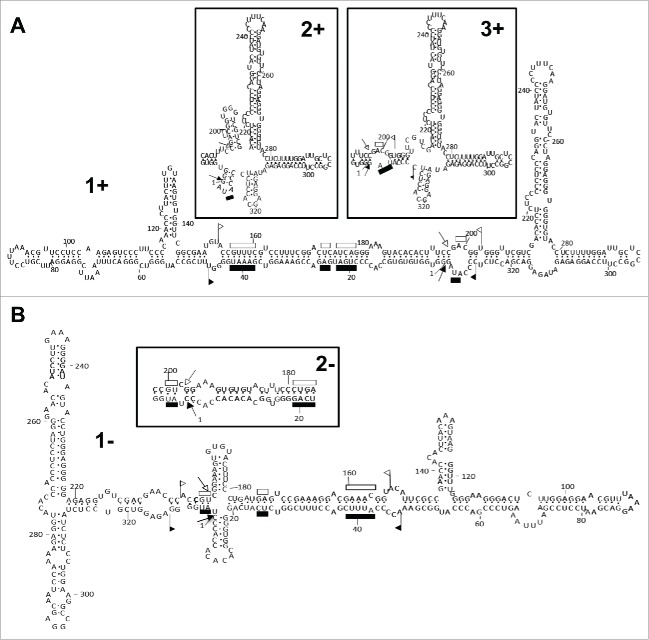
In the first report on the molecular properties of ELVd, application of the Mfold software for RNA folding22 to the (+) strand of 4 representative variants of this viroid generated a quasi-rod-like conformation with 68–70% of the residues paired —including those corresponding to the hammerhead ribozymes of both polarities that appear faced each other in the central domain— and bifurcations at both terminal domains.21 Moreover, within a 10% interval of minimal free energy, none of the predicted secondary structures were of the rod-like class typical of the family Pospiviroidae.21 Re-examination of this question with a more recent version of the software23 resulted in the same conformation (Fig. 1A, structure 1+), plus 3 other less stable structures with slightly different right terminal bifurcations, while 2 alternative softwares, RNAfold24 and RNAstructure,25 produced the most stable structure of Mfold or/and 2 other conformations with minor modifications also affecting the right terminal bifurcation (Fig. 1A, structures 2+ and 3+). Regarding the ELVd (−) strand, the 3 softwares predicted a bifurcated conformation for the reference variant ELVd-221 mimicking that of the (+) strand (Fig. 1B, structure 2-), with RNAstructure additionally predicting a slightly more stable conformation with a small cruciform motif in the central domain (Fig. 1B, structure 1-). Besides these 2 most stable structures, Mfold and RNAstructure generated variations thereof with small differences.
Figure 1.
Conformations of minimum free energy predicted for both strands of ELVd (reference variant). (A) Structure 1+ generated for ELVd (+) RNA by the Mfold and RNAstructure softwares for RNA folding. RNAstructure and RNAfold additionally produced structures 2+ and 3+ with minor modifications affecting the right terminal bifurcation (insets). (B) Structure 2- generated for ELVd (−) RNA by the 3 softwares. RNAstructure additionally produced structure 1- with a small cruciform motif in the central domain. Sequences forming the hammerhead structures are delimited by flags, motifs conserved in natural hammerhead structures are denoted by bars, and self-cleavage sites are marked by arrows. Solid and open symbols refer to plus and minus polarities, respectively. The same numbers are used for both polarities.
In summary, thermodynamics-based predictions were consistent among the 3 approaches, leading to similar bifurcated conformations (or variations thereof) for both ELVd strands.
Non-denaturing PAGE shows differences between the secondary structures adopted in vitro by ELVd (+) and (−) RNAs
Next we examined if, despite the close similarity of the bifurcated conformations predicted in silico for both ELVd strands, a physical distinction between them could be established in vitro. To this end, the monomeric linear (+) and (−) forms of ELVd resulting from hammerhead-mediated self-cleavage in vitro of their corresponding dimeric transcripts were analyzed by non-denaturing PAGE. Prior to electrophoresis, 2 aliquots of each gel-eluted RNAs were heated at 98°C for 2 min and either snap-cooled on ice or gradually-cooled at 25°C along 15 min, with a third aliquot remaining untreated serving as control. Three other aliquots of the 2 gel-eluted RNAs were treated similarly, but in the presence of 5 mM Mg2+. The treatments did not affect the electrophoretic mobility of ELVd RNAs, although the presence of Mg2+ led to an attenuation of the signal of those samples heated at 98°C, most likely resulting from RNA degradation catalyzed by this cation (Fig. 2A). Two additional observations are worthy of note: i) both ELVd strands migrated as prominent single bands accompanied by close satellite bands of much lower intensity, and ii) compared with the size markers, the ELVd (+) RNA displayed faster mobility than its (−) counterpart. This latter observation was confirmed by co-electrophoresis of mixed aliquots applied to the same well in the presence and absence of Mg2+ (Fig. 2B).
Figure 2.
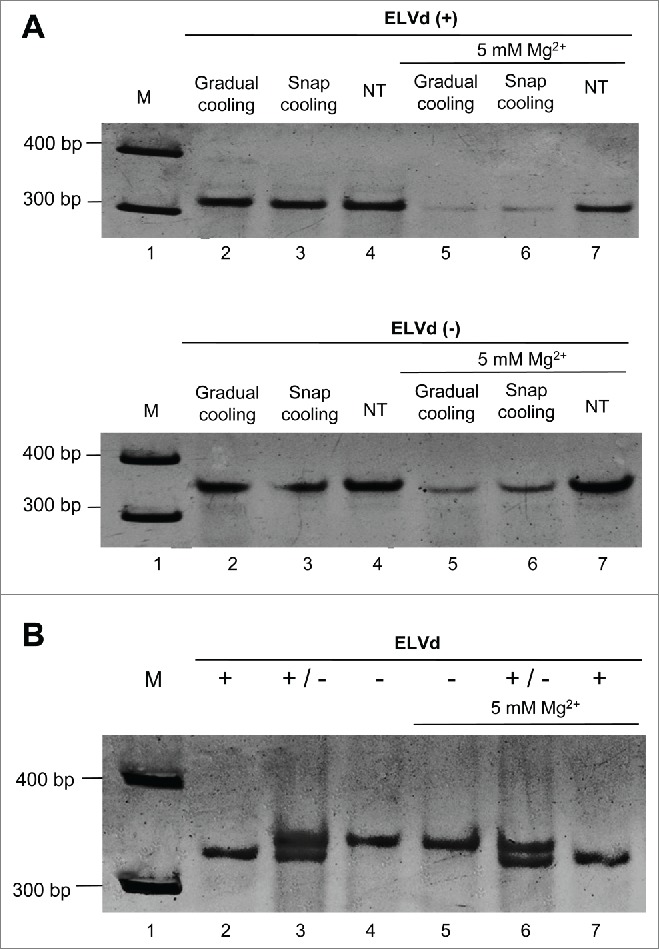
The monomeric linear (+) and (−) RNAs of ELVd display a different mobility in non-denaturing PAGE. (A) Before electrophoresis, aliquots of the gel-eluted ELVd (+) and (−) forms (upper and lower panels, respectively) were heated at 98°C for 2 min and gradually-cooled at 35°C along 15 min (lane 2), snap-cooled on ice (lane 3), or applied directly with no thermal treatment (NT) (lane 4). Three other aliquots of the same RNAs were processed similarly, but in the presence of 5 mM Mg2+ (lanes 5, 6 and 7, respectively). (B) Aliquots of untreated ELVd (+) and (−) RNAs were applied individually (lanes 2 and 4, respectively) or concurrently (lane 3). Three other aliquots of the same RNAs were processed similarly, but in the presence of 5 mM Mg2+ (lanes 5, 6 and 7, respectively). M refers to DNA markers with their size (in base pairs) indicated on the left (lanes 1). Gels were stained with ethidium bromide and are shown in the inverted option to facilitate visualization.
Altogether these results indicated that each ELVd strand adopts one dominant conformation (or a set of closely-related conformations not separable individually by PAGE), with that of the (+) strand being comparatively more compact. The lack of detectable Mg2+ effects does not support the existence of tertiary interactions like the kissing loops present in PLMVd and CChMVd (see below). Moreover, in contrast with these 2 viroid RNAs that are insoluble in 2 M LiCl as a likely consequence of their multibranched conformations,20 ELVd (+) and (−) RNAs were soluble under these high-salt conditions (data not shown), thus suggesting that they fold into less complex structures.
SHAPE analysis coupled to computer-assisted prediction corroborates the ELVd conformations derived from in silico approaches
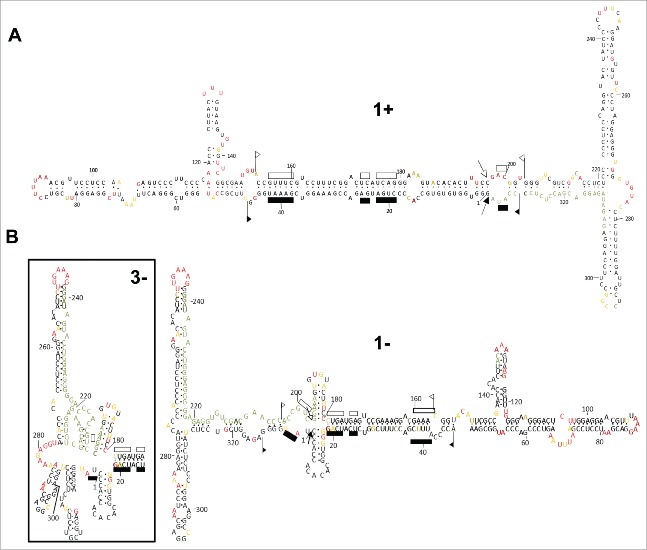
The finding that each ELVd strand folds in vitro into dominant conformations made feasible their finer dissection at the nucleotide level with SHAPE (using NMIA) coupled to computer-assisted prediction.30 The obtained resolution allowed determination of SHAPE reactivity of most positions (Fig. S1). The conformation of minimal free energy derived in vitro for the ELVd (+) RNA was consistent, with minor rearrangements, with that predicted in silico initially21 and in the present work (Fig. 1A, structure 1+): a central domain, resulting from base-pairing of the residues forming the hammerhead ribozymes of both polarities, flanked by bifurcations at both terminal domains (Fig. 3A). On the other hand, the conformations obtained in vitro for the ELVd (−) RNA were also consistent to a very good extent with those predicted in silico (Fig. 1A, structure 1- and 3-): a secondary structure resembling that of the (+) strand but with a small cruciform motif in the central domain (only partially supported because some positions correspond to the primer binding site), and a rearranged left terminal domain for accommodating the high SHAPE reactivity detected for nucleotides around position 280 (Fig. 3B). When this reactivity was not computed considering the compensatory mutations detected in vivo (see below), the conformation adopted was that with a simple left terminal bifurcation (Fig. 3B). Again, no elements of tertiary structure were apparent.
Figure 3.
SHAPE analysis results in ELVd conformations with a central domain, wherein most of the nucleotides that can potentially form the hammerheads are base-paired, flanked by terminal domains with bifurcations. (A) ELVd (+) RNA folding in vitro shows minor differences in some loops with respect to one of the structures predicted in silico. (B) ELVd (−) RNA folding in vitro is also consistent with the structures predicted in silico, including 2 variations with a cruciform motif in the central domain and a rearrangement in one of the arms forming the left terminal bifurcation. Nucleotides in red, yellow and black displayed high (more that 0.85), intermediate (0.85–0.40) and low (less than 0.40) SHAPE-reactivity. Nucleotides in gray, for which the SHAPE-reactivity could not be determined, correspond to the primer binding region and adjacent positions, and to blurry positions. Other details as in the legend to Fig. 1.
The natural variability found in ELVd supports that both strands adopt in vivo conformations very similar to those proposed by in silico and in vitro approaches
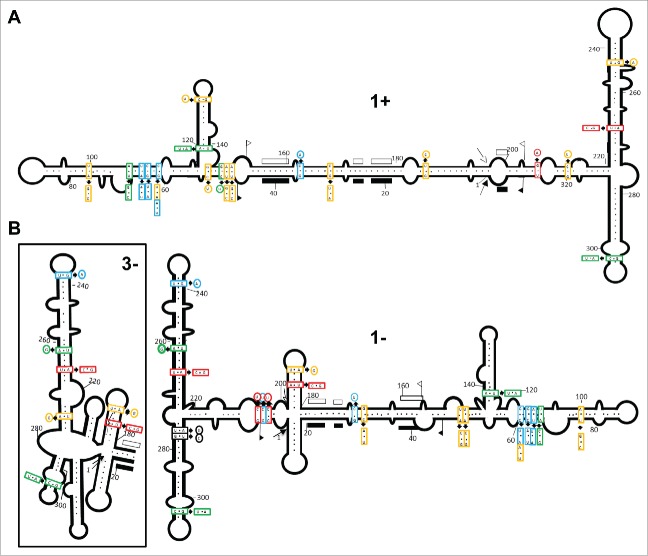
There are several reasons explaining why in vitro-derived RNA conformations may not reflect the situation existing in vivo, chief among which is that RNAs interact with proteins in their natural habitat (see Discussion). To overcome this limitation, we examined the natural variability of 100 ELVd full-length progeny variants from the parental variant ELVd-221 obtained by RT-PCR amplification (50 with a pair of adjacent primers and 50 with another pair in order to cover the complete molecule), cloning and sequencing (Table 1). First, we search for co-variations or compensatory mutations (converting canonical into wobble pairs or vice versa) preserving the stems predicted by the in silico and in vitro approaches. Regarding the ELVd (+) strand, the conformation with a central domain flanked by 2 bifurcations at both terminal domains (Fig. 1A and 3A, structure 1+), is strongly supported because of the presence of co-variations and compensatory mutations in most of the stems that do not disrupt their base-pairing (Fig. 4A) (Table S1). The variability found in the 10 original variants21 support also the same conformation. Thus, this is a good example of coherence among results generated by the 3 experimental approaches.
Table 1.
Primers for RT-PCR amplification, SHAPE and RLM-RACE.
| ELVd-specific primers (and polaritya) | Sequence (5′→ 3′) | Positionsb | Use |
|---|---|---|---|
| RF-1024 (c) | TATGGGGAGAGGTCGTC | 333–317 | SHAPE (+) and 5′-RACE (RT) |
| RF-701 (c) | AGAGGTCGTCCTCTATCTCTCCTGGAAGG | 326–298 | 5′-RACE (+) (PCR) |
| RF-1070 (c) | GAGCAATCCTTTTGAGGTACACCC | 294–271 | 5′-RACE (+) (nested PCR) |
| RF-975 (h) | GGGTGGGTGTACCTCTTTTGGATTGCT | 267–293 | SHAPE (−) and 5′-RACE (RT) |
| RF-1071 (h) | CCTTCCAGGAGAGATAGAGGACGACCTCT | 298–326 | 5′-RACE (−) (PCR) |
| RF-1313 (h) | GGGTGGTGTGTGCCACCCCT | 1–20 | 5′-RACE (−) (nested PCR) |
| RF-1331 (h) | GACGGTGGGTTCGTCGACACC | 198–218 | SHAPE (−) |
| RF-676 (h) | CCTTWAAWCGTTCCTCCAAG | 86–105 | RT-PCR (−) (Progeny) |
| RF-677 (c) | WCGAATCCTCCGAATTTA | 85–68 | PCR (+) (Progeny) |
| RF-1299 (h) | GAGGACGACCTCTCCCCATA | 314–333 | RT-PCR (−) (Progeny) |
| RF-1298 (c) | TATCTCTCCTGGAAGRCCGG | 313–294 | PCR (+) (Progeny) |
| Adaptors and adaptor-specific primers | Sequence (5′→ 3′) | Positions | Use |
| Adaptor A | CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA | ||
| RF-553 (h) | TGGAGCACGAGGACACTGACATG | 5–27 | 5′-RACE (+) (PCR) |
| RF-554 (h) | GACACTGACATGGACTGAAGGAGTAG | 16–41 | 5′-RACE (+) (nested PCR) |
| Adaptor B | CUCAAAAGUUUCGCCGUAUCUCAACGGCUCAUCAGUGGGCUUAGCCCAGACUUUUGAGAGAAGUGGCG | ||
| RF-37 (h) | CGGCCTCATCAGTGGGCTTAG | 25–44 | 5′-RACE (−) (PCR) |
| RF-1086 (h) | CCCAGACTTTTGAGAGAAG | 45–63 | 5′-RACE (−) (nested PCR) |
h, homologous; c, complementary
Numbering refers to plus polarity
Figure 4.
The natural sequence heterogeneity is consistent with the conformations adopted by ELVd strands in vivo being very similar to those proposed in silico and in vitro. (A) Co-variations and mutations resulting in conversion of canonical into wobble pairs or vice versa do not disrupt base-pairing in most of the stems of ELVd (+) RNA folding. (B) For similar reasons, the conformation with the cruciform motif in the central domain and an unbranched lower arm in the terminal left bifurcation appear favored in ELVd (−) RNA folding. Mutation frequencies detected in the ELVd variants sequenced are denoted with different color: red (in more than 30 variants), blue (in 20–30 variants), green (in 10–20 variants), yellow (in 5–10 variants) and black (in 2 of the original variants).21 The schemes representing ELVd (+) and (−) conformations are derived from those obtained by SHAPE analyses. Other details as in the legend to Fig. 1.
What about the ELVd (−) strand for which 2 structures were derived from in silico and in vitro analyses (Fig. 1A and Fig. 3B)? On the one hand the conformation with the small cruciform motif in the central domain appears favored, because the upper hairpin stem of this motif is supported by the co-variation A-U→C-G (positions 183 and 194) in 35 variants, and by the substitution A→G (position 186) converting a canonical into a wobble pair in 6 variants (Fig. 4B) (Table S2). On the other hand the bifurcated, rather than the multibranched structure of the left terminal domain, is supported by: i) 3 consecutive substitutions C→U (positions 204, 205 and 206) in 35, 20 and 35 variants, respectively, converting canonical into wobble pairs, and ii) 2 substitutions U→C and A→G (positions 311 and 312) in 2 of the original variants,21 converting a wobble into a canonical pair and vice versa, respectively (Fig. 4B) (Table S2). Overall, these results bolster the adoption by ELVd (+) and (−) strands of slightly different foldings in vivo, which could also be adopted in vitro, thus explaining the different mobility displayed by both RNAs in non-denaturing PAGE (Fig. 2). The in vivo conformations of ELVd (+) and (−) strands are those favoring ligation, and not self-cleavage, because the nucleotides potentially forming the hammerhead structures are involved in alternative catalytically-inactive foldings.55,56 As expected, the nucleotides forming the catalytic core of the hammerhead structures are conserved as well as the base-pairing of the flanking stems (Fig. S2).
The transcription initiation site of ELVd (+) strands maps at an internal loop of a hairpin
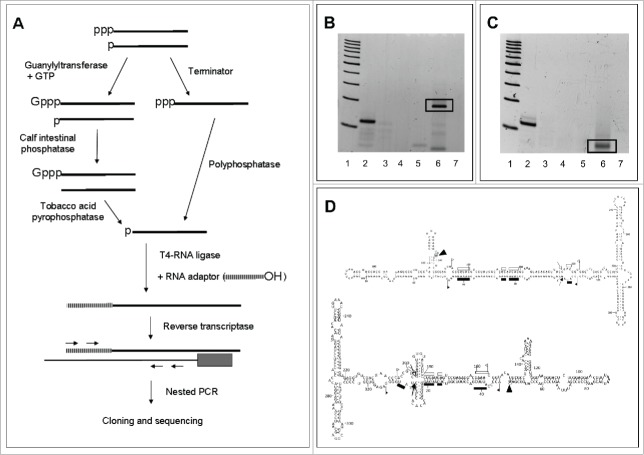
Once reliably determined the in vivo conformations of the 2 ELVd strands, we then moved to map their transcription initiation sites. We first began with that of (+) strands, the most abundant in vivo, using the approach developed previously to address a similar issue in PLMVd.50 In brief, this RLM-RACE approach is a derivation of a previous one set up for mapping the genuine 5′ termini of eukaryotic mRNAs, which are capped.57,58 Because, there is evidence indicating that ELVd accumulates59 (our unpublished in situ hybridization data) and most likely replicates in chloroplasts, wherein capping does not occur, we assumed that the primary ELVd transcripts, like other chloroplastic RNA transcripts, should be characteristically tagged by a 5′-triphosphorylated group. Only RNAs with such a group are specifically capped in vitro with guanylyltransferase and GTP, while other RNAs with 5′-monophosphorylated or -hydroxylated groups are not.60,61 After in vitro capping, ensuing treatment with calf intestinal phosphatase (CIP) removes the 5′-monophosphorylated groups, thus hindering ligation of these RNAs to an RNA adaptor. Subsequent digestion with tobacco acid pyrophosphatase (TAP) removes the cap structure of the genuine 5′ termini, leaving a free 5′-monophosphorylated group suitable for ligation to the RNA adaptor with T4-RNA ligase. Following reverse transcription of the resulting product with a viroid-specific primer, the cDNA obtained was amplified by nested PCR (with primers specific for the viroid and the RNA adaptor), cloned, and sequenced (Fig. 5A).
Figure 5.
Identification of the transcription initiation sites of ELVd strands. (A) Scheme of the 2 RLM-RACE approaches used to exclusively amplify RNAs with a 5′-triphosphorylated end. (B) Non-denaturing PAGE of the RT-PCR products resulting from applying in vitro capping and RLM-RACE to ELVd (+) strands. Lane 1, ladder of 100-bp DNA multimers. Lane 2, artificial reaction mixture containing an in vitro ELVd (+) transcript starting at position 210 combined with excesses of a CF11-fractionated RNA preparation from mock-inoculated eggplant. Lanes 3 and 4, reaction mixtures in which in vitro capping and addition of the RNA adaptor were omitted, respectively. Lanes 5 and 6, RNA preparations from mock-inoculated and ELVd-infected eggplant, respectively. Lane 7, reaction mixture in which the RNA template was omitted. Gel was stained with ethidium bromide. (C) Non-denaturing PAGE of the RT-PCR products resulting from applying RLM-RACE to ELVd (−) strands after their pretreatment with Terminator™ exonuclease and Polyphosphatase™. Lane 1, ladder of 100-bp DNA multimers. Lane 2, artificial reaction mixture containing an in vitro ELVd (−) transcript starting at position 113 combined with excesses of a CF11-fractionated RNA preparation from mock-inoculated eggplant. Lanes 3 and 4, reaction mixtures in which the treatment order with Terminator™ and Polyphosphatase™ was reversed, or the addition of the RNA adaptor was omitted, respectively. Lanes 5 and 6, RNA preparations from mock-inoculated and ELVd-infected eggplant, respectively. Lane 7, reaction mixture in which the RNA template was omitted. Gel was stained with ethidium bromide. (D) Arrowheads indicate the transcription initiation sites in the proposed in vivo conformations of ELVd (+) and (−) RNAs (upper and lower panels, respectively). Other details as in the legends to Figs. 1 and 2.
This methodology led to consistent results. First of all, when applied to an artificial mixture —made up of 10 ng of an in vitro ELVd (+) transcript with a known 5′-triphosphorylated end, combined with an excess of a nucleic acid preparation from mock-inoculated eggplants— cloning and sequencing of the resulting RT-PCR product of about 114 bp corroborated the expected terminus (Fig. 5B, lane 2). Two control reaction mixtures, wherein the treatments with guanylyltransferase or the addition of the RNA adaptor were omitted, failed to generate the same RT-PCR product (Fig. 5B, lanes 3 and 4, respectively), as also failed a third control in which the RNA template was omitted (Fig. 5B, lane 7). Application of this approach to RNA preparations from mock-inoculated and ELVd-infected eggplants produced a prominent RT-PCR amplicon of approximately 200 bp only in the last case (Fig. 5B, compare lanes 5 and 6). Cloning and sequencing of the amplified product characteristic of the ELVd-infected preparations identified position U138 as the transcription initiation site of ELVd (+) strands, located in an asymmetric loop forming part of a hairpin (Fig. 5D). This result was further confirmed using, instead of RNA preparations from ELVd-infected leaves, the ELVd monomeric linear forms and an ELVd subgenomic RNA of approximately 200 nt purified from these preparations. Application of RLM-RACE, cloning and sequencing resulted in the same site, and primer-extension experiments revealed a population of molecules with their 5′-end mapping at position U138 (data not shown). However, the substitutions U138→A or U138→C were detected in some of the full-length sequenced variants and the first one also in some RLM-RACE clones, thus indicating that the nucleotide at this position is not strictly conserved.
The transcription initiation site of ELVd (−) strands maps at a structural motif different from that of their (+) counterparts
Next we extended the same RLM-RACE methodology to the ELVd (−) strands but the results obtained were erratic, possibly due to the lower accumulation of these strands. Therefore, we modified the methodology substituting the in vitro capping and subsequent treatments with CIP and TAP, by 2 consecutive digestions with: i) Terminator™ exonuclease, which degrades RNAs with a 5′ monophosphorylated group, and ii) Polyphosphatase™, which transforms a 5′ triphosphorylated group into a 5′ monophosphorylated group (Fig. 5A). When first tested with an artificial control —consisting of 10 ng of an in vitro ELVd (−) transcript with a known 5′-triphosphorylated end mixed with an excess of a nucleic acid preparation from mock-inoculated eggplants— this approach resulted in an amplification product of about 115 bp, the cloning and sequencing of which confirmed the expected terminus (Fig. 5C, lane 2). Two control reaction mixtures, in which the order of treatments with Terminator™ and Polyphosphatase™ was reversed, or the addition of the RNA adaptor was omitted, failed to produce the same RT-PCR amplicon (Fig. 5C, lanes 3 and 4, respectively), as also failed a third control in which the RNA template was omitted (Fig. 5C, lane 7). Subsequent application of this approach to RNA preparations from mock-inoculated and ELVd-infected eggplants produced an RT-PCR amplicon of approximately 70 bp present only in the last case (Fig. 5C, compare lanes 5 and 6). The same RT-PCR product was observed when the RNA preparation from ELVd-infected eggplant was replaced by ELVd monomeric linear forms purified from this preparation (data not shown). Cloning and sequencing of the amplification product, only generated when using RNA preparations from ELVd-infected material, mapped the transcription initiation site of ELVd (−) strands at position A48 (Fig. 5D). However, some of the full-length sequenced variants presented the substitution A48→G indicating that, like in the ELVd (+) strands, the nucleotide at this position is not strictly conserved. The transcription initiation site of ELVd (−) strands does not map at the hairpin equivalent to that containing the initiation site of ELVd (+) strands, but at the end of a proximal double-stranded segment adjacent to an internal asymmetric loop (Fig. 5D). Moreover, no sequence conservation between the regions flanking both transcription initiation sites was observed (Fig. 5D).
Discussion
Initial thermodynamics-based analyses suggested the presence of terminal bifurcations in the secondary structure predicted for ELVd (+) RNA,21 a conformation lying between the rod-like structure proposed for ASBVd strands62,63 and the clearly branched structures —stabilized by a kissing-loop interaction— proposed for PLMVd19,26,33 and CChMVd RNAs.20,26,53 Because the predicted secondary structure of ELVd (+) RNA21 does not contain the same sequence/structural motifs where the initiation sites of ASBVd and PLMVd RNAs have been previously mapped,49-51 we assumed the existence of novel ELVd-specific motifs recruiting the NEP or some associated transcription factor. Before mapping these sites, we decided to obtain firmer evidence on the secondary structures proposed for both ELVd RNAs.
First we assessed the structures of minimal free energy predicted by Mfold with an updated version thereof and with 2 additional softwares, which corroborated the conformation proposed initially for the ELVd (+) strand21 and suggested a similar conformation for the complementary strand. Moreover, analyses by non-denaturing PAGE revealed, irrespective of the denaturing/renaturing treatments, a predominant band for each polarity strand, consistent with the adoption of a single major conformation in each case. Coupling SHAPE data to computer-assisted prediction resulted in conformations to a good extent in agreement with those generated by Mfold and RNAstructure, and to that proposed for the ELVd (+) RNA by a recent SHAPE analysis63 (with some differences in the hairpins delimited by positions 119–141 and 259–310). However, with respect to this latter analysis,63 we detected a cruciform motif in the central domain of ELVd (−) structure. These differences could result from the use in the 2 SHAPE analyses of distinct ELVd variants (89% sequence similarity), acylating agents (NMIA and benzoyl cyanide) and folding conditions. Most importantly, the existence of our SHAPE-predicted structures in vivo was validated by the presence in natural ELVd sequence variants of co-variations and conversions of canonical into wobble base-pairs or vice versa, thus preserving the internal pairing of most double-stranded stems. The more frequently occurrence of co-variations than the corresponding single mutations, which are more likely to appear from a statistical viewpoint, strongly supports the role of selection in purging variants non-viable in vivo because of their destabilized stems. The in vivo conformation is the one that really matters because, being viroids non-protein-coding RNAs, they must rely on RNA sequence and structural motifs for specific interactions with the host proteins involved in their replication, movement, pathogenesis and overcoming of the host defensive response.8
This good concordance between the in silico, in vitro and in vivo structures of ELVd strands, however, should not be taken for granted a priori. While free-energy minimization analyses predict correctly the structure of most small RNA motifs, they become less precise as the RNA size increases due to the lack of accurate thermodynamic parameters for some motifs and higher-order interactions.64 On the other hand, leaving aside that SHAPE analysis interrogates local backbone RNA flexibility in a protein-free solution —with an ionic composition that in some cases includes high concentration of Mg+2 not reflecting the physiological habitat— this approach, despite representing a great step forward in the analysis of RNA structure in vitro, has intrinsic limitations partly resulting from the initial thermal denaturation/renaturation applied to RNA.27 As pointed out before,32 a good deal of RNA biochemistry has been (and still is) done with renatured RNA, but how likely is that an RNA will renature into a unique conformation and how likely is it that this conformation will be the biologically relevant species remains unknown in most instances. Because of the high folding free energy of even small RNA hairpins, non-native states of RNA are often quite stable and pose a serious kinetic folding problem for RNA that has been denatured. Even non-denatured in vitro transcripts, frequently used because they are affordable and easy to prepare, may not fold into the physiological conformation. Moreover, certain RNAs may need a specific binding protein to maintain its active conformation, which may differ when this protein is removed. Therefore, it is critical to have independent genetic or phylogenetic evidence supporting the biochemical data.32,65
Several non-exhaustive examples illustrate the limitations of in vitro approaches for viroids. First, early studies revealed that, after thermal denaturation, the PSTVd RNA did not recover its initial conformation even following slow cooling.9 Although more refined folding conditions were subsequently developed,13 full reversibility to the original “native” structure cannot be always guaranteed. Second, when applied by independent groups, SHAPE predicted for ASBVd strands different conformations, rod-like or branched, and with or without a kissing-loop interaction in that of (−) polarity.63,66 An third, the SHAPE-derived structure for the CChMVd (+) RNA63 does not agree with that supported by numerous data obtained with natural and artificial mutants combined with bioassays and progeny analysis, particularly in the region proximal to one of the loops involved in a kissing-loop interaction critical for in vitro folding and in vivo viability.53 A similar kissing-loop interaction in the PLMVd (+) RNA has been detected by in vitro analyses (nuclease probing and SHAPE).26,63 However, the secondary structure proposed for the (+) strand of a viroid-like RNA from grapevine, which appears also stabilized by a similar kissing-loop interaction,67 is not present in the SHAPE-predicted structure.63
A well-supported secondary structure, as that reported here for both ELVd strands, is crucial for searching RNA sequence or structural motifs that could direct initiation of transcription, which as indicated above, may influence the structure of nascent viroid RNAs that start folding co-transcriptionally. Indeed, the co- and post-transcriptional self-cleavage of both ELVd hammerheads differs in vitro and possibly in vivo.68 Because the transcription initiation sites of the (+) and (−) strands of 2 members of the family Avsunviroidae map at sites similar in sequence and structure —those of ASBVd at (A+U)-rich terminal loops in the rod-like secondary structure,49 and those of PLMVd at short double-stranded RNA stems containing also the hammerhead self-cleavage sites—50,51 we anticipated a sort of similar situation for ELVd (+) and (−) initiations sites. However, our data indicate that this is not the case. After checking with appropriate controls the consistency of the RLM-RACE approaches adapted to eggplant tissues, we identified position U138 (mapping at an asymmetric loop forming part of a hairpin) as the transcription initiation site of ELVd (+) strands, and position A48 (mapping at a double-stranded segment adjacent to an internal asymmetric loop) as the transcription initiation site of ELVd (−) strands. Because no sequence conservation was found in the regions flanking the 2 sites, the possibility that initiation of transcription could be determined by a sequence rather than by structural motifs appears unlikely. Therefore, we need to conclude that the eggplant NEP involved (or some auxiliary transcription factor), is recruited to initiate transcription of both ELVd strands at sites unrelated in sequence but sharing some common structural feature (e.g. mapping at, or adjacent to, asymmetric internal loops).
Materials and methods
Extraction, fractionation, and analysis of RNA
Total nucleic acids of young leaves from mock-inoculated and ELVd-infected eggplants (Solanun melongena cv. ‘Redonda morada’) were extracted either with phenol-saturated buffer69 or with a method avoiding organic solvents,70 and then partitioned on non-ionic cellulose (CF11; Whatman) with STE (50 mM Tris-HCl, pH 7.2, 100 mM NaCl, 1 mM EDTA) containing 35% ethanol. The preparations were further clarified by removing polysaccharides with methoxyethanol.71 RNAs were examined by non-denaturing PAGE in 5% gels with 1X TAE (Tris-acetate-EDTA), and by denaturing PAGE in 5% gels with 1X TBE (Tris-borate-EDTA) and 8 M urea.72 For preparative purposes, following double electrophoresis first in non-denaturing and then in denaturing conditions,69 the second gel was stained with ethidium bromide and the bands of interest, identified with appropriate markers, were excised and the corresponding RNAs eluted.
Computer-assisted prediction of RNA structure
The structures of minimal free energy for the linear (+) and (−) forms of ELVd were searched with 3 softwares: Mfold version 3.5,23 using the circular version and default parameters, and RNAfold version 2.1.924 and RNAstructure version 5.725 using the default parameters.
SHAPE analysis
The substrates for this analysis were the unit-length self-cleavage products resulting from in vitro transcription driven by the T7 or T3 promoters,72 of a recombinant plasmids containing a dimeric head-to-tail ELVd-cDNA insert of the reference variant ELVd-2 (GenBank AJ536613). The resulting monomeric linear (+) and (−) forms of ELVd were purified by denaturing PAGE and subsequent elution. SHAPE was performed with N-methylisatoic anhydride (NMIA) essentially as reported.28 Briefly, the RNA (3 pmol) was denatured at 95°C for 3 min, transferred to ice for 15 min, and renatured at 37°C for 5 min in the folding buffer (100 mM HEPES pH 8.0, 100 mM NaCl, 6 mM MgCl2). 2′-acylation was initiated by adding NMIA (6 mM in dimethyl-sulfoxide), while only dimethyl-sulfoxide was incorporated to the control. Reactions proceeded for 15 min at 37°C and formation of 2′-O-adducts was stopped by adding 3 volumes of ethanol. RNA was recovered by centrifugation, washed 3 times with 70% ethanol, and subjected to primer extension.
Fluorescently-labeled DNA oligonucleotides (Applied Biosystems) were purified by denaturing PAGE in 20% gels. After adding 4 pmol of the appropriate primer (Table 1) tagged with the VIC fluorophore, the RNAs corresponding to the (+) and (−) NMIA reactions were heated to 95°C for 3 min and snap-cooled on ice for 15 min. Extensions were performed at 52°C for 45 min (in a 20 μl reaction volume) with 100 U SuperScript III RT (Invitrogen) and 0.5 mM dNTPs in the buffer recommended by the supplier. Sequencing reactions used to identify the peaks were prepared similarly but adding 10 mM ddGTP to the primer extension mix and using the appropriate primer (Table 1) tagged with the NED fluorophore and non-modified RNA. The resulting cDNAs were ethanol-precipitated, collected by centrifugation, resuspended in deionized formamide and resolved by capillary electrophoresis in an ABI 3130 XL Genetic Analyzer (Applied Biosystems) as previously described.28 Electrophoregrams were analyzed using the QuShape software,73 which also normalized the reactivity data. Between 4 to 6 replicas were performed for each sample, and the mean and standard deviation of the reactivity of each nucleotide was calculated.
Cloning and sequencing
ELVd circular forms purified from infected tissue were reverse transcribed with SuperScript II RT (Invitrogen) and primer RF-677 complementary to positions 85 to 68 of the ELVd reference sequence, and PCR-amplified with Pfu DNA polymerase (Agilent) and primers RF-677 and RF-676 homologous to positions 86 to 105 of the same sequence.21 To cover the complete ELVd sequence a second amplification was performed using for RT primer RF-1298 complementary to positions 313 to 294 of the ELVd reference sequence, and for PCR primers RF-1298 and RF-1299 homologous to positions 314 to 333 of the same sequence (Table 1). The resulting products were separated by non-denaturing PAGE in 5% gels and the ELVd DNAs of the expected full-length were eluted and cloned into the EcoRV restriction site of plasmid pBS II KS (+) (Stratagene). Fifty plasmid inserts from each amplification were sequenced automatically by capillary electrophoresis (see above) using a Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems). The nucleotide sequences obtained in this study have been deposited in GenBank (accession numbers KT901835 to KT901928 corresponding to 94 of the 100 plasmid inserts sequenced; the other 6 were incomplete or presented cloning artifacts).
In vitro capping
After adding N-terminal histidine tags to the 2 subunits of the vaccinia virus guanylyltransferase (mRNA-capping enzyme) they were coexpressed in bacteria and purified by affinity chromatography in Ni-agarose columns.74 RNA preparations (3 µg) from mock-inoculated and ELVd-infected eggplant leaves and, when indicated, from individual RNAs eluted from denaturing gels were subjected to capping in vitro in reaction mixtures (30 µl final volume) containing 50 mM Tris-HCl, pH 7.9, 1.25 mM MgCl2, 6 mM KCl, 2.5 mM DTT, 10 U of human placental RNase inhibitor (HPRI) (Roche Applied Science), 0.3 mM GTP, and 2.5 µl of a purified guanylyltransferase preparation, which in previous experiments was sufficient to efficiently label with [α-32P]GTP 200 ng of a synthetic transcript. Following incubation at 37°C for 45 min, addition of the same amount of fresh enzyme and incubation for another 45 min at 37°C, the RNAs were extracted with phenol-chloroform, recovered by ethanol precipitation, and resuspended in sterile distilled water.50
RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE)
After in vitro capping, the RNAs were sequentially treated with calf intestinal phosphatase (CIP) (Roche Applied Science), tobacco acid pyrophosphatase (TAP) (Epicenter Technologies), and ligated to an RNA adaptor using T4-RNA ligase (Roche Applied Science).50 Reverse transcription of the resulting products and nested PCR using the Expand High Fidelity DNA polymerase (Roche Applied Science) and appropriate primers (Table 1) were also as reported previously.50
Alternatively to in vitro capping, eggplant RNA preparations (3 μg in 20 μl final volume) were treated for 1 h at 60°C with 1 U of Terminator™ exonuclease (Epicentre) in the buffer recommended by the supplier complemented with 10 U of HPRI and, subsequently, with 20 U of RNA 5′ Polyphosphatase™ (Epicentre Technologies) for 30 min at 37°C (in 20 μl final volume) containing 50 mM HEPES-KOH (pH 7.5), 100 mM NaCl, 1 mM EDTA, 0.1% Triton X-100 and 10 U of HPRI. Ligation, reverse transcription and PCR amplification were performed as described above.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. S. Shuman for kindly providing the recombinant plasmid for coexpression of the 2 subunits of the vaccinia virus mRNA capping enzyme, to Dr. C. Romero and Dr. A. Barroso for their help in initial SHAPE experiments, and to A. Ahuir for excellent technical assistance.
Funding
This work was supported by grants BFU2011–28443 and BFU2014–56812-P (to R.F.) from Ministerio de Economía y Competitividad (MINECO) of Spain. A.L.C. and S.M. were recipients of predoctoral fellowships from MINECO and SD of a postdoctoral contract from the same organism.
References
- 1.Diener TO. Discovering viroids—a personal perspective. Nature Rev Microbiol 2003; 1:75-80; http://dx.doi.org/ 10.1038/nrmicro736. [DOI] [PubMed] [Google Scholar]
- 2.Flores R, Hernández C, Martínez de Alba E, Darós JA, Di Serio F. Viroids and viroid-host interactions. Annu Rev Phytopathol 2005; 43:117-39; PMID:16078879; http://dx.doi.org/ 10.1146/annurev.phyto.43.040204.140243. [DOI] [PubMed] [Google Scholar]
- 3.Tsagris EM, Martínez de Alba AE, Gozmanova M, Kalantidis K. Viroids. Cell Microbiol 2008; 10:2168-79; PMID:18764915; http://dx.doi.org/ 10.1111/j.1462-5822.2008.01231.x. [DOI] [PubMed] [Google Scholar]
- 4.Ding B. The biology of viroid-host interactions. Annu Rev Phytopathol 2009; 47:105-31; PMID:19400635; http://dx.doi.org/ 10.1146/annurev-phyto-080508-081927 [DOI] [PubMed] [Google Scholar]
- 5.Kovalskaya N, Hammond RW. Molecular biology of viroid-host interactions and disease control strategies. Plant Sci 2014; 228:48-60; PMID:25438785; http://dx.doi.org/ 10.1016/j.plantsci.2014.05.006 [DOI] [PubMed] [Google Scholar]
- 6.Palukaitis P. What has been happening with viroids? Virus Genes 2014; 49:175-84; PMID:25164861; http://dx.doi.org/ 10.1007/s11262-014-1110-8 [DOI] [PubMed] [Google Scholar]
- 7.Ding B. Viroids: self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip Rev RNA; 2010; 1:362-75 [DOI] [PubMed] [Google Scholar]
- 8.Flores R, Serra P, Minoia S, Di Serio F, Navarro B. Viroids: from genotype to phenotype just relying on RNA sequence and structural motifs. Front Microbiol 2012; 3:217; PMID:22719735; http://dx.doi: 10.3389/fmicb.2012.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diener TO. Potato spindle tuber viroid VIII. Correlation of infectivity with a UV-absorbing component and thermal denaturation properties of the RNA. Virology 1972; 50:606-9; PMID:4636118; http://dx.doi.org/ 10.1016/0042-6822(72)90412-6 [DOI] [PubMed] [Google Scholar]
- 10.Gross HJ, Domdey H, Lossow C, Jank P, Raba M, Alberty H, Sänger HL. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 1978; 273:203-8; PMID:643081; http://dx.doi.org/ 10.1038/273203a0 [DOI] [PubMed] [Google Scholar]
- 11.Gast FU, Kempe D, Spieker RL, Sänger HL. Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J Mol Biol 1996; 262:652-70; PMID:8876645; http://dx.doi.org/ 10.1006/jmbi.1996.0543 [DOI] [PubMed] [Google Scholar]
- 12.Sänger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt A. Viroids are single-stranded covalently-closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852-6; http://dx.doi.org/ 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riesner D, Henco K, Rokohl U, Klotz G, Kleinschmidt AK, Domdey H, Jank P, Gross HJ, Sänger HL. Structure and structure formation of viroids. J Mol Biol 1979; 133:85-115; PMID:529284; http://dx.doi.org/ 10.1016/0022-2836(79)90252-3 [DOI] [PubMed] [Google Scholar]
- 14.Sogo JM, Koller T, Diener TO. Potato spindle tuber viroid X. Visualization and size determination by electron microscopy. Virology 1973; 55:70-80; PMID:4728831; http://dx.doi.org/ 10.1016/S0042-6822(73)81009-8 [DOI] [PubMed] [Google Scholar]
- 15.Hernández C, Elena SF, Moya A, Flores R. Pear blister canker viroid is a member of the apple scar skin viroid subgroup (apscaviroids) and also has sequence homologies with viroids from other subgroups. J Gen Virol 1992; 73:2503-7; http://dx.doi.org/ 10.1099/0022-1317-73-10-2503 [DOI] [PubMed] [Google Scholar]
- 16.Giguère T, Raj Adkar-Purushothama C, Perreault J-P. Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae. PLoS ONE 2014; 9:e98655; http://dx.doi.org/ 10.1371/journal.pone.0098655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores R, Gago-Zachert S, Serra P, Sanjuán R, Elena SF. Viroids: survivors from the RNA world? Annu Rev Microbiol 2014; 68:395-414; PMID:25002087; http://dx.doi.org/ 10.1146/annurev-micro-091313-103416 [DOI] [PubMed] [Google Scholar]
- 18.Flores R, Darós JA, Hernández C. The Avsunviroidae family: viroids with hammerhead ribozymes. Adv Virus Res 2000; 55:271-323; PMID:11050945; http://dx.doi.org/ 10.1016/S0065-3527(00)55006-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernández C, Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro via hammerhead structures. Proc Natl Acad Sci U S A 1992; 89:3711-5; http://dx.doi.org/ 10.1073/pnas.89.9.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navarro B, Flores R. Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of viroids with hammerhead ribozymes. Proc Natl Acad Sci U S A 1997; 94:11262-7; PMID:9326597; http://dx.doi.org/ 10.1073/pnas.94.21.11262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fadda Z, Darós JA, Fagoaga C, Flores R, Duran-Vila N. Eggplant latent viroid (ELVd): candidate type species for a new genus within family Avsunviroidae (hammerhead viroids). J Virol 2003; 77:6528-32; PMID:12743309; http://dx.doi.org/ 10.1128/JVI.77.11.6528-6532.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M, Mathews DH, Turner DH. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide In RNA Biochemistry and Biotechnology NATO ASI Series (Barciszewski J. & Clark BFC., eds), p. 11-43, Kluwer Academic Publishers, Boston, Mass. 1999 [Google Scholar]
- 23.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz R, Bernhart SH, Hoener zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 2.0. Algorith Mol Biol 2011; 6:26; http://dx.doi.org/ 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuter JS, Mathews DH. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinform 2010; 11:129; http://dx.doi.org/ 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bussière F, Ouellet J, Côté F, Lévesque D, Perreault J-P. Mapping in solution shows the peach latent mosaic viroid to possess a new pseudoknot in a complex, branched secondary structure. J Virol 2000; 74:2647-54; http://dx.doi.org/ 10.1128/JVI.74.6.2647-2654.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 2005; 127:4223-31; PMID:15783204; http://dx.doi.org/ 10.1021/ja043822v [DOI] [PubMed] [Google Scholar]
- 28.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 2009; 106:97-102; PMID:19109441; http://dx.doi.org/ 10.1073/pnas.0806929106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res 2011; 44:1280-91; PMID:21615079; http://dx.doi.org/ 10.1021/ar200051h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A 2013; 110:5498-503; PMID:23503844; http://dx.doi.org/ 10.1073/pnas.1219988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutell RR, Larsen N, Woese CR. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev 1994; 58:10-26; PMID:8177168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uhlenbeck OC. Keeping RNA happy. RNA 1995; 1:4-6; PMID:7489487 [PMC free article] [PubMed] [Google Scholar]
- 33.Ambrós S, Hernández C, Desvignes JC, Flores R. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J Virol 1998; 72:7397-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De la Peña M, Navarro B, Flores R. Mapping the molecular determinant of pathogenicity in a hammerhead viroid: a tetraloop within the in vivo branched RNA conformation. Proc Natl Acad Sci U S A 1999; 96:9960-5; http://dx.doi.org/ 10.1073/pnas.96.17.9960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leontis NB, Stombaugh J, Westhof E. Motif prediction in ribosomal RNAs: lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie 2002; 84:961-73; PMID:12458088; http://dx.doi.org/ 10.1016/S0300-9084(02)01463-3 [DOI] [PubMed] [Google Scholar]
- 36.Zhong X, Leontis N, Qiang S, Itaya A, Qi Y, Boris-Lawrie K, Ding B. Tertiary structural and functional analysis of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J Virol 2006; 80:8566-81; PMID:16912306; http://dx.doi.org/ 10.1128/JVI.00837-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gago S, Elena SF, Flores R, Sanjuán R. Extremely high variability of a hammerhead viroid. Science 2009; 323:1308; PMID:19265013; http://dx.doi.org/ 10.1126/science.1169202 [DOI] [PubMed] [Google Scholar]
- 38.Ambrós S, Hernández C, Flores R. Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J Gen Virol 1999; 80:2239-52; http://dx.doi.org/ 10.1099/0022-1317-80-8-2239 [DOI] [PubMed] [Google Scholar]
- 39.Dubé A, Bolduc F, Bisaillon M, Perreault J-P. Mapping studies of the peach latent mosaic viroid reveal novel structural features. Mol Plant Pathol 2011; 12:688-701; http://dx.doi.org/ 10.1111/j.1364-3703.2010.00703.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polivka H, Staub U, Gross HJ. Variation of viroid profiles in individual grapevine plants: novel grapevine yellow speckle viroid 1 mutants show alterations of hairpin I. J Gen Virol 1996; 77:155-61; PMID:8558124; http://dx.doi.org/ 10.1099/0022-1317-77-1-155 [DOI] [PubMed] [Google Scholar]
- 41.Visvader JE, Forster AC, Symons RH. Infectivity and in vitro mutagenesis of monomeric cDNA clones of citrus exocortis viroid indicates the site of processing of viroid precursors. Nucleic Acids Res 1985; 13: 5843-56; PMID:2994014; http://dx.doi.org/ 10.1093/nar/13.16.5843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flores R, Di Serio F, Hernández C. Viroids: the noncoding genomes. Semin Virol 1997; 8:65-73; http://dx.doi.org/ 10.1006/smvy.1997.0107 [DOI] [Google Scholar]
- 43.Loss P, Schmitz M, Steger G, Riesner D. Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J 1991; 10:719-27; PMID:2001685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu F, Heinrich C, Loss P, Steger G, Tien P, Riesner D. Multiple pathways of reversion in viroids for conservation of structural domains. EMBO J 1995; 12:2129-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Candresse T, Góra-Sochacka A, Zagórski W. Restoration of secondary hairpin II is associated with restoration of infectivity of a non-viable recombinant viroid. Virus Res 2001; 75:29-34; PMID:11311425; http://dx.doi.org/ 10.1016/S0168-1702(00)00255-0 [DOI] [PubMed] [Google Scholar]
- 46.Gas ME, Hernández C, Flores R, Darós JA. Processing of nuclear viroids in vivo: an interplay between RNA conformations. PLoS Pathog 2007; 3:1813-26; http://dx.doi.org/ 10.1371/journal.ppat.0030182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarro JA, Vera A, Flores R. A chloroplastic RNA polymerase resistant to tagetitoxin is involved in replication of avocado sunblotch viroid. Virology 2000; 268:218-25; PMID:10683343 ; http://dx.doi.org/ 10.1006/viro.1999.0161 [DOI] [PubMed] [Google Scholar]
- 48.Rodio ME, Delgado S, De Stradis AE, Gómez MD, Flores R, Di Serio F. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell 2007; 19:3610-26; PMID:18055612; http://dx.doi.org/ 10.1105/tpc.106.049775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarro JA, Flores R. Characterization of the initiation sites of both polarity strands of a viroid RNA reveals a motif conserved in sequence and structure. EMBO J 2000; 19:2662-70; PMID:10835363; http://dx.doi.org/ 10.1093/emboj/19.11.2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delgado S, Martínez de Alba E, Hernández C, Flores R. A short double-stranded RNA motif of peach latent mosaic viroid contains the initiation and the self-cleavage sites of both polarity strands. J Virol 2005; 79:12934-43; PMID:16188995; http://dx.doi.org/ 10.1128/JVI.79.20.12934-12943.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motard J, Bolduc F, Thompson D, Perreault J-P. The peach latent mosaic viroid replication initiation site is located at a universal position that appears to be defined by a conserved sequence. Virology 2008; 373:362-75; PMID:18190946; http://dx.doi.org/ 10.1016/j.virol.2007.12.010 [DOI] [PubMed] [Google Scholar]
- 52.Lai D, Proctor JR, Meyer IM. On the importance of cotranscriptional RNA structure formation. RNA 2013; 19:1461-73; PMID:24131802; http://dx.doi.org/ 10.1261/rna.037390.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gago S, De la Peña M, Flores R. A kissing-loop interaction in a hammerhead viroid RNA critical for its in vitro folding and in vivo viability. RNA 2005; 11:1073-83; PMID:15928342; http://dx.doi.org/ 10.1261/rna.2230605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Repsilber D, Wiese S, Rachen M, Schröder AW, Riesner D, Steger G. Formation of metastable RNA structures by sequential folding during transcription: time-resolved structural analysis of potato spindle tuber viroid (−)-stranded RNA by temperature-gradient gel electrophoresis. RNA 1999; 5:574-84; PMID:10199573; http://dx.doi.org/ 10.1017/S1355838299982018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martínez F, Marqués J, Salvador ML, Darós JA. Mutational analysis of eggplant latent viroid RNA processing in Chlamydomonas reinhardtii chloroplast. J Gen Virol 2009; 90:3057-65; http://dx.doi.org/ 10.1099/vir.0.013425-0 [DOI] [PubMed] [Google Scholar]
- 56.Nohales MA, Molina-Serrano D, Flores R, Darós JA. Involvement of the chloroplastic isoform of tRNA ligase in the replication of the viroids belonging to the family Avsunviroidae. J Virol 2012; 86:8269-76; PMID:22623792; http://dx.doi.org/ 10.1128/JVI.00629-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 1994; 138:171-4; PMID:8125298; http://dx.doi.org/ 10.1016/0378-1119(94)90802-8 [DOI] [PubMed] [Google Scholar]
- 58.Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem 1995; 227:255-73; PMID:7573945; http://dx.doi.org/ 10.1006/abio.1995.1279 [DOI] [PubMed] [Google Scholar]
- 59.Gómez G, Pallás V. A pathogenic non coding RNA that replicates and accumulates in chloroplasts traffics to this organelle through a nuclear-dependent step. Plant Signal Behav 2012; 7:882-4; http://dx.doi.org/ 10.4161/psb.20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desai NA, Shankar V. Single-strand-specific nucleases. FEMS Microbiol Rev 2003; 26:457-91; PMID:12586391; http://dx.doi.org/ 10.1111/j.1574-6976.2003.tb00626.x [DOI] [PubMed] [Google Scholar]
- 61.Mishra NC. Nucleases: molecular biology and applications John Wiley and Sons, Chichester, UK: 2002 [Google Scholar]
- 62.Symons RH. Avocado sunblotch viroid: primary sequence and proposed secondary structure. Nucleic Acids Res 1981; 9:6527-37; PMID:7322921; http://dx.doi.org/ 10.1093/nar/9.23.6527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giguère T, Adkar-Purushothama CR, Bolduc F, Perreault J-P. Elucidation of the structures of all members of the Avsunviroidae family. Mol Plant Pathol 2014; 15:767-779; http://dx.doi.org/ 10.1111/mpp.12130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathews DH, Turner DH. Prediction of RNA secondary structure by free energy minimization. Curr Opin Struct Biol 2006; 16:270-8; PMID:16713706; http://dx.doi.org/ 10.1016/j.sbi.2006.05.010 [DOI] [PubMed] [Google Scholar]
- 65.Cech TR. RNA World research-still evolving. RNA 2015; 21:474-5; PMID:25780099; http://dx.doi.org/ 10.1261/rna.049965.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delan-Forino C, Deforges J, Benard L, Sargueil B, Maurel MC, Torchet C. Structural analyses of avocado sunblotch viroid reveal differences in the folding of plus and minus RNA strands. Viruses 2014; 6:489-506; PMID:24481250; http://dx.doi.org/ 10.3390/v6020489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu Q, Wang Y, Cao M, Pantaleo V, Burgyan J, Li W-X, Ding S-W. Homology-independent discovery of replicating pathogenic circular RNAs by deep sequencing and a new computational algorithm. Proc Natl Acad Sci U S A 2012; 109:3938-43; PMID:22345560; http://dx.doi.org/ 10.1073/pnas.1117815109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carbonell A, De la Peña M, Flores R, Gago S. Effects of the trinucleotide preceding the self-cleavage site on eggplant latent viroid hammerheads: differences in co- and post-transcriptional self-cleavage may explain the lack of trinucleotide AUC in most natural hammerheads. Nucleic Acids Res 2006; 34:5613-22; PMID:17028097; http://dx.doi.org/ 10.1093/nar/gkl717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pallás V, Navarro A, Flores R. Isolation of a viroid-like RNA from hop different from hop stunt viroid. J Gen Virol 1987; 68:3201-5; http://dx.doi.org/ 10.1099/0022-1317-68-12-3201 [DOI] [Google Scholar]
- 70.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep 1983; 1:19-21; http://dx.doi.org/ 10.1007/BF02712670 [DOI] [Google Scholar]
- 71.Bellamy AR, Ralph RK. Recovery and purification of nucleic acids by means of cetyltrimethylammonium bromide. Methods Enzymol 1968; XII:156-60; http://dx.doi.org/ 10.1016/0076-6879(67)12125-3 [DOI] [Google Scholar]
- 72.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor (NY: ): Cold Spring Harbor Laboratory Press; 1989 [Google Scholar]
- 73.Karabiber F, McGinnis JL, Favorov OV, Weeks KM. QuShape: Rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 2013; 19:63-73; PMID:23188808; http://dx.doi.org/ 10.1261/rna.036327.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Luo Y, Mao X, Deng L, Cong P, Shuman S. The D1 and D12 subunits are both essential for the transcription termination factor activity of vaccinia virus capping enzyme. J Virol 1995; 69:3852-6; PMID:7745734 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.