Abstract
Cancer/testis antigen HCA587/MAGE-C2 has been considered as a tumor specific target for immunotherapy. It has been reported that HCA587/MAGE-C2 plays an active role in tumorigenesis by promoting the growth and survival of tumor cells. However, the regulation of HCA587/MAGE-C2 expression in cancer cells remains largely unknown. MicroRNAs (miRNAs), a large family of gene regulators, have been shown to negatively regulate the expression of important cancer-related genes and contribute to the initiation and development of cancers. In this study, we conducted searches of miRNAs that regulate HCA587/MAGE-C2 expression. We combined bioinformatics tools with biological validation assays to demonstrate that HCA587/MAGE-C2 is a direct target of microRNA-874 (miR-874). Furthermore, we investigated the expression levels of miR-874 in human hepatocellular carcinoma tissues and paired adjacent normal tissues by stem-loop reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The results revealed a significant downregulation of miR-874 expression in tumor tissues compared to adjacent normal tissues. Finally, we demonstrated that overexpression of miR-874, as well as HCA587/MAGE-C2 silencing, resulted in suppression of tumor cell proliferation and invasion. Moreover, the inhibition effects of miR-874 on cell proliferation and invasion were reversed by co-expression of HCA587/MAGE-C2 in A375 cells. Taken together, our data demonstrated that HCA587/MAGE-C2 is a direct target of miR-874, and miR-874 may function as a tumor suppressive miRNA, at least in part, by negatively regulating HCA587/MAGE-C2 expression in cancer cells.
Keywords: Cancer/testis antigen, HCA587/MAGE-C2, MiRNA-874, Cell proliferation, Cell invasion.
Introduction
Cancer/testis antigens are characterized by a unique pattern of tissue expression, aberrantly expressed in a wide variety of tumors but absent from normal adult tissues other than immunoprivileged germ-line tissues 1. This tissue restriction expression makes them ideal targets for cancer immunotherapy. The list of cancer/testis antigens has grown to include more than 100 different gene families during the past two decades, with additional novel cancer/testis antigens continuing to be discovered 2. HCA587/MAGE-C2, a member of MAGE family, is a cancer/testis antigen expressed in a number of malignancies, including hepatocellular carcinoma, melanoma, bladder cancer, prostate cancer, lung cancer and breast cancer, etc 3-5. Recent studies revealed that HCA587/MAGE-C2 forms complexes with KAP1, a known co-repressor of p53, to suppress p53-dependent apoptosis, which indicated that HCA587/MAGE-C2 may play a role in the development of malignancies by promoting tumor cells survival 6, 7. In addition, HCA587/MAGE-C2 expression in primary melanoma has been identified as a potent predictor of sentinel lymph node metastasis 8. However, the regulation of HCA587/MAGE-C2 expression in cancer cells has been relatively understudied and remains a mystery.
MicroRNAs (miRNAs), a family of small non-coding RNAs of ~22 nucleotides, are a novel class of gene regulators. It is estimated that miRNAs potentially control the expression of about one-third of human mRNAs 9, 10. MiRNAs have been shown to negatively regulate the expression of important cancer-related genes and contribute to the initiation and development of various types of cancer 11-14. In this study, we conducted searches of miRNAs that regulate HCA587/MAGE-C2 expression. We identified MicroRNA-874 (miR-874) as a negative regulator of HCA587/MAGE-C2 and validated that HCA587/MAGE-C2 is a direct target of miR-874. Furthermore, we found that miR-874 was significantly downregulated in hepatocellular carcinoma tissues, and overexpression of miR-874 resulted in suppression of tumor cell proliferation and invasion.
Materials and methods
Tissue specimens
Hepatocellular carcinoma specimens and adjacent normal tissues were obtained from the PLA General Hospital. Tumor and adjacent normal tissues were flash frozen in liquid nitrogen for miR-874 analysis. Written informed consents were obtained from all the participants before collection of specimens. The study protocol was performed strictly according to the regulation of Peking University Health Science Center.
Cell culture and transfection
All the tumor cell lines were cultured in incubator supplemented with 5% CO2 at 37ºC. Human embryo kidney cell (HEK293T), human hepatocellular carcinoma cell (HLE) and human melanoma cell A375 were maintained in DMEM medium (ATCC, USA) with 10% fetal bovine serum (FBS, Invitrogen). Cells were transfected with miR-mimic, miR-inhibitor or siRNA (Ribobio, China) by using Lipofectamine 2000 (Invitrogen), and co-transfected with miR-mimic and HCA587 expression vector by using jetPRIME (Polyplus, France) according to the instructions. The siRNA sequence for HCA587/MAGE-C2 is: 5'-CAAUUGAUACCGCAGAUGA-3'.
Reverse-transcription quantitative PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 2μg RNA was reverse transcribed into cDNA using Reverse Transcription System kit (Promega, USA) according to the manufacture's protocol. Subsequently, real-time quantitative PCR was performed on ECO real-time polymerase chain reaction instruments (Illumina, USA). All the primers used for testing miR-874 expression were designed and synthesized by Ribobio Company. The specific primers for HCA587/MAGE-C2 used in quantitative PCR were: forward 5'-GGCCCTGAGGAAGAACTGAG-3', reverse 5'-TGAGATCCAACAGGCCTTGAC-3'. All experiments were performed independently in triplicate.
Western blot
Total protein from cell lines was extracted in RIPA lysis buffer, and then size-fractionated using SDS polyacrylamide gel electrophoresis. After that, protein was transferred to NC membrane. After blocking, the membrane was incubated with specific testing antibodies for HCA587/MAGE-C2, MAGE-A1 (Bioworld, USA), or MAGE-A3 (Origene, USA) and GAPDH (Bioworld). After blotting overnight at 4ºC, the membrane was incubated with HRP-conjugated anti-mouse or anti-rabbit IgG antibodies for 1h at room temperature. The expression of proteins was then tested by ECL system. GAPDH was used as internal reference.
3'-UTR luciferase plasmid construct and analysis
3'-UTR of HCA587/MAGE-C2 was cloned from the genome of A375 cells and mutated the putative sites which can bind miR-874. The mutant and wild type sequences were then cloned into pmiR-RB-Report vector (Ribobio) respectively. HEK293T cells were co-transfected with the constructed vector and miR-874 mimic. After 36 h of transfection, the firefly and Renilla dual-luciferase testing system were used to measure and calculate the luciferase activities.
MTS cell proliferation assay
HLE or A375 cells were transfected with miR-874 mimic and were seeded into 96-well plate at 36 h post transfection (5×103/well cells), respectively. After additional 24 h, 48 h, or 72 h, cells were added with 10 μl MTS (Promega, USA) per 100 μl medium and then proliferation of cells was assessed by recording the absorbance at 490 nm (OD values). A375 cells were transfected with HCA587/MAGE-C2 siRNA and proliferation of cells was measured at 48 h and 72 h post transfection with the method mentioned above.
Cell apoptosis assay—Annexin V/PI
A375 cells were transfected with miR-874 mimic. After 36 h, cells were harvested, washed and resuspended with binding buffer (Sungene, China). Cells were incubated with Annexin V-FITC for 20 min and propidium iodide (PI) for 5 min at room temperature in darkness before analyzing with flow cytometer.
Cell apoptosis assay—JC-1
The transfected A375 cells were harvested at 36 h post-transfection and washed, and then incubated with JC-1 (5 μl JC-1 into 500 μl assay buffer). After washing and resuspending, the apoptotic cells were analyzed via flow cytometer.
Cell invasion assay
HLE or A375 cells were transfected with miR-874 mimic, inhibitor or siRNA, and after 48 h of transfection, cells were seeded into the chambers (8 µm, Corning) containing 60 µg matrigel. For co-transfection of miRNA mimic and expression vector in A375 cells, HCA587 expression vector was first transfected into cells for 20 h, and then miR-874 mimic was transduced, and after 30 h, cells were seeded into the chambers. HLE cells transfected with miR-874 mimic or inhibitor were measured after seeding for 12 h or 8 h, while A375 cells were assessed after 16-18 h.
Statistical analysis
The data were expressed as mean ± standard deviation (SD), and statistical analysis was measured by SPSS 11.0. The relationship between two groups was analyzed via t-test. The relationship among two more variables was analyzed by One-way ANOVA.
Results
HCA587/MAGE-C2 is a direct target of miR-874
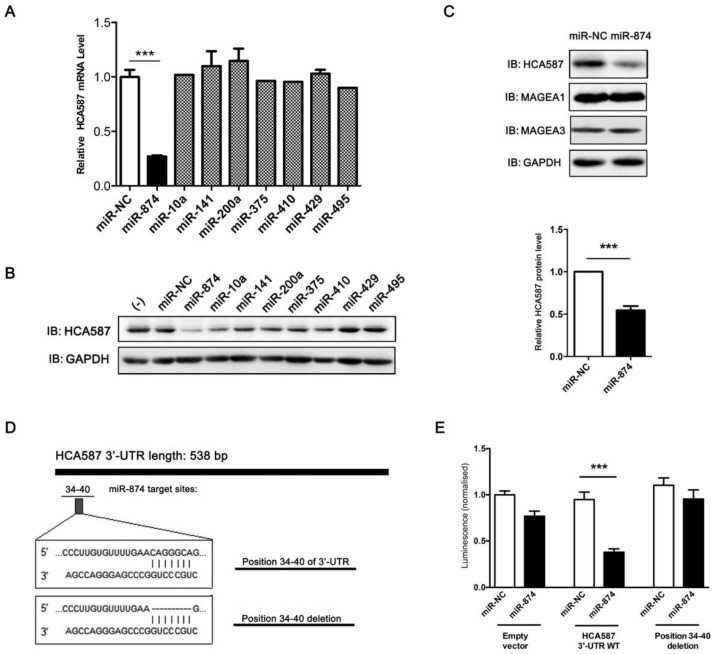
To explore the candidate miRNAs regulating HCA587/MAGE-C2, we first predicated the putative target sites of miRNAs in 3'-UTR of HCA587/MAGE-C2 using target prediction algorithms, TargetScan, miRanda and PicTar. Based on the data from different prediction software, 8 conserved miRNAs (miR-10a, miR-141, miR-200a, miR-375, miR-410, miR-429, miR-495 and miR-874) were selected, and in vitro gain-of-function analyses were performed using an overexpression strategy in human melanoma cell line A375, which endogenously expresses HCA587/MAGE-C2. As shown in Fig. 1A and 1B, overexpression of miR-874 mimic in A375 cells led to significant reduction of HCA587/MAGE-C2 at both mRNA and protein levels. We also detected the expression of other members of MAGE family, MAGE-A1 and MAGE-A3, in miR-874 mimic-transduced A375 cells, and no change was observed in both MAGE-A1 and MAGE-A3 expression compared with control cells (Fig. 1C).
Figure 1.
Prediction and validation of HCA587/MAGE-C2 as the target of miR-874. (A) Quantitative RT-PCR analysis of the relative HCA587 mRNA expression in A375 cells transfected with each mimic of 8 candidate miRNAs for 48 h. (B) Western blot analysis of the endogenous HCA587 protein level in A375 cells transfected with each mimic of 8 candidate miRNAs for 48 h. The quantitative analysis is shown in the right panel. (C) Western blot analysis of the endogenous HCA587, MAGE-A1 and MAGE-A3 protein levels in A375 cells transfected with miR-874 mimic for 48 h. (D) Schematic of putative binding site of miR-874 in HCA587 3'-UTR. A mutant HCA587 3'-UTR construct (position 34-40 deletion) was tested in parallel. (E) Analysis of luciferase activity. Renilla luciferase reporters containing either the wild-type or mutant form of HCA587 3'-UTR were co-transfected into HEK293T cells with miR-874 mimic or miR-NC. At 48 h post-transfection, Renilla luciferase activity was measured. The results were normalized against firefly luciferase values. (mean ± S.D.; *** p < 0.001).
To determine whether HCA587/MAGE-C2 is a direct target of miR-874, we performed luciferase reporter assay with a vector encoding the total sequence of the 3'-UTR of HCA587/MAGE-C2 mRNA, or a vector encoding the mutant 3'-UTR of HCA587/MAGE-C2 mRNA lacking the predicted miR-874 target site (positions 34-40). The putative target site of miR-874 in the 3'-UTR of HCA587/MAGE-C2 is illustrated in Fig. 1D. The results indicated that the luciferase activity was significantly reduced by co-transfection of the entire 3'-UTR of HCA587/MAGE-C2 and miR-874 mimic, whereas deletion of the miR-874 target site blocked the decrease in luciferase activity (Fig. 1E). These results clearly indicate that HCA587 is a direct target of miR-874.
MiR-874 is downregulated in human hepatocellular carcinoma tissues
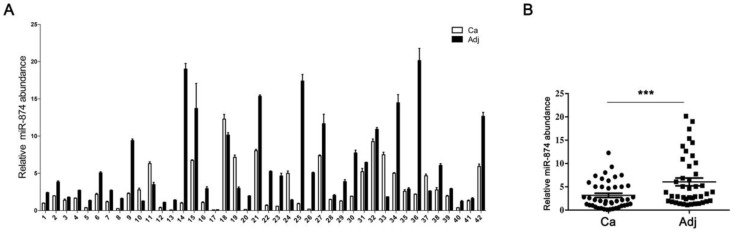
Based on the reports that HCA587/MAGE-C2 was highly expressed in human hepatocellular carcinoma (HCC) tissues 3, 15, we attempted to evaluate the expression level of miR-874 in 42 paired HCC and adjacent normal tissues. Stem-loop RT-qPCR showed that the expression level of miR-874 was significantly lower in 38 HCC specimens compared with their corresponding adjacent normal tissues (Fig. 2A and B), suggesting that miR-874 may function as a tumor suppressive miRNA in hepatocellular carcinoma.
Figure 2.
Expression of miR-874 in human hepatocellular carcinoma (HCC) tissues. (A) Expression levels of miR-874 in 42 HCC and corresponding adjacent normal tissues were measured by stem-loop quantitative RT-PCR. U6 snRNA was used as an internal control for miR-874. (B) Analysis of miR-874 expression in HCC and corresponding adjacent normal tissues. (mean ± S.D.; *** p < 0.001).
To explore the potential correlation between miR-874 and HCA587/MAGE-C2 expression in HCC tissues, we also detected HCA587/MAGE-C2 expression in the above mentioned 42 HCC tissues by PCR (Fig. S1A). We observed the expression of miR-874 in HCA587-positive samples is decreased than that in HCA587-negative groups (Fig. S1B). However, no statistically significant difference was found, probably owing to the limited number of samples.
MiR-874 inhibits cancer cell growth and invasion
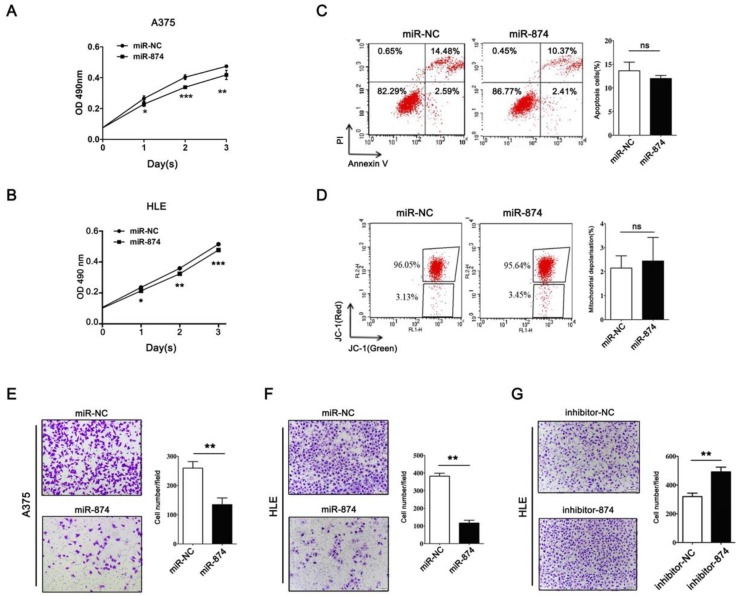
To investigate the biological roles of miR-874 in tumor cells, hepatocellular carcinoma cell line HLE and A375 cells were transfected with miR-874 mimic or miR-NC (negative control). MTS assay showed significant inhibition of cell proliferation in the miR-874 mimic-transfected cells compared with the miR-NC-transfected cells (Fig. 3A and B). To further address whether the observed changes in proliferation were due to cell apoptosis, both annexin-V/PI and JC-1 staining assays were used to detect the effect of miR-874 on cell apoptosis. The proportion of apoptotic cells in miR-874-transfected A375 cells showed no significant difference with that in control groups (Fig. 3C and 3D). Taken together, miR-874 inhibits the proliferation of cancer cells without affecting cell apoptosis.
Figure 3.
Effects of miR-874 on tumor cell proliferation and invasion. (A) MTS assays were performed at days 1, 2 and 3 after transfection of HLE cells with miR-874 mimic (transfection of 36 h is defined as day 0). (B) MTS assays were performed at days 1, 2 and 3 after transfection of A375 cells with miR-874 mimic (transfection of 36 h is defined as day 0). (C) Apoptosis profiles for A375 cells transfected with miR-874 mimic or miR-NC staining for Annexin V/PI. The quantitative analysis is shown in the right panel. (D) Apoptosis profiles for A375 cells transfected with miR-874 mimic or miR-NC staining for JC-1. The quantitative analysis is shown in the right panel. (E) Transwell invasion analysis of A375 cells treated with miR-874 mimic or miR-NC. The quantitative analysis is shown in the right panel. (F) Transwell invasion analysis of HLE cells treated with miR-874 mimic or miR-NC. (G) Transwell invasion analysis of HLE cells treated with miR-874 inhibitor or inhibitor-NC. (mean ± S.D.; * p < 0.05; ** p < 0.01; *** p < 0.001).
Matrigel invasion assay indicated that invading cell numbers were significantly decreased in miR-874 mimic-transfected A375 and HLE cells in comparison with the controls (Fig. 3E and 3F). Consistently, transfection with miR-874 inhibitor in HLE cells resulted in an increase in invasion ability (Fig. 3G).
HCA587/MAGE-C2 is involved in miR-874-regulated cell proliferation and invasion in A375 cells
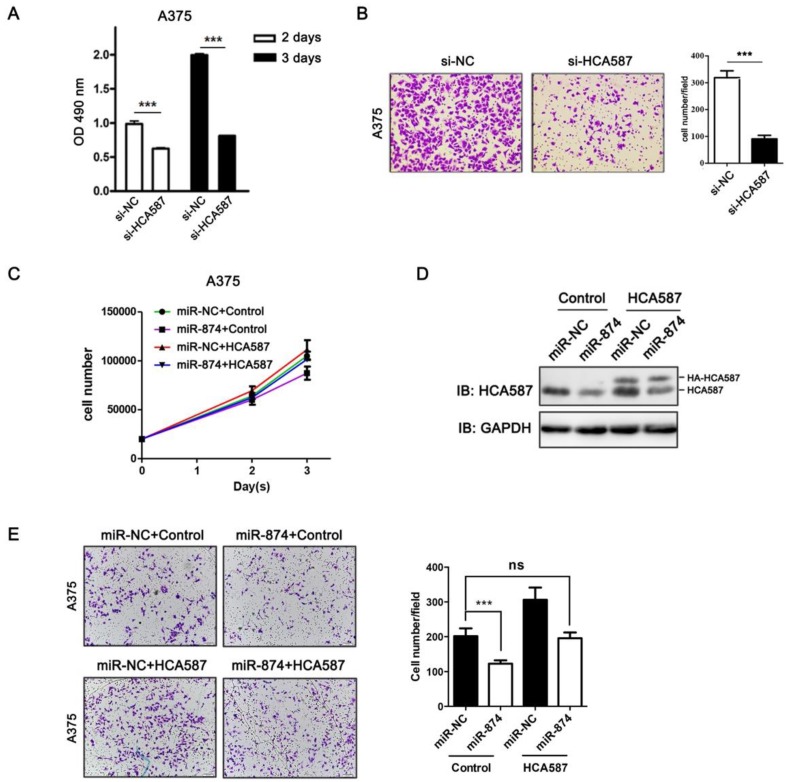
To examine the biological functions of HCA587/MAGE-C2, we performed loss-of-function studies by transfecting HCA587/MAGE-C2 siRNA into A375 cells. The protein and mRNA levels of HCA587/MAGE-C2 were markedly reduced by HCA587/MAGE-C2 siRNA transfection (Fig. S2A and S2B). MTS assay showed a significant inhibition of cell proliferation in HCA587/MAGE-C2 siRNA-transfected A375 cells compared with si-NC-transfected cells (Fig. 4A). Matrigel invasion assay demonstrated that the number of invading cells was significantly decreased in HCA587/MAGE-C2 siRNA-transfected cells in comparison with control (Fig. 4B). Furthermore, the inhibition effects of miR-874 on cell proliferation and invasion were rescued by co-expression of HCA587 vector in A375 cells (Fig.4C and 4E). The expression level of HCA587 in A375 cells transfected with miR-874 and HCA587 or control vectors was detected (Fig. 4D). These findings indicated that miR-874 inhibits cell proliferation and invasion by regulating HCA587 expression in A375 cells.
Figure 4.
HCA587/MAGE-C2 is involved in miR-874-regulated tumor cell proliferation and invasion. (A) MTS assays were performed at days 2 and 3 after transfection with si-HCA587/MAGE-C2 or si-NC. (B) Transwell invasion analysis of A375 cells treated with si-HCA587 or si-NC, and the quantitative analysis is shown in the right panel. (C) Cells counting analysis of A375 cells transfected with miR-874 or miR-NC plus HA-HCA587 expression vector or control at indicated times. (D) Western blot analyses of HCA587 expression in A375 cells transfected with miR-874 or miR-NC plus HA-HCA587 expression vector or control. (E) Transwell analysis of A375 cells treated with miR-874 or miR-NC plus HA-HCA587 expression vector or control, and the quantitative analysis is shown in the right panel. (mean ± S.D.; *** p < 0.001).
Discussion
HCA587/MAGE-C2, as a cancer/testis antigen, has been widely investigated as a potential target for vaccine in cancer 16-18. To date, 9 cytotoxic T lymphocyte (CTL) and 4 help T cell (Th) epitopes have been identified by our group and other investigators 19-24. In addition, it has been reported that HCA587/MAGE-C2 is an important prognostic factor and is associated with metastases as well as clinical relapse in melanoma and prostate cancer 8, 25. Recent evidence indicates that HCA587/MAGE-C2 plays an active role in tumorigenesis by promoting the growth and survival of tumor cells 6, 7. However, the regulation of HCA587/MAGE-C2 expression in cancer cells remains largely unknown to date. Here, we demonstrated that HCA587/MAGE-C2 is a direct target of miR-874, and miR-874 may function as a tumor suppressor by negatively regulating the expression of HCA587/MAGE-C2 in cancer cells.
MiRNAs have been known as a large family of gene regulators, and they negatively regulate their targets and are involved in the modulation of many physiological or pathological biological processes 9, 26. Bioinformatic analysis indicated that miRNAs regulate more than 30% of the protein coding genes 9. Accumulating evidence has indicated that miRNA expression correlated with various types of cancers 11. Some highly expressed microRNAs in tumor cells could function as oncogenes by repressing tumor suppressor genes, whereas downregulated microRNAs could act as tumor suppressors by negatively regulating oncogenes 9.
Based on bioinformatic analysis and biological validation, we found that miR-874 negatively regulated the expression of HCA587/MAGE-C2 in cancer cells, and further dual-luciferase reporter assay showed that HCA587/MAGE-C2 is a direct target of miR-874. MiR-874 was first identified based on small RNA library sequencing 27. MiR-874 is located on chromosome 5q31.2, a frequent fragile site of human genome which is often deleted in cancers 28, 29, implying that miR-874 may function as a tumor suppressor. Consistently, recent studies revealed that miR-874 was downregulated in maxillary sinus squamous cell carcinoma, non-small cell lung cancer, and breast cancer, and furthermore, tumor suppressive roles of miR-874 in inhibiting tumor growth and invasion in these cancer cells support that miR-874 is a tumor suppressor 30-32.
Our group and others have reported that HCA587/MAGE-C2 was highly expressed in hepatocellular carcinoma tissues 3, 15. In this study, we detected the expression of miR-874 by stem-loop RT-qPCR in hepatocellular carcinoma tissues, and downregulation of miR-874 in tumor tissues was discovered comparing with adjacent non-tumorous liver tissues. Furthermore, we found that overexpression of miR-874 in A375 cells decreased tumor cell proliferation and invasion, and knockdown of HCA587/MAGE-C2 with siRNA also resulted in reduced tumor cell invasion which is in consistence with overexpression of miR-874. In addition, overexpression of HCA587 rescued the inhibition effects of miR-874 on cell proliferation and invasion in A375 cells. By evaluating the proliferation capacity of A375 cells while performing invasion assay, we excluded the possibility that the inhibitory effect of miR-874 on cell invasion was associated with proliferation inhibition (data not shown). Our findings suggest the potential tumor suppressive roles of miR-874 in hepatocellular carcinoma tissues.
It was reported that miR-874 promotes apoptosis in gastric and breast cancer cells by targeting aquaporin-3 or CDK9, respectively, and stimulates necrosis of myocardial cells by regulating caspase-8 expression 32-34. However, we did not observe the effect of miR-874 on apoptosis in A375 cells, which is consistent with the results from knockdown of HCA587 with siRNAs in A375 cells (data not shown). It has been well known that microRNA can regulate multiple targets, and the protein targets are usually cell type-dependent, for example, SPRY2 is a target of miR-21 in colon cancer cells, but not in B16 cells 35. Whether CDK9, aquaporin-3, or caspase-8 is the target of miR-874 in A375 cells needs further investigation.
In summary, our study demonstrated that HCA587/MAGE-C2 is a direct target of miR-874, and miR-874 may function as a tumor suppressive microRNA, at least in part, by negatively regulating HCA587/MAGE-C2 in cancer cells.
Supplementary materials
Figures S1-S2.
Acknowledgments
This study was supported by grants from National Natural Science Foundation of China (No. 81472645) and Beijing Natural Science Foundation (7142087).
References
- 1.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer science. 2009;100(11):2014–21. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T. et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic acids research. 2009;37(Database issue):D816–9. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li B, Qian X-P, Pang X-W, Zou W-Z, Wang Y-P, Wu H-Y. et al. HCA587 Antigen Expression in Normal Tissues and Cancers: Correlation with Tumor Differentiation in Hepatocellular Carcinoma. Laboratory Investigation. 2003;83(8):1185–92. doi: 10.1097/01.lab.0000080605.73839.96. [DOI] [PubMed] [Google Scholar]
- 4.Lucas S, De Plaen E, Boon T. MAGE-B5, MAGE-B6, MAGE-C2, and MAGE-C3: four new members of the MAGE family with tumor-specific expression. International journal of cancer Journal international du cancer. 2000;87(1):55–60. [PubMed] [Google Scholar]
- 5.Wang Y, Han KJ, Pang XW, Vaughan HA, Qu W, Dong XY. et al. Large scale identification of human hepatocellular carcinoma-associated antigens by autoantibodies. Journal of immunology. 2002;169(2):1102–9. doi: 10.4049/jimmunol.169.2.1102. [DOI] [PubMed] [Google Scholar]
- 6.Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM. et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer research. 2007;67(20):9954–62. doi: 10.1158/0008-5472.CAN-07-1478. [DOI] [PubMed] [Google Scholar]
- 7.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Molecular cell. 2010;39(6):963–74. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curioni-Fontecedro A, Nuber N, Mihic-Probst D, Seifert B, Soldini D, Dummer R. et al. Expression of MAGE-C1/CT7 and MAGE-C2/CT10 predicts lymph node metastasis in melanoma patients. PloS one. 2011;6(6):e21418. doi: 10.1371/journal.pone.0021418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 10.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews Genetics. 2008;9(2):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(39):13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng JR, Chen HS, Mou DC, Cao J, Cong X, Qin LL. et al. Expression of cancer/testis (CT) antigens in Chinese hepatocellular carcinoma and its correlation with clinical parameters. Cancer letters. 2005;219(2):223–32. doi: 10.1016/j.canlet.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 16.Li B, Wu HY, Qian XP, Li Y, Chen WF. Expression, purification and serological analysis of hepatocellular carcinoma associated antigen HCA587 in insect cells. World J Gastroenterol. 2003;9(4):678–82. doi: 10.3748/wjg.v9.i4.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Germeau C, Ma W, Schiavetti F, Lurquin C, Henry E, Vigneron N. et al. High frequency of antitumor T cells in the blood of melanoma patients before and after vaccination with tumor antigens. The Journal of experimental medicine. 2005;201(2):241–8. doi: 10.1084/jem.20041379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, He X, Pang X, Zhang H, Chen J, Chen W. Elicitation of both CD4 and CD8 T-cell-mediated specific immune responses to HCA587 protein by autologous dendritic cells. Scand J Immunol. 2004;60(5):506–13. doi: 10.1111/j.0300-9475.2004.01503.x. [DOI] [PubMed] [Google Scholar]
- 19.Li B, Wang Y, Chen J, Wu H, Chen W. Identification of a new HLA-A*0201-restricted CD8+ T cell epitope from hepatocellular carcinoma-associated antigen HCA587. Clinical and experimental immunology. 2005;140(2):310–9. doi: 10.1111/j.1365-2249.2005.02786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xing Q, Pang XW, Peng JR, Yin YH, Li Y, Yu X. et al. Identification of new cytotoxic T-lymphocyte epitopes from cancer testis antigen HCA587. Biochemical and biophysical research communications. 2008;372(2):331–5. doi: 10.1016/j.bbrc.2008.05.049. [DOI] [PubMed] [Google Scholar]
- 21.Godelaine D, Carrasco J, Brasseur F, Neyns B, Thielemans K, Boon T. et al. A new tumor-specific antigen encoded by MAGE-C2 and presented to cytolytic T lymphocytes by HLA-B44. Cancer immunology, immunotherapy: CII. 2007;56(6):753–9. doi: 10.1007/s00262-006-0244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma W, Germeau C, Vigneron N, Maernoudt AS, Morel S, Boon T. et al. Two new tumor-specific antigenic peptides encoded by gene MAGE-C2 and presented to cytolytic T lymphocytes by HLA-A2. International journal of cancer Journal international du cancer. 2004;109(5):698–702. doi: 10.1002/ijc.20038. [DOI] [PubMed] [Google Scholar]
- 23.Ma W, Vigneron N, Chapiro J, Stroobant V, Germeau C, Boon T. et al. A MAGE-C2 antigenic peptide processed by the immunoproteasome is recognized by cytolytic T cells isolated from a melanoma patient after successful immunotherapy. International journal of cancer Journal international du cancer. 2011;129(10):2427–34. doi: 10.1002/ijc.25911. [DOI] [PubMed] [Google Scholar]
- 24.Wen W, Zhang L, Peng J, Chen J, Hao J, Li X. et al. Identification of promiscuous HLA-DR-restricted CD4(+) T-cell epitopes on the cancer-testis antigen HCA587. Cancer science. 2011;102(8):1455–61. doi: 10.1111/j.1349-7006.2011.01986.x. [DOI] [PubMed] [Google Scholar]
- 25.von Boehmer L, Keller L, Mortezavi A, Provenzano M, Sais G, Hermanns T. et al. MAGE-C2/CT10 protein expression is an independent predictor of recurrence in prostate cancer. PloS one. 2011;6(7):e21366. doi: 10.1371/journal.pone.0021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fundia AF, Gorla NB, Bonduel MM, Azpilicueta O, Lejarraga H, Muriel FS. et al. Increased expression of 5q31 fragile site in a Bloom syndrome family. Human genetics. 1992;89(5):569–72. doi: 10.1007/BF00219187. [DOI] [PubMed] [Google Scholar]
- 29.Thorland EC, Myers SL, Gostout BS, Smith DI. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene. 2003;22(8):1225–37. doi: 10.1038/sj.onc.1206170. [DOI] [PubMed] [Google Scholar]
- 30.Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T. et al. Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. British journal of cancer. 2011;105(6):833–41. doi: 10.1038/bjc.2011.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kesanakurti D, Maddirela DR, Chittivelu S, Rao JS, Chetty C. Suppression of tumor cell invasiveness and in vivo tumor growth by microRNA-874 in non-small cell lung cancer. Biochemical and biophysical research communications. 2013;434(3):627–33. doi: 10.1016/j.bbrc.2013.03.132. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Gao W, Hu F, Xu Z, Wang F. MicroRNA-874 inhibits cell proliferation and induces apoptosis in human breast cancer by targeting CDK9. FEBS Lett. 2014;588(24):4527–35. doi: 10.1016/j.febslet.2014.09.035. [DOI] [PubMed] [Google Scholar]
- 33.Jiang B, Li Z, Zhang W, Wang H, Zhi X, Feng J, Chen Z, Zhu Y, Yang L, Xu H, Xu Z. miR-874 Inhibits cell proliferation, migration and invasion through targeting aquaporin-3 in gastric cancer. J Gastroenterol. 2014;49(6):1011–25. doi: 10.1007/s00535-013-0851-9. [DOI] [PubMed] [Google Scholar]
- 34.Wang K, Liu F, Zhou LY, Ding SL, Long B, Liu CY, Sun T, Fan YY, Sun L, Li PF. miR-874 regulates myocardial necrosis by targeting caspase-8. Cell Death Dis. 2013;4:e709. doi: 10.1038/cddis.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011;286(45):39172–8. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S2.