Abstract
Male infertility is a worldwide medical problem. Asthenozoospermia is a common cause of infertility. Epigenetic modifications of DNA and histones have been shown to influence human infertility, but no research has explored whether N6-methyladenosine (m6A) level in RNA is associated with asthenozoospermia. Here, we collected a total of 52 semen samples, including 20 asthenozoospermia patients and 32 healthy controls. An LC-ESI-MS/MS method was used to detect m6A contents in sperm RNA, and real-time PCR was performed to determine the mRNA expression of demethylase (FTO, ALKBH5), methyltransferase (METTL3, METTL14, WTAP) and an m6A-selective-binding protein (YTHDF2). We found that m6A content (p = 0.033) and the mRNA expression of METTL3 (p = 0.016) and METTL14 (p = 0.025) in asthenozoospermia patients were significantly higher than those of controls. Increased m6A content was a risk factor for asthenozoospermia (odds ratio (OR) 3.229, 95% confidence interval (CI) 1.178 – 8.853, p = 0.023). Moreover, m6A content was correlated with the expression of METTL3 (r = 0.303, p = 0.032) and with sperm motility (progressive motility: r = −0.288, p = 0.038; non-progressive motility: r = −0.293, p = 0.037; immotility: r = 0.387, p = 0.005). Our data suggest that increased m6A content is a risk factor for asthenozoospermia and affects sperm motility. Methyltransferases, particularly METTL3, play key roles in increasing m6A contents in sperm RNA.
Human infertility is a prevalent medical problem, which affects approximately 15% of couples worldwide1,2. Tragically, the infertility rates in some developing countries can reach 30%3. Male infertility contributes to more than 50% of all cases of infertility4. Optimal male fertility depends on an adequate number of normal, motile, mature and physiologically functional sperm5, which may be influenced by many factors, such as genetic disorders, infections of the genital tract, medical intervention and environmental degradation1. Decreased semen quality and defective sperm function appear to be common causes of male infertility1. Many current studies have probed the pathogenesis of infertility, but the interference of different epigenetic alterations in infertility is not fully understood.
Epigenetics is defined as a heritable change affecting gene expression that is not caused by DNA sequence alterations6. Epigenetic modifications of DNA and histones have been confirmed as having significant roles throughout spermatogenesis2. There are two characteristic features of sperm epigenetic programming. The first feature is a major removal of epigenetic markers occurring in the primordial germ cells (PGCs). The second feature is the reorganization and condensation of the germ cell genome during postmeiotic maturation, including DNA methylation and histone modifications during spermiogenesis2. Abnormal sperm DNA methylation levels are related to altered semen parameters7. Moreover, many specific RNAs, mRNAs, miRNAs, and Piwi-interacting RNAs (piRNAs) in sperm are vital for male fertilization8,9,10,11. In addition to DNA and histone modifications, epigenetic modifications of RNA have been proposed to be another layer of epigenetic regulation. Up to date, more than 100 types of RNA modifications, occurring in mRNA, tRNA, rRNA and small nuclear RNA (snRNA), have been identified12. Among these modifications, N6-methyladenosine (m6A) modification is the most prevalent in mammalian mRNA13. Despite m6A modification being first reported in early 1970s14, its biological function and significance in sperm RNA are largely unknown.
In vivo, the dynamic regulation of m6A modification plays important roles through the functional interplay among m6A methyltransferases and demethylases15. The methyltransferase complex, which is composed of methyltransferase like 3 (METTL3), methyltransferase like 14 (METTL14) and Wilms tumor 1 associated protein (WTAP), has been identified as a crucial factor catalyzing the formation of m6A with S-adenosyl-L-methionine (SAM) as the methyl donor16. In contrast to methyltransferases, two demethylases, the alpha-ketoglutarate- and Fe2+ -dependent dioxygenase fat mass and obesity associated protein (FTO) and AlkB family member 5 protein (ALKBH5), have similar functions as the TET enzyme in DNA demethylation17; i.e., both FTO and ALKBH5 can remove the methyl group from m6A in RNA18,19. In this dynamic modification, methyltransferases act as ‘writers’, demethylases serve as ‘erasers’, and m6A-selective-binding proteins(YTHDF) represent ‘readers’ of m6A in mRNA. More recently, the YTHDF domain family 2 protein (YTHDF2) has been shown to regulate RNA stability, translation, splicing, transport and localization through selective recognition of methylated RNA20,21.
Unlike DNA methylation, very few studies directly focused on the role of RNA methylation in human disease, particularly m6A. Recent advances have demonstrated that FTO expression and m6A levels are inversely correlated during adipogenesis22 and the FTO gene encoding an m6A demethylase is also associated with human obesity23. We have reported that the increased mRNA expression of FTO contributed to the reduction of m6A in type 2 diabetes mellitus24. However, the association of m6A with human fertility can be traced back to 1997, when Bokar and colleagues first revealed using Northern blot analysis that the mRNA of METTL3 (MT-A70) was expressed in a wide variety of human tissues, with the highest levels being in testis25. In addition, a recent animal study documented that Alkbh5-deficient male mice showed increased m6A in mRNA and impaired fertility due to compromised spermatogenesis19. Based on these experimental results, we hypothesize that m6A modification is associated with human asthenozoospermia and plays a role in male infertility.
To verify our hypothesis, in the current study, we first detected the m6A contents in sperm RNA from asthenozoospermia patients and healthy controls. Compared with the controls, m6A content in asthenozoospermia patients was significantly increased, which is a risk factor for asthenozoospermia. To better understand this phenomenon, we performed real-time PCR to examine the mRNA expression of key genes involved in m6A regulation. Our results indicated that the mRNA expression of METTL3 and METTL14 was higher than that of controls. In addition, m6A content was closely related to the mRNA expression of METTL3 and clinical parameters of sperm.
Results
Clinical parameters of semen
The clinical parameters of semen from 20 asthenozoospermia patients and 32 healthy controls are given in Table 1. Student’s t test indicated that there was no significant difference in semen pH and teratozoospermia index (TZI) between the two groups; nevertheless, asthenozoospermia patients showed significantly lower sperm concentration (39.2 × 106/mL vs. 96.6 × 106/mL), progressive motility (8.6% vs. 24.7%), non-progressive motility (19.8% vs. 44.5%) and normal morphology (5.1% vs. 8.2%) and higher sperm immotility (72.4% vs.30.8%) and sperm deformity index (1.1% vs. 1.0%) compared with the controls.
Table 1. Clinical parameters of semen in controls and patients.
| Patients (n = 20) | Controls (n = 32) | p values | |
|---|---|---|---|
| pH value | 7.1 ± 0.2 | 7.1 ± 0.1 | 0.906 |
| Concentration (106/mL) | 39.2 ± 23.9 | 96.6 ± 74.0 | 0.000 |
| Progressive motility (PR, %) | 8.6 ± 6.0 | 24.7 ± 10.2 | 0.000 |
| Non-progressive motility (NP, %) | 19.8 ± 7.5 | 44.5 ± 14.2 | 0.000 |
| Immotility (IM, %) | 72.4 ± 10.5 | 30.8 ± 13.2 | 0.000 |
| Normal morphology (%) | 5.1 ± 3.2 | 8.2 ± 4.7 | 0.017 |
| Teratozoospermia index (TZI, %) | 1.1 ± 0.1 | 1.1 ± 0.1 | 0.291 |
| Sperm deformity index (SDI, %) | 1.1 ± 0.1 | 1.0 ± 0.1 | 0.029 |
LC-ESI-MS/MS method development
The linearity of this method was investigated using 40 pmol rA standard supplemented with m6A at different amounts ranging from 8 fmol to 800 fmol (Table S1). The calibration curves were constructed by plotting the mean peak area ratio of m6A/rA versus the mean molar ratio of m6A/rA based on data obtained from triplicate measurements. The results showed linearity within the range of 0.02–2% (molar ratio of m6A/rA) with the coefficient of determination (R2) higher than 0.9977 (Table S1). Limits of detection (LOD) and limits of quantification (LOQ) for m6A were calculated as the amounts of the analytes at signal/noise ratios of 3 and 10, respectively. The LOD and LOQ were 1.2 fmol and 4.0 fmol for m6A, respectively (Table S1). Validation of the method was accomplished using the synthesized m6A-containing oligonucleotide by comparing the measured m6A content to the theoretical m6A content (Table S2). m6A was determined from RNA hydrolysis products with RSDs being 1.8–10.6% and REs being 2.7–12.1% (Table S3), indicating high accuracy and precision for the determination of m6A by LC-ESI-MS/MS.
m6A contents increased in asthenozoospermia
To investigate whether m6A modification differs in asthenozoospermia patients and controls, the m6A contents of 52 sperm RNA samples, including 20 asthenozoospermia patients and 32 controls, were analyzed by LC-ESI-MS/MS in multiple reaction monitoring (MRM) mode (Table S4). The MRM chromatograms of 9 standard nucleosides and the hydrolysis products of 50 ng sperm RNA from an asthenozoospermia patient are shown in Fig. 1A,B. The average contents of m6A in asthenozoospermia patients and healthy controls were 0.18 ± 0.02% and 0.13 ± 0.01%, respectively. Student’s t test indicated asthenozoospermia patients had higher m6A contents (p = 0.033), as shown in Fig. 1C. Importantly, logistic regression analysis revealed that increased m6A content in sperm RNA was a risk factor for asthenozoospermia (odds ratio (OR) 3.229, 95% confidence interval (CI) 1.178 – 8.853, p = 0.023).
Figure 1. The MRM chromatograms of nucleosides and difference of m6A contents between the two groups.
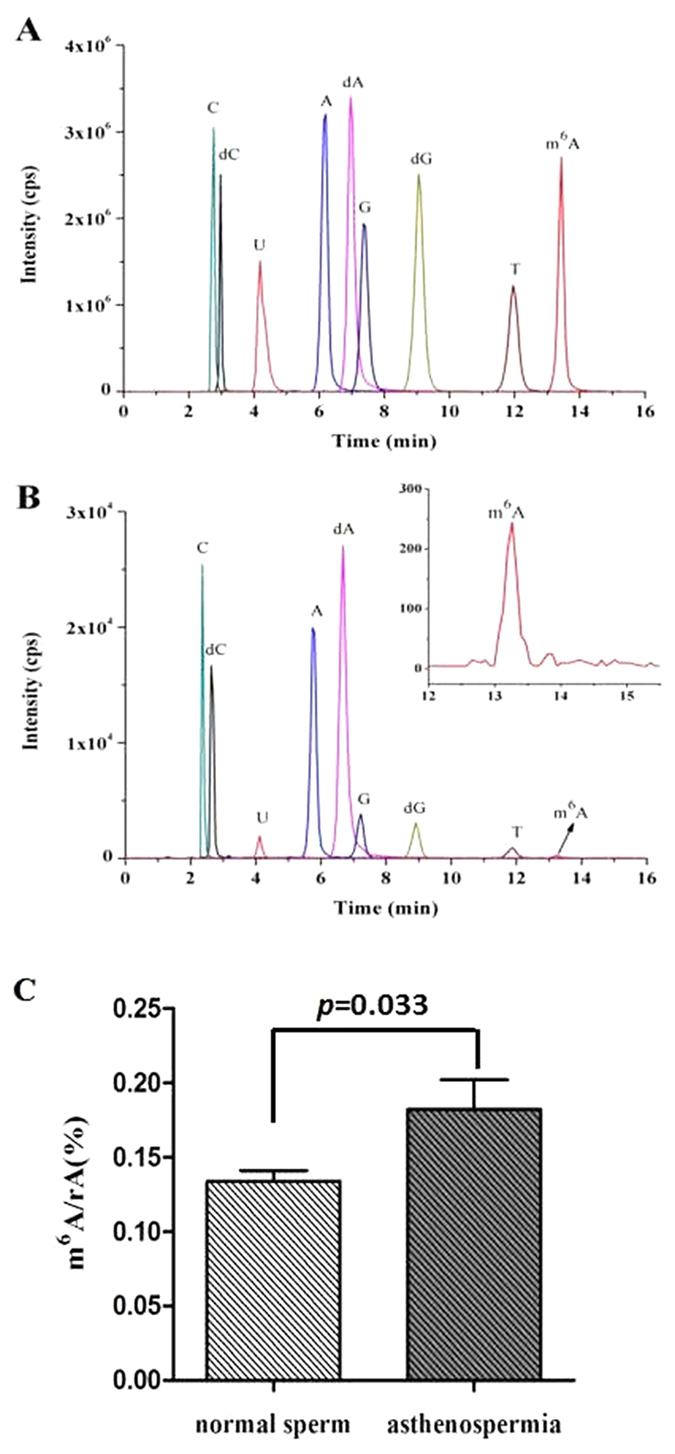
(A) Standard nucleosides. (B) 50 ng sperm RNA from an asthenozoospermia patient. Shown in inset is the enlargement chromatogram of m6A. (C) Comparative analysis of m6A contents in sperm RNA from asthenozoospermia patients (n = 20) and controls (n = 32).
mRNA expression of the regulatory genes of dynamic RNA methylation and its correlation with m6A contents
To address why m6A contents increased in asthenozoospermia patients, we utilized real-time PCR to determine the mRNA expression levels of the core regulatory genes that would be engaged in dynamic m6A modification of RNA (Fig. S1). We used Student’s t test to compare the differences in ALKBH5 and METTL14 between the two groups, and the Mann-Whitney U test for FTO, METTL3, WTAP and YTHDF2 comparisons. Asthenozoospermia patients showed significantly higher mRNA expression levels of the methyltransferases, including METTL3 (p = 0.016) and METTL14 (p = 0.025) compared to the healthy controls (Fig. 2B). The average mRNA expression levels of METTL3 and METTL14 were 2.4-fold (p = 0.016) and 2.5-fold (p = 0.025) higher, respectively, than those of controls (Fig. 2B). However, there was no significant difference in FTO, ALKBH5, WTAP or YTHDF2 mRNA levels between asthenozoospermia patients and controls (Fig. 2A,B). Interestingly, Spearman correlation analysis indicated that the mRNA expression of METTL3 (r = 0.303, p = 0.032) (Fig. 2C), WTAP (r = 0.324, p = 0.023) and YTHDF2 (r = 0.338, p = 0.014) (Fig. S2) associated with the contents of m6A in sperm RNA; while no correlation was found between FTO expression and m6A contents (r = 0.268, p = 0.057); Pearson correlation analysis revealed neither ALKBH5 (r = 0.055, p = 0.700) nor METTL14 (r = 0.194, p = 0.172) was correlated with the contents of m6A (Fig. S2). Our data suggested that the increased m6A contents in asthenozoospermia patients might be caused by the upregulation of methyltransferase encoding genes, particularly METTL3.
Figure 2. mRNA expression of the regulatory genes of dynamic RNA methylation and the correlation of METTL3 expression with m6A contents.
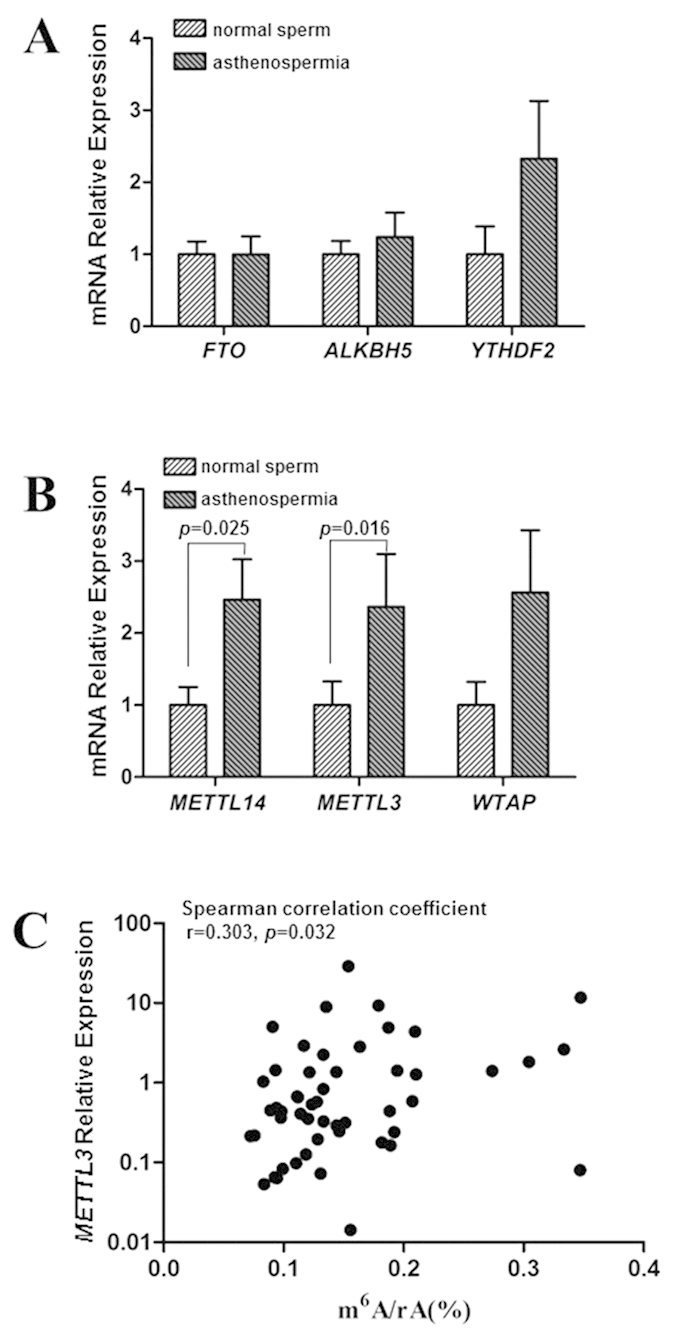
(A,B) Gene expression levels of the regulatory genes of dynamic RNA methylation, normalized to GAPDH, were examined by real-time PCR. Data are expressed as means ± SEM. (C) Spearman correlation analysis between m6A contents and mRNA expression of METTL3 in all subjects.
m6A content associated with clinical parameters of sperm
To test whether m6A content is related to the clinical parameters of sperm, we performed Pearson correlation analysis to evaluate the correlation between m6A content and several clinical parameters. m6A content was negatively correlated with sperm progressive motility (r = −0.288, p = 0.038) and non-progressive motility (r = −0.293, p = 0.037), and positively correlated with sperm immotility (r = 0.387, p = 0.005) (Table 2). No correlation was found between m6A contents and other clinical parameters.
Table 2. Pearson correlation analysis of m6A content with clinical parameters of semen.
| Pearson correlation coefficient | p values | |
|---|---|---|
| pH value | 0.043 | 0.760 |
| Concentration (106/mL) | −0.238 | 0.089 |
| Progressive motility (PR, %) | −0.288 | 0.038 |
| Non−progressive motility (NP, %) | −0.293 | 0.037 |
| Immotility (IM, %) | 0.387 | 0.005 |
| Normal morphology (%) | −0.039 | 0.792 |
| Teratozoospermia index (TZI, %) | −0.033 | 0.823 |
| Sperm deformity index (SDI, %) | 0.001 | 0.996 |
Discussion
The majority of male infertility results from asthenozoospermia. Despite tremendous efforts to characterize the genetic basis for this disorder, almost half of male infertility cases are of unknown etiology26. So far, few reports described RNA epigenetic alterations in human asthenozoospermia. Given the importance of m6A in RNA stability, splicing regulation, miRNA attenuation, RNA editing prevention, disease acceleration and epigenome control27, we first established a highly sensitive LC-ESI-MS/MS method to measure the m6A content in sperm RNA.
Even though there are many methods to detect m6A in RNA, such as MeRIP-Seq (m6A-specific methylated RNA immunoprecipitation combined with next-generation sequencing)28, the facile enzymatic ligation-based method29, the selective Polymerase method30, SMRT reverse transcription31 and the LC-ESI-MS/MS method24. We chose the LC-ESI-MS/MS method to determine m6A contents in the semen samples as its high sensitivity, relative low cost and satisfying qualitative and quantitative ability.
One of the most obvious and distinctive features of asthenozoospermia is weak sperm motility, which is defined as the proportion of progressively motile sperm. In our study, correlations existed between m6A contents and sperm progressive motility, non-progressive motility and immotility, suggesting that increased m6A in sperm RNA from asthenozoospermia patients might be a leading cause of the reduction of sperm motility. Previous studies have demonstrated that the nucleus of mature sperm contains a complex population of RNAs8, and differential expression of sperm mRNA is correlated with sperm motility and male reproduction32. We also found that m6A was present in human sperm RNA, and asthenozoospermia patients had higher m6A contents. Thus, we propose that the abnormal m6A modification in sperm RNA may affect the steady mRNA expression of certain genes related to sperm motility, such as cysteine-rich secretory protein 2 (CRISP2)33,34 and human β-defensin 1 (DEFB1)35. However, determining which genes are responsible will require further exploration.
The discovery and characterization of m6A demethylases and methyltransferases and an m6A-selective-binding protein reveals the functional and reversible regulatory mechanism of m6A modification. Theoretically, the altered mRNA expression levels of demethylases, methyltransferases and m6A-selective-binding protein encoding genes may lead to abnormal m6A modification and further impact on the fundamental biological process and development of disease36. Our results demonstrated that METTL3 played a major role in altering the contents of m6A in asthenozoospermia patients and influenced fertility. METTL3 (MT-A70) was confirmed as one component of the N6-adenosine methyltransferase complex. Bokar and colleagues25 isolated the methyltransferase complex from HeLa cell nuclear extracts and named the 70 kDa protein, which exhibited methyltransferase activity, MT-A70 or METTL3. Together with METTL14, another methyltransferase, METTL3 forms a stable heterodimer to mediate mammalian nuclear m6A methylation16. Interestingly, WTAP itself has no methyltransferase activity, but it can affect cellular m6A by interacting with the METTL3-METTL14 complex16. Recent studies have shown that METTL3 participates in the regulation of a variety of life processes by catalyzing the formation of m6A, including stem cell self-renewal and differentiation37, controlling the circadian clock38 and marking primary microRNAs for recognition and processing39.
It is worth noting that ALKBH5 deficiency can result in the elevation of m6A in male mice and further impair fertility19, whereas our data revealed that METTL3 but not ALKBH5 played a key role in human male infertility, especially in asthenozoospermia. The possible reason for this difference between humans and mice might be the diversity of the gene expression model.
Indeed, the isolation of sperm RNA is a challenging work owing to the intrinsic heterogeneous cells exist in the ejaculate and the low quantity of RNA present in Spermatozoon. Although we cannot absolutely ensure that no RNA isolated from other cellular structures in sperm purified by a typical Percoll gradient centrifugation, the potential influences are likely limited.
In summary, we first reported that methyltransferases (METTL3 and METTL14), particularly METTL3, play key roles in the dynamic modification of m6A in asthenozoospermia patients and that increased m6A can impair sperm motility. According to our results, we deduce a process of dynamic modification of m6A in asthenozoospermia patients (Fig. 3). The underlying mechanisms of m6A modification in asthenozoospermia still need further investigation.
Figure 3. A process of the dynamic m6A modification in asthenozoospermia.
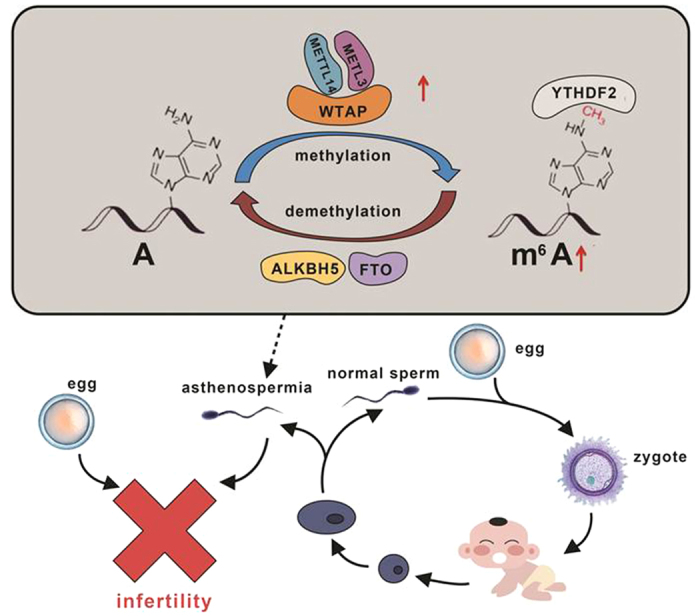
Increased mRNA expression of METTL3 and METTL14 contributes to the elevation of m6A content which affects sperm motility and further results in male infertility.
Materials and Methods
Chemicals and reagents
Adenosine (rA), uridine (rU), cytidine (rC), guanosine (rG), 2′-deoxyadenosine (dA), thymidine (T), 2′-deoxycytidine (dC), 2′-deoxyguanosine (dG) were purchased from Sigma-Aldrich (Beijing, China). N6-methyladenosine (m6A) was from Hanhong Chemical Co., Ltd. (Shanghai, China). S1 nuclease and alkaline phosphatase (CIAP) were purchased from Takara Biotechnology Co., Ltd. (Dalian, China). Phosphodiesterase I was from Sigma-Aldrich (St. Louis, MO, USA). Chloroform and formic acid were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Chromatographic grade methanol was purchased from Merck (Darmstadt, Germany). All water was purified by a Milli-Q apparatus (Millipore, Bedford, MA). Stock solutions of the ribonucleosides and 2′-deoxynucleosides were prepared in Milli-Q water at a concentration of 5 mmol/L.
Semen sample collection
The ethics committee of Zhongnan Hospital of Wuhan University has approved this study. The study was carried out in accordance with the approved guidelines of the ethics committee of Zhongnan Hospital of Wuhan University and informed consent was obtained from each participant at the time of semen sample collection. According to the 5th edition of the WHO laboratory manual for the examination and processing of human semen26, 20 asthenozoospermia patients and 32 healthy controls from the Productive Medicine Center of Zhongnan Hospital donated semen samples by masturbation after 2–7 days of abstinence. All participants were without leukocytospermia or reproduction tract infection. To eliminate the influence of age on the results, in subjects selection we ensured no significant difference in age between healthy controls and asthenozoospermia patients (32.2 ± 5.5 years VS. 32.4 ± 4.5 years, p = 0.901). We defined asthenozoospermia as forward progression (progressive motility and non-progressive motility) sperm <40% and healthy control as forward progression sperm ≥40% or progressive motility sperm ≥32%. Fresh semen samples were incubated at 37 °C for 30 min to allow for liquefaction before further processing.
Sperm purification and RNA extraction
Human sperm was purified from 2–3 mL of liquefied semen samples using density gradient centrifugation26. Briefly, the sample was transferred to a 15 mL conical tube (RNase-Free), and then 6–10 mL (0.01 mol/L) phosphate-buffered saline (PBS) was added and the tube inverted several times to wash the sperm. After centrifugation at 300 × g for 15 min at 4 °C, the supernatant was discarded and the sediment was resuspended in 1 mL of PBS. The sample was carefully overlaid onto a two-layer (40–80%) Percoll gradient to avoid disturbing the interface. Following another centrifugation at 400 × g for 20 min, the supernatant was discarded and the sperm pellet was resuspended in 2 mL of PBS. To confirm the purity of spermatozoa preparation, the sperms were examined by microscope(Olympus, Japan) after purification, and no lymphocytes, epithelial cells or bacteria was found under 5 microscopic vision fields (400×) (Fig. S3).Then the sperm RNA was extracted from the purified sperm suspension in a biosafety cabinet using a commercial RNAprep pure Cell/Bacteria Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. RNA was quantified by spectrophotometry (ND-2000, NanoDrop Inc., USA).
Oligonucleotides
A 15-mer m6A-containing oligonucleotide was synthesized according to a previously described method40. The 10-mer RNA (5′-AUCUAUAUGC-3′) was purchased from Takara Biotechnology Co., Ltd. (Dalian, China).
Enzymatic digestion of RNA
RNA (200 ng) was first denatured by heating at 95 °C for 5 min and then chilling on ice for 2 min. After adding 1/10 volume of S1 nuclease buffer (30 mM CH3COONa, pH 4.6; 280 mM NaCl; 1 mM ZnSO4) and 150 units of S1 nuclease, the mixture (20 μL) was incubated at 37 °C for 4 h. The solution was subsequently added to 1/10 volume of alkaline phosphatase buffer (50 mM Tris-HCl, 10 mM MgCl2, pH 9.0), 0.002 units of venom phosphodiesterase I and 15 units of alkaline phosphatase. Then, the incubation was continued at 37 °C for an additional 2 h followed by extraction with an equal volume of chloroform twice. The resulting aqueous layer was collected and lyophilized to dryness and reconstituted in 100 μL of water. Next, 30 μL of the obtained samples was subjected to liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) analysis.
Measurement of m6A content by LC-ESI-MS/MS
Analysis of nucleosides was performed on the LC-ESI-MS/MS system consisting of an AB 3200 QTRAP mass spectrometer (Applied Biosystems, Foster City, CA, USA) with an electrospray ionization source (TurboIonSpray) and a Shimadzu LC-20AD HPLC (Tokyo, Japan) with two LC-20AD pumps, a SIL-20A autosampler, a CTO-20AC thermostatic column compartment, and a DGU-20A3 degasser. Data acquisition and processing were performed using AB SCIEX Analyst 1.5 Software (Applied Biosystems, Foster City, CA). The HPLC separation was performed on a Hisep C18-T column (150 mm × 2.1 mm i.d., 5 μm, Weltech Co., Ltd., Wuhan, China) with a flow rate of 0.2 mL/min at 35 °C. Formic acid in water (0.1%, v/v, solvent A) and formic acid in methanol (0.1%, v/v, solvent B) were employed as the mobile phase. A gradient of 5 min 5% B, 10 min 5–30% B, 5 min 30–50% B, 3 min 50% B-5% B and 17 min 5% B was used.
The mass spectrometry detection was performed under positive electrospray ionization mode. The target nucleosides were monitored by multiple reaction monitoring (MRM) mode using the mass transitions (precursor ions → product ions) of m6A (282.2 →150.1), rA (268.4 → 136.2), rU (245.4 → 113.1), rC (244.4 → 112.2), rG (284.5 → 152.2), dA (252.4 → 136.2), T (243.3 → 127.2), dC (228.4 → 112.2), dG (268.4 → 152.4). The MRM parameters of all nucleosides were optimized to achieve maximal detection sensitivity.
Quantification of mRNA expression by real-time PCR
For mRNA quantifications of FTO, ALKBH5, METTL3, METTL14, WTAP and YTHDF2, cDNA was synthesized by DNase treatment and reverse transcription (TOYOBO, Osaka, Japan) and real-time PCR was performed on a CFX96 Touch TM Real-Time PCR Detection System (BioRad) with iTaq TM Universal Supermixes (BioRad) and intron-spanning primers, which are listed in Table S5. The mRNA levels of the target genes were normalized to that of the reference gene GAPDH, and the results are expressed as the means ± SEM.
Statistical analyses
All statistical analyses were performed using SPSS 19.0 software (SPSS Inc., Chicago, USA). A normality test was used to explore the data distribution, p > 0.05 was consider to be normal distributed. Two-tailed Student’s t test and Mann-Whitney U test were used to compare the differences in normal and non-normal distributed data between the two groups, respectively. The correlations of m6A contents with gene expression and semen parameters were assessed by Pearson correlation coefficient for normal distributed data and Spearman correlation coefficient for non-normal distributed data. Logistic regression analysis was used to evaluate whether m6A contents associated with asthenozoospermia risk. p < 0.05 was considered to be statistically significant.
Additional Information
How to cite this article: Yang, Y. et al. Increased N6-methyladenosine in Human Sperm RNA as a Risk Factor for Asthenozoospermia. Sci. Rep. 6, 24345; doi: 10.1038/srep24345 (2016).
Supplementary Material
Acknowledgments
We thank Professor Chuanhua Yu (School of Public Health, Global Health Institute, Wuhan University) for statistical guidance. We also thank the financial support from the National Basic Research Program of China (973 Program) (2012CB720600, 2012CB720601, 2012CB720603, 2012CB720605), the National Natural Science Foundation of China (81271919, 81472023).
Footnotes
Author Contributions Y.Y., Y.-Q.F., B.-F.Y. and S.-M.L. designed research; Y.Y., W.H., J.-T.H., F.S. and E.-F.Y. conducted research; M.Z., J.-T.H. and S.S.-Q. collected the semen samples and clinical informations; W.H. and J.X. detected m6A contents; Y.Y., W.H., B.-F.Y. and S.-M.L. prepared the initial manuscript draft; Y.Q.-F., B.-F.Y. and S.-M.L. edited and revised subsequent drafts; S.-M.L. had primary responsibility for final content.
References
- Urdinguio R. G. et al. Aberrant DNA methylation patterns of spermatozoa in men with unexplained infertility. Human reproduction, 10.1093/humrep/dev053 (2015). [DOI] [PubMed] [Google Scholar]
- Boissonnas C. C., Jouannet P. & Jammes H. Epigenetic disorders and male subfertility. Fertil Steril 99, 624–631, 10.1016/j.fertnstert.2013.01.124 (2013). [DOI] [PubMed] [Google Scholar]
- Ombelet W., Cooke I., Dyer S., Serour G. & Devroey P. Infertility and the provision of infertility medical services in developing countries. Human reproduction update 14, 605–621, 10.1093/humupd/dmn042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inhorn M. C. & Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Human reproduction update, 10.1093/humupd/dmv016 (2015). [DOI] [PubMed] [Google Scholar]
- Garrido N., Garcia-Herrero S. & Meseguer M. Assessment of sperm using mRNA microarray technology. Fertil Steril 99, 1008–1022, 10.1016/j.fertnstert.2013.02.006 (2013). [DOI] [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science 238, 163–170 (1987). [DOI] [PubMed] [Google Scholar]
- Aston K. I., Punj V., Liu L. & Carrell D. T. Genome-wide sperm deoxyribonucleic acid methylation is altered in some men with abnormal chromatin packaging or poor in vitro fertilization embryogenesis. Fertil Steril 97, 285–292, 10.1016/j.fertnstert.2011.11.008 (2012). [DOI] [PubMed] [Google Scholar]
- Hamatani T. Human spermatozoal RNAs. Fertil Steril 97, 275–281, 10.1016/j.fertnstert.2011.12.035 (2012). [DOI] [PubMed] [Google Scholar]
- Sendler E. et al. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic acids research 41, 4104–4117, 10.1093/nar/gkt132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. M. et al. Sperm-borne microRNA-34c is required for the first cleavage division in mouse. Proceedings of the National Academy of Sciences of the United States of America 109, 490–494, 10.1073/pnas.1110368109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G. D., Mackie P., Jodar M., Moskovtsev S. & Krawetz S. A. Chromatin and extracellular vesicle associated sperm RNAs. Nucleic acids research 43, 6847–6859, 10.1093/nar/gkv591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W. J. et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic acids research 44, D259–265, 10.1093/nar/gkv1036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu J. & He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes & development 29, 1343–1355, 10.1101/gad.262766.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R., Friderici K. & Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America 71, 3971–3975 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A. & Dahl J. A. Dynamic RNA modifications in disease. Current opinion in genetics & development 26, 47–52, 10.1016/j.gde.2014.05.006 (2014). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature chemical biology 10, 93–95, 10.1038/nchembio.1432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. & Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends in genetics : TIG 30, 464–474, 10.1016/j.tig.2014.07.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology 7, 885–887, 10.1038/nchembio.687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell 49, 18–29, 10.1016/j.molcel.2012.10.015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120, 10.1038/nature12730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. S. & He C. Fate by RNA methylation: m6A steers stem cell pluripotency. Genome biology 16, 43, 10.1186/s13059-015-0609-1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell research 24, 1403–1419, 10.1038/Cr.2014.151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim M. S., Moshitch-Moshkovitz S. & Rechavi G. FTO: linking m6A demethylation to adipogenesis. Cell research 25, 3–4, 10.1038/cr.2014.162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F. et al. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. The Journal of clinical endocrinology and metabolism 100, E148–154, 10.1210/jc.2014-1893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar J. A., Shambaugh M. E., Polayes D., Matera A. G. & Rottman F. M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. Rna 3, 1233–1247 (1997). [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. WHO laboratory manual for the examination and processing of human semen. 5th edn, (World Health Organization, 2010). [Google Scholar]
- Saletore Y., Chen-Kiang S. & Mason C. E. Novel RNA regulatory mechanisms revealed in the epitranscriptome. RNA biology 10, 342–346, 10.4161/rna.23812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646, 10.1016/j.cell.2012.05.003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q. et al. Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic acids research 35, 6322–6329, 10.1093/nar/gkm657 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt E. M., Ehrenschwender T., Batista P. J., Chang H. Y. & Kool E. T. Identification of a selective polymerase enables detection of N(6)-methyladenosine in RNA. Journal of the American Chemical Society 135, 19079–19082, 10.1021/ja4105792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilfan I. D. et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. Journal of nanobiotechnology 11, 8, 10.1186/1477-3155-11-8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambard S. et al. Analysis and significance of mRNA in human ejaculated sperm from normozoospermic donors: relationship to sperm motility and capacitation. Molecular human reproduction 10, 535–541, 10.1093/molehr/gah064 (2004). [DOI] [PubMed] [Google Scholar]
- Zhou J. et al. [Expression of cysteine-rich secretory protein 2 in patients with asthenozoospermia and its clinical significance]. Nan fang yi ke da xue xue bao = Journal of Southern Medical University 34, 1528–1533 (2014). [PubMed] [Google Scholar]
- Zhou J. H. et al. The expression of cysteine-rich secretory protein 2 (CRISP2) and its specific regulator miR-27b in the spermatozoa of patients with asthenozoospermia. Biology of reproduction 92, 28, 10.1095/biolreprod.114.124487 (2015). [DOI] [PubMed] [Google Scholar]
- Diao R. et al. Deficient human beta-defensin 1 underlies male infertility associated with poor sperm motility and genital tract infection. Science translational medicine 6, 249ra108, 10.1126/scitranslmed.3009071 (2014). [DOI] [PubMed] [Google Scholar]
- Jia G., Fu Y. & He C. Reversible RNA adenosine methylation in biological regulation. Trends in genetics : TIG 29, 108–115, 10.1016/j.tig.2012.11.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006, 10.1126/science.1261417 (2015). [DOI] [PubMed] [Google Scholar]
- Fustin J. M. et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806, 10.1016/j.cell.2013.10.026 (2013). [DOI] [PubMed] [Google Scholar]
- Alarcon C. R., Lee H., Goodarzi H., Halberg N. & Tavazoie S. F. N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485, 10.1038/nature14281 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun 4, 1798, 10.1038/ncomms2822 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


