Abstract
In this study, we report the contribution of a PDI-like gene from wheat wild relative Haynaldia villosa in combating powdery mildew. PDI-V protein contains two conserved thioredoxin (TRX) active domains (a and a′) and an inactive domain (b). PDI-V interacted with E3 ligase CMPG1-V protein, which is a positive regulator of powdery mildew response. PDI-V was mono-ubiquitinated by CMPG1-V without degradation being detected. PDI-V was located on H. villosa chromosome 5V and encoded for a protein located in the endoplasmic reticulum. Bgt infection in leaves of H. villosa induced PDI-V expression. Virus induced gene silencing of PDIs in a T. durum-H. villosa amphiploid compromised the resistance. Single cell transient over-expression of PDI-V or a truncated version containing the active TXR domain a decreased the haustorial index in moderately susceptible wheat cultivar Yangmai 158. Stable transgenic lines over-expressing PDI-V in Yangmai 158 displayed improved powdery mildew resistance at both the seedling and adult stages. By contrast over-expression of point-mutated PDI-VC57A did not increase the level of resistance in Yangmai 158. The above results indicate a pivotal role of PDI-V in powdery mildew resistance and showed that conserved TRX domain a is critical for its function.
Every year plant diseases are responsible for billions of dollars worth of crop losses worldwide. A comprehensive understanding of how plants coordinate their defense systems will facilitate exploitation of the most effective protection strategies. Powdery mildew, caused by the fungus Blumeria graminis (DC) f. sp. tritici (Bgt), is one of the most important foliar diseases of wheat worldwide1. Yield losses caused by the disease may be as high as 50%2. Plants possess remarkably sophisticated mechanisms to fend off attack by pathogens and pests. The first line of defense by the plant immune system against pathogens involves basal defense responses that are triggered upon detection of pathogen associated molecular patterns (PAMPs)3,4. To suppress PAMP-triggered immunity (PTI) pathogens secrete small unique protein molecules known as effectors. Upon recognition, host plants trigger a second line of defense, effector triggered immunity (ETI)5,6.
The identification of genes related to non-host resistance and regulators of PTI will help breeders to access durable resistance. Powdery mildew resistance gene Pm21 from Haynaldia villosa (Syn. Dasypyrum villosum, 2n = 2x = 14, genome VV), confers durable broad-spectrum resistance (BSR) to Bgt7. The Bgt-H. villosa provides a useful pathosystem for elucidating the mechanism of BSR. In a previous study the resistance pathway was analyzed by comparing transcription patterns of H. villosa before and after Bgt infection by microarray analysis using the Barley1 Genechip (Affymetrix). Based on the microarray results two genes, a serine/threonine kinase gene Stpk-V and an E3 ligase gene CMPG1-V were cloned and both positively contributed to BSR when over-expressed in wheat1,2. Further clarification of resistance signaling pathways and identification of more genes related to disease response will facilitate an understanding of the mechanism of Pm21-mediated BSR.
The ubiquitin proteasome system (UPS) is a highly regulated mechanism of intracellular protein degradation in eukaryotes. Many studies over the last decade, have shown that E3 ligase genes are involved in regulation of innate immunity in plants8,9. The functions of a dozen E3 ligase genes in plant immunity were recently well characterized in Arabidopsis10. An in vitro ubiquitination activity study showed that CMPG1-V is a positive regulator of the powdery mildew resistance. CMPG1-V-mediated resistance involves ROS and SA pathways1. To further elucidate the molecular pathway of CMPG-V-mediated powdery mildew resistance, a yeast two hybrid cDNA library of H. villosa leaves was constructed and screened for interacting cDNA-expressed genes. One of the positive cDNA clones encoded a disulphide isomerase (PDI)-like protein.
PDI (EC 5.3.4.1), a ubiquitious sulfydryl oxidoreductase found in abundance in the lumen of the endoplasmic reticulum (ER) of all eukaryotic cells, is an important cellular protein with multiple biological functions, displays versatile redox behavior, is highly interactive with other proteins and has an implied role in various diseases11,12. The fundamental role of PDI-like proteins and their wide range of substrates make it very difficult to understand their function in vivo, especially in plants13,14. However, specific functions of plant PDIs have been postulated and supported with accumulating evidence. Arabidopsis thaliana encodes 12 PDI-like proteins. Among them PDI5 regulates the timing of programmed cell death (PCD) in endothelial cell15, while PDIL-2;1 plays a critical role in the development of the embryo sac16. Typical wheat PDIs are involved in the assembly of storage protein within the ER and also participate in quality control by regulation of unfolded proteins14,17,18,19,20. The barley variant HvPDIL5-1 confers BSR against many strains of Bymoviruses21.
PDI is one of the redox proteins that regulate reactive oxygen species (ROS) production by the Nox enzyme family, as well as changes in the redox status of cells to activate the defense system22. Calcium influx and nitric oxide (NO) production results in the S-nitrosylation of PDI (SNO-PDI), which increases the level of polyubiquitinated proteins and triggers cell death23 and inhibition of mitochondria, leading to the generation of ROS and NO24. Antioxidant properties of PDI could help limit potential cell damage by ROS generated during pathogen infection. PDI may be integral to the repertoire of mechanisms that host plants have evolved to suppress the highly destructive and energy-consuming processes accompanying hypersensitive responses25. Plant defense systems produce ROS which not only causes damage to hydrolytic enzymes in pathogens but also regulate various cellular pathways during infection26. Some studies in plants have suggested the role of cell surface PDI in transportation of defense-signaling cascades i.e. movement of NO between cells27,28,29. ER-quality control components (which also involve PDI) directly take part in PTI as any disturbance of ER-localized processing of the PAMP receptor EFR1 disrupts PTI response and results in enhanced susceptibility to bacterial and fungal pathogens30,31.
Only one study in wheat has reported involvement of a PDI gene in defense; this was against the fungal pathogen Mycosphaerella graminicola25. Here, we report the cloning and functional analysis of a PDI-like gene from H. villosa in combating powdery mildew. H. villosa, a wild diploid grass, belongs to the tertiary wheat gene pool and possesses high levels of resistance to abiotic and biotic stresses, including resistance to powdery mildew. The present research aimed to investigate the interaction of PDI-V with CMPG1-V, to examine possible involvement of PDI-V during powdery mildew infection using molecular approaches, and to fully characterize the PDI-V gene in response to different phytohormone and abiotic stress treatments.
Results
PDI-V was cloned from H. villosa by screening a Y2H cDNA library using CMPG1-V as bait
CMPG1-V positively regulates powdery mildew resistance in common wheat1. To identify proteins interacting with CMPG1-V, a Y2H cDNA library of H. villosa was constructed and used to elucidate the resistance pathway mediated by CMPG1-V32. Using CMPG1-V as bait, 17 putative cDNA clones interacting with CMPG1-V were identified (Supplementary Table S1); one of them encoded a protein disulphide isomerase (PDI). Based on the cDNA sequence, the 1,615 bp full-length PDI gene was isolated from H. villosa. The gene has an ORF of 1,323 bp, encoding a protein with 440 amino acids (Fig. 1A,B). BLASTn analysis showed that the sequence was highly similar to the T. aestivum PDI-like genes TaPDIL5-1a & 1b, and hence was designated PDI-V.
Figure 1. Structural features of the PDI-V cDNA sequence and its translation product.
(A) Structure deduced from the full-length cDNA sequence of PDI-V. The size of each domain, signal peptide and 3′ and 5′- UTR sizes and position of the ATG and TAG stop codons are shown. (B) Graphical representation of domain organization as provided by the NCBI conserved domain database. (C) Deduced amino acid sequence of PDI-V. Thioredoxin catalytic motifs and ER retention signals are shown in red.
PDI-V has two tandem thioredoxin active domains (a and a′), each containing a typical tetra peptide motif (CGHC) at the N terminus, and an inactive thioredoxin b domain at the C-terminus. Signal P analysis predicted the presence of a signal peptide from the start of the N terminus to the 30th amino acid, and a modified NDEL signal for retention in the ER at the C-terminus (Fig. 1C). Alignment of PDI-V with homologous proteins from other grass species and Arabidopsis indicated that PDIs are well conserved across mono- and dicotyledonous plants (Supplementary Fig. S1). Phylogenetic analysis showed that PDI-V homologs from monocots clustered into one clade, and PDI-V was closest to PDIs from Hordeum vulgare, T. aestivum, T. urartu and Aegilops tauschii (Supplementary Fig. S2A).
PDI-V interacted with, and is ubiquitinated, by CMPG1-V without detectable degradation
Y2H confirmed the interaction of PDI-V with CMPG1-V in yeast (Fig. 2A). To test their physical interaction in vivo, bimolecular fluorescence complementation (BiFC) with split yellow fluorescent protein (YFP) was employed. Co-expression of nYFP-PDI-V and cYFP-CMPG1-V in onion epidermal cells resulted in complementation of YFP in the cytoplasm, proving their interaction in vivo (Fig. 2B).
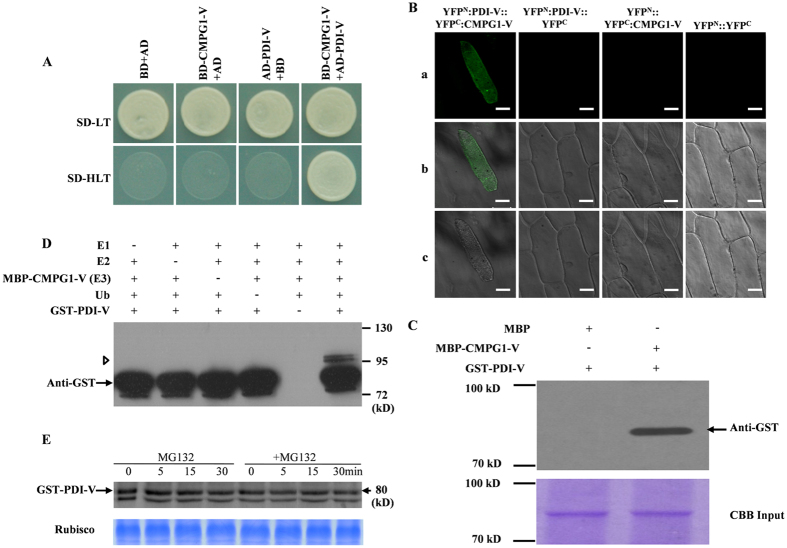
Figure 2. Interaction of PDI-V with CMPG1-V.
(A) Y2H assay showing interaction of PDI-V and CMPG1-V. Yeast growth on SD-Leu/-Trp/-His confirms protein: protein interaction. (B) In vivo interaction of YFPN:PDI-V with YFPC:CMPG1-V in the BiFC assay in onion epidermal cells. Epiflourescence (a) bright field (b) and light images (c) of YFPN:PDI-V::YFPC:CMPG1-V, YFPN:PDI-V, YFPC:CMPG1-V and empty vectors YFPN:YFPC (C) In vitro interaction of MBP-CMPG1-V with GST-PDI-V by pull-down assay. GST protein loading is shown by Coomassie brilliant (CBB) blue staining (bottom panel). The position and size of the molecular marker are shown to the left of the blot (D) In vitro ubiquitination assay of recombinant GST-PDI-V fusion protein by MBP-CMPG1-V. The recombinant protein was assayed for E3 activity in the presence of E1, E2, biotinylated ubiquitin (Ub) and GST-PDI-V. + and – represent the presence and the absence of the corresponding protein in the reaction mixture respectively. The position and size of the molecular marker are shown to the right of the blot. (E) Cell-free degradation assay of recombinant GST-PDI-V. Total protein extracted from wheat leaves was incubated with MBP-CMPG1 and GST-PDI-V at 30 °C for 90 min with or without MG132, with sampling at different time points. GST-PDI-V protein was detected by western blotting using anti-GST antibody.
To further confirm the physical interaction revealed by Y2H and BiFC assays, a pull-down assay was performed. A specific 80 kDa band for GST-PDI-V was obtained only when using MBP-CMPG1-V (Fig. 2C, Lane 2), but not when using MBP protein alone (Fig. 2C, Lane 1), further confirming the existence of physical interaction between CMPG1-V and PDI-V. CMPG1-V acts as an U-box/ARM E3 ubiquitin ligase. Ubiquitination of PDI-V by CMPG1-V was demonstrated by an in vitro ubiquitination assay. In the presence of all requisite components for ubiquitination, GST-PDI-V was monoubiquitinated by MBP-CMPG1-V, as evidenced by increased size of the target band (Fig. 2D, Lane 6). A cell-free degradation assay showed that, regardless of the presence or absence of 26S proteasome inhibitor reagent MG132, the amount of GST-tagged PDI-V showed no significant change when co-incubated with MBP-tagged CMPG1-V, indicating GST-tagged PDI-V could not be degraded by CMPG1-V and hence should not be its degradation target (Fig. 1E).
PDI-V gene is located on long arm of chromosome 5V of H. villosa, and PDI-V subcelluarly localized in the ER
A specific primer pair (PDIV-40F/R, Supplementary Table S2) was designed based on alignment of genomic sequences of PDI-V and its wheat genome A, B and D homologs. PCR using H. villosa (VV), T. durum-H. villosa amphiploid (AABBVV), a complete set of Chinese Spring-H. villosa addition lines (DA1V-7V) and Chinese Spring as templates showed that the 600 bp band specific for PDI-V was amplified only from H. villosa, AABBVV and DA5V (Supplementary Fig. S2B), indicating the location of PDI-V on chromosome 5V. Further amplification using translocation lines CINAU-61 and CINAU-158 showed that the PDI-V band was amplified only by CINAU-158 (Supplementary Fig. S2B), indicating the PDI-V was located on 5VL. All identified PDI orthologs from the sub-genomes of wheat (Chinese Spring A, B and D) and related species barley, rice and Brachypodium were located in syntenic regions of group 5L chromosomes (Supplementary Table S3).
Vectors of PDI-V-GFP and endoplasmic reticulum (ER) marker gene (RFP) were transiently co-expressed in onion epidermal cells to detect subcellular location. GFP-tagged PDI-V protein signals were localized in the ER (Fig. 3A). A complete overlap of GFP and RFP signals was observed in transiently over-expressing cells, confirming that PDI-V-GFP was localized to the lumens of endoplasmic reticulum (Fig. 3A).
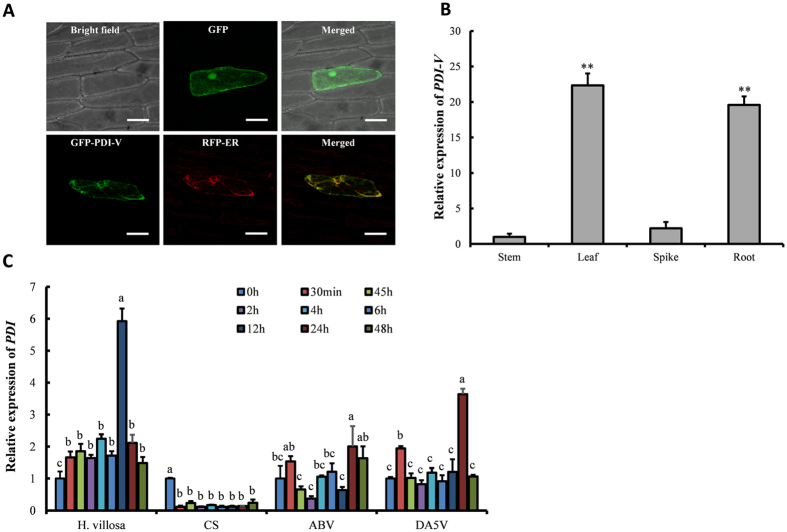
Figure 3. Subcellular localization of PDI-V and expression profiling of PDI-V.
(A) Subcellular localization of GFP-PDI-V co-expressed with RFP in onion epidermal cells. Localization of green fluorescence protein (GFP) as control is shown in the upper panel. The first image in the lower panel shows the green GFP-PDI-V signal localized in the endoplasmic reticulum and the second image shows localization of the RFP ER marker. The third image in this panel shows the co-localization of GFP-PDI-V and RFP-ER. Scale bar = 100 μm. (B) Expression pattern of PDI-V in tissues (stem, leaf, immature spike and root) of H. villosa (C) Expression pattern of PDI in H. villosa and wheat lines i.e. Chinese Spring (CS), T. turgidum-H. villosa amphiploid (AABBVV), and the 5V addition line (DA5V) after inoculation with Bgt. Each result is the mean of three independent biological repeats while bars show standard deviations. The Tubulin gene was used as an internal control to normalize qRT-PCR values. The lower script alphabet letters represent the statistically significant differences at p = 0.05 (one way ANOVA test).
PDI-V plays a positive role in powdery mildew resistance of wheat and domain a is involved in restricting Bgt haustorial formation
Quantitative reverse transcription PCR (qRT-PCR) analysis in different tissues of H. villosa showed that expression of PDI-V was 19- and 22-fold higher in roots and leaves than in stems. PDI-V expression in spikes was lower than in leaves and roots, but slightly higher than in stems (Fig. 3B). When two-week-old H. villosa seedlings were inoculated with Bgt, PDI-V expression in leaves increased slightly at 0.5 hours post inoculation (hpi), and reached a 6-fold level compared to the control at 24 hpi (Fig. 3C). In Chinese Spring PDI remained at constant levels after inoculation, whereas expression in the AABBVV amphiploid and DA5V increased at 0.5 hpi, but no obvious change was observed from 1 hpi to 12 hpi. At 24 hpi expression in the amphiploid and DA5V was twice that at 0 hpi, but expression levels for both lines were significantly lower than for H. villosa. At 48 hpi the PDI-V expression remained high in the amphiploid, but had fallen to a pre-treatment level in DA5V (Fig. 3C).
To further characterize the role of PDI-V in powdery mildew response, a single-cell transient over-expression assay (TOA) was conducted using the moderate susceptible wheat cultivar (cv) Yangmai 158 as receptor. The haustorial index (HI) for Yangmai 158 was 59.9% when transformed with GUS alone, but was significantly decreased to 38.3% when co-transformed with GUS and pBI220-PDI-V (Fig. 4A). This demonstrated an involvement of PDI-V in response to Bgt. To determine the PDI-V domain involved three vectors, viz pBI220-PDI-Va, pBI220-PDI-Va′ and pBI220-PDI-Vc, were constructed and used in single-cell TOA. Only the domain a construct significantly reduced the HI (34.36%) to a similar level as the full-length PDI-V (Fig. 4A). This proved that domain a of PDI-V was involved in restraining the haustorium formation of Bgt and restricted their further penetration.
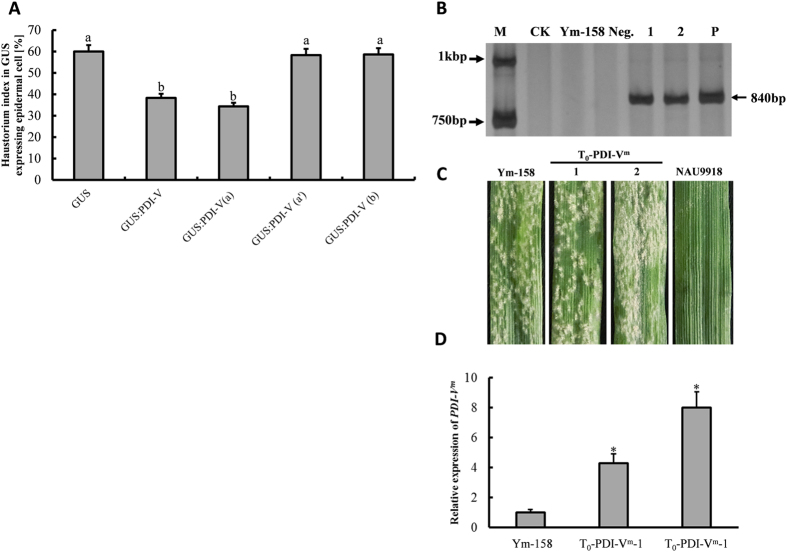
Figure 4. Gene function analysis of PDI-V.
(A) Functional analysis of PDI-V by a single cell transient expression assay in wheat leaves by the particle bombardment method. As a control treatment, the GUS-reporting empty vector pWMB002 was transformed into the epidermal layer of susceptible variety Yangmai 158. To assess the function of PDI-V and its truncated domains a, a′ and b, GUS was co-transformed with each construct. The experiment was repeated in three biological repeats and mean data with standard deviations are displayed. Lowercase letters indicate statistically significance differences at p = 0.05 (one way ANOVA). (B) PCR of positive point mutated (PDI-Vm) T0 transgenic plants. M, DNA marker DL2kb; CK, ddH2O; Ym-158, Yangmai 158; Neg., negative T2 transgenic plant 1 & 2, T0-PDI-Vm-1 & T0-PDI-Vm-2 plants; P, plasmid of pBI220:PDI-V. (C) Interaction of Bgt with T0-PDI-Vm evaluated at 7 dpi using inoculated detached leaves of Yangmai 158 and Nannong-9918 (NAU 9918) as susceptible and resistant controls (D) Over-expression of PDI-Vm in T0-transgenic plants assayed by qRT-PCR using Tubulin as an internal control to normalize the quantification data. Significance was determined by paired sample t-tests. *p < 0.05.
To confirm the essential role of domain a in chaperon involvement in Bgt response, a point mutation PDI-VC57A (PDI-Vm) was created in the catalytic motif of domain a. When PDI-Vm was transformed into Yangmai 158, 13 positive T0 transformants overexpressing PDI-Vm were identified (Fig. 4B,D). All showed similar levels of powdery mildew susceptibility to that of non-transformed Yangmai 158 (Fig. 4C), confirming an essential role of domain a in function of PDI-V in powdery mildew response in wheat.
Stable transgenic plants overexpressing PDI-V in Yangmai 158 were generated by particle bombardment. Transgenic plants T0 to T2 generation plants were identified. PCR using a combined primer pair CaMV35S-F & PDI-V-R (Supplementary Table S2) confirmed that 11 regenerated plants were positively transformed. Stable transgenic lines PDI-V-T2-1 and PDI-V-T2-2, showed enhanced seedling and adult resistance to Bgt compared to non-transformed Yangmai 158 (Fig. 5A). At seedling stage both over-expressing lines had infection type (IT) 0, whereas Yangmai 158 was fully susceptible (IT 4). At the adult stage, a few Bgt pustules were observed on the PDI-V over-expressing lines but far less than on the Yangmai 158 control. In a qRT-PCR assay the expression levels of the PDI-V transgene in the two transgenic lines were increased by 2- and 9-fold relative to the non-transgenic control (Fig. 5C).
Figure 5. Powdery mildew evaluation and PCR identification of PDI-V over-expressing T2 plants.
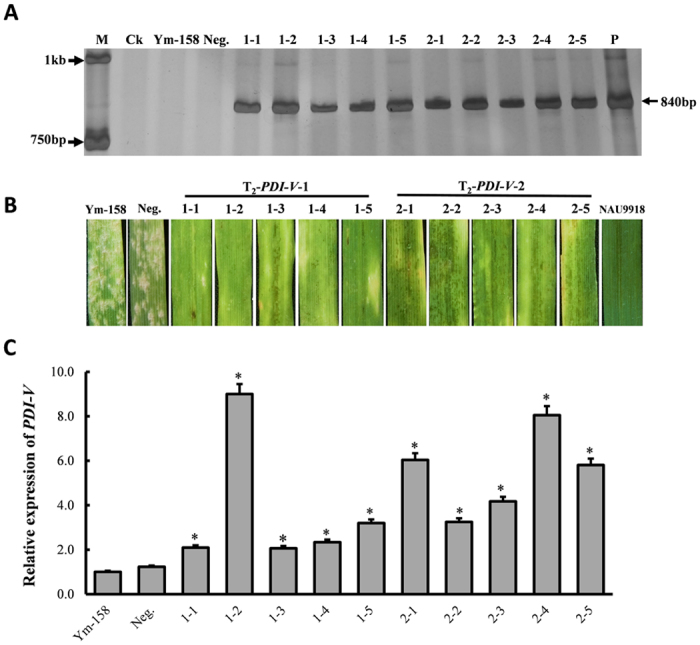
(A) Detection of positive T2 transgenic plants by PCR amplification of PDI-V using pBI220:PDI-V as positive control. M, DNA marker DL2kb; CK, ddH2O; Ym-158, Yangmai 158; Neg., negative T2 transgenic plant, P, pBI220:PDI-V plasmid. Arrows on the left side of the image indicate the size of the two bands of the 2 kb DNA marker; arrow on the right side shows the size and amplification of the PDI-V-specific band. (B) Disease responses of T2 plants evaluated at the seedling stage following inoculation of detached leaves, using Yangmai 158 and Nannong-9918 (NAU 9918) as susceptible and resistant controls. (C) Over-expression of PDI-V in T2-transgenic plants assayed by qRT-PCR using Tubulin as internal control to normalize the quantification data. Significance was determined according to paired sample t-tests. *p < 0.05.
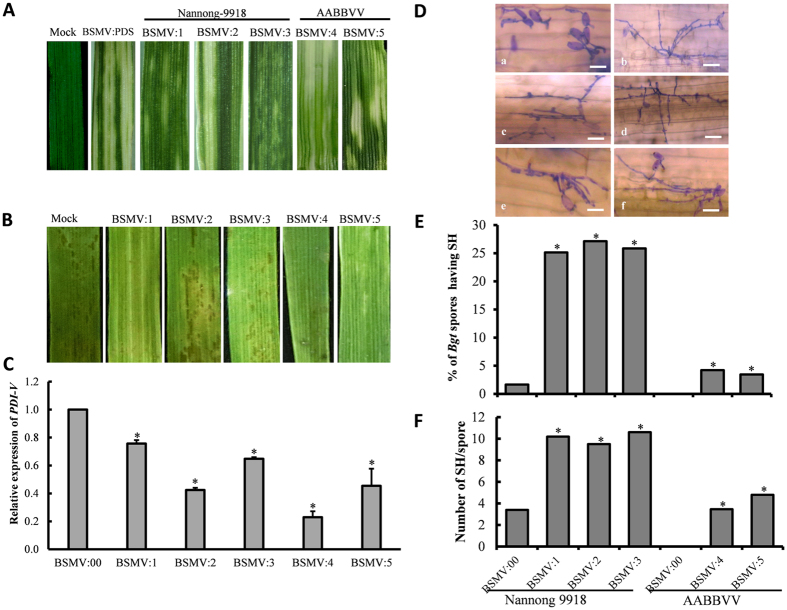
VIGS was used to silence PDI-V and its wheat orthologs in the T. durum-H. villosa amphiploid and wheat cv Nannong 9918. Typical mild chlorotic mosaic symptoms appeared on the 4th leaves of all the BSMV infected plants at 9 dpi, and photobleaching was observed only in plants infected with BSMV:PDS (Fig. 6A). Leaves of plants infected with BSMV:PDI-V sampled at 15 dpi were placed on media containing 6-benzylaminopurine (6-BA) and inoculated with Bgt; qRT-PCR showed that PDI-V transcripts were significantly decreased in silenced plants compared to the control (Fig. 6C). At 7 dpi no visible powdery mildew symptoms were present on the leaves of both resistant lines (Fig. 6B). However, there were differences in fungal development between the lines with respect to the percentages of Bgt conidia producing secondary hyphae (SH) and numbers of SH produced by individual conidia. The percentage of Bgt conidia producing SH was significantly higher in PDI-silenced leaves of Nannong 9918 than in the amphiploid (Fig. 6E). Similarly, the number of SH produced by individual conidia in PDI-V-silenced leaves was also higher (Fig. 6F). These results indicated that PDI- silencing reduced the level of resistance in these lines. The haustorial indices were higher in Nannong 9918 than in the T. durum-H. villosa amphiploid. We speculate this may due to other genes affecting powdery mildew response in the amphiploid. To obtain a better understanding of the relationships of PDI-V, CMPG1-V and Pm 21, PDI expression levels were profiled in plants over-expressing CMPG1-V and Stpk-V plants (T2-generation). The results showed that the expression of PDI was greatly induced in both over-expressing lines after Bgt inoculation (Supplementary Fig. S4) supporting our hypothesis that decreased resistance in silenced plants was associated with lack of PDI expression.
Figure 6. Functional characterization of PDI-V in Bgt-infected Nannong 9918 and H. villosa-T. turgidum amphiploid (AABBVV) by BSMV-mediated VIGS.
(A) Typical photo bleaching of BSMV:PDS-infected was first observed on the third leaf at 9 dpi. Photographs show the fourth leaves at 15 dpi. (B) Disease responses at 7 dpi of Bgt inoculation on the fourth leaves of BSMV:PDI-V-silenced plants. (C) Assessment of silencing efficiency of BSMV:PDI-V by qRT-PCR assay. The fifth leaves of PDI-V-silenced plants were sampled; BSMV:00 infected plant were used as the control. Data were normalized using the Tubulin gene as an internal control. Each point represents the mean of three replicates. Bars indicate SD, *p < 0.05. (D) Microscopic observation of fungal growth in BSMV:00 (a) and BSMV:PDI-V infected plants (b–d from Nannong 9918; e,f from the AABBVV amphiploid). Scale bar = 30 μm (E) Comparison of Bgt spores producing SH between BSMV:00- and BSMV:PDI- infected plants (BSMV:1 to BSMV:3, Nannong 9918; BSMV:4 and BSMV:5, AABBVV). (F) Numbers of individual fungal spores producing secondary hyphae were counted from whole detached leaves of each genotype and compared with BSMV:00. *P < 0.05 according to Student’s t-test.
Enhanced H2O2 production may contribute to inhibited Bgt development
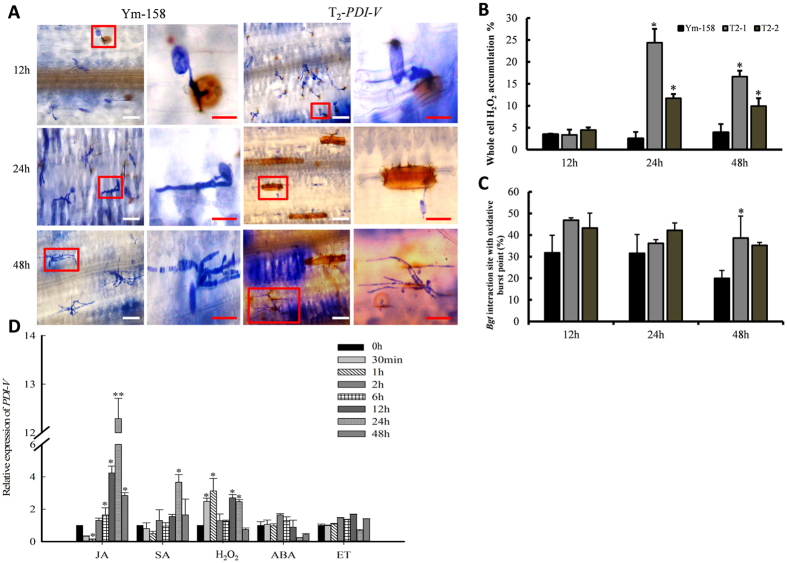
H2O2 is the key driver of ROS production and signaling that regulates interaction between the SA, JA and ethylene defense pathways in many plants and also in powdery mildew resistance33,34. H2O2 treatment triggered immediate expression of PDI-V; increased expression was detected at 30 min and 1 h, and significant up-regulation continued at 24 h and 48 h (Fig. 7D). T2 PDI-V transgenic plants and Yangmai 158 were DAB-stained to measure H2O2 production and accumulation at different time points within and around Bgt-infected cells (Fig. 7A). At 24 and 48 hpi the percentages of H2O2 accumulation in PDI-V-T2-2 were significantly higher than in Yangmai 158. While no significant difference was observed at 12 hpi (Fig. 7B), the percentages of H2O2 accumulation at 48 hpi were much higher in T2-trangenic plants than Yangmai 158 (Fig. 7C). We speculate that overexpression of PDI-V induced delayed cell death following pathogen penetration, resulting in improved powdery mildew resistance.
Figure 7. Accumulation of H2O2 at the Bgt infection sites in wheat epidermal cells.
(A) Microscopic view of H2O2 production in wheat leaf epidermal cells. Two-week-old seedlings were inoculated with Bgt and sampled at different time points as indicated. H2O2 accumulation was detected by in situ histochemical staining using DAB and observed in bright field under an Olympus microscope. Enlarged views of marked red boxes in lanes 1 and 3 (scale bar = 30 μm) are shown in lanes 2 and 4. Scale bar = 20 μm. (B,C) Percentage of cells with H2O2 accumulation throughout the entire cell, and with accumulation only around the infection sites in Yangmai 158 and PDI-V-over-expressing plants. (D) Expression pattern of PDI-V in response to exogenous hormones. Each result is the mean of three independent biological repeats; bars show standard deviations. The Tubulin gene was used as an internal control to normalize qRT-PCR values. Asterisks indicate a significant difference (P < 0.05) from the control (0 hpi) based on Student’s t-tests. JA, methyl jasmonate; SA, salicylic acid; H2O2, hydrogen peroxide; ET, ethylene.
PDI-V expression is up-regulated by different stresses and phytohormone treatments
Expression profiling of PDI-V in response to salt (200 mM NaCl), cold (4 °C) and heat shock (40 °C) were assayed by qRT-PCR. All three stresses induced PDI-V expression changes (Supplementary Fig. S3). NaCl treatment induced an almost twofold increase in expression of PDI-V in H. villosa leaves after 3 h. Expression then decreased slightly to a constant level of about 1.75-fold until 12 h, decreased somewhat at 24 h and it was again up-regulated to 1.76-fold at 48 h (Supplementary Fig. S3A). After 24 h exposure of H. villlosa at 4 °C, PDI-V expression was sharply up-regulated and reached a maximum level of about 3-fold higher than the control (Supplementary Fig. S3C). Heat shock induced a 3-fold up-regulation at 1 h, and maximum 11-fold level at 6 h, before decreasing to the pre-treatment level at 48 h (Supplementary Fig. S3D).
Expression patterns of PDI-V in response to treatments by phytohormones (MethJA, ethylene, ABA, SA) were investigated by qRT-PCR. With SA treatment, no significant change was observed until 24 h post treatment when the gene expression reached a maximum level, then decreased and finally back to the pre-treatment level. PDI-V in response of MeJA treatment was significantly down-regulated at 30 min, then increased gradually from 2 h and reached a maximum level (12-fold) at 24 h and was maintained at relatively high level of up-regulation for the remainder of the experiment. Ethylene and ABA treatments induced no significant changes in PDI-V expression level (Fig. 3D).
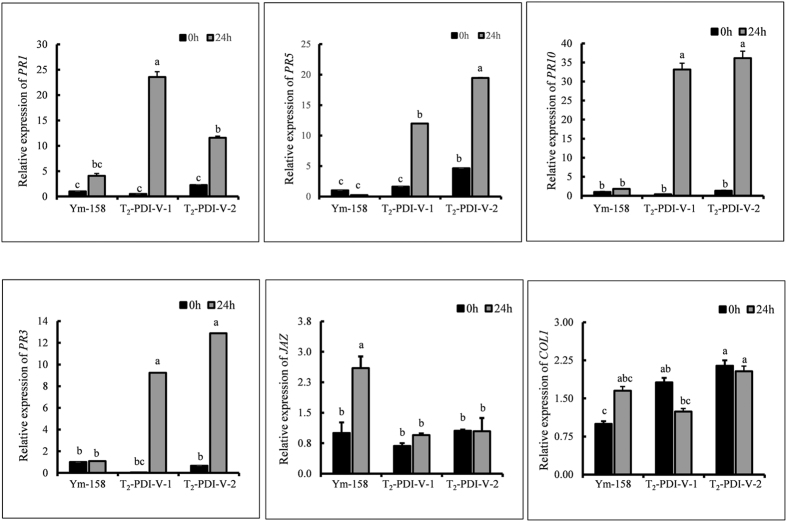
Expression of some PR genes was studied in over-expressing T2 plants to investigate the mechanism of resistance conferred by PDI-V. The results showed that expression of PR1, PR5 and PR10 were significantly up-regulated at 24 hpi relative to the non-transgenic Yangmai 158 control (Fig. 8). Expression of PR1, the key marker for SA-systemic acquire resistance (SAR) was also significantly higher in Yangmai 158 after inoculation, however in PDI-V transgenic lines it was much higher. The expression pattern of PR3, the marker for JA-signaling pathway was also up-regulated in PDI-V over-expressing lines, but expressions of JAZ and COL1 were unchanged (Fig. 8). From these results we speculated that PDI-V might use SA to produce resistance responses to powdery mildew.
Figure 8. Expression patterns of pathogenesis-related (PR) genes in Bgt-inoculated leaf tissues of Yangmai 158 and PDI-V over-expressing T2 lines at 0 and 24 hpi.
Each result is the mean of three independent biological repeats; bars shows standard deviations. The Tubulin gene was used as an internal control to normalize qRT-PCR values. Lowercase letters represent statistically significant differences at p = 0.05 (one way ANOVA).
Discussion
Very little is reported in literature about the participation of PDI genes in plant disease response. Some studies suggest a role of cell surface PDIs in molecular transportation in defense-signaling cascades, for example the movement of nitric oxide across cell walls27,28,29. Recent work with barley proved that an HvPDIL5-1 variant conferred broad spectrum resistance to many Bymoviruses strains21. Only one study in wheat reported involvement of a PDI gene in defense against the hemibiotrophic fungal pathogen (M. graminicola). It found immediate induction of PDI in resistant wheat cultivars challenged with the fungus and suggested that PDI may be integral to the repertoire of mechanisms that host plants have evolved to suppress highly destructive, energy-consuming processes accompanying hypersensitive responses25. Here we report a functional analysis of PDI from the wheat wild relative H. villosa in combating wheat powdery mildew.
A Y2H assay and BiFC proved that PDI-V interacted with CMPG1-V, reported previously as a positive regulator of powdery mildew resistance1. As an E3 ligase, one possibility was that CMPG-mediated powdery mildew resistance involved triggering of degradation of one or more target proteins35. However, we found that PDI-V resides in the ER and is mono-ubiquitinated by CMPG1-V without detectable degradation. Thus PDI-V is an interaction protein of CMPG-V but is not its degradation target. In the host-pathogen interaction systems in animals, PDI not only processes antigens but it is also involved in transferring pathogen antigens from the ER to the cytosol by an ER-associated degradation system for further degradation by proteasomes22. We therefore suggest that CMPG1-V might be processed by PDI-V in the ER lumen before being secreted to the cytoplasm as the ER localization of CMPG1-V was already proved1.
PDI-V is located in chromosome 5VL. Bioinformatic analysis showed high syntenic conservation of the gene location across homologues in monocots (rice, Brachypodium and barley) including the A, B and D sub-genomes of hexaploid wheat36,37. d’Aloisio et al.19 characterized the PDI genes in bread wheat and grouped them into eight clades. PDI-V has the highest similarity with group V (containing two genes designated as TaPDIL5-1a and TaPDIL5-1b), which were also located on homeologous group 5 wheat chromosomes.
The protein structure of PDI-V is also highly conserved across the monocots in terms of structure, domain number and active thioredoxin sites which are characterized by presence of three domains (a, a′ and b), two thioredoxin-active sites and a modified ER retention NDEL signal at the C-terminus. The PDIs having such structural features are known as PDI-a-P5 due to close homology with human P5 PDI. Domain a is present on the N-terminus while a′ is located almost in the middle of PDI-V protein. Studies on the evolution of PDI domains suggested that the C-terminus TRX domain evolved from duplication of the N-terminus TRX domain present in all classes of PDI enzymes38.
Single-cell TOA of full-length of PDI-V clearly demonstrated that HI, which represents a measure of the compatible interaction between the host and Bgt, was reduced significantly in transformed plants compared to Yangmai 158. The multilobe shape of the Bgt haustorium not only increases the surface area but also facilitates nutrient absorption from the host cell39. Thus a decrease in HI shows that PDI-V restricts Bgt in establishing contact with the host, ultimately leading to a lower disease level. Our results from single-cell TOA showed that domain a of PDI-V is critical in restraining Bgt growth. Over-expression of domain a mutant gene PDI-Vm did not improve resistance compared to over-expression of the wild type gene. This implies that catalytic site in TXR domains present in PDI-V has chaperon activity that regulates the physiological function of cells in a complex way.
To gain further insight into PDI-V functions in the defense pathway of Bgt, we knocked down the gene in two different lines; one having a complete VV genome i.e. H. villosa-T. turgidum amphiploid (AABBVV) and wheat cv Nannong 9918 having translocation chromosome 6VS/6AL containing Pm21. Pm21 confers broad spectrum resistance to Bgt in China2. The purpose of silencing PDI in Nannong 9918 was to investigate a possible functional relationship between the two genes. PDI-V silencing was insufficient to produce visible disease symptoms on knockdown leaves of both genotypes, but microscopic observations on numbers of germinating Bgt conidiospores producing SH per unit area and numbers of SH produced per spore were much higher in Nannong 9918 than in the H. villosa-T. turgidum amphiploid. One explanation for this is the presence of additional genes affecting powdery mildew response in the amphiploid than in Nannong 9918. Moreover, high expression of PDI in CMPG1-V and Stpk-V transgenic wheat plants provided preliminary indications that Pm21 might mediate the PDIs and CMPGs to regulate defense responses. During pathogen infection, ROS production provides the hallmark for successful recognition of pathogen and activation of defense mechanism in plants40. The first and quickest response is an oxidative burst to inhibit pathogen attack mainly by virtue of H2O2 production and consequently act as a signal molecule for the induction of many defensive genes41,42. Our results confirmed rapid accumulation of H2O2 at Bgt infection sites in transgenic plants as well as in the receptor Yangmai 158. Although initially there was no statistically significant difference in H2O2 production between PDI-V transgenic plants and Yangmai 158 but still the accumulation of H2O2 in transgenic plants was higher than in Yangmai 158. The whole cell oxidative burst in response to Bgt attack was significantly higher in transgenic plants than in Yangmai 158 at 24 hai and thereafter. From these results we supposed that H2O2 initially functioned as a signal to trigger PDI-V induction, which upon activation produced H2O2 to reduce the Bgt growth. A previous study also detected rapid H2O2 accumulation in wheat mesophyll cells during incompatible Bgt interactions, suggesting signaling behavior in defense against Bgt43. Like the powdery mildew resistance gene Stpk-V, PDI-V was also triggered by exogenous H2O2 application and Bgt inoculation2. Expression of PR1, PR3, PR5 and PR10 were up-regulated in PDI-V transgenic plants, suggesting that PDI-V may use SA signaling to inhibit fungal growth. It has been reported that high expression of PR1, PR2, PR5 and PR10 are consequences of SA-dependent signaling responses in resistance to biotrophic pathogens in many plants species including wheat and Arabidopsis44,45,46. Although expression of PR1, the key marker for SA-systemic acquired resistance (SAR), has also increased significantly in Yangmai 158 after inoculation but it was much higher in PDI-V transgenic lines. This is in accordance with previous findings, as it is a well understood phenomenon that expression of PR1, and even PR5, is much higher in incompatible than in compatible interaction47. In PDI-V over-expressing plants, high expression of PR3 was observed. In many plants, PR3 has inhibitory role against invading fungal pathogens and can increase the PR1 expression level, which lead to enhanced disease resistance45,48.
PDI-V was also up-regulated under abiotic stresses salinity, cold and heat shock. It was shown previously that both pathogen infection and abiotic stresses produced unfolded protein stress conditions49, which results in triggering of an ER-quality control (ER-QC) mechanism50. The ER-QC system comprised an SDF2-ERdj3b-BIP complex, the calreticulin/calnexin cycle, and disulfide isomerase (PDI)51. However, the degree to which PDI-V can elevate abiotic stress needs further study.
In conclusion, the present study reports for the first time function-based evidence of involvement of PDIs in disease resistance in wheat. The catalytic site of thioredoxin domains in PDI-V provides chaperon activity in restricting Bgt penetration of host cells in a complex regulatory mechanism. PDI-V may function coordinately with Stpk-V and CMPG1-V, and its activity involves the H2O2 pathway. Future studies will be focused on the functional analysis of the TXR domain and to determine the exact connection between PDI-V and CMPG1-V during host infection.
Materials and Methods
Plant materials, growth conditions and chemical treatments
Haynaldia villosa (genome VV, accession no. 91C43) was introduced from Cambrige, UK. A Triticum durum-H. villosa amphiploid (genome AABBVV, accession no. NAU 201), a set of T. aestivum-H. villosa addition lines (DA1V to DA7V, accession no. NAU307 to NAU313, each containing one pair of chromosomes from H. villosa in Chinese Spring wheat, and two T. aestivum-H. villosa whole arm translocation lines T5VS/5DL and T5VL/W (CINAU-61 and CINAU-158) were developed by the Cytogenetics Institute, Nanjing Agricultural University (CINAU). Wheat cv. Yangmai 158 (moderately susceptible to Bgt) and Nannong 9918 (containing Pm21, and immune to Bgt) were used as receptors for transient and/or stable transformation and virus-induced gene silencing (VIGS). The conditions for plant growth and Bgt inoculations were as described in Xing et al.52.
To study the expression level of PDI-V in response to different treatments, H. villosa seedlings at the two-leaf stages were either inoculated with Bgt, or moved to 4 °C or 40 °C conditions for cold and heat shock stress treatments, or dipped into 200 mM NaCl for salinity stress. Leaf tissue samples were collected at 0 h, 30 min, 45 min, 1 h, 2 h, 6 h, 12 h, 24 h, 48 h and 72 h post treatments. Parallel mock-inoculated controls were treated with sterile water. Treatments with exogenous hormones or signal molecules were performed as described in Xing et al.52. Three biological repeats were used for each assay. All samples were rapidly frozen in liquid nitrogen and stored in an ultra-freezer (−80 °C) until use.
Total RNA extraction and quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) following the instructions of the manufacturer. Primer oligo (dT)18 was used to reversely transcribe 2 μg of total RNA into cDNA using AMV reverse transcriptase (Takara) in RT-PCR. The expression pattern of PDI-V was analyzed using a SYBR Green/Fluorescent qPCR master mix (Takara) on a Roche-480 system (Roche), and the wheat Tubulin gene was used as the internal control. The qRT-PCR program started with denaturation at 95 °C for 1 min, followed by 40 amplification cycles programmed as 95 °C 5 sec, 57 °C 30 sec, and 72 °C 30 sec. The CT values for target and standard control genes were retrieved and the comparative threshold 2−∆∆CT method was applied to quantify the relative expression of given genes53. All reactions were conducted in three biological replicates with three technical repeats for each replication. Double distilled H2O was used as template for negative controls. All primers used in the study are listed in Supplementary Table S2.
Yeast two-hybrid protein-protein interaction
A yeast two-hybrid (Y2H) H. villosa cDNA library was constructed by Li32. To investigate protein to protein interaction between CMPG1-V and PDI-V, GAL-4-based Y2H was performed in the HF7c strain of S. cerevisiae that harbored reporter genes lacZ and HIS3. The complete ORF of CMPG1-V was fused with the GAL4 DNA binding domain (DNA-BD) of protein expression vector pGBKT7 as a bait while the PDI-V cDNA sequence was inserted as prey into a pGADT7 vector harboring an activation domain (AD) using the SmaI and BamHI cloning sites. The resulting constructs and empty vectors were transformed into yeast strain HF7c as described by Xie et al.54. The transformation mixtures were plated on yeast drop-down selection media (SD-LTH) deprived of leucine (Leu), tryptophan (Trp) and histidine (His) but supplemented with 10 mM 3-amino-1,2,4,triazole (3-AT) to reduce the appearance of false positive colonies. In addition the same combination of transformation mixtures were plated on SD-Leu-Trp medium (SD-LT) as a control to check for normal growth of the yeast colonies. Proteins interactions were evaluated on the basis of growth status of yeast cells on SD-LTH media after incubation at 30 °C for 3 days.
Bimolecular fluorescence complementation (BiFC) assay
A BiFC assay was performed as described by Walter et al.55. In short, coding regions of CMPG1-V and PDI-V were amplified by PCR using specific primers (Supplementary Table S1) and cloned into pSPYNE-35S and pSPYCE-35S, respectively. The resulting constructs (CMPG1-V-nYFP and PDI-V-cYFP) were transformed into onion epidermal cells with a BIOLISTIC-PDS-1000/He particle delivery system (Bio-Rad) at a pressure of 1100 p.s.i. The transformed onion epidermal layers were incubated in darkness at 22 °C for 16 h before detection of yellow florescence signals by confocal microscopy (Leica Microsystem SP2).
In vitro pull-down assay
To purify GST-PDI-V protein, the full-length ORF of PDI-V was cloned into a pGEX4 vector using the restriction sites BamHI and SmaI. Resultant GST-tagged PDI-V protein was expressed in the Escherichia coli strain BL21 (DE3) and purified by affinity chromatography using glutathione sepharose beads. Generation of the MBP-CMPG1-V construct and protein purification were performed as described by Zhu et al.1. For the pull-down assay about 10 μg of the MBP-PDI-V and MBP alone (control) proteins were incubated with 10 μL of pre-washed amylose beads into different microfuge tubes containing 1 mL of pull down buffer (20 mM TRIS-HCl, pH 7.5, 1 mM β-mercaptoethanol, 3 mM EDTA, 150 mM NaCl, and 1% NP-40) for 1 h at 4 °C with gentle shaking. Incubated beads were centrifuged at 2000 rpm for 1 min before second incubation with 10 μg of GST-PDI-V protein in 1 mL of pull down buffer for 30 min at room temperature with constant gentle shaking. After incubation the beads were harvested, washed once with PBS buffer containing 500 mM NaCl and subsequently washed five times with the same buffer containing 135 mM of NaCl. The bound protein complex retained on the beads was extracted by boiling the beads in 10 μL of 2 × SDS-PAGE loading buffer and finally analyzed by western blotting using α-GST antibody.
Chromosome location of PDI-V
To determine the chromosome location of PDI-V, H. villosa, T. turgidum-H. villosa amphiploid, a set of all seven addition lines (DA1V to DA7V), and Chinese Spring were used for PCR using primer pair PDI-V-40F/R, designed from the PDI homolog from the sequenced D-genome of Ae. tauschii (Supplementary Table S2). PCR were performed in 25 μL reaction volumes including 1 × PCR buffer, 2 mmol/l MgCl2, 0.15 mmol/l dNTPs, 20 ng of each primer, 2 μL template and 1U r-Taq DNA polymerase (Takara, Japan). Conditions for thermal cycling were: denaturing at 94 °C for 5 min, followed by 94 °C 45 s, 59 °C 45 s, 72 °C for 1 min for 35 cycles, and final extension at 72 °C for 10 min. The amplified products were visualized in 1% agrose gels containing ethidum bromide.
Sub-cellular localization of PDI-V
The coding sequence of PDI-V without a stop codon was amplified with specific primers (PDI-V-580F & R) and cloned into the SpeI and BamHI sites in front of green fluorescent protein (GFP) under control of the Cauliflower mosaic virus (CaMV) 35S promoter in transient expression vector -pAN580. The resultant recombinant pAN580-PDI-V-GFP vector was confirmed by DNA sequencing. For onion bombardment, gold particles (1 mm in diameter) were coated separately with 5 mg of plasmid DNA with the empty vector pAN580 and pAN580-PDI-V-GFP and bombarded with a BIOLISTIC-PDS-1000/He particle delivery system (Bio-Rad) at a pressure of 1100 p.s.i. Onion epidermal peels were placed on solidified half-strength Murashige and Skoog (MS) medium and kept at 22 °C in darkness for 24 h before imaging with a Zeiss LSM 730 microscope.
In vitro ubiquitination and cell-free degradation assays
The purified proteins (MBP-CMPG1-V and GST-PDI-V) were used for in vitro ubiquitination assay according to Xie et al.56 with minor modifications. A final volume of 30 μL was used for each reaction, containing 40 mM Tris-HCl pH 7.4, 5 mM MgCl2, 50 mM KCl, 2 mM ATP, 1 mM DTT, 10% glycerol, 200 ng E1, 200 ng E2, 200 ng MBP-CMPG1-V and 600 ng of GST-PDI-V. Reactions were incubated at 30 °C for 1.5 h. Samples were separated on 10% SDS-PAGE gels and ubiquitination was detected by western blotting using anti-GST antibody (Roche).
Cell-free degradation assays were carried out according to García-Cano et al.57. Briefly, total protein was extracted by grinding wheat leaves in 1 × PBS protein extraction buffer. Sequential centrifugations at 12,000 g for 5 min yielded clear supernatant. Equal concentrations of GST-PDI-V and MBP-CMPG1-V were added to 200 μL of total protein extract supplemented with, or without, the 26S proteasome inhibitor MG132 (40 μM). Samples were taken from both treatments at time intervals of 0, 5, 15 and 30 min and analyzed by western blotting using anti-GST antibody (Roche).
VIGS assay
To silence PDI-V, barley stripe mosaic virus (BSMV)-based infectious vectors were constructed as documented by Wang et al.58. The partial sequence of PDI-V (200 bp) was cloned by specific primer pair PDIV-VIGS-F/R (Supplementary Table S2) and inserted in RNAγ of BSMV. About 2.5 μL of each BSMV RNA i.e., α, β and genetically modified γ mixed with 42.5 μL of 2 × GKP buffer, was inoculated to two-leaf seedlings by gently rubbing the leaves with gloved fingers. BSMV:PDS and BSMV:00 were used as controls. Each assay consisted of 20 seedlings and repeated at least three times. Detached fourth leaves from 10 plants in each treatment were placed on 6-BA media and inoculated with high densities of freshly collected Bgt conidiospores. The phenotypes of inoculated leaves were observed and photographed at 10 dpi. To determine the silencing efficiency RNA extracted from fourth leaves of remaining plants was used to measure PDI-V expression levels by qRT-PCR.
Single-cell transient overexpression assay (TOA)
Single-cell TOA is a highly reliable, quick and powerful approach to study defense functions of foreign genes against Bgt due to the ectoparasitic nature of pathogen and synchronous restriction of the fungus to penetrated cell only59. The TOA of PDI-V was performed using a Bio-Rad He/1000 particle delivery system to wheat leaves as described by Shirasu et al.60. Equal volumes (1 μg/μL) of plasmid DNA (pBI220:PDI-V) and pWMB002 vector (containing the GUS gene) were mixed before coating tungsten particles. Mixed tungsten particles and plasmids were prepared for 15 shots. Second leaves of one-week-old Yangmai 158 seedlings were firmly placed on the plated media for delivery of mixed plasmids into host cells by gene gun. Delivery of pWMB002 alone was used as control. Bombarded leaves were incubated at 25 °C for 4 to 6 h in darkness, then infected with fresh Bgt inoculum and placed in a growth chamber with a 14 h light/10 h darkness photoperiod for 40 h61. Leaves were stained for determination of GUS activity, bleached with 90% ethanol, and observed under a microscope to record the haustorial index (HI, percentage of GUS-staining cells with haustoria among all GUS-stained cells invaded by Bgt). The assay was repeated twice and significant differences between treatments were analyzed by paired sample t-tests using SPSS software.
Generation and characterization of PDI-V transgenic wheat plants
A point mutation PDI-Vm was created in the catalytic motif of domain a, using the Mut Express II Fast Mutagenesis Kit (Vazyme, Nanjing) according to the manufacturer’s recommendations. The coding regions of PDI-V and PDI-Vm were amplified by PCR and cloned into the over- expression vector pBI220-HA between the BamHI and StuI cloning sites. The resulting constructs pBI220-HA-PDI-V and pBI220-HA-PDI-Vm were co-transformed along with pAHC20 (having a herbicide tolerance marker gene) into immature-embryo derived calli of wheat cv. Yangmai 158 by particle bombardment. Transformed calli were cultured and regenerated into fully developed plants as prescribed by Xing et al.16. Total genomic DNA of T0 transgenic and subsequent generation (T1 and T2) plants were used to detect the positive plants by amplification with a combined specific primer flanking the CaMV35S promoter and PDI-V, CaMV35S-F and PDI-V-R (Supplementary Table S2).
For characterizing the function of PDI-V, detached young leaves from all plants in each generation (T0, T1 and T2) were inoculated with Bgt to check the infection types according to Sheng et al.62. Two T2 transgenic lines derived from positive T0 plants showing high levels of powdery mildew resistance were selected and grown in the field and greenhouse. Five rows of each T2 transgenic line were planted along with the negative control Yangmai 158 (receptor variety). Two rows of highly susceptible cv. Sumai 3 were planted every six rows for generation of Bgt inoculum. Disease responses of adult T2 plants was recorded using a 0–9 disease scale64.
Detection of ROS
Accumulation of H2O2 in plant cells was detected by 3, 3′-diaminobenzidine (DAB) staining as described by Xing et al.52. Leaves of two T2-PDI-V transgenic lines and Yangmai 158 were soaked in DAB solution (Bio Basic Inc., Shanghai) for 8 h for uptake and then incubated at 25 °C for an additional 8 h. At 12, 24 and 48 hpi, 1.5 cm leaf segments from each host line were sampled, and boiled in 95% ethanol for 5 min. The cleared leaf segments were mounted in 50% glyercerol and observed under a microscope (Olympus, Japan). Randomly selected 5 leaf segments from each line and at least 100 infection sites on each leaf were examined. Standard deviations were determined and pair sample t-tests were used for statistical analysis.
In silico analysis
The BLAST tool (http://www.ncbi.nlm.nih.gov/blast/) was used to analyze conserved motifs and domain structure of PDI-V. The full ORF was predicted by online ORF finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Retrieved sequences were aligned by the Clustal X 1.83 program using the Gonnet series as protein weight matrix along with other parameters set to a 30% divergent sequence delay and 10 gap open penalty. The phylogenetic tree was constructed using the neighbour-joining method in MEGA 4.0 software. For this all the gaps in the alignment were deleted prior to estimation of distance matrices calculated according to a Poisson correction amino acid model. Statistical significance of phylogenetic tree was tested with 1,000 bootstrap replicates.
Additional Information
How to cite this article: Faheem, M. et al. A disulphide isomerase gene (PDI-V) from Haynaldia villosa contributes to powdery mildew resistance in common wheat. Sci. Rep. 6, 24227; doi: 10.1038/srep24227 (2016).
Supplementary Material
Acknowledgments
This work was supported by the grants from Higher Education Commission Pakistan, the National Science Foundation of China (No. 31471490), the Important National Science & Technology Specific Projects in Transgenic Research (No. 2013ZX08002001-007, 2011ZX08002-001 and 2014ZX0800907B), the Chinese High Tech Program of China (No. 2011AA1001), the Program of Introducing Talents of Discipline to Universities (No.B08025), High-level Talent in Six Industries of Jiangsu province, and the 333 Talent Project of Jiangsu Province. The funders had no role in study design, data collection, analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions X.W., M.F., Y.L. and A.C. designed the research project. M.F., Y.L. and M.A. did all the molecular experiments. C.J., Z.J., Z.W. and F.Y. participated in pull down and in vitro ubiquitination assays. J.X., H.W., L.X. and R.Z. took part in bioinformatics and phenotypic analysis. M.F., X.W. and Q.X. drafted the manuscript. All authors read and approved the final manuscript.
References
- Zhu Y. et al. E3 ubiquitin ligase gene CMPG1–V from Haynaldia villosa L. contributes to powdery mildew resistance in common wheat (Triticum aestivum L.). Plant J. 84, 154–168 (2015). [DOI] [PubMed] [Google Scholar]
- Cao A. et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. 108, 7727–7732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P. N. & Rathjen J. P. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010). [DOI] [PubMed] [Google Scholar]
- Beck M., Heard W., Mbengue M. & Robatzek S. The INs and OUTs of pattern recognition receptors at the cell surface. Curr. Opin. Plant Biol. 15, 367–374 (2012). [DOI] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Banfield M. J. & Kamoun S. Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483–492 (2012). [DOI] [PubMed] [Google Scholar]
- Rafiqi M., Ellis J. G., Ludowici V. A., Hardham A. R. & Dodds P. N. Challenges and progress towards understanding the role of effectors in plant–fungal interactions. Curr. Opin. Plant Biol. 15, 477–482 (2012). [DOI] [PubMed] [Google Scholar]
- Chen P., Qi L., Zhou B., Zhang S. & Liu D. Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 91, 1125–1128 (1995). [DOI] [PubMed] [Google Scholar]
- Chen L. & Hellmann H. Plant E3 ligases: flexible enzymes in a sessile world. Mol. Plant 6, 1388–1404 (2013). [DOI] [PubMed] [Google Scholar]
- Liu J. et al. The U-box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol. 160, 28–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furniss J. J. & Spoel S. H. Cullin-RING ubiquitin ligases in salicylic acid-mediated plant immune signaling. Front. Plant Sci. 6, 154; 10.3389/fpls.2015.00154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman R. B., Hirst T. R. & Tuite M. F. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem. Sci. 19, 331–336 (1994). [DOI] [PubMed] [Google Scholar]
- Hatahet F. & Ruddock L. W. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid. Redox signal. 11, 2807–2850 (2009). [DOI] [PubMed] [Google Scholar]
- Ciaffi M., Paolacci A., Dominici L., Tanzarella O. & Porceddu E. Molecular characterization of gene sequences coding for protein disulfide isomerase (PDI) in durum wheat (Triticum turgidum ssp. durum). Gene 265, 147–156 (2001). [DOI] [PubMed] [Google Scholar]
- Dhanapal A. P. & Porceddu E. Funtional and evolutionary genomics of protein disulphide isomerase (PDI) gene family in wheat and its wild relatives: A review. Annu. Rev. Res. Biol. 3, 935–958 (2013). [Google Scholar]
- Ondzighi C. A., Christopher D. A., Cho E. J., Chang S.-C. & Staehelin L. A. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell Online 20, 2205–2220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Boavida L. C., Ron M. & McCormick S. Truncation of a protein disulfide isomerase, PDIL2-1, delays embryo sac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell 20, 3300–3311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y., Zhu X., Levanony H., Segal G. & Galili G. Purification, characterization, and intracellular localization of glycosylated protein disulfide isomerase from wheat grains. Plant Physiol. 108, 327–335 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaffi M., Dominici L., Tanzarella O. & Porceddu E. Chromosomal assignment of gene sequences coding for protein disulphide isomerase (PDI) in wheat. Theor. Appl. Genet. 98, 405–410 (1999). [Google Scholar]
- d’Aloisio E. et al. The protein disulfide isomerase gene family in bread wheat (T. aestivum L.). BMC Plant Biol. 10, 101; 10.1186/1471-2229-10-101 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Higashino Y., Kitao Y., Masuda T. & Urade R. Expression and characterization of protein disulfide isomerase family proteins in bread wheat. BMC Plant Biol. 15, 73; 10.1186/s12870-015-0460-2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P. et al. PROTEIN DISULFIDE ISOMERASE LIKE 5-1 is a susceptibility factor to plant viruses. Proc. Natl. Acad. Sci. 111, 2104–2109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolf B. S. et al. Protein disulfide isomerase and host-pathogen interaction. Sci. World J. 11, 1749–1761 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester M. T., Benhar M. & Stamler J. S. Nitrosative stress in the ER: a new role for S-nitrosylation in neurodegenerative diseases. ACS Chem. Biol. 1, 355–358 (2006). [DOI] [PubMed] [Google Scholar]
- Halloran M., Parakh S. & Atkin J. The role of s-nitrosylation and s-glutathionylation of protein disulphide isomerase in protein misfolding and neurodegeneration. Int. J. Cell Biol. 2013, 10.1155/2013/797914 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S., Anderson J. M., Urmeev F. I. & Goodwin S. B. Rapid induction of a protein disulfide isomerase and defense-related genes in wheat in response to the hemibiotrophic fungal pathogen Mycosphaerella graminicola. Plant Mol. Biol. 53, 741–754 (2003). [DOI] [PubMed] [Google Scholar]
- Giraldo M. C. & Valent B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11, 800–814 (2013). [DOI] [PubMed] [Google Scholar]
- Durner J., Wendehenne D. & Klessig D. F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. 95, 10328–10333 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M., Xia Y., Dixon R. A. & Lamb C. Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588 (1998). [DOI] [PubMed] [Google Scholar]
- Zai A., Rudd M. A., Scribner A. W. & Loscalzo J. Cell-surface protein disulfide isomerase catalyzes transnitrosation and regulates intracellular transfer of nitric oxide. J. Clin. Invest. 103, 393–399 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V. et al. Control of the pattern‐recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y. et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28, 3439–3449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Identification of a CMPG1-V interaction protein HvHIPP1 from Haynaldia villosa L. and its functional analysis in powdery mildew resistance. Ph.D Thesis, Nanjing Agricultural Univeristy (2014).
- Overmyer K., Brosché M. & Kangasjärvi J. Reactive oxygen species and hormonal control of cell death. Trends Plant Sci. 8, 335–342 (2003). [DOI] [PubMed] [Google Scholar]
- Bouchez O., Huard C., Lorrain S., Roby D. & Balagué C. Ethylene is one of the key elements for cell death and defense response control in the Arabidopsis lesion mimic mutant vad1. Plant Physiol. 145, 465–477 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy E. M. et al. CMPG1‐dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytol. 190, 653–666 (2011). [DOI] [PubMed] [Google Scholar]
- Wu H., Dorse S. & Bhave M. In silico identification and analysis of the protein disulphide isomerases in wheat and rice. Biologia 67, 48–60 (2012). [Google Scholar]
- Zhu C. et al. Molecular characterization and expression profiling of the protein disulfide isomerase gene family in Brachypodium distachyon L. PLos ONE 9, e94704; 10.1371/journal.pone.0094704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai S., Toh H., Hayano T. & Kikuchi M. Molecular evolution of the domain structures of protein disulfide isomerases. J. Mol. Evol. 47, 200–210 (1998). [DOI] [PubMed] [Google Scholar]
- Braun U. et al. The taxonomy of the powdery mildew fungi. The powdery mildews: a comprehensive treatise, (edsm Belanger R. R. et al.) 13–55 (APS press, 2002). [Google Scholar]
- Li A. et al. Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive fatty acid desaturase gene participating in powdery mildew resistance. PLos ONE 6, e28810; 10.1371/journal.pone.0028810 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M. A., Jones J. D. & Dangl J. L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenucci M. S., Piro G. & Dalessandro G. In muro feruloylation and oxidative coupling in monocots: a possible role in plant defense against pathogen attacks. Plant Signal. Behav. 4, 228–230 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. et al. Comparative analysis of early H2O2 accumulation in compatible and incompatible wheat–powdery mildew interactions. Plant Pathol. 54, 308–316 (2005). [Google Scholar]
- Niu J. S., Liu R. & Zheng L. Expression analysis of wheat PR-1, PR-2, PR-5 activated by Bgt and SA, and powdery mildew resistance. Wheat Crop 27, 1132–1137 (2007). [Google Scholar]
- Hamamouch N., Li C., Seo P. J., Park C. M. & Davis E. L. Expression of Arabidopsis pathogenesis‐related genes during nematode infection. Mol. Plant Pathol. 12, 355–364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M. et al. Transcriptome comparison of susceptible and resistant wheat in response to powdery mildew infection. Genomics Proteomics Bioinformatics 10, 94–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritsch C., Muehlbauer G. J., Bushnell W. R., Somers D. A. & Vance C. P. Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol. Plant Microbe Interact. 13, 159–169 (2000). [DOI] [PubMed] [Google Scholar]
- Fiocchetti F. et al. Over-expression of a pathogenesis-related protein gene in transgenic tomato alters the transcription patterns of other defence genes. J. Hortic. Sci. Biotechnol. 81, 27–32 (2006). [Google Scholar]
- Urade R. The endoplasmic reticulum stress signaling pathways in plants. Biofactors 35, 326–331 (2009). [DOI] [PubMed] [Google Scholar]
- Malhotra J. D. & Kaufman R. J. The endoplasmic reticulum and the unfolded protein respnse. In Seminars in Cell & Developmental Biology. 716–731 (Elsevier, 2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli T. & Sitia R. Protein quality control in the early secretory pathway. EMBO J. 27, 315–327 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L. et al. The Hv-SGT1 gene from Haynaldia villosa contributes to resistances towards both biotrophic and hemi-biotrophic pathogens in common wheat (Triticum aestivum L.). PLos ONE 8, e72571; 10.1371/journal.pone.0072571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Xie Q., Sanz-Burgos A. P., Guo H., García J. A. & Gutiérrez C. GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39, 647–656 (1999). [DOI] [PubMed] [Google Scholar]
- Walter M. et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 (2004). [DOI] [PubMed] [Google Scholar]
- Xie Q. et al. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170 (2002). [DOI] [PubMed] [Google Scholar]
- García-Cano E., Zaltsman A. & Citovsky V. Assaying proteasomal degradation in a cell-free system in plants. J. Vis. Exp. 85, 10.3791/51293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Establishment of an effective virus induced gene silencing system with BSMV in Haynaldia villosa. Mol. Biol. Rep. 37, 967–972 (2010). [DOI] [PubMed] [Google Scholar]
- Schweizer P., Pokorny J., Abderhalden O. & Dudler R. A transient assay system for the functional assessment of defense-related genes in wheat. Mol. Plant Microbe Interact. 12, 647–654 (1999). [Google Scholar]
- Shirasu K., Nielsen K., Piffanelli P., Oliver R. & Schulze‐Lefert P. Cell‐autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 17, 293–299 (1999). [Google Scholar]
- Ai-li L. A transient expression system for the functional assessment of early response genes on the powdery mildew infected barley or wheat leaves. Agric. Sci. China 2, 1061–1068 (2003). [Google Scholar]
- Xing L. et al. Transformation of wheat thaumatin-like protein gene and diseases resistance analysis of transgenic plants. Acta Agron. Sin. 34, 349–354 (2008). [Google Scholar]
- Sheng B. Grades of resistance to powdery mildew classified by different phenotypes of response in the seeding stage of wheat. Plant Prot. 1, 49 (1988). [Google Scholar]
- Jakobson I., Peusha H., Timofejeva L. & Järve K. Adult plant and seedling resistance to powdery mildew in a Triticum aestivum × Triticum militinae hybrid line. Theor. Appl. Genet. 112, 760–769 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.