Abstract
Background
The exon junction complex (EJC), which contains four core components, eukaryotic initiation factor 4AIII (eIF4AIII), MAGO/NASHI (MAGO), Y14/Tsunagi/RNA-binding protein 8A, and Barentsz/Metastatic lymph node 51, is formed in both nucleus and cytoplasm, and plays important roles in gene expression. Genes encoding core EJC components have been found in plants, including rice. Currently, the functional characterizations of MAGO and Y14 homologs have been demonstrated in rice. However, it is still unknown whether eIF4AIII is essential for the functional EJC in rice.
Results
This study investigated two DEAD box RNA helicases, OsRH2 and OsRH34, which are homologous to eIF4AIII, in rice. Amino acid sequence analysis indicated that OsRH2 and OsRH34 had 99 % identity and 100 % similarity, and their gene expression patterns were similar in various rice tissues, but the level of OsRH2 mRNA was about 58-fold higher than that of OsRH34 mRNA in seedlings. From bimolecular fluorescence complementation results, OsRH2 and OsRH34 interacted physically with OsMAGO1 and OsY14b, respectively, which indicated that both of OsRH2 and OsRH34 were core components of the EJC in rice. To study the biological roles of OsRH2 and OsRH34 in rice, transgenic rice plants were generated by RNA interference. The phenotypes of three independent OsRH2 and OsRH34 double-knockdown transgenic lines included dwarfism, a short internode distance, reproductive delay, defective embryonic development, and a low seed setting rate. These phenotypes resembled those of mutants with gibberellin-related developmental defects. In addition, the OsRH2 and OsRH34 double-knockdown transgenic lines exhibited the accumulation of unspliced rice UNDEVELOPED TAPETUM 1 mRNA.
Conclusions
Rice contains two eIF4AIII paralogous genes, OsRH2 and OsRH34. The abundance of OsRH2 mRNA was about 58-fold higher than that of OsRH34 mRNA in seedlings, suggesting that the OsRH2 is major eIF4AIII in rice. Both OsRH2 and OsRH34 are core components of the EJC, and participate in regulating of plant height, pollen, and seed development in rice.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0769-5) contains supplementary material, which is available to authorized users.
Keywords: DEAD box RNA helicase, Eukaryotic initiation factor 4AIII (eIF4AIII), Exon junction complex (EJC), Rice (Oryza sativa)
Background
The DEAD box RNA helicase family, the largest family of RNA helicases, belongs to helicase superfamily 2. Each DEAD box RNA helicase contains nine conserved amino acid motifs that constitute the helicase core domain. Besides these conserved motifs within DEAD box proteins, there are also N- and C-terminal extension sequences in each DEAD box RNA family member that varies in terms of their length and composition; they have been proposed to provide substrate binding specificity, and to act as signals for subcellular localization or as domains that interact with accessory components [1–3]. DEAD box proteins are found in most prokaryotes and all eukaryotes, including plants [4–10]. Rice is an important staple food crop and is also valuable as a model plant for studies in cereal functional genomics. Although predicted protein sequences in the rice genome database as determined by silico analysis to indicate that there are at least 51 DEAD box proteins in rice [10], the functional characterizations of most of them remain unknown.
Eukaryotic initiation factor 4AIII (eIF4AIII), a DEAD box RNA helicase, is a core component of the exon junction complex (EJC) that also contains MAGO/NASHI (MAGO), Y14/Tsunagi/RNA-binding protein 8A, and Barentsz/Metastatic lymph node 51 [11–16]. The EJC is formed in both the nucleus and the cytoplasm, and plays important roles in gene expression, including the following: (1) It assembles 20–24 bases upstream of each exon of pre-mRNA for its involvement in mRNA splicing [17]. (2) It is involved in nonsense-mediated decay, a surveillance mechanism that degrades mRNA containing premature termination codons [18]. (3) It is involved in the regulation of gene expression at the translational level [19]. (4) It has a role in mRNA subcellular localization [20, 21].
Although most research has been undertaken in mammals, genes encoding core EJC components have been found in plants [22], suggesting that there is structural and functional conservation in the EJC complex among plant and mammalian. However, only limited evidence has been reported on the physiological role of the EJC in plants. In Arabidopsis, eIF4AIII interacts with an EJC component, ALY/Ref, and colocalizes with other EJC components, such as Mago, Y14, and RNPS1 [23]. In O. sativa, two forms of MAGO, OsMAGO1 and OsMAGO2, and two forms of Y14, OsY14a and OsY14b, were analyzed [24–26]. OsMAGO1 and OsMAGO2 double-knockdown rice plants displayed dwarfism and abnormal flowers in which the endothecium and tapetum of the stamen were maintained [24]. OsY14b may function in embryogenesis, while the down-regulation of OsY14b resulted in a failure to induce plantlets [24]. OsY14a knockdown plants also displayed phenotypes similar to those of OsMAGO1 and OsMAGO2 double-knockdown rice plants [24]. Moreover, OsMAGO1 and OsMAGO2 double-knockdown, and OsY14a knockdown transgenic plants showed abnormal accumulation of the pre-mRNA of UNDEVELOPED TAPETUM 1 (OsUDT1), a key regulator of stamen development [24]. These findings indicate that the EJC participates in the regulation of pre-mRNA splicing in rice.
Despite the fact that the functions of homologs of MAGO and Y14 have been demonstrated in rice, it is still unknown whether eIF4AIII is essential for EJC function in rice. In this study, two putative rice DEAD box RNA helicase genes, OsRH2 (Os01g0639100) and OsRH34 (Os03g0566800), were therefore characterized. Both OsRH2 and OsRH34 are homologous to eIF4AIII, which is a member of the eIF4A family, and their gene expression patterns were similar in various rice tissues, but the level of OsRH2 mRNA was about 58-fold higher than that of OsRH34 mRNA in seedlings. The results from bimolecular fluorescence complementation (BiFC) analysis showed that both OsRH2 and OsRH34 can interact with OsMAGO1 and OsY14b. Transgenic plants with both OsRH2 and OsRH34 knocked down by RNA interference displayed phenotypes that resembled those of mutants with gibberellin-related developmental defects. Moreover, these OsRH2 and OsRH34 double-knockdown plants exhibited severe defects in terms of pollen and seed development. The accumulation of OsUDT1 pre-mRNA was also detected in the OsRH2 and OsRH34 double-knockdown transgenic lines. Our data demonstrate that both OsRH2 and OsRH34 are core components of the EJC and play critical roles in regulation of plant height, pollen, and seed development in rice.
Results
OsRH2 and OsRH34 are putative DEAD box RNA helicases
To identify rice eIF4AIII homologs, human eIF4AIII protein sequences were used as queries to search protein databases at phytozome and National Center for Biotechnology Information (NCBI). Two eIF4AIII-like putative proteins, encoded by OsRH2 (Os01g0639100) and OsRH34 (Os03g0566800) were identified in rice (Additional file 1). The OsRH2 is located on rice chromosome 1 and has eight exons. The deduced amino acid sequence of OsRH2 cDNA consists of nine conserved RNA helicase domains (Fig. 1) and the characteristic amino acid residues D-E-A-D in motif II. Besides, the OsRH34 gene has eight exons and is located on chromosome 3. The levels of identity between OsRH2 and OsRH34 in terms of the DNA sequence and the deduced amino acid sequence were found to be 97 and 99 %, respectively. Phylogenetic relationships were established using amino acid sequences from the eIF4A families of dicots, monocots, green algae, vertebrates, invertebrates, and yeast (Additional file 2), which showed that OsRH2 and OsRH34 are closely related to eIF4AIII and can be clustered into the monocot group (Fig. 2).
Fig. 1.

Amino acid sequences and domain structures of the OsRH2 and OsRH34 proteins. A. The amino acid sequences of OsRH2 and OsRH34 were compared using the CLUSTAL W program. Identical amino acid residues are labeled in black. Different amino acid residues are marked by asterisks. The conserved helicase motif is highlighted by a line above it and includes motifs Q, I, Ia, Ib, II, III, IV, V, and VI
Fig. 2.
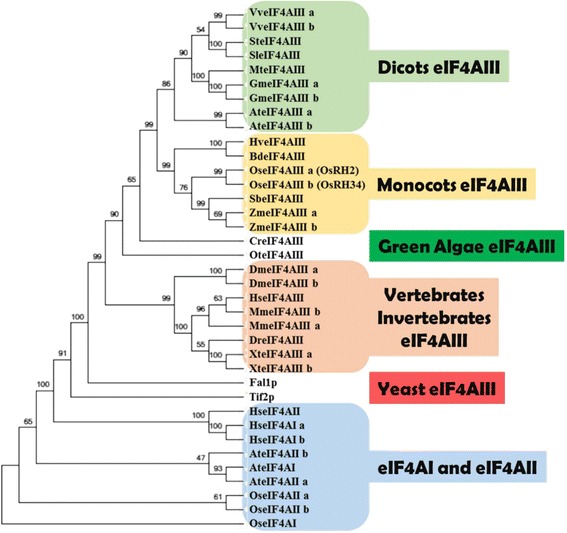
Phylogenetic relationships of eIF4AIII family members. A phylogenetic tree for eIF4AIII in dicots, monocots, green algae, vertebrates, invertebrates, and yeast was generated using MEGA 5. eIF4AIII members from rice, maize, sorghum, and Brachypodium are categorized into the monocot group with at least 50 % bootstrap support. Accession numbers of the genes listed here are shown in Additional file 2
Expression patterns of OsRH2 and OsRH34
To determine the relative expression levels of OsRH2 and OsRH34 in rice, total RNA was isolated from a variety of vegetative and reproductive tissues and was subjected to qRT-PCR with specific primers (Additional file 1). The OsRH2 transcript was expressed in all selected tissues and organs, including roots, stems, leaves, sheaths, panicles, and seedlings (Fig. 3a). Relatively high levels of OsRH2 mRNA were detected in vegetative leaf blades, flag leaves, and panicles before heading (Fig. 3a). Expression of OsRH34 was relatively abundant in vegetative leaf blades, flag leaves, and seedlings, whereas its expression was rarely detected in roots, stems, and panicles (Fig. 3a). These results indicate that these two paralogous genes are coexpressed in most selected tissues and organs in rice. To compare the levels of OsRH2 and OsRH34 mRNA in rice plants, absolute qRT-PCR was performed. Standard curves were used with a serial dilution of either OsRH2 cDNA- or OsRH34 cDNA-containing plasmids. As shown in Fig. 3b, the level of OsRH2 mRNA was 58-fold higher than that of OsRH34 mRNA in rice seedlings at the three-leaf stage.
Fig. 3.
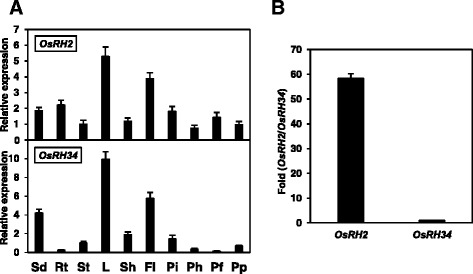
Expression of OsRH2 and OsRH34. a qRT-PCR analysis of OsRH2 and OsRH34 gene expression in rice. Total RNA was isolated from seedlings (Sd), roots (Rt), stems (St), leaves (L), sheaths (Sh), flag leaves (Fl), booting panicles (Pi), heading panicles (Ph), flowering panicles (Pf), and pollinated panicles (Pp). The rice Act1 gene was used as an internal control. b Absolute quantitative RT-PCR analysis of OsRH2 and OsRH34, in which plasmid DNA was applied as a control to compare the mRNA levels of OsRH2 and OsRH34
OsRH2 and OsRH34 were colocalized in nucleus and cytoplasm
To determine the subcellular localization of OsRH2 and OsRH34, plasmids containing an OsRH2–GFP fusion gene and OsRH34–GFP under the control of the CaMV 35S promoter were generated and introduced into onion epidermal cells. Fluorescent signals were emitted from both OsRH2–GFP (Fig. 4a) and OsRH34–GFP (Fig. 4c) in both the nucleus and the cytoplasm. Similar results were obtained in onion cells for the expression of either GFP–OsRH2 (Fig. 4b) or mCherry–OsRH34 (Fig. 4d). To confirm the subcellular localization of OsRH2 and OsRH34, onion cells were cotransformed with GFP–OsRH2 and mCherry–OsRH34. GFP and mCherry signals were colocalized in the nucleus and the cytoplasm (Fig. 4e). These results suggest that the OsRH2 and OsRH34 proteins are localized in both the nucleus and the cytoplasm.
Fig. 4.
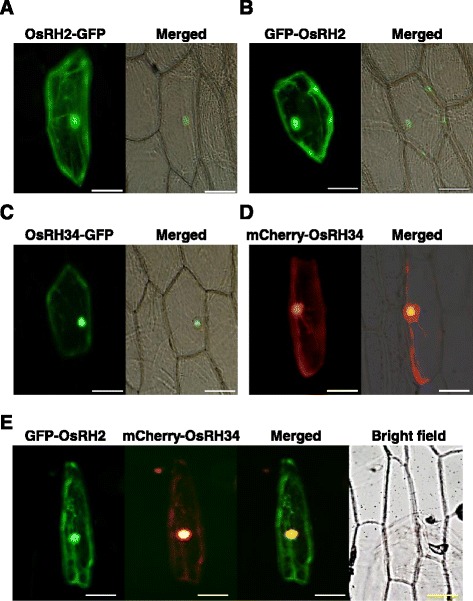
Subcellular localization of OsRH2 and OsRH34. a and b OsRH2 fluorescence fusion protein was localized in the nucleus and the cytoplasm. Onion epidermal cells were transformed with either 35S::OsRH2–GFP (a) or 35S::GFP–OsRH2 (b). c and d Onion epidermal cells were transformed with either 35S::OsRH34–GFP (c) or 35S::mCherry–OsRH34 (d). e Colocalization of GFP–OsRH2 and mCherry–OsRH34 in the nucleus and the cytoplasm. Onion epidermal cells were cotransformed with 35S::GFP–OsRH2 and 35S::mCherry–OsRH34. Bars = 100 μm
Both OsRH2 and OsRH34 are components of the EJC core complex
eIF4AIII can interact with Y14 and MAGO to form the EJC core complex in eukaryotic cells [27, 28]. Gong and He [24] have also reported that rice MAGO and Y14 can form heterodimers. To determine whether OsRH2 and OsRH34 were components of the EJC in rice, interactions among rice MAGO, Y14, and eIF4AIII were examined by BiFC. The N-terminus (YN) of yellow fluorescent protein (YFP) was fused at the downstream end of OsRH2 and OsRH34. The C-terminus (YC) of YFP was fused at the downstream end of OsY14b and OsMAGO1. Coexpression of OsRH2-YN and YC, OsRH34-YN and YC, YN and OsMAGO1-YC, YN and OsY14b-YC in onion epidermal cells were used as negative controls for interaction tests among OsRH2, OsMAGO1, and OsY14, and no fluorescent signals were detected (Fig. 5a). The interaction between OsMAGO1 and OsY14b was used as a positive control that exhibited remarkable fluorescent signals in onion cells (Fig. 5b). These two fusion proteins, OsRH2-YN and OsY14b-YC, were coexpressed in onion cells and the YFP fluorescence was observed (Fig. 5c). OsRH2-YN and OsMAGO1-YC coexpressed in onion cells also displayed the YFP signal (Fig. 5d). Meanwhile, YFP fluorescence was also detected upon the coexpression of OsRH34-YN with OsY14b-YC (Fig. 5e) and OsRH34-YN with OsMAGO1-YC (Fig. 5f), respectively. These results indicate that both OsRH2 and OsRH34 directly interact with OsY14b and OsMAGO1, demonstrating that they are indeed a component of the EJC core complex in rice.
Fig. 5.
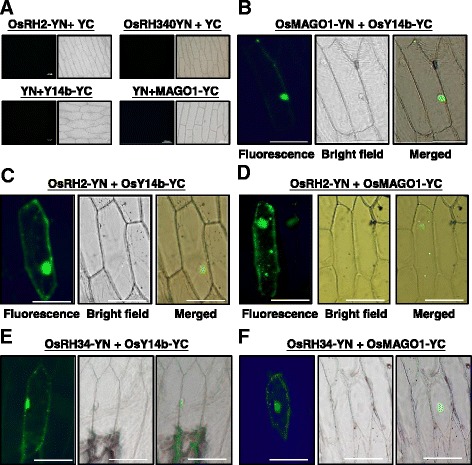
BiFC analysis of the interaction among rice MAGO, Y14, and eIF4AIII in onion epidermal cells. N- and C-terminal fragments of YFP (YN and YC) were fused to the C-terminus of OsRH2, OsRH34, OsMAGO1, and OsY14b, respectively. Onion epidermal cells were cotransformed with combinations of 35S:: OsRH2–YN and 35S::YC, 35S::OsRH34–YN and 35S::YC, 35S::YN and 35S::Y14b–YC, and 35S::YN and 35S::MAGO1–YC as negative controls (a) Onion epidermal cells were cotransformed with 35S::OsMAGO1–YN and 35S::OsY14b–YC (b), 35S::OsRH2–YN and 35S::OsY14b–YC (c), 35S::OsRH2–YN and 35S::OsMAGO1–YC (d), 35S::OsRH34–YN and 35S::OsY14b–YC (e), 35S::OsRH34–YN and 35S::OsMAGO1–YC. (e) Bars = 100 μm
The OsRH2 and the OsRH34 were colocalized (Fig. 4), so protein interaction between these two isoforms was further examined by the BiFC analysis. The YFP fluorescent signals were not be observed in onion cells coexpressed with either combinations of OsRH2-YN and OsRH2-YC, OsRH34-YN and OsRH34-YC, OsRH2-YN and OsRH34-YC, or OsRH34-YN and OsRH2-YC (Additional file 3). These results indicated that proteins of OsRH2 and OsRH34 were not able to interact to form homomer or heteromer.
Characterization of double knockdown of OsRH2 and OsRH34 transgenic lines
To unravel the physiological functions of OsRH2 and OsRH34, a RNA interference mediated genes silencing approach was performed. Because OsRH2 and OsRH34 shared extremely high sequence identity, it was difficult to achieve specific gene silencing. Thus, double knockdown of OsRH2 and OsRH34 was carried out in rice. To minimize the potential off-target gene silencing, the sequences of 271-bp RNAi designed region at the 3´ end of OsRH2 cDNA and OsRH34 cDNA were used as queries to search rice mRNA databases at NCBI. None of region identical of around or more than 16 nucleotides was obtained. Further, a public web-based computational tool developed for identification of potential off-targets, siRNA Scan [29], was applied to search rice mRNA databases, and no potential off-target was detected in the RNAi designed region. Inverted repeat of the 271-bp region was fused at the up- and downstream ends of a GFP coding sequence, and the fusion construct was expressed under the control of the maize ubiquitin gene (Ubi) promoter (Fig. 6a) in transgenic rice. Several independent T1 transgenic plants were obtained, and the levels of OsRH2 mRNA and OsRH34 were determined by qRT-PCR. As results showed in Fig. 6b, both OsRH2 mRNA and OsRH34 mRNA were barely detectable in three independent T1 transgenic lines, RH2Ri 2b, RH2Ri 4, and RH2Ri 14b, indicating that both OsRH2 and OsRH34 were knocked down. Therefore, RH2Ri 2b, RH2Ri 4, and RH2Ri 14b lines were selected to address roles of OsRH2 and OsRH34 in rice.
Fig. 6.
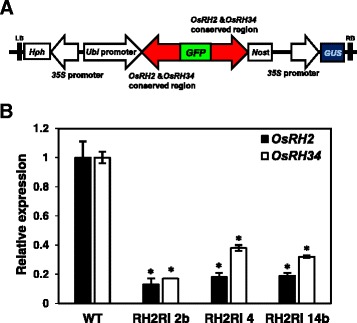
Characterization of OsRH2 and OsRH34 double-knockdown transgenic lines. a Schematic presentation of the double silencing of OsRH2 and OsRH34 of the RNA interference construct. A 271-bp fragment at the 3′ end of OsRH2 and OsRH34 conserved region was ligated in sense and antisense orientations to the GFP cDNA and fused downstream of the Ubi promoter. b Expression of OsRH2 and OsRH34 in T1 transgenic rice seedlings. Total RNA was isolated from 14-day-old seedlings and subjected to qRT-PCR using OsRH2- and OsRH34-specific primers. Rice Act1 was used as an internal control. Error bars indicate the standard deviations (SD) of triplicate experiments. Gene expression was related to wild-type plants, as 1. * is significantly different from the wild-type plants (Student’s t test: p <0.05). OsRH2 and OsRH34 double-knockdown lines are named as RH2Ri 2b, 4, and 14b. Wild-type line is indicated by WT
Reduced plant height in transgenic rice double knockdown of OsRH2 and OsRH34
Significant differences in the height of plants in the T1 transgenic lines were observed. RH2Ri 2b, RH2Ri 4, and RH2Ri 14b showed a dwarf phenotype; their seedlings were 27 to 44 % shorter than those of wild-type plants at 2 weeks old (Fig. 7a and b). Moreover, RH2Ri transgenic plants were shorter than wild-type plants at following growth stage. One example was shown in Fig. 7c, the plant height of RH2Ri 2b T1 plant was 20 and 26 % shorter than wild-type plants at 78-day-old and 147-day-old stages, respectively. Plant height was further compared between wild-type and RH2Ri transgenic plants at the reproductive stage. The culm of wild-type plants contained five internodes, named I to V from top to bottom. Culm lengths of the RH2Ri transgenic plants also appeared to be reduced in each internode region compared with those in the wild-type plants (Fig. 7d and e). The dwarf phenotype of RH2Ri transgenic plants was also observed in a paddy field. Significant differences in plant heights between wild-type plants and RH2Ri plants of the three transgenic T1-T3 generation were observed (Table 1). In addition, the leaves of the RH2Ri transgenic plants were a deeper green and they had a greater number of tillers than the wild-type plants (Fig. 7c).
Fig. 7.
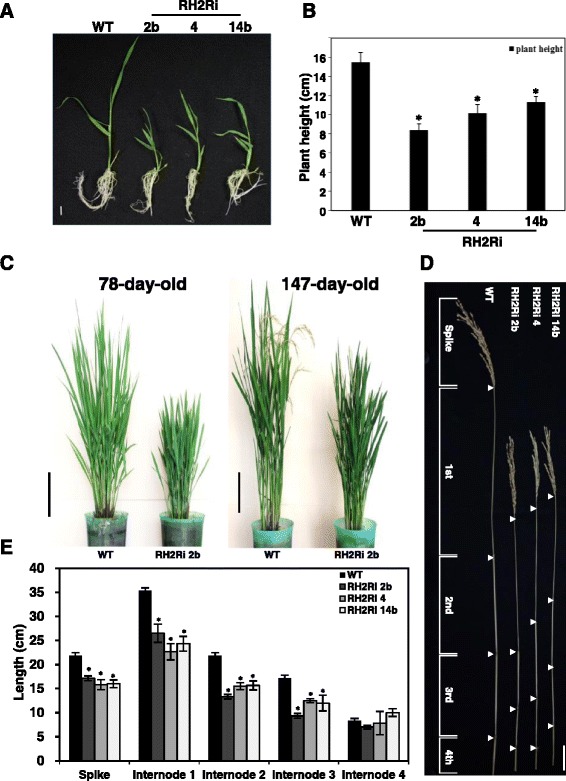
Phenotype of OsRH2 and OsRH34 double-knockdown T1 transgenic rice. a WT and three independent OsRH2 and OsRH34 double-knockdown lines, RH2Ri 2b, RH2Ri 4, and RH2Ri 14b, seedlings were grown on ½ MS agar medium for 10 days and transferred to hydroponic cultures for 7 days. Bar = 1 cm. b Quantification of plant height at seedling stages. The plant height of 17-day-old seedlings was measured. Error bars indicate the SD of ten individual plants for each line. * is significantly different from the wild-type plants (Student’s t test: p <0.05). c Comparison of plant height between WT and RH2Ri 2b in 78-day-old plants and 147-day-old plants. Bars = 19 cm. d Comparison of internode distance of 4-month-old rice plants among WT, RH2Ri 2b, RH2Ri 4, and RH2Ri 14b. Bars = 5 cm. e Determination of internode distance of RH2Ri 2b, RH2Ri 4, RH2Ri 14b, and wild-type plants. Error bars show ± SD (n = 20), * is significantly different from the wild-type plants (Student’s t test: p <0.05)
Table 1.
Heights (cm) of RH2Ri transgenic plants
| Line | ||||
|---|---|---|---|---|
| Generation | WT | RH2i-2b | RH2i-4 | RH2i-14b |
| T1 | 98 ± 3.2 | 78 ± 3.0 * | 87 ± 2.5 * | 89.4 ± 3.8 * |
| T2 | 115 ± 4.2 | 88 ± 5.2 * | 95 ± 6.2 * | 97 ± 4.1 * |
| T3 | 102 ± 5.5 | 80 ± 2.2 * | 85 ± 2.4 * | 87 ± 3.7 * |
± indicates standard deviation, n = 20 for each line
* is significantly different from the wild-type plants (Student’s t test: p <0.05)
Severe defects in pollen and seed development in double knockdowns of OsRH2 and OsRH34
The RH2Ri transgenic plants had 30 ~ 40 % fewer seeds than the wild-type plants (Fig. 8a and b). This marked reduction in the number of seeds suggested that double knockdown of OsRH2 and OsRH34 may cause defects in fertilization or seed development. Aborted pollen was previously identified in OsMAGO1 and OsMAGO2 double-knockdown plants and OsY14a knockdown plants [24]. To address whether OsRH2 and OsRH34 function in the male gametophyte development, pollen viability of RH2Ri transgenic plants was determined by the Alexander staining. In Fig. 8c, aborted pollens were more in RH2Ri transgenic plants than that in wild type, suggesting that double knockdowns of OsRH2 and OsRH34 affected male gametophyte development.
Fig. 8.
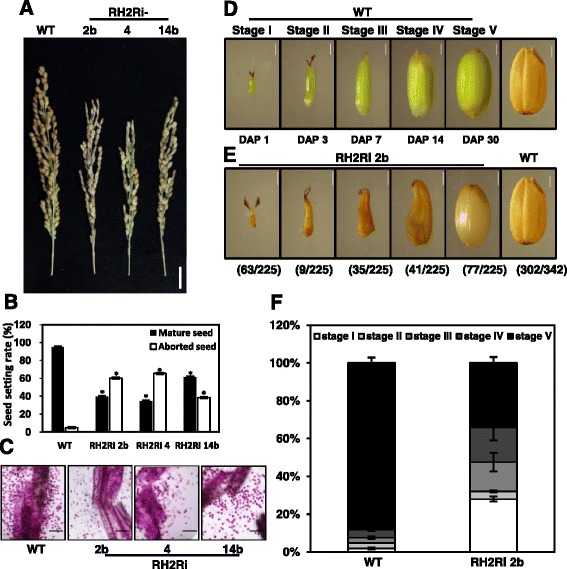
Seed setting rate and seed development in OsRH2 and OsRH34 double-knockdown transgenic rice. a and b A low seed setting rate was observed in OsRH2 and OsRH34 double-knockdown plants. a Spikelet phenotype of three independent OsRH2 and OsRH34 double-knockdown lines, RH2Ri 2b, RH2Ri 4, and RH2Ri 14b. Bars = 5 cm. b Determination of the numbers of mature and aborted seeds in OsRH2 and OsRH34 double-knockdown lines. Error bars show ± SD (n = 20), * is significantly different from the wild-type plants (Student’s t test: p <0.05). c The OsRH2 and OsRH34 double-knockdown lines showed defects in pollen development. Bars = 200 μm. d–f The OsRH2 and OsRH34 double-knockdown lines showed defects in embryonic development. d Micrographs of husked of wild-type rice seeds at various developmental stages. Rice seeds were harvested at 1, 3, 7, 14, and 30 days after pollination (DAP). e The internal seed stages of the RH2Ri 2b line at 30 DAP. f Determination of the numbers of the seeds at different stages in the RH2Ri 2b transgenic and wild-type plants
On the other hand, the levels of seed development normally seen at 1, 3, 7, 14, and 30 days after pollination (DAP) were set as stage I to stage V, respectively (Fig. 8d). Most seeds in the wild-type plants had developed to stage V at 30 DAP (Fig. 8d). However, in the RH2Ri 2b transgenic plants, the level of seed development at 30 DAP varied, from stage I to V; about one-third of the plants remained at stage I, one-third were at stages II, III, or IV, and one-third formed mature seeds (stage V) (Fig. 8e and f). These phenotypes suggested that the OsRH2 and OsRH34 genes play critical roles in the development of rice seeds.
Exogenous gibberellic acid (GA) partially rescues the phenotype of RH2Ri transgenic plants and double knockdown of OsRH2 and OsRH34 influences on GA biosynthesis and GA signaling genes
Phenotypes of the OsRH2 RNAi transgenic plants included dwarf, reduced internode length, deep green in the leaf color, increased tiller number, abnormal seed development and reduced seed germination rate, are similar to mutants deficient in GA biosynthesis or GA signaling pathway. To investigate whether OsRH2 and OsRH34 are involved in the GA biosynthesis or signaling pathway, rice seedlings were treated with 0.1 and 1 μM GA3. Elongation of the dwarf phenotype of 10-day-old RH2Ri seedlings was recovered partially by GA3 treatment (Fig. 9a). To further characterize of GA sensing in RH2Ri transgenic plants, starch plate assay for activity of α-amylase from aleurone layer cells was conducted. The embryoless half-seeds were placed on starch plates with or without 1 μM GA3 for 2 days, and then starch plates were stained with iodine. Activity of α-amylase was not detected in RH2Ri 2b and wild-type embryoless half seeds without treatment of GA3 (Fig. 9b). Cleared zone was detected both in GA3 treatment of half seeds, and no difference in cleared zone size was observed between wild-type and RH2Ri 2b transgenic lines (Fig. 9b and c). These results demonstrated that RH2Ri transgenic plants were responsive to exogenously supplied GA3.
Fig. 9.
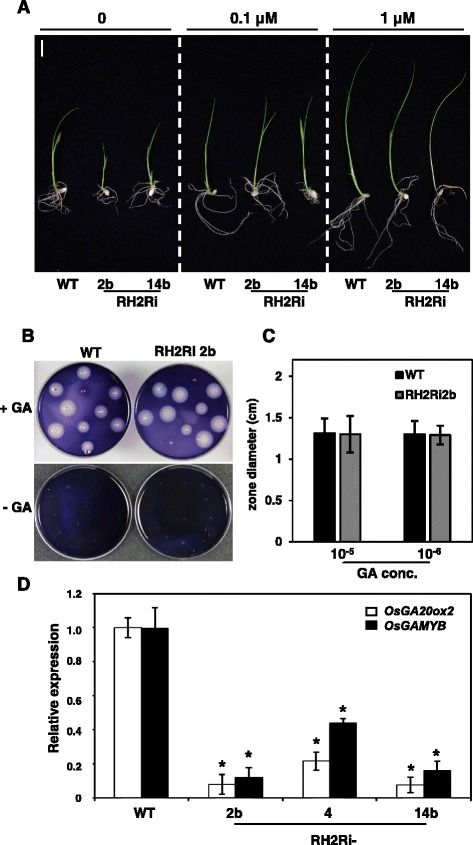
Effect of exogenous GA on the OsRH2 and OsRH34 double-knockdown T1 transgenic rice. a Three-day-old rice seedlings were incubated in water containing 0, 0.1, and 1 μM GA3 for 7 days. Bars = 1 cm. b A starch plate assay of α-amylase activity. Embryoless half seeds were incubated on starch plates with 10−6 M GA3 for 2 days. c Quantification of clear zone diameter on starch plates with 10−5 and 10−6 M GA3. Error bars show ± SD (n = 40). d Expression of OsGA20ox2 and OsGAMYB in the OsRH2 and OsRH34 double-knockdown seedlings. Total RNAs were isolated from three-leaf-stage seedling and subjected to qRT-PCR. Rice Act1 as an internal control. Error bars indicate the SD of four replicate experiments with two biological replicates. Gene expression was related to wild-type plants, as 1. * is significantly different from the wild-type plants (Student’s t test: p <0.05)
To investigate the role of OsRH2 and OsRH34 in GA biosynthesis and GA signaling, the expression levels of the OsGA20ox2, a gene encoded for GA biosynthesis, and the OsGAMYB, a transcription factor in GA signaling, were determined. Total RNAs were isolated from three-leaf-stage of RH2Ri transgenic seedlings and subjected to qRT-PCR analyses. The mRNA levels of the OsGA20ox2 and the OsGAMYB were significantly decreased in various RH2Ri lines, compared to the wild type (Fig. 9d). This result suggested that OsRH2 and OsRH34 participate the regulation of GA biosynthesis and GA signaling pathways.
Double knockdown of OsRH2 and OsRH34 transgenic plants exhibit accumulation of unspliced OsUDT1 mRNA
It has been demonstrated that OsMAGO1, OsMAGO2, and OsY14b are involved in the splicing of OsUDT1 mRNA [24]. Both OsRH2 and OsRH34 are one component of the EJC core complex, suggesting that double knockdowns of OsRH2 and OsRH34 may affect OsUDT1 mRNA maturation. Total RNA was isolated from inflorescence of plants and subjected to RT-PCR using specific primers (Fig. 10a, Additional file 1) for amplifying fragments of OsUDT1 mRNA. Four fragments, namely type I, mature, type II, and type III, were amplified using UDT1R and UDT1F primers (Fig. 10b). The accumulated levels of the type I, type II, and type III were higher in three independent OsRH2 and OsRH34 double-knockdown lines than wild type (Fig. 10b and c). Using the UDTIn1F and UDTIn1R primer pair to specifically amplify the type I fragments (Fig. 10b), more accumulated unspliced type I OsUDT1 mRNAs were detected in these three independent OsRH2 and OsRH34 double-knockdown transgenic lines, as compared to wild type (Fig. 10b and c). These results indicate that OsRH2 and OsRH34 play critical roles in the accurate splicing of OsUDT1 pre-mRNA.
Fig. 10.
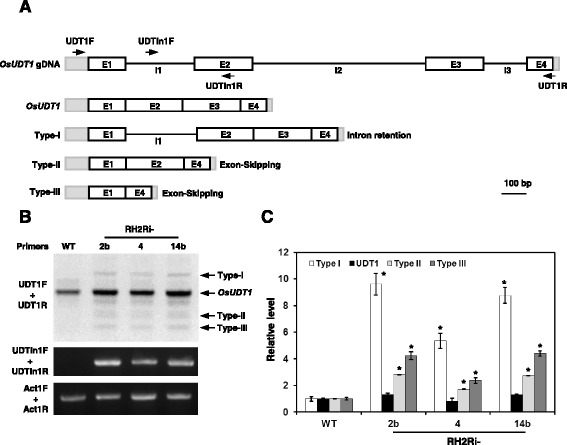
Accumulation of abnormal OsUDT1 transcripts in OsRH2 and OsRH34 double-knockdown plants. a Illustration of gene structure and abnormal transcript structures of OsUDT1 [24], and the positions of primers used for RT-PCR analysis. Gray rectangles, UTRs; white rectangles, exons (E); lines, introns (I). b and c Accumulation of OsUDT1 abnormal transcripts. Total RNAs were isolated from inflorescence of WT, RH2Ri 2b, RH2Ri 4, and RH2Ri 14b plants at vegetative stage. Unspliced OsUDT1 pre-mRNAs were detected by RT-PCR analysis with specific primers (Additional file 1). Act1 mRNAs were used as internal control. c Relative level of abnormal (type I, II and III) and mature OsUDT1 mRNAs were determined by Image J with normalization relative to the WT. Error bars indicated the SD of four replicate experiments with two biological replicates. Level of DNA fragment was related to wild-type plants, as 1. * is significantly different from the wild-type plants (Student’s t test: p <0.05)
Discussion
In this study, two DEAD box RNA helicase genes, OsRH2 and OsRH34, were characterized in rice. Amino acid sequence analysis indicated that OsRH2 and OsRH34 share 99 % identity and 100 % similarity, suggesting that these two DEAD box RNA helicases might have similar biochemical properties in rice. Both OsRH2 and OsRH34 are homologous to eIF4AIII, which is a member of the eIF4A family. eIF4AIII is a core component of the EJC, which is one of the fundamental factors involved in post-transcriptional processes in eukaryotes [17, 30]. Besides eIF4AIII, the EJC also contains three other subunits, MAGO, Y14, and Btz [28]. The results obtained in the present study demonstrate that both OsRH2 and OsRH34 interact physically with OsMAGO1 and OsY14b. Three independent OsRH2 and OsRH34 double-knockdown transgenic lines showed phenotypes that were similar to those of plants in which the OsY14a gene had been knocked down or both OsMAGO1 and OsMAGO2 had been knocked down, namely, reduced plant height and abnormal endothecium and tapetum in flowers [24]. Thus, OsRH2 and OsRH34 are a core component of the EJC in rice.
Immunofluorescence microscopy indicated that eIF4AIII was localized to the nucleoplasm [28] in HeLa cells; a similar localization pattern of eIF4AIII was observed for transiently expressed myc-eIF4AIII [28]. However, excessive eIF4AIII were found in the cytoplasm by subcellular fractionation analysis [28, 31]. These studies indicated that eIF4AIII is localized in the nucleus and the cytoplasm. In terms of the results of the subcellular localization of the OsRH2–GFP and GFP–OsRH2 fusion protein, fluorescence was detected in the nucleus and the cytoplasm. Similarly, the OsRH34–GFP and mCherry–OsRH34 fluorescence was detected in the nucleus and cytoplasm. These results indicate that both OsRH2 and OsRH34 proteins are localized in the nucleus and the cytoplasm, and also suggest that they can shuttle between these two locations. Indeed, Arabidopsis eIF4AIII is mainly localized in the nucleoplasm under normal growth conditions, but is located in the nucleolus and forms splicing speckles under hypoxic stress [23].
The function of the EJC in rice is poorly understood. However, recently, the EJC core subunits OsMAGO1, OsMAGO2, OsY14a, and OsY14b were identified. It has been identified that there are different types of MAGO-Y14 complex, and variation in their specific functions has been proposed [25]. Knockdown of a single MAGO did not lead to any visible phenotype, while the double knockdown of MAGO genes in rice plants led to dwarfism with abnormal flowers [24], suggesting that OsMAGO1 and OsMAGO2 are functionally redundant. The phenotype of OsY14a knockdown rice plants matched that of OsMAGO1 and OsMAGO2 double-knockdown plants, while the knockdown of OsY14b led to failure of the induction of plantlets [24], suggesting the functional specialization of OsY14b in embryogenesis. The amino acid sequences of OsRH2 and OsRH34 were found to be highly conserved, and their gene expression patterns were also similar in various rice tissues. However, the abundance of OsRH2 mRNA was about 58-fold higher than that of OsRH34 mRNA in seedlings. These results suggest that OsRH2 and OsRH34 may be functionally redundant, and that OsRH2 plays a major role in rice. Since DNA sequence of OsRH2 and OsRH34 are too similar to make specific gene silencing in rice, we therefore cannot rule out a possibility whether each of them has specific functions in rice.
We introduced OsRH2 mRNA-based interfering RNA into rice and knocked down both the OsRH2 and the OsRH34 genes in the same transgenic line. The OsRH2 and OsRH34 double-knockdown transgenic lines showed a dwarf phenotype. The number of nodes and the internode distance in OsRH2 and OsRH34 double-knockdown lines were less than in the wild type. In addition, the leaves of the OsRH2 and OsRH34 double-knockdown transgenic plants were a deeper green and they had a greater number of tillers than the wild-type plants. These phenotypes are similar to those of mutants with defects in gibberellin signaling [32–35] or gibberellin biosynthesis [36–40]. Exogenous GA was able to partially rescue the dwarf phenotype and induce α-amylase activities in aleurone layers of the OsRH2 and OsRH34 double-knockdown transgenic lines. The expression levels of the OsGA20ox2 were decreased in the OsRH2 and OsRH34 double-knockdown transgenic lines as compared to WT. These results suggested that the dwarf phenotype of the OsRH2 and OsRH34 double-knockdown transgenic lines may due to the decreased level of GA. However, low levels of OsGAMYB mRNA were detected in the OsRH2 and OsRH34 double-knockdown transgenic lines. In a previous study, OsY14a knockdown rice plants and those with double knockdown with MAGO genes also exhibited dwarfism with abnormal flowers, and low level of the OsGA20ox2 and the OsGAMYB mRNA [24]. Thus, the function of EJC was suggested to be strongly correlated with the gibberellin action in rice.
There was a discrepancy in the seed setting rate between the wild-type and OsRH2 and OsRH34 double-knockdown transgenic lines. The ratio of mature seeds to total seeds in the OsRH2 and OsRH34 double-knockdown transgenic lines was lower than in the wild type. The aborted pollen phenotype observed in the OsRH2 and OsRH34 double-knockdown plants was consistent with previously identified in OsMAGO1 and OsMAGO2 double-knockdown plants and OsY14a knockdown plants [24]. These results suggest that the low seed setting rate may be caused by a defective EJC, which affects pollen development. However, it is necessary to address whether the EJC is also involved in female gametophyte development. Alternatively, seed development was compared between the wild type and the OsRH2 and OsRH34 double-knockdown transgenic lines. After pollination, 90 % of the seeds of the wild type developed to the mature stage. In contrast, in the transgenic lines, one-third of the seeds developed to maturity, one-third remained at an intermediate stage, and one-third did not progress beyond a very early stage. These results suggested that the double knockdown of OsRH2 and OsRH34 impaired seed development. Taking these findings together, the double knockdown of the OsRH2 and OsRH34 genes may cause defects in pollen and seed development.
In eukaryotic cells, nonsense-mediated mRNA decay (NMD), a surveillance mechanism, eliminates mRNA that contains nonsense mutations or has acquired premature termination codons because of aberrant splicing [18]. It is thus an effective safeguard for eliminating aberrant gene expression [18, 41–44]. The EJC has been demonstrated to be involved in the post-transcriptional processing of mRNA, including mRNA splicing and NMD, in eukaryotes [45]. In rice, OsMAGO1 and OsMAGO2 double-knockdown plants and OsY14a knockdown plants exhibited abnormal splicing of OsUDT1 transcripts. Multiple types of OsUDT1 mRNA were detected in OsMAGO1 and OsMAGO2 double-knockdown plants and OsY14a knockdown plants [24]. In the present study, double knockdown of the OsRH2 and OsRH34 genes also led to abnormal OsUDT1 pre-mRNA splicing accumulation. Thus, the knockdown of one component of the EJC causes defects of EJC function, which is strongly correlated to the accumulation of certain abnormal pre-mRNA. However, this type of intron retained pre-mRNA accumulation may be due to an EJC dependent-splicing defect or an EJC dependent-NMD defect. Future studies to identify the proteins that interact with OsRH2 or OsRH34 might provide more information on the specificity of the function of EJC in rice.
Conclusion
The EJC contains four core components, eukaryotic initiation factor 4AIII (eIF4AIII), MAGO/NASHI, Y14/Tsunagi/RNA-binding protein 8A, and Barentsz/Metastatic lymph node 51, and plays important roles in gene regulation. Genes encoding core EJC components have been found in rice, and currently.the functional characterizations of MAGO and Y14 homologs have been demonstrated in economically important crop, rice. However, little is known about how important of eIF4AIII in rice. In this study, two rice eIF4AIII homologous genes, OsRH2 and OsRH34, were identified. Deduced amino acid sequence of OsRH2 and OsRH34 share 99 % identity and 100 % similarity. Both rice eIF4AIII fluorescent fusion proteins were localized in the cytoplasm and the nucleus. Moreover, OsRH2 and OsRH34 can interact with rice MAGO and Y14, indicating that OsRH2 and OsRH34 are core components of the EJC. OsRH2 and OsRH34 may be functionally redundant, but the abundantly expressed OsRH2 may play a major role in rice. Double-knockdown of OsRH2 and OsRH34 exhibited severe defects in terms of plant height, pollen, and seed development. Moreover, double knockdown of the OsRH2 and OsRH34 genes led to decrease in expression levels of OsGA20ox2 and the OsGAMYB and abnormal accumulation of OsUDT1 pre-mRNA. These visible and molecular phenotypes caused by OsRH2 and OsRH34 double-knockdown are similar to OsMAGO1 and OsMAGO2 double-knockdown plants and OsY14a knockdown plants. Collectively, our findings demonstrate the eIF4AIII proteins, OsRH2 and OsRH34, play critical roles in the functional rice EJC.
Methods
Plant materials and growth conditions
The rice cultivar Oryza sativa L. cv Tainung 67 (TNG67) was collected from the Taiwan Agricultural Research Institute and used in this study. Transgenic rice plants were cultivated at the Agricultural Experiment Station, National Chung-Hsing University (Taichung, Taiwan). For seed germination, seeds were de-hulled, sterilized with 3 % NaOCl for 30 min, and washed extensively with sterile water. Sterilized seeds were placed on ½ Murashige Skoog (MS) agar medium, and then cultivated in a growth chamber at 28 °C under constant light. Seedlings at the three-leaf stage were transferred into hydroponic culture medium (Kimura B solution) for 2 days and then used for various treatments.
Primers
The nucleotide sequences of all primers used for plasmid construction, PCR, RT-PCR, and qRT-PCR analyses are listed in Additional file 1.
Plasmids
Plasmid pMDC43 [46] was used for fusion of the OsRH2–GFP chimeric protein. Plasmids pSAT4-DEST-nEYFP-C1 and pSAT5-DEST-cEYFP-C1, were used as gateway vectors for the BiFC assay, and were obtained from the Arabidopsis Biological Resource Center. The pCAMBIA vectors were obtained from CAMBIA.
Plasmid construction
The OsRH2 and OsRH34 coding regions were amplified with specific primers (Additional file 1) by Phusion High-Fidelity DNA Polymerase (NEB, Ipswich, MA, USA) using the cDNA of seedlings at the three-leaf stage as templates. The PCR products were cloned into the yT&A cloning vector (Yeastern, Taipei, Taiwan) to generate pOsRH2 and pOsRH34, respectively. To investigate the subcellular localization of OsRH2 and OsRH34, their full-length cDNA fragments were excised from pOsRH2 and pOsRH34 with AscI and NotI, and then ligated into the same sites of pENTR-TOPO vector to generate pOsRH2-ENTR and pOsRH34-ENTR vectors. Using LR clonase (Invitrogen, Carlsbad, CA), recombination was carried out to transfer OsRH2 and OsRH34 DNA fragments from entry clones to the destination vector, pMDC43, to generate the GFP-OsRH2 and GFP-OsRH34, respectively. The OsRH2 and OsRH34 DNA fragments were also constructed into the pMDC85 to generate the OsRH2-GFP and OsRH34-GFP plasmids, respectively. To construct the OsRH34-mCherry expression vector, a mCherry destination vector, pMDC43m, was generated by replacing the GFP with mCherry in pMDC43. The expression plasmid of mCherry-OsRH34 was generated by LR clonase.
For the BiFC assay, full-length coding regions of OsRH2, OsMAGO1, and OsY14b were amplified with specific primers and then subcloned into the pDonor221 binary vector between the attL1 and attL2 sites using BP clonase (Invitrogen). Each fragment was subcloned into pSAT4-DEST-nEYFP-C1 and pSAT5-DEST-cEYFP-C1 (B) binary vectors using LR clonase to generate OsRH2–, OsMAGO1–, and OsY14b–YFP (n), and OsRH2–, OsMAGO1–, and OsY14b–YFP (c) fusion genes.
For construction of the OsRH2 interfering RNA vector, a 271-bp DNA fragment containing 153 bp of the coding region and 117 bp of the 3′’ UTR of OsRH2 was amplified using specific primers (Additional file 1). This DNA fragment was cloned into the yT&A cloning vector, generating pRH2Ri. Green fluorescent protein (GFP) cDNA was amplified by PCR using a forward primer and a reverse primer (Additional file 1), and was then subcloned into the yT&A cloning vector, generating pGFPRI. The OsRH2 RNAi DNA fragment was isolated from pRH2Ri by digestion with EcoRI and BamHI, the GFP DNA fragment was isolated from pGFPRI by digestion with EcoRI, and these two fragments were ligated into the BamHI site of the pAHC18 expression vector, generating pAHC18-OsRH2-Ri. This RNA silencing construct was linearized by digestion with HindIII and inserted into the HindIII site of the pCAMBIA1301 binary vector for Agrobacterium-mediated gene transformation.
RT-PCR and qRT-PCR analyses
Total RNA was isolated from whole seedlings and various tissues of mature plants using Trizol reagent (Invitrogen) and then treated with RNase-free DNase I (NEB) to remove genomic DNA contamination. First-strand cDNA was synthesized using RTace reverse transcriptase (Toyobo, Osaka, Japan) with oligo-dT primers. A 20-fold dilution of the resultant first-strand cDNA was subjected to PCR (22–35 reaction cycles) with gene-specific primers (Additional file 1). For the qRT-PCR reaction, first-strand cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen). A 10-fold dilution of the first-strand cDNA was subjected to qRT-PCR using FastStart Essential DNA Green Master (Roche, Basel, Switzerland) and an iQ5 RT-PCR machine (Bio-Rad, Hercules, CA, USA), in accordance with the manufacturers’ instructions. The PCR procedure was independently repeated at least three times. The relative gene expression levels are expressed as ratios of the abundance of the target gene’s mRNA to that of Act1 mRNA. Data were analyzed using the iQ5 2.1 software provided by the manufacturer. The gene-specific primers used for qRT-PCR are listed in Additional file 1.
Plant transformation
Rice embryonic calli were induced from germinated seeds on N6 solid medium with 9 μM 2,4-dichlorophenoxy. Agrobacterium tumefaciens strain EHA105 was used to perform rice transformation, as previously described [47]. Transformed calli were selected on N6 medium containing 25 mg/L hygromycin B.
Subcellular localization analysis and BiFC assay
The onion bulb epidermis was prepared and particle bombardment was carried out as described previously [48, 49] with a PDS-1000 biolistic device (Bio-Rad) at 1100 psi. To introduce the plasmid DNA, the bombarded material was cultured in MS medium for 24 h, and then observed and imaged with an Olympus IX71 inverted fluorescence microscope (Olympus, Tokyo, Japan) with a digital camera. The Olympus UMWIBA3 and the Olympus U-MWIGA3 filters were used to obtain GFP and mCherry images, respectively, and images were merged by the DP Manager program.
For BiFC analysis, various combinations of expression vector carriers with YFPN- and YFPC-fused genes were coexpressed in epidermal cells of onion bulb epidermis by particle bombardment. The YFP signal was observed using an Olympus IX71 inverted fluorescence microscope with the Olympus UMWIBA3 filter.
GA treatment
Sterilized seeds were placed on ½ MS agar medium, and then cultivated in a growth chamber at 28 °C under constant light for 3 days. Seedlings were transferred into hydroponic culture medium (Kimura B solution) with various GA concentrations for 7 days.
α-amylase activity assay
Embryoless half seeds (endosperms) were sterilized with 3 % NaOCl for 30 min, washed extensively with sterile water. Each plate contained 16 half seeds that were arranged in a small circle. The plates were incubated in the dark for 1–3 days at 30 °C and then stained with iodine solution. The sizes of colorless zone were measured.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files.
Acknowledgements
This work was supported by grants “103-231-B-008-001-“and “104-2321-B-008-001-“from the Ministry of Science and Technology of the Republic of China, Taiwan.
Abbreviations
- Act
actin
- ALY/Ref
Aly/REF export factor
- BiFC
bimolecular fluorescence complementation
- DAP
days after pollination
- eIF4A
eukaryotic initiation factor 4A
- eIF4AIII
eukaryotic initiation factor 4AIII
- EJC
exon junction complex
- GA
gibberellic acid
- GA20ox2
gibberellin 20 oxidase 2
- GAMYB
GAMYB transcription factor
- GFP
green fluorescent protein
- MAGO
exon junction complex mago nashi
- mCherry
mCherry red fluorescent protein
- MS medium
Murashige Skoog medium
- NCBI
National Center for Biotechnology Information
- NMD
nonsense-mediated mRNA decay
- qRT-PCR
quantitative real time reverse transcription polymerase chain reaction
- RH
RNA helicase
- RNPS1
RNA-binding protein S1
- Ubi
maize ubiquitin gene
- UDT1
undeveloped tapetum 1
- Y14
exon junction complex protein Y14
- YFP
yellow fluorescent protein
Additional files
Primers used in the study. (DOCX 17 kb)
Accession numbers and proteins homologous to eIF4A. (DOCX 18 kb)
BiFC analysis of the interaction between OsRH2 and OsRH34. (PPTX 122 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CKH and CAL participated in the design of the study. CKH, YSS, and YFC carried out the bioinformatics analysis, gene cloning, real-time RT-PCR, subcellular localization, and BiFC. CKH, YSS, and CAL carried out data analysis, and wrote manuscript. YSS, YFC, and TSH assisted in collected the tissues for gene expression analysis. All authors read and approved the final manuscript.
Contributor Information
Chun-Kai Huang, Email: hck0903@gate.sinica.edu.tw.
Yi-Syuan Sie, Email: syuan0714@gmail.com.
Yu-Fu Chen, Email: vicky_vic0609@hotmail.com.
Tian-Sheng Huang, Email: joeson0617@yahoo.com.tw.
Chung-An Lu, Phone: 886-3-4227151, Email: chungan@cc.ncu.edu.tw.
References
- 1.Byrd AK, Raney KD. Superfamily 2 helicases. Front Biosci (Landmark Ed) 2012;17:2070–88. doi: 10.2741/4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 3.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20(3):313–24. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubourg S, Kreis M, Lecharny A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 1999;27(2):628–36. doi: 10.1093/nar/27.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okanami M, Meshi T, Iwabuchi M. Characterization of a DEAD box ATPase/RNA helicase protein of Arabidopsis thaliana. Nucleic Acids Res. 1998;26(11):2638–43. doi: 10.1093/nar/26.11.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owttrim GW. RNA helicases: diverse roles in prokaryotic response to abiotic stress. RNA Biol. 2013;10(1):96–110. doi: 10.4161/rna.22638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owttrim GW. RNA helicases and abiotic stress. Nucleic Acids Res. 2006;34(11):3220–30. doi: 10.1093/nar/gkl408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocak S, Linder P. Dead-box proteins: The driving forces behind RNA metabolism. Nat Rev Mol Cell Bio. 2004;5(3):232–41. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 9.Umate P, Tuteja R, Tuteja N. Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol Biol. 2010;73(4–5):449–65. doi: 10.1007/s11103-010-9632-5. [DOI] [PubMed] [Google Scholar]
- 10.Xu R, Zhang S, Huang J, Zheng C. Genome-wide comparative in silico analysis of the RNA helicase gene family in Zea mays and Glycine max: a comparison with Arabidopsis and Oryza sativa. PLoS One. 2013;8(11):e78982. doi: 10.1371/journal.pone.0078982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293(5536):1832–6. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 12.Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2(12):1119–24. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohr SE, Dillon ST, Boswell RE. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Gene Dev. 2001;15(21):2886–99. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427(6976):753–7. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 15.Park N-I, Yeung EC, Muench DG. Mago Nashi is involved in meristem organization, pollen formation, and seed development in Arabidopsis. Plant Sci. 2009;176(4):461–9. doi: 10.1016/j.plantsci.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X-F, Nowak NJ, Shows TB, Aplan PD. MAGOH interacts with a novel RNA-binding protein. Genomics. 2000;63(1):145–8. doi: 10.1006/geno.1999.6064. [DOI] [PubMed] [Google Scholar]
- 17.Tange TØ, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16(3):279–84. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 19.Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Gene Dev. 2004;18(2):210–22. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh S, Marchand V, Gaspar I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Struct Mol Biol. 2012;19(4):441–9. doi: 10.1038/nsmb.2257. [DOI] [PubMed] [Google Scholar]
- 21.Simon B, Masiewicz P, Ephrussi A, Carlomagno T. The structure of the SOLE element of oskar mRNA. RNA. 2015;21(8):1444–53. doi: 10.1261/rna.049601.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, et al. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell. 2005;16(1):260–9. doi: 10.1091/mbc.E04-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Simpson CG, et al. Dynamic behavior of Arabidopsis eIF4A-III, putative core protein of exon junction complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell. 2009;21(5):1592–606. doi: 10.1105/tpc.108.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong P, He C. Uncovering divergence of rice exon junction complex core heterodimer gene duplication reveals their essential role in growth, development, and reproduction. Plant Physiol. 2014;165(3):1047–61. doi: 10.1104/pp.114.237958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong P, Quan H, He C. Targeting MAGO proteins with a peptide aptamer reinforces their essential roles in multiple rice developmental pathways. Plant J. 2014;80(5):905–14. doi: 10.1111/tpj.12672. [DOI] [PubMed] [Google Scholar]
- 26.Gong P, Zhao M, He C. Slow co-evolution of the MAGO and Y14 protein families is required for the maintenance of their obligate heterodimerization mode. PLoS One. 2014;9(1):e84842. doi: 10.1371/journal.pone.0084842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12(10):861–9. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 28.Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, et al. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10(2):200–9. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P, Zhang YJ, Kang L, Roossinck MJ, Mysore KS. Computational estimation and experimental verification of off-target silencing during posttranscriptional gene sikencing in plants. Plant Physiol. 2006;142:429–440. doi: 10.1104/pp.106.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, et al. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005;17(10):2705–22. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mufarrege EF, Gonzalez DH, Curi GC. Functional interconnections of Arabidopsis exon junction complex proteins and genes at multiple steps of gene expression. J Exp Bot. 2011;62(14):5025–36. doi: 10.1093/jxb/err202. [DOI] [PubMed] [Google Scholar]
- 32.Holzmann K, Gerner C, Pöltl A, Schäfer R, Obrist P, Ensinger C, et al. A human common nuclear matrix protein homologous to eukaryotic translation initiation factor 4A. Biochem Biophys Res Commun. 2000;267(1):339–44. doi: 10.1006/bbrc.1999.1973. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, et al. slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13(5):999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, et al. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 2003;35(1):104–15. doi: 10.1046/j.1365-313X.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- 35.Ueguchi-Tanaka M, Fujisawa Y, Kobayashi M, Ashikari M, Iwasaki Y, Kitano H, et al. Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc Natl Acad Sci U S A. 2000;97(21):11638–43. doi: 10.1073/pnas.97.21.11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chhun T, Aya K, Asano K, Yamamoto E, Morinaka Y, Watanabe M, et al. Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell. 2007;19(12):3876–88. doi: 10.1105/tpc.107.054759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiao F, Zhao KJ. The influence of RNAi targeting of OsGA20ox2 gene on plant height in rice. Plant Mol Biol Rep. 2011;29(4):952–60. doi: 10.1007/s11105-011-0309-2. [DOI] [Google Scholar]
- 38.Sakai M, Sakamoto T, Saito T, Matsuoka M, Tanaka H, Kobayashi M. Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. J Plant Res. 2003;116(2):161–4. doi: 10.1007/s10265-003-0080-z. [DOI] [PubMed] [Google Scholar]
- 39.Sakamoto T, Kobayashi M, Itoh H, Tagiri A, Kayano T, Tanaka H, et al. Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiol. 2001;125(3):1508–16. doi: 10.1104/pp.125.3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, et al. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 2004;134(4):1642–53. doi: 10.1104/pp.103.033696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo SF, Yang SY, Chen KT, Hsing YI, Zeevaart JA, Chen LJ, et al. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 2008;20(10):2603–18. doi: 10.1105/tpc.108.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chuang T-W, Chang W-L, Lee K-M, Tarn W-Y. The RNA-binding protein Y14 inhibits mRNA decapping and modulates processing body formation. Mol Biol Cell. 2013;24(1):1–13. doi: 10.1091/mbc.E12-03-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012;40(6):2454–69. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Gene Dev. 2006;20(3):355–67. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyikó T, Kerényi F, Szabadkai L, Benkovics AH, Major P, Sonkoly B, et al. Plant nonsense-mediated mRNA decay is controlled by different autoregulatory circuits and can be induced by an EJC-like complex. Nucleic Acids Res. 2013;41(13):6715–28. doi: 10.1093/nar/gkt366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Gene Dev. 2007;21(15):1833–3856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 47.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133(2):462–9. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang CK, Yu SM, Lu CA. A rice DEAD-box protein, OsRH36, can complement an Arabidopsis atrh36 mutant, but cannot functionally replace its yeast homolog Dbp8p. Plant Mol Biol. 2010;74(1–2):119–28. doi: 10.1007/s11103-010-9659-7. [DOI] [PubMed] [Google Scholar]
- 49.Scott A, Wyatt S, Tsou P, Robertson D, Allen NS. Model system for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques. 1999;26(6):1125. doi: 10.2144/99266st04. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are included within the article and its additional files.


