Abstract
The polarization and migration of neural progenitor cells (NPCs) are critical for embryonic brain development and neurogenesis after brain injury. Although stromal-derived factor-1α (SDF-1α, CXCL12) and its receptor CXCR4 are well-known to mediate the migration of NPCs in the developing brain, the dynamic cellular processes and structure-related molecular events remain elusive. Transwell and microfluidic-based assays are classical assays to effectively study cellular migration. However, both of them have limitations in the analysis of a single cell. In this study, we modified the stripe assay and extended its applications in the study of NPC polarization and intracellular molecular events associated with CXCL12-mediated migration. In response to localized CXCL12, NPCs formed lamellipodia in the stripe assay. Furthermore, CXCR4 and Rac1 quickly re-distributed to the area of lamellipodia, indicating their roles in NPC polarization upon CXCL12 stimulation. Although the chemokine stripes in the assay provided concentration gradients that can be best used to study cellular polarization and migration through immunocytochemistry, they can also generate live imaging data with comparable quality. In conclusion, stripe assay is a visual, dynamic and economical tool to study cellular mobility and its related molecule mechanisms.
Keywords: PDMS, Stripe assay, Polarization, Migration, CXCL12, CXCR4
1. Introduction
The migration and polarization of neural progenitor cells (NPCs) are critical processes for neural development and neural repair during diseases in the central nervous system (CNS). Understanding the detailed cellular events and intracellular signaling intermediates through gene expressions and protein analyses may provide important information on the mechanisms of neural development and repair [1-3]. Stromal-derived factor-1α (SDF-1α, CXCL12) is a member of the CXC chemokine family that plays an essential role on hematopoiesis, neoangiogenesis, tumorigenesis, immune response and the maintenance of tissue stem cells [4-8]. Notably, CXCL12, its traditional receptor CXCR4 and newly identified receptor CXCR7 are widely express in the CNS, regulating neurogenesis and neural regeneration during the development and pathogenic processes of brain diseases, respectively [9-12]. Although CXCL12/CXCR4 plays crucial roles in various aspects of neurogenesis by regulating NPC migration, proliferation and formation of neuronal circuits, the cellular events and associated molecular mechanisms remain unclear.
Transwell assays, either the Boyden or modified Boyden chamber assay [13], are commonly used to study the migration of cells [14]. In these assays, adherent or nonadherent cells migrate through a permeable filter after triggered by very low chemotactic inducers. Therefore, these assays are appropriate for a variety of cells and quite sensitive. However, these assays are also traditionally being end-point assays, making it difficult to assess cells during migration. Microfluidic-based assays are more recently developed migration assays that connect two chambers with an internal channel allowing for cell migration. These assays are generally sensitive and effective. However, they often come with considerable cost for the kits.
Stripe assays was originally developed to investigate the fundamental axonal guidance in vitro by F. Bonhoeffer and co-workers [15, 16] in the late 1980s [17-20]. By forming stripes containing different factors under investigation, this assay gains its popularity in the research of process outgrowth involving multiple cells, including neural crest cells [21], oligodendrocytes [22, 23], and tumor cells [24]. We have previously successfully used stripe assay to explore the surprising role of CXCR7, a newly discovered atypical chemokine receptor, on CXCL12-mediated migration and polarization [12]. The paper generated considerable interest. Therefore, we intend to further characterize the stripe assay in studying cellular migration, polarization, and their associated signaling intermediates in the current paper. We demonstrated that the stripe assay can generate remarkable migration and polarization data with comparable quality comparing with live cell imaging data. In addition, we determined that this assay could be used to investigate signaling intermediates involved in cellular migration and polarization.
2. Materials and Methods
2.1. Reagents
Epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) were obtained from Life Technologies. Recombinant mouse CXCL12 was obtained from R&D systems. Primary antibodies included monoclonal rat anti-CXCR4 (R&D Systems, 1:200), polyclonal rabbit anti-Rac1 (Sigma, 1:100), and monoclonal mouse anti-Nestin (Millipore, 1:100). Secondary antibodies included goat anti-mouse IgG (coupled with Alexa Flour 568, Invitrogen), goat anti-rat IgG (coupled with Alexa Fluor 488, Invitrogen), goat anti-rabbit IgG (coupled with Alexa Fluor 647, Invitrogen). AMD3100 was obtained from Sigma-Aldrich.
2.2. Mouse NPCs collection and cell culture
All mice were housed and bred in the Comparative Medicine facilities of the University of Tongji Medical College, China or the University of Nebraska Medical Center, USA. All procedures were conducted according to protocols approved by the Institutional Animal Care and Use Committee of the University of Tongji Medical College and the University of Nebraska Medical Center.
The mouse forebrain of each embryo at gestational day E13.5 was dissected and dissociated into single cells by triturating the tissue with a 1 ml pipette. Cells from each forebrain were seeded into a 100 mm Petri dish separately at a density of 2 × 105 cells/ml in 10 ml of mouse NeuroCult NSC Proliferation Medium (Stem Cell Technologies) supplemented with epidermal growth factor (20 ng/ml) and basic fibroblast growth factor (10 ng/ml) for neurosphere cultures. Neurospheres were passaged when they reached 100 - 150 μm in diameter.
2.3. Transwell assay
NPC migration was evaluated using an 8-mm pore size transwell system (Costar) coated with fibronectin (Sigma-Aldirich) at 5 ng/ml and poly-D-lysine (PDL, Sigma-Aldirich) at 100 μg/ml in PBS overnight. Briefly, NPCs were dissociated into single cells and resuspended in Mouse NeuroCult Proliferation Medium at a density of 105 cells/ml. The top chamber of the transwell was loaded with 100 μl of cell suspension and cells were cultured for 12 hours to form an adherent monolayer culture. CXCL12 was added to the bottom chamber. After 12 hours, the membrane of the transwell inserts was fixed with 4% PFA in PBS, and cells on top of the membrane were removed with a cotton swab. Cells that migrated to the bottom of the membrane were stained with DAPI (Sigma) in PBS at 10 ng/ml. For each insert, 10 fields were randomly selected to be captured under a microscope at 20×, and cell numbers were quantified using Image-Pro Plus 6.0. The cell number of each treated group was normalized to the cell number of the control group to calculate the migration index.
2.4. PDMS stripe device fabrication
The stripe device was fabricated using mold etched by soft lithography with polydimethylsiloxane (PDMS), modified from the earlier work [25]. To fabricate the PDMS stripe device, the mold was first exposed to chlorotrimethylsilane (Alfa Aesar, Lancs, England) vapor for 3 minutes in order to promote elastomer release after carrying out the baking steps. A mixture of PDMS was vacuumed for 10 minutes and then poured onto the mold. After degassing (5 minutes), the mold was baked for 60 minutes at 85 °C, after which the PDMS flow layer structure was then peeled from the mold.
2.5. Stripe assay
A silicon wafer was used to generate a template for the PDMS mold and substrates were patterned in parallel stripes of 60 μm width separated by 90 μm gaps. Briefly, PDMS mold was reversibly sealed on PDL-coated glass bottom dishes (35 mm dish with 20 mm bottom well), and microchannels were formed between the PDMS mold and well. The mixture of BSA and CXCL12 or BSA alone was added to one end of the microchannels, and vacuum was applied to the other end of the microchannels to ensure BSA/CXCL12 or BSA filled in all the microchannels. PDMS molds were removed after drying overnight. The stripe-coated dishes were planted with dissociated NPCs to study cell migration or polarization. Cells were then fixed on cover glasses and stained with Nestin and DAPI. Images were taken by a Zeiss 710 confocal microscope and quantified by Image Pro Plus. The percent of cells on stripe versus total cell number was calculated to evaluate cell migration.
2.6. Immunofluorescence and confocal microscopy
For immunofluorescence staining, mNPCs were fixed using 4% paraformaldehyde (PFA), and permeabilized with 0.4% triton-X in PBS. After blocked by 1% BSA in PBS, mNPCs were incubated with primary antibodies overnight. Cultures were washed and then incubated with phalloidin (coupled with rhodamine, Invitrogen) and corresponding secondary antibodies for one hour at room temperature. Nuclear DNA was labeled with 4’, 6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) for 2 minutes after the secondary antibody at room temperature. Cover slips were mounted on glass slides with mounting medium (Sigma-Aldrich). Triple immunostaining was examined by a Zeiss META 710 confocal microscope.
2.7. Transfectiion with LiveAct-RFP plasmid
Lipofectamine® LTX and PLUS™ Reagents (Life Technologies) was utilized for the transfection of LiveAct-RFP plasmid (ibidi). The experiment was performed according to the manufacturer's instruction. Briefly, NPCs were transfected with LiveAct-RFP plasmid in a 6-well format after optimizing the transfection efficiency. NPCs were incubated at 37°C in a CO2 incubator for 18 - 48 hours prior to stripe assays.
2.8. 3D reconstruction of immunofluorescence
After dissociation, mix single NPCs (after transfection of LifeAct-RFP and stain with Hoechst33342) with MatriGel (BD) properly on ice (liquid state) and inoculate into the stripe-coated dishes. Incubate the dishes at 37°C in a CO2 incubator for 30 minutes (becoming a solid state), add 1 ml mouse NeuroCult NSC Proliferation Medium (Stem Cell Technologies) with EGF and FGF. After 2 hours, fix mouse NPCs. Cultures were washed. After 3 times wash, images were taken by Z-stack scanning and 3 d reconstruction of Zeiss 710 confocal microscope.
2.9. Statistics analysis
Statistical analysis was performed on all quantitative assays. The appropriate one-way ANOVA were run to determine statistical significance with p < 0.05 considered significant (GraphPad Prism).
3. Results
3.1. Generation of PDMS stamp
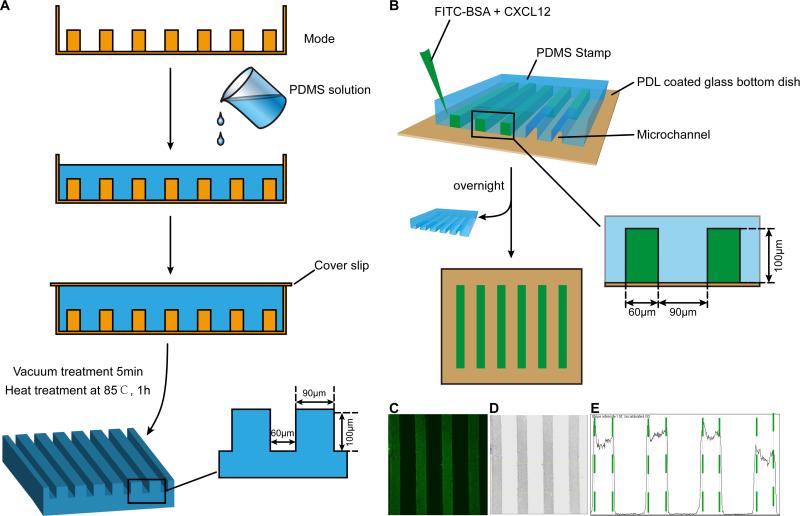
The CXCL12 and control stripes were generated by embedding protein mix of Fluorescein isothiocyanate (FITC) -BSA and CXCL12 in the PDMS stamp. The stamp was fabricated using mold etched by soft lithography with PDMS. To fabricate the PDMS stamp, the mold was first exposed to chlorotrimethylsilane vapor for 3 minutes in order to promote elastomer release after the baking steps. A mixture of PDMS was degassed for 10 minutes and then poured onto the mold to form the PDMS stamp (Fig. 1A).
Figure 1. Schematic illustration of stripe assay.
The PDMS stamp was fabricated using mold etched by soft lithography with PDMS. A: To fabricate the PDMS stamp, the mold was first exposed to chlorotrimethylsilane vapor for 3 minutes in order to promote elastomer release after the baking steps. A mixture of PDMS was degassed for 10 minutes and then poured onto the mold. After degassing (5 minutes), the mold was baked for 60 min at 85 °C. Subsequently, the PDMS flow layer structure was then peeled from the mold. B: Schematic illustration of the procedures for fabricating stripe. C: Fluorescent image of FITC-conjugated BSA diluted in PBS (0.1 mg/ml) and infused into micro channels sealed by a PDMS stamp on a glass slide to create BSA density. D: After inverted and discolored, the picture in B turned to C. E: Pixel intensity of the channel was quantified using Image J software.
3.2. Procedures and quantification of fluorescent stripe assay
Using PDMS as the matrix stamp, a PDL-coated glass bottom dish (35mm dish with 20mm bottom well) as the foundation, and a protein solution of FITC-BSA or FITC-BSA CXCL12 (100 μg/ml) infused into the microchannels within the PDMS stamp on the dish, we created stripes on glass bottom dishes for the stripe assays (Fig. 1B). The PDMS molds were removed after the protein stripes were dried overnight. We used FITC-BSA as a control protein to FITC-BSA CXCL12 stripes. The FITC in the stripes helped visualize the stripes. Under fluorescent confocal microscopy, the FITC stripes appeared as well-defined and parallel green stripes (Fig. 1C). To quantify the fluorescent intensities of FITC-BSA, we converted the images into grey scales (Fig. 1D) and analyzed the pixel intensities by Image J software (Fig. 1E). This mapping of signal intensities of the FITC-BSA stripes suggested that a gradient of CXCL12 may exist from the high plateau of the stripes towards intermediate space between stripes.
3.3. Quantification of CXCL12-mediated NPCs migration in stripe assays
NPCs derived from E13.5 mouse cortices were first characterized through immunocytochemistry (see Figure 1 in Ref [32]). After initial enrichments in mouse NeuroCult NSC Proliferation Medium containing EGF and bFGF, the NPCs cultures expanded in the form of neurosphere. Notably, both neurospheres and adherent cultures derived form neurospheres had specific immunoreactivity with both nestin and SOX2 antibody (see Figure 1 in Ref [32]). Nestin and SOX2 are NPC markers [26, 27] and therefore the prevalence of nestin and SOX2 double positive cells suggested that cells used in the stripe assays are indeed NPCs.
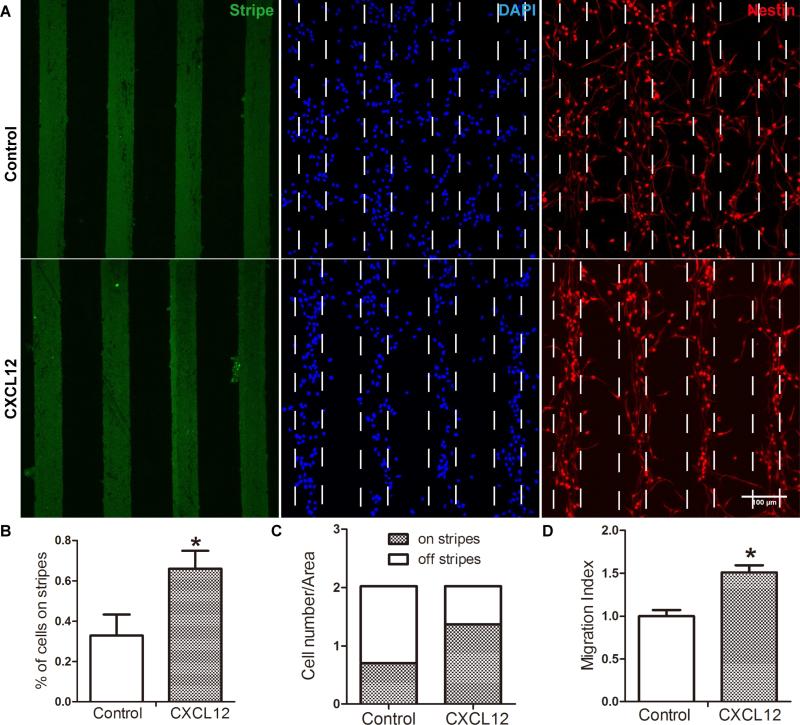
To study CXCL12-mediated mouse NPC migration, both stripe assay and transwell assay were used. In stripe assays, mouse NPCs were seeded and incubated for 48 hours on PDL-coated glass bottom dishes printed with FITC-conjugated BSA stripes (green) with or without CXCL12 (Fig. 2 A). Importantly, cells on glass bottom dishes remained nestin (red) positive. On the dishes printed with BSA stripe without CXCL12, NPCs distributed evenly on and off the stripes, suggesting a random distribution pattern (control group) at 48 hours after seeding. In contrast, on the dishes printed with BSA CXCL12 stripes, more cells were aligned on the BSA/CXCL12 stripes, forming a pattern of stripes (CXCL12 group) in two days. The percentage of cells on stripes to total cells was 33% and 66% in the control group and CXCL12 group, respectively (Fig. 2 B-C), indicating a two fold increase of migration in CXCL12 group over BSA control. As a comparison, we also plated NPCs in transwell assays. Quantification of CXCL12-mediated NPC migration in the transwell assays suggested migration increased 1.5 fold (p < 0.05) compared with BSA control (Fig. 2 D).
Figure 2. CXCL12 induce NPC migration in stripe assay.
NPC migration was assessed by stripe assay. A: Mouse NPCs were seeded on cover glasses coated with BSA stripes (green) without CXCL12 or with CXCL12. After 48 h, cells were fixed and stained with nestin (red) and DAPI (blue). The quantification of stripe migration assay were shown as B and C: The percent of cells on stripes to total cells was 33% and 66%, the control group and CXCL12 group respectively. D NPCs on transwell insert were incubated in the presence of CXCL12 (100 ng/ml) and quantified. * denotes p<0.05. Scale bar: D, 100 μm.
3.4. Observation of CXCL12-mediated NPC polarization in 2D and 3D environments
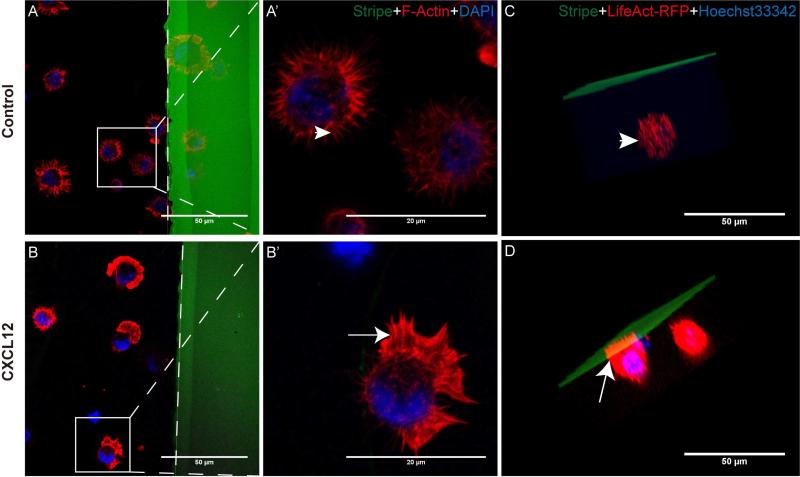
To determine whether stripe assays can be used to study NPC polarization, we first cultured NPCs in the presence of CXCL12 stripes or control BSA stripes. In the BSA control group in 2D cultures, NPCs formed random filopodia and no lamellipodia towards BSA stripes (Fig. 3 A, A’, arrowhead), suggesting that cells were not polarized for migration. In contrast, the changes of cell polarization and the formation of lamellipodia were evident with CXCL12 stripes. NPCs were polarized with the feature of lamellipodia (Fig. 3 B, B’, arrow) formation at the leading edge toward BSA/CXCL12 stripes at as early as 10 minutes after seeding on BSA/CXCL12 stripes, suggesting that the cells were polarized for migration towards CXCL12 stripes. NPCs in both the control group and CXCL12 group continued to express high levels of nestin (Fig. 4 D, H). Next, we used 3D culture of NPCs in the stripe assays and found a similar spatial relationship between NPCs and the CXCL12 stripes: NPCs polarized to CXCL12 stripe and formed lamellipodia (Fig. 3 D), while NPCs in the control group formed random filopodia (Fig. 3 C) after 3D reconstruction. Together, these data strongly suggest that the stripe assays can be used to study cellular polarization.
Figure 3. CXCL12 induce NPC polarization in 2D and 3D cultures.
NPCs polarizations both in 2D and 3D cultures mediated by CXCL12 were determined by stripe assay. A-A’, B-B’: Mouse NPCs were seeded on cover glasses coated with BSA stripes (green) without CXCL12 (A-A’, control group) or with CXCL12 (B-B’). After 10 min, cells were fixed and stained with phalloidin-rhodamine (red) and DAPI (blue). The dotted line displayed the boundary of the stripe. C, D: After dissociation, mix single NPCs (after transfection of LifeAct-RFP and stain with Hoechst33342) with MatriGel (BD) properly on ice (liquid state) and inoculate into the stripe-coated dishes. Incubate the dishes at 37°C in a CO2 incubator for 30 minutes (becoming a solid state), add 1 ml mouse NeuroCult NSC Proliferation Medium (Stem Cell Technologies) with EGF and FGF. After 2 hours, fix mouse NPCs. Cultures were washed. After 3 times wash, images were taken by Z-stack scanning and 3 d reconstruction of Zeiss 710 confocal microscope. Scale bar:A-D, F 50 μm; A’-B’, 20 μm.
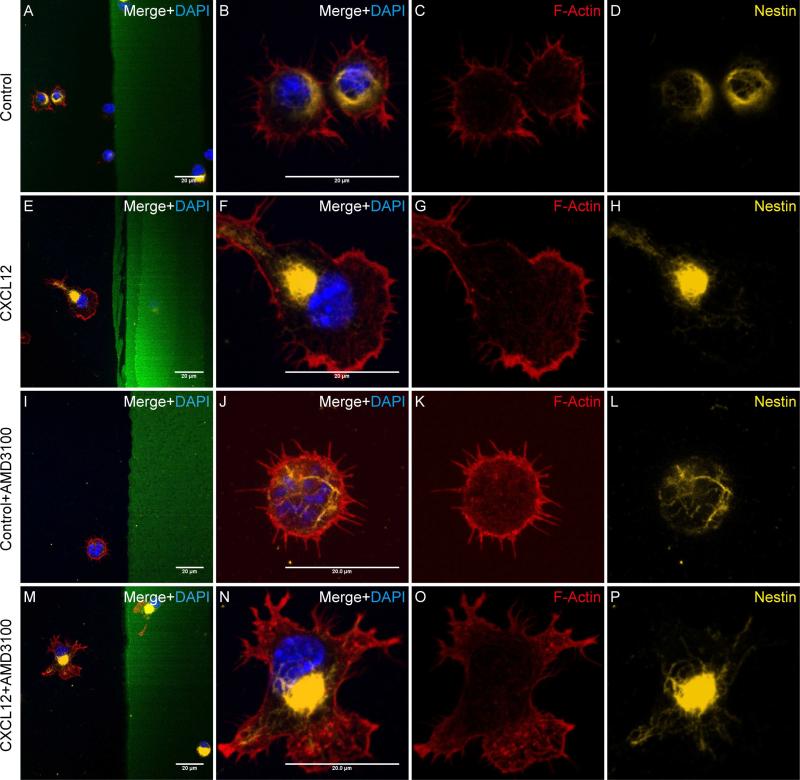
Figure 4. CXCL12-mediated NPC polarization is dependent on CXCR4.
A-H, The polarized NPCs in CXCL12 group (E-H) and unpolarized NPCs in the control group (A-D) expressed nestin. I-P, The NPCs’ polarization induced by CXCL12 was blocked by AMD3100. After treating mouse NPCs with AMD3100 (1 μg/ml, 1h), the NPCs were seeded on cover glasses coated with BSA stripes without CXCL12 (I-L Control group) or with CXCL12 (M-P). After 10 min, cells were fixed and stained with phalloidin-rhodamine (red), Nestin (yellow) and DAPI (blue). The cells in the control group appeared no evidence change in morphology (I-H), while the cells in the CXCL12 group no longer polarized towards the CXCL12 stripe (M-P).Scale bar: 20 μm.
3.5. CXCL12-mediated NPC polarization is dependent on CXCR4
To determine the receptor involved in CXCL12-mediated NPC polarization, we treated mouse NPCs without (Fig. 4A-H) or with AMD3100 (1 μg/ml, Fig. 4I-P), a CXCR4 antagonist, for 1 hour and seeded the cells on dishes coated with control BSA stripes or CXCL12 strips. In the BSA control groups, with (Fig. 4I-L) or without AMD3100 (Fig. 4A-D), the cells showed little evidence of polarization. In contrast, the presence of CXCL12 stripes induced NPC polarization (Fig. 4E-H). As expected, the addition of AMD3100 in the culture blocked CXCL12-mediated NPC polarization, suggesting that CXCL12-mediated NPC polarization is dependent on CXCR4.
3.6. CXCL12-mediated proteins translocation at the NPC leading edge in stripe assays
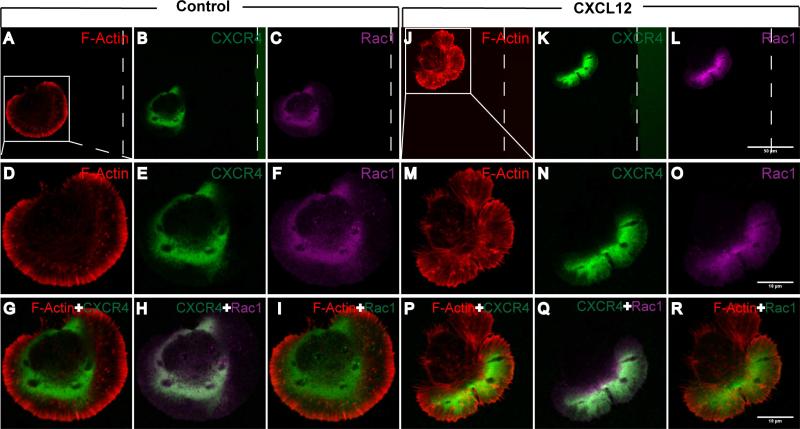
To determine the molecular mechanism(s) of CXCL12-mediated NPC polarization and migration in the stripe assays, we used immunostaining to study the distribution of CXCR4 and the expression of signaling intermediates in mouse NPCs. In the absence of CXCL12 stripes, CXCR4 distributed uniformly around the plasma membrane (Fig. 5A-I). However, within 10 minutes of seeding on BSA/CXCL12 stripe, CXCR4 re-distributed within the cells accumulated along the edge towards the CXCL12 stripe (Fig. 5 J-R). Receptor activation is likely first step of cell polarization in the migration event [28, 29]. During this process, small G proteins such as Rac1 play a key role in orchestrating the cytoskeleton necessary for the migration. Activated Rac1 is typically at the leading edge of the polarized cells, and is essential for cytoskeleton reorganization [7] and the protrusion of lamellipodia [30, 31]. In our stripe assay, Rac1 appeared to have polarized distribution (Fig. 5 L, O), which co-localized with CXCR4 and F-Actin at 10 minutes (Fig. 5 Q) in the CXCL12 group. In contrast, Rac1 was not polarized in the control BSA group (Fig. 5 C, F). The co-localized of Rac1 and CXCR4 at the lamellipodia of polarized cells suggested their involvement in the cytoskeleton reorganization during cellular polarization and migration.
Figure 5. CXCL12 induces protein translocation at the leading edge.
NPCs were seeded on cover glasses coated with BSA stripes (the control group) and BSA stripes plus CXCL12 (CXCL12 group). 10 minutes later, cells were fixed and stained with F-Actin (phalloidin-rhodamine, red), CXCR4 (green) and Rac1 (purple). The polar localization and co-localization of CXCR4 and Rac1 showed in NPC seeded on BSA/CXCL12 stripe preprinted and PDL coated cover glasses after 10 min, while CXCR4 and Rac1 in the NPC seeded on BSA stripe stayed in silent or random state. The dotted line displayed the boundary of the stripe. Scale bar: A-C, J-L 50 μm; D-I, M-R 10 μm.
3.7. Stripe assays generate migration and polarization data comparable to live cell imaging data
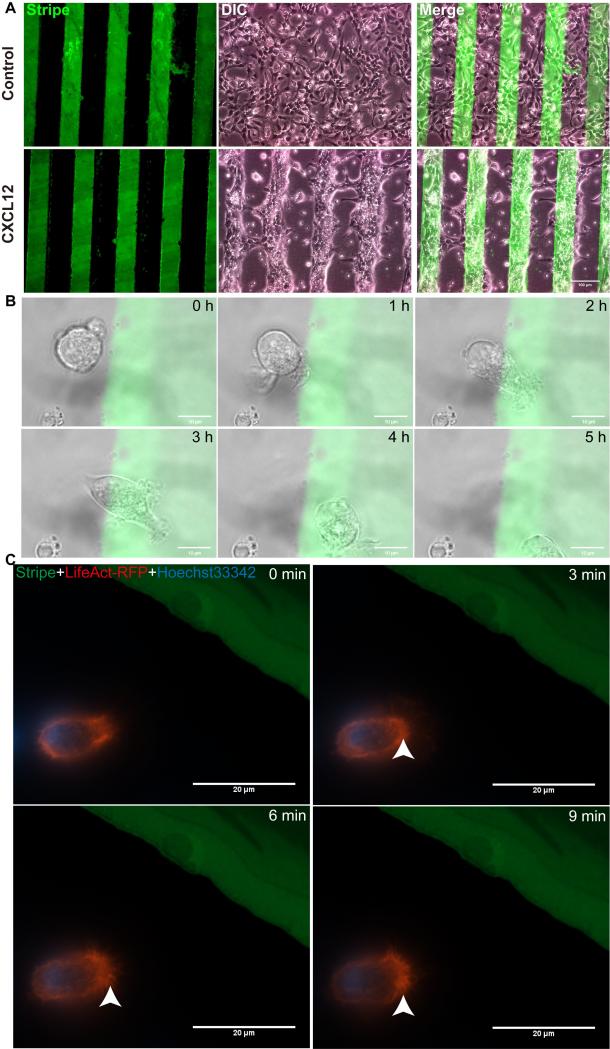
Live cell imaging, despite its sophisticated setup and instrument requirements, is often more desirable compared with various migration assays. We next compared the stripe assays with live cell imaging in the cellular migration and polarization assays. Mouse NPCs (1×107/ml) were seeded on PDL dishes printed with BSA stripes (green) with or without CXCL12. NPCs in the BSA control group showed a pattern of random migration, while those in BSA/CXCL12 group displayed a pattern of directional migration (Fig. 6 A). In the live cell imaging data obtained from NPCs in the stripe assays, NPC migrated towards CXCL12 stripes during 5 hours of recording (Fig. 6 B, see Video 1 in Ref [32]). To visualize the polarization of NPCs, we transfected cells with LifeAct-RFP (Ibidi). These plasmids encode a 17 amino acid peptide that binds filamentous actin (F-actin) structures without affecting actin dynamics. NPCs transfected with LifeAct-RFP were polarizing towards CXCL12 stripes and forming lamellipodia (Fig. 6 C, see Video 2 in Ref [32]) but not the control BSA stripe (see Figure 3 and Video 3 in Ref [32],). Together, these data suggested that stripe assays can generate migration and polarization data comparable to live cell imaging data.
Figure 6. NPCs polarize and migrate towards CXCL12 stripes in live cell imaging experiments.
A: Mouse NPCs (1×107/ml) were seeded on plastic Petri dishes (first coated with PDL) preprinted with BSA stripes (green) without CXCL12 (the control group) or with CXCL12 (CXCL12 group). After proper time, mouse NPC migrated directionally to the BSA/CXCL12 stripe. NPCs in control group showed a shape of random migration, while those in CXCL12 group displayed a shape of directional migration. B, The process of NPCs’ migration induced by CXCL12 on stripes was captured during 5 hours. C, NPCs transfected with LifeAct-RFP were polarizing towards CXCL12 stripe and forming lamellipodia. Scale bar: A, 100μm, B, 10μm, C, 20 μm.
Interestingly, NPCs in the CXCL12 or control BSA groups could be harvested to obtain total protein and RNA. The quality of protein was sufficient to process molecular experiments, including western blot (data not shown), co-immunoprecipitation [12], and small GTPase pull down [12]. Additionally, Rac1-pull down followed by immunoblotting with Rac1-GTP, the activated form of Rac1, suggested that showed Rac1 is activated upon exposure to CXCL12 stripes for mere 2 minutes (see Figure 2 in Ref [32]). Total RNA extracted from the stripe assays was also in good quality. The RNA was tested in Bioanalyzer and RNA gel for RNA integrity and showed appropriate RNA quality for subsequent applications such as mRNA Microarray and real-time PCR (data not shown).
4. Discussions
In our study, we modified the stripe assay to study CXCL12-mediated NPCs migration, polarization, and the associated intracellular signaling intermediates during these processes. Using this stripe assay, we found that localized CXCL12 on stripes effectively induced NPC migration. The NPC migration was also preceded by polarization of NPCs towards CXCL12 stripes at the initial stage. Furthermore, a quick polarized distribution of CXCR4 and Rac1 in the area of lamellipodia was strongly associated with cellular polarization and migration. Most interestingly, the migration and polarization processes in the stripe assay could also be visualized though live cell imaging setup.
Our group was among the first to the apply stripe assay in the research of NPCs’ polarity during migration and the associated molecular mechanism(s). Although transwell have been widely used for studying the cell chemotaxis, we have found that the stripe assay have several advantages. First, stripe assay is easier to visualize cellular polarized and migration in a dish. It is quite easy to observe the movement of a cell in time-dependent manner in the stripe assays. Second, we can specifically detect the change of related molecules or markers in the polarization process. Third, large amounts of live and migrating cells can be collected to extract mRNA or protein for further analysis. Fourth, it is convenient to alter the stamps, including the size, shape and stripe spaces to fit for multi-culture conditions. Lastly, CXCL12 can be substituted with other protein of chemoattractants for similar migration and polarization studies. Thus, the stripe assay is a promising method to analyze cell migration and polarization.
In this report, we found that CXCL12-mediated migration and polarization of NPCs is strongly associated with activation of Rac1 via CXCR4. The underlying molecular mechanism(s) between G protein-coupled receptors and the small G protein, such as Rac1 remain unclear. Future protein analysis from the stripe assay should shed light on the possible molecular connections.
Although stripe assay data is straightforward for interpretation, several questions need to be considered for the use of assay. It is unknown whether CXCL12 on the stripes can form a stable concentration gradient. The concentration gradient is the key to simulate cellular migration in more physiologically relevant microenvironment. Therefore, more validation and future modification may be required to improve the assay. Nonetheless, the stripe assays often an economic solution for the laboratories without live cell imaging setup and costly migration chips to study cellular migration and polarization.
5. Conclusions
In summary, we developed a functional assay to study the migration and polarization of NPCs. Stripe assay not only visualized cell migration and polarization, but it also helps detect the related protein changes during these processes. We expect the stripe assay to be fitted with other chemoattractants and serve as one of the regular migration and polarization assays to help discover important biological functions.
Acknowledgments
This work was supported by grants from National Key Basic Research Program of China (973 Program Grant No. 2014CB965000, project 1 No. 2014CB965001 and project 3 No. 2014CB965003) and National Natural Science Foundation of China (#81271420 and #81471936 to DX, #31271139 to AS), Innovative Research Groups of the National Natural Science Foundation of China (#81221001 to JZ), and Joint Research Fund for Overseas Chinese, Hong Kong and Macao Young Scientists of the National Natural Science Foundation of China (#81329002 to JZ); National Institutes of Health: R01 NS 41858-01, 2R56NS041858 - 15A1 (JZ), and R03 NS094071-01 (YH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contribution: MZ, AS, SL performed experiment. MZ, DX and JZ designed experimental. MZ, AS, SL,YH, DX, JZ wrote the manuscript.
References
- 1.Armstrong RJ, Rosser AE, Dunnett SB, Barker RA. Neural stem cell technology as a novel treatment for Parkinson's disease. Methods Mol Med. 2001;62:289–307. doi: 10.1385/1-59259-142-6:289. [DOI] [PubMed] [Google Scholar]
- 2.Honmou O, Uede T, Hashi K. [Neural stem cells derived from adult human brain: implications for a cell therapy for CNS diseases]. No Shinkei Geka. 2001;29(4):293–304. [PubMed] [Google Scholar]
- 3.Sukhikh GT, Malaitsev VV. Neural stem cell: biology and prospects of neurotransplantation. Bull Exp Biol Med. 2001;131(3):203–12. doi: 10.1023/a:1017503325833. [DOI] [PubMed] [Google Scholar]
- 4.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393(6685):595–9. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 5.Klein RS, Rubin JB, Gibson HD, DeHaan EN, Alvarez-Hernandez X, Segal RA, et al. SDF-1 alpha induces chemotaxis and enhances Sonic hedgehog-induced proliferation of cerebellar granule cells. Development. 2001;128(11):1971–81. doi: 10.1242/dev.128.11.1971. [DOI] [PubMed] [Google Scholar]
- 6.Lu M, Grove EA, Miller RJ. Abnormal development of the hippocampal dentate gyrus in mice lacking the CXCR4 chemokine receptor. Proc Natl Acad Sci U S A. 2002;99(10):7090–5. doi: 10.1073/pnas.092013799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imitola J, Raddassi K, Park KI, Mueller FJ, Nieto M, Teng YD, et al. Directed migration of neural stem cells to sites of CNS injury by the stromal cell-derived factor 1alpha/CXC chemokine receptor 4 pathway. Proc Natl Acad Sci U S A. 2004;101(52):18117–22. doi: 10.1073/pnas.0408258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 2006;7(12):243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banisadr G, Skrzydelski D, Kitabgi P, Rostene W, Parsadaniantz SM. Highly regionalized distribution of stromal cell-derived factor-1/CXCL12 in adult rat brain: constitutive expression in cholinergic, dopaminergic and vasopressinergic neurons. Eur J Neurosci. 2003;18(6):1593–606. doi: 10.1046/j.1460-9568.2003.02893.x. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Ransohoff RM. Multiple roles of chemokine CXCL12 in the central nervous system: a migration from immunology to neurobiology. Prog Neurobiol. 2008;84(2):116–31. doi: 10.1016/j.pneurobio.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu B, Xu D, Deng X, Chen Q, Huang Y, Peng H, et al. CXCL12 enhances human neural progenitor cell survival through a CXCR7- and CXCR4-mediated endocytotic signaling pathway. Stem Cells. 2012;30(11):2571–83. doi: 10.1002/stem.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q, Zhang M, Li Y, Xu D, Wang Y, Song A, et al. CXCR7 Mediates Neural Progenitor Cells Migration to CXCL12 Independent of CXCR4. Stem Cells. 2015;33(8):2574–85. doi: 10.1002/stem.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyden S. The chemotactic effect of mixtures of antibody and antigen on polymorphonuclear leucocytes. J Exp Med. 1962;115:453–66. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Senger DR, Perruzzi CA, Streit M, Koteliansky VE, de Fougerolles AR, Detmar M. The alpha(1)beta(1) and alpha(2)beta(1) integrins provide critical support for vascular endothelial growth factor signaling, endothelial cell migration, and tumor angiogenesis. Am J Pathol. 2002;160(1):195–204. doi: 10.1016/s0002-9440(10)64363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter J, Kern-Veits B, Huf J, Stolze B, Bonhoeffer F. Recognition of position-specific properties of tectal cell membranes by retinal axons in vitro. Development. 1987;101(4):685–96. doi: 10.1242/dev.101.4.685. [DOI] [PubMed] [Google Scholar]
- 16.Walter J, Henke-Fahle S, Bonhoeffer F. Avoidance of posterior tectal membranes by temporal retinal axons. Development. 1987;101(4):909–13. doi: 10.1242/dev.101.4.909. [DOI] [PubMed] [Google Scholar]
- 17.Knoll B, Kretz O, Fiedler C, Alberti S, Schutz G, Frotscher M, et al. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat Neurosci. 2006;9(2):195–204. doi: 10.1038/nn1627. [DOI] [PubMed] [Google Scholar]
- 18.Knoll B, Zarbalis K, Wurst W, Drescher U. A role for the EphA family in the topographic targeting of vomeronasal axons. Development. 2001;128(6):895–906. doi: 10.1242/dev.128.6.895. [DOI] [PubMed] [Google Scholar]
- 19.Mann F, Zhukareva V, Pimenta A, Levitt P, Bolz J. Membrane-associated molecules guide limbic and nonlimbic thalamocortical projections. J Neurosci. 1998;18(22):9409–19. doi: 10.1523/JNEUROSCI.18-22-09409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savaskan NE, Plaschke M, Ninnemann O, Spillmann AA, Schwab ME, Nitsch R, et al. Myelin does not influence the choice behaviour of entorhinal axons but strongly inhibits their outgrowth length in vitro. Eur J Neurosci. 1999;11(1):316–26. doi: 10.1046/j.1460-9568.1999.00430.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang HU, Anderson DJ. Eph family transmembrane ligands can mediate repulsive guidance of trunk neural crest migration and motor axon outgrowth. Neuron. 1997;18(3):383–96. doi: 10.1016/s0896-6273(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 22.Cohen RI, Rottkamp DM, Maric D, Barker JL, Hudson LD. A role for semaphorins and neuropilins in oligodendrocyte guidance. J Neurochem. 2003;85(5):1262–78. doi: 10.1046/j.1471-4159.2003.01722.x. [DOI] [PubMed] [Google Scholar]
- 23.Moreau-Fauvarque C, Kumanogoh A, Camand E, Jaillard C, Barbin G, Boquet I, et al. The transmembrane semaphorin Sema4D/CD100, an inhibitor of axonal growth, is expressed on oligodendrocytes and upregulated after CNS lesion. J Neurosci. 2003;23(27):9229–39. doi: 10.1523/JNEUROSCI.23-27-09229.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasarre P, Kusy S, Constantin B, Castellani V, Drabkin HA, Bagnard D, et al. Semaphorin SEMA3F has a repulsing activity on breast cancer cells and inhibits E-cadherin-mediated cell adhesion. Neoplasia. 2005;7(2):180–9. doi: 10.1593/neo.04481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi YS, Vincent LG, Lee AR, Kretchmer KC, Chirasatitsin S, Dobke MK, et al. The alignment and fusion assembly of adipose-derived stem cells on mechanically patterned matrices. Biomaterials. 2012;33(29):6943–51. doi: 10.1016/j.biomaterials.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60(4):585–95. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- 27.Messam CA, Hou J, Berman JW, Major EO. Analysis of the temporal expression of nestin in human fetal brain derived neuronal and glial progenitor cells. Brain Res Dev Brain Res. 2002;134(1-2):87–92. doi: 10.1016/s0165-3806(01)00325-x. [DOI] [PubMed] [Google Scholar]
- 28.Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284(5415):765–70. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 29.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–9. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 30.Balabanian K, Lagane B, Infantino S, Chow KY, Harriague J, Moepps B, et al. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280(42):35760–6. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- 31.Sierro F, Biben C, Martinez-Munoz L, Mellado M, Ransohoff RM, Li M, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104(37):14759–64. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Song AH, Lai SQ, Qiu LS, Huang YL, Chen Q, et al. Living Cell Imaging and Rac1-GTP Levels of CXCL12-treated Migrating Neural Progenitor Cells in Stripe Assay. Biomaterials Data in Brief “submitted”. doi: 10.1016/j.dib.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]