Abstract
Anti-allergic effects of dietary polyphenols were extensively studied in numerous allergic disease models, but the molecular mechanisms of anti-allergic effects by polyphenols remain poorly understood. In the present study, we show that the release of granular cargo molecules, contained in distinct subsets of granules of mast cells, is specifically mediated by two sets of SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins, and that various polyphenols differentially inhibit the formation of those SNARE complexes. Expression analysis of RBL-2H3 cells for 11 SNARE genes and a lipid mixing assay of 24 possible combinations of reconstituted SNAREs indicated that the only two active SNARE complexes involved in mast cell degranulation are Syn (syntaxin) 4/SNAP (23 kDa synaptosome-associated protein)-23/VAMP (vesicle-associated membrane protein) 2 and Syn4/SNAP-23/VAMP8. Various polyphenols selectively or commonly interfered with ternary complex formation of these two SNARE complexes, thereby stopping membrane fusion between granules and plasma membrane. This led to the differential effect of polyphenols on degranulation of three distinct subsets of granules. These results suggest the possibility that formation of a variety of SNARE complexes in numerous cell types is controlled by polyphenols which, in turn, might regulate corresponding membrane trafficking.
Keywords: degranulation, β-hexosaminidase, histamine, membrane fusion, polyphenol, soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)
INTRODUCTION
The incidence of allergic disorders has dramatically increased over the last few decades. A wide variety of factors, including those of the environment, genetics, hygiene and diet, are known to influence the development of related diseases such as atopy, asthma and rhinitis. Although several therapeutic options such as corticosteroids and antihistamines are available as an anti-allergic therapy, polyphenols are now gaining special interest because adverse effects associated with chronic use are relatively rare with the abundant evidence of their efficacy. Furthermore, polyphenols are found in abundance within our daily diet including vegetables, fruit, tea, cocoa and coffee. The efficacy of polyphenols was extensively investigated using numerous disease models, and the safety profile of the dietary polyphenols points to the potential of the polyphenols as anti-allergy agents [1–6]. In spite of those clear beneficial effects of polyphenols, the mechanism of their specific function as anti-allergic agents remains surprisingly an open question. Polyphenols are known to exhibit antiallergic activity by scavenging free radicals [4,7–9], or through binding to allergenic proteins [10–12], affinity receptors [13] and intracellular signalling enzymes [14–17].
Mast cells (specialized secretory cells) are recognized as the major effector cells of type 1 hypersensitivity reactions owing to FcεRI (high-affinity receptor for IgE) [18]. They are known to play a pivotal role in host defence by discharging a variety of biologically active inflammatory mediators, including histamine, serotonin, heparin, serine proteases, chemotactic factors and specific immune-regulatory cytokines. They are also closely related to allergic diseases including atopic rhinitis, asthma and atopic dermatitis. Mast cells acutely respond to specific allergens or other stimuli by releasing various internal components [19]. Following stimulation, cross-linking of FcεRI on the cell surface by multivalent antigens activates intracellular signalling cascades, which leads to degranulation [18]. Three types of granules in mast cells have been reported [20,21]. Type I and type II granules were both labelled by MHC class II and lysosomal markers, such as β-hexosaminidase. Serotonin (amine) was localized to type II and III granules, of which the latter type did not contain MHC class II. It was suggested that types I (amine-free lysosome) and III (secretory granules) might form type II granules (lysosomal amine containing secretory granule) by a fusion event [20].
Exocytosis of these granular contents requires the fusion of the granular membrane and the plasma membrane. It is well established that the degranulation of mast cells is mediated by membrane-anchored SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) proteins [22,23]. SNARE proteins promote membrane fusion between the vesicle and the plasma membrane, and open a fusion pore through which allergy mediators are secreted. On stimulation of FcεRI, IκB (inhibitor of nuclear factor κB) kinase 2 phosphorylates SNAP-23 (23 kDa synaptosome-associated protein), and in turn, the phosphorylated SNAP-23 recruits Syn (syntaxin) 4 and VAMP (vesicle-associated membrane protein) 2 to form the ternary SNARE complex [24]. Several isoforms of SNARE proteins working in mast cells have been identified, which are different from neuronal SNAREs [23,25,26]. Previous studies have suggested Syn3, Syn4 and SNAP-23 as t-SNAREs (target SNAREs) on mast cell plasma membrane. On the granule membrane, VAMP2, VAMP3, VAMP7 (TI-VAMP; tetanus toxin-insensitive VAMP) and VAMP8 have been reported as v-SNAREs (vesicle SNAREs) [26,27]. Although these SNARE proteins are thought to be involved in membrane fusion in mast cells, the functional ternary SNARE complex mediating mast cell degranulation has not been unequivocally characterized. Several studies through immunoprecipitation using recombinant SNARE proteins or antibodies by permeabilizing mast cells with streptolysin-O, or RNA interference technology aimed at characterizing the ternary SNARE complex in mast cells, have yielded the controversial results that suggest different SNARE complex models [23,28–32].
Our previous study demonstrated that some polyphenols could stop neuronal SNARE zippering [33]. ‘SNARE zippering’ is a term applied to the process of directional SNARE complex formation, starting from the membrane-distal N-terminus to the membrane-proximal C-terminus. Membrane fusion intermediates such as hemifusion and partially zipped SNARE complexes, which are otherwise transient, could be captured and dissected by the identified polyphenols [33]. The polyphenols did indeed impair neuroexocytosis by inhibiting SNARE complex formation in neuron and paralysed muscles [34].On the basis of these results, we hypothesized that some polyphenols interfere with SNARE complex formation in cells other than neuronal cells, although the functional SNARE complex-containing proteins might be different. Mast cells were selected to demonstrate our hypothesis, as the secretory behaviours of mast cells have been relatively well characterized. The major challenge of this hypothesis was, however, to explain how polyphenols differentially regulate the degranulation of distinct subsets of secretory granules, of which exocytosis is controlled by different v-SNAREs. On the basis of our observations, it is suggested that SNARE complexes are common targets of many polyphenols.
MATERIALS AND METHODS
Materials
POPC (1-palmitoyl-2-dioleoyl-sn-glycero-3-phosphatidylcholine), DOPS (1,2-dioleoyl-sn-glycero-3-phosphatidylserine), cholesterol, NBD-PS [1,2–dioleoyl-sn-glycero-3-phosphoserine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl)] and rhodamine–PE [1,2– dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl)] were obtained from Avanti Polar Lipids. All other chemicals were purchased from Sigma–Aldrich.
Antibodies
Anti-SNAP-23 (ab4114), anti-VAMP8 (ab6186) and anti-Syn4 (ab101879) polyclonal sera and anti-SNAP-23 monoclonal antibody (ab57961) were purchased from Abcam. Anti-VAMP2 (V1389) and anti-β-actin (A1978) antibodies were purchased from Sigma.
Cell culture
The RBL-2H3 cells from the A.T.C.C. (CRL-2256) were cultured in DMEM (Dulbecco’s modified Eagle’s medium) with high glucose supplemented with 10% heat-inactivated fetal bovine serum and antibiotics, at 37°C in an atmosphere of 5% CO2.
Mast cell stimulation and degranulation assay
Degranulation assays, described in detail elsewhere [35,36], were performed by determining the amount of released β-hexosaminidase or histamine into the supernatant. Briefly, RBL-2H3 mast cells were plated overnight in 500 µl of culture media in 24-well plates (2 × 105 cells/well). After cells were washed with the Hepes-buffered saline solution [140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.6 mM MgCl2, 0.1% glucose, 0.1% BSA and 10 mM Hepes (pH 7.4)], they were sensitized by anti-DNP (dinitrophenyl)-IgE (100 ng/ml; Sigma) for 3 h, and then further incubated with or without polyphenols (10 µM) for 30 min. After incubation, cells were stimulated with DNP groups that had been conjugated with BSA (200 ng/ml, DNP-BSA) for antigen-induced degranulation, or 10 µMA23187 for ionophore-induced degranulation. Degranulation was carried out in Hepesbuffered saline solution for 30 min at 37°C and was quantified by measurement of β-hexosaminidase or histamine activity via colorimetric biochemical assay. Aliquots of supernatant were transferred to a 96-well plate (50 µl/well) and incubated with 50 µl of substrate solution [(1 mM p-nitrophenyl-N-acetyl-β-d-glucosaminide in 100 mM citrate buffer (pH 4.5)] for 3 h at 37°C. After the reaction was terminated with 100 µl NaCO3 and NaHCO3 buffer [1:1 mixture, 100 mM NaCO3/NaHCO3 (pH 10.0)], absorbance at 405 nm was measured using a microplate reader. The release activity relative to the total β-hexosaminidase content of the cells was calculated. Total β-hexosaminidase content (lysate) was determined by dissolving cells with 0.1% Triton X-100. The supernatant was also used to measure the release of histamine, by following themanufacturer’s protocol for the EIA (enzyme immunoassay) system (Oxford Biomedical Research). The percentage of β-hexosaminidase or histamine release was calculated using eqn (1) [37]:
T (test): DNP-BSA (+), polyphenol (+)/B (blank): DNPBSA (−), polyphenol (+)/C (control): DNP-BSA (+), polyphenol (−)/N (normal): DNP-BSA (−), polyphenol (−).
Co-immunoprecipitation and immunoblotting analysis
A total of (0.5–1) × 106 cells were lysed with 1 ml of lysis buffer [50 mM Tris/HCl, 150 mM NaCl, 1% (v/v) Triton X-100 and 1 mM EDTA (pH 7.5)] containing a protease inhibitor cocktail (Calbiochem). The lysates were centrifuged at 13000 g for 15 min at 4°C to remove cell debris, and pre-cleared with 100 µl of Protein G–Sepharose (GE Healthcare) for 2 h at 4°C. Antibodies against SNARE proteins (SNAP-23, Syn4, VAMP2 and VAMP8) were added to the pre-cleared lysates and allowed to bind for 2 h at 4°C. Antibody–antigen complexes were precipitated by the addition of Protein G slurry for overnight. After extensive washing with lysis buffer, the precipitates were subjected to immunoblotting analysis. Immunoprecipitated proteins and cell lysates were analysed by separation on SDS/12% PAGE gels, transferred on to nitrocellulose membranes (Whatman). After blocking with TBST [10 mMTris/HCl, 150 mMNaCl and 0.05% Tween 20 (pH 8.0)] containing 5%(w/v) non-fat dried skimmed milk powder and 0.5% BSA for 1 h at room temperature (25 °C), blots were probed with the primary antibody against SNAP-23 or Syn4 (1:1000 dilution) overnight. After being washed with TBST, the membrane was treated with HRP (horseradish peroxidase)-conjugated secondary anti-(mouse IgG) antibody (A4416; Sigma), and developed using an ECL (enhanced chemiluminescence) solution.
RNA isolation and reverse transcription–PCR
Total RNA was isolated from RBL-2H3 cells using the TRIzol® reagent (Qiagen) according to the manufacturer’s instructions. Briefly, 106 cells were homogenized by the addition of 1 ml of TRIzol®. Chloroform (200 µl) was added to the TRIzol® lysate, which was then mixed and incubated for 3 min at room temperature. The solution was then centrifuged at 13000 g for 10 min at 4°C in a microcentrifuge. Following centrifugation, the upper aqueous phase was transferred to a new tube, and the same volume of propan-2-ol was added, after which RNA purification was performed as described in the manufacturer’s instructions. RNA was quantified using a spectrophotometer at 260 nm and the quality was determined by A260/A280 ratios. Reverse transcription was performed using MMLV (Moloney murine leukaemia virus) reverse transcriptase (Promega) using 4 µg of total RNA. Semi-quantitative PCR of the expression of SNARE genes was performed under the conditions detailed in Supplementary Table S1 at http://www.biochemj.org/bj/450/bj4500537add.htm.
Cloning, expression and purification of recombinant protein preparation
Total RNA obtained from RBL-2H3 cells was reverse-transcribed into cDNA. The generated cDNAs were used to obtain genes for SNAP23, SYNTAXIN4, VAMP2, VAMP4, VAMP7 and VAMP8. The PCR products were cloned into the pGEX-4T-1 vector in order to purify as GST (glutathione transferase)-tagged proteins. Protein expression and purification procedures for mast cell SNARE proteins have been described previously [33]. Briefly, transformed Escherichia coli BL21(DE3) Codon(+) cells were grown in LB (Luria–Bertani) medium to a D600 of 0.8, and protein expression was induced by 0.3 mM IPTG (isopropyl β-d-thiogalactopyranoside) treatment. GST–SNAREs were purified with glutathione–agarose beads, and GST was cleaved by thrombin. The purified proteins were examined by SDS/12.5% PAGE, and the purity was at least 90% for all proteins.
Reconstitution of SNARE proteins into membranes
A total of 100 nm LUVs (large unilamellar vesicles) were prepared by extrusion through polycarbonate filters (Avanti Polar Lipids). The lipid molar ratios for the FRET (fluorescence resonance energy transfer) reactions were 65:35 (v/v) POPC/DOPS for the t-vesicles (vesicles that contain t-SNARE protein) and 62:35:1.5:1.5 (by vol.) POPC/DOPS/NBD/rhodamine B for the v-vesicles (vesicles that contain v-SNARE protein). Binary t-SNARE complex and VAMP2 or VAMP8 were reconstituted into t-vesicles and v-vesicles with a 50:1 lipid/protein ratio as described previously [33,38]. The final lipid concentration of each t- and v-vesicles was 1 mM.
FRET-based lipid mixing assay
To measure total lipid mixing, v-SNARE liposomes were mixed with t-SNARE liposomes at a ratio of 1:9. Fluorescence was measured at excitation and emission wavelengths of 465 and 530 nm respectively. Changes in fluorescence were recorded with a SpectraMax M2 (Molecular Devices) fluorescence spectrophotometer. The maximum fluorescence intensity was determined by adding 0.1% Triton X-100. All lipid mixing experiments were carried out at 37°C as described previously [39].
UV absorbance measurement
The UV absorption spectra of the polyphenols were recorded on a SpectraMax M2 spectrophotometer at a concentration of 20 µM in buffer solution [50 mM Tris/HCl, 150 mM NaCl and 2.5 mM CaCl2 (pH 8.0)]. The absorption spectra of the SNARE complex/polyphenol mixtures were measured from 250 to 700 nm at a polyphenol–SNARE complex molar ratio of 0.8 [40].
RESULTS
Differential effects of polyphenols on histamine and β-hexosaminidase release
Mast cells contain enzymes and inflammatory mediators including histamine and β-hexosaminidase. β-Hexosaminidase, stored in the secretory granules of the mast cells, is released concomitantly with histamine when the mast cells are immunologically activated [41,42]. Many polyphenols are known to be anti-allergic because they inhibit β-hexosaminidase and/or histamine release frommast cells [1]. Thus we tested whether all polyphenols concomitantly inhibit histamine and β-hexosaminidase release. A total of 32 polyphenolic compounds representative of 11 subclasses of polyphenols (Supplementary Figure S1a at http://www.biochemj.org/bj/450/bj4500537add.htm) were used to treat RBL-2H3 cells. We also tested LY (Ly294002), a PI3K (phosphoinositide 3-kinase) inhibitor, as a positive control. After stimulating degranulation with IgE and DNP-BSA, the released β-hexosaminidase and histamine were analysed (Supplementary Figures S1b and S1c). Surprisingly, the inhibitory activity of various polyphenols was quite selective to β-hexosaminidase or histamine release. For example, CY (cyanidin) and GN (gosmisin N) did not inhibit β-hexosaminidase release, but markedly inhibited histamine release (in red circle). In sharp contrast, some polyphenols such as FA (ferulic acid), DL (delphinidin) and CA (caffeic acid) inhibited 40–60% histamine release, whereas they did not affect β-hexosaminidase release (in blue circle). Other polyphenols such as MR (myricetin), LT (luteolin) and AP (apigenin) commonly and almost equally inhibited β-hexosaminidase and histamine release comparable with the effects by LY (in yellow circle). In general, flavonols (labelled in cyan) strongly inhibited β-hexosaminidase release, but their effect on histamine release varied with a wide range of 10–80%. Similarly, when the mast cell was degranulated by calcium ionophore A23187, the effect of polyphenols on β-hexosaminidase release was almost the same as that induced by IgE + DNPBSA (Supplementary Figure S2 at http://www.biochemj.org/bj/450/bj4500537add.htm). This result excludes the possibility that the differential effect of polyphenols on mast cell degranulation is dependent on the stimulation method. On the basis of our observation of the selective inhibition of histamine or β-hexosaminidase release, we conclude that the inhibitory effect of polyphenols on mast cell degranulation is granule-specific.
Functional ternary complexes involved in mast cell degranulation
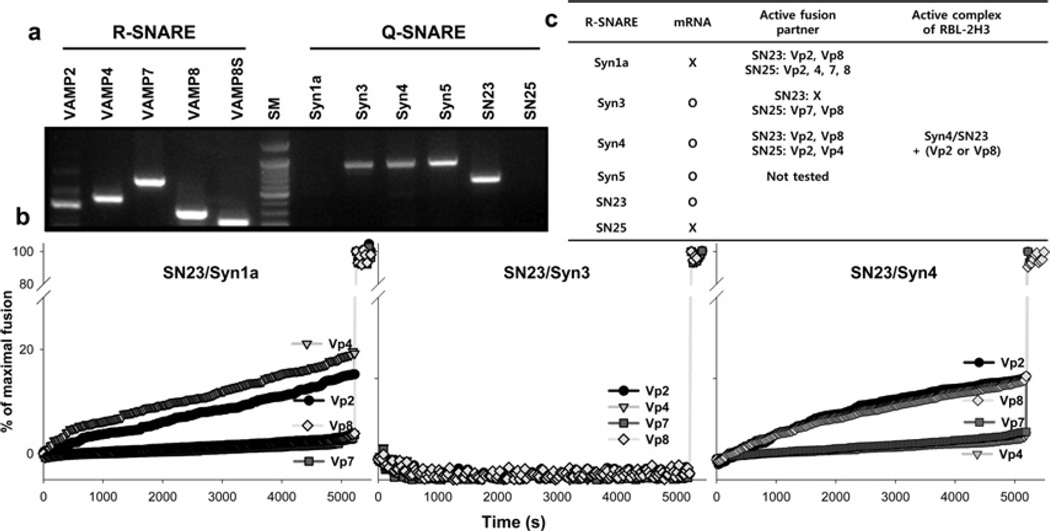
Previous studies have identified several isoforms of SNARE in mast cells [23,32]. However, our understanding of the functional SNARE complexes responsible for mast cell exocytosis is still far from complete, and the processes involved in the regulation of mast cell degranulation remain largely elusive. To resolve this issue, we employed a SNARE-driven lipid mixing assay, which was first developed in order to show that SNARE complex is minimal fusion machinery [39], and is now widely employed to elucidate various features of neuronal exocytosis [43]. Before performing the lipid mixing assay, we analysed expression of SNARE genes using reverse transcription–PCR (Figure 1a). Consistent with previous findings [23,32,44], Syn1a and neuron-specific SNAP-25 (25 kDa synaptosome-associated protein) were not expressed in RBL-2H3 cells, whereas Syn3, Syn4, Syn5, SNAP-23, VAMP2, VAMP4, VAMP7 and VAMP8 were expressed. We then reconstituted purified SNARE proteins into LUVs and measured the lipid mixing ability of various combinations (Figure 1b). Even though SNAP-25 was able to induce lipid mixing in combination with various SNARE proteins (Supplementary Figure S3 at http://www.biochemj.org/bj/450/bj4500537add.htm), SNAP-25-containing ternary complexes cannot be the mediators of mast cell degranulation, as SNAP-25 is not expressed inmast cells [23,32] (Figure 1a). For the same reason, Syn1a-containing ternary complexes were excluded regardless of whether they were fusogenic or not. When the t-SNARE complex was composed of SNAP-23 and Syn3, it was not able to induce membrane fusion with any of the tested v-SNARE proteins. However, the t-SNARE complex composed of SNAP-23 and Syn4 was able to induce lipid mixing with VAMP2 and VAMP8, but not with VAMP4 and VAMP7. Although Syn5 was not tested because it could not be recovered in a soluble form, a previous study excluded the involvement of Syn5 for fusion in mast cells [23]. Therefore we concluded that SNAP- 23/Syn4/VAMP2 and SNAP-23/Syn4/VAMP8 were the only two functional ternary SNARE complexes that are validated in mast cells (Figure 1c).
Figure 1. Identification of active SNARE complexes in the mast cells.
(a) mRNA expression of various SNAREs was analysed by reverse transcription–PCR. VAMP8 was repeatedly analysed with another primer set covering only the soluble SNARE motif (denoted by VAMP8S). (b) The lipid mixing ability of various SNARE complexes. Fusion traces for SNAP-25-containg complexes are shown in Supplementary Figure S3 at http://www.biochemj.org/bj/450/bj4500537add.htm. (c) Summary of SNARE protein expression in mast cell and lipid mixing ability of SNARE complexes.
Differential inhibition of SNARE-driven membrane fusion by polyphenols
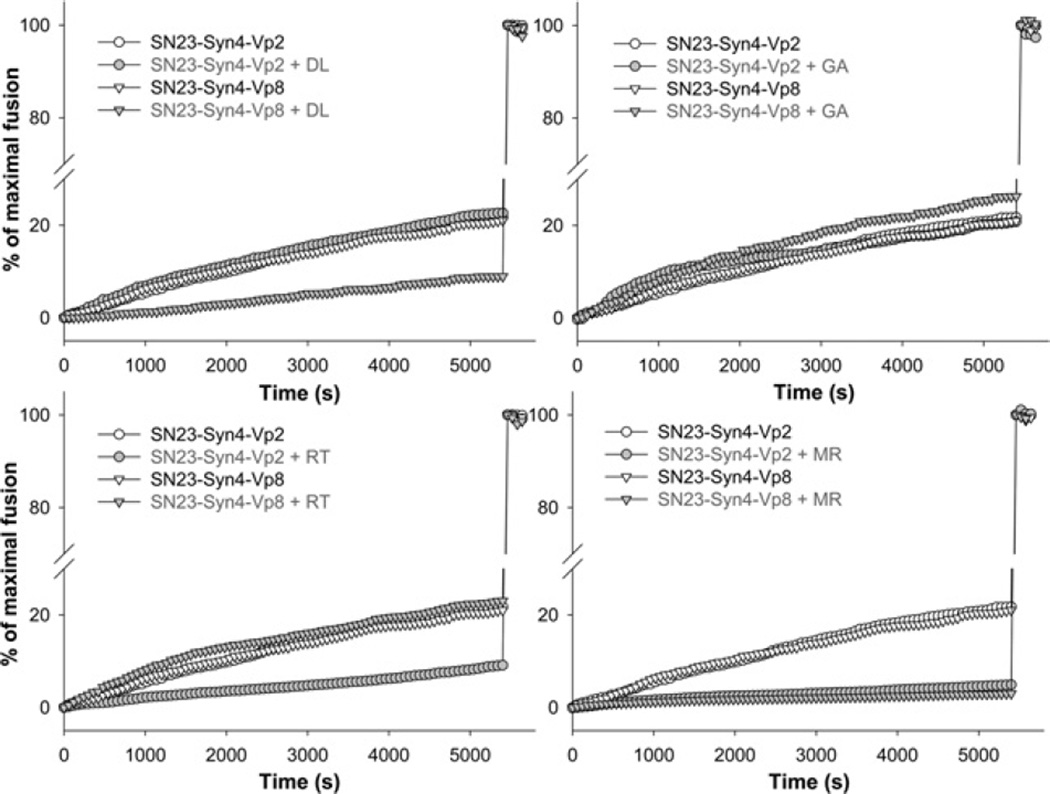
The lipid mixing assay was performed using the two validated combinations of SNARE proteins (SNAP-23/Syn4/VAMP2 and SNAP-23/Syn4/VAMP8) in the presence of polyphenols. Several fusion traces clearly indicated that polyphenols could differentially inhibit the lipid mixing driven by SNAP- 23/Syn4/VAMP2 or SNAP-23/Syn4/VAMP8 (Figure 2). When DL was present, lipid mixing driven by the combination of SNAP-23/Syn4/VAMP8 was strongly inhibited, but SNAP- 23/Syn4/VAMP2-driven lipid mixing was not affected. In contrast, RT (rutin) did not inhibit SNAP-23/Syn4/VAMP8- driven fusion, but did inhibit SNAP-23/Syn4/VAMP2-driven fusion. GA (gomisin A) affected fusion little regardless of SNARE complexes, whereas MR inhibited fusion by both SNARE complexes (Figure 2). These results suggest that the inhibitory effect of polyphenol on SNARE-driven membrane fusion is dependent on the combination of SNARE proteins rather than on the individual SNARE protein, giving us a clear insight into the way polyphenols differentially regulate mast cell degranulation.
Figure 2. Differential inhibitory effects of polyphenols DL, RT, GA and MR on the membrane fusion driven by different SNARE complexes.
Percentage of maximal fusion was plotted as a function of time in the presence or absence of polyphenols. Polyphenols were treated at 10 µM concentration. The maximum fluorescence intensity was obtained by adding 0.1% Triton X-100.
In order to fully understand the inhibitory activity of polyphenols on SNARE-driven membrane fusion, all 32 compounds were subjected to the lipid mixing assay. For this assay, three SNARE complexes were prepared: two for mast cells complexes (SNAP-23/Syn4/VAMP2 and SNAP-23/Syn4/VAMP8) and one for neuronal complex (SNAP-25/Syn1/VAMP2). After reading all 96 fusion traces, the inhibition percentage of polyphenols relative to that of the control was calculated. Scatter plots of these values clearly show that each polyphenolic compound differentially affected the SNARE complexes (Supplementary Figure S4 at http://www.biochemj.org/bj/450/bj4500537add.htm). For example, DL inhibited ~90% SNAP-25/Syn1/ VAMP2-driven fusion, and ~60% SNAP-23/Syn4/VAMP8- driven fusion, but had little effect on SNAP-23/Syn4/VAMP2- driven fusion. AP inhibited little SNAP-25/Syn1/VAMP2-driven fusion, but inhibited ~90% SNAP-23/Syn4/VAMP8-driven and ~40% SNAP-23/Syn4/VAMP2-driven fusion. MR commonly inhibited all three SNARE complex-driven fusion events with 75– 90% efficiency. GA, SC (schizandrin), FA and PH (phloridzin) did not affect any of the SNARE-driven fusions. In the absence of SNARE protein reconstitution, no polyphenol induced lipid mixing, excluding the possibility of SNARE-independent membrane fusion by polyphenols. Furthermore, there was little turbidity change at the tested polyphenol concentration, indicative of no liposome aggregation by polyphenols.
Next, we tested whether these differential inhibitions of membrane fusion occurred as a direct consequence of the differential binding of polyphenols to different SNARE complexes. Although the binding of several polyphenols to the inner layer of neuronal SNARE complex (Syn1/SNAP-25/VAMP2) could be visualized by Nitro Blue Tetrazolium staining [33], the same in-gel visualization method was inapplicable to mast cell SNAREs, as neither of the mast cell SNARE complexes are SDS-resistant. As an alternative approach, we therefore measured the bathochromic shift of the polyphenols, which occurs when a light-absorbing molecule binds to macromolecules. After preparing soluble SNARE motifs of the SNARE proteins, absorption spectra of the polyphenols were measured in the presence or absence of the SNARE proteins (Figure 3). Three equimolar SNARE proteins were added to each of the polyphenols at a polyphenol/each SNARE protein molar ratio of 0.8. DL, QT (quercetin) and FS (fisetin) exhibited an apparent bathochromic shift only when they were mixed with Syn4S, SNAP-23 and VAMP8S, clearly indicating that these three polyphenols exclusively bind to the Syn4/SNAP-23/VAMP8 complex, and not to the Syn4/SNAP-23/VAMP2 complex. In contrast, CY and RT did not bind to the Syn4/SNAP- 23/VAMP8 complex, but bound to Syn4/SNAP-23/VAMP2. MR and LT showed a bathochromic shift with both SNARE complexes, whereas KP (kaempferol) did not exhibit such a shift with any complex. Free VAMP2S, VAMP8S or a Syn4/SNAP-23 binary complex induced much less bathochromic shift of polyphenols compared with corresponding ternary complexes (Supplementary Figure S5 at http://www.biochemj.org/bj/450/bj4500537add.htm), suggesting that polyphenol binding to SNARE complex is specific to the ternary complex, but not to a single SNARE protein. The effect of polyphenols on bathochromic shift was perfectly consistent with the SNARE-driven lipid mixing assay, indicating that polyphenols differentially inhibited SNARE-driven membrane fusion by specifically binding to different SNARE complexes.
Figure 3. Differential binding of polyphenols to different SNARE complexes observed by bathochromic shift.
UV absorption spectra of polyphenols before and after mixing with soluble SNARE proteins were overlapped. (a–c) Bathochromic shifts of DL, QT and FS are induced only by Syn4/SNAP-23/VAMP8 complex, but not by Syn4/SNAP-23/VAMP2 complex; (d–e) bathochromic shifts of CY and RT are induced by only Syn4/SNAP-23/VAMP2 complex; (f) KP does not show any bathochromic shift with SNARE proteins; (g and h) bathochromic shifts of MR and LT by both complexes; (i) scheme of panel array.
Differential inhibition of SNARE-mediated exocytosis of distinct subsets of granules by polyphenols
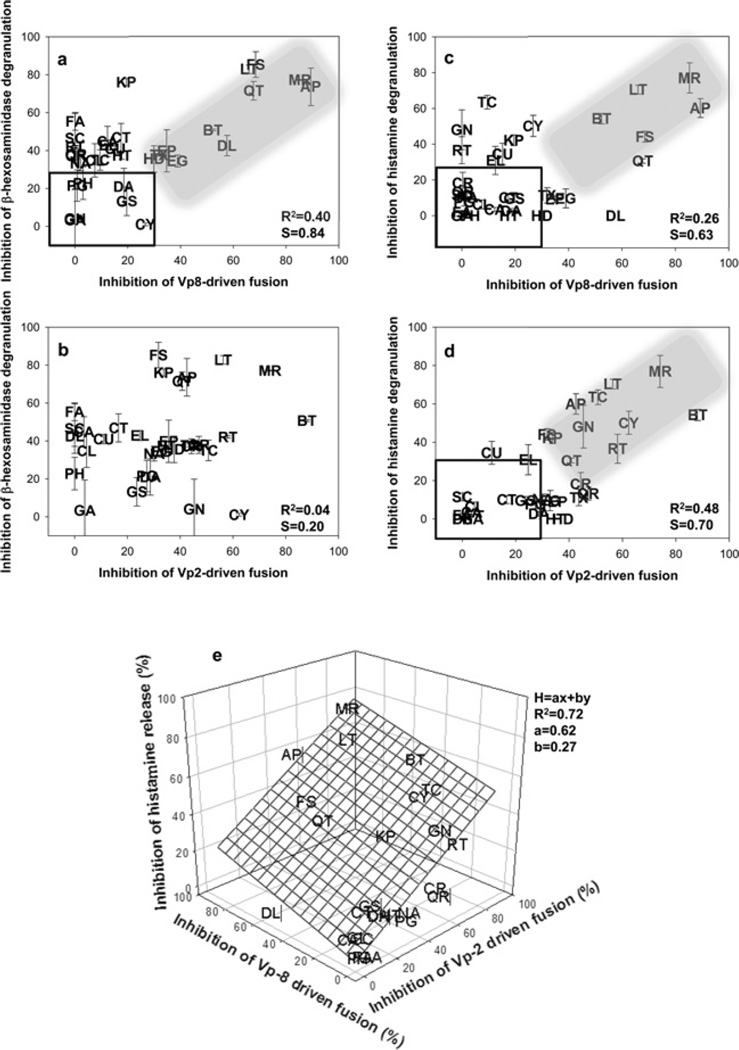
In order to establish the relationship between mast cell degranulation and SNARE-driven membrane fusion, both of which are affected by polyphenols, the inhibition percentage of degranulation of Supplementary Figure S1 and percentage of inhibition of SNARE-driven fusion of Supplementary Figure S4 by each polyphenol were scattered on planes (Figure 4, and Supplementary Figure S6 at http://www.biochemj.org/bj/450/bj4500537add.htm). We plotted four planes for SNAP- 23/Syn4/VAMP8-driven fusion compared with inhibition of β-hexosaminidase release (Figure 4a); SNAP-23/Syn4/VAMP2- driven fusion compared with inhibition of β-hexosaminidase release (Figure 4b); SNAP-23/Syn4/VAMP8-driven fusion compared with inhibition of histamine release (Figure 4c); and SNAP-23/Syn4/VAMP2-driven fusion compared with inhibition of histamine release (Figure 4d). None of the four plots exhibited a clear linear relationship between SNARE-driven fusion and degranulation when all of the polyphenols were taken into account (R2 =0.04–0.48). However, the inhibition of degranulation was linearly dependent on the inhibition of SNARE-driven fusion for some of the polyphenols (shaded area in Figures 4a, 4b and 4d). For example, when only 11 polyphenols (in the shaded area in Figure 4a) were taken into account, inhibition of β-hexosaminidase release was found to be linearly dependent on the inhibition of SNAP-23/Syn4/VAMP8-driven fusion (R2 =0.85 and slope=0.90). Conversely, the release of β-hexosaminidase was found to occur independently of the inhibition of SNAP-23/Syn4/VAMP2-driven fusion, with no observable correlation between the two processes (R2 =0.04) (Figure 4b), consistent with several previous reports [29,32]. Thus it seems that many polyphenols, but not all, inhibit β-hexosaminidase release through the inhibition of SNAP-23/Syn4/VAMP8-driven fusion. It is further noted that some polyphenols such as KP and FA strongly inhibited β-hexosaminidase release, whereas they did not affect SNARE-driven fusion, perhaps through inhibiting signalling pathways [14–17].
Figure 4. Correlation between mast cell degranulation and SNARE-driven membrane fusion affected by polyphenols.
(a–d) Scatter plots for inhibition of Syn4/SNAP-23/VAMP2-or Syn4/SNAP-23/VAMP8-driven fusion compared with β-hexosaminidase or histamine release. Dense areas are shown enlarged in Supplementary Figure S6 at http://www.biochemj.org/bj/450/bj4500537add.htm for better resolution between polyphenolic compounds. R2 and slope (S) shown in the inset are the values considering all polyphenols. A linear relationship between the inhibition of SNARE-driven fusion and inhibition of degranulation by some polyphenols is highlighted in the shaded area. Polyphenols with relatively small inhibitory effect (<30%) are included in the box. (e) Inhibition of histamine release by polyphenols was extrapolated by using the simultaneous equation H=ax + by, where H is the inhibition percentage of histamine release by a polyphenol, × is the inhibition percentage of VAMP2-driven fusion by the polyphenol, y is the inhibition percentage of VAMP8-driven fusion by the polyphenol, and a and b are extrapolated constants.
Although β-hexosaminidase release was found to be dependent only on the presence of VAMP8, histamine release was found to be dependent on both VAMP2 and VAMP8 (Figures 4c and 4d). Some polyphenols (in the shaded areas) showed an apparent and direct relationship between SNAP-23/Syn4/VAMP2- or VAMP8-driven fusion and histamine release. However, there were also apparent outliers such as GN, RT and TC (trans-cinnamic acid), which did not inhibit SNAP-23/Syn4/VAMP8-driven fusion, but strongly inhibited histamine release (Figure 4c). Interestingly, these polyphenols robustly inhibited SNAP-23/Syn4/VAMP2- driven fusion (Figure 4d), indicating how they reduced the release of histamine. Of the three types of granules in mast cells, type I granules are amine-free and contain the lysosomal marker β-hexosaminidase [20,21]. Type III granules are known to contain histamine, whereas type II granules contain both β-hexosaminidase and histamine. On the basis of this suggested classification, VAMP8 controls type I (containing β-hexosaminidase) and type II (containing both β-hexosaminidase and histamine) granules, whereas VAMP2 controls type III (containing histamine) granules. If histamine release is controlled by both VAMP2 and VAMP8, which are located on type III and type II granules respectively, inhibition of histamine release should be a simultaneous equation of the inhibitions of SNAP-23/Syn4/VAMP2- and SNAP-23/Syn4/VAMP8-driven fusion. We therefore applied the simple spontaneous equation H=ax + by, where H is the inhibition percentage of histamine release by a compound, × is the inhibition percentage of VAMP2-driven fusion by the same compound, and y is the inhibition percentage of VAMP8-driven fusion by the same compound. The constants a and b determined from the regression process would be indicative of the leverage of each SNARE protein on histamine release (Figure 4e). After removing six polyphenols {CU (p-coumaric acid), EL (ellagic acid), HD (hesperedin), TX (taxifolin), EGCG (epigallocatechin gallate) and EP [(-)-epicatechin]}, which did not fall within any of the shaded areas and boxes (Figures 4c and 4d) from the dataset, 26 polyphenols were fitted into the equation. The extrapolated plane resulted in 0.62 and 0.27 for a and b respectively, with an R2 value of 0.72, suggesting that ~62% of histamine release is mediated by VAMP2, whereas VAMP8 is responsible for ~27% of histamine release. This result is consistent with the previous study that an siRNA (small interfering RNA) of VAMP8 decreased ~25% of antigen-induced histamine release [32]. On the basis of these findings, we expect that type III granules contain ~62% of total histamine and the remaining histamines are contained in type II granules, as VAMP2 is solely associated with type III granules.
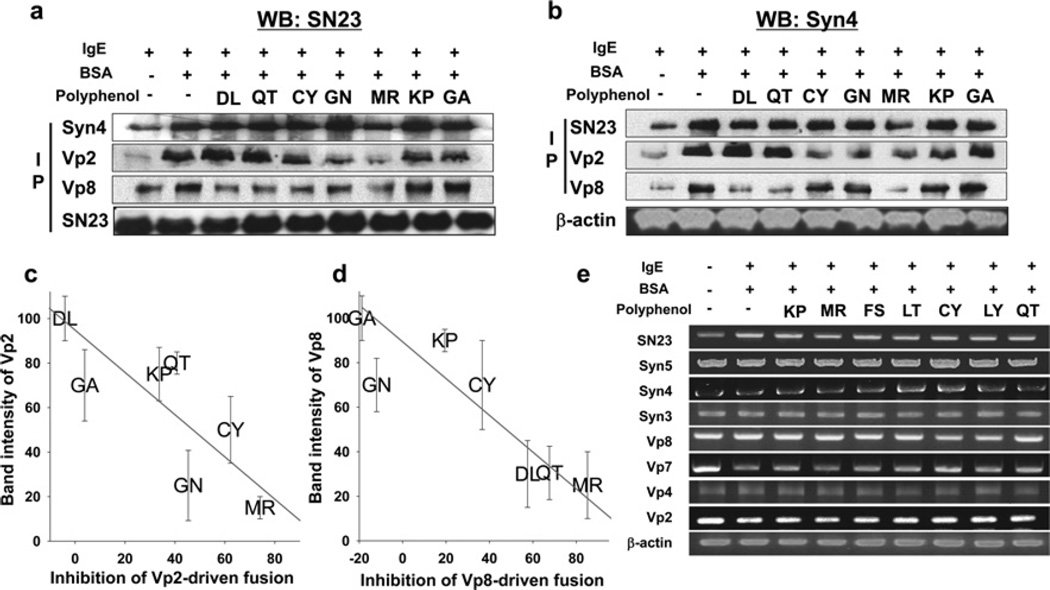
We further investigated whether polyphenols selectively inhibit SNARE complex formation in mast cells using co-immunoprecipitation. Western blot analysis was performed using anti-SNAP-23 antibody after immunoprecipitating SNARE proteins with anti-Syn4 antibody, anti-VAMP2 antibody or anti-VAMP8 antibody (Figure 5a). In addition, co-immunoprecipitated Syn4 protein was evaluated after immunoprecipitating other SNARE proteins (Figure 5b). The first columns of Figures 5(a) and 5(b) clearly show that none of the polyphenols inhibited the binary t-SNARE complex formation of SNAP-23 and Syn4, with the exception of MR. Approximately 20% of t-SNARE complex was inhibited by MR. Also, the result shows that IgE stimulation induces t-SNARE complex formation, as well as ternary complex formationwithVAMP2 orVAMP8.As expected, polyphenols differentially inhibited SNARE complex formation. MR inhibited ternary complex formation of both Syn4/SNAP-23/VAMP2 and Syn4/SNAP-23/VAMP8 complex, whereas DL and QT reduced only Syn4/SNAP-23/VAMP8 complex formation. CY and GN reduced only Syn4/SNAP-23/VAMP2 complex formation, whereas KP and GA did not inhibit the formation of any complex (Figures 5a and 5b). There was an apparent linear relationship between the complex formation in the cell (averaged from relative band intensities of Figures 5a and 5b) and in vitro fusion inhibitory activity (Supplementary Figure S4a), indicating that polyphenols differentially inhibited specific SNARE-driven fusion by inhibiting complex formation, which eventually leads to differential effects on distinct granules of mast cells (Figures 5c and 5d). None of the polyphenols repressed the expression of SNARE proteins (Figure 5e).
Figure 5. Co-immunoprecipitation assay of different SNARE complexes in mast cells.
Western blot analysis was performed using anti-SNAP-23 antibody after immunoprecipitating with several SNARE-specific antibodies. (b) Western blot analysis was performed using anti-Syn4 antibody after immunoprecipitating with several SNARE-specific antibodies. (c and d) Correlation between SNARE-mediated membrane fusion (lipid mixing assay) and SNARE complex formation in RBL-2H3 cells (co-immunoprecipitation). (e) Effect of polyphenols on the mRNA expression of SNARE genes in RBL-2H3 cells.
Finally, half-maximal inhibitory concentrations of selected polyphenols on histamine and β-hexosaminidase were determined (Table 1, and Supplementary Figure S7 at http://www.biochemj.org/bj/450/bj4500537add.htm). IC50 values of CY and GN against histamine release were 8.1 and 14.1 µM respectively, whereas those polyphenols inhibited little β-hexosaminidase release even at much higher concentrations. IC50 values of DL and QT against histamine release were >50 µM, whereas those against β-hexosaminidase release were 13.6 and 5.9 µM respectively. This result confirms SNARE complex-specific inhibition of polyphenols, which eventually leads to the inhibition of corresponding exocytosis.
Table 1.
IC50 values of some polyphenols against β-hexosaminidase and histamine release
| Compound | IC50 against β-hexosaminidase release (µM) |
IC50 against histamine release (µM) |
|---|---|---|
| LY | 6.5 | 5.1 |
| CY | >50 | 8.1 |
| GN | >50 | 14.1 |
| DL | 13.6 | >50 |
| QT | 5.9 | >50 |
| MR | 4.9 | 3.5 |
| KP | 4.6 | 11.4 |
DISCUSSION
In spite of the abundance of polyphenols in our diet, the general belief that diet is closely related to allergic responses and the well-documented efficacies of polyphenols on various allergic models, the mechanism of how they function as antiallergic agents surprisingly remains an open question. One most popular hypothesis is that the endogenous antioxidant ability of polyphenols limits the extent of cellular injury from free radicals during the allergic insult [4,7–9]. A second hypothesis is that each polyphenolic compound has its cognate binding partner. For example, QT, a flavonol, has been postulated to inhibit allergen– IgE complex formation, or hinder the binding of the complex to its receptor FcεRI [42,45,46]. Also, EGCG is known to have an affinity for CD4 [13]. One of best-known examples of a polyphenol binding target is oestrogen receptors, which have been shown to interact with isoflavones [47]. In addition to receptors, enzymes involved in various signalling pathways seem to be affected by some polyphenols [14–17]. A third possibility is that the reduced allergenicity of common food allergens is mediated by polyphenol–protein interactions. Proteins complexed with polyphenols may be rendered less allergenic either by changing the structure of the allergenic protein or by becoming less bioavailable [10–12]. However, even with these plausible postulations, there seems to be an unknown common pathway through which various polyphenols reduce allergic responses. First, only small numbers of polyphenols have been known to have their own binding partner, whereas most polyphenols more or less inhibit the release of allergy mediators from mast cells, macrophages, etc. Secondly, FcεRI stimulation of mast cells induces the release of lysosomal hydrolases, such as β-hexosaminidase and cathepsin D, in addition to the well-known cargo molecules of serotonin and histamine [19]. However, none of the above hypotheses explain clearly the reason some polyphenols inhibit histamine release, but do not inhibit β-hexosaminidase release, and vice versa. Thirdly, even with complexity of multiple interactions of various signalling cascades, a simple question remains unanswered: how do the signalling cascades differentially affect the release of allergy mediators. In general, little is known about the target of polyphenols, and their specific mode of action is still elusive, whereas information relating to the effectiveness of various polyphenols on numerous allergic disease models is plentiful.
In a previous study, we observed that some polyphenols intercalated into the inner layer of neuronal SNARE complex leading to the inhibition of neurotransmitter release from neuronal cells [33]. On the basis of these observations, we postulated that SNARE complexes in the cells related to allergy responses might be regulated by polyphenols. In fact, polyphenols regulated SNARE complex formation in mast cells (Figure 5), resulting in degranulation inhibition (Supplementary Figure S1). Some polyphenols specifically bound to Syn4/SNAP-23/VAMP2 or Syn4/SNAP-23/VAMP8 ternary complexes, whereas some polyphenols concomitantly bound to both complexes (Figure 3 and Supplementary Figure S5). This SNARE complex-specific binding of polyphenols coincides very well with the inhibitory effect of the polyphenol on in vitro SNARE-driven membrane fusion results (Figure 2 and Supplementary Figure S4). As a result of this selective or common binding of polyphenols to two different SNARE complexes (Figure 3), membrane fusion of the corresponding granules with plasma membrane was selectively or commonly inhibited (Figure 2 and Supplementary Figure S4), leading to polyphenol-mediated differential inhibition of degranulation of distinct subsets of granules (Figures 4 and 5). Nevertheless, our results do not exclude the possibility that polyphenols’ anti-allergic effects are also attributed to the well-known function as antioxidants or as regulators of many signalling pathways. Binding of polyphenols to SNARE complexes and to other proteins are not necessarily mutually exclusive. Thus we conclude that polyphenols regulate allergenic responses by inhibiting SNARE complex formation in mast cells.
The present study also suggests the possibility that SNARE complexes found in other types of cells are controlled by polyphenols. All intracellular membrane fusion events are mediated by SNARE family members. SNARE proteins are distributed on nearly every membrane of all eukaryotic cells, and are implicated at each step of membrane fusion in intracellular trafficking pathways. In the cells of the immune system, SNARE-driven membrane fusion mediates the delivery of receptors to and from the cell surface, as well as the constitutive secretion of immune mediators, phagocytosis and endocytosis, and the release of stored inflammatory mediators and products from secretory granules. Neutrophils, eosinophils, platelets and mast cells utilize SNARE-driven membrane fusion in order to release stored inflammatory granular contents on stimulation [12]. In macrophages, SNAREs transport cytokines between intracellular compartments and, eventually, to the cell surface for release [48]. Each cell type expresses different combinations of SNARE proteins, for which currently 38 SNARE family members are known to be distributed within organelles. Thus it is highly probable that polyphenols directly control intracellular trafficking pathways by interfering with the formation of various SNARE complexes in various cell types.
The inhibitory effects of polyphenols on degranulation of mast cells can be extrapolated into human diet and health. Studies on the bioavailability of polyphenols in humans [48,49] indicate that plasma concentrations of some polyphenols could reach up to severalmicromolar in 1–2 h after ingestion. For example, ingested EGCG as a form of green tea extract was as high as 4.4 µM in human plasma. Ingestion of derivatives of QT and naringin led to 6.0–7.6 µMplasma concentration. On the basis of current studies of IC50 values of polyphenol compared with β-hexosaminidase and histamine release, these plasma concentrations of polyphenols derived from human diets seem to be high enough to regulate human physiology including mast cell degranulation.
Supplementary Material
Acknowledgments
FUNDING
This study was supported by a grant from the Korea Healthcare technology R&D Project, Ministry of Health & Welfare, Republic of Korea [grant number A103017] and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology [grant numbers 2011- 0006268 and 2011-0004054].
Abbreviations used
- AP
apigenin
- CY
cyanidin
- DL
delphinidin
- DNP
dinitrophenyl
- DOPS
1, 2-dioleoyl-sn-glycero-3-phosphatidylserine
- EGCG
epigallocatechin gallate
- FA
ferulic acid
- FcεRI
high-affinity receptor for IgE
- FRET
fluorescence resonance energy transfer
- FS
fisetin
- GA
gomisin A
- GN
gosmisin N
- GST
glutathione transferase
- KP
kaempferol
- LT
luteolin
- LUV
large unilamellar vesicle
- LY
Ly294002
- MR
myricetin
- NBD
1, 2-dioleoyl-sn-glycero-3-phosphoserine-N-(7-nitro-2-1, 3-benzoxadiazol-4-yl)
- POPC
1-palmitoyl-2-dioleoyl-sn-glycero-3-phosphatidylcholine
- QT
quercetin
- RT
rutin
- SC
schizandrin
- SNAP-23
23 kDa synaptosome-associated protein
- SNAP-25
25 kDa synaptosome-associated protein
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- Syn
syntaxin
- t-SNARE
target SNARE
- VAMP
vesicle-associated membrane protein
- v-SNARE
vesicle SNARE
Footnotes
AUTHOR CONTRIBUTION
Dae-Hyuk Kweon and Yeon-Kyun Shin co-ordinated the research design.Yoosoo Yang, Jung-Mi Oh, Paul Heo, Byoungjae Kong, Jonghyeok Shin and Ji-Chun Lee performed the experiments and analysed the data. Jae Yoon Shin and Choong Hwan Lee contributed to the discussion. Dae-Hyuk Kweon and Yoosoo Yang wrote the paper, with contributions to writing and editing prior to submission from Kye Won Park and Jeong Su Oh.
REFERENCES
- 1.Park HH, Lee S, Son HY, Park SB, Kim MS, Choi EJ, Singh TS, Ha JH, Lee MG, Kim JE, et al. Flavonoids inhibit histamine release and expression of proinflammatory cytokines in mast cells. Arch. Pharm. Res. 2008;31:1303–1311. doi: 10.1007/s12272-001-2110-5. [DOI] [PubMed] [Google Scholar]
- 2.Hounsome N, Hounsome B, Tomos D, Edwards-Jones G. Plant metabolites and nutritional quality of vegetables. J. Food Sci. 2008;73:R48–R65. doi: 10.1111/j.1750-3841.2008.00716.x. [DOI] [PubMed] [Google Scholar]
- 3.Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. Polyphenols: food sources and bioavailability. Am. J. Clin. Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 4.Mennen LI, Walker R, Bennetau-Pelissero C, Scalbert A. Risks and safety of polyphenol consumption. Am. J. Clin. Nutr. 2005;81:S326–S329. doi: 10.1093/ajcn/81.1.326S. [DOI] [PubMed] [Google Scholar]
- 5.Akazome Y, Kametani N, Kanda T, Shimasaki H, Kobayashi S. Evaluation of safety of excessive intake and efficacy of long-term intake of beverages containing apple polyphenols. J. Oleo Sci. 2010;59:321–338. doi: 10.5650/jos.59.321. [DOI] [PubMed] [Google Scholar]
- 6.Ross JA, Kasum CM. Dietary flavonoids: bioavailability, metabolic effects, and safety. Annu. Rev. Nutr. 2002;22:19–34. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- 7.Noguchi Y, Fukuda K, Matsushima A, Haishi D, Hiroto M, Kodera Y, Nishimura H, Inada Y. Inhibition of Df-protease associated with allergic diseases by polyphenol. J. Agric. Food Chem. 1999;47:2969–2972. doi: 10.1021/jf9812073. [DOI] [PubMed] [Google Scholar]
- 8.Garcia V, Arts IC, Sterne JA, Thompson RL, Shaheen SO. Dietary intake of flavonoids and asthma in adults. Eur. Respir. J. 2005;26:449–452. doi: 10.1183/09031936.05.00142104. [DOI] [PubMed] [Google Scholar]
- 9.Seifried HE, Anderson DE, Fisher EI, Milner JA. A review of the interaction among dietary antioxidants and reactive oxygen species. J. Nutr. Biochem. 2007;18:567–579. doi: 10.1016/j.jnutbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Chung S-Y, Champagne ET. Reducing the allergenic capacity of peanut extracts and liquid peanut butter by phenolic compounds. Food Chem. 2009;115:1345–1349. [Google Scholar]
- 11.Nozaki A, Hori M, Kimura T, Ito H, Hatano T. Interaction of polyphenols with proteins: binding of (−)-epigallocatechin gallate to serum albumin, estimated by induced circular dichroism. Chem. Pharm. Bull. 2009;57:224–228. doi: 10.1248/cpb.57.224. [DOI] [PubMed] [Google Scholar]
- 12.Labuckas DO, Maestri DM, Perelló M, Martínez ML, Lamarque AL. Phenolics from walnut (Juglans regia L.) kernels: antioxidant activity and interactions with proteins. Food Chem. 2008;107:607–612. [Google Scholar]
- 13.Williamson MP, McCormick TG, Nance CL, Shearer WT. Epigallocatechin gallate, the main polyphenol in green tea, binds to the T-cell receptor, CD4: potential for HIV-1 therapy. J. Allergy Clin. Immunol. 2006;118:1369–1374. doi: 10.1016/j.jaci.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Targeting multiple signaling pathways by green tea polyphenol (−)-epigallocatechin-3-gallate. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 15.Biesalski HK. Polyphenols and inflammation: basic interactions. Curr. Opin. Clin. Nutr. Metab. Care. 2007;10:724–728. doi: 10.1097/MCO.0b013e3282f0cef2. [DOI] [PubMed] [Google Scholar]
- 16.Santangelo C, Vari R, Scazzocchio B, Di Benedetto R, Filesi C, Masella R. Polyphenols, intracellular signalling and inflammation. Ann. Ist. Super. Sanita. 2007;43:394–405. [PubMed] [Google Scholar]
- 17.Kanda T, Akiyama H, Yanagida A, Tanabe M, Goda Y, Toyoda M, Teshima R, Saito Y. Inhibitory effects of apple polyphenol on induced histamine release from RBL-2H3 cells and rat mast cells. Biosci., Biotechnol., Biochem. 1998;62:1284–1289. doi: 10.1271/bbb.62.1284. [DOI] [PubMed] [Google Scholar]
- 18.Abraham SN, Malaviya R. Mast cells in infection and immunity. Infect. Immun. 1997;65:3501–3508. doi: 10.1128/iai.65.9.3501-3508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metcalfe DD, Baram D, Mekori YA. Mast cells. Physiol. Rev. 1997;77:1033–1079. doi: 10.1152/physrev.1997.77.4.1033. [DOI] [PubMed] [Google Scholar]
- 20.Raposo G, Tenza D, Mecheri S, Peronet R, Bonnerot C, Desaymard C. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol. Biol. Cell. 1997;8:2631–2645. doi: 10.1091/mbc.8.12.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baram D, Adachi R, Medalia O, Tuvim M, Dickey BF, Mekori YA, Sagi-Eisenberg R. Synaptotagmin II negatively regulates Ca2+ -triggered exocytosis of lysosomes in mast cells. J. Exp. Med. 1999;189:1649–1658. doi: 10.1084/jem.189.10.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 23.Blank U, Cyprien B, Martin-Verdeaux S, Paumet F, Pombo I, Rivera J, Roa M, Varin-Blank N. SNAREs and associated regulators in the control of exocytosis in the RBL-2H3 mast cell line. Mol. Immunol. 2002;38:1341–1345. doi: 10.1016/s0161-5890(02)00085-8. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki K, Verma IM. Phosphorylation of SNAP-23 by IκB kinase 2 regulates mast cell degranulation. Cell. 2008;134:485–495. doi: 10.1016/j.cell.2008.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stow JL, Manderson AP, Murray RZ. SNAREing immunity: the role of SNAREs in the immune system. Nat. Rev. Immunol. 2006;6:919–929. doi: 10.1038/nri1980. [DOI] [PubMed] [Google Scholar]
- 26.Guo Z, Turner C, Castle D. Relocation of the t-SNARE SNAP-23 from lamellipodia-like cell surface projections regulates compound exocytosis in mast cells. Cell. 1998;94:537–548. doi: 10.1016/s0092-8674(00)81594-9. [DOI] [PubMed] [Google Scholar]
- 27.Puri N, Roche PA. Ternary SNARE complexes are enriched in lipid rafts during mast cell exocytosis. Traffic. 2006;7:1482–1494. doi: 10.1111/j.1600-0854.2006.00490.x. [DOI] [PubMed] [Google Scholar]
- 28.Pombo I, Rivera J, Blank U. Munc18–2/syntaxin3 complexes are spatially separated from syntaxin3-containing SNARE complexes. FEBS Lett. 2003;550:144–148. doi: 10.1016/s0014-5793(03)00864-0. [DOI] [PubMed] [Google Scholar]
- 29.Lippert U, Ferrari DM, Jahn R. Endobrevin/VAMP8 mediates exocytotic release of hexosaminidase from rat basophilic leukaemia cells. FEBS Lett. 2007;581:3479–3484. doi: 10.1016/j.febslet.2007.06.057. [DOI] [PubMed] [Google Scholar]
- 30.Salinas E, Quintanar-Stephano A, Cordova LE, Ouintanar JL. Allergen-sensitization increases mast-cell expression of the exocytotic proteins SNAP-23 and syntaxin 4, which are involved in histamine secretion. J. Invest. Allergol. Clin. Immunol. 2008;18:366–371. [PubMed] [Google Scholar]
- 31.Puri N, Roche PA. Mast cells possess distinct secretory granule subsets whose exocytosis is regulated by different SNARE isoforms. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2580–2585. doi: 10.1073/pnas.0707854105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woska JR, Jr, Gillespie ME. Small-interfering RNA-mediated identification and regulation of the ternary SNARE complex mediating RBL-2H3 mast cell degranulation. Scand. J. Immunol. 2011;73:8–17. doi: 10.1111/j.1365-3083.2010.02471.x. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Shin JY, Oh JM, Jung CH, Hwang Y, Kim S, Kim JS, Yoon KJ, Ryu JY, Shin J, et al. Dissection of SNARE-driven membrane fusion and neuroexocytosis by wedging small hydrophobic molecules into the SNARE zipper. Proc. Natl. Acad. Sci. U.S.A. 2010;107:22145–22150. doi: 10.1073/pnas.1006899108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Choi JK, Jung CH, Koh HJ, Heo P, Shin JY, Kim S, Park WS, Shin HJ, Kweon DH. SNARE-wedging polyphenols as small molecular botox. Planta Med. 2012;78:233–236. doi: 10.1055/s-0031-1280385. [DOI] [PubMed] [Google Scholar]
- 35.Schwartz LB, Austen KF, Wasserman SI. Immunologic release of β-hexosaminidase and β-glucuronidase from purified rat serosal mast cells. J. Immunol. 1979;123:1445–1450. [PubMed] [Google Scholar]
- 36.Kawasaki Y, Saitoh T, Okabe T, Kumakura K, Ohara-Imaizumi M. Visualization of exocytotic secretory processes of mast cells by fluorescence techniques. Biochim. Biophys. Acta. 1991;1067:71–80. doi: 10.1016/0005-2736(91)90027-6. [DOI] [PubMed] [Google Scholar]
- 37.Tewtrakul S, Itharat A. Anti-allergic substances from the rhizomes of Dioscorea membranacea. Bioorg. Med. Chem. 2006;14:8707–8711. doi: 10.1016/j.bmc.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Jung CH, Yang YS, Kim JS, Shin JI, Jin YS, Shin JY, Lee JH, Chung KM, Hwang JS, Oh JM, et al. A search for synthetic peptides that inhibit soluble N-ethylmaleimide sensitive-factor attachment receptor-mediated membrane fusion. FEBS J. 2008;275:3051–3063. doi: 10.1111/j.1742-4658.2008.06458.x. [DOI] [PubMed] [Google Scholar]
- 39.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 40.Qin C, Xie MX, Liu Y. Characterization of the myricetin-human serum albumin complex by spectroscopic and molecular modeling approaches. Biomacromolecules. 2007;8:2182–2189. doi: 10.1021/bm070319c. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz LB, Lewis RA, Seldin D, Austen KF. Acid hydrolases and tryptase from secretory granules of dispersed human lung mast cells. J. Immunol. 1981;126:1290–1294. [PubMed] [Google Scholar]
- 42.Fujimura Y, Tachibana H, Maeda-Yamamoto M, Miyase T, Sano M, Yamada K. Antiallergic tea catechin, (−)-epigallocatechin-3-O-(3-O-methyl)-gallate, suppresses FcεRI expression in human basophilic KU812 cells. J. Agric. Food Chem. 2002;50:5729–5734. doi: 10.1021/jf025680z. [DOI] [PubMed] [Google Scholar]
- 43.Tong J, Borbat PP, Freed JH, Shin YK. A scissors mechanism for stimulation of SNARE-mediated lipid mixing by cholesterol. Proc. Natl. Acad. Sci. U.S. A. 2009;106:5141–5146. doi: 10.1073/pnas.0813138106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puri N, Kruhlak MJ, Whiteheart SW, Roche PA. Mast cell degranulation requires N-ethylmaleimide-sensitive factor-mediated SNARE disassembly. J. Immunol. 2003;171:5345–5352. doi: 10.4049/jimmunol.171.10.5345. [DOI] [PubMed] [Google Scholar]
- 45.Fujimura Y, Tachibana H, Yamada K. A tea catechin suppresses the expression of the high-affinity IgE receptor FcεRI in human basophilic KU812 cells. J. Agric. Food Chem. 2001;49:2527–2531. doi: 10.1021/jf001392w. [DOI] [PubMed] [Google Scholar]
- 46.Yano S, Tachibana H, Yamada K. Flavones suppress the expression of the high-affinity IgE receptor FcεRI in human basophilic KU812 cells. J. Agric. Food. Chem. 2005;53:1812–1817. doi: 10.1021/jf047929d. [DOI] [PubMed] [Google Scholar]
- 47.Morabito N, Crisafulli A, Vergara C, Gaudio A, Lasco A, Frisina N, D’Anna R, Corrado F, Pizzoleo MA, Cincotta M, et al. Effects of genistein and hormone-replacement therapy on bone loss in early postmenopausal women: a randomized double-blind placebo-controlled study. J. Bone Miner. Res. 2002;17:1904–1912. doi: 10.1359/jbmr.2002.17.10.1904. [DOI] [PubMed] [Google Scholar]
- 48.Scalbert A, Williamson G. Dietary intake and bioavailability of polyphenols. J. Nutr. 2000;130:S2073–S2085. doi: 10.1093/jn/130.8.2073S. [DOI] [PubMed] [Google Scholar]
- 49.Manach C, Williamson G, Morand C, Scalbert A, Rémésy C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005;81:S230–S242. doi: 10.1093/ajcn/81.1.230S. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.