Abstract
Since its first isolation in 1996 in Guangdong, China, the highly pathogenic avian influenza virus (HPAIV) H5N1 has circulated in avian hosts for almost two decades and spread to more than 60 countries worldwide. The role of different avian hosts and the domestic-wild bird interface has been critical in shaping the complex HPAIV H5N1 disease ecology, but remains difficult to ascertain. To shed light on the large-scale H5N1 transmission patterns and disentangle the contributions of different avian hosts on the tempo and mode of HPAIV H5N1 dispersal, we apply Bayesian evolutionary inference techniques to comprehensive sets of hemagglutinin and neuraminidase gene sequences sampled between 1996 and 2011 throughout Asia and Russia. Our analyses demonstrate that the large-scale H5N1 transmission dynamics are structured according to different avian flyways, and that the incursion of the Central Asian flyway specifically was driven by Anatidae hosts coinciding with rapid rate of spread and an epidemic wavefront acceleration. This also resulted in long-distance dispersal that is likely to be explained by wild bird migration. We identify a significant degree of asymmetry in the large-scale transmission dynamics between Anatidae and Phasianidae, with the latter largely representing poultry as an evolutionary sink. A joint analysis of host dynamics and continuous spatial diffusion demonstrates that the rate of viral dispersal and host diffusivity is significantly higher for Anatidae compared with Phasianidae. These findings complement risk modeling studies and satellite tracking of wild birds in demonstrating a continental-scale structuring into areas of H5N1 persistence that are connected through migratory waterfowl.
Keywords: H5N1, phylogeography, Bayesian inference, viral evolution, disease ecology
Introduction
Disentangling the complex contributions of various avian hosts to the spread of highly pathogenic avian influenza virus (HPAIV) H5N1 has been termed as one of the most challenging tasks of HPAI disease ecology (Xiao et al. 2007). HPAIV H5N1 is a strain from the Influenza virus A genus, which belongs to the Orthomyxoviridae family, and is characterized by a high mortality in bird populations (Webster et al. 2006), thus imposing a very high economic burden. HPAIV H5N1 was detected for the first time in 1996 in geese from Guangdong, China (Xu et al. 1999) and has since spread across Asia, Europe, and north African countries. The extensive spread of HPAIV H5N1 in avian populations, which took place despite several control measures, such as culling, stamping out and cleaning or disinfection, had raised the fear early on for human pandemic spread. Consequently, poultry and wild bird vaccination has been taken into consideration as a preventive measure, but only a few countries have adopted it efficiently.
HPAIV H5N1 shows a considerable capacity for xenospecific transmission, including to human hosts, and can lead to infection through the fecal–oral route, oral–oral route, consumption of raw infected birds, and by fomites (Webster et al. 1992; Songserm et al. 2006; Gilbert et al. 2010; Pfeiffer et al. 2013; Poovorawan et al. 2013; Bett et al. 2014). For human infections, mortality rates of approximately 60% have been reported (WHO 2013) although the real mortality rates are undoubtedly lower (Li et al. 2008). Experimental studies in ferrets, which are used as models for human transmission, indicate that there is only a relatively small genetic barrier for HPAIV H5N1 to acquire the capacity to efficiently transmit by droplets or aerosols (Herfst et al. 2012). However, some of the required substitutions are likely to be negatively selected in birds, and viral variants transmissible by respiratory droplets may not sufficiently contribute to the within-host viral population to transmit successfully (Russell et al. 2012).
The threat of pathogenic influenza viruses emerging in avian populations has recently also been stressed by an outbreak of the novel H7N9 influenza virus in China around February 2013 (Lam et al. 2013). Although less pathogenic in avian species, the H7N9 virus resulted in 571 confirmed cases and 212 deaths in humans over a 2-year span whereas 19 years of HPAIV H5N1 spread amounted to about 844 cases (WHO 2015; Wang et al. 2014). The rapid emergence and relatively high incidence in the human population make H7N9 an additional threat for triggering a human pandemic, and it may shift the attention away from HPAIV H5N1 (To et al. 2013).
Although evolutionary analyses suggest that influenza A virus may have been a more mammalian generalist in the distant past (Worobey et al. 2014), wild waterfowl belonging to the Anatidae family (including ducks, geese, and swans) are now considered to be the natural reservoir because all known subtypes (16 hemagglutinin [HA] and 9 neuraminidase [NA] (Fouchier et al. 2005), except for HA17 and H18 which were only detected in bats [Tong et al. 2012, 2013]) have been isolated from aquatic birds. Viruses isolated from other hosts in recent decades also find close relatives in waterfowl (Webster et al. 1992).
HPAIV H5N1 has rapidly evolved to evade host immunity and achieved efficient transmission in new host species, such as domestic poultry (Phasianidae, which includes pheasants, chickens and quails among others) and the remaining modern birds (Neoaves, which encompass the remaining avian diversity, such as sparrows, pigeons and falcons, with the exception of ratites) (Nelson and Holmes 2007). Despite the importance for predicting and preventing viral spread, transmission pathways and host-specific contributions to spread between countries have been difficult to establish and two competing views on the spread of HPAIV H5N1 have been put forward. On the one hand, Anatidae are hypothesized to be the main spreaders of the virus in mainly two ways. First, the long distance migration routes expose the domestic or resident bird population at multiple stopover sites to the virus, as observed in 2005 during the rapid viral spread from Russia and Kazakhstan to Turkey, Romania and Ukraine, or in the western European 2006 outbreaks, where migration or unusual movement of wild birds implicated their role as vectors to areas that had no previous record of HPAIV H5N1 presence (Songserm et al. 2006; Kim et al. 2009; Hill et al. 2012). Second, they may represent silent spreaders, because often HPAIV H5N1 causes asymptomatic infection in some species of domestic and wild ducks, as demonstrated by the 2004 case study of ten flocks of grazing ducks in Thailand, of which few were found to be symptomatic despite shedding the virus for 5–10 days (Songserm et al. 2006; Keawcharoen et al. 2008). Surveillance studies on the other hand have suggested that HPAIV H5N1 transmission is maintained by Phasianidae, largely due to the intensive movement of poultry, and that transmission within poultry is the major mechanism for sustaining viral endemicity rather than by continuous reintroduction through migrating birds (Chen et al. 2006; Fournié et al. 2013).
Because of the commensurate time-scale of evolutionary and spatial dispersal dynamics, genetic data may offer a valuable source of information to reconstruct transmission for rapidly evolving pathogens (Holmes 2008; Pybus and Rambaut 2009). As a consequence, the field of viral phylodynamics has witnessed a rich development of quantitative approaches to effectively infer epidemic processes from viral evolutionary histories (Pybus and Rambaut 2009). HPAIV H5N1 represents in fact a prime example of viral phylogeographic reconstructions that stimulated methodological developments, first within a parsimony context (Wallace et al. 2007; Wallace and Fitch 2008), and more recently, in a Bayesian statistical framework (Lemey et al. 2009). These analyses focused on the location of HPAIV H5N1 emergence, and provided descriptive analyses of source-sink dynamics and restrictions to viral migration. Two stochastic models of phylogenetic diffusion have been proposed to perform spatiotemporal reconstructions in a Bayesian framework (Bloomquist et al. 2010; Faria et al. 2011): A continuous-time Markov chain (CTMC) process to model transitioning among discrete location states throughout evolutionary history (Lemey et al. 2009) and a Brownian random walk process to model diffusion in continuous space (the change in longitude and latitude along phylogenetic branches) (Lemey et al. 2010). The discrete model has recently been extended to test and quantify the contribution of potential predictors of viral diffusion (Lemey et al. 2014). By evaluating competing hypotheses of spatiotemporal origins (Bouckaert et al. 2012) and by providing estimates for key spatial epidemiological variables (Pybus et al. 2012), the continuous diffusion model has also proven to be a powerful hypothesis testing tool. The latter work was performed on the West Nile virus (WNV) invasion in North America, for which phylogeographic estimates of the diffusion coefficient and the variation in the spatial spread revealed that this invasion was not characterized by a steady front of east-to-west dissemination, but heavily impacted by rare long-range movements. Although rooted in phylogeographic applications, Bayesian phylogenetic diffusion models are not limited to geography and can be applied in various studies that aim at jointly reconstructing evolutionary history and discrete or continuous trait evolution, including, for example, viral host switching (Faria et al. 2013) and phenotypic evolution (Bedford et al. 2014; Vrancken et al. 2014).
To gain further insight into the large-scale transmission patterns of HPAIV H5N1, from its emergence in Guangdong to its maintenance across Asia and Russia, and to clarify the contribution of different avian host populations, we here undertake a detailed phylogeographic analysis of large data sets of HA and NA gene sequences from Russian and Asian HPAIV H5N1 viruses. We apply discrete phylogeographic inference techniques and extend a recently developed generalized linear model (GLM) approach to incorporate random effects that allow identify spatial transmission patterns that deviate from regular distance-based dispersal dynamics. We extend a continuous diffusion approach to distinguish between different host populations when quantifying spatial dynamics. Specifically, we jointly reconstruct both spatial and host independent processes upon a posterior sample of trees in a single probabilistic analysis and apply this to large sets of HPAIV H5N1 sequence data sampled over 15 years to disentangle the contribution of the Anatidae and Phasianidae hosts to the tempo and mode of HPAIV H5N1 epidemic expansion.
Results
We analyze data sets comprising 806 HA and NA gene sequences sampled between 1996 and 2011 from two avian families, Anatidae and Phasianidae, and the Neoaves avian superorder (supplementary fig. S1 and tables S1 and S2, Supplementary Material online). Different data sets were produced by different subsampling strategies to mitigate sampling bias, but we mainly report the results for randomly downsampling overrepresented locations (based on comparisons of phylogeny-trait association and reconstruction uncertainty; supplementary table S3, Supplementary Material online), and mirror them with estimates for the other data sets in the supplementary material, Supplementary Material online. Our probabilistic analysis approach encompasses model components of temporal sequence evolution, geographic dispersal, and host switching. Estimates of evolutionary rates and phylogenetic divergence times are in agreement with previous studies (supplementary table S3, Supplementary Material online) (Lemey et al. 2010). We start by presenting spatial dispersal estimates in both discrete and continuous spaces and subsequently introduce the discrete host transmission to finally arrive at a joint analysis of both processes.
Spatial Expansion
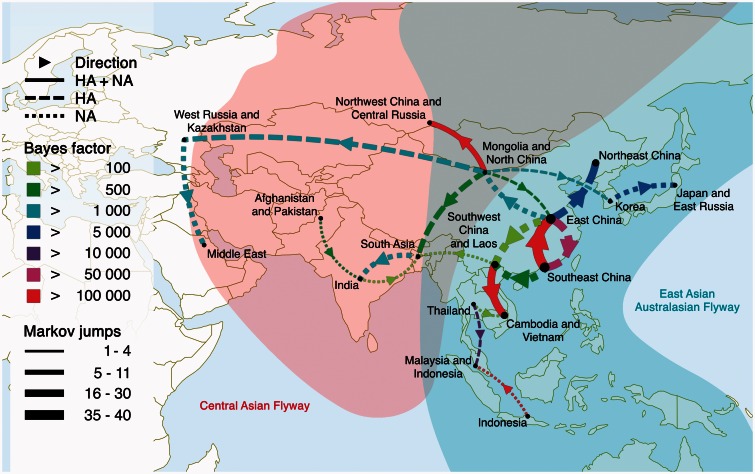
To capture the main spatial diffusion dynamics underlying HPAIV H5N1 spread across Eurasia, we summarize the best supported rates of discrete location transitioning among all pairs of 19 locations as inferred using Bayesian stochastic search variable selection (BSSVS) in figure 1. These dynamics can be largely discriminated by two Asian migratory bird flyways, the East Asian–Australasian flyway and the Central Asian flyway, with a central connecting role for Mongolia and North China where these flyways overlap. These patterns also largely recover the gene flow structures recently estimated for HPAIV H5N1 clade 2.3.2 (Tian et al. 2015), which were shown to follow bird migration networks along these two flyways. Taken together, these diffusion pathways represent 83% and 79% of the location state transitions in the HA and NA evolutionary histories, respectively. As expected, root state probabilities provide support for Southeast China (mostly represented by samples from Guangdong and Hong Kong) as the origin of HPAIV H5N1 emergence (supplementary fig. S2, Supplementary Material online).
Fig. 1.
BF support for nonzero rates in HPAIV H5N1 HA and NA. Rates are represented for a BF > 100 in either gene (dashed and dotted lines for HA and NA, respectively) or both genes (full lines). The line color (in the online color figures) represents the relative strength by which the rates are supported: green and red reflect relatively weak and strong support, respectively. When the rate is supported by both HA and NA, the color represents the lowest support for the rate. The thickness of the lines representing the rates is proportional to the number of Markov jumps (MJ): thin and thick reflect a relatively small and large number of MJ, respectively. When the rate is supported for both HA and NA, we set the thickness according to the smallest number of MJ. The representation of the East Asian–Australasian flyway and the Central Asian flyway is adapted from data from the East Asian–Australasian Flyway Partnership (www.eaaflyway.net/the-flyway, last accessed September 15, 2015). The well-supported rates for differently downsampled data sets are shown in supplementary figure S3, Supplementary Material online.
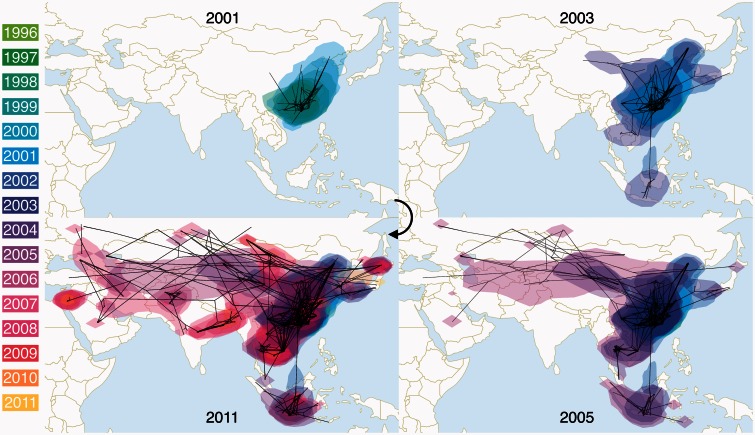
A more detailed picture of the HPAIV H5N1 spread through time along this transmission network is provided by a phylogeographic reconstruction in continuous space (fig. 2 and supplementary fig. S4, Supplementary Material online). This depicts that HPAIV H5N1 spread remained largely restricted to East and Southeast China up 2001, but migration events to North China and Japan as well as to Vietnam are starting to emerge from this time. These migrations became more pronounced by 2003 and include the seeding of Thailand and Indonesia, with spread between different islands within the latter. The dynamics remain largely restricted to the area encompassing the East Asian–Australasian flyway, but westward movements from North and East China into the area where both flyways overlap are emerging. More phylogenetic branches have bridged these flyways by 2005 in line with the multiple well-supported corresponding rates in the discrete analysis (fig. 1), but also spread over a large geographic area to the west has now been established. By 2011, the patterns of spread also encompass South Asia (e.g., India, Nepal and Bangladesh) (Chen et al. 2006; Webster et al. 2006; Pfeiffer et al. 2013).
Fig. 2.
Spatiotemporal dispersal of HPAIV H5N1 in Eurasia reconstructed using continuous phylogeographic analysis of HA. Dispersal patterns are shown up to four different years: 2001, 2003, 2005, and 2011. The black lines project the part of the MCC tree up to each of those times, whereas the contours represent statistical uncertainty of the estimated locations at the internal nodes (95% credible contours based on kernel density estimates). The dispersal patterns for the differently downsampled data sets are shown in supplementary figure S4, Supplementary Material online.
The extensive transmission within China (fig. 2), despite that vaccination efforts that have been unrolled, is also reflected in discrete diffusion estimates showing that the virus remains circulating throughout a large proportion of the evolutionary history in these areas (Southeast, East, and to a lesser extent also Southwest China; supplementary figs. S2 and S5, Supplementary Material online), a persistence that may be attributed to intense duck production in terms of flock size and movement (Tian et al. 2005; Gilbert et al. 2010).
The spatial expansion in continuous space is characterized by an average invasion velocity of about 700 km/year, but considerable diffusion rate heterogeneity underlies these dynamics as formal testing (using log marginal likelihood estimation techniques; Baele et al. 2012, 2013; Baele and Lemey 2013) rejects a phylogenetic Brownian diffusion model in favor of more flexible relaxed random walk (RRW) models (supplementary table S4, Supplementary Material online). Using a weighted average estimate for the diffusion coefficient (a measure of the diffusivity of the viral transmission, cfr. Materials and Methods), we arrive at estimates of 584 (503–665) and 712 (558–884) km2/year for HA and NA, respectively, which is substantially higher than the comparable estimate of 210.27 (174.36–25317) km2/year for WNV in North America.
For the discrete partitioning of the large geographic scale we focus on here, many of the well-supported diffusion pathways occur between locations that are in relative close proximity from each other (fig. 1). To formally evaluate this pattern, we perform an analysis using a recently developed GLM diffusion model (Lemey et al. 2014), in which we include geographic distance as a potential predictor, but that we also extend here with random effects for the diffusion rates (cfr. Materials and Methods). For all data sets, this analysis yields a maximal posterior inclusion probability for geographic distance, providing evidence that distance helps to explain the diffusion dynamics. Because diffusion rates are parameterized as a function of geographic distance in this GLM, the random effects may help to identify exceptions to relatively regular distance-based diffusion patterns. To identify those exceptions, we use a statistic that summarizes the probability that a random effect is the highest on an absolute scale. When random effects are ranked according to this statistic, its value rapidly declines (supplementary fig. S7, Supplementary Material online), and only the rate between Mongolia/North China and West Russia/Kazakhstan is accompanied by a high random effect with a probability of 0.4 or 0.5 for being the top-ranked effect in the analysis of HA and NA, respectively. This is indeed the rate that connects the two most distant locations among all the well-supported diffusion rates (fig. 1). When recording the number of transitions along this connection in the evolutionary history using Markov jumps, we infer an average of 8.18 and 13.76 transitions for HA and NA, respectively, suggesting multiple migration events over long distance.
Host Transmission Patterns
We investigated the coarse-grain host switching dynamics in our analysis by including a discrete diffusion process among the avian families, Anatidae (n = 299) and Phasianidae (n = 428), and the Neoaves superorder (n = 74), from which our samples were obtained (supplementary table S6, Supplementary Material online). To characterize the overall dynamics between the most abundantly sampled Anatidae and Phasianidae hosts, we estimated model fit for four different models: A bidirectional symmetric and asymmetric model and two different unidirectional models (table 1). This identifies a directional asymmetric model as the best-fitting model for both HA and NA, suggesting transmission in both directions but with a relatively strong preference for one of the directions.
Table 1.
Log Marginal Likelihoods Estimated by Stepping Stone Sampling for Four Discrete Host Transmission Scenarios.
| HA | NA | |
|---|---|---|
| Bidirectional Asymmetric | −551.50 | −599.47 |
| Bidirectional Symmetric | −558.91 | −603.75 |
| Unidirectional Ana-to-Pha | −650.02 | −682.37 |
| Unidirectional Pha-to-Ana | −638.12 | −695.69 |
To quantify the host transitions in the ancestral history of our sample under this model, as well as the time spent in each of these avian hosts, we summarized Markov jumps and Markov reward estimates in table 2. These revealed about three times as many Anatidae to Phasianidae jumps as vice versa. The estimates are unlikely to be impacted by sampling bias because more samples were included from Phasianidae, and accordingly, more time appears to be spent in this Phasianidae host (table 2). The same differences in transmission preference can also be deduced from the host transition rate estimates in an asymmetric diffusion model that considers all three host orders (supplementary table S7, Supplementary Material online). The overall preference of Anatidae to Phasianidae transmission is also apparent from the ancestral host reconstructions in the HA and NA phylogenies, which reveals a general backbone associated with Anatidae (supplementary fig. S8, Supplementary Material online).
Table 2.
Host Transitions and Host Reward Time under the Bidirectional Asymmetric Model.
| HA | NA | |
|---|---|---|
| Ana-to-Pha jumps | 122 (105–139) | 131 (99–154) |
| Pha-to-Ana jumps | 43 (30–57) | 48 (30–74) |
| Ana reward time (years) | 352 (321–385) | 382 (329–434) |
| Pha reward time (years) | 400 (370–431) | 424 (377–480) |
Note.—Values in brackets represent 95% HPD intervals.
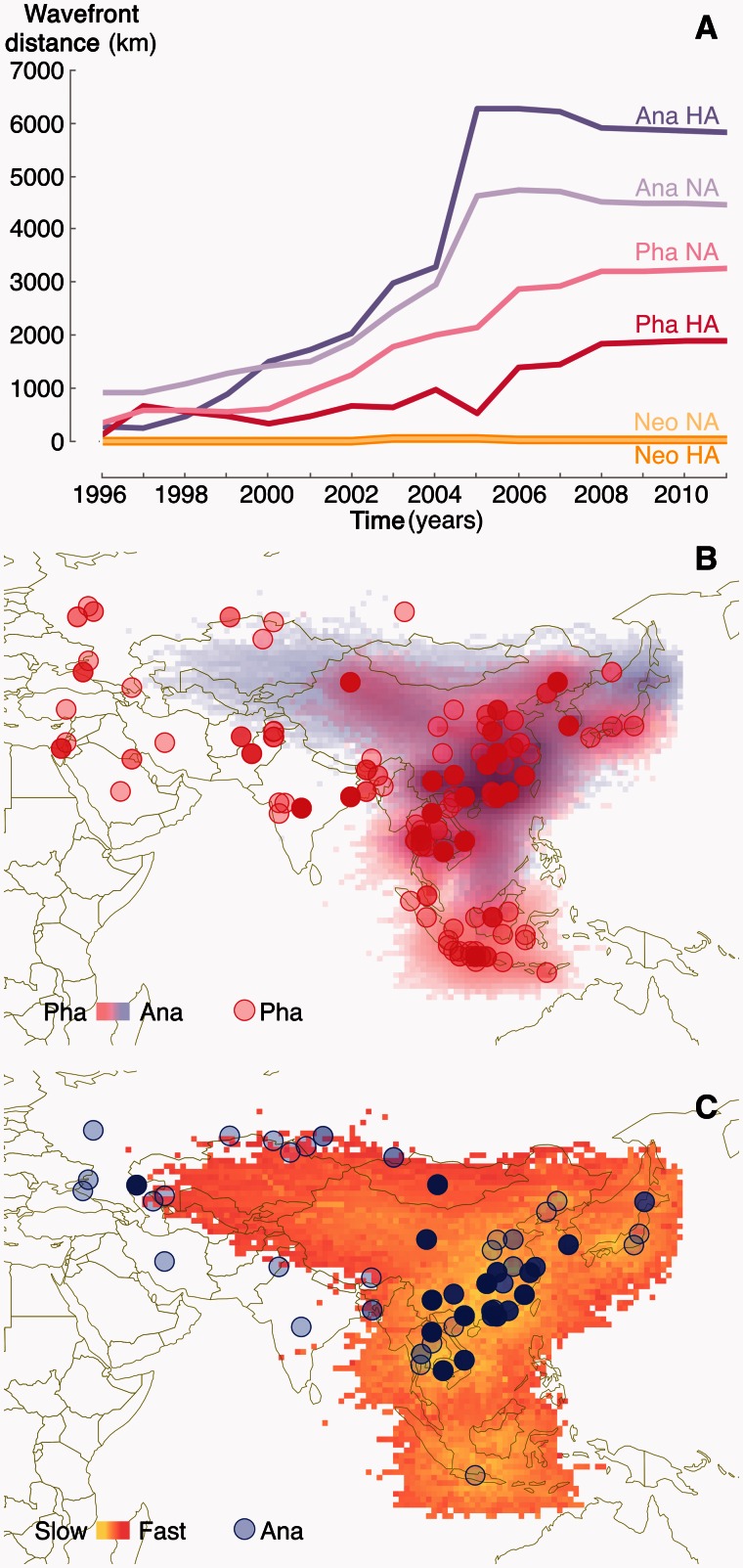
To further disentangle the host contributions in the spatial expansion of HPAIV H5N1, we summarized specific estimates from a joint analysis of discrete host and continuous spatial diffusion (cfr. Materials and Methods). In accordance with the Anatidae phylogenetic backbone, this reveals a large overall contribution of this host to the epidemic wavefront (fig. 3A), summarized by the largest great-circle distance traveled from the root location estimate divided by the time since the most recent common ancestor. In the HA history, a comparatively high contribution of Phasianidae can be noted in the early wavefront around 1997, but this subsequently decreases, perhaps due to intensive poultry culling (Yee et al. 2009). The higher contribution for Anatidae is particularly noticeable during the HPAIV H5N1 epidemic wave dynamics between 2004 and 2005 (fig. 3A).
Fig. 3.
Host-specific wavefront distance estimates for HA and NA (A). These estimates summarize for each host (Anatidae—Ana, Phasianidae—Pha, and Neoaves—Neo), the fraction of estimated amount of great circle distance from the phylogeographic origin to the wavefront that can be associated with that host according to the host ancestral reconstruction. Host-specific wavefront distance estimates for HAD and NAD are shown in supplementary figure S9, Supplementary Material online. Host-specific densities (B) and diffusion rates (C) summarized for the year 2004 from a joint spatial and host diffusion analysis for HA. The Ana and Pha samples in this analysis are separately represented in the rate (C) and density (B) plot for clarity. In the density plot (B), Ana and Pha densities are represented by a transparent blue and red color (in the online color figures), respectively. High and low dispersal rates are represented by a red-yellow color gradient in the rate plot (in the online color figures) (C).
To demonstrate how the Anatidae-driven expansion spatially manifests in the invasion dynamics, we summarize phylogeographic realizations by host in the posterior distribution of trees on a fine spatial grid to visualize both host-specific densities and dispersal rates through time and space (cfr. Materials and Methods). In figure 3B and C, we focus on 2004 in the HA analysis, which marks the acceleration in wavefront for both HA and NA (fig. 3A). In agreement with the host-specific wavefront contribution (fig. 3A), this demonstrates that the forefront of the westward expansion appears to be dominated by the Anatidae host. This finding does not reflect a biased sampling to the west of Qinghai lake, which includes 53 sequences from Anatidae, 46 from Phasianidae, and 21 samples from Neoaves. In agreement with accelerated spread, the westward expansion through Anatidae is also characterized by relatively high rates of dispersal (fig. 3C).
In Indonesia, the almost exclusive sampling of Phasianidae explains the highly specific host density in this region. Despite this however, the seeding from South China is still at least equally likely to have occurred in association with the Anatidae host (cfr. the connecting density in fig. 3B). A high density of Phasianidae samples generally coincides with areas of relatively low dispersal rates (fig. 3B), which may point to more localized persistence dynamics in poultry.
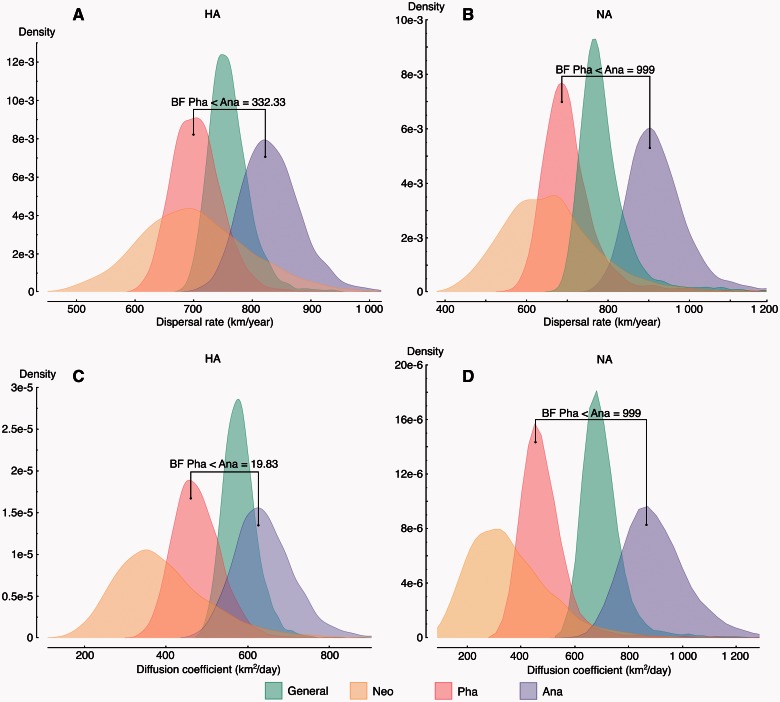
Host-specific diffusion statistics also allow to quantify the overall rate of viral spread and the intrinsic diffusivity of the infected host through the diffusion coefficient (Pybus et al. 2012) (fig. 4). These estimates reveal a significantly higher rate of spread and diffusivity in Anatidae as compared with Phasianidae. Due to a relatively large degree of uncertainty, the posterior distributions for these statistics overlap, most notably for the rate of spread. Therefore, to formally asses the difference between Anatidae and Phasianidae, we compute Bayes factor (BF) support based on the prior and posterior expectation that a statistic is larger for one host compared with the other (cfr. Materials and Methods). For both the diffusion rate (BF = 332 and 999 for HA and NA, respectively) and the diffusion coefficient (BF = 20 and 999 for HA and NA, respectively), this provides strong support for higher estimates for Anatidae. Rates of diffusion and intrinsic diffusivity in the Neoaves host appear as low or lower than that for Phasianidae. However, due to the sparsity of sampling from this host order, we caution against drawing strong conclusions for the contribution of Neoaves to HPAIV H5N1 spread from this analysis.
Fig. 4.
Posterior dispersal rate (top) and diffusion coefficient (bottom) distributions for each host (from left to right: yellow—Neoaves, red—Phasianidae, greengeneral, and blue—Anatidae, in the online color figures) obtained by the joint host analysis of HA (left) and NA (right). Posterior dispersal rate and diffusion coefficient distributions for differently downsampled data sets are shown in supplementary figure S10, Supplementary Material online.
Discussion
In this phylodynamic study, we combine spatial and host dynamics to disentangle the contributions of different avian hosts to the viral invasion dynamics. We first reconstructed the spatial diffusion dynamics using different Bayesian phylogenetic diffusion models and demonstrated that the spread of HPAIV H5N1 is a heterogeneous process that is structured according to avian flyways in Asia and Russia. Together with dispersal pathways that bridge large geographic distances (Mongolia/North China to West Russia/Kazakhstan), this suggests a role for wild bird migration in the expansion dynamics of H5N1. In a second part, we infer host transmission dynamics to demonstrate that highly asymmetric jumping between Anatidae and Phasianidae underlies the patterns of spread. Anatidae have driven the epidemic wavefront, in particular for the incursion of the area coinciding with the Central Asian flyway. These findings complement risk modeling and satellite-tracked wild waterfowl studies in suggesting that the continental-scale dynamics of HPAIV H5N1 are structured into persistence areas delineated by domestic ducks and connected by relatively rare transmission through migratory waterfowl (Gilbert et al. 2010).
Although we identify a wavefront dominated by Anatidae and a faster and more diffusive viral spread in this avian family, it remains difficult to determine the exact contributions of domestic and wild ducks to these dynamics because this information was unavailable for many of the isolates we investigated. The Anatidae isolates for which this information was available appeared to be a roughly equal mix between wild and domestic, whereas most Phasianidae isolates were known to be sampled from domestic hosts. Long-distance dispersal events, like the ones we identify between the region of Mongolia and North China and the region of West Russia and Kazakhstan, can be attributed to wild migratory birds, but the tempo of spatial spread in domestic ducks may also be higher than in chickens because of differences in farming practices. In southeast Asian countries in particular, duck flocks are allowed to free-graze nearby wet lands or rice cultivations, not only because of pest control and fertilization advantage but also because it cuts down food consumption by up to 50% (Gilbert et al. 2007; Pfeiffer et al. 2013). Here, domestic ducks may become infected as silent carriers of HPAIV H5N1, and subsequently spread the disease to other poultry flocks (Phasianidae) when returning to their farms (Songserm et al. 2006; Kim et al. 2009; Cappelle et al. 2014). Transmission in the opposite direction may also sustain spillover from domestic poultry to wild birds, which is considered to be a unique feature of HPAIV H5N1 (Gilbert et al. 2010). If a distinction between domesticated and wild birds could be made for the currently available sequence data, it may be possible to further quantify differences in spatial spread within the avian hosts families. However, given the potentially close contact between domesticated and wild ducks as suggested above, disentangling these transmission patterns is also likely to require a denser sampling of the different hosts in space and time and a better differentiation between domestic and wild ducks in the sequence data annotation.
Our analyses do not argue against movement of poultry as playing an important role for HPAIV H5N1 spread, but this is likely to be less important for connecting more distant areas of persistence on the geographic scale that we consider here. The importance of domesticated ducks, as also recognized by HPAIV H5N1 risk mapping (Hulse-Post et al. 2005), explains why controlling this virus has been particularly challenging. Compared with chickens, domestic ducks not only exhibit fewer clinical signs of disease while shedding virus up to 17 days (Hulse-Post et al. 2005), but vaccination is difficult to implement for ducks because they have longer production cycles and show lower vaccine efficiency, requiring repeated injections throughout the animal’s life (Tian et al. 2005).
The impact of bird migration on HPAIV H5N1 spread has very recently been highlighted using a combination of HPAIV H5N1 outbreak records, whole-genome data, and satellite tracking data (Tian et al. 2015). This demonstrated that virus migration across six regions in Asia coincided well with bird migration networks in both the East Asian–Australasian flyway and the Central Asian flyway for HPAIV H5N1 clade 2.3.2. We expand the scale of this study and recover similar gene flow dynamics with a central connecting role for Mongolia and North China, including lake Qinghai. Lake Qinghai is an important breeding place for migratory bird species in East Asia, and wild waterfowl were also likely to be responsible for viral spread through Mongolia because it has very few poultry production areas (Tian et al. 2005; Gilbert et al. 2012). As predicted earlier by satellite-tracked waterfowl (Gilbert et al. 2010), and confirmed by recent satellite tracking and phylogeographic analysis (Tian et al. 2015), we also identified a link between this northern area and South Asia (Bangladesh, and connected with this, India; fig. 1). We further complement these findings by demonstrating relatively frequent long-distance dispersal to West Russia and Kazakhstan. This is likely to be associated with wild bird migration to the west Siberian lowlands, which is an important breeding area and by far the largest wetlands in the world (Gilbert et al. 2006).
As an extension of the GLM diffusion model (Lemey et al. 2014), we here introduced random effects to identify exceptions to diffusion modelled by a specific predictor. The rationale of this approach is that if a predictor does not explain the intensity of viral migration very well between a specific pair of locations, then an additional effect for the corresponding rate may be needed to adequately explain the location transitions. As for a standard discrete phylogeographic analysis, this requires a large number of parameters to be estimated from a single observation of location states at the tip of the tree, and as a consequence, random effects estimates will be associated with high uncertainties. For this reason, we only consider these effects to be important if the estimates yield consistently high absolute effects on a log scale, which may be a fairly conservative approach. In the future, it may be useful to explore search variable selection approaches that try to identify a restricted number of nonzero effects, but current prior specification on the random effects complicates such strategies.
Similar to a recent WNV analysis (Pybus et al. 2012), our continuous phylogeographic reconstruction indicates a considerable heterogeneity in the spatial HPAIV H5N1 diffusion dynamics. In fact, the diffusion coefficient we estimate for HPAIV H5N1 is higher than for WNV (supplementary fig. S6, Supplementary Material online), suggesting an even stronger impact of long-range movements on avian influenza spread. Here, we hypothesized that the host dynamics are responsible for a large degree of the heterogeneity in spatial spread and we attempt to quantify this based on summaries of a joint analysis of continuous spatial and discrete host traits. We rely on stochastic mapping techniques to obtain realizations of the host jumping process throughout the posterior distribution of trees and summarize the continuous spatial diffusion process according to host-specific trajectories in these jump histories. The ability to find clear differences in spatial statistics (e.g., fig. 4), and in spatial visualizations (e.g., fig. 3), by host using this approach is remarkable because the current analysis integrates host transmission and spatial diffusion as conditionally independent processes. Ancestral trait reconstruction processes are inherently uncertain, and the statistics that incorporate time inherit also the uncertainty of the phylogenetic divergence time estimation. This hints at the potential of future approaches that would aim at introducing an estimable dependency between the two trait processes, such that cross talk is allowed between the different processes and different ancestral reconstructions can borrow strength from each other.
Despite efforts to downsample particular locations to arrive at equitable numbers of sequences, we acknowledge that sample sizes for both location and host have an important impact on ancestral reconstruction approaches, and the results we obtained for the discrete location estimation are specific to the spatial partitioning we applied. In addition, we note that the sequence data we examined generally lacked detailed annotation on the specific sampling date (day and month), and not all isolates could be assigned to a specific city or region within a country. Sampling time uncertainty can now readily be taken into account in recent versions of the BEAST software and there are also possibilities to accommodate the location uncertainty in continuous spatial diffusion (Bouckaert et al. 2012; Nylinder et al. 2014). However, given the time-scale and geographic scale of our analysis, we do not expect that these aspects will considerably impact our estimates and the main point of attention probably remains representative spatial and host sampling, and specific knowledge about the host (domestic or wild hosts, as argued above).
The framework we introduce here and the model extensions we hint at may find applications to various pathogen systems for which the spatial dynamics involve a multiplicity of host species. The approach is also not restricted to spatial applications and readily generalizes to any trait of interest. Staying close to the current application however, the emergence and continuing spread of avian influenza A H7N9 in multiple waves is now receiving considerable attention (Lam et al. 2013; Gilbert et al. 2014). Despite the pandemic potential and high mortality rate of these viruses in humans, low pathogenic avian influenza virus H7N9 seems to mainly affect poultry in live markets. Although the importance of live poultry market networks has also been demonstrated for the persistence and spread of HPAIV H5N1, the relationship between disease and markets appears to be even stronger for H7N9 (Cowling et al. 2013; Han et al. 2013; Li et al. 2014). However, H7N9 spread has remained difficult to contain along the poultry market chain, and as pointed out by Gilbert et al. (2014), together with the persistence between subsequent waves, this may indicate H7N9 spread beyond the distribution of human cases. Surveillance may need to take this into account as well as possible spillover at the domestic–wild bird interface.
In conclusion, we have introduced a framework to simultaneously reconstruct the spatial dynamics and transmission patterns among different hosts in time-calibrated phylogenies. By applying this to a comprehensive data set of H5N1 sequences sampled throughout the HPAIV H5N1 expansion across Asia and Russia, we demonstrate differences in the tempo and mode of spread between different avian hosts and a key role for Anatidae in the invasion dynamics. Such phylodynamic analyses contribute to our understanding of the impact of specific hosts on pathogen dispersal and may assist disease surveillance and pandemic preparedness.
Materials and Methods
Data Collection
To assemble comprehensive genetic data sets representative of the HPAIV H5N1 expansion, we first retrieved the HA and NA gene sequences for all available () Russian and Asian HPAIV H5N1 avian isolates from the public Influenza Research Database (www.fludb.org accessed on January 11, 2012). From this sequence set, we selected the strains with known sampling location, sampling year (ranging from 1996 to 2011) and host, for which both an HA and NA sequences were available. In case identical HA or NA sequences were present, we retained only a single isolate and we also excluded sequences with less than 75% of the total length of the segment. The sequences were grouped by country of sampling, and also by Province for China, to investigate which discrete partitioning scheme (according to geography and administrative borders) would enable downsampling to roughly equitable numbers of sequences per location. This led us to condition on 19 geographic regions to subsample the HA and NA sequence data to obviate some of the biases in surveillance intensity in the different geographic regions (supplementary table S1, Supplementary Material online). More detail on the subsampling procedure is provided in the supplementary materials and methods, Supplementary Material online. In the section describing the discrete phylogeographic diffusion model we extend in this study, we describe how we partition the subsampled data in a geographically more coherent way, arriving at the data sets summarized in supplementary figure S1 and file S1, Supplementary Material online.
The sequences of each genome segment were aligned using the Multiple Alignment using Fast Fourier Transform (MAFFT) program version 6.864b (Katoh and Toh 2008) and manual editing was carried out to delete indels occurring in more than 50% of the sequences.
Bayesian Evolutionary Inference
Our interest lies in integrating spatial diffusion and host jumping processes with sequence evolution in a phylogenetic context. Although our Bayesian framework allows tackling such problems using a joint inference approach, it remains computationally challenging for the data set sizes we examine here, in particular when model testing needs to be performed (Baele et al. 2012, 2013; Baele and Lemey 2013). Because of this and the fact that both diffusion processes are modelled independently from the substitution process throughout evolutionary history, we split up the inference problem into two steps: We first focus on the sequence evolution process to generate an empirical distribution of trees, and then condition on this set of posterior trees to fit several discrete or continuous trait diffusion processes. In the supplementary materials and methods, Supplementary Material online, we describe how we model the more standard sequence evolutionary process.
All Markov chain Monte Carlo (MCMC) sampling analyses were performed using BEAST in conjunction with the Broad-platform Evolutionary Analysis General Likelihood Evaluator library to enhance computation speed (Suchard and Rambaut 2009; Ayres et al. 2012). A subset of 500 trees were selected from the combined posterior distribution and used as an empirical distribution in the subsequent spatial and host diffusion inference. Following Pagel et al. (2004), we achieved this by incorporating a proposal mechanism that randomly draws a new tree from the empirical distribution (Lemey et al. 2014). We used TreeAnnotator to summarize a maximum clade credibility (MCC) tree and FigTree version 1.4.1 to visualize annotated trees.
Discrete Geography
We estimated spatial diffusion dynamics among a set of 19 geographic regions using a Bayesian discrete phylogeographic approach (Lemey et al. 2009). This approach conditions on geographic locations recorded at the tips of the (empirical) HA and NA phylogenies and models the transition history among those locations as a CTMC process, allowing the inference of unobserved locations at the ancestral nodes in each tree of the posterior distribution. We used a nonreversible CTMC model (Edwards et al. 2011) and incorporated BSSVS to identify a sparse set of transition rates that adequately summarizes the epidemiological connectivity (Lemey et al. 2009). As part of these analyses, we also incorporated posterior inference of the complete Markov jump history through time (Minin and Suchard 2008; Lemey et al. 2014), allowing to quantify state transitions and the time spent in a particular location state along each phylogenetic branch.
The discrete diffusion model has recently been extended to test and quantify potential predictors of the diffusion process (Lemey et al. 2014). This has been achieved by adopting a GLM approach that parameterizes the log of the rates in the CTMC matrix as a log-linear function of several potential predictors of viral diffusion. Here, we extend this approach to identify long-distance dispersal dynamics that significantly stand out from a more regular, distance-based diffusion process by introducing random effects (supplementary materials and methods, Supplementary Material online).
Continuous Geography
As a complementary approach to discrete phylogeographic inference, we also estimated the HPAIV H5N1 diffusion dynamics in continuous space using a Brownian motion random walk mode (Lemey et al. 2010). We relax the constant-variance assumption of the standard Brownian diffusion model by allowing for branch-specific scaling factors that are drawn from an underlying distribution. We evaluate different probability distributions (Cauchy, gamma, lognormal) as RRW model and compare it with the homogenous process (Lemey et al. 2010) using log marginal likelihoods obtained by path sampling and stepping stone sampling (Baele et al. 2012, 2013; Baele and Lemey 2013).
To draw inference under these models, we employ a Bayesian inference method that incorporates a dynamic programming approach to achieve an analytic solution for the trait likelihood (integrating over all possible realizations of the unobserved traits; Pybus et al. 2012). To visualize the dispersal process through time, we use the Spatial Phylogenetic Reconstruction of Evolutionary Dynamics application (Bielejec et al. 2011) to complement an MCC tree with location kernel density estimates through time and to convert these estimates to the keyhole markup language format.
The spatiotemporal realizations of the continuous diffusion process can be used to draw different statistics about the spatial epidemic dynamics without the need to account for autocorrelation as is required for mathematical models in spatial ecology (Pybus et al. 2012). Here, we use several such statistics, either for the entire evolutionary history or for a particular time interval, to characterize the dispersal dynamics of HPAIV H5N1, including the dispersal rate or the phylogenetic great-circle distance travelled per unit time (km/year) and the wavefront distance, which tracks the largest great-circle distance between each phylogenetic lineage and the estimated origin of the epidemic at a particular time.
We also employ an estimator of the diffusion coefficient, , a fundamental ecological measure of the intrinsic diffusivity of infected hosts. Pybus et al. (2012) showed that this measure can be estimated using
| (1) |
where di represents the great-circle distance covered along a branch, ti represents the time elapsed on the branch, and n represents the number of branches in the phylogeny. This estimator can however be sensitive to uncertain diffusion and time estimates for very short branches and hence suffer from high variances. Therefore, we here explore a weighted average alternative that divides the total squared phylogenetic displacement by four times the total phylogenetic time in the evolutionary history:
| (2) |
This statistic arrives at consistently lower estimates with considerable less variance (supplementary fig. S9, Supplementary Material online).
Combining Host and Geography
Phylogenetic diffusion models are general trait evolutionary models and are therefore not restricted to spatial locations (Faria et al. 2013; Bedford et al. 2014). We exploit this generality to examine HPAIV H5N1 host transmission throughout the viral evolutionary history. Given the multitude of avian hosts from which HPAIV H5N1 viruses were samples, we follow a coarse-grained approach and consider transmission among Anatidae, Phasianidae, and Neoaves hosts using a nonreversible CTMC model. As with the discrete location inference, we also map the complete Markov jump history for the host trait in the posterior tree distribution and summarize the number of Markov jumps as well as the time spent in a particular host populations (Markov rewards).
To formally assess the directionality and symmetry in viral jumps between the two largest host populations (Anatidae and Phasianidae), we follow Mather et al. (2013) in setting up four different models: Two unidirectional models for Anatidae to Phasianidae jumps and vice versa, by fixing one of the rates in a nonreversible model to zero, and two bidirectional models, including one that enforces symmetric transition rates and another that allows for different intensities in host jumping. We compare the different models using log marginal likelihoods obtained through stepping stone sampling (Baele et al. 2012, 2013; Baele and Lemey 2013).
To help elucidate the role of different avian host populations in the expansion dynamics of HPAIV H5N1, we incorporate both a continuous spatial diffusion process and a discrete host transmission process in a single Bayesian analysis. Although both processes are modelled independently, the joint inference allows us to summarize host-specific contributions to the spatial dispersal dynamics. To this end, we map the complete host trait history in the posterior tree distribution and condition on this to delineate host-specific trajectories in the phylogeographic history. If branches would be entirely associated with one specific host population, then the host-specific summaries described above (e.g., the dispersal rate or diffusion coefficient) can be obtained by conditioning on the relevant branch set identified by the host mapping. However, a CTMC process realization involves transmission among (potentially multiple) discrete states at specific time points along a branch. In this case, we impute the bivariate location at these time points and draw our statistics from the compound host-specific trajectories.
We provide new statistics (implemented as of BEAST v1.8.1) to obtain such estimates. To evaluate differences in spatial statistics between two different hosts, we conduct a BF test (Suchard et al. 2005) that expresses the posterior odds over the prior odds that the statistic for host i is larger than for host j. To determine the posterior odds, we set up an indicator function that expresses this difference and note that its MCMC sample average converges to the posterior probability of that model.
In addition to host-specific statistics, we developed a novel visualization approach of continuous phylogeographic dispersal on a two-dimensional grid (described in detail in supplementary materials and methods, Supplementary Material online) that integrates three dimensions (time, space, and host).
Supplementary Material
Supplementary file S1, tables S1–S7, figures S1–S11, and materials and methods are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under Grant Agreement No. 278433-PREDEMICS and ERC Grant Agreement No. 260864, the National Institute of Health under R01 AI107034 and R01 HG006139, and the National Science Foundation under DMS 1264153.
References
- Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT, Lewis PO, Huelsenbeck JP, Ronquist F, Swofford DL, Cummings MP, et al. 2012. Beagle: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol. 61(1):170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Lemey P. 2013. Bayesian evolutionary model testing in the phylogenomics era: matching model complexity with computational efficiency. Bioinformatics 29(16):1970–1979. [DOI] [PubMed] [Google Scholar]
- Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 29(9):2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Lemey P, Vansteelandt S. 2013. Make the most of your samples: Bayes factor estimators for high-dimensional models of sequence evolution. BMC Bioinformatics 14:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, McCauley JW, Russell CA, Smith DJ, Rambaut A. 2014. Integrating influenza antigenic dynamics with molecular evolution. Elife 3:e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bett B, Henning J, Abdu P, Okike I, Poole J, Young J, Randolph TF, Perry BD. 2014. Transmission rate and reproductive number of the H5N1 highly pathogenic avian influenza virus during the December 2005-July 2008 epidemic in Nigeria. Transbound Emerg Dis. 61(1):60–68. [DOI] [PubMed] [Google Scholar]
- Bielejec F, Rambaut A, Suchard MA, Lemey P. 2011. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27(20):2910–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist EW, Lemey P, Suchard MA. 2010. Three roads diverged? Routes to phylogeographic inference. Trends Ecol Evol. 25(11):626–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Lemey P, Dunn M, Greenhill SJ, Alekseyenko AV, Drummond AJ, Gray RD, Suchard MA, Atkinson QD. 2012. Mapping the origins and expansion of the Indo-European language family. Science 337(6097):957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappelle J, Zhao D, Gilbert M, Nelson MI, Newman SH, Takekawa JY, Gaidet N, Prosser DJ, Liu Y, Li P, et al. 2014. Risks of avian influenza transmission in areas of intensive free-ranging duck production with wild waterfowl. Ecohealth 11:109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith GJD, Li KS, Wang J, Fan XH, Rayner JM, Vijaykrishna D, Zhang JX, Zhang LJ, Guo CT, et al. 2006. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc Natl Acad Sci U S A. 103(8):2845–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling BJ, Jin L, Lau EHY, Liao Q, Wu P, Jiang H, Tsang TK, Zheng J, Fang VJ, Chang Z, et al. 2013. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet 382(9887):129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CJ, Suchard MA, Lemey P, Welch JJ, Barnes I, Fulton TL, Barnett R, O’Connell TC, Coxon P, Monaghan N, et al. 2011. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol. 21(15):1251–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Suchard MA, Rambaut A, Lemey P. 2011. Toward a quantitative understanding of viral phylogeography. Curr Opin Virol. 1(5):423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria NR, Suchard MA, Rambaut A, Streicker DG, Lemey P. 2013. Simultaneously reconstructing viral cross-species transmission history and identifying the underlying constraints. Philos Trans R Soc Lond B Biol Sci. 368(1614):20120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RAM, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus ADME. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 79(5):2814–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournié G, Guitian J, Desvaux S, Cuong VC, Dung DH, Pfeiffer DU, Mangtani P, Ghani AC. 2013. Interventions for avian influenza A (H5N1) risk management in live bird market networks. Proc Natl Acad Sci U S A. 110(22):9177–9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Golding N, Zhou H, Wint GRW, Robinson TP, Tatem AJ, Lai S, Zhou S, Jiang H, Guo D, et al. 2014. Predicting the risk of avian influenza A H7N9 infection in live-poultry markets across Asia. Nat Commun. 5:4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Jambal L, Karesh WB, Fine A, Shiilegdamba E, Dulam P, Sodnomdarjaa R, Ganzorig K, Batchuluun D, Tseveenmyadag N, et al. 2012. Highly pathogenic avian influenza virus among wild birds in Mongolia. PLoS One 7(9):e44097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Newman SH, Takekawa JY, Loth L, Biradar C, Prosser DJ, Balachandran S, SubbaRao MV, Mundkur T, Yan B, et al. 2010. Flying over an infected landscape: distribution of highly pathogenic avian influenza H5N1 risk in South Asia and satellite tracking of wild waterfowl. Ecohealth 7(4):448–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Xiao X, Chaitaweesub P, Kalpravidh W, Premashthira S, Boles S, Slingenbergh J. 2007. Avian influenza, domestic ducks and rice agriculture in Thailand. Agric Ecosyst Environ. 119:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M, Xiao X, Domenech J, Lubroth J, Martin V, Slingenbergh J. 2006. Anatidae migration in the western Palearctic and spread of highly pathogenic avian influenza H5NI virus. Emerg Infect Dis. 12(11):1650–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Jin M, Zhang P, Liu J, Wang L, Wen D, Wu X, Liu G, Zou Y, Lv X, et al. 2013. Epidemiological link between exposure to poultry and all influenza A(H7N9) confirmed cases in Huzhou city, China, March to May 2013. Euro Surveill. 18(20). [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, deWit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, et al. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336(6088):1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill NJ, Takekawa JY, Ackerman JT, Hobson KA, Herring G, Cardona CJ, Runstadler JA, Boyce WM. 2012. Migration strategy affects avian influenza dynamics in mallards (Anas platyrhynchos). Mol Ecol. 21(24):5986–5999. [DOI] [PubMed] [Google Scholar]
- Holmes EC. 2008. Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol. 62:307–328. [DOI] [PubMed] [Google Scholar]
- Hulse-Post DJ, Sturm-Ramirez KM, Humberd J, Seiler P, Govorkova EA, Krauss S, Scholtissek C, Puthavathana P, Buranathai C, Nguyen TD, et al. 2005. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc Natl Acad Sci U S A. 102(30):10682–10687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9(4):286–298. [DOI] [PubMed] [Google Scholar]
- Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyer WE, van Lavieren R, Osterhaus ADME, Fouchier RAM, Kuiken T. 2008. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1). Emerg Infect Dis. 14(4):600–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-K, Negovetich NJ, Forrest HL, Webster RG. 2009. Ducks: the ”Trojan horses” of H5N1 influenza. Influenza Other Respir Viruses. 3(4):121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT-Y, Wang J, Shen Y, Zhou B, Duan L, Cheung C-L, Ma C, Lycett SJ, Leung CY-H, Chen X, et al. 2013. The genesis and source of the H7N9 influenza viruses causing human infections in china. Nature 502(7470):241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, Baele G, Russell CA, Smith DJ, Pybus OG, Brockmann D, et al. 2014. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 10(2):e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput Biol. 5(9):e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Welch JJ, Suchard MA. 2010. Phylogeography takes a relaxed random walk in continuous space and time. Mol Biol Evol. 27(8):1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FCK, Choi BCK, Sly T, Pak AWP. 2008. Finding the real case-fatality rate of H5N1 avian influenza. J Epidemiol Community Health. 62(6):555–559. [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, et al. 2014. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med. 370:520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, et al. 2013. Distinguishable epidemics of multidrug-resistant Salmonella typhimurium dt104 in different hosts. Science 341(6153):1514–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin VN, Suchard MA. 2008. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol. 56(3):391–412. [DOI] [PubMed] [Google Scholar]
- Nelson MI, Holmes EC. 2007. The evolution of epidemic influenza. Nat Rev Genet. 8(3):196–205. [DOI] [PubMed] [Google Scholar]
- Nylinder S, Lemey P, DeBruyn M, Suchard MA, Pfeil BE, Walsh N, Anderberg AA. 2014. On the biogeography of Centipeda: a species-tree diffusion approach. Syst Biol. 63(2):178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M, Meade A, Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 53(5):673–684. [DOI] [PubMed] [Google Scholar]
- Pfeiffer DU, Otte MJ, Roland-Holst D, Zilberman D. 2013. A one health perspective on HPAI H5N1 in the Greater Mekong sub-region. Comp Immunol Microbiol Infect Dis. 36:309–319. [DOI] [PubMed] [Google Scholar]
- Poovorawan Y, Pyungporn S, Prachayangprecha S, Makkoch J. 2013. Global alert to avian influenza virus infection: from H5N1 to H7N9. Pathog Glob Health. 107(5):217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A. 2009. Evolutionary analysis of the dynamics of viral infectious disease. Nat Rev Genet. 10(8):540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Suchard MA, Lemey P, Bernardin FJ, Rambaut A, Crawford FW, Gray RR, Arinaminpathy N, Stramer SL, Busch MP, et al. 2012. Unifying the spatial epidemiology and molecular evolution of emerging epidemics. Proc Natl Acad Sci U S A. 109(37):15066–15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell CA, Fonville JM, Brown AEX, Burke DF, Smith DL, James SL, Herfst S, van Boheemen S, Linster M, Schrauwen EJ, et al. 2012. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science 336(6088):1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songserm T, Jam-on R, Sae-Heng N, Meemak N, Hulse-Post DJ, Sturm-Ramirez KM, Webster RG. 2006. Domestic ducks and H5N1 influenza epidemic, Thailand. Emerg Infect Dis. 12(4):575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Rambaut A. 2009. Many-core algorithms for statistical phylogenetics. Bioinformatics 25(11):1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard MA, Weiss RE, Sinsheimer JS. 2005. Models for estimating Bayes factors with applications to phylogeny and tests of monophyly. Biometrics 61(3):665–673. [DOI] [PubMed] [Google Scholar]
- Tian G, Zhang S, Li Y, Bu Z, Liu P, Zhou J, Li C, Shi J, Yu K, Chen H. 2005. Protective efficacy in chickens, geese and ducks of an H5N1-inactivated vaccine developed by reverse genetics. Virology 341(1):153–162. [DOI] [PubMed] [Google Scholar]
- Tian H, Zhou S, Dong L, VanBoeckel TP, Cui Y, Wu Y, Cazelles B, Huang S, Yang R, Grenfell BT, et al. 2015. Avian influenza H5N1 viral and bird migration networks in Asia. Proc Natl Acad Sci U S A. 112(1):172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To KKW, Chan JFW, Chen H, Li L, Yuen K-Y. 2013. The emergence of influenza A H7N9 in human beings 16 years after influenza A H5N1: a tale of two cities. Lancet Infect Dis. 13(9):809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DAA, Chen L-M, Recuenco S, Ellison JA, Davis CT, York IA, et al. 2012. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 109(11):4269–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog. 9(10):e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrancken B, Rambaut A, Suchard MA, Drummond A, Baele G, Derdelinckx I, VanWijngaerden E, Vandamme A-M, VanLaethem K, Lemey P. 2014. The genealogical population dynamics of HIV-1 in a large transmission chain: bridging within and among host evolutionary rates. PLoS Comput Biol. 10(4):e1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RG, Fitch WM. 2008. Influenza A H5N1 immigration is filtered out at some international borders. PLoS One 3(2):e1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RG, Hodac H, Lathrop RH, Fitch WM. 2007. A statistical phylogeography of influenza A H5N1. Proc Natl Acad Sci U S A. 104(11):4473–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Luo J, Wang J, Su W, Gao S, Zhang M, Xie L, Ding H, Liu S, Liu X, et al. 2014. Novel human H7N9 influenza virus in China. Integr Zool. 9(3):372–375. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev. 56(1):152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Peiris M, Chen H, Guan Y. 2006. H5N1 outbreaks and enzootic influenza. Emerg Infect Dis. 12(1):3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2015. Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO. Geneva (Switzerland): World Health Organization. [Google Scholar]
- Worobey M, Han G-Z, Rambaut A. 2014. A synchronized global sweep of the internal genes of modern avian influenza virus. Nature. 508:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Gilbert M, Slingenbergh J, Lei F, Boles S. 2007. Remote sensing, ecological variables, and wild bird migration related to outbreaks of highly pathogenic H5N1 avian influenza. J Wildl Dis. 43(1):40–47. [PMC free article] [PubMed] [Google Scholar]
- Xu X, Subbarao, Cox NJ, Guo Y. 1999. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 261(1):15–19. [DOI] [PubMed] [Google Scholar]
- Yee KS, Carpenter TE, Cardona CJ. 2009. Epidemiology of H5N1 avian influenza. Comp Immunol Microbiol Infect Dis. 32(4):325–340 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.