Abstract
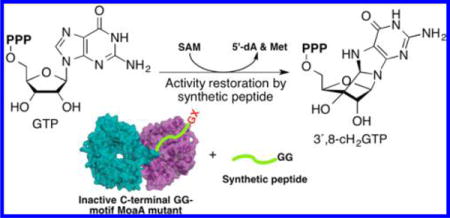
The molybdenum cofactor (Moco) is an essential redox cofactor found in all kingdoms of life. Genetic mutations in the human Moco biosynthetic enzymes lead to a fatal metabolic disorder, Moco deficiency (MoCD). Greater than 50% of all human MoCD patients have mutations in MOCS1A, a radical S-adenosyl-L-methionine (SAM) enzyme involved in the conversion of guanosine 5′-triphosphate (GTP) into cyclic pyranopterin monophosphate. In MOCS1A, one of the frequently affected locations is the GG motif constituted of two consecutive Gly at the C-terminus. The GG motif is conserved among all MOCS1A homologues, but its role in catalysis or the mechanism by which its mutation causes MoCD was unknown. Here, we report the functional characterization of the GG motif using MoaA, a bacterial homologue of MOCS1A, as a model. Our study elucidated that the GG motif is essential for the activity of MoaA to produce 3′,8-cH2GTP from GTP (GTP 3′,8-cyclase), and that synthetic peptides corresponding to the C-terminal region of wt-MoaA rescue the GTP 3′,8-cyclase activity of the GG-motif mutants. Further biochemical characterization suggested that the C-terminal tail containing the GG motif interacts with the SAM-binding pocket of MoaA, and is essential for the binding of SAM and subsequent radical initiation. In sum, these observations suggest that the C-terminal tail of MoaA provides an essential mechanism to trigger the free radical reaction, impairment of which results in the complete loss of catalytic function of the enzyme, and causes MoCD.
Graphical abstract
INTRODUCTION
The molybdenum cofactor (Moco) is an essential cofactor that mediates redox reactions in the active sites of enzymes found in all kingdoms of life.1,2 In all organisms, including humans, Moco requires de novo biosynthesis (Figure 1A).3 In humans, Moco-dependent enzymes are involved in various detoxification and catabolic pathways. Failure to produce Moco de novo due to a genetic disorder causes a fatal disease, Moco deficiency (MoCD), characterized by severe neurological symptoms such as progressive seizures and microcephaly.
Figure 1.
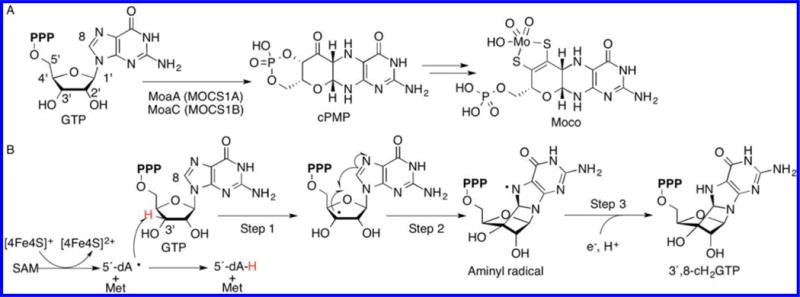
Moco biosynthesis. (A) Moco biosynthetic pathway in bacteria and humans. The human enzymes are indicated in parentheses. (B) Proposed reaction catalyzed by MoaA.11 MoaA was also proposed to perform rearrangement to pyranopterin triphosphate.12,13
In the reported MoCD patients studied, more than 50% of the mutations occur in molybdenum cofactor synthesis enzyme 1A (MOCS1A),4,5 which belongs to the radical S-adenosyl-L-methionine (SAM) superfamily.6 Enzymes in this superfamily catalyze the reductive cleavage of SAM using oxygen-sensitive 4Fe-4S clusters, and transiently generate a 5′-deoxyadenosyl radical (5′-dA•), which then abstracts a H-atom from the substrate to initiate radical reactions.7 During the Moco biosynthesis, MOCS1A, together with a product of the MOCS1B gene, is involved in the formation of cyclic pyranopterin monophosphate (cPMP) from guanosine 5′-triphosphate (GTP).8–10 The specific functions of MOCS1A and MOCS1B are currently under extensive investigation by multiple groups. The current consensus is that MOCS1A catalyzes an H-atom abstraction from the 3′ position of GTP using 5′-dA•, followed by a radical cyclization reaction to produce a unique cyclic nucleotide, 3′,8-cH2GTP (Figure 1B).11,12 Whether the subsequent rearrangement is catalyzed also by MOCS1A12,13 or by MOCS1B11 is the subject of extensive study. In this report, we will focus on the MOCS1A function where the consensus has already been achieved: the transformation of GTP into 3′,8-cH2GTP (GTP 3′,8-cyclase activity).
Of the MoCD-causing mutations frequently found in MOCS1A, the large majority occurs in three locations. Two of these hot spots, Cys ligands of the two 4Fe-4S clusters and the conserved Arg residues, are located in the active site (Figure 2A).10,14 On the other hand, the third hot spot, two consecutive Gly’s at the C-terminus (GG motif), was disordered in the reported X-ray crystal structures (Figure 2B), and it is unknown how their mutations cause MoCD. The GG motif is conserved in all MOCS1A homologues in prokaryotes and eukaryotes (highlighted in yellow in Figure 2C). The essential role of the GG motif for the function of MOCS1A was shown by heterologous expression of MOCS1A in Escherichia coli lacking the moaA gene (MOCS1A homologue).9 While wt-MOCS1A complemented the Moco biosynthesis in E. coli ΔmoaA strain, MOCS1A with mutations in the GG motif did not. Although these studies demonstrated the importance of the GG motif, the function of the motif and the mechanism by which its mutations cause MoCD have remained elusive.
Figure 2.
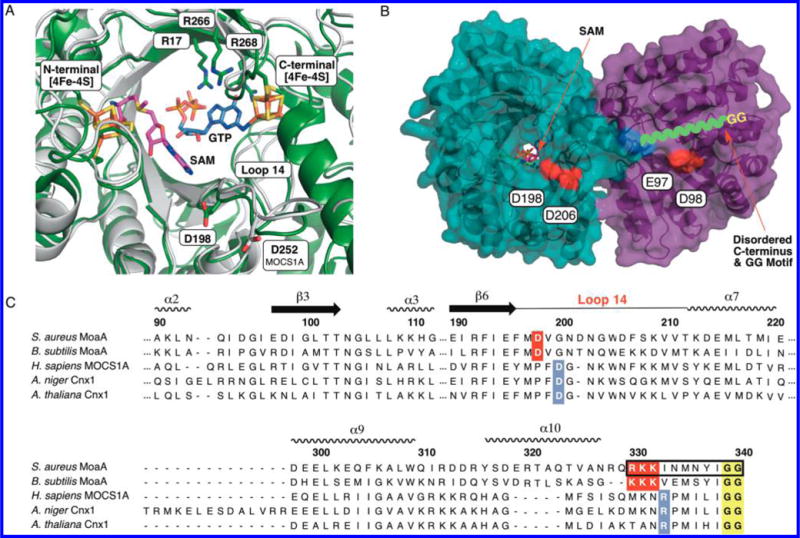
C-terminal GG motif in MoaA and MOCS1A. (A) Overlay of the crystal structure of MoaA in complex with GTP (blue sticks, PDB ID: 2FB3),14 and a homology model structure of MOCS1A (gray). The position of SAM (pink sticks) is based on the crystal structure of MoaA in complex with SAM (PDB ID: 1TV8).10 The homology model of MOCS1A was prepared on the basis of the reported MoaA structure (PDB ID: 1TV8) using SWISS-MODEL.15 Shown in sticks are the active-site residues frequently found mutated in MoCD patients (“hot spots”; Cys ligands of the two 4Fe-4S clusters and three active-site Arg). Also shown are D198, which is essential for the C-terminal tail interaction in S. aureus MoaA, and D252, its putative counterpart in human MOCS1A. (B) S. aureus MoaA homodimer in complex with SAM.10 The disordered C-terminal tail is drawn as a green wavy line, the last ordered residue, Q329, is highlighted in blue, and the acidic residues investigated for the C-terminal tail binding are highlighted in red. (C) Sequence alignment of MoaA/MOCS1A homologues. The GG motif is highlighted in yellow, the C-terminal tail/Loop 14 interface residues in bacteria are highlighted in red, and those predicted for eukaryotes are highlighted in blue. The C-terminal tail sequence corresponding to the 11-mer peptide is indicated in a black square.
C-terminal GG motifs have also been found in ubiquitin and ubiquitin-like proteins, in which the GG motif binds to the active site of partner enzymes and receives post-translational modifications at the C-terminal carboxylate.16,17 However, such modification is not known for MoaA/MOCS1A, and therefore a novel function for the GG motif was expected. To elucidate the role of the GG motif, we used Staphylococcus aureus MoaA as a model (36% sequence identity to human MOCS1A), and characterized the function of the GG motif by a combination of site-directed mutagenesis and peptide rescue. Our work demonstrated that synthetic peptides corresponding to the C-terminal amino acid sequence of wt-MoaA containing the GG motif rescue the catalytic activity of MoaA variants with mutations in the GG motif. Investigation of the peptide binding site using a combination of the MoaA variants and the peptide rescue assay revealed that the peptide and the C-terminal tail share the interaction interface in Loop 14 that constitutes a part of the SAM-binding pocket. Further biochemical and functional characterization of the GG-motif mutants using the peptide rescue assay suggested that the GG motif is critical for the binding of SAM and the subsequent radical initiation process. On the basis of these observations, we propose that the GG motif interacts with the active site of MoaA directly or through surrounded loops, and provides a gating mechanism to control the SAM binding and the reaction of 5′-dA• with GTP. These findings are in line with the emerging notion that protein conformational dynamics play a key role in controlling the radical initiation process in radical SAM enzymes.18 Furthermore, our demonstration of the peptide rescue of the MoCD-causing mutations highlights the possibility of developing novel therapy against human MoCD.
RESULTS
Functional Characterization of MoaA GG-Motif Mutants
To investigate the function of GG motif in vitro, we prepared and characterized MoaA variants with single-point mutations replacing Gly at the 339 or 340 position with Ala, Ser, or Val, or with 5 or 11 amino acid truncations at the C-terminus, producing Δ336–340 and Δ330–340 MoaA, respectively. All mutants were heterologously expressed in E. coli similar to the wild-type (wt) enzyme, and subsequently purified and anaerobically reconstituted to give holo-proteins with >90% purity. All of these mutants were reconstituted with 1.3–2.0 4Fe-4S clusters per monomer (Supporting Information, Figure S1b), comparable to that found in wt-MoaA (2.1 ± 0.03 4Fe-4S clusters per monomer), suggesting that the mutations in the GG motif have little effect on the cluster loading. Similarly, using anaerobic size exclusion chromatography in the presences of SAM and GTP, the single-point mutants migrated identical to that of wt-MoaA, indicating that the GG mutations are also likely not affecting the overall oligomeric state of MoaA (Figure S1c,d).
The catalytic functions of MoaA variants were investigated using a coupled assay with MoaC, which monitors the conversion of GTP into cPMP. Since cPMP is the known physiological biosynthetic intermediate in humans as well as bacteria,8 this assay likely represents the physiological catalytic function of MoaA. Based on this assay, no cPMP formation was observed in any of the tested mutants (Figure 3A, the lower limit of detection is 0.3% of the wt activity). We also investigated the GTP 3′,8-cyclase activity of MoaA (Figure 1A) in the absence of MoaC using the previously described HPLC method.11 No detectable amounts of 3′,8-cH2GTP (Figure 3B, the lower limit of detection is 0.04% of the wt activity) was observed in any of the mutants. These results suggest that even a minimal perturbation to the GG motif, such as an introduction of a methyl group, completely abolishes the GTP 3′,8-cyclase activity without significantly affecting the cofactor loading or the overall oligomeric states of MoaA.
Figure 3.
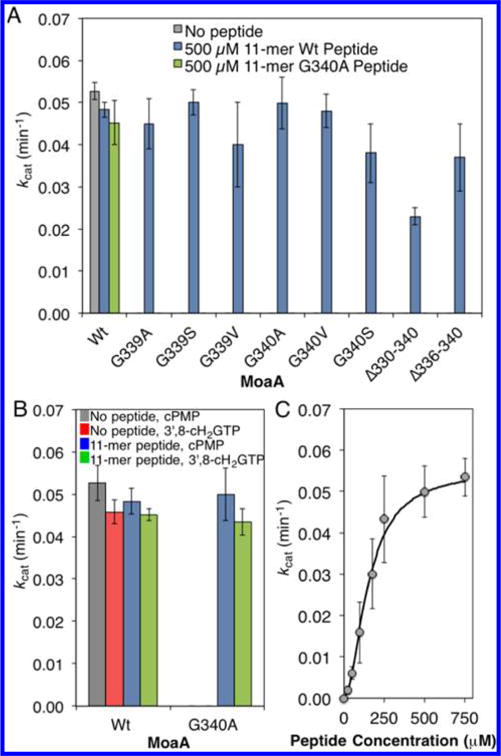
Peptide rescue of MoaA GG-motif mutant activity. (A) Catalytic activity of wt-MoaA and MoaA GG-motif mutants. The kcat values were determined on the basis of formation of cPMP in the MoaA/MoaC coupled assay in the absence (gray bar), or in the presence of the 11-mer wt peptide (500 μM, blue bars) or the 11-mer G340A peptide (500 μM, green bar). (B) Catalytic activity of wt- and G340A-MoaA in the absence (gray and red bars) or presence (blue and green bars) of the 11-mer wt peptide. The kcat values are based on the formation of cPMP in the MoaA/MoaC coupled assay (gray and blue bars) and those of 3′,8-cH2GTP in the absence of MoaC (red and green bars). (C) Activity of G340A-MoaA in the presence of specified concentrations of the 11-mer wt peptide. The activity was determined by the MoaA/MoaC coupled assay. The solid line is the nonlinear fit to the Hill equation,19 with kcat = 0.055 min−1, KD = 151 μM, and Hill coefficient = 1.9. All experiments were at least triplicated, and error bars represent standard deviation.
Peptide Rescue of MoaA GG-Motif Mutant Activity
The C-terminal tail of MoaA, including the GG motif (330RKKINMNYIGG340), is disordered in the reported X-ray crystal structures (Figure 2B), suggesting that the C-terminal tail may be conformationally flexible and interact with other parts of MoaA. Such transient interaction of the flexible C-terminal tail may be mimicked by synthetic peptides that correspond to MoaA’s C-terminal amino acid sequence. This hypothesis can be tested by investigating the ability of synthetic peptides to rescue the catalytic activity of the MoaA GG-motif mutants. Therefore, we synthesized peptides with amino acid sequence corresponding to the disordered region of the MoaA C-terminus, and tested for their ability to restore the in vitro catalytic function of the inactive MoaA GG-motif mutants. The MoaA/MoaC coupled assay revealed that the presence of the 11-mer peptide (RKKINMNYIGG, Figure 2C) restored the catalytic activity of all the tested GG-motif mutants (Figure 3A). The activity restoration was also observed in the absence of MoaC (Figure 3B), where MoaA was assayed by monitoring the formation of 3′,8-cH2GTP. The rate of 3′,8-cH2GTP formation by G340A-MoaA in the presence of the 11-mer wt peptide was comparable to that of cPMP formation, which corresponds to the increase in kcat values by >2500-fold. Thus, the 11-mer peptide rescues the GTP 3′,8-cyclase activity of the MoaA GG-motif mutants.
The observed activity restoration was dependent on the concentration of the peptide, on the basis of which the apparent Kd value for the interaction was determined (Figure 3C). The peptide titration curve exhibited an apparent cooperatively (Hill coefficient = 2.0 ± 0.2) with an apparent Kd value of 0.15 ± .04 mM, and a maximum activity (kcat = 0.054 ± 0.005 min−1) comparable to that of the wt-MoaA (Table 1, kcat = 0.042–0.045 min−1). An 11-mer peptide with a mutation in the GG motif (RKKINMNYIGA) did not restore the MoaA activity, even at 1.0 mM peptide concentration (Figure 3A), suggesting the critical role of the GG motif in the peptide. These results demonstrate the importance of the GG motif for the restoration of the GTP 3′,8-cyclase activity, suggesting that the activity restoration is mediated by a specific interaction between the synthetic peptide and MoaA.
Table 1.
Steady-State Kinetic Parameters for MoaA GG-Motif Mutants
| enzyme | peptide (μM)a | substrate | Km (μM)b | kcat (min−1)b | Kd (μM)b |
|---|---|---|---|---|---|
| MoaA Wt | 0 | GTP | 3.1 ± 0.67 | 0.042 ± 0.005 | 5.0 ± 3.0 |
| 0 | SAM | 5.1 ± 1.4 | 0.045 ± 0.007 | 1.7 ± 0.6 | |
| MoaA G339A | 0 | GTP | N.D.c | N.D.c | 0.46 ± 0.01 |
| 500 | 19 ± 1.8 | 0.045 ± 0.006 | −d | ||
| 0 | SAM | N.D.c | N.D.c | >90e | |
| 500 | 34 ± 2.7 | 0.046 ± 0.004 | −d | ||
| MoaA G340A | 0 | GTP | N.D.c | N.D.c | 5.2 ± 2.3 |
| 500 | 7.7 ± 3.8 | 0.047 ± 0.003 | −d | ||
| 0 | SAM | N.D.c | N.D.c | >90e | |
| 500 | 37 ± 14 | 0.037 ± 0.006 | −d | ||
| D198A-MoaA | 0 | GTP | 6.9 ± 0.73 | 0.0046 ± 0.0015 | −d |
| 0 | SAM | 36 ± 6.9 | 0.0039 ± 0.00074 | −d |
11-mer wt peptide was used.
Km and kcat were determined by the coupled assay with MoaC and HPLC quantitation of cPMP. Kd was determined by anaerobic ITC.
Not detected. The amount of cPMP formation was below the limit of detection.
Not tested.
Kd values were above the upper limit of detection for ITC analysis, 90 μM.
Site of the Peptide Binding
To obtain further insights about the peptide binding specificity, we investigated the activity restoration ability of shorter peptides, 8-mer (INMNYIGG) and 5-mer (NYIGG), truncated at the N-terminus. When these peptides were used in the activity assay of G340A-MoaA, both peptides exhibited activity restoration, but to an extent 10- and 100-fold lower than the 11-mer peptide, respectively (Figure 4A).
Figure 4.
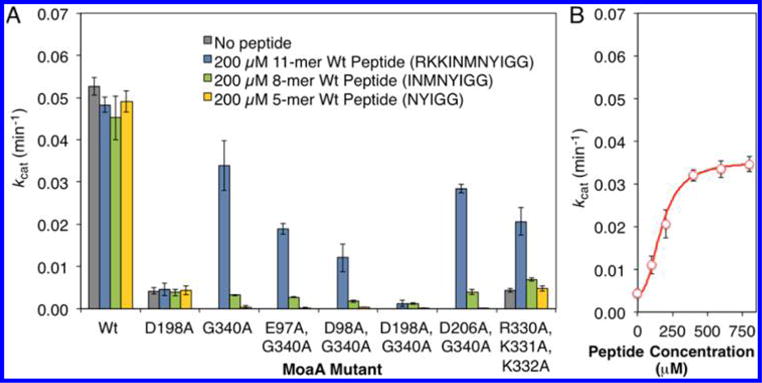
Investigation of the peptide binding site. (A) Activity of MoaA variants with mutations in the putative C-terminal tail interaction interface residues. The activity was determined by the MoaA/MoaC coupled assay in the absence (gray bar), or in the presence of the 11-mer (blue bars), 8-mer (green), or 5-mer (yellow) wt peptide (200 μM). (B) Activity of R330A/K331A/K332A-MoaA in the presence of specified concentrations of the 11-mer wt peptide. The activity was determined by the MoaA/MoaC coupled assay. The solid line is the nonlinear fit to Hill equation19 with kcat = 0.035 min−1, Kd = 172 μM, and Hill coefficient = 2.46. Rescue of the R330A/K331A/K332A-MoaA activity by the 11-mer peptide with Kd comparable to the rescue of G340A-MoaA provides strong evidence that the R330/K331/K332 residues are critical for the binding of the peptide as well as the C-terminal tail. All experiments were at least triplicated, and error bars represent standard deviation.
The difference in the activity restoration abilities between 11-and 8-mer peptides suggested that the three cationic residues (RKK) at the N-terminus of the 11-mer peptide may be important for the peptide binding to MoaA. This hypothesis predicted the presence of electrostatic interaction between the three cationic residues of the peptide and surface exposed acidic residue(s) of MoaA. In the reported crystal structures,10,14 four acidic residues (E97, D98, D198, and D206) were found within <20 Å from the last ordered residue at the C-terminus (Q329, Figure 2B). D198 and D206 are in Loop 14, which is located adjacent to the SAM binding site (Figure 2A,B) and contains F196 and M197 that directly interact with SAM. E97 and D98 are closer to the GTP binding site (Figure 2B). To investigate the potential interaction of these residues with the 11-mer peptide, we prepared MoaA double mutants harboring G340A and an additional Ala mutation in one of the acidic residues. The effects of the mutations were evaluated in the presence or absence of the peptides. If the mutated acidic residues are important for the peptide binding, significant loss of activity restoration is expected. Among the four mutants, only D198A/G340A-MoaA exhibited low-level activity restoration by the 11-mer peptide (2.6% of wt-MoaA, Figure 4A). In this mutant, the activities restored by the 11- and 8-mer peptides were within error (0.0012 ± 0.0007 and 0.0012 ± 0.0002, respectively). This observation suggests that in the absence of D198, the presence of three cationic residues (RKK) on the peptide no longer has any effect on the activity restoration. On the other hand, the activities of the other double mutants were restored by the 11-mer peptide to the extent comparable to that of wt-MoaA, and >5-fold greater than those by the 8-mer peptide. These observations demonstrate that D198 and the three cationic residues in the peptide (RKK) are both critical for the activity restoration of the MoaA GG-motif mutants by the synthetic peptide.
To investigate the relevance of the observed interaction between the peptide and Loop 14 to the function of C-terminal tail in the intact MoaA, we characterized D198A-MoaA and R330A/K331A/K332A-MoaA mutants. If the interaction between 11-mer peptide and MoaA mimics the interaction of the C-terminal tail in the intact MoaA, D198 and R330/K331/K332 are also important for the function of C-terminal tail in the interact MoaA. Thus, we prepared and tested the activity of D198A-MoaA and R330A/K331A/K332A-MoaA in the absence or presence of exogenous peptides. Without the peptide, the two mutants exhibited only 8% the activity of wt-MoaA (Supporting Information, Table S1, and Figure 4A). The observed activities of the two mutants were within errors of each other, and were also comparable to that observed for G340A-MoaA complemented with the 8-mer peptide without the three cationic residues (RKK). This observation may suggest that the loss of interaction between D198 and the three cationic residues results in activity ~8% that of wt-MoaA. Importantly, the activity of the R330A/K331A/K332A-MoaA triple mutant was restored by the 11-mer peptide (Figure 4A). The peptide titration curve (Figure 4B) showed a sigmoidal relation with an apparent Kd = 0.17 ± 0.030 mM and Hill coefficient = 2.5 ± 0.6, comparable to those observed in the peptide titration experiments for G340A-MoaA (Figure 3C, Table S1). The activity at the saturating concentration of the peptide was comparable to that of wt-MoaA (kcat = 0.035 ± 0.0022 min−1). No significant activity restoration was observed by the 8- and 5-mer peptides (Figure 4A). The activity of D198A-MoaA was not restored by any of the peptides (Figure 4A). These observations suggest that D198 and R330/K331/K332 are critical for the function of the C-terminal tail in the intact MoaA, and provide strong support that the synthetic 11-mer peptide binds to the location identical to that for the C-terminal tail binding, and the peptide rescue assay mimics the function of the C-terminal tail of wt-MoaC. Furthermore, the observation suggested that the C-terminal tail of MoaA interacts with D198 in Loop 14 that constitutes a part of the SAM binding pocket (Figure 2A).
The Role of MoaA GG Motif in Substrate Binding
With the peptide rescue assay established, we performed a detailed steady-state kinetic characterization of the peptide-rescued activities of the five GG-motif mutants (four single mutants at the 339 or 340 position, and one Δ330–340 truncated mutant) (Table 1, Table S1). In these assays, the 11-mer peptide was used at the saturating concentration (500 μM). In all tested mutants, in the presence of the peptide, kcat values (0.021–0.055 min−1) were comparable to that for wt-MoaA (0.045 ±0.007 min−1). On the other hand, increased Km values were observed for both substrates; 7–18-fold for SAM and 2–17-fold for GTP. The most striking effects were observed for SAM binding determined on the basis of the anaerobic isothermal titration calorimetry (ITC); no thermal binding was observed when SAM was titrated to mutant MoaA’s, suggesting that Kd > 90 μM (Table 1, Figure S2). wt-MoaA exhibited clear exothermic binding events with Kd = 1.7 ± 0.6 μM (Table 1, Figure S2). In contrast, no effects were detectable in the Kd values for GTP (0.5–5.2 μM in mutants vs 5.0 ± 3.0 μM in wt-MoaA). The observed decreases in the affinity for SAM are consistent with perturbation in the SAM binding in these GG-motif mutants. This observation parallels the interaction between the C-terminal tail and Loop 14 that constitutes a part of the SAM binding pocket.
The Role of MoaA GG Motif in H-Atom Transfer
We further investigated the potential involvement of the C-terminal tail in the radical initiation process. In the MoaA catalysis, SAM is reductively cleaved to transiently generate 5′-dA•, which then abstracts a H-atom from the 3′-position of GTP (Figure 1B).11,12 To investigate the perturbation in the H-atom-transfer step, we studied kinetic isotope effects (KIE) using [3′-D]GTP or GTP as substrate. In our previous study, we reported a small KIE for wt-MoaA reaction (1.28 ± 0.05),11 suggesting the kinetic masking of the H-atom transfer. In contrast, when G340A-MoaA was assayed in the presence the 11-mer wt peptide, a significantly greater KIE of 3.0 ± 0.30 was observed (Figure 5A, Figure S3). The significant increase in the KIE suggests that the H-atom abstraction is more rate-limiting in G340A-MoaA compared to the wt enzyme, suggesting the perturbation in the H-atom-transfer step.
Figure 5.
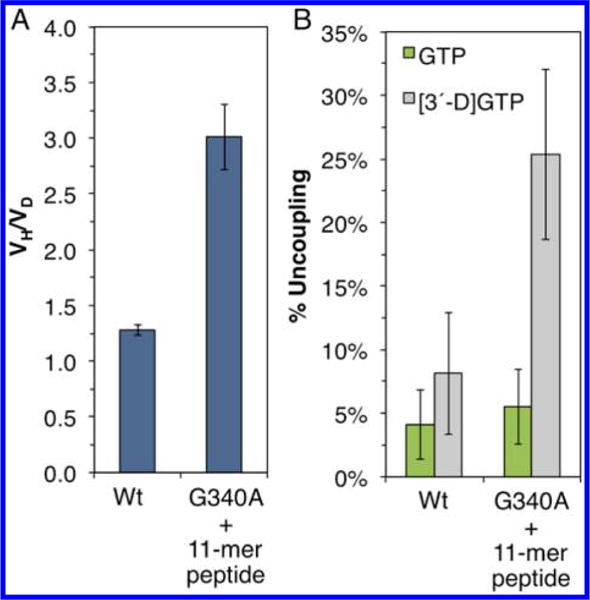
Effects of GG-motif mutations on the H-atom abstraction. (A) KIE (VH/VD) observed in the assays of wt-MoaA and G340A-MoaA (with 11-mer peptide). The catalytic rates were determined by the coupled assay in the presence of MoaA (3 μM), MoaC (10 μM), SAM (1 mM), and GTP or [3′-D]GTP (1 mM). The 11-mer peptide (200 μM) was used in the G340A-MoaA mutant assays. KIE was determined on the basis of the ratio of kcat values for the reactions with GTP and [3′-D]GTP. (B) The amount of abortive SAM cleavage relative to cPMP formation by wt-MoaA and G340A-MoaA with 11-mer peptide. Assays were performed as in A using GTP (green bars) or [3′-D]GTP (gray bars) as substrate. The percent uncoupling was determined on the basis of the amounts of 5′-dA and cPMP formation. All reactions were triplicated, and error bars represent standard deviation.
Perturbation of the H-atom-transfer step was also suggested by the increased amounts of the abortive cleavage of SAM. Abortive SAM cleavage is frequently observed in radical SAM enzymes when the formation of 5′-dA• by the reductive cleavage of SAM is uncoupled from the H-atom abstraction from substrate.20,21 In our in vitro assays of the wt enzyme, MoaA reaction is very well coupled and only minimal amount of uncoupling is detectable (4.1 ± 2.7 and 5.5 ± 3.0% for GTP and [3′-D]GTP, respectively; Figure 5B). Comparable uncoupling (8.2 ± 4.8%) was observed for G340A-MoaA in the presence of the 11-mer wt peptide when assayed using GTP with natural isotope abundance. In contrast, when [3′-D]GTP was used, a significantly greater value (25 ± 6.7%, Figure 5B) was observed. Together with the increased KIE in G340A-MoaA, these observations suggest that the perturbation in the GG motif significantly slows the rate of H-atom abstraction.
DISCUSSION
Human MoCD is caused by genetic mutations in Moco biosynthetic enzymes. One of the hot spots for the MoCD-causing mutations is the GG motif of MOCS1A. However, the exact mechanism by which the mutations in the GG motif cause MoCD was unknown. The studies described here provide the first in vitro functional characterization of the GG motif using a bacterial homologue (MoaA) as a model, and the first demonstration that the mutations can be rescued by synthetic peptides. Our initial characterization of MoaA variants with mutations in the GG motif revealed no significant structural perturbation, but complete depletion of the GTP 3′,8-cyclase activity. Even the minimal structural perturbation by mutation of Gly into Ala resulted in the complete loss of activity. Intriguingly, the GTP 3′,8-cyclase activities of these GG-motif mutants were restored by supplementation of synthetic peptides corresponding to the C-terminal tail of wt-MoaA. The peptides’ ability to restore activity was specific to their amino acid sequence or their lengths. Studies using MoaA variants with mutations in the putative binding interface revealed specific interaction of three cationic residues in the peptide with D198, a partially conserved residue in Loop 14 locating adjacent to the SAM binding site (Figure 2A). The observed interaction was also critical for the function of the C-terminal tail in the intact MoaA, suggesting that the synthetic peptide likely binds to the location identical to the binding site of C-terminal tail in the intact enzyme. These observations demonstrated the relevance of the peptide rescue assay to study the role of C-terminal tail in MoaA catalysis.
Functional characterizations of the GG-motif mutants using the peptide rescue assay suggested that the GG motif is essential for the SAM binding and the H-atom transfer from GTP to 5′-dA•. Although they were not described in the Results, we also investigated the previously discussed possibilities for the functions of the GG motif; post-translational modification and glycyl radical formation.9,14 All of our data suggest the absence of such mechanisms (see Figures S4 and S5 for details). These observations in sum suggest that the physical interaction of the GG motif with the active site or surrounding loops is critical for SAM binding and subsequent radical reactions.
Comparison of our biochemical observations and the previously reported crystal structures of MoaA suggest potential function of the C-terminal tail. In the reported structures, MoaA active site is highly exposed to solvent (Figure 6A,B),10 a unique feature compared to the active-site structures of other radical SAM enzymes.22 For example, in pyruvate formate-lyase activating enzymes (PFL-AE, see Figure 6C,D), SAM is bound to the active site through extensive H-bonding and hydrophobic interactions.18 Particularly, the adenine base interacts with the surrounding amino acid residues through H-bonding and π-stacking interactions with two His side chains (Figure 6D).18 Analogously, SAM is also deeply buried in the active site in other radical SAM enzymes (5–10 Å from the nearest protein surface) with extensive interactions with surrounding amino acid residues (Figure S6). On the other hand, in the MoaA structure, SAM is exposed to solvent (Figure 6A), and few interactions were observed between the adenine base and the surrounding amino acid residues. While in some radical SAM enzymes, substrate itself provides the shielding,22 such mechanism is unlikely for MoaA because its substrate, GTP, is small and buried more deeply in the active site than SAM (Figure 2A). In many radical SAM enzymes, the active-site closure is considered as an important mechanism to provide hydrophobic environment to overcome the large apparent free-energy barrier (~0.39 V) for SAM cleavage.23–25 Thus, MoaA requires a specific mechanism to close and isolate the active site from the solvent. Since our data suggest that the interaction of C-terminal tail and Loop 14 is essential for the SAM binding and subsequent H-atom-abstraction step, C-terminal tail of MoaA likely plays a key role in the active-site closure and the initiation of the radical catalysis.
Figure 6.
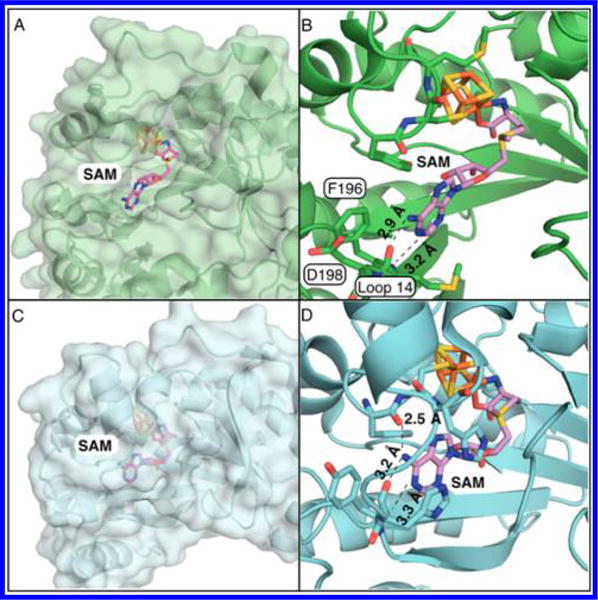
Comparison of active-site structures of MoaA and pyruvate formate-lyase activating enzyme (PFL-AE). (A,B) The active site of S. aureus MoaA in surface representation (A) and in stick and cartoon representation (B).10 (C,D) The active site of PFL-AE in surface representation (C) and in stick and cartoon representation (D).18 (B) and (D) show H-bonding distances (Å) between the adenine of SAM and amino acid residues.
On the basis of these considerations, we propose that the C-terminal tail serves as a gate-keeper of the MoaA active site essential for the initiation of the catalysis (Figure 7). The binding of the C-terminal tail to Loop 14 likely occurs in concert with SAM binding, which shields the active site from solvent. In this process, the GG motif may be inserted into the active site and directly interact with SAM or the active-site amino acid residues. The small sizes of the two Gly residues may be important for their insertion into the spatially limited active site, which parallels the function of the GG motifs in the ubiquitin-like proteins.16,26 It is also possible that the C-terminal tail does not directly interact with the active-site residues or substrates, but interacts with the loops around the SAM binding pocket (Figure S7). In this case, the C-terminal tail would stabilize the closed conformation of the loops to isolate the active site from solvent. In either case, the interaction of C-terminal tail with the active site is critical for SAM binding and subsequent H-atom-abstraction step. Therefore, the C-terminal tail of MoaA likely plays a crucial role in the active-site closure to initiate the free radical reaction. Because the activity of the MoaA GG-motif mutants can be triggered by the exogenous addition of the peptide and the H-atom transfer is more rate-determining, further study of this system may provide important insights into the mechanism of radical SAM enzymes in general.
Figure 7.
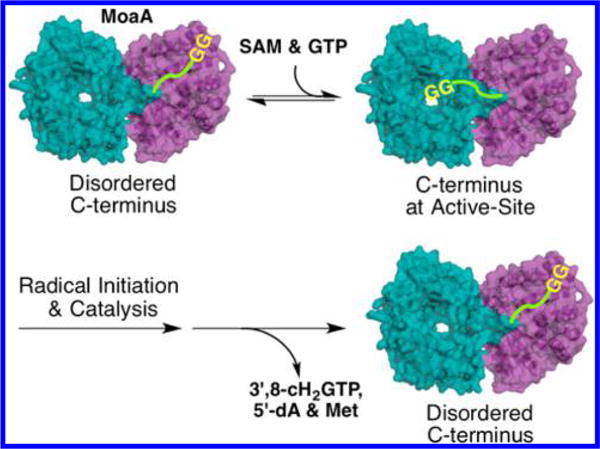
Model for the C-terminal tail dynamics and the initiation of MoaA catalysis. C-terminal tail was indicated with green line with GG motif at the C-terminus.
While the GG motif is conserved among all the prokaryotic and eukaryotic MoaA/MOCS1A homologues, the residues responsible for the interaction between the C-terminal tail and Loop 14 vary between bacteria and eukaryotes, which likely represent differences in their interaction interface. The interface determined in this study for S. aureus MoaA involves D198 in Loop 14 and the three cationic residues in the C-terminal tail, which are conserved among many of the Gram-positive bacteria, but not in eukaryotes. Instead, the eukaryotic homologues (mammals, plants, and fungi) harbor a uniquely conserved Asp in the Loop 14 (D251 in humans, Figure 2A) and Arg in the C-terminal tail (R378 in humans, Figure 2C). Thus, in eukaryotes, these residues may provide an important affinity for the C-terminal tail binding. These analyses suggest that the interaction interface of C-terminal tail is likely specific to evolutionary related organisms. On the other hand, the GG motif is evolutionary conserved, and its mutation causes impairment in initiation of catalysis. In humans, this results in the pleiotropic loss of all the Moco-dependent enzyme functions, and causes the fatal MoCD.
Overall, we demonstrated that synthetic peptides corresponding to the C-terminal tail of MoaA rescues mutations in the catalytically essential GG motif. Biochemical characterization using the peptide rescue assay revealed that the human MoCD associated with the mutations in the GG motif is caused by impairment in the SAM binding and the radical initiation process of the MOCS1A catalysis. These results suggest the possibility that the rescue of the MoCD-causing mutations may be possible using chemical complementation of enzyme activity, which may lead to development of novel and sustainable therapy for MoCD patients.
METHODS
General
Guanosine 5′-triphosphate (GTP), S-adenosyl-L-methionine (SAM), dithiothreitol (DTT), and sodium dithionite were purchased from Sigma-Aldrich. [3′-D]GTP was prepared as described previously.11 Nonlinear least-squares fitting of kinetic data was carried out using KaleidaGraph software (Synergy Software, Reading, PA). Anaerobic experiments were carried out in an UNIlab workstation glovebox (MBaun, Stratham, NH) maintained at 10 ± 2 °C with an O2 concentration <0.1 ppm. All HPLC experiments were performed on a Hitachi L-2130 pump equipped with an L-2455 diode array detector, an L-2485 fluorescence detector, an L-2200 autosampler, and an ODS Hypersil C18 column (Thermo Scientific) housed in an L-2300 column oven maintained at 40 °C. UV–vis absorption spectra were determined using the U-3900 UV–vis ratio recording double-beam spectrometer (Hitachi). Synthetic peptides were purchased from Genscript.
Expression and Purification of wt-MoaA and MoaA Variants
N-terminally His6-tagged S. aureus wt-MoaA and mutant MoaA was expressed in E. coli, purified, and anaerobically reconstituted as previously described.11 MoaA variants with single-point mutations were prepared by following the Stratagene’s QuikChange site-directed mutagenesis protocol using the primers shown in Table S2 and pET-HisMoaA11 as a template. The triple mutant, R330A/K331A/K332A-MoaA, was prepared using pET-HisMoaA-R330A as a template.
In Vitro Characterization of MoaA
For the coupled assay, purified wt-MoaA or mutant MoaA (1 μM) was anaerobically incubated with MoaC (3 μM), GTP (1 mM), SAM (1 mM), and sodium dithionite (1 mM) in the assay buffer (50 mM Tris-HCl pH 7.6, 1 mM MgCl2, 2 mM DTT, and 0.3 M NaCl) at 25 °C. The reaction was quenched at specified time with 0.1 volume of 25% trichloroacetic acid (TCA), and cPMP was quantified by HPLC after its conversion to a fluorescent derivative, compound Z.11 Peptide rescue assays of GG-motif mutants were performed in an identical manner in the presence of synthetic peptides at specified concentrations. To determine the steady state kinetic parameters, GTP or SAM concentration was varied (0, 1, 2, 5, 10, 50, or 100 μM). To determine the KIE values, the assays were performed in the presence of MoaA (3 μM) and MoaC (10 μM) as descried above using GTP or [3′-D]GTP as substrate (1 mM). KIE values were determined by comparing rate of reaction with GTP vs [3′-D]GTP. To determine the abortive SAM cleavage, 20 μM MoaA (wt- and G340A-MoaA with or without 11-mer peptide) was incubated with 40 μM MoaC as above for 20 min at 25 °C.
Anaerobic ITC
Using a GE MicroCal VP-ITC instrument under the flow of argon, anaerobic ITC was performed on wt-MoaA and MoaA GG-motif mutants. A solution containing GTP or SAM (350 μM in 0.5 mL) or degassed assay buffer was titrated into 1.5 mL of 35 μM MoaA in 20 mM Tris, pH 7.6, 150 mM NaCl, 5 mM DTT over 29 injections, at 24 °C, 307 rpm.
Supplementary Material
Acknowledgments
This work was supported by start-up funds from the Duke University Medical Center (to K.Y.). We thank George R. Dubay (Duke University, Department of Chemistry) for assistance with the MS measurements.
Footnotes
Supporting Information
Additional experimental details, supplementary methods, and extended data. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Mendel RR, Schwarz G. Coord Chem Rev. 2011;255:1145–1158. [Google Scholar]
- 2.Leimkuhler S, Wuebbens MM, Rajagopalan KV. Coord Chem Rev. 2011;255:1129–1144. doi: 10.1016/j.ccr.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson JL, Waud WR, Rajagopalan KV, Duran M, Beemer FA, Wadman SK. Proc Natl Acad Sci USA. 1980;77:3715–3719. doi: 10.1073/pnas.77.6.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss J. Hum Genet. 2000;106:157–163. doi: 10.1007/s004390051023. [DOI] [PubMed] [Google Scholar]
- 5.Reiss J, Hahnewald R. Hum Mutat. 2011;32:10–18. doi: 10.1002/humu.21390. [DOI] [PubMed] [Google Scholar]
- 6.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey PA, Hegeman AD, Ruzicka FJ. Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 8.Wuebbens MM, Rajagopalan KV. J Biol Chem. 1993;268:13493–13498. [PubMed] [Google Scholar]
- 9.Hänzelmann P, Schwarz G, Mendel RR. J Biol Chem. 2002;277:18303–18312. doi: 10.1074/jbc.M200947200. [DOI] [PubMed] [Google Scholar]
- 10.Hänzelmann P, Schindelin H. Proc Natl Acad Sci USA. 2004;101:12870–12875. doi: 10.1073/pnas.0404624101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hover BM, Loksztejn A, Ribeiro AA, Yokoyama K. J Am Chem Soc. 2013;135:7019–7032. doi: 10.1021/ja401781t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mehta AP, Hanes JW, Abdelwahed SH, Hilmey DG, Hänzelmann P, Begley TP. Biochemistry. 2013;52:1134–1136. doi: 10.1021/bi3016026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehta AP, Abdelwahed SH, Xu H, Begley TP. J Am Chem Soc. 2014;136:10609–10614. doi: 10.1021/ja502663k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hänzelmann P, Schindelin H. Proc Natl Acad Sci USA. 2006;103:6829–6834. doi: 10.1073/pnas.0510711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold K, Bordoli L, Kopp J, Schwede T. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 16.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Nature. 2001;414:325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 17.van der Veen AG, Ploegh HL. Annu Rev Biochem. 2012;81:323–357. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 18.Vey JL, Yang J, Li M, Broderick WE, Broderick JB, Drennan CL. Proc Natl Acad Sci USA. 2008;105:16137–16141. doi: 10.1073/pnas.0806640105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill AV. Biochemical Journal. 1913;7:471–480. doi: 10.1042/bj0070471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove TL, Lee KH, St Clair J, Krebs C, Booker SJ. Biochemistry. 2008;47:7523–7538. doi: 10.1021/bi8004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarty RM, Krebs C, Bandarian V. Biochemistry. 2013;52:188–198. doi: 10.1021/bi301156w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vey JL, Drennan CL. Chem Rev. 2011;111:2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang SC, Frey PA. Biochemistry. 2007;46:12889–12895. doi: 10.1021/bi701745h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shisler KA, Broderick JB. Curr Opin Struct Biol. 2012;22:701–710. doi: 10.1016/j.sbi.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dey A, Peng Y, Broderick WE, Hedman B, Hodgson KO, Broderick JB, Solomon EI. J Am Chem Soc. 2011;133:18656–18662. doi: 10.1021/ja203780t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmitz J, Wuebbens MM, Rajagopalan KV, Leimkuhler S. Biochemistry. 2007;46:909–916. doi: 10.1021/bi062011w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


