Abstract
Pancreatic cancer is the fourth leading cause of cancer death in the U.S. Once diagnosed, prognosis is poor with a 5-year survival rate of less than 5%. Exposure to carcinogenic heterocyclic amines (HCAs) derived from cooked meat has been shown to be positively associated with pancreatic cancer risk. To evaluate the processes that determines the carcinogenic potential of HCAs for human pancreas, 14-carbon labeled 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx), a putative human carcinogenic HCA found in well-done cooked meat, was administered at a dietary relevant dose to human volunteers diagnosed with pancreatic cancer undergoing partial pancreatectomy and healthy control volunteers. After 14C-MeIQx exposure, blood and urine was collected for pharmacokinetic and metabolite analysis. MeIQx-DNA adducts levels were quantified by accelerator mass spectrometry from pancreatic tissue excised during surgery from the cancer patient group. Pharmacokinetic analysis of plasma revealed a rapid distribution of MeIQx with a plasma elimination half-life of approximately 3.5 hr in 50% of the cancer patients and all of the control volunteers. In 2 of the 4 cancer patients very low levels of MeIQx were detected in plasma and urine suggesting low absorption from the gut into the plasma. Urinary metabolite analysis revealed five MeIQx metabolites with 2-amino-3-methylimidazo[4,5-f]quinoxaline-8-carboxylic acid being the most abundant accounting for 25%–50% of the recovered 14-carbon/ml urine. There was no discernable difference in metabolite levels between the cancer patient volunteers and the control group. MeIQx-DNA adduct analysis of pancreas and duodenum tissue revealed adduct levels indistinguishable from background levels. Although other meat-derived HCA mutagens have been shown to bind DNA in pancreatic tissue, indicating that exposure to HCAs from cooked meat cannot be discounted as a risk factor for pancreatic cancer, the results from this current study show that exposure to a single dietary dose of MeIQx does not readily form measurable DNA adducts under the conditions of the experiment.
TOC Graphic
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer death in the United States.1 It is generally diagnosed at a late stage and does not respond well to therapies. The 5-year survival rate is less than 5%.2 The etiology of pancreatic cancer is not well understood, but diet has been implicated as an important environmental risk factor.3–5 Epidemiology studies have shown a positive relationship between the consumption of red meat and pancreatic cancer incidence, however, results have been inconsistent.6–12 Meat cooked at high temperatures is a source of carcinogen exposure from the formation of heterocyclic amines (HCAs) and polycyclic aromatic hydrocarbons (PAHs). Several HCAs found in cooked meat have been shown to be carcinogenic in multiple tissues, including pancreas, in animal models.13,14 While laboratory studies and observational data in humans support the hypothesis that HCAs are carcinogenic for humans, the biological evidence needed to determine which human organs are susceptible to HCA-induced cancers is lacking. A better understanding of the pancreas-relevant pathways that detoxify HCAs, activate them to DNA-binding species, and repair DNA damage is needed so that strategies can be developed to reduce the risk from exposure to these compounds. A causal relationship between meat-derived HCA mutagens and pancreatic cancer has not been established, nor do we fully understand how individual differences in metabolism may impact risk. Thus, the rationale for this research is to identify the mechanisms that contribute to the carcinogenic potential of HCAs for human pancreas.
2-Amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx) is one of the most mutagenic HCAs found in cooked meat and has been shown to be carcinogenic in rodents. Chronic administration in rats and mice results in tumors primarily in the liver, although tumors in the lung, and skin,13,14 as well as, mouse lymphomas and leukemias have also been documented. Although no pancreatic tumors have been detected in experimental animals after MeIQx exposure, the compound was detected at relatively high levels in the pancreas of mice after a single oral dose.15 In humans, epidemiology studies have shown a positive association between MeIQx exposure and pancreatic cancer in some but not all studies.7–9, 12,16 These studies suggest that MeIQx, or other HCAs may be causative agents for human pancreatic cancer, however studies to determine the extent of MeIQx as a risk factor for pancreatic cancer are limited.
The objective of the current study is to compare MeIQx metabolism and DNA adduct formation in a small population of cancer patients and controls as a first step towards understanding how exposure to MeIQx may impact pancreatic cancer risk. The relationship between MeIQx-DNA adduct formation, metabolism, and exposure has been primarily established at high MeIQx doses using animal models, mostly due to limitations in assay sensitivity and the difficulties associated with human in vivo studies. In the present study, these limitations have been overcome by using accelerator mass spectrometry (AMS), which is capable of accurately measuring attomole (10−18) quantities of radiolabeled compound.17–18
Human volunteers diagnosed with pancreatic cancer and healthy control volunteers were exposed to a dietary equivalent dose of MeIQx, labeled with a very low level of 14-carbon. AMS was used to quantify plasma MeIQx concentrations and urinary MeIQx metabolite levels. Comparisons were made between the two populations to determine if pharmacokinetic parameters and urinary metabolite profiles differ between the two volunteer groups. MeIQx-DNA adducts in pancreatic tissue were measured by AMS in the cancer patients from tissue obtained after partial pancreatectomy. This pilot study serves as a first step in identifying potential mechanisms that determine the carcinogenic potential of HCAs for human pancreas.
MATERIAL AND METHODS
Chemicals
14C-MeIQx was obtained from Toronto Research Chemicals (Ontario, Canada). Radio-chemical purity was assessed by HPLC and was determined to be >98% pure. MeIQx metabolite standards were kindly provided by Dr. Robert Turesky (University of Minnesota, MN). All reagents were of analytical grade or better.
Human Study
The human study protocol was independently reviewed and approved by the Institutional Review Boards for Human Subjects at the Lawrence Livermore National Laboratory (LLNL) (Livermore, CA) and the University of Minnesota (UMN) (Minneapolis, MN). An Investigational New Drug (IND) application was submitted to the FDA and an exploratory IND (no. 113501) was prepared for this study. All human volunteers participating in the study gave informed consent prior to enrollment. Eight volunteers were recruited for the study, four were pancreatic cancer patients scheduled for partial pancreatectomy (depicted as P1, P2, P3, P4) and four were healthy control volunteers (depicted as C1, C2, C3, C4).
Eligible subjects were required to have adequate hepatic function within 4 weeks of study enrollment which was defined as total bilirubin ≤ 2.0 mg/dL and alkaline phosphatase, aspartate aminotransferase and alanine transaminase ≤ 2 × under the normal limit. Subjects were not eligible if they had chronic conditions such as: cardiovascular disease, hypertension, angina, chronic obstructive pulmonary disease (COPD) or other conditions that may alter metabolism, other than diabetes. Subjects were also excluded if known to be pregnant or lactating. To reduce the risk of recruiting cancer patients who were later found to be unresectable, patients were excluded if they met any of the following criteria: tumor size ≥ 3 cm, CA-19-9 > 400 or ascites present.
14C-MeIQx was packaged in gelatin capsules containing lactose filler. The capsules were administered orally with a glass of water and each volunteer received a dose of 21 μg of 14C-MeIQx (specific activity, 43.5 mCi/mmol). The radioactive dose received was 0.002 mSv, which is the energy equivalent of 1/29 of the energy received from an average chest X-ray. At 0, 0.5, 1, 2, 4, 6, and 8 h after dosing, blood samples were collected and plasma was isolated by centrifugation. Urine was collected at various time intervals up to 24 h post dose. All samples were stored at −80° C until analysis. For the cancer cases only, the subjects underwent surgery for pancreatectomy in which uninvolved resected tissue from surgery was collected between six to eight hours after dosing. The tissue was subsequently stored by the Tissue Procurement Facility at the UMN, then shipped to LLNL and stored at −80° C until DNA adduct analysis.
Plasma pharmacokinetics
Whole blood was collected from each volunteer at pre-dose, 0.5, 1, 2, 4, 6, and 8 hr post 14C-MeIQx administration. Plasma was separated from whole blood by centrifugation and stored at −80°C until it was shipped to LLNL for analysis by AMS to quantify the levels of 14-carbon.
The pharmacokinetic parameters of MeIQx from each volunteer were calculated by noncompartmental analysis using PK Solutions software (Summit Research Services, Montrose, CO). The half-life (t1/2) and the maximum concentration observed in plasma (Cmax) were determined by observations from the concentration-versus-time data. The area under the curve (AUC) was calculated for the intervals from time zero to time t (AUC0–t), where t is the time of the last measurable concentration (8 h), and for time zero to infinity (AUC0–inf), using the linear trapezoidal method. The volume of distribution (V) was determined on the basis of the AUC determination and reflects the V during the elimination phase. The clearance (CL) calculation is based on the AUC0–t.
Separation of urinary MeIQx metabolites
Urine was collected at time intervals of 0–4 h, 4–8 h and 8–24 h post-ingestion of the 14C-MeIQx dose, and immediately frozen and stored at −80°C. Prior to HPLC analysis, each urine sample was thawed and a 1.0 ml aliquot from each fraction was analyzed by liquid scintillation counting to determine the 14-carbon content. Each sample was then analyzed by reversed-phase HPLC for MeIQx and MeIQx metabolites. Approximately 1000–4000 disintegrations per minute (dpm) of each urine sample was filtered using a 0.45 μm nylon centrifuge filter (MSI, Westborough, MA), concentrated under vacuum centrifugation to 100 μl, then directly injected into an Alliance HPLC system (Waters, Milford, MA) equipped with a 4 μm, 4.6 × 250 mm Synergi Max-RP 80A column (Phenomenex, Torrance, CA), and monitored at 273 nm. Following the methods of Turesky etal.,19 metabolites were eluted at 1.0 ml/min initially using a solvent of 89% (v/v) 50 mM ammonium acetate/11% methanol for 10 min. This was followed by a gradient to 80% (v/v) ammonium acetate/20% methanol at 40 min, followed by a final gradient to 100% (v/v) methanol at 51 min. The methanol concentration was maintained at 100% (v/v) from 51 to 60 min.19 The column eluent was collected at 1 min intervals and radioactivity was quantified by scintillation counting (Perkin Elmer Packard). Approximately 90% of the radioactivity was recovered after sample preparation and analysis.
DNA Extraction from tissue
All tissue samples were processed in a biosafety cabinet until DNA was extracted. To homogenize the tissue samples, between 30–50 mgs of tissue were weighed from stock samples and transferred to sealed O-ring style bead beating tubes (Lysis Matrix A, MB Biomedicals, Santa Ana, CA). Approximately 350 μl of Qiagen ATL buffer was added to the tube along with 1.75 μl of anti-foam reagent (0.5%v/v). The samples were vortexed for several minutes and then run on a Mini-Beadbeater (Biospec Products, Bartlesville, OK) with 4 sets of 20-second intervals while cooling on ice for 30 seconds in between intervals. After bead beating, the samples were centrifuged at 12,000 RPM for 3 minutes. The supernatants were transferred to a new tube for DNA extraction. RNA-free genomic DNA was isolated using the Qiagen DNEASY Blood and Tissue Mini Kit (Qiagen, Redwood City, CA) following the kit protocol. DNA concentration was determined by measuring the absorbance at 260 nm assuming an absorbance value of 1.0 is equal to 50 μg/ml DNA. Purity was assessed by measuring the 260/280 absorbance ratio. DNA with a ratio of 1.8 was considered pure. Following quantification, 15 μg of pure DNA was then added to quartz tubes for analysis by AMS.
AMS analysis
Preparation of the samples for radiocarbon analysis by AMS requires conversion of the samples to graphite. This procedure has been described previously.20 All the samples and reagents were handled carefully to avoid radiocarbon cross-contamination, including using disposable materials for any item that might come into contact with the samples. For the plasma samples, a 30 μl aliquot of each sample was pipetted into a 6- by 55-mm quartz tube using aerosol-resistant tips and subsequently dried under vacuum centrifugation. For the DNA analysis, 15 μg of each DNA sample was pipetted into a quartz tube followed by 1 μg of tributyrin, to provide the carbon content necessary for efficient graphitization. The samples were subsequently dried under vacuum centrifugation. The dried plasma and DNA samples were then converted to graphite by a two-step process using published methods.21 Briefly, the dried samples were oxidized to CO2 by heating at 900°C for 4 h in the presence of copper oxide. The CO2 was then cryogenically transferred to a septum-sealed vial under vacuum and reduced to filamentous graphite in the presence of cobalt, titanium hydride, and zinc powder.
The total radiocarbon content of the samples was quantified by AMS as described previously.17,21,22 The 14C/12C ratios from the graphitized samples obtained by AMS were converted to either ng MeIQx per ml of plasma or pg MeIQx per μg of DNA after subtraction of the background carbon contribution from any added tributyrin and correction for the specific activity of the 14C-labeled MeIQx dosing material.18 The precision of the AMS measurements is based on the standard deviation of a number of measurements for equivalent samples and is a nominal 3%. Accuracy is determined by normalization to well-defined standards and with high-accuracy counting measurements and is ~1%.18
RESULTS
Plasma Pharmacokinetics
Plasma was collected at the time points designated above and analyzed for 14C-MeIQx content by quantifying the 14-carbon at each time point using AMS. The pharmacokinetics for each volunteer were calculated by noncompartmental analysis. The plasma concentration vs. time curve was similar in all four of the control volunteers (C1–C4) and two of the cancer patient volunteers following first order kinetics (Figure 1). In these six individuals the observed Cmax was attained within 1 h post 14C-MeIQx administration. There was some inter-individual variation in the Cmax between the subjects with a range between 0.3–0.9 ng/ml of plasma. The terminal half-life between the six subjects was similar except in control subject C3 where it was approximately 2-fold higher (7.5 hr) indicating a slower elimination from the plasma (Table 1). This was not, however, reflected in the clearance rate. The apparent volume of distribution (V) ranged from 31.5 L to 119.6 L suggesting rapid and extensive distribution beyond the plasma compartment. Total clearance was variable, ranging between 7.9–24.1 L/hr. Due to the small sample size (4 controls, 2 cases) no inferences could be made from the data. In the remaining 2 cancer cases (P2, P3) there was significantly less 14-carbon in the plasma compared to the other 6 volunteers. This could be due to a lack of absorption of the 14C-MeIQx dose from the gut into the plasma. The plasma concentration time curves for these two patients did not follow the traditional profile that was observed in the other six volunteers. Due to the low levels of 14-carbon and non-traditional curves it was not possible to determine accurate PK parameters for these subjects.
Figure 1.
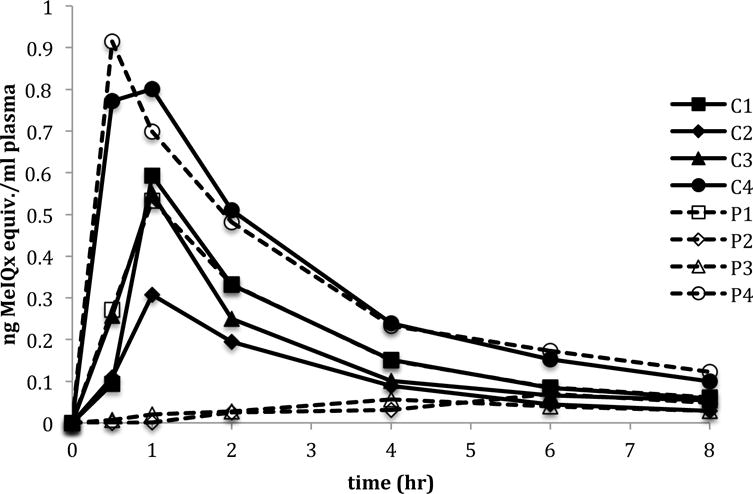
Plasma concentration versus time of MeIQx following a single oral dose of 14C-MeIQx (21 μg) in four healthy human volunteers (C1–C4) and four pancreatic cancer patient volunteers (P1–P4).
Table 1.
Pharmacokinetic parameters of MeIQx following a single oral administration of 21 μg 14C-MeIQx to humans
| Subject | T1/2 elim. (hr) |
Tmax (hr) |
Cmax (ng/ml) |
AUC0–t (ng-hr/ml) |
Vd (ml) |
Cl (ml/hr) |
|---|---|---|---|---|---|---|
| C1 | 3.9 | 1.0 | 0.59 | 1.5 | 63844 | 13762 |
| C2 | 3.2 | 1.0 | 0.31 | 0.9 | 97495 | 24062 |
| C3 | 7.5 | 1.0 | 0.55 | 1.3 | 119585 | 16052 |
| C4 | 3.2 | 1.0 | 0.80 | 2.6 | 31476 | 7933 |
| P1 | 3.4 | 1.0 | 0.53 | 1.6 | 55194 | 13301 |
| P4 | 4.0 | 0.5 | 0.91 | 2.6 | 36370 | 7933 |
MeIQx Levels in Urine
Consistent with the plasma data, the majority of the urinary 14-carbon was detected in the first 4 hours after 14C-MeIQx administration, indicating rapid clearance. MeIQx levels were similar across the non-cancer control volunteers with a range of 21–33 ng MeIQx equivalents/ml urine in the first 4 hr post dose (Figure 2). This accounted for 64%–85% of the recovered radioactivity/ml of urine (Table 2). Cancer patient P4 had the highest level of MeIQx in the first 4 hours of collection at 68 ng MeIQx equivalents/ml of urine were detected, accounting for over 90% of the recovered dose/ml of urine. In cancer cases P2 and P3, very low levels of 14-carbon were detected in the urine compared to the rest of the study group and control group. This finding is in accordance with the low 14-carbon levels observed in the plasma samples from these same two individuals. Only a total of 8.7 and 6.5 ng MeIQx/ml of urine was detected in P2 and P3, respectively, over the 24 hr course of the study (Figure 2), whereas the levels of MeIQx for the other volunteers ranged from 26 ng–75 ng MeIQx/ml urine for the same time period. Interestingly a relatively higher level of 14-carbon was detected in the 4–8 hr samples of volunteers P2 and P3 (65% and 43% of recovered dose/ml, respectively) compared to the 0–4 hr samples, suggesting a slower elimination rate compared to the other 2 cancer cases and 4 control individuals.
Figure 2.
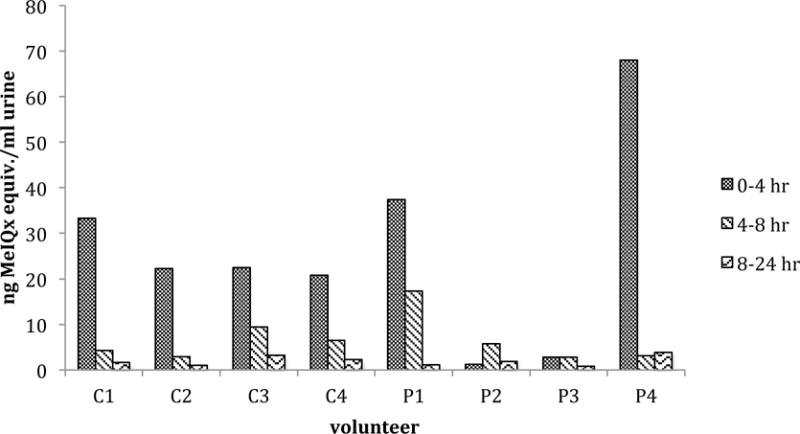
MeIQx levels in urine fractions collected over time following a single oral dose of 14C-MeIQx (21 μg) in four healthy human volunteers (C1–C4) and four pancreatic cancer patient volunteers (P1–P4). Data is expressed as ng MeIQx equivalents/ml of urine.
Table 2.
Percent of recovered 14-carbon in urine after administration of 14C-MeIQx
| Collection time (hr) | C1 | C2 | C3 | C4 | P1 | P2 | P3 | P4 |
|---|---|---|---|---|---|---|---|---|
| 0–4 | 84.8 | 85.2 | 63.9 | 70.2 | 66.9 | 13.5 | 43.9 | 90.7 |
| 4–8 | 10.8 | 11.1 | 26.7 | 22.1 | 31.0 | 65.1 | 43.2 | 4.1 |
| 8–24 | 4.3 | 3.7 | 9.3 | 7.7 | 2.0 | 21.5 | 12.9 | 5.2 |
Data are expressed as percent of recovered carbon-14/ml of urine
Chromatographic analysis of the 0–4 hr urine samples revealed six major radioactive peaks in all eight of the volunteers (Figure 3). Based on HPLC co-elution with authentic MeIQx and MeIQx metabolite standards 5 radioactive peaks were identified as MeIQx metabolites and one was determined to be unchanged MeIQx. All five metabolites have been previously identified in humans.19,23 There was some inter-individual variation among the volunteers in the relative quantities of urinary metabolites excreted. 2-Amino-3-methylimidazo[4,5-f]quinoxaline-8-carboxylic acid (IQx-8-COOH) was the most abundant metabolite in all 8 volunteers accounting for 25–50% of the recovered radioactivity (Table 3). The phase II conjugates N2-(3,8-dimethylimidazo[4,5-f]quinoxaline-2-yl)sulfamic acid (MeIQx-N2-SO3) and N2-(β-1-glucosiduronyl)-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx-N2-Gl) accounted for 5.1%–17.7% and 5.6%–13.4% of the recovered 14-carbon signal, respectively, across all volunteers. The urinary levels of the cytochrome P450 oxidation products N2-(β-1-glucosiduronyl)-N-hydroxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (HON-MeIQx-N2-Gl) and 2-amino-8-(hydoxymethyl)-3-methylimidazo[4,5-f]quinoxaline (8-CH2OH-IQx), ranged from 12.9%–22.2% and 2.7%–9.2%, respectively. Unchanged MeIQx was the least abundant urinary product at 1.5%–5.6% of the recovered dose/ml of urine. The unidentified minor metabolites are presumed to be conjugates of MeIQx and OH-MeIQx.23 Overall there was no observable difference in the concentrations of metabolites between the non-cancer controls and the cancer subjects.
Figure 3.

A representative HPLC radio-profile of urinary MeIQx metabolites. The chromatograph was derived from a 0–4 hr urine fraction from volunteer P1 following a single oral dose of 14C-MeIQx (21 μg). Metabolites were identified based on co-elution with authentic MeIQx metabolite standards.
Table 3.
Percent of MeIQx metabolites recovered in urine after administration of 14C-MeIQx
| Metabolite | Subject
|
|||||||
|---|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | P1 | P2 | P3 | P4 | |
| IQx-8-COOH | 34.8 | 35.5 | 25 | 28 | 42.1 | 26.9 | 29.6 | 50.3 |
| MeIQx-N2-Gl | 9.2 | 7.6 | 8.8 | 6.3 | 5.6 | 5.8 | 13.4 | 6.2 |
| HON-MeIQx-N2-Gl | 13.9 | 18 | 19.8 | 15.8 | 12.9 | 15.1 | 22.2 | 13.7 |
| MeIQx-N2-SO3 | 12.8 | 12.6 | 17.7 | 12 | 10.3 | 8.4 | 5.1 | 7.7 |
| 8-CH2OH-IQx | 6.5 | 5.5 | 7 | 3.9 | 9.2 | 2.7 | 4.1 | 3.8 |
| MeIQx | 3.2 | 2.3 | 2.4 | 5.6 | 4.1 | 4.7 | 1.5 | 2.6 |
Data are expressed as percent of recovered carbon-14/ml of urine
Tissue DNA adducts
The ability of 14C-MeIQx to covalently bind DNA was assessed in the 4 pancreatic cancer volunteers. DNA was isolated from normal uninvolved pancreas and duodenum tissue that was excised during surgery and analyzed for 14C-MeIQx adduct formation by AMS. No duodenum tissue was collected from subject P1. The duration between 14C-MeIQx administration and tissue excision varied between the individuals as follows: subject P1: 7.75 hr, subject P2: 7.0 hr, subject P3: 7.5 hr, subject P4: 6.3 hr. Adduct levels in all tissues analyzed were very low and there was no observable difference between the two tissue types (Figure 4). Most measurements were barely distinguishable from natural 14-carbon background levels indicating that, under the study conditions, MeIQx-DNA adducts cannot readily be detected in these tissues.
Figure 4.
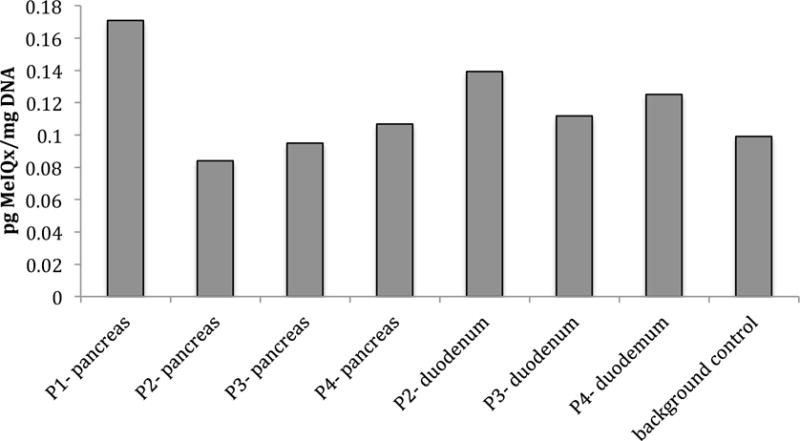
MeIQx-DNA adduct levels in pancreatic and duodenum tissue from pancreatic cancer patients (P1–P4) following a single oral dose of 14C-MeIQx (21 μg). No duodenum tissue was available for analysis for subject P1.
DISCUSSION
MeIQx is well established as a mutagen and potential human carcinogen.24–26 Epidemiologic evidence supports cooked-meat mutagens such as MeIQx as important risk factors for the development of colon cancer.27–31 Diet has also been implicated as a risk factor for pancreatic cancer, however studies investigating a relationship between meat-derived HCA mutagens and pancreatic cancer have been inconsistent.6–9,12,16
In this current pilot study, MeIQx pharmacokinetics and metabolism was assessed in both healthy human volunteers and volunteers diagnosed with pancreatic cancer after exposure to a defined dietary relevant dose of 14C-MeIQx. Additionally, MeIQx-DNA adducts in pancreatic tissue were assessed in the cancer patients from tissue obtained after partial pancreatectomy. This is the first study to investigate the formation of MeIQx-DNA adducts in human pancreas at a dietary relevant dose, and to compare pharmacokinetic response between healthy and cancer compromised individuals. The goal of the study was to gain insight into the role of MeIQx in human pancreatic cancer and to determine if cancer compromised individual respond differently than healthy individuals in regards to PK and metabolism of MeIQx. Interpretation of the results from this study must take into account the small size of the sample population and the health compromised state of the cancer patients. The data presented shows that urinary MeIQx metabolite profiles are similar between pancreatic cancer patients and healthy individuals at dose levels that are typical of dietary human exposure conditions. Plasma pharmacokinetic profiles revealed some differences between the two study groups. DNA isolated from pancreatic tissue from the cancer patients revealed low levels of MeIQx adducts.
Plasma pharmacokinetics of MeIQx revealed first order elimination in all four of the controls and 2 of the 4 cancer patients. The results showed rapid absorption into the plasma with the Cmax occurring within 1 hr post 14C-MeIQx dose. The large apparent volume of distribution suggests rapid and extensive distribution beyond the plasma compartment. The low absorption of MeIQx into the plasma from two of the cancer patient volunteers was unexpected. The low plasma MeIQx levels could be due to poor absorption (disturbed transport through the portal vein), which has been shown to occur in pancreatic cancer cases.32 In a study investigating malabsorption in patients with pancreatic cancer, 76% of cancer patients displayed abnormalities in absorption using the pancreatic function diagnostic test and the 5 g D-xylose absorption test.32 Differences in body mass index (BMI) could have also contributed to low plasma levels in the 2 cancer patients. The BMI for these patients (subjects P2 an P3) was 31 and 28, respectively. The BMI for the 2 cancer patients with higher levels of MeIQx in the plasma was 25 and 17. The subject with the BMI of 17 had the highest level of plasma MeIQx levels within the study group. These findings suggest that BMI could affect the rate of MeIQx absorption with higher BMI relating to slower absorption and clearance. BMI was not reported for the control subjects. Because of the low levels of MeIQx in the plasma of the 2 cancer patients, an increase in MeIQx concentration in excreta could be expected. The urine, however, did not show any increase in MeIQx levels in these two volunteers over the course of the experiment. The urinary levels of 14C-MeIQx were similar to the plasma with much lower MeIQx levels when compared to the six other volunteers. Since the exposure was oral, the lack of absorption into the plasma and low urinary levels could infer an increase in MeIQx concentration in the feces; however, fecal samples were not collected in this study. The wide variation of 14-carbon in the 0–4 h urine fractions of the cancer patients again suggests differences in urinary excretion rates between these individuals that could reflect malabsorption.32 In an effort to explain the poor absorption of the compound in two of the cases, the medical records for all cases were reviewed by a physician. None of the patients were documented to have pancreas insufficiency prior to surgery and none were on pancreas enzyme replacement prior to surgery. Thus, we were not able to positively identify the reason for the poor absorption in the two cancer patients, but the data suggests it could be related to BMI.
The five principle metabolites detected in the urine of all individuals are in accordance with what has been reported previously for human metabolism of MeIQx, with IQx-8-COOH being the most abundant detoxification product.19,23 In the two cancer case subjects with low urinary excretion of 14-carbon the relative amounts of individual metabolites were similar to the other six individuals in the study.
A major focus of this study was to determine the level of MeIQx-DNA adducts in pancreatic tissue from a population of pancreatic cancer patients. MeIQX-adducts have been primarily detected in the liver and colon of rodent models and in colon tissue of humans.33,34 This is the first study to investigate whether adducts are formed in human pancreatic tissue when exposed to a human dietary relevant dose of MeIQx. The results revealed no discernable difference in adduct levels among the samples. In fact, the adduct levels in all the cancer cases, including those with plasma and urine profiles similar to the controls, were extremely low suggesting that, under the conditions of the experiment, MeIQx-DNA adducts cannot readily be detected in these tissues. The lack of detectable adducts could possibly be due to a non-optimal sampling time with regards to the kinetics of adduct formation and/or DNA adduct stability and repair since previous studies assessing MeIQx adducts in rodents have reported the presence of adducts in pancreatic tissues after chronic and acute oral exposure to MeIQx.15,35,36,37 The lack of detectable MeIQx-DNA adducts in the current study cannot definitively discount an association of HCA exposure and pancreatic cancer. Several other mutagenic HCAs are formed during the cooking of meat that could also contribute to pancreatic cancer incidence. 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), the most abundant HCA found in cooked beef and chicken, was detected in pancreatic tissue at significant levels in mice exposed orally to a human dietary equivalent dose of 14C-PhIP38, and PhIP-DNA adducts were observed in the pancreas of rats exposed to PhIP through the diet.35,39 PhIP-DNA adducts were also detected in human pancreatic tissue from both normal tissue and tissue from patients with pancreatic adenocarcinoma suggesting this HCA may contribute to pancreatic cancer development.40
The results from the current study show that MeIQx is rapidly cleared from the plasma, and is extensively metabolized with less than 5% of unchanged MeIQx remaining in the urine. These results also suggest that persons with pancreatic cancer could have absorption insufficiencies leading to a reduced amount of MeIQx being absorbed from the gut into the blood stream. Additionally, individual BMI could have affected MeIQx absorption as well. Under the experimental conditions MeIQx DNA adducts were detected at or near back ground levels suggesting either adducts did not readily form in this tissue or that the sampling time was sub-optimal due to the kinetics of adduct formation and repair. These results suggest that exposure to MeIQx alone may not be a significant risk factor for pancreatic cancer. However, other meat-derived HCA mutagens have been shown to covalently bind DNA in pancreatic tissue indicating that exposure to HCAs from cooked meat cannot be discounted as a risk factor for pancreatic cancer.40 This pilot study serves as a starting point for additional studies with larger populations to fully understand the contribution of HCA exposure to pancreatic cancer incidence.
Acknowledgments
The Authors wish to thank the following individuals for their contributions to this research: Gerri Anderson and Danielle Jin, for assistance in study logistics and subject recruitment; Bruce Lindgren for statistical advice, Dr. Myron Gross for sample storage; Dr. Robert Turesky for providing the MeIQx metabolite standards, and Dr. Ted Ognibene and Elise Hill for AMS sample analysis. The late Dr. Fred Kadlubar contributed to the design and planning of this project.
FUNDING SOURCES
This work was performed under the auspices of the U.S. Department of Energy by, Lawrence Livermore National Laboratory at the Research Resource for Biomedical AMS under contract DE-AC52-07NA27344, and supported by a grant from the National Institute of General Medical Sciences (2P41GM103483-16). Support was also provided through a pilot funding mechanism from UAB/UMN SPORE in Pancreatic Cancer (NIH/NCI P50 CA101955-07).
ABBREVIATIONS
- HCAs
heterocyclic amines
- PAHs
polycyclic aromatic hydrocarbons
- MeIQx
2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- AMS
accelerator mass spectrometry
- LLNL
Livermore National Laboratory
- UMN
University of Minnesota
- IND
Investigational New Drug
- COPD
Chronic obstructive pulmonary disease
- AUC
area under the curve
- V
volume of distribution
- CL
clearance
- dpm
disintegrations per minute
- IQx-8-COOH
2-Amino-3-methylimidazo[4,5-f]quinoxaline-8-carboxylic acid
- MeIQx-N2-SO3
N2-(3,8-dimethylimidazo[4,5-f]quinoxaline-2-yl)sulfamic acid
- MeIQx-N2-Gl
N2-(β-1-glucosiduronyl)-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- HON-MeIQx-N2-Gl
N2-(β-1-glucosiduronyl)-N-hydroxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- 8-CH2OH-IQx
2-amino-8-(hydoxymethyl)-3-methylimidazo[4,5-f]quinoxaline
- BMI
body mass index
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
References
- 1.Siegel R, Ma JM, Zou ZH, Jemal A. Cancer Statistics. Ca-Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2011. National Cancer Institute; Bethesda, MD: http://seer.cancer.gov/csr/1975_2011/, based on November 2013 SEER data submission. [Google Scholar]
- 3.Nothlings U, Wilkens LR, Murphy SP, Hankin JH, Henderson BE, Kolonel LN. Meat and fat intake as risk factors for pancreatic cancer: the multiethnic cohort study. J Natl Cancer Inst. 2005;97:1458–1465. doi: 10.1093/jnci/dji292. [DOI] [PubMed] [Google Scholar]
- 4.Pericleous M, Rossi RE, Mandair D, Whyand T, Caplin ME. Nutrition and pancreatic cancer. Anticancer Res. 2014;34:9–21. [PubMed] [Google Scholar]
- 5.Rohrmann S, Linseisen J, Nöthlings U, Overvad K, Egeberg R, Tjønneland A, Boutron-Ruault MC, Clavel-Chapelon F, Cottet V, Pala V, Tumino R, Palli D, Panico S, Vineis P, Boeing H, Pischon T, Grote V, Teucher B, Khaw KT, Wareham NJ, Crowe FL, Goufa I, Orfanos P, Trichopoulou A, Jeurnink SM, Siersema PD, Peeters PHM, Brustad M, Engeset D, Skeie G, Duell EJ, Amiano P, Barricarte A, Molina-Montes E, Rodríguez L, Tormo M-J, Sund M, Ye W, Lindkvist B, Johansen D, Ferrari P, Jenab M, Slimani N, Ward H, Riboli E, Norat T, Bueno-de-Mesquita HB. Meat and fish consumption and risk of pancreatic cancer: Results from the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2013;132:617–624. doi: 10.1002/ijc.27637. [DOI] [PubMed] [Google Scholar]
- 6.Anderson KE, Sinha R, Kulldorff M, Gross M, Lang NP, Barber C, Harnack L, DiMagno E, Bliss R, Kadlubar FF. Meat intake and cooking techniques: associations with pancreatic cancer. Mutat Res. 2002;506–507:225–231. doi: 10.1016/s0027-5107(02)00169-0. [DOI] [PubMed] [Google Scholar]
- 7.Anderson KE, Kadlubar FF, Kulldorff M, Harnack L, Gross M, Lang NP, Barber C, Rothman N, Sinha R. Dietary Intake of Heterocyclic Amines and Benzo(a)Pyrene: Associations with Pancreatic Cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(9):2261–2265. doi: 10.1158/1055-9965.EPI-04-0514. [DOI] [PubMed] [Google Scholar]
- 8.Li D, Day RS, Bondy ML, Sinha R, Nguyen NT, Evans DB, Abbruzzese JL, Hassan MM. Dietary Mutagen Exposure and Risk of Pancreatic Cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(4):655–661. doi: 10.1158/1055-9965.EPI-06-0993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stolzenberg-Solomon RZ, Cross AJ, Silverman DT, Schairer C, Thompson FE, Kipnis V, Subar AF, Hollenbeck A, Schatzkin A, Sinha R. Meat and meat-mutagen intake and pancreatic cancer risk in the NIH-AARP cohort. Cancer Epidemiol Biomarkers Prev. 2007;16:2664–2675. doi: 10.1158/1055-9965.EPI-07-0378. [DOI] [PubMed] [Google Scholar]
- 10.Cross AJ, Peters U, Kirsh VA, Andriole GL, Reding D, Hayes RB, Sinha R. A prospective study of meat and meat mutagens and prostate cancer risk. Cancer Res. 2005;65(24):11779–11784. doi: 10.1158/0008-5472.CAN-05-2191. [DOI] [PubMed] [Google Scholar]
- 11.Zheng W, Lee SA. Well-done meat intake, heterocyclic amine exposure, and cancer risk. Nutr Cancer. 2009;61(4):437–446. doi: 10.1080/01635580802710741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson KE, Mongin SJ, Sinha R, Stolzenberg-Solomon R, Gross MD, Ziegler RG, Mabie JE, Risch A, Kazin SS, Church TR. Pancreatic cancer risk: Associations with meat-derived carcinogen intake in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) cohort. Mol Carcinog. 2012;51(1):128–137. doi: 10.1002/mc.20794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato T, Ohagaki H, Hasegawa H, Sato S, Takayama S, Sugimura T. Induction of tumours in the Zymbal gland, oral cavity, colon, skin and mammary gland of F344 rats by a mutagenic compound 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline. Carcinogenesis. 1988;9:71–73. doi: 10.1093/carcin/9.1.71. [DOI] [PubMed] [Google Scholar]
- 14.Ohagaki H, Hasegawa H, Suenaga M, Sato S, Takayama S, Sugimura T. Carcinogenicity in mice of a mutagenic compound 2-amino-3,8-dimethylimidazo[4,5-b]quinoxaline. Carcinogenesis. 1997;8:665–668. doi: 10.1093/carcin/8.5.665. [DOI] [PubMed] [Google Scholar]
- 15.Turteltaub KW, Mauthe RJ, Dingley KH, Vogel JS, Frantz CE, Garner RC, Shen N. MeIQx-DNA adduct formation in rodent and human tissues at low doses. Mutat Res. 1997;376:243–252. doi: 10.1016/s0027-5107(97)00049-3. [DOI] [PubMed] [Google Scholar]
- 16.Jansen RJ, Robinson DP, Frank RD, Stolzenberg-Solomon RZ, Bamlet WR, Oberg AL, Rabe KG, Olson JE, Petersen GM, Sinha R, Anderson KE. Meat-related mutagens and pancreatic cancer: Null results from a clinic-based case-control study. Cancer Epidemiol Biomarkers Prev. 2013;22(7):1336–1339. doi: 10.1158/1055-9965.EPI-13-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turteltaub KW, Felton JS, Gledhill BL, Vogel JS, Southon JR, Caffee MW, Finkel RC, Nelson DE, Proctor ID, Davis JC. Accelerator mass spectrometry in biomedical dosimetry: relationship between low-level exposure and covalent binding of heterocyclic amine carcinogens to DNA. Proc Natl Acad Sci U S A. 1990;87:5288–5292. doi: 10.1073/pnas.87.14.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel JS, Turteltaub KW, Finkel RC, Nelson DE. Accelerator mass spectrometry—isotope quantification at attomole sensitivity. Anal Chem. 1995;67:A353–A359. doi: 10.1021/ac00107a001. [DOI] [PubMed] [Google Scholar]
- 19.Turesky RJ, Garner C, Welti DH, Richoz J, Leveson SH, Dingley KH, Turteltaub KW, Fay LB. Metabolism of the Food-Borne Mutagen 2-Amino-3,8-dimethylimidazo[4,5-f]quinoxaline in Humans. Chem Res Toxicol. 1998;11:217–225. doi: 10.1021/tx9701891. [DOI] [PubMed] [Google Scholar]
- 20.Creek MR, Mani C, Vogel JS, Turteltaub KW. Tissue distribution and macromolecular binding of extremely low doses of [14C]-benzene in B6C3F1 mice. Carcinogenesis. 1997;18:2421–2427. doi: 10.1093/carcin/18.12.2421. [DOI] [PubMed] [Google Scholar]
- 21.Ognibene TJ, Bench G, Vogel JS, Peaslee GF, Murov S. A high-throughput method for the conversion of CO2 obtained from biochemical samples to graphite in septa-sealed vials for quantification of 14C via accelerator mass spectrometry. Anal Chem. 2003;75:2192–2196. doi: 10.1021/ac026334j. [DOI] [PubMed] [Google Scholar]
- 22.Vogel JS. Rapid production of graphite without contamination for bio medical AMS. Radiocarbon. 1992;34:333–350. [Google Scholar]
- 23.Langouët S, Welti DH, Kerriguy N, Fay LB, Huynh-Ba T, Markovic J, Guengerich FP, Guillouzo A, Turesky RJ. Metabolism of 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in human hepatocytes: 2-amino-3-methylimidazo[4,5-f]quinoxaline-8-carboxylic acid is a major detoxification pathway catalyzed by cytochrome P450 1A2. Chem Res Toxicol. 2001;14(2):211–221. doi: 10.1021/tx000176e. [DOI] [PubMed] [Google Scholar]
- 24.Wakabayashi K, Nagao M, Esumi H, Sugimura T. Food-derived mutagens and carcinogens. Cancer Res. 1992;52:2092s–2098s. [PubMed] [Google Scholar]
- 25.Lynch AM, Knize MG, Boobis AR, Gooderham NJ, Davies DS, Murray S. Intra-and interindividual variability in systemic exposure in humans to 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline and 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine, carcinogens present in cooked beef. Cancer Res. 1992;52:6216–6223. [PubMed] [Google Scholar]
- 26.Stillwell WG, Kidd LC, Wishnok JS, Tannenbaum SR, Sinha R. Urinary excretion of unmetabolized and phase II conjugates of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in humans: relationship to cyto-chrome P4501A2 and N-acetyltransferase activity. Cancer Res. 1997;57:3457–3464. [PubMed] [Google Scholar]
- 27.Helmus DS, Thompson CL, Zelenskiy S, Tucker TC, Li L. Red meat-derived heterocyclic amines increase risk of colon cancer: a population-based case-control study. Nutr Cancer. 2013;65(8):1141–1150. doi: 10.1080/01635581.2013.834945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler LM, Sinha R, Millikan RC, Martin CF, Newman B, Gammon MD, Ammerman AS, Sandler RS. Heterocyclic amines, meat intake, and association with colon cancer in a population-based study. Am J Epidemiol. 2003;157(5):434–445. doi: 10.1093/aje/kwf221. [DOI] [PubMed] [Google Scholar]
- 29.Cotterchio M, Boucher BA, Manno M, Gallinger S, Okey AB, Harper PA. Red meat intake, doneness, polymorphisms in genes that encode carcinogen-metabolizing enzymes, and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17(11):3098–3107. doi: 10.1158/1055-9965.EPI-08-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowell S, Coles B, Sinha R, MacLeod S, Ratnasinghe D, Stotts C, Kadlubar FF, Ambrosone CB, Lang NP. Analysis of total meat intake and exposure to individual heterocyclic amines in a case-control study of colorectal cancer: contribution of metabolic variation to risk. Mutat Res. 2002;506–507:175–785. doi: 10.1016/s0027-5107(02)00164-1. [DOI] [PubMed] [Google Scholar]
- 31.Gilsing AM, Berndt SI, Ruder EH, Graubard BI, Ferrucci LM, Burdett L, Weissfeld JL, Cross AJ, Sinha R. Meat-related mutagen exposure, xenobiotic metabolizing gene polymorphisms and the risk of advanced colorectal adenoma and cancer. Carcinogenesis. 2012;33(7):1332–1339. doi: 10.1093/carcin/bgs158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakasugi H, Hara Y, Abe M. A study of malabsorption in pancreatic cancer. J Gastroenterol. 1996;31:81–85. doi: 10.1007/BF01211191. [DOI] [PubMed] [Google Scholar]
- 33.Yamashita K, Adachi M, Kato S, Nakagama H, Ochiai M, Wakabayashi K, Sato S, Nagao M, Sugimura T. DNA adducts formed by 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in rat liver: dose-response on chronic administration. Jpn J Cancer Res. 1990;81(5):470–476. doi: 10.1111/j.1349-7006.1990.tb02593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mauthe RJ, Dingley KH, Leveson SH, Freeman SP, Turesky RJ, Garner RC, Turteltaub KW. Comparison of DNA-adduct and tissue-available dose levels of MeIQx in human and rodent colon following administration of a very low dose. Int J Cancer. 1999;80(4):539–545. doi: 10.1002/(sici)1097-0215(19990209)80:4<539::aid-ijc10>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 35.Takayama K, Yamashita K, Wakabayashi K, Sugimura T, Nagao M. DNA modification by 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in rats. Jpn J Cancer Res. 1989;80:1145–1148. doi: 10.1111/j.1349-7006.1989.tb01644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metry KJ, Neale JR, Bendaly J, Smith NB, Pierce WM, Jr, Hein DW. Effect of N-Acetyltransferase 2 Polymorphism on Tumor Target Tissue DNA Adduct Levels in Rapid and Slow Acetylator Congenic Rats Administered 2-Amino-1-methyl-6-phenylimidazo[4,5-b] pyridine or 2-Amino-3,8-dimethylimidazo-[4,5-f]quinoxaline. Drug Metab Dispos. 2009;37:2123–2126. doi: 10.1124/dmd.109.029512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turesky RJ, Bessette EE, Dunbar D, Liberman RG, Skipper PL. Cytochrome P450-mediated metabolism and DNA binding of 2-amino-1,7-dimethylimidazo[4,5-g]quinoxaline and its carcinogenic isomer 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline in mice. Chem Res Toxicol. 2012;25(2):410–421. doi: 10.1021/tx2004536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turteltaub KW, Vogel JS, Frantz CE, Shen N. Fate and distribution of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine in mice at a human dietary equivalent dose. Cancer Res. 1992;52(17):4682–4687. [PubMed] [Google Scholar]
- 39.Pfau W, Brockstedt U, Shirai T, Ito N, Marquardt H. Pancreatic DNA adducts formed in vitro and in vivo by the food mutagens 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and 2-amino-3-methyl-9H-pyrido[2,3-b]indole (MeA alphaC) Mutat Res. 1997;378(1–2):13–22. doi: 10.1016/s0027-5107(97)00093-6. [DOI] [PubMed] [Google Scholar]
- 40.Zhu J, Rashid A, Cleary K, Abbruzzese JL, Friess H, Takahashi S, Shirai T, Li D. Detection of 2-amino-1-methyl-6-phenylimidazo [4,5-b]-pyridine (PhIP)-DNA adducts in human pancreatic tissues. Biomarkers. 2006;11(4):319–328. doi: 10.1080/13547500600667911. [DOI] [PMC free article] [PubMed] [Google Scholar]


