Abstract
Type 2 diabetes (T2D) is a common, multifactorial disease that is influenced by genetic and environmental factors and their interactions. However, common variants identified by genome wide association studies (GWAS) explain only about 10% of the total trait variance for T2D and less than 5% of the variance for obesity, indicating that a large proportion of heritability is still unexplained. The transcriptomic approach described here uses quantitative gene expression and disease-related physiological data (deep phenotyping) to measure the direct correlation between the expression of specific genes and physiological traits. Transcriptomic analysis bridges the gulf between GWAS and physiological studies. Recent GWAS studies have utilized very large population samples, numbering in the tens of thousands (or even hundreds of thousands) of individuals, yet establishing causal functional relationships between strongly associated genetic variants and disease remains elusive. In light of the findings described below, it is appropriate to consider how and why transcriptomic approaches in small samples might be capable of identifying complex disease-related genes which are not apparent using GWAS in large samples.
Keywords: Type 2 diabetes, Insulin resistance, Obesity, Gene expression, Mexican Americans, VAGES, ADH1B, Transcriptomics, GWAS, eQTL
1. Introduction
1.1. The value of transcriptomic analysis in genetic studies
The transcriptome is the complete set of expression products transcribed from the genome in a specified tissue or population of cells. It is commonly used to refer to the expression of genome-wide protein coding genes and we follow that convention here, although our thesis applies equally to the expression of non-protein coding genes. In this review we discuss the use of whole genome gene expression approaches (referred to here as transcriptomics) in searching for the genes underlying complex genetic diseases. For brevity and clarity we focus on one such metabolic disease, type 2 diabetes (T2D), and its effect in Mexican Americans, as a typical example of a well-studied complex genetic disease. While we focus on this as a single complex genetic disease, it should be noted that T2D is strongly correlated with other complex disorders such as obesity, insulin resistance, the metabolic syndrome, dyslipidemia, cardiovascular disease, and a variety of diabetic complications. Thus, this discussion has wide relevance to other complex genetic diseases. It is our contention that transcriptomic approaches can provide a valuable bridge between the genotype and the phenotype. Recent studies have highlighted several issues in connecting genotypic approaches, such as (1) genome-wide association studies (GWAS), (2) genome-wide sequencing (GWS), and (3) expression quantitative trait locus (eQTL) analysis, with physiological disease phenotypes. We propose that the gap between these genotypic approaches and resulting phenotypes may be bridged or narrowed by the use of transcriptomic approaches. The reason for this is that transcriptomics occupy a unique position between the genomic or genotypic level and the physiologic or phenotypic level. We note that eQTL also provide a potential link between genotype and phenotype but eQTL typically rely on the connection between genotypes and gene expression and thus may be considered an enhanced form of GWAS. Gene expression as RNA provides the direct raw material for protein expression, which acts on and within cells and tissues, influencing so-called endo-phenotypes, and intermediate phenotypes and which eventually are measurable as physiological changes at the whole body level
In this review we also address potentially confounding factors such as the effect of the phenotype, including the disease phenotype, on transcription and on the myriad mechanisms by which proteins undergo post-translational modification. Nevertheless, these factors do not seriously affect the underlying rationale. A second issue is that transcriptomic analysis does not provide a direct causal link between specific nucleotides in the genome and downstream phenotypes such as disease-related clinical traits. This is also a limitation of GWAS and eQTL studies. However, transcriptomic analysis provides a link between RNA transcripts, which are copied directly from specific regions of the genome, and downstream measurable phenotypes. In this sense they provide a bridge between genotype and phenotype. Causal evidence requires careful follow-up experiments in model cells, tissues, organisms and eventually in humans. Transcriptomic experiments provide valuable initial correlational evidence to guide these follow-up studies.
2. Background
2.1. The diabetes pandemic and the burden of diabetes in Mexican Americans
Type 2 diabetes (T2D) is a major public health issue in the US, particularly in minority groups including Mexican Americans, the major sub-group of the US Hispanic population. Over the past 20 years, we have examined the genetics of T2D, obesity, metabolic syndrome and their related traits in low-income Mexican American families in San Antonio, Texas. T2D is a complex cardio-metabolic disorder characterized by both insulin resistance and pancreatic β-cell dysfunction [17] and is associated with significant morbidity from the micro- and macro-vascular complications of the disease. The compound burden of an increasing global T2D epidemic together with its co-morbid conditions including obesity, dyslipidemia and hypertension makes this a major world-wide public health problem [3]. The International Diabetes Federation estimates that globally 382 million people had diabetes in 2013, and that about 592 million people will be afflicted with diabetes by the year 2035. In the US, according to the Centers for Disease Control and Prevention [76], it is estimated that approximately 29.1 million people were afflicted with diabetes during the years 2010–2012, of whom about 21.1 million had diagnosed diabetes, while the remaining 8.1 million were unaware of their disease. The estimated prevalence of diagnosed diabetes based on age-adjusted data for individuals aged 20 years or older exhibits remarkable ethnic disparities (CDC, 2010–2012): non-Hispanic European Americans = 7.6%, African Americans = 13.2%, Asians = 9.0%, and Hispanics = 12.8% (of whom Mexican Americans = 13.9%).
2.2. Genetic variation in T2D and other complex diseases
T2D is a common, multifactorial disease that is influenced by genetic and environmental factors and their interactions. There are many large-scale ongoing efforts to localize and characterize T2D susceptibility genes using genome-wide association study (GWAS) approaches. To date, the GWAS method has achieved substantial success in localizing novel T2D susceptibility loci and loci for T2D-related glycemic traits (about 90 loci), obesity loci (~ 90), and loci for metabolic syndrome or its components (~ 50 loci), e.g. reviews: [4], [20], [28], [29], [41], [47], [51], [64], [65], [67]. However, common variants identified by GWAS explain only about 10% of the total trait variance for T2D [29] and less than 5% of the trait variance for obesity [4], indicating that a large proportion of trait variance, and of heritability, is still unexplained [48]. Therefore, there has been increased interest in the potential role of rare variants in the etiology of common complex diseases (rare variant/common disease hypothesis). These rare variants are postulated to have larger penetrance with potential functional consequences and could contribute to missing heritability [13], [26], [48]. Recent advances in next generation sequencing (NGS) technologies have made it possible to obtain complete information on rare and common variants across the whole exome, genome or within a targeted region [25] and new information on low frequency or rare variants, including exome chip based data, that are associated with T2D is emerging [16], [24], [45], [66], [73].
2.3. Functional translation of T2D association signals and the case for deep phenotyping
A large number of identified T2D loci has been associated specifically with defects in insulin secretion, highlighting the importance of pancreatic β-cell development and dysfunction. Beta cell function is commonly measured by the oral disposition index and the hyperglycemic clamp, which measures insulin secretory capacity. In contrast, relatively few loci have been discovered relating to insulin resistance which is commonly assayed by measuring levels of insulin and glucose during the oral glucose tolerance test (OGTT) and the gold standard euglycemic hyperinsulinemic clamp. However, recent studies in nondiabetic subjects have focused on loci associated with insulin resistance, e.g. reviews by [29], [47], [55], [68]. Since the overlap between the GWAS association signals with T2D and fasting glucose is partial, differential genetic influences on physiological and pathophysiological variation in glucose homeostasis are possible [29], [39]. These localization signals have, so far, yielded few causal gene identifications and the functional relevance of a majority of the implicated genetic variants has yet to be established. More than 85% of GWAS-identified variants, including those for T2D, fall in non-coding regions of the genome, highlighting their potential role in gene regulation [21], [22], [34], [39]. Efforts to functionally assess GWAS susceptibility loci are challenging. To enhance our knowledge of biological mechanisms and translational possibilities corresponding to specific genetic findings, the genotype-driven approach, aided with deep phenotyping, appears to be a more appealing and powerful strategy [52], [69]. Together with more precise characterization of clinical and pathophysiologic phenotypes, a range of deep physiological phenotypes (sometimes referred to as “endophenotypes” or intermediate phenotypes) and “omic-metrics” such as epigenomics, transcriptomics, proteomics and metabolomics can be examined in an integrated fashion to understand the molecular mechanisms underlying the phenotypic expression of diseases such as T2D [31], [40], [43], [63]. It is hoped that these more functionally-specific phenotypes will exhibit higher genetic signal-to-noise ratios and speed causal gene identification.
2.4. Role of transcriptomics in disease gene identification
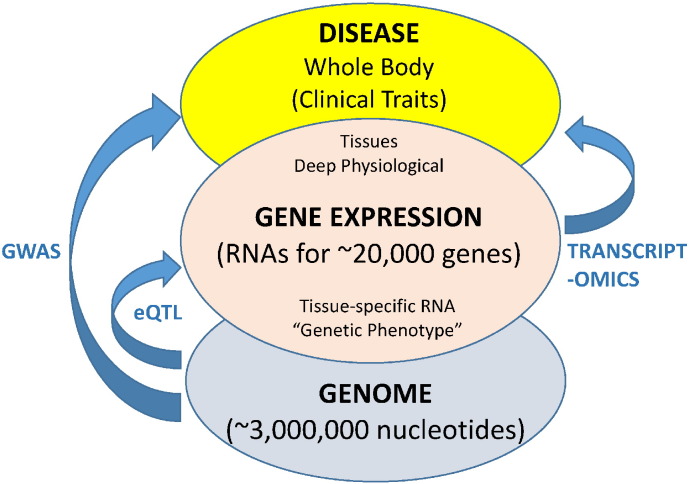
Transcriptomic analysis, the measurement of gene expression at the genome-wide level, bridges the gulf between GWAS and physiological studies (Fig. 1). This is because of the causal relationships between the genome, the transcriptome and physiological traits. Information normally flows from the genome to traits of interest via the transcriptome. Thus, the information residing in genes predisposing to disease is often differentially expressed as RNA whose concentration can be correlated with disease-related traits at the physiological level. Recent GWAS studies have utilized very large population samples, numbering in the tens of thousands (or even hundreds of thousands) of individuals, yet establishing causal functional relationships between strongly associated genetic variants and disease remains elusive [4]. Finally, with the advent of inexpensive and accurate Whole Genome Sequencing (WGS) we are now on the cusp of a revolution in genomic association studies, involving genetic datasets of trillions of nucleotide measurements in combination with multiple physiological measurements [40]. As we enter this exciting era of genetic research, it may be an appropriate time to reassess some of the fundamental assumptions underlying these types of studies.
Fig. 1.
Simplified depiction of the relationship between complex diseases, gene expression and the genome. Both GWAS and transcriptomics seek to deduce the underlying causal genetic relationships between DNA sequence variation and physiological variation. Transcript profiling analyzes a more direct and greatly simplified connection between the genome and complex disease pathology than GWAS. eQTL explicitly analyzes the relationship between DNA sequence variation and gene expression although their extension to phenotypic analysis can be performed in a similar manner to GWAS. It should be noted that the relationships in each case, between nucleotides and clinical traits, are correlative rather than causal. For clarity, the role of active non-coding RNAs is not included although their role is implicit. Their incompletely understood function would presumably mimic that of the relatively well-understood protein coding RNAs. Furthermore, their concentration can be accurately assayed by the same transcriptomic approaches used for protein-coding RNAs and their similar transcript variants.
2.4.1. GWAS vs physiological studies
In striking contrast to recent GWAS results is the remarkable paradox that many physiological correlative studies can be well-powered to detect clinically meaningful and scientifically generalizable information with modest sample sizes, often including less than 100 individuals, e.g. [11], [14], [15], [46], [54], [61]. The paradox lies in the fact that the physiological traits under investigation are often highly heritable and strongly genetically determined. The majority (> 85%) of GWAS signals are located in non-protein-coding DNA sequences, which collectively comprise 98.5% of the genome, and it is likely that their effect is due to their location within or near to regulatory regions which modulate gene expression. It is also likely that, given the GWAS data and what is currently known about the regulation of gene expression, multiple variants can act in concert to regulate the expression of genes which may be involved in key physiological processes, thereby contributing to complex multifactorial traits/diseases. Transcriptomics provides a method to detect these combinatorial effects by measuring changes in the “genetic phenotype” (i.e. gene expression) which is functionally intermediate between the genome sequence and clinically important traits. This biological gap is further reduced when deep physiological traits are used. By definition, these traits lie closer to gene effects than truncated clinical traits, such as the presence or absence of T2D or obesity, which are commonly used in large-scale genetic epidemiological investigations.
2.4.2. Complexity of gene regulation
Gene regulation is a complex multi-layered process involving numerous proteins and non-coding RNAs which may act at a great distance from their target gene. Elaborate multi-protein/RNA complexes must be assembled at the site of regulation. The regulatory mechanism may be intricate and variable, potentially involving transcript rearrangement and mRNA degradation. It is now clear that RNA has a diverse set of functions and is more than just a messenger between gene and protein. The mammalian genome is extensively transcribed, giving rise to thousands of RNA transcripts that are never translated into proteins. Whether all of these transcripts are functional is currently debatable, but it is evident that these include families of RNA molecules with a regulatory function [34]. The presence of a gene expression change, which is strongly correlated with relevant physiological changes, in the absence of proximate significant GWAS signals, suggests that relatively distant regulatory variants (and potentially many such variants) may act in combination to regulate the expression of the target gene of interest. Such putative gene expression-modulating variants could potentially act upon target gene expression through the mediation of non-protein-coding regulatory RNAs. For example, recent studies have shown that the expression of many genes is modulated by small interfering RNAs (siRNAs) and micro-RNAs, e.g. reviews by [10], [30], which do not encode proteins. In addition to microRNAs, many non-protein-coding RNA species (or “RNA genes”), such as long non-coding RNAs [42], are transcribed from the genome. Thus, there is compelling evidence that most of the genome may be transcribed [5], [6], [9], [19], [38], [53], [58], [59], [62] and the potential role of non-protein coding RNA genes in the modulation of protein-coding gene expression remains to be fully evaluated.
2.4.3. Mode of action of expression regulatory variants
The putative gene-expression-modulating variants could, in principle, act either through non-coding RNAs or through protein coding RNAs (via proteins). siRNAs have been shown to operate by binding to mRNAs, leading to premature mRNA degradation and hence down-regulation of gene expression [10]. Those proteins known to be involved in gene expression modulation operate by binding, in a coordinated sequential fashion, to enhanceosomes, repressosomes, spliceosomes and transcriptosomes (for review see [2]). In some cases the regulatory binding site has been shown to reside far from the putative target coding sequence, in regions between genes — so-called “gene deserts” [60], [72]. Regulatory binding sites thus integrate the effect of multiple signals and the effect of individual proteins or RNAs may be multiplicative, by analogy with the amplification effects of enzyme regulatory complexes found in intermediary metabolism and other cellular processes. Thus, such putative genetic mechanisms could evade GWAS approaches designed to detect the association of common or rare variants with whole-body physiological traits such as the presence or absence of T2D or obesity. In contrast, transcriptomic techniques measure gene action directly and can bypass the relatively weak signals of widely scattered variants within the genome.
2.5. Transcriptomics vs expression quantitative trait loci (eQTL) approaches
The transcriptomic approach uses quantitative gene expression data and disease-related physiological data to measure the direct correlation between the expression of specific genes and physiological traits. A related but conceptually distinct technique measures the correlation of genetic variants with gene expression. This approach seeks to identify genomic regions, termed expression quantitative trait loci (eQTL) that are significantly associated with the expression of specific genes. Thus, eQTL analysis lies between GWAS and the transcriptomic gene profiling described here. In some respects it may be considered a subtype of GWAS or an enhanced form of GWAS. However, unlike GWAS, eQTL analysis tends to require relatively smaller sample sizes. The power for a given sample size depends on the effect size and it is reasonable to expect larger genetic effects on transcription than on typical GWAS signals at the nucleotide level. Thus, sample size requirements may often be smaller than for GWAS. The number of statistical tests also affects power, since it determines the false positive error rate. In particular, the GWAS approach can be problematic because the massive number of statistical tests, between genetic variants and traits of interest, presents an enormous potential for false positive results. Whether considering genotyped variants of several hundred thousand or several million, including imputed variants, the scale of the problem is vast. In the case of whole genome sequencing, the problem is at least an order of magnitude greater. This also presents a problem for eQTL analyses, which measure the correlation between a similar number of genetic variants and the expression of transcripts derived from microarray or RNAseq data. In contrast, multiple testing presents much less of a problem for transcriptomic analysis, which measures the correlation between the expression of a comparatively small number of transcripts and physiological traits of interest.
Since eQTL signals involve a necessary intermediate step between genomic variants and physiological traits, they would be expected to provide support for some GWAS signals. This has been shown empirically to be the case with many loci for T2D [32]. The great majority of eQTL identified to date have been found to act in cis with the gene of interest rather than in trans [27]. Part of the explanation is the greatly reduced power to detect trans effects given the enormous multiple testing issues related to searching for trans eQTL, where most of the 3 billion bp of the genome must be searched, rather than a few hundred kbp for cis eQTL. The situation may be worse if there are multiple trans eQTL for a given gene of interest, scattered throughout the genome, and they act combinatorially and multiplicatively on gene expression (see Section 2.6 below). This may provide a partial explanation for the fact that many GWAS signals appear to act in trans. This is because a significant fraction of variants cannot, with certainty, be ascribed to act upon any particular gene because they tend to lie at some distance from any protein coding sequence and, therefore, their mode of action is less likely to occur in cis with respect to genes and potentially more likely to act in trans. A large proportion (43%) of GWAS hits are located in intergenic regions [33]. The lack of close proximity of many potential regulatory variants to coding sequences may in part be due to the fact that protein coding sequence accounts only for approximately 1.5% of the genome [35]. Nevertheless this assumption of major trans effects remains to be validated by rigorous laboratory testing of the effect of potential regulatory variants on the expression of hypothetical target genes. In general, such tests are not currently feasible on a large scale.
2.6. Gene expression variants and GWAS
In light of the findings described below (Section 2.7), it is appropriate to consider how and why transcriptomic approaches in small samples might be capable of identifying complex disease-related genes which are not apparent using GWAS in very large samples. With regard to the considerations discussed above for gene identification in complex diseases, we may consider a hypothetical simplified example. If two genetic variants exist which act combinatorially and multiplicatively to strongly regulate the expression of a protein-coding gene “G” but are located sufficiently far apart within the genome, such that the effect of one or both of the variants is operating “in trans” with respect to gene “G”, then the association of the two variants with a clinical trait of interest might be effectively invisible to detection by GWAS, even if both variants could be effectively targeted by specific oligonucleotides. This is because the association of each variant with the trait of interest may be too weak to reach significance in association testing even though the combined and multiplicative effects of the variants have a strong effect on the expression of “G”, which in turn influences the clinical trait. As previously noted, many GWAS signals are in fact located in non-exomic regions of the genome and are suspected of being regulatory variants or of tagging local regulatory sequences which are often non-canonical, such that a clear functional sequence pattern is difficult to discern. The situation becomes even more difficult if multiple genetic variants are involved (see below for a more complex hypothetical example) and if many or all of the variants are rare. In such a scenario it is not unreasonable to propose that association of gene expression signals with a clinical trait of interest may tend to be stronger than the association of individual nucleotide variants. As one example of such a putative mechanism, in a recent study of eQTL contributing to the risk for coronary artery disease, multiple cis and trans eQTL were identified, none of which contributed to the well-known 9p21.3 risk locus, containing the “first common variant associated with coronary artery disease to yield to the GWAS approach” [12]. Similarly, other studies have shown the presence of multiple trans eQTL which “are far from any GWAS association signals and thus cannot be identified from the GWAS alone” [32]. This concept finds additional support in the empirical example described below (Section 2.7).
2.6.1. Multiple distributed regulatory expression variants
Here we expand upon the theoretical example from the previous section to a more complex example. Instead of a single regulatory SNP acting in cis, relative to “G”, the regulatory region might instead, and more realistically, be hypothetically distributed over perhaps 10 genetic variants throughout the genome, all acting in trans or in combination with one or more cis variants. If all 10 variants are involved in the regulation of the “G” transcript, then the potential GWAS signal, that might otherwise have resided in a single SNP, could be diluted by several orders of magnitude. This effect would be worsened if truncated high-level phenotypes are used (e.g. the presence or absence of T2D or obesity) rather than quantitative deep physiological phenotypes (e.g. measurements from the oral glucose tolerance test [OGTT] and the euglycemic hyperinsulinemic clamp). In effect, the use of an intermediate “genetic phenotype”, gene expression, which is closer to both clinically relevant clinical phenotypes and to the immediate effect of gene action, further increases the power to detect a clinical effect of gene action. This is because the gap between the genomic measurement and the end-phenotype is greatly decreased and multiple genetic effects from individual nucleotides converge on a single gene expression phenotype. Thus, this “concentrating” effect suggests that the design of many GWAS experiments may tend to dilute the effects they seek to elucidate. Another factor enhancing transcriptomic gene identification compared with GWAS is that it isolates and effectively concentrates gene activity within a specific tissue and metabolic state, whereas GWAS necessarily targets the whole genome at all times and states.
2.7. Empirical example of gene discovery by transcriptomics
For illustrative purposes, an example of the transcriptomic approach described herein is provided by the recent identification of a gene, alcohol dehydrogenase 1B (ADH1B) with unexpectedly robust gene expression correlations with multiple deep phenotypes related to T2D. Detailed results are provided elsewhere [74]. RNA was extracted from abdominal subcutaneous adipose tissue biopsies from 75 unrelated non-diabetic Mexican American subjects and genome-wide gene expression measured on the Illumina HumanHT-12v4 Expression BeadChip platform, containing 47,324 oligonucleotide probes per array (derived from RefSeq release 38 and other sources), providing genome-wide transcriptional coverage of well-characterized genes, gene candidates, and splice variants. Microarray data were deposited in the Gene Expression Omnibus (GEO) database under ID GSE64567. ADH1B expression was inversely correlated with all obesity/insulin resistance (OB/IR) related variables at the tissue and whole body levels: waist circumference (P = 2.8 × 10− 9); BMI (P < 0.0001); fasting plasma insulin (P < 0.001); the Homeostasis Model Assessment of Insulin Resistance, HOMA-IR [50] (P < 0.01); Matsuda Insulin Sensitivity Index [49] (P < 0.01); β-cell function (Oral Disposition Index [71] across 120 min of the oral glucose tolerance test (ODI1–120; OGTT) (P = 8.6 × 10− 4), and pre-T2D (impaired glucose tolerance and fasting glucose during the OGTT) (P = 8.6 × 10− 5); and from the euglycemic hyperinsulinemic clamp technique [18]: total glucose disposal (TGD; P = 8.6 × 10− 10); hepatic glucose production (HGP, P = 8.6 × 10− 4); and free fatty acid IR index (FFA-IRI; P = 1.3 × 10− 4). The TGD rate mainly represents skeletal muscle IR; HGP indicates hepatic IR and FFA IR indicates adipose IR. As expected, ADH1B protein expression was decreased with high BMI (P < 0.05, N = 12). Transcription results for ADH1B were confirmed using quantitative PCR. In summary, decreased adipose ADH1B expression was associated with increased obesity, whole body IR, liver IR, skeletal muscle IR, adipose tissue IR, and declining β-cell function. While proof of causality remains to be established, the finding of strong consistent correlations across an array of phenotypes, including several deep physiological phenotypes, in a modest sample size, increases the prior probability that ADH1B is involved in T2D causation and helps prioritize functional testing. A cautionary note is that some of these physiological traits are, to some extent, correlated with each other and the apparent correlation between gene expression and these physiological traits could be partially explained by a reduced number of physiological variables.
In addition to the transcriptomic data obtained from 75 individuals, described above, the ADH1B chromosomal region (4q22) contains GWAS and eQTL data from approximately 69,000 and 1569 individuals, respectively. There were no significant GWAS signals found for association with T2D or related phenotypes (BMI, FPG, FPI) within at least 200 kb of ADH1B using GWAS SNP data (http://www.type2diabetesgenetics.org/accessed 4/30/2015). GWAS T2D and related trait data were obtained from a meta-analysis of 69,000 individuals of European ancestry and covered a total of 3.1 million SNPs. Similarly, no significant eQTL were found for ADH1B in subcutaneous adipose or any of more than 60 tissues examined (http://www.gtexportal.org/accessed 4/30/2015). The eQTL data were derived from 1569 RNAseq samples and contained 8930 significant genes. As previously noted, the lack of localized GWAS and eQTL signals should not be surprising and provides some support for the proposed model of gene action.
2.7.1. Relative gene expression and tissue availability for transcriptomic studies
The case for transcriptomic studies is likely to be strengthened when the gene of interest is highly expressed in the tissue(s) of interest and can, therefore, provide a potentially stronger and more accurate signal. According to the EMBL-EBI Expression Atlas (http://www-test.ebi.ac.uk) using data obtained by microarray assay of 76 normal human tissues (http://BioGPS.org; accessed 5/12/2015), ADH1B was most highly expressed in adipose tissue, with almost double the expression level found in the next highest tissue, liver. This finding was confirmed independently by high resolution sequencing of RNA (RNASeq) from 16 human tissues (Illumina Body Map 2.0; accessed 5/12/2015). In the RNAseq study, ADHIB had substantially higher expression in adipose tissue than in 15 other major tissues examined. Its expression, measured by RNAseq, was more than 3-fold higher in adipose tissue than in the next highest tissue, liver. On the other hand, transcriptomic studies in general are dependent on a limited source of available tissue which may not always be optimal for the studies' objective. In the present example, the tissue under study was highly relevant to the disease of interest and the results were not confounded by the presence of disease [27].
2.7.2. Some advantages and disadvantages of transcriptomic studies
A key advantage of transcriptomic studies is the ability to detect causative genes for complex genetic traits due to the proximity of transcripts and phenotypes of interest. A second potential advantage is the opportunity to measure environmental effects on transcription. Many transcriptomic studies tend to have case/control designs because of the difficulty of obtaining repeated samples of solid tissue, with the notable exception of liquid blood samples which can be easily sampled repeatedly over time. In either case, it is possible to incorporate environmental impacts in the study design, either by measuring differential environmental effects on the study groups or by measuring environmental effects over time and measuring the correlation of transcript expression with environmental exposure.
A limitation of transcriptomic studies is that relevant tissues are often not available and this difficulty is not easily overcome. One promising approach is the use of induced pluripotent stem cells (iPSCs) derived, for example, from human lymphoblastoid cell lines, although this too currently has limitations, including the accurate recapitulation of the target tissue phenotype. Secondly, some important genetic mechanisms may not be mediated through an effect on transcript levels, or protein levels, and thus may not be detectable by transcriptomic methods. Such mechanisms include nonsynonymous variants that may not change RNA or protein levels but may alter protein function. In this regard, transcriptomic studies can be complementary to genetic association or whole genome sequencing approaches.
2.7.3. Methods for dealing with confounding effects
Transcription is a dynamic process and it can be challenging to separate confounding effects on gene expression. One commonly used approach, used in the empirical example described above, in which disease was not a factor, is to use covariates such as age and sex in multiple regression analysis. Causal models have been developed to address issues of potential confounding. To take a hypothetical example, we may observe a correlation between a trait, such as BMI, and a gene's expression level. The obvious question to ask is whether this correlation, regardless of significance, is causal or not. If it is causal, then the gene's expression level will influence BMI, perhaps with some time lag. Conversely, there could be reverse causality, with the effect being reversed, such as the influence of BMI on a gene's expression level. More generally, there could be confounding of apparent causality by other unknown factors. As a hypothetical example, people with higher BMI might tend to come from socioeconomically deprived backgrounds that predispose them to eat low quality calorically dense foods more often, and this diet might also influence the expression of the gene(s) in question, even if the gene's expression itself does not lead to obesity
To disentangle such complex relationships, analytical methods have been developed, such as mediation analysis [44] and Mendelian randomization [8]. For example, the latter method works because genetic variants are randomized among us, by the rules of Mendelian inheritance (and some additional assumptions such as non-assortative mating) and are generally not correlated with possible confounders, such as someone's socioeconomic status and/or diet. Mendelian randomization is one of the valuable features that distinguish genetic epidemiology from most other fields of epidemiology. In the present case, if we have a genetic variant that is known to influence the expression level of the gene(s) in question, such as a cis-regulatory variant, then we could test whether this variant is also associated with BMI. If the gene's expression level causally influences BMI, then we should be able to observe an association of the cis-eQTL(s) with BMI as well. Alternatively, if causality is reversed, i.e. BMI influences gene expression, then there should be no such association between cis-eQTL(s) and BMI. This can be carried further by looking at effect sizes, e.g. if a doubling of gene expression correlates with a BMI increase of 10 units, then a cis-eQTL increasing gene expression by 50% should lead to a BMI increase of roughly 5 units. In any case, the use of this analytical strategy requires that we have cis-eQTLs for the gene(s) in question, or trans eQTLs, but this is much more unlikely. These eQTLs should come from the same tissue and preferably the same ethnicity.
2.8. Epigenomics, metabolomics and proteomics
In the context of gene expression modulation, three other related mechanisms should also be considered. (1) epigenomics/chromatin remodeling, (2) metabolomics, and (3) proteomics. Epigenomics and chromatin remodeling refer, respectively, to reversible heritable changes in DNA sequence and changes in chromosome structure. These occur most commonly via nucleotide and histone methylation and acetylation which affect localized chromosomal regions and gene expression via availability of DNA sequence and potential steric hindrance of transcriptosome formation. Epigenomics and associated chromosomal changes must be considered a potential mechanism of action in gene expression modulation. Epigenomic regulation of gene expression represents a sub-class of gene expression modulation and is, in principle, subject to the same kinds of mechanisms and processes as the modulation of RNA concentrations which are measured by transcriptomic profiling. However, as with GWAS's, whole genome epigenetic changes also occur at the level of the genome sequence and suffer from a similar difficulty in quantitation at the genome-wide level, and for similar reasons. Regions of epigenetic changes are influenced by multiple extrinsic and intrinsic factors and effectively represent changes at the genomic DNA level which can be considered analogous to DNA sequence differences similar to those measured by GWAS but with a greater variability, making epidemiological-scale measurements difficult to interpret [1], [70]
2.8.1. Complexity of proteomic and metabolomic analysis
Proteomics holds conceptually greater promise as an additional measure of disease-related genetic change given that it takes the “genetic phenotype” of RNA expression at least one step closer to the ultimate clinical phenotype of interest. However, with proteins and their multiple derivatives an enormous amount of complexity is introduced and this currently is difficult to resolve [75]. Proteomic and metabolomic measurements can be exquisitely sensitive to changes in the concentrations of specific proteins and metabolites [23]. However, the task of de novo identification of novel protein products and metabolites is more daunting than that of RNA profiling. In comparison, transcriptomic measurements, which rely on the identification of simple four-nucleotide polymers, whether measured by oligonucleotide arrays or RNA sequencing, are currently more amenable to disease gene identification. Thus, genome-wide analyses of very large numbers of proteins and metabolites, potentially orders of magnitude greater than RNA species, currently are limited by their inability to simultaneously identify and measure all possible species encoded by the genome. This is a technical issue which eventually should be resolved by better high-throughput high-resolution technology and likely will become a valuable adjunct to future transcriptomic analysis
It should be noted that there is not a simple 1:1 relationship between RNA expression and protein expression (including expression of covalently modified proteins) and this situation is even more complex for the biosynthesis and use of metabolites. This lack of a simple relationship between RNA expression and protein/metabolite expression makes interpretation of proteomic and metabolomic data challenging. In this regard, one lesson that may be drawn from GWAS analyses is that larger datasets may not always provide simple answers in multifactorial diseases where the underlying structure is very complex. For example, a small proportion of a given protein may become highly active after phosphorylation and become capable of initiating an exponential intracellular catalytic cascade of activity out of proportion to the expression of the original non-phosphorylated protein. Thus, the concentrations of multiple phosphorylated and dephosphorylated proteins may need to be measured (not to mention the concentrations of numerous other types of covalently modified proteins). As with GWAS, eQTL and transcriptomic correlations, the measurable proteomic and metabolomic changes are ultimately encoded within the genome and transferred via expression of RNAs to eventually interact with the intracellular and extra-cellular environment.
2.9. Identification of accessible chromosomal regions for gene expression regulation
Newer epigenomics-related approaches can aid the search for meaningful non-coding variation by focusing on regions of the genome where regulatory protein complexes are likely to be bound, so-called nuclease hypersensitivity sites. Such methods are based on the location of genome-wide sterically accessible open chromatin regions. These techniques exploit the power of massively parallel next generation sequencing (NGS) coupled with nuclease digestion to elucidate the genome-wide location and sequences of gene expression regulatory sites. Current methods include: (1) the assay for transposase-accessible chromatin using sequencing (ATAC-seq) [7], which locates putative sites for all DNA-binding proteins, and (2) chromatin immunoprecipitation with sequencing (ChIP-seq) [36], [37] which locates the binding sites of known or suspected DNA-binding proteins. We have previously used ChIP-seq to elucidate genome-wide putative binding sites for the TCF7L2 transcription factor protein and identified binding sites for TCF7L2 regulation of expression of hepatocyte gluconeogenic genes which appear to be involved in hepatic insulin resistance [56], [57]. Although ATAC-seq and ChIP-seq analyses can be performed independently of transcriptomic analysis, they represent powerful synergistic approaches to identify specific sites of regulation of gene expression uncovered by transcriptomic studies.
3. Conclusion
In this review we have presented evidence that transcriptomic studies may offer insights into physiological mechanisms that are not easily obtained from GWAS or eQTL. We propose that the effects of multiple genetic variants may “converge” on a single genetic mechanism that can be assayed by calculating the correlation between the levels of a particular transcript and phenotype, whereas the effects of individual genetic variants are comparatively small and difficult to detect. This approach could allow transcriptomic studies to yield insights from much smaller sample sizes than have hitherto proven to be necessary for GWAS, particularly if “deep” phenotypes, that are closer to the physiological traits of interest, are analyzed. We discussed an empirical example where highly significant associations were detected between the levels of the transcript for alcohol dehydrogenase 1B, measured in adipose tissue, and multiple measures of obesity and insulin resistance. We conclude that transcriptomic analysis can provide a more convenient and direct path bridging the genotype and phenotype than is currently available using either GWAS or eQTL approaches.
Conflict of interest statement
The authors warrant that they have no conflict of interest with regard to the material in this manuscript.
Author agreement
All authors have seen and approved the manuscript being submitted. They warrant that the article is the authors' original work, has not received prior publication and is not under consideration for publication elsewhere.
References
- 1.Almen M.S., Jacobsson J.A., Moschonis G., Benedict C., Chrousos G.P., Fredriksson R., Schioth H.B. Genome wide analysis reveals association of a FTO gene variant with epigenetic changes. Genomics. 2012;99:132–137. doi: 10.1016/j.ygeno.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez M., Rhodes S.J., Bidwell J.P. Context-dependent transcription: all politics is local. Gene. 2003;313:43–57. doi: 10.1016/s0378-1119(03)00627-9. [DOI] [PubMed] [Google Scholar]
- 3.Apovian C.M. The causes, prevalence, and treatment of obesity revisited in 2009: what have we learned so far? Am. J. Clin. Nutr. 2010;91:277S–279S. doi: 10.3945/ajcn.2009.28473A. [DOI] [PubMed] [Google Scholar]
- 4.Arya R., Puppala S., Farook V.S., Chittoor G., Jenkinson C.P., Blangero J., Hale D.E., Duggirala R., Almasy L. Mapping of susceptibility genes for obesity, type 2 diabetes, and the metabolic syndrome in human populations. In: Almasy L., Duggirala R., Williams-Blangero S., Paul S.F.D., Kole C., editors. Genome Mapping and Genomics in Animals. Springer-Verlag; Berlin Heidelberg: 2015. [Google Scholar]
- 5.Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., Gerstein M., Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 6.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E., Kuehn M.S., Taylor C.M., Neph S., Koch C.M., Asthana S., Malhotra A., Adzhubei I., Greenbaum J.A., Andrews R.M., Flicek P., Boyle P.J., Cao H., Carter N.P., Clelland G.K., Davis S., Day N., Dhami P., Dillon S.C., Dorschner M.O., Fiegler H., Giresi P.G., Goldy J., Hawrylycz M., Haydock A., Humbert R., James K.D., Johnson B.E., Johnson E.M., Frum T.T., Rosenzweig E.R., Karnani N., Lee K., Lefebvre G.C., Navas P.A., Neri F., Parker S.C., Sabo P.J., Sandstrom R., Shafer A., Vetrie D., Weaver M., Wilcox S., Yu M., Collins F.S., Dekker J., Lieb J.D., Tullius T.D., Crawford G.E., Sunyaev S., Noble W.S., Dunham I., Denoeud F., Reymond A., Kapranov P., Rozowsky J., Zheng D., Castelo R., Frankish A., Harrow J., Ghosh S., Sandelin A., Hofacker I.L., Baertsch R., Keefe D., Dike S., Cheng J., Hirsch H.A., Sekinger E.A., Lagarde J., Abril J.F., Shahab A., Flamm C., Fried C., Hackermuller J., Hertel J., Lindemeyer M., Missal K., Tanzer A., Washietl S., Korbel J., Emanuelsson O., Pedersen J.S., Holroyd N., Taylor R., Swarbreck D., Matthews N., Dickson M.C., Thomas D.J., Weirauch M.T., Gilbert J., Drenkow J., Bell I., Zhao X., Srinivasan K.G., Sung W.K., Ooi H.S., Chiu K.P., Foissac S., Alioto T., Brent M., Pachter L., Tress M.L., Valencia A., Choo S.W., Choo C.Y., Ucla C., Manzano C., Wyss C., Cheung E., Clark T.G., Brown J.B., Ganesh M., Patel S., Tammana H., Chrast J., Henrichsen C.N., Kai C., Kawai J., Nagalakshmi U., Wu J., Lian Z., Lian J., Newburger P., Zhang X., Bickel P., Mattick J.S., Carninci P., Hayashizaki Y., Weissman S., Hubbard T., Myers R.M., Rogers J., Stadler P.F., Lowe T.M., Wei C.L., Ruan Y., Struhl K., Gerstein M., Antonarakis S.E., Fu Y., Green E.D., Karaoz U., Siepel A., Taylor J., Liefer L.A., Wetterstrand K.A., Good P.J., Feingold E.A., Guyer M.S., Cooper G.M., Asimenos G., Dewey C.N., Hou M., Nikolaev S., Montoya-Burgos J.I., Loytynoja A., Whelan S., Pardi F., Massingham T., Huang H., Zhang N.R., Holmes I., Mullikin J.C., Ureta-Vidal A., Paten B., Seringhaus M., Church D., Rosenbloom K., Kent W.J., Stone E.A., Nisc Comparative Sequencing Program, Center Baylor College of Medicine Human Genome Sequencing, Center Washington University Genome Sequencing, Institute Broad, Institute Children's Hospital Oakland Research, Batzoglou S., Goldman N., Hardison R.C., Haussler D., Miller W., Sidow A., Trinklein N.D., Zhang Z.D., Barrera L., Stuart R., King D.C., Ameur A., Enroth S., Bieda M.C., Kim J., Bhinge A.A., Jiang N., Liu J., Yao F., Vega V.B., Lee C.W., Ng P., Shahab A., Yang A., Moqtaderi Z., Zhu Z., Xu X., Squazzo S., Oberley M.J., Inman D., Singer M.A., Richmond T.A., Munn K.J., Rada-Iglesias A., Wallerman O., Komorowski J., Fowler J.C., Couttet P., Bruce A.W., Dovey O.M., Ellis P.D., Langford C.F., Nix D.A., Euskirchen G., Hartman S., Urban A.E., Kraus P., Van Calcar S., Heintzman N., Kim T.H., Wang K., Qu C., Hon G., Luna R., Glass C.K., Rosenfeld M.G., Aldred S.F., Cooper S.J., Halees A., Lin J.M., Shulha H.P., Zhang X., Xu M., Haidar J.N., Yu Y., Ruan Y., Iyer V.R., Green R.D., Wadelius C., Farnham P.J., Ren B., Harte R.A., Hinrichs A.S., Trumbower H., Clawson H., Hillman-Jackson J., Zweig A.S., Smith K., Thakkapallayil A., Barber G., Kuhn R.M., Karolchik D., Armengol L., Bird C.P., de Bakker P.I., Kern A.D., Lopez-Bigas N., Martin J.D., Stranger B.E., Woodroffe A., Davydov E., Dimas A., Eyras E., Hallgrimsdottir I.B., Huppert J., Zody M.C., Abecasis G.R., Estivill X., Bouffard G.G., Guan X., Hansen N.F., Idol J.R., Maduro V.V., Maskeri B., McDowell J.C., Park M., Thomas P.J., Young A.C., Blakesley R.W., Muzny D.M., Sodergren E., Wheeler D.A., Worley K.C., Jiang H., Weinstock G.M., Gibbs R.A., Graves T., Fulton R., Mardis E.R., Wilson R.K., Clamp M., Cuff J., Gnerre S., Jaffe D.B., Chang J.L., Lindblad-Toh K., Lander E.S., Koriabine M., Nefedov M., Osoegawa K., Yoshinaga Y., Zhu B., de Jong P.J., Encode Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess S., Thompson S.G. CRC Press; New York: 2015. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. [Google Scholar]
- 9.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., Kodzius R., Shimokawa K., Bajic V.B., Brenner S.E., Batalov S., Forrest A.R., Zavolan M., Davis M.J., Wilming L.G., Aidinis V., Allen J.E., Ambesi-Impiombato A., Apweiler R., Aturaliya R.N., Bailey T.L., Bansal M., Baxter L., Beisel K.W., Bersano T., Bono H., Chalk A.M., Chiu K.P., Choudhary V., Christoffels A., Clutterbuck D.R., Crowe M.L., Dalla E., Dalrymple B.P., de Bono B., Della Gatta G., di Bernardo D., Down T., Engstrom P., Fagiolini M., Faulkner G., Fletcher C.F., Fukushima T., Furuno M., Futaki S., Gariboldi M., Georgii-Hemming P., Gingeras T.R., Gojobori T., Green R.E., Gustincich S., Harbers M., Hayashi Y., Hensch T.K., Hirokawa N., Hill D., Huminiecki L., Iacono M., Ikeo K., Iwama A., Ishikawa T., Jakt M., Kanapin A., Katoh M., Kawasawa Y., Kelso J., Kitamura H., Kitano H., Kollias G., Krishnan S.P., Kruger A., Kummerfeld S.K., Kurochkin I.V., Lareau L.F., Lazarevic D., Lipovich L., Liu J., Liuni S., McWilliam S., Madan Babu M., Madera M., Marchionni L., Matsuda H., Matsuzawa S., Miki H., Mignone F., Miyake S., Morris K., Mottagui-Tabar S., Mulder N., Nakano N., Nakauchi H., Ng P., Nilsson R., Nishiguchi S., Nishikawa S., Nori F., Ohara O., Okazaki Y., Orlando V., Pang K.C., Pavan W.J., Pavesi G., Pesole G., Petrovsky N., Piazza S., Reed J., Reid J.F., Ring B.Z., Ringwald M., Rost B., Ruan Y., Salzberg S.L., Sandelin A., Schneider C., Schonbach C., Sekiguchi K., Semple C.A., Seno S., Sessa L., Sheng Y., Shibata Y., Shimada H., Shimada K., Silva D., Sinclair B., Sperling S., Stupka E., Sugiura K., Sultana R., Takenaka Y., Taki K., Tammoja K., Tan S.L., Tang S., Taylor M.S., Tegner J., Teichmann S.A., Ueda H.R., van Nimwegen E., Verardo R., Wei C.L., Yagi K., Yamanishi H., Zabarovsky E., Zhu S., Zimmer A., Hide W., Bult C., Grimmond S.M., Teasdale R.D., Liu E.T., Brusic V., Quackenbush J., Wahlestedt C., Mattick J.S., Hume D.A., Kai C., Sasaki D., Tomaru Y., Fukuda S., Kanamori-Katayama M., Suzuki M., Aoki J., Arakawa T., Iida J., Imamura K., Itoh M., Kato T., Kawaji H., Kawagashira N., Kawashima T., Kojima M., Kondo S., Konno H., Nakano K., Ninomiya N., Nishio T., Okada M., Plessy C., Shibata K., Shiraki T., Suzuki S., Tagami M., Waki K., Watahiki A., Okamura-Oho Y., Suzuki H., Kawai J., Hayashizaki Y., Fantom Consortium, Riken Genome Exploration Research Group, Group Genome Science The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 10.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chavez A.O., Kamath S., Jani R., Sharma L.K., Monroy A., Abdul-Ghani M.A., Centonze V.E., Sathyanarayana P., Coletta D.K., Jenkinson C.P., Bai Y., Folli F., Defronzo R.A., Tripathy D. Effect of short-term free fatty acids elevation on mitochondrial function in skeletal muscle of healthy individuals. J. Clin. Endocrinol. Metab. 2010;95:422–429. doi: 10.1210/jc.2009-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H.H., Stewart A.F. Making sense of genome-wide association studies: integrating genetic variation with gene expression to derive functional mechanisms underlying disease risk. Circulation. 2015;131:519–521. doi: 10.1161/CIRCULATIONAHA.114.014634. [DOI] [PubMed] [Google Scholar]
- 13.Cirulli E.T., Goldstein D.B. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 14.Coletta D.K., Sriwijitkamol A., Wajcberg E., Tantiwong P., Li M., Prentki M., Madiraju M., Jenkinson C.P., Cersosimo E., Musi N., Defronzo R.A. Pioglitazone stimulates AMP-activated protein kinase signalling and increases the expression of genes involved in adiponectin signalling, mitochondrial function and fat oxidation in human skeletal muscle in vivo: a randomised trial. Diabetologia. 2009;52:723–732. doi: 10.1007/s00125-008-1256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coletta D.K., Fernandez M., Cersosimo E., Gastaldelli A., Musi N., DeFronzo R.A. The effect of muraglitazar on adiponectin signalling, mitochondrial function and fat oxidation genes in human skeletal muscle in vivo. Diabet. Med. 2015;32:657–664. doi: 10.1111/dme.12664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornes B.K., Brody J.A., Nikpoor N., Morrison A.C., Dang H.C., Ahn B.S., Wang S., Dauriz M., Barzilay J.I., Dupuis J., Florez J.C., Coresh J., Gibbs R.A., Kao W.H., Liu C.T., McKnight B., Muzny D., Pankow J.S., Reid J.G., White C.C., Johnson A.D., Wong T.Y., Psaty B.M., Boerwinkle E., Rotter J.I., Siscovick D.S., Sladek R., Meigs J.B. Association of levels of fasting glucose and insulin with rare variants at the chromosome 11p11.2-MADD locus: Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Targeted Sequencing Study. Circ. Cardiovasc. Genet. 2014;7:374–382. doi: 10.1161/CIRCGENETICS.113.000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeFronzo R.A. Banting lecture. From the triumvirate to the ominous octet: a new paradigm for the treatment of type 2 diabetes mellitus. Diabetes. 2009;58:773–795. doi: 10.2337/db09-9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeFronzo R.A., Tobin J.D., Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am. J. Physiol. 1979;237:E214–E223. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 19.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G.K., Khatun J., Williams B.A., Zaleski C., Rozowsky J., Roder M., Kokocinski F., Abdelhamid R.F., Alioto T., Antoshechkin I., Baer M.T., Bar N.S., Batut P., Bell K., Bell I., Chakrabortty S., Chen X., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Falconnet E., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M.J., Gao H., Gonzalez D., Gordon A., Gunawardena H., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Luo O.J., Park E., Persaud K., Preall J.B., Ribeca P., Risk B., Robyr D., Sammeth M., Schaffer L., See L.H., Shahab A., Skancke J., Suzuki A.M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Wrobel J., Yu Y., Ruan X., Hayashizaki Y., Harrow J., Gerstein M., Hubbard T., Reymond A., Antonarakis S.E., Hannon G., Giddings M.C., Ruan Y., Wold B., Carninci P., Guigo R., Gingeras T.R. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., Lindgren C.M., Magi R., Morris A.P., Randall J., Johnson T., Elliott P., Rybin D., Thorleifsson G., Steinthorsdottir V., Henneman P., Grallert H., Dehghan A., Hottenga J.J., Franklin C.S., Navarro P., Song K., Goel A., Perry J.R., Egan J.M., Lajunen T., Grarup N., Sparso T., Doney A., Voight B.F., Stringham H.M., Li M., Kanoni S., Shrader P., Cavalcanti-Proenca C., Kumari M., Qi L., Timpson N.J., Gieger C., Zabena C., Rocheleau G., Ingelsson E., An P., O'Connell J., Luan J., Elliott A., McCarroll S.A., Payne F., Roccasecca R.M., Pattou F., Sethupathy P., Ardlie K., Ariyurek Y., Balkau B., Barter P., Beilby J.P., Ben-Shlomo Y., Benediktsson R., Bennett A.J., Bergmann S., Bochud M., Boerwinkle E., Bonnefond A., Bonnycastle L.L., Borch-Johnsen K., Bottcher Y., Brunner E., Bumpstead S.J., Charpentier G., Chen Y.D., Chines P., Clarke R., Coin L.J., Cooper M.N., Cornelis M., Crawford G., Crisponi L., Day I.N., de Geus E.J., Delplanque J., Dina C., Erdos M.R., Fedson A.C., Fischer-Rosinsky A., Forouhi N.G., Fox C.S., Frants R., Franzosi M.G., Galan P., Goodarzi M.O., Graessler J., Groves C.J., Grundy S., Gwilliam R., Gyllensten U., Hadjadj S., Hallmans G., Hammond N., Han X., Hartikainen A.L., Hassanali N., Hayward C., Heath S.C., Hercberg S., Herder C., Hicks A.A., Hillman D.R., Hingorani A.D., Hofman A., Hui J., Hung J., Isomaa B., Johnson P.R., Jorgensen T., Jula A., Kaakinen M., Kaprio J., Kesaniemi Y.A., Kivimaki M., Knight B., Koskinen S., Kovacs P., Kyvik K.O., Lathrop G.M., Lawlor D.A., Le Bacquer O., Lecoeur C., Li Y., Lyssenko V., Mahley R., Mangino M., Manning A.K., Martinez-Larrad M.T., McAteer J.B., McCulloch L.J., McPherson R., Meisinger C., Melzer D., Meyre D., Mitchell B.D., Morken M.A., Mukherjee S., Naitza S., Narisu N., Neville M.J., Oostra B.A., Orru M., Pakyz R., Palmer C.N., Paolisso G., Pattaro C., Pearson D., Peden J.F., Pedersen N.L., Perola M., Pfeiffer A.F., Pichler I., Polasek O., Posthuma D., Potter S.C., Pouta A., Province M.A., Psaty B.M., Rathmann W., Rayner N.W., Rice K., Ripatti S., Rivadeneira F., Roden M., Rolandsson O., Sandbaek A., Sandhu M., Sanna S., Sayer A.A., Scheet P., Scott L.J., Seedorf U., Sharp S.J., Shields B., Sigurethsson G., Sijbrands E.J., Silveira A., Simpson L., Singleton A., Smith N.L., Sovio U., Swift A., Syddall H., Syvanen A.C., Tanaka T., Thorand B., Tichet J., Tonjes A., Tuomi T., Uitterlinden A.G., van Dijk K.W., van Hoek M., Varma D., Visvikis-Siest S., Vitart V., Vogelzangs N., Waeber G., Wagner P.J., Walley A., Walters G.B., Ward K.L., Watkins H., Weedon M.N., Wild S.H., Willemsen G., Witteman J.C., Yarnell J.W., Zeggini E., Zelenika D., Zethelius B., Zhai G., Zhao J.H., Zillikens M.C., Diagram Consortium, Giant Consortium, BPgen Consortium Global, Borecki I.B., Loos R.J., Meneton P., Magnusson P.K., Nathan D.M., Williams G.H., Hattersley A.T., Silander K., Salomaa V., Smith G.D., Bornstein S.R., Schwarz P., Spranger J., Karpe F., Shuldiner A.R., Cooper C., Dedoussis G.V., Serrano-Rios M., Morris A.D., Lind L., Palmer L.J., Hu F.B., Franks P.W., Ebrahim S., Marmot M., Kao W.H., Pankow J.S., Sampson M.J., Kuusisto J., Laakso M., Hansen T., Pedersen O., Pramstaller P.P., Wichmann H.E., Illig T., Rudan I., Wright A.F., Stumvoll M., Campbell H., Wilson J.F., Consortium Anders Hamsten on behalf of Procardis, Magic investigators, Bergman R.N., Buchanan T.A., Collins F.S., Mohlke K.L., Tuomilehto J., Valle T.T., Altshuler D., Rotter J.I., Siscovick D.S., Penninx B.W., Boomsma D.I., Deloukas P., Spector T.D., Frayling T.M., Ferrucci L., Kong A., Thorsteinsdottir U., Stefansson K., van Duijn C.M., Aulchenko Y.S., Cao A., Scuteri A., Schlessinger D., Uda M., Ruokonen A., Jarvelin M.R., Waterworth D.M., Vollenweider P., Peltonen L., Mooser V., Abecasis G.R., Wareham N.J., Sladek R., Froguel P., Watanabe R.M., Meigs J.B., Groop L., Boehnke M., McCarthy M.I., Florez J.C., Barroso I. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elbein S.C., Gamazon E.R., Das S.K., Rasouli N., Kern P.A., Cox N.J. Genetic risk factors for type 2 diabetes: a trans-regulatory genetic architecture? Am. J. Hum. Genet. 2012;91:466–477. doi: 10.1016/j.ajhg.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farook V.S., Reddivari L., Chittoor G., Puppala S., Arya R., Fowler S.P., Hunt K.J., Curran J.E., Comuzzie A.G., Lehman D.M., Jenkinson C.P., Lynch J.L., DeFronzo R.A., Blangero J., Hale D.E., Duggirala R., Vanamala J. Metabolites as novel biomarkers for childhood obesity-related traits in Mexican-American children. Pediatr. Obes. 2014 doi: 10.1111/ijpo.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flannick J., Thorleifsson G., Beer N.L., Jacobs S.B., Grarup N., Burtt N.P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R., Blangero J., Bowden D.W., Brandslund I., Brosnan J., Burslem F., Chambers J., Cho Y.S., Christensen C., Douglas D.A., Duggirala R., Dymek Z., Farjoun Y., Fennell T., Fontanillas P., Forsen T., Gabriel S., Glaser B., Gudbjartsson D.F., Hanis C., Hansen T., Hreidarsson A.B., Hveem K., Ingelsson E., Isomaa B., Johansson S., Jorgensen T., Jorgensen M.E., Kathiresan S., Kong A., Kooner J., Kravic J., Laakso M., Lee J.Y., Lind L., Lindgren C.M., Linneberg A., Masson G., Meitinger T., Mohlke K.L., Molven A., Morris A.P., Potluri S., Rauramaa R., Ribel-Madsen R., Richard A.M., Rolph T., Salomaa V., Segre A.V., Skarstrand H., Steinthorsdottir V., Stringham H.M., Sulem P., Tai E.S., Teo Y.Y., Teslovich T., Thorsteinsdottir U., Trimmer J.K., Tuomi T., Tuomilehto J., Vaziri-Sani F., Voight B.F., Wilson J.G., Boehnke M., McCarthy M.I., Njolstad P.R., Pedersen O., Consortium Go T.D., Genes Consortium T.D., Groop L., Cox D.R., Stefansson K., Altshuler D. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genomes Project, Consortium, Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Göring H.H. Gene expression studies and complex diseases. In: Duggirala R., Almasy L., Williams-Blangero S., Paul S.F.D., Kole C., editors. Genome Mapping and Genomics in Human and Non-human Primates. Springer-Verlag; Heidelberg, New York, Dordrecht, London: 2015. [Google Scholar]
- 28.Grant S.F., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A., Styrkarsdottir U., Magnusson K.P., Walters G.B., Palsdottir E., Jonsdottir T., Gudmundsdottir T., Gylfason A., Saemundsdottir J., Wilensky R.L., Reilly M.P., Rader D.J., Bagger Y., Christiansen C., Gudnason V., Sigurdsson G., Thorsteinsdottir U., Gulcher J.R., Kong A., Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 29.Grarup N., Sandholt C.H., Hansen T., Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014;57:1528–1541. doi: 10.1007/s00125-014-3270-4. [DOI] [PubMed] [Google Scholar]
- 30.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 31.Haring R., Wallaschofski H. Diving through the “-omics”: the case for deep phenotyping and systems epidemiology. OMICS. 2012;16:231–234. doi: 10.1089/omi.2011.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He X., Fuller C.K., Song Y., Meng Q., Zhang B., Yang X., Li H. Sherlock: detecting gene-disease associations by matching patterns of expression QTL and GWAS. Am. J. Hum. Genet. 2013;92:667–680. doi: 10.1016/j.ajhg.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hrdlickova B., de Almeida R.C., Borek Z., Withoff S. Genetic variation in the non-coding genome: involvement of micro-RNAs and long non-coding RNAs in disease. Biochim. Biophys. Acta. 2014;1842:1910–1922. doi: 10.1016/j.bbadis.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 35.International Human Geenome Sequencing, Consortium, Adekoya E., Ait-Zahra M., Allen N., Anderson M., Anderson S., Anufriev F., Ambruster J., Ayele K., Baker J., Baldwin J., Barna N., Bastien V., Batzoglou S., Beckerly R., Beda F., Bernard J., Birren B., Blumensteil B., Boguslavsky L., Boukghalter B., Brown A., Burkett G., Camarata J., Campopiano A., Carneiro H., Chen Z., Choephal Y., Colangelo M., Collins S., Collymore A., Cooke P., Davis C., Dawoe T., DeArellano K., Devon K., Dewar K., Diaz J.S., Dodge S., Donelan E., Dorjee K., Doyle M., Dube A., Dupes A., Endrizzi M., Farina A., Faro S., Ferguson D., Ferriera P., Fischer H., Fitzhugh W., Flaherty K., Foley K., Funke R., Gage D., Galagan J., Gardyna S., Gilbert D., Ginde S., Gomes A., Goyette M., Graham J., Graham L., Grandbois A., Grand-Pierre N., Grant G., Gregoire D., Guerrero R., Hagos B., Harris K., Hart D., Hatcher B., Heaford A., Horton L., Hosage-Norman C., Howland J., Hulme B., Iliev I., Johnson R., Jones C., Joseph M., Judd M., Kann L., Karatas A., Kelley D., Kelly M., Lama D., Lamazares J., Lander E.S., Landers T., Lane A., LaRocque K., LeBlanc H., Leger J.P., Lehoczky J., LeVine R., Lewis D., Lewis T., Lieu C., Linton L., Liu G., Liu X., Locke K., Lokyitsang Y., Macdonald P., Martinez R., Maru K., McCarthy M., McEwan P., McGhee T., McGing B., McGurk A., McKernan K., McLaughlin J., McPheeters R., Meneus L., Mesirov J., Mihova T., Meldrim J., Meneus L., Mesirov J., Mihova T., Miranda C., Mlenga V., Modeski M., Montello G., Morris W., Morrow J., Mulrain L., Murphy T., Mychaleckyj J., Naylor J., Newes C., Ngodup T., Nguyen C., Nguyen T., Norbu C.D., Norbu N., Nusbaum C., O'Connor T., O'Donnell P., Okaf Y., O'Neil D., O'Shea J., Osman S., Paresi M., Pavlin B., Peterson K.M., Phunkang P., Pierre N., Pollara V., Raymond C., Rieback M., Riley B., Rise C., Rogov P., Roman J., Roman M., Rosetti M., Rothman D., Roy A., Roycraft K., Santos R., Schauer S., Schupbach R., Seaman S., Sheridan A., Smith C., Sougnez C., Speece T., Spencer B., Stange-Thomann N., Stojanovic N., Stone C., Strauss N., Subramanian A., Talamas J., Tchuinga P., Temelko M., Tenzin P., Tesfaye S., Theodore J., Tirrell A., Torruella-Miller I., Trac T., Travers M., Travis N., Trigilio J., Tsao E., Vassiliev H., Veil R., Vo A., Wagner A., Walsh J., Wangdi T., Wierzbowski J., Wilson B., Wu X., Abdellah Z., Ahamdi A., Ahmed S., Aimable M., Ainscough R., Almeida J., Ambler A., Ambrose K., Ambrose K., Andrews D., Andrews N., Arbery H., Archer B., Ash G., Ashcroft K., Ashurst J., Ashwell R., Atkin D., Atkinson A., Atwood J., Aubin K., Avis T., Babbage A., Bacon J., Bagguley C., Bailey J., Baker A., Bardill S., Barker D., Barlow K., Baron L., Barrett A., Bartlett R., Basham D., Basham V., Bateman A., Bates K., Baynes C., Beard L., Beard L., Beard S., Beare D., Beasley A., Beasley O., Beck S., Bell E., Bellerby D., Bellerby T., Bemrose R., Bennett J., Bentley D., Berks M., Berks M., Bethel G., Bird C., Birney E., Bissell H., Blackburne-Maze S., Blakey S., Bonnett R., Bonnett R., Border R., Brady N., Bray J., Bray-Allen S., Bridgeman A., Brook J., Brooking S., Brown A., Brown C., Brown J., Brown M., Brown M., Bruskiewich R., Bryant J., Buck D., Buckle V., Budd C., Burberry J., Burford D., Burgess J., Burrill W., Burrows C., Burton J., Butcher P., Butler A., Cairns M., Caning B., Carder C., Carder P., Carter N., Cavanna T., Chan K., Chapman J., Charles R., Clothia T., Chui C., Clamp M., Clark A., Clark G., Clark G., Clark K., Clark S., Clark S., Clarke B., Clarke E., Clarke K., Clee C., Clegg S., Clifford K., Coates J., Cobley V., Coffey A., Coggill P., Lotte C., Collier R., Collings S., Collins J., Collins P., Connor P., Conquer J., Conroy D., Constance D., Cook L., Cooper J., Cooper R., Cooper R., Copsey T., Corby N., Cornell L., Cornell R., Cottage A., Coulson A., Coville G., Cox A., Cox T., Coxhill R., Craig M., Crane T., Crawley M., Crew V., Cuff J., Culley K., Cummings A., Cummings K., Cummings P., Curran A., Curwen V., Cutts J., Daniels R., Davidson L., Davies J., Davies J., Davies N., Davies R., Davies J., Dawson E., Deadman R., Dean P., Dear S., Dearden F., Delgado M., Deloukas P., Dennis J., Dhami P., Dibling C., Dobbs R., Dobson R., Dockree C., Doddington D., Dodsworth S., Doggett N., Dunham A., Dunham I., Dunn A., Dunn M., Durbin R., Durham J., Dwyer R., Earthrowl M., Eastham T., Edwards C., Edwards K., Ellington A., Ellwood M., Emberson B., Errington H., Evans G., Evans J., Evans K., Evans R., Feltwell T., Fennell S., Finn R., Flack T., Fleming K., Flint J., Flint M., Floyd Y., Footman S., Fowler J., Frame D., Francis M., Francis S., Frankland J., Fraser A., Fraser D., French L., Frost D., Frost J., Frost L., Frost C., Fuller L., Fullerton K., Gardner A., Garner P., Garnett J., Gatland L., Gatland L., Ghori J., Gibbs B., Gibson D., Gilbert J., Gilby L., Gilson C., Gorton M., Grafham D., Grant M., Grant S., Gray I., Green L., Greenhalgh J., Greenhill J., Gregg P., Gregory S., Griffiths E., Griffiths M., Guthrie I., Gwilliam R., Hall R., Halls K., Hall-Tamlyn G., Hamlett J., Hammond S., Hancock J., Harding A., Harley J., Harper D., Harper G., Harradence G., Harrison C.L., Harrison R., Hassan D., Hawkins N., Hawley K., Hayes K., Heath P., Heathcott R., Hembry C., Herd T., Hewitt S., Higgs D., Hillyard G., Hinkins R., Ho S.J., Hodgson D., Hoffs M., Holden J., Holdgate J., Holloway E., Holmes I., Holmes S., Holroyd S., Hoooper A., Hopewell L., Hopkins B., Hornett G., Hornsby G., Hornsby T., Horsley S., Horton R., Howard P., Howden P., Howell G., Hubbard T., Huckle E., Hughes J., Hughes J., Hull L., Hummerich H., Humphray S., Humphries M., Hunt A., Hunt P., Hunt S., Hyde D., Ince M., Isherwood J., Izatt J., Izmajlowicz M., Jareborg N., Jassal B., Jeffery G., Jeffery K., Jeffery C., Jekosch K., Jenkins L., Johansen T., Johnson C., Johnson C., Johnson D., Jolley K., Jones A., Jones C., Jones J., Jones M., Jones M., Jones S., Joseph S., Joy A., Joy L., Joy V., Joyce G., Jubb M., Karunaratne K., Kay M., Kaye D., Kearney L., Kelley S., Kershaw J., Kettleborough R., Kidd C., Kierstan P., Kimberley A., King A., Kingsley S., Klingle G., Knights A., Krogh A., Laidlaw P., Laing M., Laird G., Lambart C., Lamble R., Lamble R., Langford C., Lau T., Lawlor S., Leather S., Lehvaslaiho M., Leonard S., Leongamornlert D., Leversha M., Lightning J., Lindsay S., Line M., Linsdell S., Little P., Lloyd C., Lloyd D., Lock V., Lock W., Lodziak A., Longden I., Loraine H., Lord R., Lovell J., Lye G., Marriott N., Marrone A., Marsden P., Marsh V., Martin M., Martin S., Maslen G., Mason D., Matthews L., Matthews P., Maynard M., McCann O., McClay J., McCollum C., McConnachie L., McDonald B., McDonald L., McDowall J., McKeown C., McLauren S., McLay K., McLean J., McMurdo J., McMurray A., McMurray D., McWilliams N., Mehta N., Menuge N., Mercer S., Miah A., Micklem G., Miles S., Milne S., Mistry D., Mistry S., Mitchell J., Mitchell J., Mohammadi M., Molina C., Mooney P., Moore M., Moreland A., Mortimore B., Mott R., Mullikan J., Munday B., Munday E., Mungall A., Murname C., Murrell K., Myers A., Negus D., Niblett D., Nicholson J., Nickerson T., Nijjar S., Ning Z., Nisbet J., Odell C., O'Donovan D., Ogbighele F., Oinn T., Oliver H., Oliver K., Orbell H., Osborn A., Osborne J., Overton-Larty E., Parkin C., Parkin K., Parry-Brown G., Patel D., Patel R., Pearce A., Pearson D., Peck A., Peck R., Peden J., Percy C., Perito A., Perrault I., Peters A., Pettett R., Phillimore B., Phillips K., Phillips S., Platt D., Playford E., Plumb B., Pocock M., Porter K., Potter C., Potter S., Powell D., Prathalingham R., Quail M., Quince C., Quareshi M., Ramsay H., Ramsey Y., Ranby S., Rance R., Rand V., Ratford J., Ratford L., Read D., Redhead D., Rees C., Reid M., Reinhardt A., Rice A., Rice C., Rice P., Richard S., Richardson S., Ridler K., Riethoven L., Robinson M., Rochford R., Rogers J., Rogers L., Ross H., Ross M., Rule A., Rule J., Russell B., Rutter J., Safdar K., Salter N., Santoyo-Lopez J., Saunders D., Scott C., Scott D., Scott I., Seager F., Searle M., Searle P., Sehra H., Shardelow J., Sharp G., Shaw T., Shaw-Smith C., Shearing J., Sheppard K., Sheppard K., Sheppard R., Sheridan E., Shownkeen R., Silk R., Sims M., Sims S., Sivadasan S., Skuce C., Smink L., Smith A., Smith L., Smith L., Smith M., Smith R., Smith S., Sneath H., Soderlund C., Solovyev V., Sonhammer E., Sotheran E., Spraggon L., Squares J., Squares S., Stables M., Stalker J., Stamford S., Stammers M., Steingruber H., Stephens Y., Steward C., Stewart A., Stewart M., Stock M., Stoppard L., Storey P., Sturdy J., Sulston J., Swainson C., Swann M., Sycamore N., Tagney M., Tan S., Tarling E., Taylor A., Taylor G., Tauylor K., Taylor R., Taylor S., Tee L., Tester J., Theaker A., Thomas C., Thomas D., Thomas K., Thomas R., Thommai R., Thorpe A., Thorpe K., Threadgold G., Tinsley E., Tracey A., Travers J., Tromans A., Tubby B., Tufarelli C., Turney K., Upson D., Vaudin M., Viknaraja R., Vine W., Voak P., Walker S., Wall M., Wallis J., Wallis M., Warren G., Warry G., Watson A., Webb A., Webb J., Wells A., Wells S., Welton R., West P., West T., Wheatley A., Wheatley C., Wheeler G., Whitaker H., White A., White A., White B., White J., White S., Whiteley M., Whittaker A., Whittaker P., Widaa S., Wild A., Wilkinson J., Wilkinson P., Willey D., Williams A., Williams B., Williams L., Williams S., Williamson H., Wilmer T., Wilming L., Wilson B., Wilson G., Wilson M., Wilson N., Wilson S., Wilson W., Window P., Winster J., Witt J., Wobus F., Wood E., Wood J., Woodeson S., Woodhouse R., Wooster R., Wray M., Wray P., Wright K., Wyatt J., Xie J., Young L., Young S., Younger R., Zhao S., Abbas S., Abbott A., Abu-Threideh J., Ahluwalia R., Alexander E., Alhawagri M., Ali J., Allen J., Ames M., Andrews S., Angell S., Antonacci P., Antonacci-Fulton L., Antoniou B., Armstrong J., Arnett C., Atkins V., Austin K., Bailey C., Baisden D., Barbazuk B., Barrett M., Bartko L., Bauer C., Bauer H., Baum D., Beck C., Becker M.C., Bedell J., Behymer K., Behymer S., Belter E., Bemis G., Bentley D., Berghoff A., Bernard K., Bevins Z., Bielicki L., Biewald T., Blackwood L., Blaine R., Blair D., Blanchard M., Blandford M., Blasiar D., Bolandis J., Bolla S., Bollinger T., Bong J., Boren-Prydydasz J., Bourne S., Bova K., Boyer E., Bradford K., Brennan S., Broy M., Buatsi D., Budnicki C., Burkett M., Burkhart J., Buss C., Butler J., Caldwell D., Caldwell R., Cardenas M., Carpenter K., Carter J., Carter T., Carter T., Casimere D., Chapman A., Chiapelli B., Chinwalla A.T., Chissoe S.L., Christy W., Cissell M., Clark B., Clark M.J., Clarke K., Clifton S.W., Cloud J., Coblitz B., Cofman M., Connell M., Conyers J., Cook L.L., Cook M., Cooper M., Coppedge V., Cordes M., Cordum H., Cotton M., Courtney W., Creason K., Crockett J.M., Crouse K., Crum T., Dante M., Davenport R., David M., Davidson S., Davidson T., Davis S., Delehaunty A., Delehaunty K.D., Dempsey S., Desai A., Despot J., Dickes M., Dickinson K., Dietrich N., Dignan G., Dixon R., Doebber A., Doerr N., Donoho M., Dotson M., Doucett J., Drone K., Du F., Du H., Du Z., Dubbelde C., Duckels G., Eddy S., Edinger S., Edwards J., Ehlman T., Eldred J., Elkin A., Elliott G., Exum E., Falk A., Farrow K., Favello A., Fedele J., Fewell G., Ficenec D., Fiedler T., Flagg L., Fleming A., Florence N., Fries J., Fronick W., Fryman J., Fuhrmann D., Fulton L.A., Fulton R.S., Gaige D., Gaige T., Garrett J., Gattung S., Geisel C., Geisel S., Gibson A., Gibson E., Giddings C., Gillam B., Gincherman Y., Gish W.R., Glaser E., Glossip D., Godfrey J., Goela D., Goins N., Gotway J., Goyea-Gbadebo E., Granderson L., Graves T., Gregory S., Grewal S., Griffin J., Grover H., Gualberto G., Gund C., Haakenson W., Haglund K., Hale P., Hale S., Hall T., Hamdan Z., Hannah C., Harkins R., Harmon G., Harper M., Harris A., Harrison M., Hart R., Haub K., Hawkins J., Hawryszko C., Heidbrink C., Hendrix K., Henkhaus J., Henley K., Henry C., Hersberger N., Heyen J., Hickenbotham M., Hill P., Hillen T., Hillier L.D., Hinds K., Hodges J., Hoefgen E., Holbrook L., Hollingsworth H., Holloway P., Holman M., Holmes A., Hotic M., Hou S., Houshmandi S., Howell C., Hoyt D., Hubbard C., Isaiah L., Isak A., Jacobs A., Jaeger S., Jeliti C., Jentes E., Johnson A., Johnson D.L., Jones B., Jones K., Jones R., Joshu C., Kang K., Kassos P., Keen K., Kellen J., Kennedy S., Keppler N., Ketterman M., Kim K., Kitchell S., Klebe D., Klinke B., Kloss J., Knight L., Koch M., Kock J., Kohlberg S., Korg I., Kovcic D., Kraemer J., Kramer J.B., Krasucki P., Krasucki P., Krauss R., Kremitzki C., Kruchowski S., Kucaba T., Lacy M., Lakanen T., Lamar E., Lane K., Langston Y., Latreille J.P., Layman D., Le T., Pham Le T.T., Le T.T., Ledwith J.J., Lee N., Lehnert L., Lennox S., Leonard S., Lesley K., Levin L., Levy A., Lewis S., Li L., Littlejohn T., Long N., Lowery P., Luxen S., Lynch T., Maas J., MacDonald J., Maggi L., Maher M., Marchetto P., Mardis E.R., Markovic C., Marquis-Homeyer C., Marra M.A., Marth G., Martin J.C., Martin J., Martinka S., Maupin R., Maxeiner K., McAdow R., Carther M., McCabe M.C., McCray Q., McDill B., McDonald K., McDonald R., McDonald T., McDonough D., McGrane R., McKinney S., McLellan M., McMahon R., McPherson J.D., McQuerrey Y., Mead K., Meininger B., Merry B., Meyer R., Meyers C., Miller K., Miller N., Miller W., Miner T.L., Minges B., Minx P.J., Mishra S., Moeller D., Mohd Nor L., Moire K., Moore B., Moore T., Morales R., Mudd N., Mullen G., Mullen M., Mulvaney E., Murray J., Myers M., Nash A., Nash W., Nelson J., Nguyen C., Nhan N., Nicol C., Niemann L., Nothaker L., Nwagbo T., Oberkfell B., O'Brien D., O'Brien D., Odunfa-Jones T., Okuka M.K., O'Malley M., Owens S., Ozersky P., Page S., Panussis D., Pape K., Parker C., Pauley A., Paulson E., Peak J., Pearman C., Peluso D., Pepin K.H., Peterson D., Pettiford J., Pfeiffer B., Phillips A., Pierce G., Pikula C., Podhrasky A., Pohl C., Ponce T., Puro S., Ralph C., Randall J., Randolph J., Reed J., Reily A., Reiniesch D., Reitz L., Reskusich J., Rhine C., Rice L., Richards M., Richey J., Rieff J., Riley J., Ritchey E., Robertson J., Robinson K., Rock S., Rohling T., Rose C., Ryan E., Ryan J., Ryan J., Ryno S., Sammons L., Sandberg B., Sandbothe T., Sander N., Sapetti L., Sasso S., Schaller M., Schaus C., Scheer D., Scheet P., Scherger E., Schneider L., Schultz B., Scott K., Scott S., Scronce D., Seim R., Sekhon M., Shafer S., Shah N., Shadid S., Shapiro K., Shelby P., Shih K., Slaughter M., Small J., Smith A., Smith A., Smith E., Smith J., Smith N., Smith R., Smoker B., Snider J., Spalding L., Spieth J., Steele P., Stellyes L., Stiziel N., Stoneking T., Strong C., Strong J., Strowmatt C., Stuebe E., Stumpf J., Suk R., Sun H., Stutterer C., Swift G., Talcherkar S., Thipkhosithkun P., Thompson J., Tin-Wollam A.M., Tomlinson C., Tonn M., Trani L., Trevaskis E., Tucci S., Twyman B., Underwood K., Ureta M., Valencia P., Van Bruntg A., Veath C., Veizer J., Wagner-McPherson C., Waligorski J., Walker C., Walker R., Wall T., Wallis J., Wamsley P., Waterston R.H., Wedgeworth P., Weihe A., Wendl M.C., Wheeler N., White S., Whitworth N., Williams D., Williamson A., Wilson R.K., Winchester K., Winkelmann M., Woessner J., Wohldmann P., Wolff J., Wollam C., Woods K., Woolley J.P., Worthington R., Wu X., Wylie K., Wylie T., Yandell M., Yang S.P., Yeh R., Yoakum M., Zerazion S., Zheng X., Zhu H.J., Zidanic M., Abrajano A., Aerts A., Alcivare D., Altherr M., Amico-Keller G., Andora J., Andredesz C.H., Andriese T., Arcaina L., Arcaina T., Archuleta R., Arellano A., Armstrong N., Ashworth L., Attix C., Avery A., Avila A., Avila J., Badri H., Bakis M., Balch M., Banda M., Beall K., Beaton D., Bercovitz J., Bergmann A., Beugelsdijk T., Bobo T., Boehm J., Bondoc M., Bonner M.P., Bowen E., Brannon W., Branscomb E., Brower A., Brown N., Brown R., Bruce D., Bruce R., Brunkhorst E., Bryant J., Buckingham J., Burkhart-Schultz K., Bursell B., Bussod M., Campbell C., Campbell E., Campbell M., Caoile C., Cardenas H., Cepeda M., Chain P., Chaparro S., Chasteen L., Chen X., Cheng J.F., Chin S.G., Chinn C., Christensen M., Chung A., Cifelli R., Clark L., Cofield J., Cohn J.R., Colayco R., Copeland A., Cordray R., Connell E., Corsetti L., Critchlow T., Critz P., Danganan L., Dean W., Deaven L., Deere K., Dehal P., Deng Z., Detter J.C., Detter S., Dias J.M., Dias V., Dickson M., DiGennaro R., Dilts K., Dimitrijevic-Bussod M., Dixon K., Do L., Doggett N., Duarte S., Elkin C., Erler A.M., Fawcitt J., Fey J., Fink M., Fischer K., Fischer L., Fitch J.P., Flowers D., Folta P., Fotopulos D., Fourcade M., Frankel K., Frazier M., Fridlyand J., Gammon S., Georgescu A., Geotina A., Gil I., Glavina T., Golvineaux K., Goodman S., Goodwin L., Gordon L., Gould K., Gray B., Green L., Griffith J., Grimwood J., Groza M., Guarin H., Gunning K., Ha C., Halsey C., Hammond S., Han C., Hawkins T., Henderson N., Hom W., Hosseini R., Huang Z., Hughes-Hull H., Humphries D., Hupman D., Hurshman J., Hutchings K., Hyatt D., Jaklevic J., Jamaca K., Jenecki T., Jett J., Jewitt P., Jiang L., Jin J., Jones M., Jung E., Kadner K., Kapur H., Kegg L., Khine S.S., Kim J., Kimball-Rojeski H., Kimmerly W., Ko C., Kobayashi A., Kolbe W., Kommander K., Krawczyk M., Kronmiller B., Krysiak V.A., Kuhn C., Lamerdin J., Land M., Larimer F., Lato B., Lee J.H., Lee M., Lehmann K., Leyba T., Lindo K., Lindquist K., Linkowski A., Litton K., Liu S.Y., Llewellyn-Silva C., Lobb R., Logan J., Longmire J., Lopez J.L., Lou Y., Lowry S., Lu X., Lucas S., Machrus M., Macht M., Madabhushi R., Mahnke R., Maltbie M., Mariano M., Marieiro L.M., Martin C., Martin J., Martinez M., McReady P., McGurn P., McMurry K., Medina C., Meier K., Meincke L., Menke J., Meyne J., Miguel T., Miller C., Milligan T., Miner S., Montgomery V., Moy D., Mundt M., Munk C., Mural R., Myers R., Narla M., Nelson D., Neunkirch J., Newman A., Nguyen H., Nguyen L., Nguyen Q., Nolan M., Oddone P., Olivas J., Olsen A., Ow D., Parang M., Parson-Quintana B., Patel B., Patel S., Peng Y., Peng Z., Peterman K., Petit B., Pfeiffer J., Phan H., Pitluck S., Pittson L., Plajzer-Frick I., Pollard M., Poundstone P., Prakash E., Predki P., Primus J., Probst L., Prusso E., Quan G., Ramirez L., Ramirez M., Randolph D., Rapier I., Regala W., Reiter C., Ren X., Richardson P., Ricke D., Robinson D., Rodriguez J., Sakaldasis G., Sanders C., Sarmiento R., Saunders E., Schmoyer D., Schmutz J., Scott D., Scott D., Shah M., Shang J., Shin M., Shreve J., Simoni J., Sims J., Sindelar L., Skowronski E., Slezak T., Smith J., Snoddy J., Stanley G., Stilwagen S., Stubbs L., Stultz J., Subramanian S., Sutherland R., Tacey K., Takenaka T., Tatum T., Terry A., Tesmer J., Thiel J., Thomas P., Thompson L., Thompson S., Thompson W., Tong G., Torney D., Tran M., Trankiem M., Trong S., Tsai M.Y., Turner H., Turner J., Turtrice J., Uberbacher E., Un C., Ung Q., Luchen R.V., Vargas M., Vartanian N.P., Velasco N.P., Velasquez O., Vertuca C., Viswanathan V.S., Wagner J., Wagner M., Wan W., Wang M., Wehri E., Weidenbach R., Wenning S., Wentz S., White C., White J., White S., Williams A., Wilson D., Winleblech-Kelly B., Wollard J.R., Woo L., Woolley J., Wright T., Wycoff-Montegro M., Yang J., Yeh M., Yu C., Yumae B., Zimmerman D.W., Adams C., Adio-Odulo B., Allen C., Allen H., Amaratunge H., Arenson A., Bailey M., Banks T., Barbaria J., Bimage K., Bodota B., Bonnin D., Bouck J., Brooks A., Brown E., Brown M.J., Bryant N., Buhay C., Burch P., Buckett C., Burrell K., Carron T., Carter K., Cavazos S., Chacko J., Chavez D., Chen G., Chen M., Chen R., Chen Z., Christopoulos C., Clerc-Blankenburg K., Cockrell R., Cox C., Coyle M., Dathorne S., David R., Davila M.L., Davis C., Davy-Carroll L., Delgado O., Denn A., DeShazo D., Ding Y., Dinh H., Douthwaite K., Draper H., Dugan-Rocha S., Durbin K.J., Earnhardt C., Edgar D., Allen C., Elhaj C., Escotto M., Falls T., Flagg N., Forcum-Tansey J., Foster P., Frantz J.P., Gabisi A., Garcia A., Garcia D.K., Garner T., Gibbs R.A., Gill R., Gorrell J.H., Gorrell L.L., Guevara W., Gunaratne P., Hamilton K., Hanak V., Harris K., Havlak P., Hawes A., Hernandez J., Hernandez O., Hodgson A., Hogues M., Hollins B., Homsi F., Hosak H., Hou X., Huber J., Hume J., Jackson L.R., Jia Y., Johnson B., Johnson R., Jolivet A., Jones M., Joudah S., Kaminsky S., Kelly J., Kelly S., Karlsson E., Khan U., LaQuisha K., Kondejewski N., Kovar C., Kratovic J., Kondejewski N., Kucherlapati R., Leal B., Lewis L.K., Lewis L., Kondejewski N., Li J., Lichtarge O., Liu J., Liu W., Logan L.Q., Logan O., Loulseged H., Lozado R., Lu J., Lucier A., Lucier R., Ly T., Ma J., Maheshwari M., Mapua P., Martin R., Martindale A., Martinez C., Martinez E., Massey E., Mawhiney S., McLeod M., Meador M., Mei G., Mercado I., Metzker M., Miner G., Mitchell T., Mo W., Mohabbat K., Montgomery B., Montgomery K., Morgan M., Morris S., Moser M., Muzny D., Nash S., Naylor S., Neal D., Nelson A., Nelson D., Newton N., Nguyen A., Nguyen B.V., Nguyen N., Nguyen N., Nickerson E., Nwokenkwo S., Oguh M., Okwuonu G., Oswal G., Oviedo R., Pace A., Parish B., Paxton S., Payton B., Perez L., Peters L., Pickens A., Pieper N., Primus E., Pu L.L., Quiles M., Quiroz J., Reiter D., Ren Y., Rives C.M., Rojas A., Ruiz S.J., Savery G., Scherer S., Scott G., Shen H., Simon M., Sisson I., Sodergren E., Sonaike T., Savage L., Sparks A., Stanley H., Stone H., Sutton H., Svatek A., Svetz L.A., Tabor P., Tamerisa K., Taylor C., Taylor T., Thomas N., Thomas S., Timms K., Tran L.T., Usmani K., Vasquez L., Vera V., Villalon D., Villasana D., Vo Q., Walker D., Walker D., Wall R., Wang J., Wang S., Ward-Moore S., Warren R., Watlington S., Watrous M., Weinstock G., Wheeler D., Williams G., Williamson A., Wleczyk R., Wooden S., Worley K., Wrensford G., Zhou J., Zhou X., Zorilla S., Aizu T., Arai R., Asahi Y., Ejima F., Fujioka A., Fujiyama A., Fukano K., Fukawa R., Gu Q., Hattori M., Hirose M., Horishima M., Ishii K., Ishizaki H., Isozaki E., Ito N., Itoh T., Kawagoe C., Kobayshi Y., Kodaka N., Kondo M., Matsumura Y., Mitani Y., Morita H., Motoyama A., Nagao S., Nakagawa S., Nakamura K., Nakano C., Nashida A., Odama Y., Omori N., Ono Y., Oshima K., Oyama Y., Ozawa R., Park H.S., Sakai R., Sakaki Y., Seki H., Shimizu H., Sun J., Tahara T., Tagaki T., Takiguchi S., Tanaka M., Tanaka R., Taylor T., Terada Y., Tochigi M., Tomioka N., Totoki Y., Toyoda A., Tsukamoto Y., Tsukuni S., Tsuzuki R., Uyama N., Wada H., Watanabe H., Yada T., Yakushiji K., Yamato N., Yamashita Y., Yokoyama S., Yonezawa M., Yoshida S., Artiguenave F., Barbe N., Besnard M., Besnard M., Boscus D., Briez S., Brottier P., Bruls T., Cattolico L., Cha N., Da Silva C., Dubois I., Gouyvenoux M., Gyapay G., Heilig R., Leclerc S., Levy M., Magdelenat G., Pelletier E., Petit J.L., Robert C., Saurin W., Vacherie B., Vico V., Weissenbach J., Wincker P., Bakis M., Bashirzadeh R., Battles J., Bodnaruk M., Breton G., Brown J., Butler C., Cahill P., Caron A., Daggett P., Dorman T., Doucette-Stamm L., Dubois J.A., Edwards N., Ezedi J., Flynn S., Freeman L., Gibson R., Gleeson D., Gryan G., Herman B., Hitti J., Ho T., Holtham K., Huynh K., Hynds C., Johnson M., Joseph P., Kadel-Garcia R., Kamath V., Kana A., Keane K., Kopcewiez K., Lach A., Lee A., Lee H.M., Little R., Lumm W., Madan D., Magararu R., Mao J.I., Mitnik-Gankin L., Munoz M., Nguyen M., Nielson W., Prabhakar S., Prescott-Roy J., Qiu D., Reinemann B., Robinson S., Roche M., Rosetti D., Rubenfield M., Russakovskaya O., Segal J., Smith D.R., Snell P., Stroika M., Turenne G.A., Walsh J., Wang Y., Weinstock K., Wheaton G., Wong L., Xu Q., Yang H., Zafiropoulos E., Zhang E., Baumgart C., Baumgart I., Blechschmidt K., Boehm E., Brunnckow C., Creutzburg N., Dette M., Drescher B., Drescher B., Eibmann P., Fabisch S., Fischer B., Foerste S., Galgoczy P., Gallert S., Glockner G., Gorlich Y., Grosser C., Hamann J., Heintze I., Jahn N., Kantowski E., Klabunde H., Kluge S., Lagemann D., Landmann S., Lehmann R., Lenk D., Ludewig H., Meier E., Menzel U., Michaelis E., Mockel K., Mortag K., Muller O., Nordsiek G., Nyakatura G., Pawelka B., Petz U., Pick U., Platzer M., Pohlmann C., Polley A., Raguschke B., Rahnis N., Reichwald K., Rosenthal A., Rosenthal S., Rothe S., Rump A., Schattevoy R., Schauer A., Schilhabel M., Schilling M., Schlenkert L., Schmid M.L., Schoemburg J., Schudy A., Schulz R., Taudien S., Tautkus B., Teuchtler M., Voigt B., Weber J., Wenderoth C., Werler D., Wiehe T., Zeise N., Zenker R., Zimmerman W.D., Bao J., Bao Q., Bao W., Bi S., Bian X., Bolund L., Cai T., Cao T., Cao Y., Chen B., Chen C., Chen J., Chen J., Chen J., Chen T., Chen Y., Chen Z., Cui H., Cui J., Cui P., Dai L., Ding H., Dong H., Dong W., Dong X., Du Y., Fan H., Fang J., Feng H., Feng J., Feng X., Fu G., Gao J., Gao Q., Gao Y., Geng J., Gong G., Gong J., Gu J., Gu W., Gu X., Guan Q., Gui Q., Guo D., He F., He J., He L., Hu J., Hu S., Huang F., Huang G., Jia J., Jia N., Jiang L., Jin Y., Jin Y., Kang N., King M.C., Kong Y., Lei M., Li C., Li C., Li E., Li G., Li J., Li J., Li L., Li L., Li M., Li N., Li R., Li S., Li S., Li S., Li S., Li T., Li W., Li W., Li Y., Li Y., Li Z., Liao J., Lin W., Liu B., Liu H., Liu K., Liu N., Liu S., Liu W., Liu X., Liu Y., Liu Y., Liu Y., Liu Z., Lu T., Lu Y., Lv G., Ma C., Ma J., Ma Q., Meng S., Mu F., Niu Y., Pan J., Qi Q., Qi X., Qian X., Qian Z., Qiang B., Qiao Z., Ren S., Rong L., Shao Y., Shen F., Shen Y., Shi Y., Shi H., Smith M., Song L., Song S., Sun J., Sun M., Sun T., Sun Y., Sun Y., Sun Y., Tan W., Tan X., Tang X., Tao R., Tian Y., Tian Y., Tong J., Tu Y., Wan M., Wang D., Wang D., Wang F., Wang G., Wang G., Wang G., Wang H., Wang H., Wang J., Wang J., Wang L., Wang L., Wang L., Wang L., Wenjun W., Xiaolei W., Wang X., Wang X., Wang Y., Wang Y., Wu C., Wu D., Wu Q., Wu X., Xi Y., Xie F., Xu R., Xu S., Xu W., Xu Y., Xue R., Xue Y., Yan C., Yan F., Yan G., Huanming Y., Yang S., Yang X., Bing Y., Yu B., Yu J., Yuan K., Zeng Y., Zhai D., Zhang B., Zhang F., Zhang G., Zhang G., Zhang H., Zhang L., Zhang L., Zhang L., Zhang M., Zhang M., Zhang M., Zhang R., Zhang W., Zhang X., Zang X., Zhang X., Zhang Y., Zhang Y., Zhang Y., Zhang Y., Zhang Y., Zhao H., Zhao L., Zhao Z., Zhen Z., Zhong M., Zhou H., Zhou N., Zhou X., Zhou Y., Zhou Y., Zhu B., Zhu B., Zhu G., Zhu N., Zhu Y., Zhu Z., Abassi N., Ahearn M.E., Baradarani L., Baskin D., Birditt B., Bloom S., Boysen C., Bumgarner R., Dickhoff R., Dors M., Fleetwood P., Friedman C., Harrison G., Hood L., James R., Kaur A., Lasky S., Lee I., Loretz C., Madan A., Madan A., Mahairas G.G., Nesbitt R., Qin S., Ratcliffe A., Rowen L., Seto J., Shaffer T., Smit A., Smith T., Swartzell S., Trask B.J., Wang K., Federspiel A., Foreman P., Glukhov S., Hansen N., Herman Z., Hyman R., Kalman S., Kurdi O., Mao J., Marathe R., Proctor M.J., Morehouse A., Oefner P., Palm C., Ramirez D., Rexan M., Dela Rosa M., Smith M., Vollrath D., Wilhelmy J., Willils T., Yu S., Bajorek E., Caoile C., Carriere J., Cox D.R., Dickson M., Dixon K., Fischer L., Flowers D., Fotopulos D., Garcia C., Gold D., Grimwood J., Haydu L., Holtzer C., Litton K., Logan J., Lopez J., Medina C., Myers R.M., Nguyen L., Ramirez L., Rodriquez A., Rogers S., Salazar A., Schmutz J., Shang J., Stone N., Tsai M., Valesquez O., Vartanian S., Vitale D., Wheeler J., Yang J., Bubb K., Daza R., Desmarais C., Duenwald S., Erickson K., Gilbert T., Hite M., Hubley R., Huges W., Iodanoto S., Jewett D., Junker C., Kas A., Kaul R., Le M., Lim R., Lytle L., Magness C., Magness Z., Maza M., McClelland E., Olson M., Passey D., Pham X.Q., Phelps K.A., Qiu R., Ramsey S., Raymond C., Richards B., Sadhegi Z., Saenphimmachak C., Sims E., Smit A., Stone M., Thomas T., Wang G.K., Wu Z., Yu J., Zhou Y., Aoki N., Asahina M., Asakwa S., Kawasaki K., Kudoh J., Minoshima S., Mitsuyama S., Sasaki T., Shibuya K., Shimizu A., Shimizu N., Shintani A., Yoshizaki Y., Aguayo P., Arenare S., Armstrong D., Athanasiou M., Basit M., Black D., Brandon J., Buettner J., Butler C., Card P., Chamblis S., Dunn J., English C., Ethridge S., Evans G.A., Federova N., Fribish A., Garza M., Gordon M., Gorman C., Grant O.D., Hahner L., Hayes S., Joslin J., Lam S., Le T., Lester T., Lewis E., Loo K.N., Loo M., Major T., McFairland J., Nguyen M., Osborne-Lawrence S., Rakoshchik I., Schageman J., Schultz R., Stimson S., Tran M., Varghese F., Wagner N., Waller K., Ward T., Wharton J., Whitaker J., Willcot J.N., Zanoni J., Ahmad M., Bodenteich A., Chen F., Chu L., Crabtree J., Deschamps S., Do A., Do T., Dolance J., Dorman A., Ducummon C., Duty A., Elraham M., Elkins W., Fang F., Fu Y., Hall G., Hartman K., Hill K., Hu P., Hu X., Hua A., Huang E., Honggui J., Jiang X., Kenton S., Khan A., Kupfer D., Lai H., Lane L., Lao H.I., Lau C., Lewis J., Lewis S., Li H., Lin S., Loh P., Malaj E., Milam J., Morales-Diaz R., Najar F., Nguyen T., Ni Y., Oommen, Pan H., Perry B., Phan S., Qi S., Qian Y., Ray L., Ren Q., Ren Q., Roe B.A., Shaull S., Sloan D., Song L., Stone J., Tian J., Tian R., Tilahun Y., Wang Q., Wang Y.P., Wang Z., White D., White J., Willingham D., Wong S., Wright H., Wu H.M., Wu H., Yang L., Yao Z., Yoon Y., Zhan M., Zhang G., Zhou L., Zhu H., Arndt S., Beck A., Borzym K., Buczek D., Chelly J., Francis F., Heitmann K., Hennig S., Hoff C., Junker E., Kioschis P., Klages S., Klein M., Kosiura A., Kube M., Langer I., Lehrach H., Lehrack S., Marquard I., McDonell N., Meindl A., Moll K., Monaco A., Nemeth A., Poustka A., Ramser J., Reinhardt R., Schuelzchen S., Seranski P., Starke P., Steffens C., Sudbrak R., Todd K., Yaspo M.L., de la Bastide M., Dedhia N., Gnoj L., Gottesman T., Granat S., Haberman K., Hameed A., Hasegawa A., Hoffman J., Huang E., Jenson K., Johnson A., Kaplan N., Lodhi M., Matero A., McCombie W.R., O'Shaughnessy A., Parnell L., Preston R., Rodgiguez M., Schutz K., See L.H., Shah R., Shekher M., Shohdy N., Spiegel L., Swaby I., Till S., Vil D., Blocker H., Brandt P., Conrad A., Dose S., Grimm M., Hornischer K., Jarke D., Kauer G., Lohnert H., Nordsiek G., Reichelt J., Scharfe M., Schon O., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birren B., Birney E., Bork P., Bouck J.B., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Chinwalla A.T., Church D., Clamp M., Collins F.S., Copley R.R., Doerks T., Durbin R., Eddy S.R., Eichler E.E., FitzHugh W., Felsenfeld A., Furey T.S., Galagan J., Gibbs R.A., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermiakob H., Hillier L.D., Hokamp K., Hubbard T., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lander E.S., Lowe T.M., McLysaght A., Mesirov J., Mikkelsen T., Moran J.V., Mulder N., Mullikn J.C., Nusbaum C., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Sulston J., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Waterson R., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Zody M.C., Agarwala R., Aravind L., Chen H.C., Church D., Jang W., Kitts P., Koonin E.V., Schuler G., Thierry-Mieg D., Thierry M.I.J., Wagner L., Wolf Y.I., Birney E., Reashci N., Sterk P., Fukami-Kobayashi K., Gojobori T., Ikeo K., Imanishi T., Myazaki S., Nishikawa K., Ohta M., Sugawara H., Tateno Y., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K., Patrinos A., Morgan M.J. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 36.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 37.Jothi R., Cuddapah S., Barski A., Cui K., Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kapranov P., Laurent G.S., Raz T., Ozsolak F., Reynolds C.P., Sorensen P.H., Reaman G., Milos P., Arceci R.J., Thompson J.F., Triche T.J. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato N. Insights into the genetic basis of type 2 diabetes. J. Diabetes Investig. 2013;4:233–244. doi: 10.1111/jdi.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohane I.S. An autism case history to review the systematic analysis of large-scale data to refine the diagnosis and treatment of neuropsychiatric disorders. Biol. Psychiatry. 2015;77:59–65. doi: 10.1016/j.biopsych.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P., Been L.F., Chia K.S., Dimas A.S., Hassanali N., Jafar T., Jowett J.B., Li X., Radha V., Rees S.D., Takeuchi F., Young R., Aung T., Basit A., Chidambaram M., Das D., Grundberg E., Hedman A.K., Hydrie Z.I., Islam M., Khor C.C., Kowlessur S., Kristensen M.M., Liju S., Lim W.Y., Matthews D.R., Liu J., Morris A.P., Nica A.C., Pinidiyapathirage J.M., Prokopenko I., Rasheed A., Samuel M., Shah N., Shera A.S., Small K.S., Suo C., Wickremasinghe A.R., Wong T.Y., Yang M., Zhang F., Diagram M.T., Abecasis G.R., Barnett A.H., Caulfield M., Deloukas P., Frayling T.M., Froguel P., Kato N., Katulanda P., Kelly M.A., Liang J., Mohan V., Sanghera D.K., Scott J., Seielstad M., Zimmet P.Z., Elliott P., Teo Y.Y., McCarthy M.I., Danesh J., Tai E.S., Chambers J.C. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kung J.T., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanktree M.B., Guo Y., Murtaza M., Glessner J.T., Bailey S.D., Onland-Moret N.C., Lettre G., Ongen H., Rajagopalan R., Johnson T., Shen H., Nelson C.P., Klopp N., Baumert J., Padmanabhan S., Pankratz N., Pankow J.S., Shah S., Taylor K., Barnard J., Peters B.J., Maloney C.M., Lobmeyer M.T., Stanton A., Zafarmand M.H., Romaine S.P., Mehta A., van Iperen E.P., Gong Y., Price T.S., Smith E.N., Kim C.E., Li Y.R., Asselbergs F.W., Atwood L.D., Bailey K.M., Bhatt D., Bauer F., Behr E.R., Bhangale T., Boer J.M., Boehm B.O., Bradfield J.P., Brown M., Braund P.S., Burton P.R., Carty C., Chandrupatla H.R., Chen W., Connell J., Dalgeorgou C., Ad B., Drenos F., Elbers C.C., Fang J.C., Fox C.S., Frackelton E.C., Fuchs B., Furlong C.E., Gibson Q., Gieger C., Goel A., Grobbee D.E., Hastie C., Howard P.J., Huang G.H., Johnson W.C., Li Q., Kleber M.E., Klein B.E., Klein R., Kooperberg C., Ky B., Lacroix A., Lanken P., Lathrop M., Li M., Marshall V., Melander O., Mentch F.D., Meyer N.J., Monda K.L., Montpetit A., Murugesan G., Nakayama K., Nondahl D., Onipinla A., Rafelt S., Newhouse S.J., Otieno F.G., Patel S.R., Putt M.E., Rodriguez S., Safa R.N., Sawyer D.B., Schreiner P.J., Simpson C., Sivapalaratnam S., Srinivasan S.R., Suver C., Swergold G., Sweitzer N.K., Thomas K.A., Thorand B., Timpson N.J., Tischfield S., Tobin M., Tomaszewski M., Verschuren W.M., Wallace C., Winkelmann B., Zhang H., Zheng D., Zhang L., Zmuda J.M., Clarke R., Balmforth A.J., Danesh J., Day I.N., Schork N.J., de Bakker P.I., Delles C., Duggan D., Hingorani A.D., Hirschhorn J.N., Hofker M.H., Humphries S.E., Kivimaki M., Lawlor D.A., Kottke-Marchant K., Mega J.L., Mitchell B.D., Morrow D.A., Palmen J., Redline S., Shields D.C., Shuldiner A.R., Sleiman P.M., Smith G.D., Farrall M., Jamshidi Y., Christiani D.C., Casas J.P., Hall A.S., Doevendans P.A., Christie J.D., Berenson G.S., Murray S.S., Illig T., Dorn G.W., 2nd, Cappola T.P., Boerwinkle E., Sever P., Rader D.J., Reilly M.P., Caulfield M., Talmud P.J., Topol E., Engert J.C., Wang K., Dominiczak A., Hamsten A., Curtis S.P., Silverstein R.L., Lange L.A., Sabatine M.S., Trip M., Saleheen D., Peden J.F., Cruickshanks K.J., Marz W., O'Connell J.R., Klungel O.H., Wijmenga C., Maitland-van der Zee A.H., Schadt E.E., Johnson J.A., Jarvik G.P., Papanicolaou G.J., Procardis Hugh Watkins on behalf of, Grant S.F., Munroe P.B., North K.E., Samani N.J., Koenig W., Gaunt T.R., Anand S.S., van der Schouw Y.T., I. I. Study Meena Kumari on behalf of the Whitehall, Whii K. Group the, Soranzo N., Fitzgerald G.A., Reiner A., Hegele R.A., Hakonarson H., Keating B.J. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am. J. Hum. Genet. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacKinnen D.P. New York; Erlbaum and Associates: 2008. Introduction to Statistical Mediation Analysis. [Google Scholar]
- 45.Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J., Ferreira T., Horikoshi M., Johnson A.D., Ng M.C., Prokopenko I., Saleheen D., Wang X., Zeggini E., Abecasis G.R., Adair L.S., Almgren P., Atalay M., Aung T., Baldassarre D., Balkau B., Bao Y., Barnett A.H., Barroso I., Basit A., Been L.F., Beilby J., Bell G.I., Benediktsson R., Bergman R.N., Boehm B.O., Boerwinkle E., Bonnycastle L.L., Burtt N., Cai Q., Campbell H., Carey J., Cauchi S., Caulfield M., Chan J.C., Chang L.C., Chang T.J., Chang Y.C., Charpentier G., Chen C.H., Chen H., Chen Y.T., Chia K.S., Chidambaram M., Chines P.S., Cho N.H., Cho Y.M., Chuang L.M., Collins F.S., Cornelis M.C., Couper D.J., Crenshaw A.T., van Dam R.M., Danesh J., Das D., de Faire U., Dedoussis G., Deloukas P., Dimas A.S., Dina C., Doney A.S., Donnelly P.J., Dorkhan M., van Duijn C., Dupuis J., Edkins S., Elliott P., Emilsson V., Erbel R., Eriksson J.G., Escobedo J., Esko T., Eury E., Florez J.C., Fontanillas P., Forouhi N.G., Forsen T., Fox C., Fraser R.M., Frayling T.M., Froguel P., Frossard P., Gao Y., Gertow K., Gieger C., Gigante B., Grallert H., Grant G.B., Grrop L.C., Groves C.J., Grundberg E., Guiducci C., Hamsten A., Han B.G., Hara K., Hassanali N., Hattersley A.T., Hayward C., Hedman A.K., Herder C., Hofman A., Holmen O.L., Hovingh K., Hreidarsson A.B., Hu C., Hu F.B., Hui J., Humphries S.E., Hunt S.E., Hunter D.J., Hveem K., Hydrie Z.I., Ikegami H., Illig T., Ingelsson E., Islam M., Isomaa B., Jackson A.U., Jafar T., James A., Jia W., Jockel K.H., Jonsson A., Jowett J.B., Kadowaki T., Kang H.M., Kanoni S., Kao W.H., Kathiresan S., Kato N., Katulanda P., Keinanen-Kiukaanniemi K.M., Kelly A.M., Khan H., Khaw K.T., Khor C.C., Kim H.L., Kim S., Kim Y.J., Kinnunen L., Klopp N., Kong A., Korpi-Hyovalti E., Kowlessur S., Kraft P., Kravic J., Kristensen M.M., Krithika S., Kumar A., Kumate J., Kuusisto J., Kwak S.H., Laakso M., Lagou V., Lakka T.A., Langenberg C., Langford C., Lawrence R., Leander K., Lee J.M., Lee N.R., Li M., Li X., Li Y., Liang J., Liju S., Lim W.Y., Lind L., Lindgren C.M., Lindholm E., Liu C.T., Liu J.J., Lobbens S., Long J., Loos R.J., Lu W., Luan J., Lyssenko V., Ma R.C., Maeda S., Magi R., Mannisto S., Matthews D.R., Meigs J.B., Melander O., Metspalu A., Meyer J., Mirza G., Mihailov E., Moebus S., Mohan V., Mohlke K.L., Morris A.D., Muhleisen T.W., Muller-Nurasyid M., Musk B., Nakamura J., Nakashima E., Navarro P., Ng P.K., Nica A.C., Nilsson P.M., Njolstad I., Nothen M.M., Ohnaka K., Ong T.H., Owen K.R., Palmer C.N., Pankow J.S., Park K.S., Parkin M., Pechlivanis S., Pedersen N.L., Peltonen L., Perry J.R., Peters A., Pinidiyapathirage J.M., Platou C.G., Potter S., Price J.F., Qi L., Radha V., Rallidis L., Rasheed A., Rathman W., Rauramaa R., Raychaudhuri S., Rayner N.W., Rees S.D., Rehnberg E., Ripatti S., Robertson N., Roden M., Rossin E.J., Rudan I., Rybin D., Saaristo T.E., Salomaa V., Saltevo J., Samuel M., Sanghera D.K., Saramies J., Scott J., Scott L.J., Scott R.A., Segre A.V., Sehmi J., Sennblad B., Shah N., Shah S., Shera A.S., Shu X.O., Shuldiner A.R., Sigurdsson G., Sijbrands E., Silveira A., Sim X., Sivapalaratnam S., Small K.S., So W.Y., Stancakova A., Stefansson K., Steinbach G., Steinthorsdottir V., Stirrups K., Strawbridge R.J., Stringham H.M., Sun Q., Suo C., Syvanen A.C., Takayanagi R., Takeuchi F., Tay W.T., Teslovich T.M., Thorand B., Thorleifsson G., Thorsteinsdottir U., Tikkanen E., Trakalo J., Tremoli E., Trip M.D., Tsai F.J., Tuomi T., Tuomilehto J., Uitterlinden A.G., Valladares-Salgado A., Vedantam S., Veglia F., Voight B.F., Wang C., Wareham N.J., Wennauer R., Wickremasinghe A.R., Wilsgaard T., Wilson J.F., Wiltshire S., Winckler W., Wong T.Y., Wood A.R., Wu J.Y., Wu Y., Yamamoto K., Yamauchi T., Yang M., Yengo L., Yokota M., Young R., Zabaneh D., Zhang F., Zhang R., Zheng W., Zimmet P.Z., Altshuler D., Bowden D.W., Cho Y.S., Cox N.J., Cruz M., Hanis C.L., Kooner J., Lee J.Y., Seielstad M., Teo Y.Y., Boehnke M., Parra E.J., Chambers J.C., Tai E.S., McCarthy M.I., Morris A.P., Meta-analysis C.D.G.R., Consortium Asian Genetic Epidemiology Network Type 2 Diabetes, Consortium South Asian Type 2 Diabetes…Consortium Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in multi-Ethnic Samples Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malin S.K., Kashyap S.R., Hammel J., Miyazaki Y., DeFronzo R.A., Kirwan J.P. Adjusting glucose-stimulated insulin secretion for adipose insulin resistance: an index of beta-cell function in obese adults. Diabetes Care. 2014;37:2940–2946. doi: 10.2337/dc13-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manning A.K., Hivert M.F., Scott R.A., Grimsby J.L., Bouatia-Naji N., Chen H., Rybin D., Liu C.T., Bielak L.F., Prokopenko I., Amin N., Barnes D., Cadby G., Hottenga J.J., Ingelsson E., Jackson A.U., Johnson T., Kanoni S., Ladenvall C., Lagou V., Lahti J., Lecoeur C., Liu Y., Martinez-Larrad M.T., Montasser M.E., Navarro P., Perry J.R., Rasmussen-Torvik L.J., Salo P., Sattar N., Shungin D., Strawbridge R.J., Tanaka T., van Duijn C.M., An P., de Andrade M., Andrews J.S., Aspelund T., Atalay M., Aulchenko Y., Balkau B., Bandinelli S., Beckmann J.S., Beilby J.P., Bellis C., Bergman R.N., Blangero J., Boban M., Boehnke M., Boerwinkle E., Bonnycastle L.L., Boomsma D.I., Borecki I.B., Bottcher Y., Bouchard C., Brunner E., Budimir D., Campbell H., Carlson O., Chines P.S., Clarke R., Collins F.S., Corbaton-Anchuelo A., Couper D., de Faire U., Dedoussis G.V., Deloukas P., Dimitriou M., Egan J.M., Eiriksdottir G., Erdos M.R., Eriksson J.G., Eury E., Ferrucci L., Ford I., Forouhi N.G., Fox C.S., Franzosi M.G., Franks P.W., Frayling T.M., Froguel P., Galan P., de Geus E., Gigante B., Glazer N.L., Goel A., Groop L., Gudnason V., Hallmans G., Hamsten A., Hansson O., Harris T.B., Hayward C., Heath S., Hercberg S., Hicks A.A., Hingorani A., Hofman A., Hui J., Hung J., Jarvelin M.R., Jhun M.A., Johnson P.C., Jukema J.W., Jula A., Kao W.H., Kaprio J., Kardia S.L., Keinanen-Kiukaanniemi S., Kivimaki M., Kolcic I., Kovacs P., Kumari M., Kuusisto J., Kyvik K.O., Laakso M., Lakka T., Lannfelt L., Lathrop G.M., Launer L.J., Leander K., Li G., Lind L., Lindstrom J., Lobbens S., Loos R.J., Luan J., Lyssenko V., Magi R., Magnusson P.K., Marmot M., Meneton P., Mohlke K.L., Mooser V., Morken M.A., Miljkovic I., Narisu N., O'Connell J., Ong K.K., Oostra B.A., Palmer L.J., Palotie A., Pankow J.S., Peden J.F., Pedersen N.L., Pehlic M., Peltonen L., Penninx B., Pericic M., Perola M., Perusse L., Peyser P.A., Polasek O., Pramstaller P.P., Province M.A., Raikkonen K., Rauramaa R., Rehnberg E., Rice K., Rotter J.I., Rudan I., Ruokonen A., Saaristo T., Sabater-Lleal M., Salomaa V., Savage D.B., Saxena R., Schwarz P., Seedorf U., Sennblad B., Serrano-Rios M., Shuldiner A.R., Sijbrands E.J., Siscovick D.S., Smit J.H., Small K.S., Smith N.L., Smith A.V., Stancakova A., Stirrups K., Stumvoll M., Sun Y.V., Swift A.J., Tonjes A., Tuomilehto J., Trompet S., Uitterlinden A.G., Uusitupa M., Vikstrom M., Vitart V., Vohl M.C., Voight B.F., Vollenweider P., Waeber G., Waterworth D.M., Watkins H., Wheeler E., Widen E., Wild S.H., Willems S.M., Willemsen G., Wilson J.F., Witteman J.C., Wright A.F., Yaghootkar H., Zelenika D., Zemunik T., Zgaga L., D. IAbetes Genetics Replication, Consortium Meta-analysis, Consortium Multiple Tissue Human Expression Resource, Wareham N.J., McCarthy M.I., Barroso I., Watanabe R.M., Florez J.C., Dupuis J., Meigs J.B., Langenberg C. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., Cho J.H., Guttmacher A.E., Kong A., Kruglyak L., Mardis E., Rotimi C.N., Slatkin M., Valle D., Whittemore A.S., Boehnke M., Clark A.G., Eichler E.E., Gibson G., Haines J.L., Mackay T.F., McCarroll S.A., Visscher P.M. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuda M., DeFronzo R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 50.Matthews D.R., Hosker J.P., Rudenski A.S., Naylor B.A., Treacher D.F., Turner R.C. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 51.McCarthy M.I., Hirschhorn J.N. Genome-wide association studies: past, present and future. Hum. Mol. Genet. 2008;17:R100–R101. doi: 10.1093/hmg/ddn298. [DOI] [PubMed] [Google Scholar]
- 52.McGuire S.E., McGuire A.L. Don't throw the baby out with the bathwater: enabling a bottom-up approach in genome-wide association studies. Genome Res. 2008;18:1683–1685. doi: 10.1101/gr.083584.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercer T.R., Gerhardt D.J., Dinger M.E., Crawford J., Trapnell C., Jeddeloh J.A., Mattick J.S., Rinn J.L. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merovci A., Mari A., Solis C., Xiong J., Daniele G., Chavez A., Tripathy D., Urban McCarthy S., Abdul-Ghani M., DeFronzo R.A. Dapagliflozin lowers plasma glucose concentration and improves beta cell function. J. Clin. Endocrinol. Metab. 2015 doi: 10.1210/jc.2014-3472. (jc20143472) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mohlke K.L., Scott L.J. What will diabetes genomes tell us? Curr. Diab. Rep. 2012;12:643–650. doi: 10.1007/s11892-012-0321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Norton L., Fourcaudot M., Abdul-Ghani M.A., Winnier D., Mehta F.F., Jenkinson C.P., Defronzo R.A. Chromatin occupancy of transcription factor 7-like 2 (TCF7L2) and its role in hepatic glucose metabolism. Diabetologia. 2011;54:3132–3142. doi: 10.1007/s00125-011-2289-z. [DOI] [PubMed] [Google Scholar]
- 57.Norton L., Chen X., Fourcaudot M., Acharya N.K., DeFronzo R.A., Heikkinen S. The mechanisms of genome-wide target gene regulation by TCF7L2 in liver cells. Nucleic Acids Res. 2014;42:13646–13661. doi: 10.1093/nar/gku1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., Yamanaka I., Kiyosawa H., Yagi K., Tomaru Y., Hasegawa Y., Nogami A., Schonbach C., Gojobori T., Baldarelli R., Hill D.P., Bult C., Hume D.A., Quackenbush J., Schriml L.M., Kanapin A., Matsuda H., Batalov S., Beisel K.W., Blake J.A., Bradt D., Brusic V., Chothia C., Corbani L.E., Cousins S., Dalla E., Dragani T.A., Fletcher C.F., Forrest A., Frazer K.S., Gaasterland T., Gariboldi M., Gissi C., Godzik A., Gough J., Grimmond S., Gustincich S., Hirokawa N., Jackson I.J., Jarvis E.D., Kanai A., Kawaji H., Kawasawa Y., Kedzierski R.M., King B.L., Konagaya A., Kurochkin I.V., Lee Y., Lenhard B., Lyons P.A., Maglott D.R., Maltais L., Marchionni L., McKenzie L., Miki H., Nagashima T., Numata K., Okido T., Pavan W.J., Pertea G., Pesole G., Petrovsky N., Pillai R., Pontius J.U., Qi D., Ramachandran S., Ravasi T., Reed J.C., Reed D.J., Reid J., Ring B.Z., Ringwald M., Sandelin A., Schneider C., Semple C.A., Setou M., Shimada K., Sultana R., Takenaka Y., Taylor M.S., Teasdale R.D., Tomita M., Verardo R., Wagner L., Wahlestedt C., Wang Y., Watanabe Y., Wells C., Wilming L.G., Wynshaw-Boris A., Yanagisawa M., Yang I., Yang L., Yuan Z., Zavolan M., Zhu Y., Zimmer A., Carninci P., Hayatsu N., Hirozane-Kishikawa T., Konno H., Nakamura M., Sakazume N., Sato K., Shiraki T., Waki K., Kawai J., Aizawa K., Arakawa T., Fukuda S., Hara A., Hashizume W., Imotani K., Ishii Y., Itoh M., Kagawa I., Miyazaki A., Sakai K., Sasaki D., Shibata K., Shinagawa A., Yasunishi A., Yoshino M., Waterston R., Lander E.S., Rogers J., Birney E., Hayashizaki Y., Fantom Consortium, Riken Genome Exploration Research Group Phase I., Team I.I. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 59.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., Wakamatsu A., Hayashi K., Sato H., Nagai K., Kimura K., Makita H., Sekine M., Obayashi M., Nishi T., Shibahara T., Tanaka T., Ishii S., Yamamoto J., Saito K., Kawai Y., Isono Y., Nakamura Y., Nagahari K., Murakami K., Yasuda T., Iwayanagi T., Wagatsuma M., Shiratori A., Sudo H., Hosoiri T., Kaku Y., Kodaira H., Kondo H., Sugawara M., Takahashi M., Kanda K., Yokoi T., Furuya T., Kikkawa E., Omura Y., Abe K., Kamihara K., Katsuta N., Sato K., Tanikawa M., Yamazaki M., Ninomiya K., Ishibashi T., Yamashita H., Murakawa K., Fujimori K., Tanai H., Kimata M., Watanabe M., Hiraoka S., Chiba Y., Ishida S., Ono Y., Takiguchi S., Watanabe S., Yosida M., Hotuta T., Kusano J., Kanehori K., Takahashi-Fujii A., Hara H., Tanase T.O., Nomura Y., Togiya S., Komai F., Hara R., Takeuchi K., Arita M., Imose N., Musashino K., Yuuki H., Oshima A., Sasaki N., Aotsuka S., Yoshikawa Y., Matsunawa H., Ichihara T., Shiohata N., Sano S., Moriya S., Momiyama H., Satoh N., Takami S., Terashima Y., Suzuki O., Nakagawa S., Senoh A., Mizoguchi H., Goto Y., Shimizu F., Wakebe H., Hishigaki H., Watanabe T., Sugiyama A., Takemoto M., Kawakami B., Yamazaki M., Watanabe K., Kumagai A., Itakura S., Fukuzumi Y., Fujimori Y., Komiyama M., Tashiro H., Tanigami A., Fujiwara T., Ono T., Yamada K., Fujii Y., Ozaki K., Hirao M., Ohmori Y., Kawabata A., Hikiji T., Kobatake N., Inagaki H., Ikema Y., Okamoto S., Okitani R., Kawakami T., Noguchi S., Itoh T., Shigeta K., Senba T., Matsumura K., Nakajima Y., Mizuno T., Morinaga M., Sasaki M., Togashi T., Oyama M., Hata H., Watanabe M., Komatsu T., Mizushima-Sugano J., Satoh T., Shirai Y., Takahashi Y., Nakagawa K., Okumura K., Nagase T., Nomura N., Kikuchi H., Masuho Y., Yamashita R., Nakai K., Yada T., Nakamura Y., Ohara O., Isogai T., Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 60.Ovcharenko I., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reyna S.M., Ghosh S., Tantiwong P., Meka C.S., Eagan P., Jenkinson C.P., Cersosimo E., Defronzo R.A., Coletta D.K., Sriwijitkamol A., Musi N. Elevated toll-like receptor 4 expression and signaling in muscle from insulin-resistant subjects. Diabetes. 2008;57:2595–2602. doi: 10.2337/db08-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., Weissman S., Snyder M. The transcriptional activity of human chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson P.N. Deep phenotyping for precision medicine. Hum. Mutat. 2012;33:777–780. doi: 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- 64.Sanghera D.K., Blackett P.R. Type 2 diabetes genetics: beyond GWAS. J. Diabetes Metab. 2012;3 doi: 10.4172/2155-6156.1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saxena R., Saleheen D., Been L.F., Garavito M.L., Braun T., Bjonnes A., Young R., Ho W.K., Rasheed A., Frossard P., Sim X., Hassanali N., Radha V., Chidambaram M., Liju S., Rees S.D., Ng D.P., Wong T.Y., Yamauchi T., Hara K., Tanaka Y., Hirose H., McCarthy M.I., Morris A.P., Diagram M.T., Agen A.B., Barnett A.H., Katulanda P., Matthews D., Mohan V., Wander G.S., Singh J.R., Mehra N.K., Ralhan S., Kamboh M.I., Mulvihill J.J., Maegawa H., Tobe K., Maeda S., Cho Y.S., Tai E.S., Kelly M.A., Chambers J.C., Kooner J.S., Kadowaki T., Deloukas P., Rader D.J., Danesh J., Sanghera D.K. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes. 2013;62:1746–1755. doi: 10.2337/db12-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steinthorsdottir V., Thorleifsson G., Sulem P., Helgason H., Grarup N., Sigurdsson A., Helgadottir H.T., Johannsdottir H., Magnusson O.T., Gudjonsson S.A., Justesen J.M., Harder M.N., Jorgensen M.E., Christensen C., Brandslund I., Sandbaek A., Lauritzen T., Vestergaard H., Linneberg A., Jorgensen T., Hansen T., Daneshpour M.S., Fallah M.S., Hreidarsson A.B., Sigurdsson G., Azizi F., Benediktsson R., Masson G., Helgason A., Kong A., Gudbjartsson D.F., Pedersen O., Thorsteinsdottir U., Stefansson K. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat. Genet. 2014;46:294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 67.Sun K., Goncalves J.P., Larminie C., Przulj N. Predicting disease associations via biological network analysis. BMC Bioinf. 2014;15:304. doi: 10.1186/1471-2105-15-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomsen S.K., Gloyn A.L. The pancreatic beta cell: recent insights from human genetics. Trends Endocrinol. Metab. 2014;25:425–434. doi: 10.1016/j.tem.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tracy R.P. Deep phenotyping: characterizing populations in the era of genomics and systems biology. Curr. Opin. Lipidol. 2008;19:151–157. doi: 10.1097/MOL.0b013e3282f73893. [DOI] [PubMed] [Google Scholar]
- 70.Ulahannan N., Greally J.M. Genome-wide assays that identify and quantify modified cytosines in human disease studies. Epigenetics Chromatin. 2015;8:5. doi: 10.1186/1756-8935-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Utzschneider K.M., Prigeon R.L., Faulenbach M.V., Tong J., Carr D.B., Boyko E.J., Leonetti D.L., McNeely M.J., Fujimoto W.Y., Kahn S.E. Oral disposition index predicts the development of future diabetes above and beyond fasting and 2-h glucose levels. Diabetes Care. 2009;32:335–341. doi: 10.2337/dc08-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Heyningen V., Bickmore W. Regulation from a distance: long-range control of gene expression in development and disease. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2013;368:20120372. doi: 10.1098/rstb.2012.0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wessel J., Chu A.Y., Willems S.M., Wang S., Yaghootkar H., Brody J.A., Dauriz M., Hivert M.F., Raghavan S., Lipovich L., Hidalgo B., Fox K., Huffman J.E., An P., Lu Y., Rasmussen-Torvik L.J., Grarup N., Ehm M.G., Li L., Baldridge A.S., Stancakova A., Abrol R., Besse C., Boland A., Bork-Jensen J., Fornage M., Freitag D.F., Garcia M.E., Guo X., Hara K., Isaacs A., Jakobsdottir J., Lange L.A., Layton J.C., Li M., Hua Zhao J., Meidtner K., Morrison A.C., Nalls M.A., Peters M.J., Sabater-Lleal M., Schurmann C., Silveira A., Smith A.V., Southam L., Stoiber M.H., Strawbridge R.J., Taylor K.D., Varga T.V., Allin K.H., Amin N., Aponte J.L., Aung T., Barbieri C., Bihlmeyer N.A., Boehnke M., Bombieri C., Bowden D.W., Burns S.M., Chen Y., Chen Y.D., Cheng C.Y., Correa A., Czajkowski J., Dehghan A., Ehret G.B., Eiriksdottir G., Escher S.A., Farmaki A.E., Franberg M., Gambaro G., Giulianini F., Goddard W.A., 3rd, Goel A., Gottesman O., Grove M.L., Gustafsson S., Hai Y., Hallmans G., Heo J., Hoffmann P., Ikram M.K., Jensen R.A., Jorgensen M.E., Jorgensen T., Karaleftheri M., Khor C.C., Kirkpatrick A., Kraja A.T., Kuusisto J., Lange E.M., Lee I.T., Lee W.J., Leong A., Liao J., Liu C., Liu Y., Lindgren C.M., Linneberg A., Malerba G., Mamakou V., Marouli E., Maruthur N.M., Matchan A., McKean-Cowdin R., McLeod O., Metcalf G.A., Mohlke K.L., Muzny D.M., Ntalla I., Palmer N.D., Pasko D., Peter A., Rayner N.W., Renstrom F., Rice K., Sala C.F., Sennblad B., Serafetinidis I., Smith J.A., Soranzo N., Speliotes E.K., Stahl E.A., Stirrups K., Tentolouris N., Thanopoulou A., Torres M., Traglia M., Tsafantakis E., Javad S., Yanek L.R., Zengini E., Becker D.M., Bis J.C., Brown J.B., Cupples L.A., Hansen T., Ingelsson E., Karter A.J., Lorenzo C., Mathias R.A., Norris J.M., Peloso G.M., Sheu W.H., Toniolo D., Vaidya D., Varma R., Wagenknecht L.E., Boeing H., Bottinger E.P., Dedoussis G., Deloukas P., Ferrannini E., Franco O.H., Franks P.W., Gibbs R.A., Gudnason V., Hamsten A., Harris T.B., Hattersley A.T., Hayward C., Hofman A., Jansson J.H., Langenberg C., Launer L.J., Levy D., Oostra B.A., O'Donnell C.J., O'Rahilly S., Padmanabhan S., Pankow J.S., Polasek O., Province M.A., Rich S.S., Ridker P.M., Rudan I., Schulze M.B., Smith B.H., Uitterlinden A.G., Walker M., Watkins H., Wong T.Y., Zeggini E., PIC-InterAct Consortium E., Laakso M., Borecki I.B., Chasman D.I., Pedersen O., Psaty B.M., Tai E.S., van Duijn C.M., Wareham N.J., Waterworth D.M., Boerwinkle E., Kao W.H., Florez J.C., Loos R.J., Wilson J.G., Frayling T.M., Siscovick D.S., Dupuis J., Rotter J.I., Meigs J.B., Scott R.A., Goodarzi M.O. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat. Commun. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winnier D.A., Fourcaudot M., Norton L., Abdul-Ghani M.A., Hu S.L., Farook V.S., Coletta D.K., Kumar S., Puppala S., Chittoor G., Dyer T.D., Arya R., Carless M., Lehman D.M., Curran J.E., Cromack D.T., Tripathy D., Blangero J., Duggirala R., Goring H.H., DeFronzo R.A., Jenkinson C.P. Transcriptomic identification of ADH1B as a novel candidate gene for obesity and insulin resistance in human adipose tissue in Mexican Americans from the Veterans Administration Genetic Epidemiology Study (VAGES) PLoS One. 2015;10 doi: 10.1371/journal.pone.0119941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi Z., Bowen B.P., Hwang H., Jenkinson C.P., Coletta D.K., Lefort N., Bajaj M., Kashyap S., Berria R., De Filippis E.A., Mandarino L.J. Global relationship between the proteome and transcriptome of human skeletal muscle. J. Proteome Res. 2008;7:3230–3241. doi: 10.1021/pr800064s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Centers for Disease Control and Prevention Diabetes Report Card 2014 Atlanta, GA: Centers for Disease Control and Prevention. US Dept. of Health and Human Services. 2015 [Google Scholar]