Abstract
Diverse cell types have unique transcriptional signatures that are best interrogated at single-cell resolution. Here we describe a novel RNA amplification approach that allows for high fidelity gene profiling of individual cells. This technique significantly diminishes the problem of 3′ bias, enabling detection of all regions of transcripts, including the recognition of mRNA with short or completely absent poly(A) tails, identification of noncoding RNAs, and discovery of the full array of splice isoforms from any given gene product. We assess this technique using statistical and bioinformatics analyses of microarray data to establish the limitations of the method. To demonstrate applicability, we profiled individual cells isolated from the mouse subventricular zone (SVZ)—a well-characterized, discrete yet highly heterogeneous neural structure involved in persistent neurogenesis. Importantly, this method revealed multiple splice variants of key germinal zone gene products within individual cells, as well as an unexpected coexpression of several mRNAs considered markers of distinct and separate SVZ cell types. These findings were independently confirmed using RNA-fluorescence in situ hybridization (RNA-FISH), contributing to the utility of this new technology that offers genomic and transcriptomic analysis of small numbers of dynamic and clinically relevant cells.
Keywords: single-cell analysis, RNA amplification, stem and progenitor cells, SVZ
Heterogeneity is a natural attribute of all living cells that allows complex systems to accommodate a wide range of environmental cues, providing a basis for a broad array of cellular responses (1–4). However, experiments performed on bulk material preclude analysis of cellular heterogeneity. Therefore, it is imperative to interrogate small cell populations, individual cells, and, in some instances, even subcellular structures (5). Some recent discoveries were only made possible by studying certain biological processes at the single-cell level (6–11).
Such studies necessitate the development of reliable methods of single-cell RNA amplification. Currently, the two most widely used methodological approaches, with numerous modifications, involve a PCR-based technique and the linear amplification procedure referred to as antisense RNA amplification (aRNA) or T7-based in vitro transcription (IVT) (12–14). The main disadvantage of the PCR-based technique is that cDNAs of different abundance, length, and composition are amplified with different efficiencies. Although similar issues must be considered with aRNA, differences with this amplification technique will be linear, rather than exponential as seen with PCR. Additionally, there is an inherent preference to amplify the 3′ end of mRNAs during the aRNA synthesis procedure (3′ bias).
Other problems with existing methods result from the limited amount of total RNA (10–20 pg) present in a single cell. Neither method provides for the nearly unlimited amplification required for transcriptome characterization. In the first case, the PCR-based method is naturally limited by the so-called “plateau effect.” Moreover, nonspecific PCR byproducts accumulate during amplification, especially during later cycles of PCR. Therefore, the number of PCR cycles that may be used for the amplification is limited. In turn, the T7-based IVT method requires two to three rounds of amplification to obtain a substantial amount of amplified RNA.
Here, we describe a new technology for interrogating small tissue and cell samples by combining exponential and linear amplification steps using a limited number of PCR cycles and T7-driven IVT. Although such combination PCR and IVT approaches for RNA amplification do exist, they were not tailored, with some exception, towards single-cell RNA amplification followed by microarray analysis (15,16), and these other methods have not solved the problem of 3′ bias for isoform detection.
Because depletion of rRNA and tRNA is not possible at the single-cell level, RNA-seq approaches are based exclusively on oligo(dT) priming. In this case, transcripts lacking poly(A) tails and a substantial amount of mRNAs longer than 3 kb (up to 36%) cannot be detected (17).
By introducing major technical modifications, we have developed an RNA amplification method that is suitable for single-cell RNA amplification and significantly diminishes the problem of 3′bias. The major modifications that distinguish this new protocol include:
Introduction of “extending primers” that contain random and semi-random sequences at the 3′ ends during PCR allows tagging of 3′ ends of cDNAs. The design of one of the extending primers includes a Kozak sequence at its 3′ ends that allows capture of 5′ ends of a gene’s coding sequence.
The combination of modified oligo(dT) and modified random primers allows a decrease of the size distribution of the resulting cDNA fragments, making the PCR step more efficient and consequently improving the preservation of relative gene abundance.
Detection of all parts of transcripts without 3′ bias, including the recognition of isoforms and mRNAs with short or completely absent poly(A) tails, as well as the identification of noncoding RNAs. The priming approach that utilizes the combination of oligo(dT) and random primers during the reverse transcription (RT) reaction is superior to any other priming strategy, and it secures full-length RNA coverage (18).
Carryover of reverse transcriptase results in PCR inhibition, which can become significantly pronounced when minute amounts of RNA are used (19–24). Our approach has greatly diminished this reverse transcriptase inhibitory effect on the following PCR step.
Our method generates 200–250 μg of amplified RNA from a single cell, sufficient to apply to virtually any RNA technique.
Cost is dramatically reduced (up to 5-fold) in comparison to commercially available kits. This is relevant because single-cell experiments usually require simultaneous study of at least 50–100 individual cells.
The limitations of this amplification approach have been rigorously tested using statistical and bioinformatics analysis of microarray data. The application of the amplification method detected various RNA isoforms with significant biological importance, as well as simultaneous expression of different cell-type specific transcripts within single cells of the neurogenic subventricular zone (SVZ).
Materials and methods
RNA amplification
Different amounts of total RNA from the LN229 glioma cell line (LN) and a collection of human neural stem/progenitor cells (Prog) were used for the amplification procedure: 20 pg, 1 ng, and 20 ng. There were 5 replicates for 1 ng and 20 ng RNA samples, and 6 replicates for 20 pg samples. In the case of single-cell amplification, the whole-cell content was used for the RT reaction. For each reaction, 0.4 μl 20 μM modified oligo(dT) primer, 0.25 μl 200 μM modified random primers, and 0.5 μl 30 μM T7-SWITCH oligonucleotide (Sigma-Aldrich, St. Louis, MO) was added to total RNA with a final volume of 6 μl. Each tube contained 100 ng of linear polyAcrylamide (LPA) (Sigma-Aldrich) as a carrier. The tubes were incubated at 65°C for 5 min and kept at 45°C thereafter. The first strand buffer (25 mM Tris-HCl, pH 8.3; 37.5 mM KCl; 1.5 mM MgCl2), 1 mM dNTP, 2 mM DTT (Life Technologies, Grand Island, NY), 3 mM MgCl2 (Sigma-Aldrich), and 100 ng of T4gp32 protein (Affymetrix, Cleveland, OH) were added to each tube (all concentrations are final). The tubes were incubated at 42°C for 1 min, 35°C for 5 min, 30°C for 10 min, and 25°C for 15 min. Superscript II (1μl) (Life Technologies) was added to the tubes, and they were incubated at 25°C for 30 min, 42°C for 2 h, 70°C for 15 min, and 25°C for 5 min. After the reactions were completed, the tubes were kept at 4°C. The final volume of reaction was 10 μl. Because the unnicked RNA:DNA hybrids do not denature completely, RT reactions were subjected to RNase H (Life Technologies) treatment at 37°C for 20 min. The tubes were allowed to stay at 4°C for 2 days prior to the PCR step.
The first-strand cDNA pool produced in the previous steps was then subjected to amplification using the Advantage 2 PCR Enzyme System (Clontech Laboratories, Mountain View, CA) in the same test tube. We added 0.3 μl 200 μM extending primers, 0.3 μl 200 μM modified random primers, 0.15 μl 200 μM amplification primer (Sigma-Aldrich), 2 μl 10 mM dNTP mix (Life Technologies), and 2 μl 50× Advantage 2 Polymerase (Clontech Laboratories) to the tube to produce double-stranded cDNA in a 100 μl reaction volume. An MJR PCR machine (Bio-Rad, Hercules, CA) was used for amplification with the following parameters: 95°C for 4 min, 25°C for 10 min, 65°C for 5 min, 68°C for 5 min; 95°C for 25 s, 58°C for 1 min, 68°C for 2 min; the next step, (95°C for 15 sec, 65°C for 30 sec, 68°C for 6 min), was run for 5–18 cycles depending on the RNA input.
Sense RNA was transcribed from double-stranded DNA using reagents from MEGAScript High Yield Transcription Kit (Life Technologies). We added 12 μl 75 mM rNTP, 3 μl 10× Buffer, and 3 μl Enzyme Mix (RNase Inhibitor and T7 RNA Polymerase) to 12 μl double-stranded DNA. The mixture was incubated for 18 h at 37°C. Upon completion, the reaction was treated with 1 μl DNase I (Life Technologies) for 30 min at 37°C. The amplified sense RNA was purified using MEGAclear Kits (Life Technologies) as described in the Ambion instruction manual. The RNA yield was evaluated using a SmartSpec 3000 spectrophotometer (Bio-Rad). This method of RNA amplification gives 200–250 μg amplified sense RNA after 1 round.
Additional material and methods information for for all experiments are supplied in the Supplementary Material.
Results and discussion
Design of the RNA amplification method
We have developed a technique that allows almost unlimited RNA amplification from a single cell without 3′ bias. Our approach involves a combination of exponential and linear amplification using a limited number of PCR cycles and T7-driven IVT.
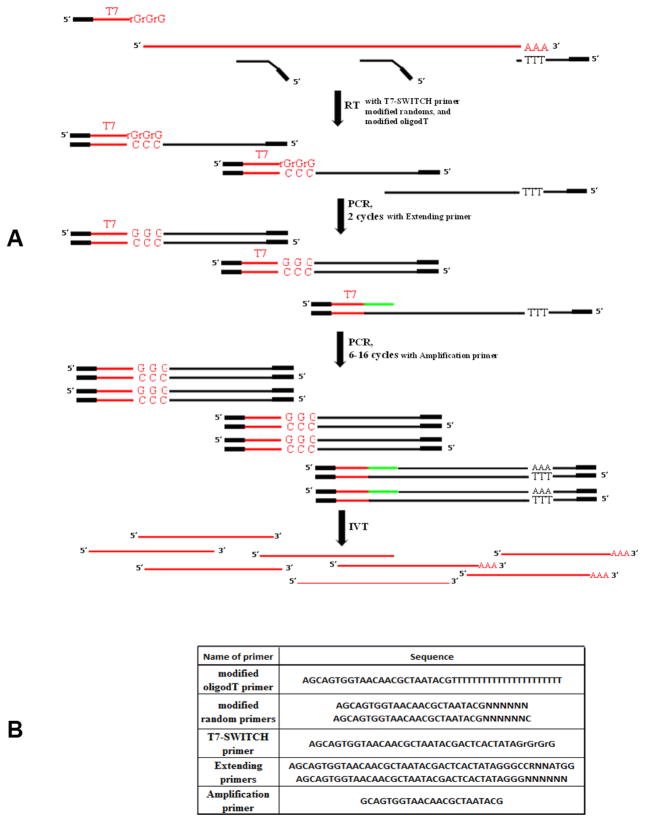
The RNA template was used in the first-strand synthesis reaction, along with a modified oligo(dT) primer, modified random primers, and a T7-SWITCH primer bearing a T7 RNA polymerase site (Figure 1A). When the reverse transcriptase reaches the 5′ end of mRNA during RT reaction, the enzyme’s terminal transferase activity adds a few additional nucleotides, primarily deoxycytidine, to the 3′ end of the cDNA (25,26). The T7-SWITCH primer comprises an oligo(rG) sequence at its 3′ end, which pairs with this deoxycytidine stretch, creating an extended template. The reverse transcriptase then switches templates and continues replicating to the end of the oligonucleotide. We also introduced extending primers that carry a specific sequence, a T7 RNA polymerase site, and either a random or Kozak semi-random sequence at the 3′ end of an oligonucleotide. This allows for the incorporation of a terminal tagging sequence at the 3′ end of cDNA molecules that were not extended by the switching mechanism. Because each of the primers has a specific sequence at its 5′ end (Figure 1B), the resulting single-stranded cDNA pool is generated with complementary sequences at their 5′ and 3′ ends. This enables the use of only one amplification primer during the following PCR cycles. Additionally, the resulting double-stranded DNA, generated during the PCR step includes a T7 RNA Polymerase site, which is subsequently used for the production of sense RNA during the T7 Polymerase amplification step.
Figure 1. RNA amplification scheme.
(A) Design of the RNA amplification method. RT step: The template for the RT reaction is polyadenylated RNA. The modified sequences of oligo(dT) and random primers are presented as black boxes. The T7-SWITCH primer contains a modified sequence, a T7 sequence, and an rGrGrG sequence (T7 and rGrGrG sequences are shown in red). Note that one of three cDNA products from the RT reaction does not have a 3′ tagging sequence because of the limited capacity of the switching effect. This cDNA is later primed with extending primers. PCR step: The PCR reaction contains all components initially, including the extending and amplification primers. The extending primers carry a specific sequence for the T7 RNA polymerase site and either random or Kozak sequences, which are are presented as green lines. The profile of two starting PCR cycles was tailored toward the preferable annealing of extending primers to the cDNA. These cycles are shown separately. The rest of the PCR cycles were modified to ensure optimal performance of the amplification primer. The product generated at the end of the PCR step was double-stranded DNA. T7 IVT step: The 5′ end localization of the T7 promoter on double-stranded DNA guarantees the synthesis of sense RNA, which is shown as red lines. Note that some RNA molecules are polyadenylated because they are the product of oligo(dT) priming. Other RNA molecules generated using random priming do not have a poly(A) stretch. (B) Primers utilized in the RNA amplification method.
It is known that carryover of reverse transcriptase inhibits PCR (19–24). The phenol/chloroform/isoamyl alcohol (PCI) procedure, followed by ethanol precipitation, removes this negative influence (24). While it is possible to utilize this method when working with nanograms of starting RNA in the amplification procedure, it was not successful for single-cell amplification. We observed a substantial loss of template after the PCI procedure; however, we did notice that the inhibition was greatly diminished when RT tubes were simply stored at 4°C for 2–3 days without any cDNA purification. Therefore, we utilized this approach to reduce PCR inhibition (24).
We minimized the number of PCR cycles and implemented the IVT step in order to avoid the excessive bias introduced by the PCR itself. We used 8, 11, and 18 PCR cycles for the amplification of 20 ng, 1 ng, and 20 pg total RNA, respectively. The yield of amplified RNA was 220–250 μg, regardless of the starting amount of total RNA. Because our RNA amplification technique is a complex multistep approach, we optimized each step of the procedure (Supplementary Material).
Analysis of the RNA amplification procedure using Agilent arrays
When sample size restrictions make RNA amplification mandatory, little information is available to the investigator to assess the probability that a distinct gene expression pattern corresponds to reality. Therefore, we compared samples before and after amplification to evaluate the performance of our approach. We isolated total RNA from LN and Prog cells. RNA amplification was applied to the equivalent of an average single cell (20 pg of total RNA), 50 cells (1 ng), or 1000 cells (20 ng) (15,27).
Two-color array analysis was performed using GeneSpring 11.5.1 (Agilent), which allowed the comparison of Prog/LN ratios for any given transcript with and without amplification. Ideally, these ratios should be the same for both samples before and after amplification. Even if RNA amplification causes some bias, this bias may be canceled out as long as it is consistent and produces comparable expression ratios between two samples, both before and after amplification (Supplementary Figure S1). The detected entities (21,589 probes) were analyzed using Volcano plots and analysis of variance (ANOVA) under different settings: P ≤ 0.05, Fold Change (FC) ≥ 2.0 and P ≤ 0.1, FC ≥ 1.5. Latter settings had less stringent conditions, which we introduced to prove that the list of outliers after amplification was limited, even in the statistically insignificant settings. We found that the Prog/LN expression ratios changed more than 2-fold compared with the samples before and after the amplification, and they were never higher than 8.1% (Table 1A). The microarray data and the protocol were deposited in the Gene Omnibus database with GEO accession no. GSE55137.
Table 1.
The characterization of the RNA amplification approach.
| A | ||||
|---|---|---|---|---|
| Total FR number out of all detected entities (%) | ||||
| p≤0.05, FC≥2.0 | p≤0.1, FC≥1.5 | |||
| ANOVA | Volcano | ANOVA | Volcano | |
| 20 ng vs w/o Amp. | 278 out of 21,589(1.3) | 132 out of 21,589(0.6) | 730 out of 21,589 (3.4) | 385 out of 21,589 (1.8) |
| 1 ng vs w/o Amp. | 618 out of 21,589 (2.9) | 253 out of 21,589(1.2) | 1.233 out of 21,589 (5.7) | 723 out of 21,589 (3.3) |
| 20 pg vs w/o Amp. | 949 out of 21,589 (4.4) | 1,025 out of 21,589 (4.7) | 1,653 out of 21,589 (7.7) | 1,744 out of 21,589 (8.1) |
| B | ||||
|---|---|---|---|---|
| % | w/o | 20ng | 1ng | 20pg |
| w/o | 96 | 90 | 70 | |
| 20ng | 96 | 76 | ||
| 1ng | 87 | |||
| 20pg | ||||
| C | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Name of Pathway’s category | p≤0.05, both WO and 20pg | p≤0.05, 20pg | p≤0.05, WO | Concordance, % | Lost after amplification, % |
| GoProcesses | 1,496 | 426 | 742 | 78 | 33 |
| GeneGoDiseases | 706 | 64 | 274 | 92 | 27 |
| GeneGoPathways | 68 | 56 | 59 | 55 | 46 |
| GeneGoProcess Networks | 19 | 3 | 20 | 86 | 51 |
(A) Prog/LN ratio comparison between the samples with and without (w/o) amplification using an ANOVA test and Volcano plot analysis. False ratio (FR) was ascribed when the absolute fold change (FC) of Prog/LN for the sample after amplification was at least 2-fold higher or lower compared with the sample without amplification. (B) Percentage of concordance between the number of differentially expressed genes detected in samples with and without amplification. Analysis was made based on Agilent array two-color data in single-color experimental settings using cutoffs of P ≤ 0.05 and FC ≥ 3.0 with the Benjamini-Hochberg FDR multiple testing correction. (C) Number of pathways that are significantly enriched for both samples, without (WO) and after 20 pg amplification, is presented in column 1. Number of pathways that are unique either for 20 pg or for WO samples is shown in columns 2 and 3 accordingly. The concordance percentage (column 4) and the percentage of overrepresented pathways lost after the amplification (column 5) were calculated for 20 pg and WO samples, where 20 pg is a primary (reference) sample. Example of concordance percentage calculation for the GOProcess category: 1496 / (1496 + 426) × 100% = 78%. Example of percentage calculation of a pathway’s loss for the GOProcess category 100 - [1496 / (1496 + 742) × 100%] = 33%.
Scatter plots were generated based on normalized log2-averaged Cy5/Cy3 ratios of signal intensities (Supplementary Figure S2). The highest correlation coefficient (r2 = 0.92) corresponded to the ratio comparison between the 20 ng amplified sample and the unamplified samples, and it decreased with lower amounts of RNA (r2 = 0.86 for 1 ng and r2 = 0.69 for 20 pg amplified samples). The correlation coefficient between 2 independently amplified samples corresponded to r2 = 0.96 ± 0.02 for 20 ng, r2 = 0.89 ± 0.02 for 1 ng, and r2 = 0.81 ± 0.04 for 20 pg samples. A correlation coefficient of 0.8–1.0 is regarded as indicative of a high correlation, and correlation coefficient of 0.6–0.8 is considered indicative of a marked degree of correlation (41).
In order to compare the number of differentially expressed genes identified before and after amplification, we analyzed Agilent two-color data with single-color experimental settings using GeneSpring. To focus on highly regulated genes, differences in gene expression between LN and Prog samples were restricted to a 3-fold change with a p-value ≤ 0.05. The concordance percentages ≥70% were retrieved by comparing lists of differentially expressed genes for samples before and after amplification (Table 1B).
Although Agilent arrays are typically 3′-biased, we were able to find genes that were represented by probes located at a minimum distance of a 3 kb away from the 3′ end of the transcript, including probes for the 5′ end as well as for the middle part of the particular gene. We focused on transcripts that were at least 6 kb in length. The number of genes that meet these criteria was 813 transcripts, with 1297 corresponding probes (Supplementary Table S1A). The number of successfully detected probes out of 1297 samples after amplification corresponded to 1180 (91%), 981 (76%), and 884 (68%) probes for 20 ng, 1 ng, and 20 pg respectively. For example, we observed the detection of 4 different isoforms of SYNE2 (transcript length of 22 kb) where probes for isoforms 1 and 5 are located at the 5′ end. Also, 2 5′-end probes revealed 2 unique isoforms of PLEC (15 kb).
We next analyzed the detection pattern of 86 probes discovered in the control sample without amplification that corresponded to non-coding RNAs. There were 84 detected probes out of 86 probes (98%) for the 20 ng sample and 75 probes (87%) each for the 1 ng and 20 pg samples after amplification (Supplementary Table S1B).
We also examined 81 probes corresponding to 71 non-polyadenylated RNAs that were detected in the sample without amplification. The number of probes identified in the samples after amplification corresponded to 79 probes (98%) for the 20 ng amplified sample and 73 probes (90%) for both the 1 ng and 20 pg samples after amplification (Supplementary Table S1C). Thus, the amplification of RNAs lacking poly(A) tails represents a significant advantage of our approach.
We analyzed the expression of 99 genes by real-time PCR after using our method of RNA amplification for 20 pg samples that utilized oligo(dT) along with random priming compared with the method that used only oligo(dT) priming. We also verified the microarray performance using real-time PCR. The genes corresponded to 3 groups: the first group included genes represented by probes located 1–3 kb from the 3′end of the transcript, the second group: 3–5 kb, and the third group: >5 kb. The results demonstrated the superiority of oligo(dT) along with random priming compared with priming without randoms. The transcripts of the second and the third groups were mostly detected after priming with the combination of oligo(dT) and random primers (91% and 82% accordingly). However, 73% of genes in the second group and only 45% in the third were detected after oligo(dT) priming. The detection of genes in the first group exhibited a similar rate regardless of mode of priming (94% with primer combination and 97% with only oligo(dT) priming). PCR analysis also demonstrated a high concordance (86%) between microarray and PCR data (Supplementary Table S2).
Pathway analysis
To translate gene expression data into functional profiles, we used the integrated knowledge database and software suite MetaCore (GeneGo), which links prior knowledge to newly generated experimental data. Our results show that the number of unique genes in a particular amplification experiment is generally smaller than the numbers of common and similar genes (Supplementary Figure S3).
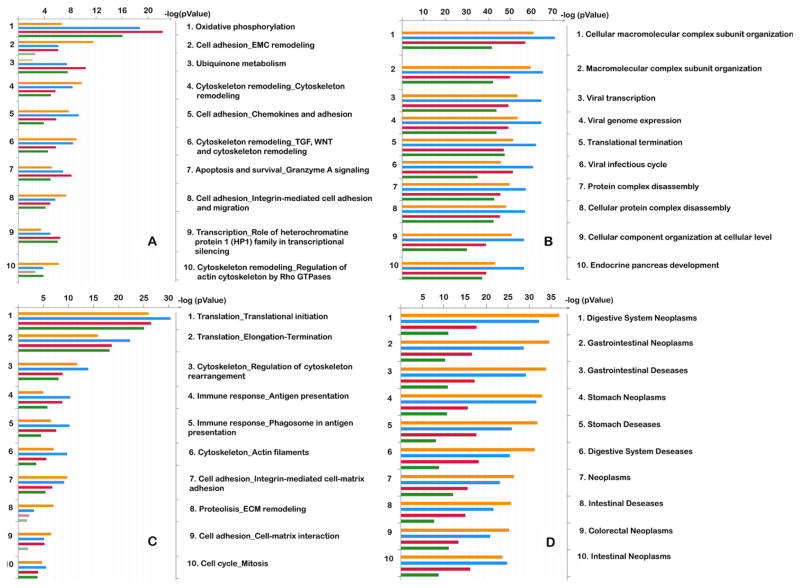
Unique genes add to dissimilarity of observed patterns of gene expression. As expected, this dissimilarity increases with reduced amounts of RNA used in the experiments. The most significantly enriched pathways for four MetaCore categories are presented in Figure 2. There is a good concordance between overrepresented pathways for each data set before and after amplification.
Figure 2. Significantly enriched pathways for four MetaCore categories.
GeneGO Pathway maps (A), GO Processes (B), GeneGO Process Networks (C), and GeneGO Diseases by Biomarkers (D) significantly enriched for differentially expressed genes are shown. Bar histograms (on the left) corresponding to the ontology terms in every section (1 to 10, listed on the right side of each diagram) are sorted in decreasing order of P-value (top to bottom) with no amplification, 20 ng, 1 ng, and 20 pg total RNA. The top terms are represented by histogram sections each having at least one longest strip regardless of which experiment it belongs to. Dimmed (semi-transparent) bars indicate marginal significance (with P-values below the 0.05 cutoff, as indicated on the logarithmic scale on top). Only the top 10 pathways are shown in every section in the form of a bar graph histogram, and the list of 50 pathways is available in Supplementary Figures S4–S7.
Since an investigator will have only single-cell information after performing a real experiment, we decided to compare 2 samples, before and after 20 pg amplification, using a 20 pg sample after amplification as a reference. We analyzed 4 pathway categories with preset P-values of ≤ 0.05 for a 20 pg sample (Table 1C). The complete pathway’s list and exemplary maps are shown in Supplementary Table S3, A–D and Supplementary Figure S8.
Three out of 4 categories have a high concordance between a 20 pg amplification sample and the sample without amplification: 78%, 92%, and 86% for GOProcesses, GeneGoDiseases, and GeneGoProcessNetworks, respectively. In contrast, the GeneGoPathway category exhibited only 55% similarity. Also, 27%–51% of pathways and networks were overrepresented only in the samples without amplification, depending on the category (Table 1C).
Thus, this analysis revealed that certain pathways or networks detected in the sample after amplification were reliably overrepresented before amplification. One limitation of this RNA amplification method is that some biologically relevant information may be lost after amplification. Only 70% of differentially expressed genes in samples without amplification were discovered after single-cell amplification. Also, certain up-regulated pathways and networks in samples without amplification were not detected after the single-cell amplification, depending on the pathway’s category.
Characterization of single cells from the SVZ: A proof of concept
The ultimate proof of the utility of our amplification technique is the ability to confirm well-established concepts from a relevant subject of interest, and generate new and significant findings that can be verified by other means.
We chose to test our method on mouse SVZ cells, because the forebrain SVZ contains several heterogeneous cell types in close proximity. Stem cells (type B cells) exhibit structural and biological markers of astrocytes. These type B cells give rise to type C cells, or transit-amplifying cells, which in turn generate immature neuroblasts (type A cells) (29–32). Other cell types populating the SVZ include astrocytes and ependymal cells. Each cell type is associated with a set of markers that presents a starting point to verify the reliability of our amplification approach (Figure 3).
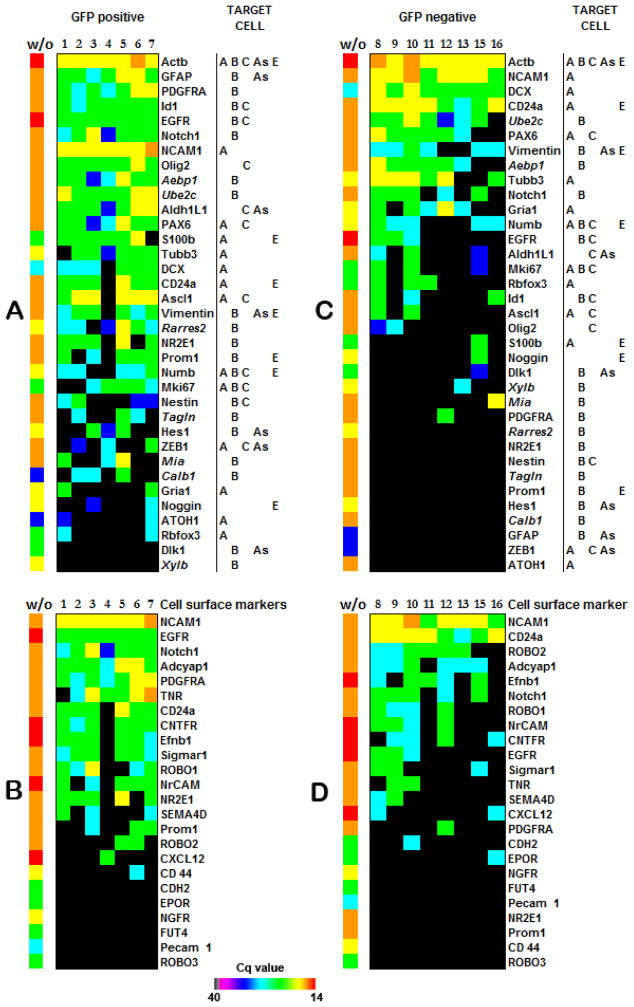
Figure 3. Expression of subventricular zone (SVZ) cell markers.
(A) SVZ markers expressed in GFP positive cells. (C) SVZ markers expressed in the GFP negative population. Markers of stem cells are defined as B type, transit-amplifying cells correspond to C type, neuroblasts correspond to A type; astrocytes correspond to As typ; and ependymal cells correspond to E type. Each cell type is identified by a combination of these markers (33). Expression of certain cell surface molecules can ensure the isolation of distinct cell types from GFP positive (B) and GFP negative (D) cell populations. Genes presented in italics were recently assigned as B cell markers using microarray data from the stem cell–enriched population (34). Heat maps reflect real-time PCR-derived Cq values for each transcript. Samples without amplification (w/o) were used as positive controls. Total RNA input of unamplified pooled SVZ cells or embryonic cells after the RT reaction corresponds to 3 μg. Cell #14 was excluded for technical reasons (low expression of ACTB and all other genes). Primer sequences are provided in Supplementary Table S4.
We studied the SVZ cell population from 6-day-old transgenic mice that express GFP under control of the GFAP promoter. Sixteen single cells from the SVZ were collected after GFP sorting: seven GFP positive cells (GFP+) and nine GFP negative cells (GFP−). The application of our method correctly confirmed GFAP expression in a GFP+ population of GFP-GFAP transgenic mice and its absence in a GFP− cell population. As expected, the mRNA expression profile of the GFP− cell population was enriched for neuronal markers (Figure 3C).
The GFP+ population was quite uniform (Figure 3, A and B). Six out of 7 cells, except for cell #4, were identical for 21 out of 44 markers, which corresponds to 48% of transcripts examined. If we consider the situation where an individual cell differs from others by 2 markers, this translates to 68% similarity. The GFP− cell population was not as uniform as the GFP+ population (30% similarity), and it exhibited the expression of mostly neuronal markers (Figure 3C).
Because GFP+ cells expressing B-type stem cell markers were also positive for transcripts characteristic of type A and C cells, we confirmed our findings with RNA-FISH probes to GFAP, Tubb3, and Olig2. Indeed, some cells simultaneously expressed all three transcripts (Supplementary Figures S9–S12).
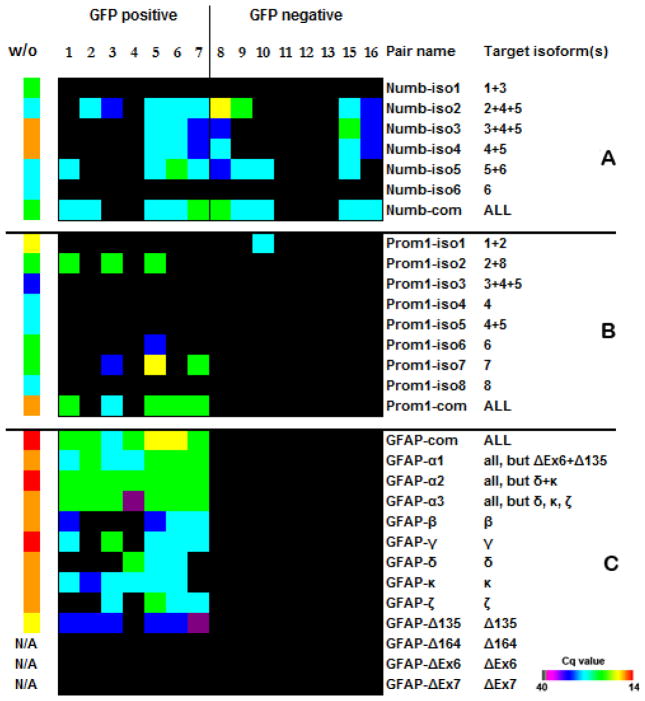
It has been shown that RNA splicing is a very pronounced process in SVZ neurogenesis (35). We were able to detect biologically important isoforms of certain transcripts in SVZ cells. For example, we observed only the expression of Numb isoforms 2 and 4, and the expression of Numb1 and Numb3 was absent in postnatal day 6 (P6) mice SVZ samples (Figure 4A). This correlates with previous findings that show a shift in numb protein expression from those isoforms containing the proline-rich region (PRR) insert (Numb1 and Numb3) in embryonic day 10 (E10) embryos, to isoforms lacking the PRR insert (Numb2 and Numb4) in P2 mice (36). We further observed that noncoding Numb isoforms also obeyed the PRR insert rule: isoform5 lacking the PRR insert was expressed in analyzed cells, and there was no expression of isoform6, which contained the PRR insert. To our knowledge, this is the first evaluation of coexpression of coding and noncoding Numb isoforms, and this finding that validates the sensitivity of our technique.
Figure 4. Expression patterns of Numb, Prominin1, and GFAP isoforms.
(A) Numb isoforms 1, 3, and 6 that were not detected contain insertions into the PRR domain, and the detected Numb isoforms 2, 4, and 5 do not have an insertion (36). Because the same pair of primers can detect different isoforms, it is not possible to assign the expression of a certain isoform to a specific cell. For example, cells 5–8 can express either the combination of isoforms 2, 4, and 5 or only isoform 5. At the same time, Numb2 is expressed in cells 2 and 3, Numb4 in cell 16, and Numb5 in all cells where it was detected. (B) The cells expressed mostly Prominin1 isoforms 2 and 7. No isoform-specific transcripts were detected in cell #6, but primers recognizing all isoforms produced a signal. Our results also show good concordance with previous studies that revealed the expression of only Prominin1 transcript isoforms 1, 2, and 6 in the subventricular zone (SVZ), and the absence of isoforms 3, 4, 5, and 8 (34,37). We also observed that isoform 1 was expressed exclusively in one of the GFP− cells, while other isoforms were present only in GFP+ population. While the expression of isoform 7 was not previously assessed, we found that this isoform together with isoform 2 is dominant in GFP+ cells. Prominin1 is strongly predisposed to alternative splicing, and there are 10 known isoforms for the human transcript. Therefore, it is likely to reveal a new murine isoform of Prominin1 amidst the eight known isoforms. (C) Glial fibrillary protein (GFAP) expression was previously detected in distinct population of astrocytes that have been identified as SVZ neural stem cells (38–40). GFAP isoforms were detected only in GFP+ cells. The cells analyzed in our experiments did not contain isoforms GFAP-ΔEx6, GFAP-ΔEx7, and GFAP-Δ164, and the GFAP-Δ135 isoform was expressed at a low level. Isoforms GFAP-ΔEx6, GFAP-Δ164 and GFAP-Δ135 were not detected in samples without amplification (w/o), but the quality of the primers was verified before (39). Primers labeled “com” are designed to anneal to the common part of all known isoforms of the transcript. Because some isoforms are homologous, certain primers detect more than one target isoform. Heat maps reflect real-time PCR-derived Cq values for each transcript. A sample without amplification was used as positive control. Total RNA input of unamplified, pooled SVZ cells or embryonic cells in the RT reaction was 3 μg.
We also were able to detect 5′ ends of the long transcripts and identify different splice isoforms of significant biological importance for genes such as Prominin1, GFAP, RbFox, Pax6, Id1, and EGFR (Figures 4 and 5).
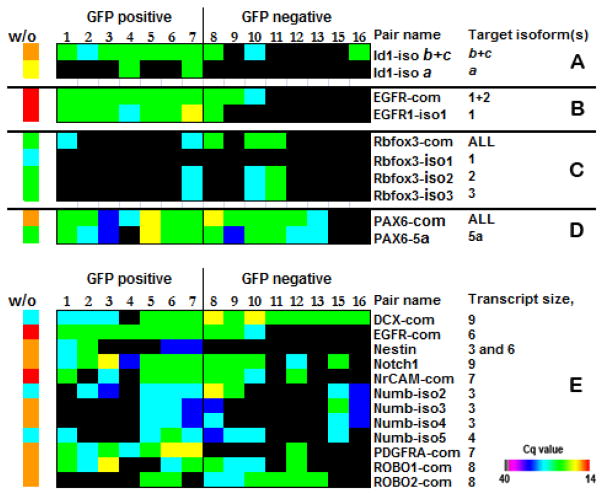
Figure 5. Expression patterns of Id1, EGFR, RbFox3, and PAX6.
(A) A high expression of DNA-binding protein inhibitor Id1 is characteristic for neural stem cells (41). It was previously established that Id1 protein expression is associated with the GFP+ cell population (41,42). This transcriptional regulator is more likely the product of Id1 isoform a, according to our results, because only this isoform is exclusively expressed in GFP+ cells. (B) Epidermal growth factor receptor (EGFR) is expressed on stem cells (B cells) and transit-amplifying progenitor cells (C cells) in the subventricular zone (SVZ), but it is absent from neuroblasts (A cells). It was proposed that EGFR regulates the balance of SVZ cell subtypes (43). All GFP+ cells express the EGFR isoform1 with a possible co-expression of isoform2. The EGFR sequence structure does not allow design of primer pairs to establish the expression pattern of EGFR isoform2 in cells 1–8 conclusively, but GFP− cells 9 and 10 definitely have this isoform. (C) It has been suggested that specific neuronal subtypes express different RbFox3 protein variants with varying nuclear/cytoplasmic ratios (44,45). It has been previously proposed that each variant of RbFox3 has its own biological target(s) and may play a key role in the regulation of neural cell differentiation. Although the protein product of RbFox3 is considered a mature neuron-specific marker, we detected only isoform 2, which is exclusively nuclear, and isoform 3 that, as was suggested, shuttles between the nucleous and cytoplasm in GFP+ as well as GFP− cells. RbFox isoform 1 was not detected in any GFP cell population. (D) It was shown that PAX6, containing the canonical form of the paired domain (PD) without the alternative exon 5a, influences cell fate and proliferation at the same time, and an exon 5a-containing PAX6 isoform inhibits cell proliferation without affecting cell fate (46). We observed that PAX6 mRNA was present in almost every cell regardless of the presence or absence of exon 5a. We speculate that the protein distribution can be different, taking in account that the PAX6 gene demonstrates translational uncoupling (34). (E) Examples of 5′ end detection of the transcripts. The primers localized at 5′ ends of mRNAs detect the transcripts of different lengths, up to 9 kb. Heat maps reflect real-time PCR–derived Cq values for each transcript. A sample without amplification (w/o) was used as a positive control. Total RNA input of unamplified pooled SVZ cells or embryonic cells to RT reaction was 3 μg.
Recent studies suggest that over 80% of all human genes undergo alternative splicing, potentially providing a molecular basis for highly complex systems (47–49). Approximately 73% of alternatively spliced exons are within coding sequences, which preferentially include internal exons. It was also observed that up to 12 isoforms may be expressed simultaneously within the same cell (50). Our method affords the detection of virtually any part of a gene sequence regardless of the length and the structure of the transcript. It also allows the detection of noncoding and non-polyadenylated RNAs. Our method also detects almost 70% of the 5′ ends of transcripts longer than 6 kb at single-cell level (Supplementary Table S1).
The application of this amplification technique uncovered an essentially uniform gene expression signature for the GFP+ cells of the SVZ. This discovery supports the notion that the SVZ micro-environment may selectively inhibit the expression of certain phenotypic genes since such uniformity was not detected at the protein level (51). This interpretation is also corroborated by several cell and molecular characterization studies of the SVZ, in which certain mRNA transcripts were identified in SVZ cells, but did not undergo subsequent translation to produce the corresponding proteins (34,40). It is worth noting that our classification of SVZ cells was based on the detection of certain RNA transcripts, while well-known A, B, and C cell markers have been mostly established at the protein level.
Therefore, it is possible that differences may emerge between the transcriptomes and proteomes of these cells. In fact, it was shown using bulk transcriptome analysis that the stem cell-enriched population was primed toward neuron-generation by transcribing, but not translating neuroblast mRNAs (e.g., PAX6, Ascl1, and CD24a) (34). We had similar results in our experiments and also found a distinct cell type, which may express some markers for all cell types populating the SVZ. These findings were made possible with this single-cell RNA amplification approach, and they were later confirmed by an independent method, RNA-FISH.
In conclusion, we suggest that our profiling results on a small number of neurogenic cells are well-supported by the literature, which speaks directly to the reliability of this new RNA amplification technology.
Supplementary Material
METHOD SUMMARY.
We present a single-cell RNA amplification approach combining exponential and linear amplification steps using a limited number of PCR cycles and T7-driven in vitro transcription (IVT). Our approach allows for unlimited RNA amplification and enables detection of gene isoforms, non-coding RNAs, and mRNAs with short or absent poly(A) tails.
Acknowledgments
We would like to thank Marda Jorgensen for her assistance in preparing tissue for RNA-FISH, Amy Poirier for her help with the epMotion robot, and Neal Benson for his assistance with cell sorting. The work supported during this study was funded by NIH grant NS055165, the McKnight Brain Institute of the University of Florida, and the Maren, McKinney, and Thompson Regeneration Fund. This paper is subject to the NIH Public Access Policy.
Footnotes
Supplementary material for this article is available at www.BioTechniques.com/article/114331.
Author contributions
O.S. conceived and designed the method, coordinated the project, performed the experiments, analyzed and interpreted the data, and wrote the manuscript. D.J.S. performed the experiments, analyzed and interpreted the data, and wrote the manuscript. F.A.S. wrote the manuscript. A.O. performed RNA-FISH experiments and wrote the manuscript. A.P. performed pathway analysis and wrote the manuscript. D.A.S. supervised the project and wrote the manuscript.
Competing interests
The authors declare no competing interests.
To purchase reprints of this article, contact: biotechniques@fosterprinting.com
References
- 1.Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat Rev Genet. 2006;7:34–44. doi: 10.1038/nrg1750. [DOI] [PubMed] [Google Scholar]
- 2.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, Barres BA. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr Opin Neurobiol. 2010;20:588–594. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, Holt CE. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McEvoy J, Flores-Otero J, Zhang J, Nemeth K, Brennan R, Bradley C, Krafcik F, Rodriguez-Galindo C, et al. Coexpression of normally incompatible developmental pathways in retinoblastoma genesis. Cancer Cell. 2011;20:260–275. doi: 10.1016/j.ccr.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eberwine J, Bartfai T. Single cell transcriptomics of hypothalamic warm sensitive neurons that control core body temperature and fever response. Pharmacol Ther. 2011;129:241–259. doi: 10.1016/j.pharmthera.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flatz L, Roychoudhuri R, Honda M, Filali-Mouhim A, Goulet JP, Kettaf N, Lin M, Roederer M, et al. Single-cell gene-expression profiling reveals qualitatively distinct CD8 T cells elicited by different gene-based vaccines. Proc Natl Acad Sci USA. 2011;108:5724–5729. doi: 10.1073/pnas.1013084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstein JA, Zeng X, Chien YH, Quake SR. Correlation of gene expression and genome mutation in single B-cells. PLoS ONE. 2013;8:e67624. doi: 10.1371/journal.pone.0067624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci USA. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brady G, Barbara M, Iscove NN. Representative in vitro cDNA amplification from individual hemopoietic cells and colonies. Methods Mol Cell Biol. 1990;2:17–25. [Google Scholar]
- 15.Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 16.Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- 17.Tang F, Kao L, Surani MA. Development and applications of single-cell transcriptome analysis. Nat Methods. 2011;8:S6–S11. doi: 10.1038/nmeth.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Resuehr D, Spiess AN. A real-time polymerase chain reaction-based evaluation of cDNA synthesis priming methods. Anal Biochem. 2003;322:287–291. doi: 10.1016/j.ab.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 19.Sellner LN, Coelen RJ, Mackenzie JS. Reverse transcriptase inhibits Taq polymerase activity. Nucleic Acids Res. 1992;20:1487–1490. doi: 10.1093/nar/20.7.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fehlmann C, Krapf R, Solioz M. Reverse transcriptase can block polymerase chain reaction. Clin Chem. 1993;39:368–369. [PubMed] [Google Scholar]
- 21.Chumakov KM. Reverse transcriptase can inhibit PCR and stimulate primer-dimer formation. PCR Methods Appl. 1994;4:62–64. doi: 10.1101/gr.4.1.62. [DOI] [PubMed] [Google Scholar]
- 22.Chandler DP, Wagnon CA, Bolton H., Jr Reverse transcriptase (RT) inhibition of PCR at low concentrations of template and its implications for quantitative RT–PCR. Appl Environ Microbiol. 1998;64:669–677. doi: 10.1128/aem.64.2.669-677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liss B. Improved quantitative real-time RT–PCR for expression profiling of individual cells. Nucleic Acids Res. 2002;30:e89. doi: 10.1093/nar/gnf088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suslov O, Steindler DA. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005;33:e181. doi: 10.1093/nar/gni176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chenchik A, Zhu YY, Diatchenko L, Li R, Hill J, Siebert PD. In: Gene Cloning and Analysis by RT–PCR. Siebert P, Larrick J, editors. Biotechniques Books; Natick, MA: 1998. pp. 305–319. [Google Scholar]
- 26.Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–1560. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subkhankulova T, Livesey FJ. Comparative evaluation of linear and exponential amplification techniques for expression profiling at the single-cell level. Genome Biol. 2006;7:R18. doi: 10.1186/gb-2006-7-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franzblau A. A primer of statistics for non-statisticians. Chapter 7. Harcourt, Brace & World, Inc; New York, NY: 1958. p. 150. [Google Scholar]
- 29.Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 30.Lois C, Alvarez-Buylla A. Long-distance neuronal migration in the adult mammalian brain. Science. 1994;264:1145–1148. doi: 10.1126/science.8178174. [DOI] [PubMed] [Google Scholar]
- 31.Thomas LB, Gates MA, Steindler DA. Young neurons from the adult subependymal zone proliferate and migrate along an astrocyte, extracellular matrix-rich pathway. Glia. 1996;17:1–14. doi: 10.1002/(SICI)1098-1136(199605)17:1<1::AID-GLIA1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 32.Sohur US, Emsley JG, Mitchell BD, Macklis JD. Adult neurogenesis and cellular brain repair with neural progenitors, precursors and stem cells. Philos Trans R Soc Lond B Biol Sci. 2006;361:1477–1497. doi: 10.1098/rstb.2006.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mamber C, Kozareva DA, Kamphuis W, Hol EM. Shades of gray: The delineation of marker expression within the adult rodent subventricular zone. Prog Neurobiol. 2013;111:1–16. doi: 10.1016/j.pneurobio.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Beckervordersandforth R, Tripathi P, Ninkovic J, Bayam E, Lepier A, Stempfhuber B, Kirchhoff F, Hirrlinger J, et al. In vivo fate mapping and expression analysis reveals molecular hallmarks of prospectively isolated adult neural stem cells. M Cell Stem Cell. 2010;7:744–758. doi: 10.1016/j.stem.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Lim DA, Muárez-Fariñas S, Naef F, Hacker CR, Menn B, Takebayashi H, Magnasco M, Patil N, et al. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol Cell Neurosci. 2006;31:131–148. doi: 10.1016/j.mcn.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Bani-Yaghoub M, Kubu CJ, Cowling R, Rochira J, Nikopoulos GN, Bellum S, Verdi JM. A switch in numb isoforms is a critical step in cortical development. Dev Dyn. 2007;236:696–705. doi: 10.1002/dvdy.21072. [DOI] [PubMed] [Google Scholar]
- 37.Walker TL, Wierick A, Sykes AM, Waldau B, Corbeil D, Carmeliet P, Kempermann G. Prominin-1 allows prospective isolation of neural stem cells from the adult murine hippocampus. J Neurosci. 2013;33:3010–3024. doi: 10.1523/JNEUROSCI.3363-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyd SE, Nair B, Ng SW, Keith JM, Orian JM. Computational characterization of 3′ splice variants in the GFAP isoform family. PLoS ONE. 2012;7:e33565. doi: 10.1371/journal.pone.0033565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamphuis W, Mamber C, Moeton M, Kooijman L, Sluijs JA, Jansen AH, Verveer M, de Groot LR, et al. GFAP isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of Alzheimer disease. PLoS ONE. 2012;7:e42823. doi: 10.1371/journal.pone.0042823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mamber C, Kamphuis W, Haring NL, Peprah N, Middeldorp J, Hol EM. GFAPδ expression in glia of the developmental and adolescent mouse brain. PLoS ONE. 2012;7:e52659. doi: 10.1371/journal.pone.0052659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niola F, Zhao X, Singh D, Castano A, Sullivan R, Lauria M, Nam H, Zhuang Y, et al. Id proteins synchronize stemness and anchorage to the niche of neural stem cells. Nat Cell Biol. 2012;14:477–487. doi: 10.1038/ncb2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y, Comte I, Szabo G, Hockberger P, Szele FG. Adult mouse subventricular zone stem and progenitor cells are sessile and epidermal growth factor receptor negatively regulates neuroblast migration. PLoS ONE. 2009;4:e8122. doi: 10.1371/journal.pone.0008122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dredge BK, Jensen KB. NeuN/Rbfox3 nuclear and cytoplasmic isoforms differentially regulate alternative splicing and nonsense-mediated decay of Rbfox2. PLoS ONE. 2011;6:e21585. doi: 10.1371/journal.pone.0021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haubst N, Berger J, Radjendirane V, Graw J, Favor J, Saunders GF, Stoykova A, Goetz M. Molecular dissection of Pax6 function: the specific roles of the paired domain and homeodomain in brain development. Development. 2004;131:6131–6140. doi: 10.1242/dev.01524. [DOI] [PubMed] [Google Scholar]
- 47.Takeda J, Suzuki Y, Nakao M, Kuroda T, Sugano S, Gojobori T, Imanishi T. H-DBAS: alternative splicing database of completely sequenced and manually annotated full-length cDNAs based on H-Invitational. Nucleic Acids Res. 2007;35(Database issue):D104–D109. doi: 10.1093/nar/gkl854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, et al. The transcriptional landscape of the Mamm. Genome Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 49.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 50.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raponi E, Agenes F, Delphin C, Assard N, Baudier J, Legraverend C, Deloulme JC. S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia. 2007;55:165–177. doi: 10.1002/glia.20445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.