SUMMARY
Dysregulation of circadian rhythms is associated with metabolic dysfunction, yet it is unclear whether enhancing clock function can ameliorate metabolic disorders. In an unbiased chemical screen using fibroblasts expressing PER2::Luc, we identified Nobiletin (NOB), a natural polymethoxylated flavone, as a Clock amplitude-Enhancing small Molecule (CEM). When administered to diet-induced obese (DIO) mice, NOB strongly counteracted metabolic syndrome and augmented energy expenditure and locomotor activity in a Clock gene-dependent manner. In db/db mutant mice, the clock is also required for the mitigating effects of NOB on metabolic disorders. In DIO mouse liver, NOB enhanced clock protein levels and elicited pronounced gene expression remodeling. We identified retinoid acid receptor-related orphan receptors (RORs) as direct targets of NOB, revealing a pharmacological intervention that enhances circadian rhythms to combat metabolic disease via the circadian gene network.
Keywords: Circadian clock, Nobiletin, natural flavonoid, retinoid acid receptor-related orphan receptors (RORs), metabolic syndrome, clock amplitude-enhancing small molecule
Graphical Abstract
INTRODUCTION
Virtually all living organisms on Earth have evolved an intrinsic timing system, the circadian clock, to anticipate and exploit daily environmental changes. In mammals, the clock system is hierarchically organized, with the central pacemaker in the hypothalamic suprachiasmatic nuclei (SCN) coordinating peripheral tissue clocks to perform physiological functions (Takahashi et al., 2008). The cell-autonomous molecular oscillator is the basic component of the clock system, composed of interlocked feedback loops (Dibner et al., 2010). The core loop, consisting of positive (transcriptional activators CLOCK/BMAL1 or NPAS2/BMAL1) and negative (PER1/2 and CRY1/2) arms, is responsible for generating molecular rhythms, whereas competing nuclear receptors REV-ERBs and RORs regulate Bmal1 expression to confer rhythm stability and robustness (Zhang and Kay, 2010). The molecular oscillators drive tissue-specific gene expression throughout the circadian cycle via both transcriptional and posttranscriptional mechanisms (Koike et al., 2012; Zhang et al., 2014).
A fundamental process tightly regulated by the clock system is metabolism (Asher and Schibler, 2011; Bass and Takahashi, 2010; Gerhart-Hines and Lazar, 2015; Green et al., 2008; Rutter et al., 2002), as both metabolites (Eckel-Mahan et al., 2012) and metabolic gene expression (Yang et al., 2006; Zhang et al., 2014) broadly exhibit circadian oscillation. In humans, circadian misalignment has been shown to cause metabolic disturbances such as glucose intolerance and hyperlipidemia (Roenneberg et al., 2012; Scheer et al., 2009). Genetic studies have also revealed overlapping metabolic deficiencies in clock-disrupted mice (Green et al., 2008). For example, the circadian mouse mutant ClockΔ19/Δ19 harboring a dominant negative allele has been found to exhibit a spectrum of metabolic disorders including obesity, hyperlipidemia, hepatic steatosis, hyperglycemia, hypoinsulinemia, and respiratory uncoupling (Marcheva et al., 2010; Shi et al., 2013; Turek et al., 2005).
Recent studies have explored the strategy of directly manipulating circadian rhythms to ameliorate the metabolic syndrome (Antoch and Kondratov, 2013; Chen et al., 2013; Farrow et al., 2012; Schroeder and Colwell, 2013). For example, time-restricted intake of high-fat diet (HFD) was shown to protect mice against metabolic disease (Hatori et al., 2012). Oscillatory amplitude of clock and metabolic gene expression was significantly enhanced in a nighttime-specific HFD regime, suggesting that the body can best expend the incoming nutrients by a concerted action of clock-associated pathways during the active period. To circumvent compliance issues inherent in behavioral interventions, a pharmacological approach involving clock-modulating small molecules was also examined (Chang et al., 2015; Chen et al., 2012; Chen et al., 2013; Hirota et al., 2012; Isojima et al., 2009; Meng et al., 2010; Solt et al., 2012; Wallach and Kramer, 2015). For example, small molecule agonists acting on the REV-ERB nuclear receptors showed beneficial metabolic effects (Solt et al., 2012), suggesting modulatory compounds can improve metabolism via clock components or clock-associated mechanisms.
We previously identified several synthetic Clock amplitude-Enhancing small Molecules (CEMs) in a high throughput chemical screen using reporter cells with highly robust rhythms (Chen et al., 2012; Chen et al., 2013). When applied to cultured heterozygous ClockΔ19/+ PER2::Luc reporter cells where reporter rhythms oscillate with a weaker amplitude (approximately 1/3) relative to wild-type (WT) Clock+/+ cells, these CEMs were able to restore the reporter rhythm amplitude to near normal levels. Here we report identification of a natural compound, Nobiletin, that enhances the clock and concordantly renders protection against metabolic syndrome in a Clock gene-dependent manner. We further identified RORs, nuclear receptors functioning in the stabilization loop of the molecular oscillator, as the direct targets of Nobiletin.
RESULTS
Identification of Nobiletin (NOB) as a clock modulator
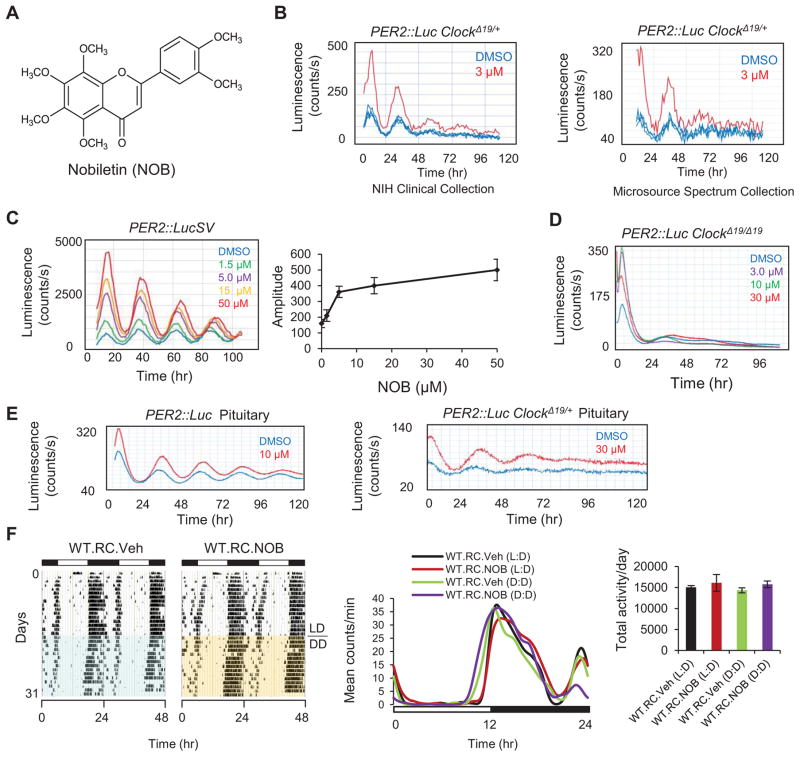
To identify additional CEMs, we screened an in-house compound collection with 5,300 small molecules using the heterozygous ClockΔ19/+ PER2::Luc reporter cells which exhibit sustained reporter rhythms at a dampened amplitude relative to WT cells (see Experimental Procedures and Supplementary Information). Interestingly, a naturally-occurring polymethoxylated flavonoid enriched in citrus peels, Nobiletin (NOB), was found independently from two sub-libraries to enhance the reporter rhythm of the ClockΔ19/+ cells (Figures 1A and 1B). Tangeretin, a close analog of NOB, similarly enhanced the PER2::Luc reporter rhythm in ClockΔ19/+ cells (Figures S1A and S1B). NOB robustly enhanced the amplitude of Per2::LucSV reporter rhythm (Chen et al., 2012) and also lengthened the period in a dose-dependent manner, with an estimated EC50 of <5.0 μM (Figure 1C). Similar to previously reported CEMs (Chen et al., 2012; Chen et al., 2013), NOB was ineffective in restoring the rhythm in clock-disrupted homozygous ClockΔ19/Δ 19 reporter cells (Figure 1D). Importantly, NOB enhanced PER2::Luc reporter rhythms in peripheral tissue explants from both ClockΔ19/+ and WT reporter knock-in mice (Figures 1E and S1C), but not in the SCN which is resistant to external perturbation due to robust inter-neuronal coupling (Figure S1D) (Buhr et al., 2010; Chen et al., 2012; Liu et al., 2007). In accordance with the resistance of the SCN to NOB manipulation, we also observed normal wheel-running activity and periodicity in WT C57BL/6J mice treated with NOB (Figure 1F).
Figure 1. Nobiletin (NOB) enhances amplitude of circadian rhythms.
(A) Nobiletin (NOB) chemical structure. (B) Primary screening of the NIH Clinical Collection (left) and Microsource Spectrum Collection (right) showing enhancement of the PER2::Luc ClockΔ19/+ reporter rhythm by NOB. (C) Left: Dose-dependent effects of NOB on reporter rhythms from PER2::LucSV cells. Right: Quantification of amplitude response to NOB doses. (D) NOB was not able to rescue reporter rhythms in PER2::Luc ClockΔ19/Δ19 fibroblast cells. (E) Clock-enhancing effects of NOB in pituitary explants from PER2::Luc WT (left) and PER2::Luc ClockΔ19/+ (right) reporter mice. (F) Left: Actograms illustrating the effect of vehicle or NOB on circadian behavior in RC-fed WT mice (n=5). Middle: Average wave plots summarizing wheel-running activity during 12–14 days of LD (12hr light, 12hr dark) or DD (Constant darkness) for indicated genotypes (n=5). Right: Daily total wheel-running activity during L:D or D:D conditions for the indicated genotypes (n=5). Data are represented as mean ± SEM.
NOB has shown a wide variety of beneficial effects (Ben-Aziz, 1967; Cui et al., 2010; Mulvihill et al., 2011; Nagase et al., 2005; Walle, 2007). However, its role as a modulator of the circadian clock was previously unknown. Whereas Per2 transcript levels were moderately altered and reduced at CT20 by NOB in PER2::LucSV cells (Figure S1E), PER2 proteins were found to accumulate to greater levels (Figures S1F and S1G), consistent with the elevated bioluminescence and suggesting a post-transcriptional mechanism for PER2 enrichment. Expression of other core clock genes was also altered by NOB (Figures S1E – S1G). In particular, CRY1, the heterodimeric partner of PERs in the negative arm of the oscillator, showed a slight trend of greater protein abundance despite markedly reduced transcript level (Figures S1E and S1F). The concordant enrichment of PER2 and CRY1 is consistent with the idea that PER2 and CRY1 proteins stabilize each other (Yagita et al., 2002), and that an improved stoichiometric ratio of the negative arm can lead to enhanced overall circadian amplitude in mouse fibroblast cells (Lee et al., 2011).
Robust Clock-dependent metabolic protection by NOB in diet-induced obese mice
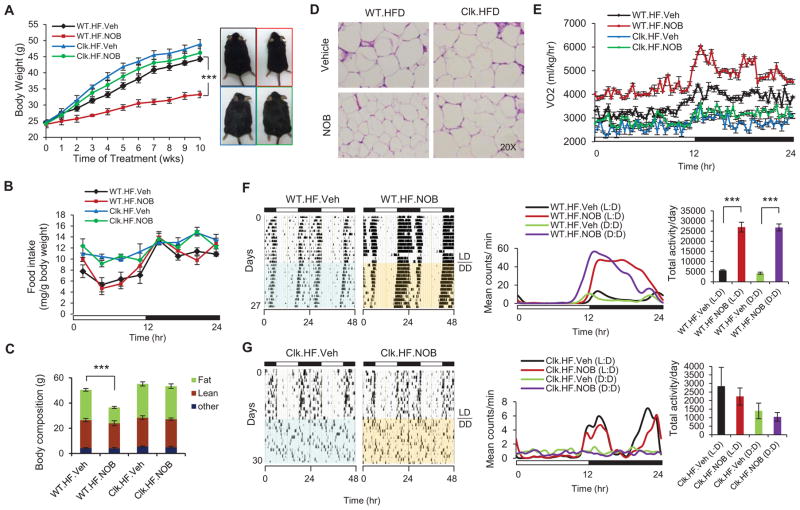
Recent studies suggested a protective role of NOB against metabolic syndrome (Kurowska and Manthey, 2004; Lee et al., 2013; Lee et al., 2010; Mulvihill et al., 2011; Roza et al., 2007). In accordance, pharmacokinetic studies revealed significant brain and systemic exposure of NOB (Figure S2A). To address whether metabolic protection by NOB depends on clock function, we first employed a diet-induced obesity (DIO) mouse model using both WT and clock-disrupted ClockΔ19/Δ19 mice. In WT C57BL/6J mice fed with high-fat diet (HFD), 10-week NOB treatment significantly attenuated body weight gain relative to the vehicle control (Figures 2A and S2B). Food intake was not significantly altered by NOB relative to the vehicle control (Figures 2B and S2C). Body mass composition analysis revealed that the body weight loss was primarily attributable to reduction in fat mass (Figure 2C) and white adipose cell size (Figures 2D and S2D). Remarkably, NOB treatment only led to very modest reduction in body weight gain in HFD-fed ClockΔ19/Δ19 mutant mice (Figures 2A and S2B), with the fat mass and adipocyte cell size essentially unchanged by NOB treatment (Figures 2C, 2D and S2D). While mutant mice consumed more food during the light phase than WT mice (Turek et al., 2005), NOB did not change food intake in the mutant mice (Figures 2B and S2C). Indicating elevated energy expenditure, NOB-treated WT mice exhibited greatly elevated oxygen consumption compared to the controls throughout the circadian cycle with the largest increase found in early dark phase (Figures 2E and S2E). Respiratory quotient was also increased in WT mice treated with NOB (Figure S2F), consistent with a switch from lipid-biased metabolism to a more balanced contribution from all major macronutrients. In contrast, ClockΔ19/Δ19 mice showed no increase in energy expenditure or respiratory quotient after NOB treatment (Figures 2E and S2F). Consistent with its effect on body weight and energy expenditure, NOB greatly increased wheel running activity levels in HFD-fed WT mice relative to control treatment (Figure 2F); in contrast, no significant difference in activity levels was detected between the treatments for ClockΔ19/Δ19 mice (Figure 2G).
Figure 2. NOB modulated energy homeostasis and behavior in diet-induced obesity (DIO) mice in a clock-dependent manner.
(A) Average body weight of WT or ClockΔ19/Δ19 mutant mice fed with high-fat diet and treated with either vehicle or NOB (WT.HF.Veh, WT.HF.NOB, Clk.HF.Veh and Clk.HF.NOB) for 10 weeks (n=8–15). (B) Daily food intake for the above 4 groups of mice (n=8–15). (C) Body mass composition as analyzed by NMR (n=3). (D) Histological analysis of white adipose fat (WAT) after 10-week treatment. WAT was subjected to H&E staining. (E) The diurnal rhythms of VO2 (volume of oxygen consumed) in the 4 groups of mice (n=8). (F) Left: Actograms illustrating the effect of vehicle or NOB on circadian behavior in HFD-fed WT mice (n=7). Middle: Average wave plots summarizing wheel-running activity during 12–14 days of LD (12hr light, 12hr dark) or DD (Constant darkness) for indicated genotypes (n=7). Right: Daily total wheel-running activity during L:D or D:D conditions for the indicated genotypes (n=7). (G) Left: Actograms illustrating the effect of vehicle or NOB on circadian behavior in HFD-fed ClockΔ19/Δ19 mutant mice (n=3). Middle: Average wave plots summarizing wheel-running activity during 10–12 days of LD (12hr light, 12hr dark) or DD (Constant darkness) for indicated genotypes (n=3). Right: Daily total wheel-running activity during L:D or D:D conditions for the indicated genotypes (n=3). Data are represented as mean ± SEM. ***p < 0.001. WT.HF.NOB vs. WT.HF.Veh.
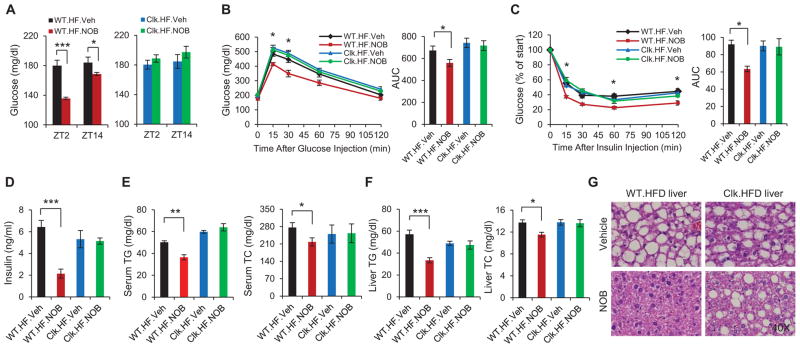
NOB also improved glucose and lipid homeostasis in WT but not ClockΔ19/Δ19 mice. NOB lowered fasting glucose levels in WT mice (Figure 3A), and significantly improved glucose tolerance and insulin sensitivity (Figures 3B and 3C). Interestingly, blood insulin levels were strongly reduced in NOB-treated WT mice relative to vehicle controls (Figure 3D). Total triglyceride (TG) and cholesterol (TC) levels in WT serum and liver were also significantly diminished by NOB (Figures 3E and 3F). Hematoxylin and eosin (H&E) and Oil Red O staining revealed that NOB strongly improved liver steatosis and essentially abolished lipid droplet formation in DIO WT liver (Figures 3G, S2H – S2J). In contrast, NOB did not significantly improve glucose and lipid homeostasis in ClockΔ19/Δ19 mice (Figure 3), and its beneficial effect on ClockΔ19/Δ19 liver steatosis was markedly attenuated compared with that in WT (Figures 3G, S2H – S2J). In contrast to HFD, regular chow (RC) feeding did not lead to metabolic disorders and NOB treatment did not show significantly beneficial effects on metabolic homeostasis (Figure S3). Together, these results demonstrate a Clock-dependent efficacy of NOB against metabolic syndrome.
Figure 3. NOB improved glucose and lipid homeostasis in diet-induced obesity (DIO) mice in a clock-dependent manner.
(A) Fasting blood glucose levels in HFD-fed WT (left) and ClockΔ19/Δ19 mutant mice (right) with Vehicle or NOB treatment at two opposite time points (n=8–15). (B) Effect of NOB on glucose tolerance in HFD-fed WT and ClockΔ19/Δ19 mutant mice as measured by glucose tolerance test (GTT) (n=8–15). Right panel: Area under curve (AUC). (C) Effect of NOB on insulin tolerance in HFD-fed WT and ClockΔ19/Δ19 mutant mice as measured by insulin tolerance test (ITT) (n=8–15). Right panel: Area under curve (AUC). (D) Blood insulin levels in HFD-fed WT mice and ClockΔ19/Δ19 mutant mice with Vehicle or NOB treatment (n=8–15). (E) Total triglyceride (TG) levels and cholesterol (TC) levels in blood after 10-week treatment (n=8–15). (F) Total triglyceride (TG) levels and cholesterol (TC) levels in liver after 10-week treatment (n=8–15). (G) H&E staining of whole livers from HFD-fed WT and ClockΔ19/Δ19 mutant mice after 10-week treatment. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. WT.HF.NOB vs. WT.HF.Veh.
Non-methoxylated flavanones such as Naringin (NAR) and its aglycone derivative Naringenin are also naturally occurring flavonoids (Figure S4A) (Assini et al., 2013), yet failed to enhance cellular circadian rhythms in our primary screen (Figures S4B – S4F). Consistent with previous studies (Mulvihill et al., 2009; Mulvihill et al., 2011), compared with NOB, NAR showed significantly attenuated effects on body weight gain and lipid/glucose homeostasis (Figure S5). Importantly, the modest effects from NAR treatment were largely indistinguishable between WT or ClockΔ19/Δ19 C57BL/6J mice.
Clock-dependent metabolic protection by NOB in db/db diabetic mice
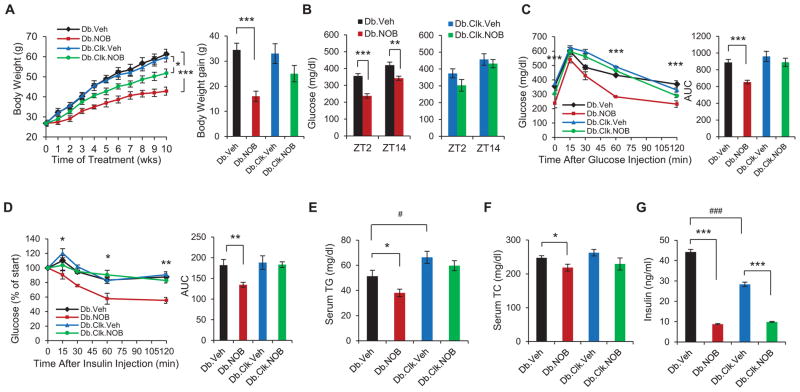
Given the improved glucose homeostasis in DIO mice treated with NOB, we next investigated effects of NOB on db/db mice, an established genetic mouse model for obesity and diabetes that lacks functional leptin receptors, and the role of the clock. NOB treatment strongly blunted body weight gain in db/db mice (Figure 4A), lowered fasting glucose levels (Figure 4B), improved glucose tolerance and insulin sensitivity (Figures 4C and 4D), and reduced serum TG and TC levels in db/db mice (Figures 4E and 4F). In contrast, db/db ClockΔ19/Δ19 double mutant mice exhibited a markedly attenuated response to NOB (Figure 4). Under Veh treatment, db/db ClockΔ19/Δ19 mutant mice showed lower insulin levels than db/db, consistent with β-cell deficits and hypoinsulinemia previously reported in ClockΔ19/Δ19 mice (Marcheva et al., 2010). NOB treatment strongly reduced circulating insulin levels in both db/db and db/db ClockΔ19/Δ19 mutant mice, with the former exhibiting a more pronounced reduction (Figure 4G). These results are consistent with lower insulin sensitivity in db/db ClockΔ19/Δ19 relative to db/db. Given the age-dependent deficits in pancreatic β-cell proliferation and insulin secretion in ClockΔ19/Δ19 (Marcheva et al., 2010), future studies will investigate the mechanistic relationship between the effects of NOB on insulin levels and β-cell function and/or insulin sensitivity.
Figure 4. NOB efficacy against metabolic syndrome in Type 2 diabetic db/db mice requires a functional circadian clock.
(A) Left panel: Average body weight of db/db or db/db ClockΔ19/Δ19 double mutant mice fed with regular chow and treated with either vehicle or NOB (Db.Veh, Db.NOB, Db.Clk.Veh and Db.Clk.NOB) for 10 weeks (n=6–8). The mice were 6–8 weeks old at the beginning of the treatment. Right panel: Average body weight gain for these 4 groups of mice. (B) Fasting blood glucose levels in db/db (left) and db/db ClockΔ19/Δ19 double mutant mice (right) at two opposite time points (n=6–8). (C) Effect of NOB on glucose tolerance in db/db and db/db ClockΔ19/Δ19 double mutant mice as measured by glucose tolerance test (GTT) (n=6–8). Right panel: Area under curve (AUC). (D) Effect of NOB on insulin tolerance in db/db and db/db ClockΔ19/Δ19 double mutant mice as measured by insulin tolerance test (ITT) (n=6–8). Right panel: Area under curve (AUC). (E and F) Blood total triglyceride (TG) levels and cholesterol (TC) levels in db/db and db/db ClockΔ19/Δ19 mice (n=6–8). (G) Effects of NOB on circulating insulin levels in both db/db and db/db ClockΔ19/Δ19 mice (n=6–8). Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. NOB vs. Veh. #p < 0.05 and ###p < 0.001. Db.Veh vs. Db.Clk.Veh.
Identification of hepatic NOB-responsive genes by microarray
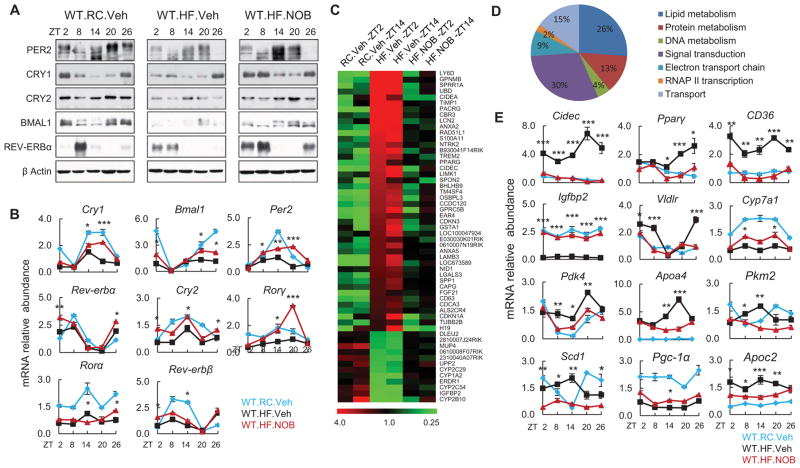
To investigate the molecular basis of NOB action in HFD-fed WT mice, we focused on a major metabolic organ, the liver, where we observed a strong protection by NOB. Consistent with previous studies (Kohsaka et al., 2007), the oscillatory amplitude of clock gene expression was generally lower in the liver of HF.Veh mice relative to lean RC.Veh mice (Figures 5A and 5B). NOB improved circadian clock transcript oscillations and largely restored clock protein rhythms (Figures 5A and 5B) (HF.NOB vs. HF.Veh), concordant with its robust physiological efficacy. To investigate metabolic output gene expression, we conducted microarray analysis. Comparative analysis was conducted to analyze gene expression changes in pair-wise comparison, namely HF.Veh vs. RC.Veh (the comparison is denoted as HF/RC) and HF.NOB vs. HF.Veh (denoted as NOB/HF), at ZT2 and ZT14 time points, which revealed altered expression of 544, 229, 915 and 243 genes, respectively (Data S1). Importantly, a total of 251 and 56 genes were identified showing altered gene expression patterns in response to HFD (HF/RC) that were reversed by NOB (NOB/HF) at one or both time points (ZT2 and/or ZT14), respectively (Figure 5C, Figure S6A and Data S2). Functional classification (Gene Ontology, GO) of these NOB-responsive genes highlighted a prominent role in metabolic regulation (Figure 5D, Data S2 and Tables S1). Real-time qPCR analysis further illustrated a broad modulatory function of NOB in metabolic output gene expression (Figures 5E). For example, transcript expression of Cidec/Fsp27, known to function in lipid droplet formation (Eckel-Mahan et al., 2013; Matsusue et al., 2008; Puri et al., 2007), was induced by >10 fold in HFD.Veh mouse liver relative to RC.Veh and reverted back to baseline levels by NOB, consistent with the NOB efficacy in mitigating hepatic steatosis (Figure 3G). NOB also altered the expression of genes involved in gluconeogenesis and glycolysis (e.g., Pdk4 and Pkm2).
Figure 5. NOB restored circadian oscillation of clock and metabolic output genes in the liver of HFD-fed WT mice.
(A and B) Liver samples were collected from HFD-fed WT mice with vehicle or NOB treatment (WT.HF.Veh and WT.HF.NOB). WT mice fed with regular chow (WT.RC.Veh) were used as controls for comparison (N=3–4). Western blotting (A) and Real-time qPCR (B) analyses were performed to determine protein and mRNA expression of clock genes. (C) Heat map of microarray gene expression data indicating that the expression patterns of 56 genes were altered by HFD, and NOB reversed, to varying degrees, their expression to approximate RC levels in WT mouse liver at both ZT2 and ZT14 time points. Color scale indicates median normalized signal intensity in relative values. (D) Functional classification of 56 genes in (C) by the Gene Ontology (GO) program. Percentages of genes sharing GO biological processes are shown. (E) Real-time qPCR analysis of mRNA expression of clock-controlled metabolic output genes in the livers from treated mice as above. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. WT.HF.NOB vs. WT.HF.Veh.
RORs as direct protein targets for NOB
Cross-examination of previous circadian ChIP-seq studies (Cho et al., 2012; Koike et al., 2012) revealed that 63% of the NOB-responsive genes showed promoter occupancy of core clock proteins (Figure S6B), particularly REV-ERBs. REV-ERBs and RORs function respectively as negative and positive transcription factors competing for binding to RORE promoter elements, playing important roles in circadian rhythms, metabolism and inflammation (Gerhart-Hines et al., 2013; Jetten et al., 2013; Kojetin and Burris, 2014). In a previous screen for inhibitors of RORγt, NOB was among a number of primary screen hits that instead activated RORγt and consequently were not validated further (Huh et al., 2011). ROR family receptors consist of α, β and γ isoforms. RORα and RORγ are more similar in tissue distribution and the ligand binding domain structure, whereas RORβ is more divergent (Kallen et al., 2002; Solt et al., 2010; Stehlin et al., 2001). We therefore focused on RORα and RORγ.
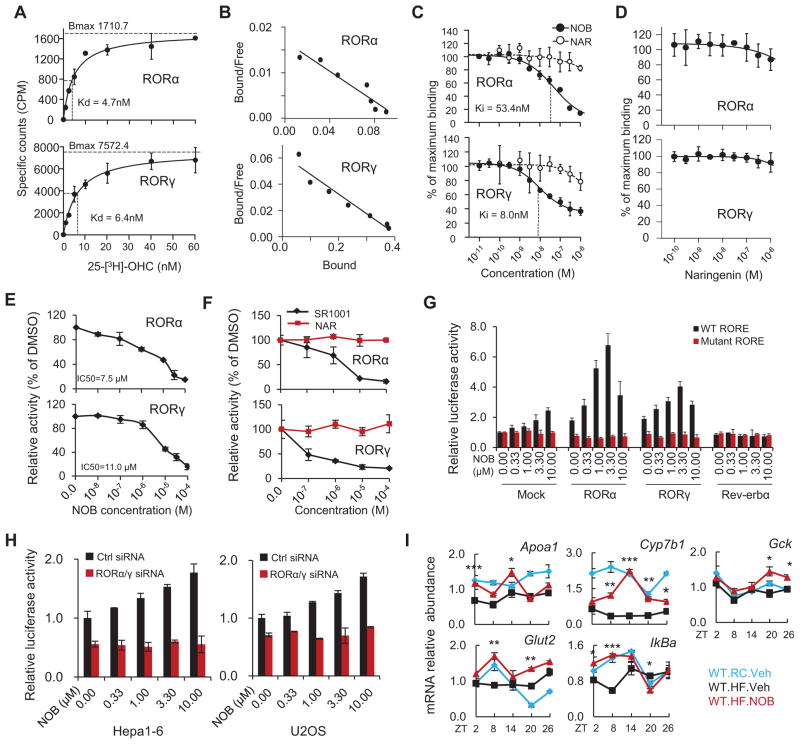
To investigate a possible direct interaction between NOB and ROR proteins, we employed a competitive radioligand binding assay for RORs using 25-[3H]-hydroxycholesterol (25-[3H]-OHC) (Kumar et al., 2010; Wang et al., 2010b). Saturation curves and Scatchard plots validated the assay, with similar Kd values to that previously reported (Figures 6A and 6B) (Kumar et al., 2010; Wang et al., 2010b). Importantly, NOB showed robust competitive binding to the ligand binding domains (LBDs) of RORα and RORγ, with higher affinity for RORγ (Figure 6C; see Ki comparison). In contrast, NAR or its aglycone derivative Naringenin showed markedly diminished binding in the same concentration range (Figures 6C and 6D). Consistent with these binding assay results, we observed robust activities of NOB and the known ROR ligand SR1001 (Solt et al., 2011) for GAL4-RORα and GAL4-RORγ chimeric receptors in mammalian one-hybrid reporter assays, and NAR showed no activities (Figures 6E and 6F). Of note, regardless of the nature of the ligand (agonist or inverse agonist), ligand interaction with these chimeric receptors has been shown to reduce transcriptional activity of these chimeric receptors (Wang et al., 2010a; Wang et al., 2010b). These results together indicate direct binding of NOB to RORα and RORγ.
Figure 6. NOB enhanced RORα/γ transcriptional activity via direct binding to RORα/γ.
(A and B) Saturation curves and Scatchard plots from filter binding assays for RORα-LBD and RORγ-LBD. (A) Saturation curves for 25-[3H]-OHC binding were generated with 100ng of RORα-LBD (top) and 200ng of RORγ-LBD (bottom) (n=3). Dissociation constant values are shown. (B) Saturation curve results in Fig. 5E were subjected to Scatchard analysis and Scatchard plots for 25-[3H]-OHC are shown for RORα-LBD (top) and RORγ-LBD (bottom) corresponding to (n=3). This analysis gave a dissociation constant (Kd) of 6.10 nM and a total number of binding sites (Bmax) of 100 fmol/mg of protein for RORα, and 6.67 nM and 410 fmol/mg of protein for RORγ. (C and D) In vitro competitive radio-ligand binding assay indicating the direct binding of NOB (C), but not NAR (C) or Naringenin (D), to RORα-LBD and RORγ-LBD within the indicated dose range. Inhibitory constant values are shown. (E and F) Mammalian one-hybrid assays showing ligand interaction with ROR-LBD. HEK293T cells were cotransfected with a GAL4 reporter construct with expression vectors for GAL4 DBD-RORα LBD or GAL4 DBD-RORγ LBD. Cells were treated with varying concentrations of NOB (E) and its non-methoxylated analog NAR (F). SR1001 served as a positive control in (F). (G) NOB dose-dependently increased Bmal1 promoter-driven luciferase reporter expression with wild-type, but not mutant, RORE in the presence of RORα or RORγ in Hepa1-6 cells. (H) Knockdown of RORα/γ expression by siRNAs abrogated NOB induction of Bmal1 promoter-driven luciferase reporter expression in both Hepa1-6 and U2OS cells. (I) Real-time qPCR analysis of RORα/γ target genes from the same mouse liver samples as in Figure 5A. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, and ***p < 0.001. WT.HF.NOB vs. WT.HF.Veh.
Next we conducted functional assays to determine the effect of NOB on RORα/γ transcriptional activity. NOB was found to dose-dependently increase Bmal1 promoter-driven luciferase reporter activity with wild-type, but not mutant, RORE elements (Preitner et al., 2002) in the presence of RORα or RORγ in Hepa1-6 cells (Figure 6G). Conversely, knockdown of the Rora/c genes encoding RORα/γ by siRNAs abrogated the NOB-mediated induction of Bmal1 promoter-driven luciferase reporter activity in both Hepa1-6 and U2OS cells (Figures 6H and S6C). Several ROR target genes (e.g., Cyp7b1, IkBa and Gck) were induced in NOB-treated DIO mouse liver relative to control treatment (Figure 6I), and Ingenuity pathway analysis (IPA) also showed an important role of RORs in the genome-wide NOB response (Figure S6D). Together, these results indicate that NOB directly binds to and activates RORs and that RORα/γ are necessary for the enhancing activity of NOB on Bmal1 transcription.
DISCUSSION
In summary, our unbiased chemical screen identified clock-enhancing polymethoxylated flavones, particularly Nobiletin. Compelling evidence from both genetic and pharmacological studies demonstrates a Clock gene-dependent efficacy of NOB in preventing metabolic syndrome in mice, providing proof in mammals that strengthening circadian amplitude is a pharmacological intervention strategy for metabolic disease and other clock-related pathologies such as age-related decline (Chen et al., 2013). The beneficial outcome of enhanced circadian amplitude could include enhanced efficiency in physiological performance, greater stimuli range and sensitized response (Hatori et al., 2012; Hogenesch and Herzog, 2011; Ikeda et al., 2013; van Ooijen and Millar, 2012). Consistent with the notion that augmented circadian amplitude enhances energy metabolism, time-restricted feeding, and thus energy expenditure, plays a dominant role determining extent of obesity from high-fat diet feeding (Hatori et al., 2012).
Polymethoxylated flavones elicit diverse benefits in mice and humans, including mitigating effects against cancer, inflammation, atherosclerosis, and more recently metabolic disorders and neurodegenerative diseases (Cui et al., 2010; Evans et al., 2012; Kurowska and Manthey, 2004; Lee et al., 2013; Mulvihill et al., 2009; Nohara et al., 2015a). Polymethoxylated flavones generally show a favorable pharmacokinetic profile (Evans et al., 2012; Saigusa et al., 2011), and no discernible toxicity was observed in chronic treatment of mice in this and previous studies (Lee et al., 2013; Mulvihill et al., 2011). Our recent studies showed a role of NOB in ammonia disposal via urea cycle regulation, and transcriptional induction of the rate-limiting Cps1 gene by NOB was impaired in ClockΔ19/Δ19 mutant mice (Nohara et al., 2015a). Providing a mechanistic basis for these functional results, the current study illustrates a direct role of NOB in the enhancement of circadian clocks and particularly the activation of ROR receptors. These findings collectively suggest a unifying circadian mechanism governing the diverse physiological effects of polymethoxylated flavones.
We identified the ROR nuclear receptors as the molecular target of NOB, which makes a direct link to the circadian gene network via the REV-ERB/ROR pathway that stabilizes the core CLOCK:BMAL1 transcriptional feedback loop (Sato et al., 2004). This function of NOB on RORs explains why there are relatively “subtle” changes in circadian gene expression, because the RORs primarily enhance Bmal1 transcription in the core loop, which in turn leads to enhancement of both activation and feedback repression (Hogenesch and Herzog, 2011). This auto-regulatory feedback limits the dynamic range of transcriptional readouts as a consequence of this homeostatic, closed-loop system, also exemplified by the finding that transgenic Clock overexpression in mice led to greater stability, but only modest amplitude increase, in Per and Cry oscillation (Antoch et al., 1997).
Genetic and pharmacological studies illustrate a complex duality of ROR functions in metabolism and immunity. On the one hand, multiple ROR-deficient mouse mutants showed metabolic benefits including resistance to obesity and improved lipid homeostasis (Lau et al., 2008), suggesting a negative role of RORs in metabolic regulation. Likewise, pharmacological studies have mainly focused on inverse agonists for RORs (Jetten et al., 2013; Solt et al., 2010), due to their therapeutic potential in Th17-dependent autoimmunity (Huh et al., 2011; Solt et al., 2011) and metabolic syndrome (Chang et al., 2015; Jetten et al., 2013). On the other hand, RORs also play important positive roles, and agonists or activating compounds of RORs can effect physiological improvements. First, genetic studies support positive metabolic and immune functions of RORs (Garidou et al., 2015; Raichur et al., 2010). For example, attenuated RORα signaling by skeletal muscle-specific expression of RORα1 ΔDE (lacking the ligand-binding domain) led to various metabolic abnormalities in mice, including hyperglycemia and glucose intolerance (Raichur et al., 2010). RORα plays an anti-inflammatory role by interfering with NF-κB signaling (Delerive et al., 2001), and homozygous Staggerer mice exhibit an enhanced susceptibility to lung inflammation (Stapleton et al., 2005). Recent studies also showed that RORγ deficiency compromises immunity to pathogens and parasites (Ohnmacht et al., 2015; Okada et al., 2015). Second, cholesterol biosynthetic intermediates (CBI) have been identified as endogenous RORγt ligand(s) that stabilize the RORγ ligand-binding domain and induced coactivator recruitment in normal physiology (Santori et al., 2015). Finally, metabolic effects in ROR-deficient mice should be considered in the broader context of overall physiological health. For example, the Staggerer mice deficient in RORα suffer from severe developmental, physiological and behavioral deficits and require special husbandry care; notably, despite reduced adiposity and apparent resistance to high-fat diet, these mice also exhibit severe atherosclerosis (Jetten, 2009; Mamontova et al., 1998). Therefore, RORs play complex roles in both immunity and metabolism, and it is possible that both agonists and antagonists can fine-tune ROR activities and exert beneficial roles. It is important to examine molecular details of NOB-ROR interaction, especially given that different inverse agonists of RORγt exhibit divergent effects on target gene expression via distinct molecular mechanisms (Xiao et al., 2014). Genetic models, such as double conditional KO and/or overexpression of functionally overlapping RORα and RORγ, will help elucidate the function of ROR ligands including NOB (Knight and Shokat, 2007).
In conclusion, our work reveals NOB as a clock-enhancing natural compound that activates RORs and protects against metabolic syndrome in a clock-dependent manner. This proof-of-principle study will spur future application of NOB and additional clock-enhancing compounds in other diseases (e.g., mood and sleep disorders) and aging known to suffer dampened circadian amplitude (Nohara et al., 2015b; Schroeder and Colwell, 2013).
EXPERIMENTAL PROCEDURES
Animals and cell lines
Animal husbandry for all the studies except tissue explant experiments was carried out under IACUC guidelines and the procedures were conducted as described in an animal protocol approved by the University of Texas Health Science Center at Houston (UTHSC-H). PER2::Luc reporter knock-in mice used for tissue explant experiments were maintained according to guidelines from IACUC at the University of Texas Southwestern Medical Center (UTSW). Adult mouse ear fibroblast and mouse embryonic fibroblast (MEF) cells were previously described (Chen et al., 2012).
High-throughput chemical screen and validation
The chemical screen for circadian clock modulators was conducted at the Chemical Genomics Core facility at the UTHSC-H. The in-house chemical library we screened consists of compounds from NIH Clinical Collection, NCI collection and Microsource Spectrum Collection. The screening was conducted largely based on the protocol previously described (Chen et al., 2012). Briefly, immortalized fibroblast cells from ClockΔ19/+ heterozygous mice expressing the PER2::Luc bioluminescence reporter were plated into 96-well plates. Upon confluency, cells were incubated with 5 μM forskolin for 1–2 h followed by the addition of chemical compounds to the plates with robotic arms (Beckman), and then subjected to continuous monitoring over several days in a temperature-controlled EnVision microplate reader (Perkin Elmer). Data analysis was carried out by using the MultiCycle software (Actimetrics) for measurement of period, phase, and amplitude.
Mouse treatment and metabolic analyses
For diet-induced obesity, male mice at 6 weeks of age were fed with HFD (D12492, Research Diets) until the end of the experimental protocol. The mice were treated with either vehicle (DMSO) or NOB (200mg/kg body weight) via oral gavage every other day, in the time window of ZT8-10, throughout the experimental period. We chose the every-other-day dosing regimen at the indicated level based on several reasons as described in Supplemental Information. Metabolic assays and energy expenditure analyses were conducted as previously described (Daniels et al., 2010; Garcia et al., 2013; He et al., 2015; Jeong et al., 2015). Please see Supplemental Information for details.
Microarray analysis
Total RNAs prepared from liver tissues from RC-fed, Vehicle-treated and HFD-fed, Vehicle or NOB-treated WT mice for 10 weeks were reverse transcribed into cDNAs, which were then biotin-UTP labeled and hybridized to the Illumina mouse WG-6v2.0 Expression BeadChip. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE78848 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE78848).
Molecular and biochemical analyses
Real-time qPCR and Western blotting analyses of circadian gene expression were conducted as previously described (Yoo et al., 2013). Primers used are listed in Table S2. For mammalian one-hybrid assays, regardless of the nature of the ligand (agonist or inverse agonist), ligand interaction with these chimeric receptors has been shown to reduce transcriptional activity of these chimeric receptors (Wang et al., 2010a; Wang et al., 2010b). For radio-ligand receptor binding assays, we adopted previously described protocols with minor modifications (Kumar et al., 2010; Wang et al., 2010b).
Statistical analysis
Data are presented as mean ± SEM. Statistical significance was determined by one-way or two-way ANOVA with Turkey’s or Dunnett’ tests for multiple group comparisons. P<0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank C. Lee for reagent, C. Stephan, E. Song, C. Ayoub, and T. M. Tran for technical support, and S. McKnight, C. Green, M. Bogdanov, G. Lee, D. Marshak, C. Wu, P. Griffin, R. Garcia-Ordonez and G. Gloston for helpful advice and/or critical reading of the manuscript. This work is in part supported by The Welch Foundation (AU-1731), AHA (11SDG7600045), and NIH/NIA (R01AG045828) to Z.C., NIH/NIGMS (R01 GM114424) to S.-H.Y., TMC-DDC P/F Awards (NIDDK Center Grant P30-DK056338) to S.-H.Y. and Z.C., U.S. Dept. of Veterans Affairs, BX000507 and CX000174; and NIH AG040583 to J.M.G., T32AG000183 to B.G., NIH/DP1 OD000895 to C.C.L., and JSPS KAKENHI (26293048) and the Uehara Memorial Foundation to N.K. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Supplemental Information contains Supplemental Experimental Procedures and Supplemental References, six Supplemental Figures, two Supplemental Tables and two Excel data files.
AUTHOR CONTRIBUTIONS
Z.C. conceived the project and wrote the manuscript; Z.C., S.-H.Y. and J.S.T. supervised research; B.H., K.N., N.P., Y.-S.P., B.G., Z.Z., S.-H.Y. and Z.C. conducted research; all authors contributed to data analysis and manuscript preparation.
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antoch MP, Kondratov RV. Pharmacological modulators of the circadian clock as potential therapeutic drugs: focus on genotoxic/anticancer therapy. Handb Exp Pharmacol. 2013:289–309. doi: 10.1007/978-3-642-25950-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Assini JM, Mulvihill EE, Huff MW. Citrus flavonoids and lipid metabolism. Curr Opin Lipidol. 2013;24:34–40. doi: 10.1097/MOL.0b013e32835c07fd. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aziz A. Nobiletin is main fungistat in tangerines resistant to mal secco. Science. 1967;155:1026–1027. doi: 10.1126/science.155.3765.1026. [DOI] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MR, He Y, Khan TM, Kuruvilla DS, Garcia-Ordonez R, Corzo CA, Unger TJ, White DW, Khan S, Lin L, et al. Antiobesity Effect of a Small Molecule Repressor of RORgamma. Mol Pharmacol. 2015;88:48–56. doi: 10.1124/mol.114.097485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, Ye ZY, Pan HL, Takahashi JS. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci U S A. 2012;109:101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yoo SH, Takahashi JS. Small molecule modifiers of circadian clocks. Cell Mol Life Sci. 2013;70:2985–2998. doi: 10.1007/s00018-012-1207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Wu J, Jung SC, Park DB, Maeng YH, Hong JY, Kim SJ, Lee SR, Eun SY. Anti-neuroinflammatory activity of nobiletin on suppression of microglial activation. Biol Pharm Bull. 2010;33:1814–1821. doi: 10.1248/bpb.33.1814. [DOI] [PubMed] [Google Scholar]
- Daniels IS, Zhang J, O’Brien WG, 3rd, Tao Z, Miki T, Zhao Z, Blackburn MR, Lee CC. A role of erythrocytes in adenosine monophosphate initiation of hypometabolism in mammals. The Journal of biological chemistry. 2010;285:20716–20723. doi: 10.1074/jbc.M109.090845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerive P, Monte D, Dubois G, Trottein F, Fruchart-Najib J, Mariani J, Fruchart JC, Staels B. The orphan nuclear receptor ROR alpha is a negative regulator of the inflammatory response. EMBO Rep. 2001;2:42–48. doi: 10.1093/embo-reports/kve007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the circadian clock by nutritional challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109:5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M, Sharma P, Guthrie N. Bioavailability of Citrus Polymethoxylated Flavones and Their Biological Role in Metabolic Syndrome and Hyperlipidemia. InTech; 2012. pp. 1–19. [Google Scholar]
- Farrow SN, Solari R, Willson TM. The importance of chronobiology to drug discovery. Expert Opin Drug Discov. 2012;7:535–541. doi: 10.1517/17460441.2012.689283. [DOI] [PubMed] [Google Scholar]
- Garcia JM, Scherer T, Chen JA, Guillory B, Nassif A, Papusha V, Smiechowska J, Asnicar M, Buettner C, Smith RG. Inhibition of cisplatin-induced lipid catabolism and weight loss by ghrelin in male mice. Endocrinology. 2013;154:3118–3129. doi: 10.1210/en.2013-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garidou L, Pomie C, Klopp P, Waget A, Charpentier J, Aloulou M, Giry A, Serino M, Stenman L, Lahtinen S, et al. The Gut Microbiota Regulates Intestinal CD4 T Cells Expressing RORgammat and Controls Metabolic Disease. Cell metabolism. 2015;22:100–112. doi: 10.1016/j.cmet.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, et al. The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature. 2013;503:410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Lazar MA. Circadian metabolism in the light of evolution. Endocr Rev. 2015;36:289–304. doi: 10.1210/er.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell metabolism. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Nohara K, Ajami NJ, Michalek RD, Tian X, Wong M, Losee-Olson SH, Petrosino JF, Yoo SH, Shimomura K, et al. Transmissible microbial and metabolomic remodeling by soluble dietary fiber improves metabolic homeostasis. Sci Rep. 2015;5:10604. doi: 10.1038/srep10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, et al. Identification of Small Molecule Activators of Cryptochrome. Science. 2012 doi: 10.1126/science.1223710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB, Herzog ED. Intracellular and intercellular processes determine robustness of the circadian clock. FEBS Lett. 2011;585:1427–1434. doi: 10.1016/j.febslet.2011.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, Manel N, Ciofani M, Kim SV, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Kumagai H, Skach A, Sato M, Yanagisawa M. Modulation of circadian glucocorticoid oscillation via adrenal opioid-CXCR7 signaling alters emotional behavior. Cell. 2013;155:1323–1336. doi: 10.1016/j.cell.2013.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, et al. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong K, He B, Nohara K, Park N, Shin Y, Kim S, Shimomura K, Koike N, Yoo SH, Chen Z. Dual attenuation of proteasomal and autophagic BMAL1 degradation in Clock(Delta19/+) mice contributes to improved glucose homeostasis. Sci Rep. 2015;5:12801. doi: 10.1038/srep12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM, Kang HS, Takeda Y. Retinoic acid-related orphan receptors alpha and gamma: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol (Lausanne) 2013;4:1. doi: 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, Fournier B. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–1707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Shokat KM. Chemical genetics: where genetics and pharmacology meet. Cell. 2007;128:425–430. doi: 10.1016/j.cell.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojetin DJ, Burris TP. REV-ERB and ROR nuclear receptors as drug targets. Nat Rev Drug Discov. 2014;13:197–216. doi: 10.1038/nrd4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, Garcia-Ordonez RD, Burris TP, Griffin PR. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol Pharmacol. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowska EM, Manthey JA. Hypolipidemic effects and absorption of citrus polymethoxylated flavones in hamsters with diet-induced hypercholesterolemia. J Agric Food Chem. 2004;52:2879–2886. doi: 10.1021/jf035354z. [DOI] [PubMed] [Google Scholar]
- Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. The Journal of biological chemistry. 2008;283:18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- Lee Y, Chen R, Lee HM, Lee C. Stoichiometric relationship among clock proteins determines robustness of circadian rhythms. The Journal of biological chemistry. 2011;286:7033–7042. doi: 10.1074/jbc.M110.207217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Cha BY, Choi SS, Choi BK, Yonezawa T, Teruya T, Nagai K, Woo JT. Nobiletin improves obesity and insulin resistance in high-fat diet-induced obese mice. J Nutr Biochem. 2013;24:156–162. doi: 10.1016/j.jnutbio.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Lee YS, Cha BY, Saito K, Yamakawa H, Choi SS, Yamaguchi K, Yonezawa T, Teruya T, Nagai K, Woo JT. Nobiletin improves hyperglycemia and insulin resistance in obese diabetic ob/ob mice. Biochem Pharmacol. 2010;79:1674–1683. doi: 10.1016/j.bcp.2010.01.034. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamontova A, Seguret-Mace S, Esposito B, Chaniale C, Bouly M, Delhaye-Bouchaud N, Luc G, Staels B, Duverger N, Mariani J, et al. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98:2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsusue K, Kusakabe T, Noguchi T, Takiguchi S, Suzuki T, Yamano S, Gonzalez FJ. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell metabolism. 2008;7:302–311. doi: 10.1016/j.cmet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A. 2010;107:15240–15245. doi: 10.1073/pnas.1005101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill EE, Allister EM, Sutherland BG, Telford DE, Sawyez CG, Edwards JY, Markle JM, Hegele RA, Huff MW. Naringenin prevents dyslipidemia, apolipoprotein B overproduction, and hyperinsulinemia in LDL receptor-null mice with diet-induced insulin resistance. Diabetes. 2009;58:2198–2210. doi: 10.2337/db09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvihill EE, Assini JM, Lee JK, Allister EM, Sutherland BG, Koppes JB, Sawyez CG, Edwards JY, Telford DE, Charbonneau A, et al. Nobiletin attenuates VLDL overproduction, dyslipidemia, and atherosclerosis in mice with diet-induced insulin resistance. Diabetes. 2011;60:1446–1457. doi: 10.2337/db10-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagase H, Yamakuni T, Matsuzaki K, Maruyama Y, Kasahara J, Hinohara Y, Kondo S, Mimaki Y, Sashida Y, Tank AW, et al. Mechanism of neurotrophic action of nobiletin in PC12D cells. Biochemistry. 2005;44:13683–13691. doi: 10.1021/bi050643x. [DOI] [PubMed] [Google Scholar]
- Nohara K, Shin Y, Park N, Jeong K, He B, Koike N, Yoo SH, Chen Z. Ammonia-lowering activities and carbamoyl phosphate synthetase 1 (Cps1) induction mechanism of a natural flavonoid. Nutr Metab (Lond) 2015a;12:23. doi: 10.1186/s12986-015-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohara K, Yoo SH, Chen ZJ. Manipulating the circadian and sleep cycles to protect against metabolic disease. Front Endocrinol (Lausanne) 2015b;6:35. doi: 10.3389/fendo.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C, Park JH, Cording S, Wing JB, Atarashi K, Obata Y, Gaboriau-Routhiau V, Marques R, Dulauroy S, Fedoseeva M, et al. MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science. 2015;349:989–993. doi: 10.1126/science.aac4263. [DOI] [PubMed] [Google Scholar]
- Okada S, Markle JG, Deenick EK, Mele F, Averbuch D, Lagos M, Alzahrani M, Al-Muhsen S, Halwani R, Ma CS, et al. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349:606–613. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Puri V, Konda S, Ranjit S, Aouadi M, Chawla A, Chouinard M, Chakladar A, Czech MP. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. The Journal of biological chemistry. 2007;282:34213–34218. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- Raichur S, Fitzsimmons RL, Myers SA, Pearen MA, Lau P, Eriksson N, Wang SM, Muscat GE. Identification and validation of the pathways and functions regulated by the orphan nuclear receptor, ROR alpha1, in skeletal muscle. Nucleic Acids Res. 2010;38:4296–4312. doi: 10.1093/nar/gkq180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roenneberg T, Allebrandt KV, Merrow M, Vetter C. Social jetlag and obesity. Curr Biol. 2012;22:939–943. doi: 10.1016/j.cub.2012.03.038. [DOI] [PubMed] [Google Scholar]
- Roza JM, Xian-Liu Z, Guthrie N. Effect of citrus flavonoids and tocotrienols on serum cholesterol levels in hypercholesterolemic subjects. Altern Ther Health Med. 2007;13:44–48. [PubMed] [Google Scholar]
- Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- Saigusa D, Shibuya M, Jinno D, Yamakoshi H, Iwabuchi Y, Yokosuka A, Mimaki Y, Naganuma A, Ohizumi Y, Tomioka Y, et al. High-performance liquid chromatography with photodiode array detection for determination of nobiletin content in the brain and serum of mice administrated the natural compound. Anal Bioanal Chem. 2011;400:3635–3641. doi: 10.1007/s00216-011-5031-2. [DOI] [PubMed] [Google Scholar]
- Santori FR, Huang P, van de Pavert SA, Douglass EF, Jr, Leaver DJ, Haubrich BA, Keber R, Lorbek G, Konijn T, Rosales BN, et al. Identification of natural RORgamma ligands that regulate the development of lymphoid cells. Cell metabolism. 2015;21:286–297. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder AM, Colwell CS. How to fix a broken clock. Trends Pharmacol Sci. 2013;34:605–619. doi: 10.1016/j.tips.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SQ, Ansari TS, McGuinness OP, Wasserman DH, Johnson CH. Circadian disruption leads to insulin resistance and obesity. Current biology: CB. 2013;23:372–381. doi: 10.1016/j.cub.2013.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Griffin PR, Burris TP. Ligand regulation of retinoic acid receptor-related orphan receptors: implications for development of novel therapeutics. Curr Opin Lipidol. 2010;21:204–211. doi: 10.1097/MOL.0b013e328338ca18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Kumar N, Nuhant P, Wang Y, Lauer JL, Liu J, Istrate MA, Kamenecka TM, Roush WR, Vidovic D, et al. Suppression of TH17 differentiation and autoimmunity by a synthetic ROR ligand. Nature. 2011;472:491–494. doi: 10.1038/nature10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton CM, Jaradat M, Dixon D, Kang HS, Kim SC, Liao G, Carey MA, Cristiano J, Moorman MP, Jetten AM. Enhanced susceptibility of staggerer (RORalphasg/sg) mice to lipopolysaccharide-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2005;289:L144–152. doi: 10.1152/ajplung.00348.2004. [DOI] [PubMed] [Google Scholar]
- Stehlin C, Wurtz JM, Steinmetz A, Greiner E, Schule R, Moras D, Renaud JP. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. Embo J. 2001;20:5822–5831. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–775. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G, Millar AJ. Non-transcriptional oscillators in circadian timekeeping. Trends Biochem Sci. 2012;37:484–492. doi: 10.1016/j.tibs.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Wallach T, Kramer A. Chemical chronobiology: Toward drugs manipulating time. FEBS Lett. 2015;589:1530–1538. doi: 10.1016/j.febslet.2015.04.059. [DOI] [PubMed] [Google Scholar]
- Walle T. Methoxylated flavones, a superior cancer chemopreventive flavonoid subclass? Semin Cancer Biol. 2007;17:354–362. doi: 10.1016/j.semcancer.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kumar N, Nuhant P, Cameron MD, Istrate MA, Roush WR, Griffin PR, Burris TP. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORalpha and RORgamma. ACS Chem Biol. 2010a;5:1029–1034. doi: 10.1021/cb100223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kumar N, Solt LA, Richardson TI, Helvering LM, Crumbley C, Garcia-Ordonez RD, Stayrook KR, Zhang X, Novick S, et al. Modulation of retinoic acid receptor-related orphan receptor alpha and gamma activity by 7-oxygenated sterol ligands. The Journal of biological chemistry. 2010b;285:5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, Wu C, Baloglu E, Schmidt D, Ramesh R, et al. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–489. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. Embo J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, Mangelsdorf DJ, Evans RM. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.