Abstract
Human brain evolution is characterized by dramatic expansion in cerebral cortex size. WDR62 (WD repeat domain 62) is one of the important gene in controlling human cortical development. Mutations in WDR62 lead to primary microcephaly, a neurodevelopmental disease characterized by three to four fold reduction in cerebral cortex size of affected individuals. This study analyzes comparative protein evolutionary rate to provide a useful insight into the molecular evolution of WDR62 and hence pinpointed human specific amino acid replacements. Comparative analysis of human WDR62 with two archaic humans (Neanderthals and Denisovans) and modern human populations revealed that five hominin specific amino acid residues (human specific amino acids shared with two archaic humans) might have been accumulated in the common ancestor of extinct archaic humans and modern humans about 550,000–765,000 years ago. Collectively, the data demonstrates an acceleration of WDR62 sequence evolution in hominin lineage and suggests that the ability of WDR62 protein to mediate the neurogenesis has been altered in the course of hominin evolution.
Keywords: Primary microcephaly, WDR62, Brain evolution, Hominin, MCPH
Graphical abstract
Highlights
-
•
We trace the evolutionary history of WDR62 and its putative paralogs.
-
•
We identify accelerated sequence evolution in human WDR62.
-
•
We pinpoint eight human specific amino acid sites that reside on the C-terminal.
-
•
Out of eight, six sites are shared with archaic humans.
1. Introduction
Homo sapiens is substantially different from other non-human primates by its unique morphological, anatomical, physiological and behavioral features, including relative brain size, craniofacial attributes, bipedalism, exquisite hairless skin, opposable elongated thumb and vocal organs (Gagneux and Varki, 2001, Carroll, 2003). The most defining feature of humans is the dramatic brain expansion, especially the forebrain and cerebral cortex, which has been implicated in the development of elevated cognitive functions such as language, intelligence and social learning (Ponting and Jackson, 2005, Roth and Dicke, 2012). Modern human brain is three fold larger than our closest extant relatives, the bonobo and chimpanzee, owing to prolonged human-specific prenatal and postnatal brain development (Sakai et al., 2012).
In genetic perspective, complex and enlarged human brain emerged by essential changes in genes and non-coding regulatory elements; however, the exact genetic basis of human brain expansion still remains enigmatic (Olson and Varki, 2003, Vallender et al., 2008). Primary microcephaly is an autosomal recessive congenital disorder defined by an occitofrontal circumference (especially the cerebral cortex) less than three standard deviation (SD) at birth with no other neuroanatomical disorders (Woods et al., 2005). The brain size of microcephaly patients is similar with that of early hominids (Mochida and Walsh, 2001). There are at least twelve autosomal recessive loci, named appropriately as MCPH1–MCPH12 which are genetically link to primary microcephaly. The underlying genes of nine loci have been identified as MCPH1 (Microcephalin), MCPH2 (WDR62), MCPH3 (CDK5RAP2), MCPH4 (CASC5), MCPH5 (ASPM), MCPH6 (CENPJ), MCPH7 (STIL), MCPH8 (CEP135) and MCPH9 (CEP152) (Thornton and Woods, 2009, Genin et al., 2012). MCPH (Microcephaly primary hereditary) genes are specific regulators of human cerebral cortex size. During neurogenesis, cortical neurons originate from the progenitor cells in the ventricular zone of the developing brain. The progenitor cells undergo a cycle of proliferative divisions before moving to neurogenic divisions. The transition from proliferative division to neurogenic division is controlled by spindle pole orientation (Thornton and Woods, 2009, Chen et al., 2014). Most of the human MCPH genes such as WDR62, ASPM, CDK5RAP2, CENPJ, and STIL have been implicated in regulating spindle pole formation and orientation and thus enabling the human specific prolonged neural proliferative divisions which is consistent with the expansion of human cerebral cortex size (Kriegstein et al., 2006, Nicholas et al., 2010, Chen et al., 2014).
Recent studies have suggested that most of the identified MCPH genes (MCPH1, ASPM, CDK5RAP2, CASC5, CEP152 and CENPJ) are under adaptive evolution in primates especially in human lineage (Wang and Su, 2004, Evans et al., 2006, Genin et al., 2012). However, the evolutionary history of WDR62 gene and its role in brain expansion during human evolution remains unknown. The newly identified MCPH2 gene WDR62 is considered as the second most common cause of primary microcephaly (Roberts et al., 1999, Nicholas et al., 2010). Recessive mutations in human WDR62 disrupt the cortical development such as primary microcephaly and some other malformation of cerebral cortex including pachygyria, lissencephaly, schizencephaly, hypoplasia of corpus callosum, and polymicrogyria (Bilguvar et al., 2010, Murdock et al., 2011). WDR62 contains 32 exons, spanning the genomic region of 50,230 bp at human chromosome 19q13.12 (Memon et al., 2013). WDR62 is a spindle pole protein with 1523 amino acids and contains multiple WD40 domains at N-terminus. Furthermore, WDR62 does not share definite sequence homology especially at the C-terminus to any known protein (Nicholas et al., 2010). Recent molecular data revealed the interaction of WDR62 protein with multiple other proteins including JNK, MKK7β1, MAPKBP1 and also with centrosomal protein CEP170 (Yu et al., 2010, Cohen-Katsenelson et al., 2011, Cohen-Katsenelson et al., 2013). Expression study of human and mouse embryonic brain implies that WDR62 is highly expressed in the forebrain particularly in ventricular and sub-ventricular zone during the neurogenesis of cerebral cortex (Bilguvar et al., 2010, Nicholas et al., 2010). WDR62 functions are indispensable for neural precursor generations as well as in neural migrations during cortical development (Cohen-Katsenelson et al., 2011). This observation suggests that human WDR62 have a starring role in human cortical development that might be implicated in immense expansion of cerebral cortex during human evolution.
The present study examines the evolutionary history of WDR62 by reconstructing the phylogenetic tree. The tree establishes an evolutionary relationship between WDR62 and its putative homologs MAPKBP1 and WDR16 in human. Furthermore, we analyze the evolutionary rate of WDR62 in various mammalian species. Human specific amino acid sites were identified through comparative sequence analysis of primates WDR62. Furthermore, in order to understand the evolutionary contribution of WDR62 in brain enlargement, human specific amino acid sites were examined in two archaic humans (Neanderthals and Denisovans) and modern human populations. In addition, variations in domain topologies were explored by comparative analysis of known functional domains of WDR62 protein.
2. Materials and methods
2.1. Sequence acquisition
The closest putative paralogs of human WDR62 are determined by paralog prediction at Ensembl 72: June 2013 (http://www.ensembl.org) (Hubbard et al., 2002). The orthologous protein sequences of human WDR62/MAPKBP1/WDR16 were extracted from protein databases available at Ensembl and NCBI (http://www.ncbi.nlm.nih.gov) by using BLASTP and Bidirectional best hit strategy BLASTP (Altschul et al., 1990, Pruitt et al., 2007). Confirmation about ancestral–descendents relationship among putative orthologs was done through clustering of homologous proteins within phylogenetic trees. We excluded sequences whose position within a tree was sharply in conflict with the uncontested animal phylogeny. The complete genomic sequences of archaic humans, the Neanderthals and Denisovans was downloaded from Max Planck Institute for Evolutionary Anthropology website (http://www.eva.mpg.de/) in binary SAM (BAM) file format with 50 × and 30 × sequence coverage, respectively (Meyer et al., 2012, Prufer et al., 2014). WDR62 gene sequence from archaic genomes was obtained by generating the consensus sequences of concerned chromosome (19) from BAM files by utilizing UGENE software (Okonechnikov et al., 2012). The sequences (protein and transcript sequence data) used in this study are provided as Supplementary Material Data 1.
Species that were used in this study included H. sapiens (human), Homo neanderthalensis (Neanderthals), Denisovans, Pan paniscus (bonobo), Pan troglodytes (chimpanzee), Gorilla gorilla (gorilla), Pongo abelii (orangutan), Macaca mulatta (macaque), Saimiri boliviensis (squirrel monkey), Mus musculus (mouse), Rattus norvegicus (rat), Equus caballus (horse), Myotis dividii, Loxdonta africana (elephant), Dasypus novemcinctus (armadillo), Monodelphis domestica (opossum), Sarcophilus hairrisii (tasmanian devil), Ornithorhynchus anatinus (platypus), Gallus gallus (chiken), Taeniopygia guttata (zebra finch), Columba livia (pigeon), Anolis carolinensis (lizard), Pelodiscus sinensis (Chinese soft-shelled turtle), Xenopus tropicalis (frog), Latimeria chalumnae (coelacanth), Takifugu rubripes (fugu), Tetraodon nigroviridis (tetraodon), Gasterosteus aculeatus (stickleback), Oryzias latipes (medaka), Denio rerio (zebra fish), Branchiostoma floridae (amphioxus), Saccoglossus kowalwvskii, Drosophila melanogaster (fruit fly), Anopheles gambiae, Apis mellifera (western honey bee), and Amphimedon queenslandica.
2.2. Sequence analysis
Amino acid sequences were aligned using CLUSTAL W with default parameters (Thompson et al., 1994). The phylogenetic tree of WDR62 and its putative paralogs was reconstructed by neighbor joining method using uncorrected p distance (Saitou and Nei, 1987). Complete deletion option was used to eliminate any position containing a gap and missing data. Maximum Likelihood tree was also reconstructed by using the Whelan and Goldman (WAG) model of amino acid replacement (Whelan and Goldman, 2001) (Supplementary Fig. 1). The topological reliability of NJ and ML trees was analyzed by bootstrap method on the basis of 1000 pseudoreplicates. Phylogenetic analyses were performed using MEGA 5.05 (Tamura et al., 2011).
To estimate the molecular evolution of WDR62 in mammals, the coding sequence of WDR62 orthologs of representative mammalian species were obtained from Ensembl and NCBI. We align these coding sequences using MUSCLE and construct the phylogenetic tree. The non-synonymous (Ka) and synonymous (Ks) substitution rates were calculated by using Pamilo–Bianchi–Li′s method in MEGA5.05 (Li, 1993).
To detect the segments under selection, the sliding window analysis of Ka/Ks ratio was performed on human and chimpanzee orthologous coding sequence of WDR62. Ka − Ks was calculated at the sliding increment of 10 codons (30 nucleotides) and the results are obtained in the graph drawn by the GNUPLOT software implemented in SWAKK (Liang et al., 2006). The non-synonymous substitutions within positively selected segments (Ka/Ks > 1) are categorized according to their physicochemical properties by using BLOSSUM 62 (Zhang, 2000).
Domains were allocated to human WDR62 as previously described (Cohen-Katsenelson et al., 2011, Cohen-Katsenelson et al., 2013). Furthermore, novel domains along human WDR62 (which were not previously known) were predicted by SMART database and MyHits tool (Schultz et al., 1998, Pagni et al., 2004). CLUSTAL W based multiple sequence alignments were used to map the putative positions of these domains in orthologs of WDR62 protein in diverse set of mammalian species and its putative paralogs in humans. To investigate if the observed patterns of variability in WDR62 sequence in human population is consistent with the neutral model, neutrality test Tajima's D (Tajima, 1989), Fu and Li′s D and Fu and Li′s F (Fu and Li, 1993) were performed on the panel of 22 validated coding SNPs downloaded from dbSNP build 137 at the National Center for Biotechnology Information. These neutrality tests were performed using the DnaSP 5.10 (Librado and Rozas, 2009).
2.3. Analyzing inter-population polymorphism data
Variation data of 1092 individuals from fourteen different human populations (CHB:97; CHS:100; JPT:89; FIN:93; GBR:89; TSI:98; IBS:14; CEU:85; CLM:60; MXL:66; PUR:55; ASW:6; LWK:97 and YRI:88) was obtained from 1000 Genomes Project in variant call format (VCF) (www.1000genomes.org) (Abecasis et al., 2012). Polymorphisms among population allele frequencies were manually calculated utilizing VCF files. The topology of modern human population's tree was depicted in accordance with previously described data (McEvoy et al., 2011). With the sense of completion, HapMap and CEPH databases were scanned to gain insights about the derived allele frequencies by exploiting the SPSmart webserver (Amigo et al., 2008).
3. Results
3.1. Phylogenetic analysis
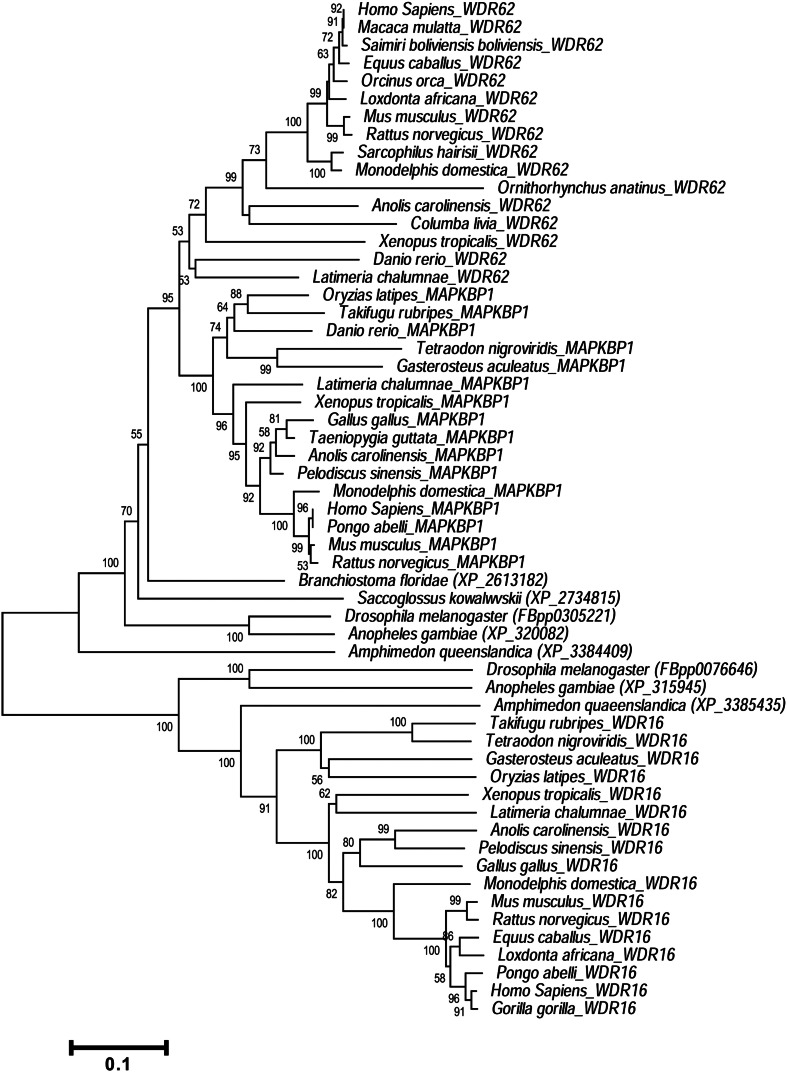
Evolutionary history of WDR62 and WD40 domain containing its putative paralogs WDR16 and MAPKBP1 was analyzed by including the protein sequences from representative members of phyla Vertebrata, Cephalochordata, Hemichordata, Arthropoda and Porifera through neighbor joining (NJ) and maximum likelihood (ML) methods (Fig. 1 and Supplementary Fig. 1). The NJ and ML yielded identical tree topologies (Fig. 1 and Supplementary Fig. 1). The tree topology infers that first duplication producing WDR16 lineage and ancestral gene of WDR62/MAPKBP1 occurred prior to bilaterian–nonbilaterian split. The second duplication splitting WDR62 and MAPKBP1 might have occurred after the divergence of vertebrate from cephalochordate and before tetrapod–teleost split.
Fig. 1.
Phylogenetic history of WDR62.
Evolutionary history of WDR62 was inferred using neighbor joining method by applying uncorrected p distance. This analysis involved 57 amino acid sequences. All gaps and missing data were eliminated by complete deletion option and there were a total 362 position in final data set. The numbers at nodes depict bootstrap value (only value ≥ 50% is shown) that was based on 1000 replicates. Scale bar represents amino acid substitution per site.
From the tree topology pattern it appears that WDR62 and MAPKBP1 are closely related genes, whereas WDR16 is very distantly related to this subgroup. The phylogeny confirms the presence of human WDR62 orthologs in all the five main classes of vertebrates, i.e. teleost fish, amphibian, reptile, bird, and mammal.
3.2. Molecular evolution of WDR62 in mammals
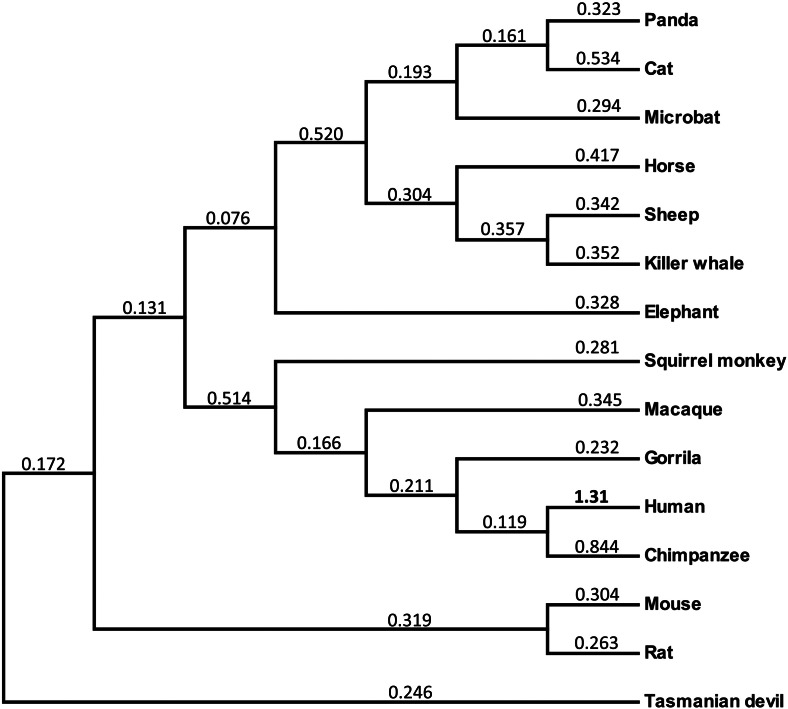
In order to identify the lineage specific Ka/Ks ratio, phylogenetic tree was constructed by using WDR62 orthologous coding sequence from representative primates and non-primate mammalian species (Fig. 2). The ratio of non-synonymous replacements to synonymous replacements was determined for each external and internal branch of phylogenetic tree. This revealed that non-synonymous substitutions outnumber the synonymous substitutions only in human terminal branch (Ka/Ks = 1.31). In contrast to human terminal branch, all other internal and terminal branches, the synonymous substitutions outnumber the non-synonymous substitutions and is suggestive of purifying selection (Fig. 2). From this analysis it appears that within mammals, the evolution of WDR62 is accelerated particularly in human terminal branch after it diverged from pan lineage.
Fig. 2.
Molecular evolution of WDR62 in mammals.
The Ka/Ks ratio for each branch of phylogenetic tree was calculated and is shown above each branch. The human terminal branch Ka/Ks score is highlighted in bold.
3.3. Human polymorphisms and signatures of selection
Molecular evolutionary rate analysis within mammals revealed different rate of WDR62 evolution among recently diverged human and pan lineage. Human WDR62 evolving slightly faster (Ka/Ks = 1.31) than its orthologous copy in pan lineage (Ka/Ks = 0.844) and hence reject neutrality. To investigate whether the pattern of variability in human WDR62 is consistent with the neutrality hypothesis, the diversity among human WDR62 is examined by exploiting human polymorphisms data from dbSNP build-137 (Sherry et al., 2001). There are total of 203 SNPs identified in the entire region of WDR62. Of these 181 SNPs are located in intronic regions and 22 in coding regions. Different statistical tests i.e. Tajima's D (Tajima, 1989), Fu and Li′s D and Fu and Li′s F (Fu and Li, 1993) (with and without using an outgroup) were employed on 22 (11 synonymous and 11 non-synonymous variations) validated polymorphisms located in coding sequence (Supplementary Table 1). Nucleotide diversity π is 0.00043 per site which is smaller than the nucleotide diversity of chromosome 19 (0.000764) (Sachidanandam et al., 2001) and Watterson's θ is 0.00129 per site. Both Tajima's test (D = − 2.5066, P < 0.001) and Fu and Li′s test without using outgroup (D* = − 4.002, P < 0.02; F* = − 4.142, P < 0.02) give significant negative values. Similarly, Fu and Li′s D and F values using chimpanzee sequence were also significantly negative (D = − 3.9448, P < 0.02; F = − 4.2124, P < 0.02). Thus significant negative values of both Tajima's D and Fu and Li′s tests reject the neutrality hypothesis and might indicate natural selection or population expansion.
3.4. Sliding window analysis of WDR62
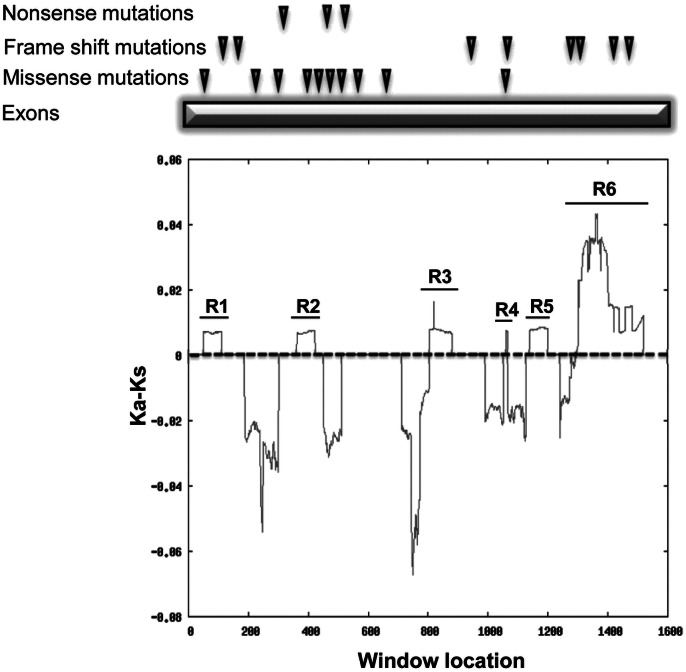
In order to pinpoint those protein regions that might be responsible in functional diversification of human WDR62 during its recent history, sliding window analysis of Ka/Ks (SWAKK) was performed. Coding sequences of human WDR62 and its chimpanzee ortholog were subjected for this analysis.
Sliding window profile revealed six regions (Fig. 3, R1–R6) with extreme peaks (Ka/Ks > 1) consistent with positive selection and many regions with valleys (Ka/Ks < 1) consistent with purifying selection (Fig. 3). The non-synonymous substitutions within positively selected (Ka/Ks > 1) regions are categorized according to their position within human WDR62 protein and their putative physicochemical impact on protein structure/function (Table 1). It appears from ML ancestral sequence reconstruction that, after the divergence from last common ancestor, eight and nine substitution fixed independently in human and chimpanzee WDR62 protein respectively. Careful comparison of these replacements with inferred human–chimpanzee ancestral residues at corresponding positions revealed that 5/8 (62%) replacements in human and 5/9 (55%) replacements in chimpanzee might have profound effect on protein structure/function (Table 1). Among protein segments with Ka/Ks > 1, regions 1 and 2 together experienced one neutral and one radical change respectively in chimpanzee within uncharacterized portion of the protein. Regions 3 and 4 collectively fixed one neutral and two radical amino acid replacements within uncharacterized medial portion of the human protein, region 5 involves one neutral and one radical replacement within an uncharacterized region and MKK7β1 binding domain (MB) of chimpanzee WDR62. Interestingly, region 6 encompasses more evolutionary non-synonymous substitutions as compared to whole gene. This region practiced three radical and two neutral amino acid replacements in both human and chimpanzee lineage, within the proline rich domain and loop helix domain. (Fig. 3) Thus, this analysis not only pinpointed the amino acid changes that fixed independently in human and chimpanzee WDR62 protein, but also discriminated the replacements that might have little or no impact on protein structure/function and the ones that are likely to be involved in altering the WDR62 protein structure/function in the course of human and chimpanzee evolution.
Fig. 3.
Sliding window analysis of WDR62.
Human and chimpanzee coding sequences were subjected for this analysis. Peaks (R1–R6) above the threshold (Ka − Ks = 0, dotted line) indicate the excess of non-synonymous substitution over neutral expectation, i.e. Ka − Ks > 0. Previously reported mutations in WDR62 that cause microcephaly are depicted on top of the plot.
Table 1.
After the divergence of human and chimpanzee, eight amino acids replacements are occurred in human lineage.
| Ka/Ks > 1 | Position | Ancestral residue | Replacement in chimpanzee | Replacement in human | Neutral/radical |
|---|---|---|---|---|---|
| Region-1 | |||||
| 81 | G | S | Neutral (0) | ||
| Region-2 | |||||
| 393 | R | G | Radical (− 2) | ||
| Region-3 | |||||
| 790 | R | H | Neutral (0) | ||
| 850 | S | L | Radical (− 2) | ||
| Region-4 | |||||
| 1091 | Y | H | Radical (2) | ||
| Region-5 | |||||
| 1169 | R | H | Neutral (0) | ||
| 1273 | T | P | Radical (− 1) | ||
| Region-6 | |||||
| 1304 | V | A | Neutral (0) | ||
| 1310 | L | Q | Radical (− 2) | ||
| 1336 | A | T | Neutral (0) | ||
| 1345 | R | H | Neutral (0) | ||
| 1369 | G | R | Radical (− 2) | ||
| 1372 | V | I | Radical (3) | ||
| 1390 | F | L | Neutral (0) | ||
| 1408 | P | S | Radical (− 1) | ||
| 1458 | R | Q | Radical (1) | ||
| 1489 | S | T | Radical (1) |
The table highlights the ancestral amino acid residues and provides a catalog of replacements occurred in each lineage since the divergence. Last column depicts the putative physicochemical impact of each replacement on protein structure/function. The numbers within bracket are the log-odds scores associated with the probability of each amino acid replacement. Positive numbers imply a preferred change, zero implies a neutral change, and negative numbers imply an unpreferred change.
3.5. Comparative analysis of WDR62 with archaic humans and modern human populations
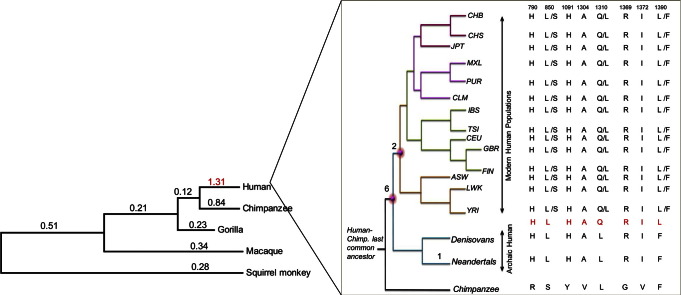
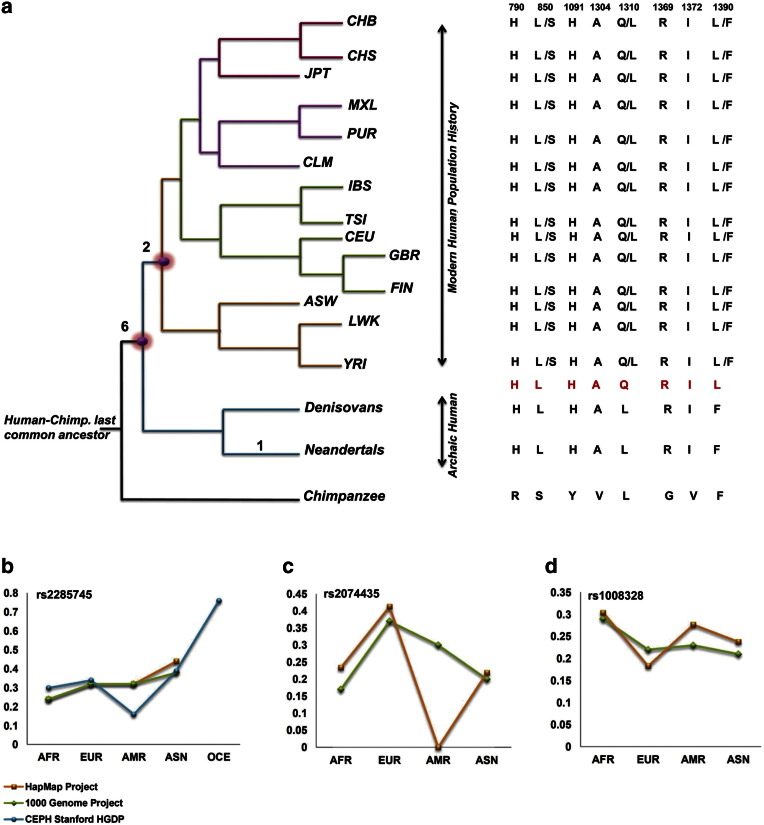
Comparative protein sequence analysis of human WDR62 with various non-human primates revealed eight human specific amino acid replacements (Fig. 2). In order to determine how many of human specific amino acid changes shared with archaic humans (Neanderthals and Denisovans) and how many of them are specific to modern humans, we compare the human WDR62 protein sequence with two archaic humans (Neanderthals and Denisovans). This analysis revealed that extinct archaic humans, the Neanderthals and Denisovans, share six amino acid replacements R790H, S850L, Y1091H, V1304A, G1369R, and V1372I with anatomically modern humans (hominin specific replacements). Two replacements L1310Q and F1390L are specific to modern humans, whereas in these sites archaic humans contain human–primate ancestral alleles (Fig. 4a).
Fig. 4.
Comparative analysis of WDR62 among human populations.
a) Tree shows the previously well-defined relationship between various modern human populations and archaic humans by using chimpanzee as outgroup (see Materials and methods). Tree illustrates six hominin specific amino acid substitutions from which five are fixed in modern human populations, while the remaining one is polymorphic in modern humans. Two amino acid substitutions are unique to modern humans and are not being shared with archaic humans. These two amino acid sites are polymorphic in modern human populations. Comparative view of modern human specific and hominin specific amino acid substitutions is illustrated on right side of the tree in modern human populations, archaic humans and chimpanzee. Human reference sequence (GRCh 37) is color coded in red. CHB; Han Chinese in Beijing, China, CHS; Han Chinese south China, JPT; Japanese in Tokyo, Japan, MXL; People with Mexican ancestry in Los Angeles, PUR; Puerto Ricans in Puerto Rico, CLM; Colombians in Medellin, Colombia, IBS; Iberian population in Spain, TSI; Toscani in Italia, CEU; Utah residents with ancestry from northern and western Europe, GBR; British from England and Scotland UK, FIN; Finnish in Finland, ASW; People with African ancestry in southwest united states, LWK; Luhya in Webuyo, Kenya, and YRI; Yoruba in Ibadan Nigeria. Three polymorphic variations are further investigated in 1000 Genomes Project, HapMap release 28 and CEPH Stanford HGDP data by SPSmart webserver. b) Derived allele frequency of SNP rs2285745 (S850L) among modern human populations in above mentioned human genomes variation projects show relatively high derived allele frequency in Oceania and Asia and low in Africa. c) SNP rs2074435 (L1310Q) show high derived allele frequency in European population as compared to Africans and Asians. American population for this SNP is not genotyped by HapMap project as depicted in graph. d) SNP rs1008328 (F1390L) demonstrated high derived allele frequency in Africa and low in Asia.
Furthermore, in order to gain insight into the status of six hominin specific and two modern human specific amino acid replacements in modern human populations, we exploited the populations' variation data from 1000 Genomes Project (Abecasis et al., 2012). These data show that, among six hominin specific amino acid replacements, five amino acid changes (R790H, Y1091H, V1304A, G1369R and V1372I) are fixed in modern human population. While the remaining one hominin specific (S850L) and two modern human specific replacements (L1310Q and F1390L) are polymorphic in modern human populations (Fig. 4a and Supplementary Tables 2, 3, and 4). These three polymorphic sites were also examined in HapMap data (International HapMap et al., 2010) and CEPH Stanford HGDP data (http://spsmart.cesga.es/). Combine analysis of 1000 Genomes Projects, HapMap data and CEPH Stanford HGDP data shows that out of three polymorphic sites, one variant S850L (human specific site shared with archaic humans), is present at relatively high derived allele frequency in non-African populations, particularly in Oceanian and Asian populations as compared to African populations (Fig. 4b). The other two polymorphic sites located in exon 30, L1310Q and F1390L show high derived allele frequency in European and African populations respectively (Fig. 4c and d).
3.6. Comparative domain analysis of WDR62 in various homologs
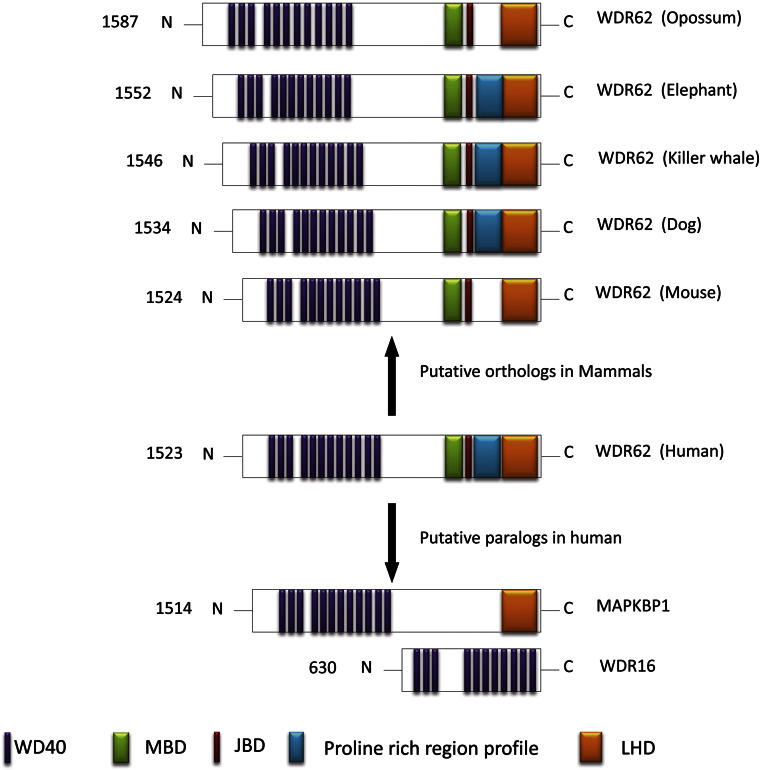
In order to gain an insight into comparative domain organization, the key functional domains of human WDR62 were analyzed and mapped on its putative paralogs in human (MAPKBP1, WDR16) and orthologs in various mammalian species including mouse, dog, elephant, killer whale, and opossum (Fig. 5).
Fig. 5.
Domains organization of WDR62 protein.
Schematic illustration of comparative organization of five key functional domains of WDR62 among human paralogous and orthologous proteins in various mammalian species. MBD; MKK7β1 binding domain, JBD; JNK binding domain, LHD; Loop helix domain.
Twelve predicted WD40 domains at the N-terminus of human WDR62 (1. 100–141, 2. 144–185, 3. 188–225, 4. 284–321, 5. 352–387, 6. 394–441, 7. 481–520, 8. 523–565, 9. 569–609, 10. 614–656, 11. 659–701, 12. 704–743) are present at conserved location in all orthologous copies analyzed and putative human paralogs except WDR16 where eleven WD40 domains were mapped instead of twelve. (Fig. 5).
MB (MKK7β1 binding) domain is responsible for the association of WDR62 with MKK7β1 through direct protein–protein interaction and is present at carboxyl terminus of human WDR62 (1212–1284) (Cohen-Katsenelson et al., 2011). Multiple sequence alignment implies the presence of MBD at conserved location across all its orthologous copies. In contrast, the paralogous comparison, suggests the absence of MB domain from MAPKBP1 and WDR16 (Fig. 5).
Human WDR62 interact with c-Jun. N terminal kinase through JBD (JNK binding domain), which is located at the carboxyl terminus of human WDR62 (1291–1301) (Cohen-Katsenelson et al., 2011). This analysis has detected the occurrence of JBD at conserved location across all orthologous counterpart. Multiple sequence alignments fail to identify JB domain in putative paralogous copies of WDR62 protein in human (Fig. 5).
Putative proline-rich domain (predicted in this study) was located at the carboxyl terminus of human WDR62 (1311–1411). Multiple sequence alignments predicted the presence of this domain across its all orthologous copies analyzed with the exception of mouse and opossum. However homology searching fails to identify putative Proline rich domain in human MAPKBP1 and WDR16 (Kay et al., 2000) (Fig. 5).
Loop helix (LH) domain is located at the carboxyl terminus of human WDR62 (1414–1520) and it is necessary for homodimerization (Cohen-Katsenelson et al., 2013). LH domain is present across all orthologous copies analyzed. LH domain also detected in human MAPKBP1 but was absent in WDR16 (Fig. 5).
For comparative analysis, the key functional domains of human WDR62 were also mapped on orthologs in various non-mammalian vertebrate species including Chinese soft-shell turtle, pigeon, frog, coelacanth, and zebrafish. This analysis revealed that all the non-mammalian vertebrates orthologs analyzed lack most of the C-terminal domains (MBD, JBD, and Proline rich region domain) (Supplementary Fig. 2).
4. Discussion
The increasing availability of whole genome sequence of extant and extinct species and advances in genomics and bioinformatics approaches have opened a new era of evolutionary study of human brain (Preuss, 2012). Homozygous mutations in human WDR62 have been reported to cause the primary microcephaly, which is characterized by severe reduction in cerebral cortex size with simplified gyral pattern (Thornton and Woods, 2009, Nicholas et al., 2010, Wasserman et al., 2010). Biochemical and molecular studies have confirmed the pivotal role of WDR62 protein in neurogenesis during embryonic development (Bilguvar et al., 2010, Yu et al., 2010). In this study, we present the phylogenetic history of human WDR62 by including representative members of vertebrate and invertebrate lineages and shed insight into the molecular evolution of WDR62 in mammals. In addition, we have selected two archaic humans (Neanderthals and Denisovans) and modern human populations' data to understand the sequence features that define the role of WDR62 in evolutionary enlargement of hominin brain.
The ML and NJ gene phylogenies (Fig. 1, and Supplementary Fig. 1) well defined by bootstrap scores, establish a distinct evolutionary relationship between WDR62/MAPKBP1 subfamily and WDR16. The tree topology indicates the diversification of WDR62 and MAPKBP1 during vertebrate history prior to tetrapod–teleost split, whereas WDR16 clade separated earlier in metazoan evolution forming the most basal branch (Fig. 1, and Supplementary Fig. 1). The close historical and sequence relationship among WDR62 and MAPKBP1 might indicate their biological similarity. This is reflected in their functional resemblance; as these vertebrate proteins are known to share the kinase activity and are involved in scaffolding functions of JNK signaling pathway (Xu et al., 2014). The most divergent phylogenetic positioning of WDR16 might account for large differences in the functional aspects of this protein and WDR62/MAPKBP1 subfamily. This is supported by the fact that this protein is responsible for proper targeting and transport of ion-channels whereas WDR62/MAPKBP1 subfamily does not perform this function (Hirschner et al., 2007). Furthermore, comparing domain features of human WDR62 with representative orthologous copies in vertebrates and its paralogous copies in human (MAPKBP1 and WDR16) revealed that N-terminal region of this protein is more preserved than the C-terminal region (Fig. 5). This suggests that N-terminus might be responsible for some fundamental ancient function. Absence of most of C-terminus domains of WDR62 in its orthologous copies in non-mammalian vertebrates and paralogous counterpart in human (MAPKBP1 and WDR16) suggests mammalian specific role for C-terminus portion of human WDR62 (Fig. 5). This is reflected in biochemical and genetic studies; as C-terminal region of WDR62 especially the MBD (MKK7β1 binding domain) and JBD (JNK binding domain) both are essential for proper neocorticogenesis through the regulation of JNK1 activity (Xu et al., 2014). These observations prompted us to propose that during early mammalian history, the C-terminal domains of WDR62 were subjected to relaxed functional constraint and accelerated evolution which might have allowed the recruitment of WDR62 for new mammalian specific function during transition from Sauropsida to Mammalia in early Jurassic era (~ 220 million years ago) (Aboitiz et al., 2002, Abdel-Mannan et al., 2008, Rakic, 2009). This time period coincides with the most important evolutionary innovation in tetrapod brain development, such as the six layered neocortex (Rakic, 2009).
Mammalian specific evolutionary rate analysis revealed the accelerated rate of WDR62 sequence evolution only in human terminal branch (Ka/Ks = 1.31) (Fig. 2). Furthermore, neutrality test (Tajima's D and Fu and Li′s tests) on human population sequence data showed significant deviation from neutrality. This deviation from neutral expectation can either be explained by natural selection (positive selection/purifying selection) or population expansion. However, in case of WDR62, the deviation from neutrality in human population cannot be attributable to purifying selection due to two reasons. First, nucleotide diversity of WDR62 (0.00043) is lower than nucleotide diversity of chromosome 19 (0.000764) and genome average diversity (0.0008–0.001) (Sachidanandam et al., 2001). Second, there is an excess of non-synonymous substitution than synonymous substitution. Functional relaxation on WDR62 is also ruled out as sliding window analysis revealed significant rate heterogeneity across WDR62 coding intervals (Fig. 3). Furthermore, careful examination of segments with Ka/Ks > 1 revealed that subset of them are functionally relevant to human brain development as mutations in these segment result in primary microcephaly and other cortical abnormalities (Fig. 3) (Bilguvar et al., 2010, Yu et al., 2010). Taken together it is argued here that the evolution of WDR62 is accelerated in human branch probably due to the collective effect of positive selection and population expansion.
Interestingly, we observed six hominin specific amino acid (human specific amino acid share with archaic humans) replacements, five of them (R790H, Y1091H, V1304A, G1369R, and V1372I) are fixed in modern human populations (Fig. 4a). This intriguing observation, prompted us to argue that these five replacements might be accumulated before the split of archaic and modern humans about 550,000–750,000 years ago (Prufer et al., 2014). The functional consequence of these replacements is yet to be understood. We assumed that these sites are potentially important in modifying the function of WDR62 during the course of hominin evolution.
Large brain and cerebral cortex are defining attributes of H. sapiens and H. neanderthalensis, and are responsible for high cognitive function including language, intelligence and social behavior (Pearce et al., 2013). Genetic and evolutionary underpinning of enlarged human brain, particularly cerebral cortex remains elusive and might be quite complex as various genes are involved in this process. This study showed that there is an acceleration of WDR62 sequence evolution only in humans terminal branch relative to other mammals and also pinpointed hominin specific amino acid substitutions that are fixed in human population. Therefore, this study set a stage for further functional and evolutionary investigations to elucidate the role of WDR62 in neurogenesis, as well in brain evolution and development.
Authors' contribution
A.A.A. conceived the project and designed the experiments. N.P. performed the experiments. A.A.A. and N.P. analyzed the data. A.A.A. and N.P. wrote the paper.
Conflict of interest statement
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the Higher Education Commission (HEC) of Pakistan (20-2562/NRPU/R&D/HEC/12/428) and the National Center for Bioinformatics, Quaid-i-Azam University, Islamabad. The authors wish to thank the Russian Unipro group especially Olga Glosova and entire UGENE team for helping in methods.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mgene.2016.02.005.
Appendix A. Supplementary data
Supplementary material.
References
- Abdel-Mannan O., Cheung A.F.P., Molnar Z. Evolution of cortical neurogenesis. Brain Res. Bull. 2008;75:398–404. doi: 10.1016/j.brainresbull.2007.10.047. [DOI] [PubMed] [Google Scholar]
- Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboitiz F., Montiel J., Morales D., Concha M. Evolutionary divergence of the reptilian and the mammalian brains: considerations on connectivity and development. Brain Res. Rev. 2002;39:141–153. doi: 10.1016/s0165-0173(02)00180-7. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amigo J., Salas A., Phillips C., Carracedo A. SPSmart: adapting population based SNP genotype databases for fast and comprehensive web access. BMC Bioinform. 2008;9:428. doi: 10.1186/1471-2105-9-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilguvar K., Ozturk A.K., Louvi A., Kwan K.Y., Choi M., Tatli B., Yalnizoglu D., Tuysuz B., Caglayan A.O., Gokben S., Kaymakcalan H., Barak T., Bakircioglu M., Yasuno K., Ho W., Sanders S., Zhu Y., Yilmaz S., Dincer A., Johnson M.H., Bronen R.A., Kocer N., Per H., Mane S., Pamir M.N., Yalcinkaya C., Kumandas S., Topcu M., Ozmen M., Sestan N., Lifton R.P., State M.W., Gunel M. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207-U93. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Zhang Y., Wilde J., Hansen K.C., Lai F., Niswander L. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat. Commun. 2014;5 doi: 10.1038/ncomms4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Katsenelson K., Wasserman T., Khateb S., Whitmarsh A.J., Aronheim A. Docking interactions of the JNK scaffold protein WDR62. Biochem. J. 2011;439:381–390. doi: 10.1042/BJ20110284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Katsenelson K., Wasserman T., Darlyuk-Saadon I., Rabner A., Glaser F., Aronheim A. Identification and analysis of a novel dimerization domain shared by various members of c-Jun N-terminal kinase (JNK) scaffold proteins. J. Biol. Chem. 2013;288:7294–7304. doi: 10.1074/jbc.M112.422055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.D., Vallender E.J., Lahn B.T. Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene. 2006;375:75–79. doi: 10.1016/j.gene.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Fu Y.X., Li W.H. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneux P., Varki A. Genetic differences between humans and great apes. Mol. Phylogenet. Evol. 2001;18:2–13. doi: 10.1006/mpev.2000.0799. [DOI] [PubMed] [Google Scholar]
- Genin A., Desir J., Lambert N., Biervliet M., Van Der Aa N., Pierquin G., Killian A., Tosi M., Urbina M., Lefort A., Libert F., Pirson I., Abramowicz M. Kinetochore KMN network gene CASC5 mutated in primary microcephaly. Hum. Mol. Genet. 2012;21:5306–5317. doi: 10.1093/hmg/dds386. [DOI] [PubMed] [Google Scholar]
- Hirschner W., Pogoda H.M., Kramer C., Thiess U., Hamprecht B., Wiesmüller K.H., Lautner M., Verleysdonk S. Biosynthesis of Wdr16, a marker protein for kinocilia-bearing cells, starts at the time of kinocilia formation in rat, and wdr16 gene knockdown causes hydrocephalus in zebrafish1. J. Neurochem. 2007;101:274–288. doi: 10.1111/j.1471-4159.2007.04500.x. [DOI] [PubMed] [Google Scholar]
- Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., Durbin R., Eyras E., Gilbert J., Hammond M., Huminiecki L., Kasprzyk A., Lehvaslaiho H., Lijnzaad P., Melsopp C., Mongin E., Pettett R., Pocock M., Potter S., Rust A., Schmidt E., Searle S., Slater G., Smith J., Spooner W., Stabenau A., Stalker J., Stupka E., Ureta-Vidal A., Vastrik I., Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap, C., Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., Dermitzakis E., Bonnen P.E., Altshuler D.M., Gibbs R.A., de Bakker P.I., Deloukas P., Gabriel S.B., Gwilliam R., Hunt S., Inouye M., Jia X., Palotie A., Parkin M., Whittaker P., Yu F., Chang K., Hawes A., Lewis L.R., Ren Y., Wheeler D., Gibbs R.A., Muzny D.M., Barnes C., Darvishi K., Hurles M., Korn J.M., Kristiansson K., Lee C., McCarrol S.A., Nemesh J., Dermitzakis E., Keinan A., Montgomery S.B., Pollack S., Price A.L., Soranzo N., Bonnen P.E., Gibbs R.A., Gonzaga-Jauregui C., Keinan A., Price A.L., Yu F., Anttila V., Brodeur W., Daly M.J., Leslie S., McVean G., Moutsianas L., Nguyen H., Schaffner S.F., Zhang Q., Ghori M.J., McGinnis R., McLaren W., Pollack S., Price A.L., Schaffner S.F., Takeuchi F., Grossman S.R., Shlyakhter I., Hostetter E.B., Sabeti P.C., Adebamowo C.A., Foster M.W., Gordon D.R., Licinio J., Manca M.C., Marshall P.A., Matsuda I., Ngare D., Wang V.O., Reddy D., Rotimi C.N., Royal C.D., Sharp R.R., Zeng C., Brooks L.D., McEwen J.E. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay B.K., Williamson M.P., Sudol P. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- Kriegstein A., Noctor S., Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Li W.H. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 1993;36:96–99. doi: 10.1007/BF02407308. [DOI] [PubMed] [Google Scholar]
- Liang H., Zhou W., Landweber L.F. SWAKK: a web server for detecting positive selection in proteins using a sliding window substitution rate analysis. Nucleic Acids Res. 2006;34:W382–W384. doi: 10.1093/nar/gkl272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- McEvoy B.P., Powell J.E., Goddard M.E., Visscher P.M. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 2011;21:821–829. doi: 10.1101/gr.119636.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memon M.M., Raza S.I., Basit S., Kousar R., Ahmad W., Ansar M. A novel WDR62 mutation causes primary microcephaly in a Pakistani family. Mol. Biol. Rep. 2013;40:591–595. doi: 10.1007/s11033-012-2097-7. [DOI] [PubMed] [Google Scholar]
- Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prufer K., de Filippo C., Sudmant P.H., Alkan C., Fu Q.M., Do R., Rohland N., Tandon A., Siebauer M., Green R.E., Bryc K., Briggs A.W., Stenzel U., Dabney J., Shendure J., Kitzman J., Hammer M.F., Shunkov M.V., Derevianko A.P., Patterson N., Andres A.M., Eichler E.E., Slatkin M., Reich D., Kelso J., Paabo S. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida G.H., Walsh C.A. Molecular genetics of human microcephaly. Curr. Opin. Neurol. 2001;14:151–156. doi: 10.1097/00019052-200104000-00003. [DOI] [PubMed] [Google Scholar]
- Murdock D.R., Clark G.D., Bainbridge M.N., Newsham I., Wu Y.Q., Muzny D.M., Cheung S.W., Gibbs R.A., Ramocki M.B. Whole-exome sequencing identifies compound heterozygous mutations in WDR62 in siblings with recurrent polymicrogyria. Am. J. Med. Genet. A. 2011;155A:2071–2077. doi: 10.1002/ajmg.a.34165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A.K., Khurshid M., Desir J., Carvalho O.P., Cox J.J., Thornton G., Kausar R., Ansar M., Ahmad W., Verloes A., Passemard S., Misson J.P., Lindsay S., Gergely F., Dobyns W.B., Roberts E., Abramowicz M., Woods C.G. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat. Genet. 2010;42:1010-U138. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Golosova O., Fursov M., Team U. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- Olson M.V., Varki A. Sequencing the chimpanzee genome: insights into human evolution and disease. Nat. Rev. Genet. 2003;4:20–28. doi: 10.1038/nrg981. [DOI] [PubMed] [Google Scholar]
- Pagni M., Ioannidis V., Cerutti L., Zahn-Zabal M., Jongeneel C.V., Falquet L. MyHits: a new interactive resource for protein annotation and domain identification. Nucleic Acids Res. 2004;32:W332–W335. doi: 10.1093/nar/gkh479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E., Stringer C., Dunbar R.I. New insights into differences in brain organization between Neanderthals and anatomically modern humans. Proc. Biol. Sci. 2013;280:20130168. doi: 10.1098/rspb.2013.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C., Jackson A.P. Evolution of primary microcephaly genes and the enlargement of primate brains. Curr. Opin. Genet. Dev. 2005;15:241–248. doi: 10.1016/j.gde.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Preuss T.M. Human brain evolution: from gene discovery to phenotype discovery. Proc. Natl. Acad. Sci. U. S. A. 2012;109(Suppl. 1):10709–10716. doi: 10.1073/pnas.1201894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C., Li H., Mallick S., Dannemann M., Fu Q.M., Kircher M., Kuhlwilm M., Lachmann M., Meyer M., Ongyerth M., Siebauer M., Theunert C., Tandon A., Moorjani P., Pickrell J., Mullikin J.C., Vohr S.H., Green R.E., Hellmann I., Johnson P.L.F., Blanche H., Cann H., Kitzman J.O., Shendure J., Eichler E.E., Lein E.S., Bakken T.E., Golovanova L.V., Doronichev V.B., Shunkov M.V., Derevianko A.P., Viola B., Slatkin M., Reich D., Kelso J., Paabo S. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43-+. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E., Jackson A.P., Carradice A.C., Deeble V.J., Mannan J., Rashid Y., Jafri H., McHale D.P., Markham A.F., Lench N.J., Woods C.G. The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1–13.2. Eur. J. Hum. Genet. 1999;7:815–820. doi: 10.1038/sj.ejhg.5200385. [DOI] [PubMed] [Google Scholar]
- Roth G., Dicke U. Evolution of the brain and intelligence in primates. Prog. Brain Res. 2012;195:413–430. doi: 10.1016/B978-0-444-53860-4.00020-9. [DOI] [PubMed] [Google Scholar]
- Sachidanandam R., Weissman D., Schmidt S.C., Kakol J.M., Stein L.D., Marth G., Sherry S., Mullikin J.C., Mortimore B.J., Willey D.L., Hunt S.E., Cole C.G., Coggill P.C., Rice C.M., Ning Z., Rogers J., Bentley D.R., Kwok P.Y., Mardis E.R., Yeh R.T., Schultz B., Cook L., Davenport R., Dante M., Fulton L., Hillier L., Waterston R.H., McPherson J.D., Gilman B., Schaffner S., Van Etten W.J., Reich D., Higgins J., Daly M.J., Blumenstiel B., Baldwin J., Stange-Thomann N., Zody M.C., Linton L., Lander E.S., Altshuler D. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method — a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai T., Hirata S., Fuwa K., Sugama K., Kusunoki K., Makishima H., Leguchi T., Yamada S., Ogihara N., Takeshita H. Fetal brain development in chimpanzees versus humans. Curr. Biol. 2012;22:R791–R792. doi: 10.1016/j.cub.2012.06.062. [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. Clustal-W — improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton G.K., Woods C.G. Primary microcephaly: do all roads lead to Rome? Trends Genet. 2009;25:501–510. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallender E.J., Mekel-Bobrov N., Lahn B.T. Genetic basis of human brain evolution. Trends Neurosci. 2008;31:637–644. doi: 10.1016/j.tins.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.Q., Su B. Molecular evolution of microcephalin, a gene determining human brain size. Hum. Mol. Genet. 2004;13:1131–1137. doi: 10.1093/hmg/ddh127. [DOI] [PubMed] [Google Scholar]
- Wasserman T., Katsenelson K., Daniliuc S., Hasin T., Choder M., Aronheim A. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell. 2010;21:117–130. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Woods C.G., Bond J., Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am. J. Hum. Genet. 2005;76:717–728. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Zhang F., Wang Y., Sun Y., Xu Z. Microcephaly-associated protein WDR62 regulates neurogenesis through JNK1 in the developing neocortex. Cell Rep. 2014;6:104–116. doi: 10.1016/j.celrep.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Yu T.W., Mochida G.H., Tischfield D.J., Sgaier S.K., Flores-Sarnat L., Sergi C.M., Topcu M., McDonald M.T., Barry B.J., Felie J.M., Sunu C., Dobyns W.B., Folkerth R.D., Barkovich A.J., Walsh C.A. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat. Genet. 2010;42:1015-U145. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Rates of conservative and radical nonsynonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 2000;50:56–68. doi: 10.1007/s002399910007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.