Significance
Caspases, a family of 12 proteases involved in irreversible cell state changes including cell death, often cleave common substrates. However, we show here by quantitative N-terminomics MS, for caspase-2 and caspase-6, that the rates of substrate cleavage vary more than 500-fold in cellular lysate. The rates of cleavage show virtually no correlation among common substrates for these two caspases, as well as for three other caspases previously studied: caspase-3, caspase-7, and caspase-8. These global and unbiased studies reveal a greater degree of substrate hierarchy and specialized functions for caspases than previously appreciated. We believe this quantitative approach is of general use to other proteases and enzymes involved in posttranslational modifications to better define their roles.
Keywords: N-terminomics, caspase, proteolysis, proteomics, apoptosis
Abstract
Proteases constitute the largest enzyme family, yet their biological roles are obscured by our rudimentary understanding of their cellular substrates. There are 12 human caspases that play crucial roles in inflammation and cell differentiation and drive the terminal stages of cell death. Recent N-terminomics technologies have begun to enumerate the diverse substrates individual caspases can cleave in complex cell lysates. It is clear that many caspases have shared substrates; however, few data exist about the catalytic efficiencies (kcat/KM) of these substrates, which is critical to understanding their true substrate preferences. In this study, we use quantitative MS to determine the catalytic efficiencies for hundreds of natural protease substrates in cellular lysate for two understudied members: caspase-2 and caspase-6. Most substrates are new, and the cleavage rates vary up to 500-fold. We compare the cleavage rates for common substrates with those found for caspase-3, caspase-7, and caspase-8, involved in apoptosis. There is little correlation in catalytic efficiencies among the five caspases, suggesting each has a unique set of preferred substrates, and thus more specialized roles than previously understood. We synthesized peptide substrates on the basis of protein cleavage sites and found similar catalytic efficiencies between the protein and peptide substrates. These data suggest the rates of proteolysis are dominated more by local primary sequence, and less by the tertiary protein fold. Our studies highlight that global quantitative rate analysis for posttranslational modification enzymes in complex milieus for native substrates is critical to better define their functions and relative sequence of events.
Caspases are cysteine-class proteases that typically cleave after aspartic acid and play critical roles in cellular remodeling during development, cell differentiation, inflammation, and cell death (reviewed in refs. 1–3). There are 12 caspases in humans alone, which have been classically grouped on the basis of sequence homology, domain architecture, and cell biology as inflammatory (caspase-1, caspase-4, caspase-5, and caspase-11), apoptotic initiators (caspase-2, caspase-8, caspase-9, and caspase-10), or executioners (caspase-3, caspase-6, and caspase-7) (4, 5). Identifying the natural protein substrates for caspases can provide important insights into their cellular roles. A series of large-scale proteomic studies from several laboratories has collectively identified nearly 2,000 native proteins cleaved after an aspartic acid during apoptosis of human cells (recently reviewed in refs. 6–8). By adding individual caspases to extracts, it has been possible to begin to identify caspase-specific substrates, ranging from just a few, in the case of caspase-4, caspase-5, and caspase-9 (9), to several hundred for caspase-1, caspase-3, caspase-7, and caspase-8 (10, 11). Some of these substrates overlap, but others are unique to specific caspases. These substrate discovery experiments provide a useful starting point, but fail to address whether a hierarchy exists for the cleavage events.
Kinetic studies have the potential to provide a much higher resolution picture by analyzing cleavage rates. For example, early work measured the rates of cleavage for several purified protein substrates by caspase-3 and caspase-7, showing significant rate differences (12). In another study, catalytic efficiency values (kcat/KM) were determined in vitro by quantitative SDS/PAGE for a sampling of eight purified caspase-3 substrates and were found to vary 100-fold (from 104 to 106 M−1⋅s−1) (13). A similar study of six caspase-1 substrates found them to vary in rate by about 200-fold, from 5 × 102 up to 1.2 × 105 M−1⋅s−1 (9). Recently, quantitative MS methods were applied to simultaneously calculate kcat/KM values for multiple substrates within cell extracts without the need to clone, express, and purify the individual proteins (10). Seven-point rate curves were measured, which allowed calculation of kcat/KM values for 180, 58, and 66 substrates for caspase-3, caspase-7, and caspase-8, respectively. The rates of cleavage varied more than 500-fold for each caspase. Together, these studies began to suggest there is a strong hierarchy in the rates at which substrates in their natural milieu are cleaved by individual caspases.
Here, we apply discovery and quantitative MS-based proteomics to caspase-2 and caspase-6, where there is far less known about their native substrates. Caspase-2 contains an initiator-like architecture with a long N-terminal caspase recruitment domain, yet it has an amino acid sequence preference for residues VDVAD↓ (P5–P1) (14), a motif that is more similar to the classical DEVD↓ motif cleaved by executioner caspase-3 and caspase-7. Protease cleavage nomenclature labels residues surrounding the cleavage site located between P1 and P1′ as Pn … P2-P1-P1′-P2′ … Pn′ (15). Caspase-6, in contrast, has closest sequence homology and architecture to caspase-3 and caspase-7, but is very inefficient at cleaving DEVD motifs and prefers hydrophobic residues at the P4 position, such as VEVD motifs seen for initiators. Caspase-6 has been implicated in neurodegenerative disease, including Huntington’s and Alzheimer’s diseases (16). Accordingly, finding small-molecule modulators of caspase-6 are of great interest (17), and knowledge of the natural substrates could be useful for inhibitor and assay design.
Using the subtiligase-based N-terminomics technology in cell extracts (18), we discovered 235 and 871 protein substrates (with P1 = D) for caspase-2 and caspase-6, respectively. We were able to carry out quantitative kinetic studies in cell extracts and measured kcat/KM values for the top 49 and 276 individual substrates cut by caspase-2 and caspase-6, respectively. This allowed us to rank cleavage sites on the basis of their roughly 500-fold variation in rate of proteolysis. The data, when combined with quantitative proteomic data from caspase-3, caspase-7, and caspase-8, allowed us to make 10 pairwise comparisons among these five caspases, which showed virtually no correlation. These data strongly suggest that these caspases have very distinct cellular functions. Last, we find the catalytic efficiencies for the protein substrates are often comparable to linear peptides, suggesting that primary sequence and not tertiary fold dominates cleavage rate and that native protein cleavage sites are as accessible as the linear peptides. Broadly, this work highlights the importance for quantitative proteomics of native proteins in complex mixtures to determine protein substrate specificity and to better understand the distinctive roles for proteases and other posttranslational modifying enzymes.
Results
Discovery of Caspase-2 and Caspase-6 Substrates in Cell Lysate Reveals Primary Sequence Specificity.
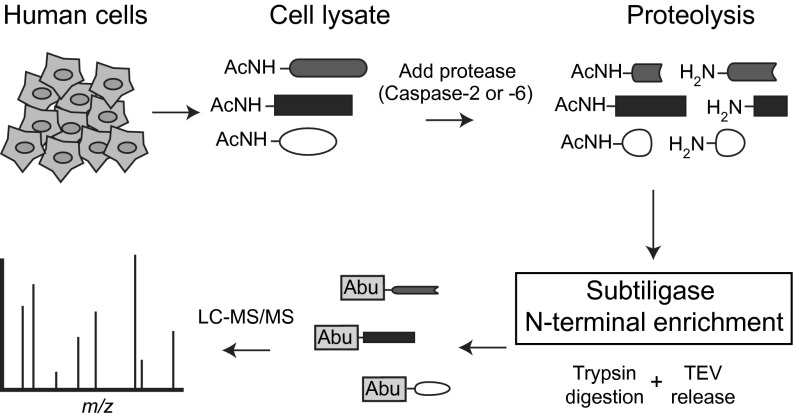
As a source of extract, we used Jurkat cells, an immortalized line of human T lymphocyte cells (19). Although no single cell line expresses all proteins, Jurkat cells have been exhaustively used for studying apoptosis and, more specifically, caspase function (3). Thus, Jurkat cells were our preferred cell line for making comparisons to published data. The protease activity for the exogenous caspase-2 and caspase-6 was the same in buffer compared with cell lysates, suggesting the absence of endogenous inhibitors or activators in the cell lysate (SI Appendix, Fig. S1). The subtiligase-based N-terminomics methodology (Fig. 1) was applied to recover cleaved products after addition of caspase-2 or caspase-6. This attaches a biotin tag onto the α-amine of the cleaved N-terminal protein fragment. Virtually all labeling occurs at sites of either endogenous or exogenous proteolysis because >80% of natural N termini for mammalian proteins are blocked by acetylation (AcNH-) during translation (20). The biotin-labeled proteins are bound to avidin beads and trypsinized. The N-terminal peptide is released by cleaving the unique tobacco etch virus (TEV) protease cleavage sequence in the tag followed by identification by LC-MS/MS. The tagging and release process generates a nonnatural remnant, aminobutyric acid (Abu), at the site of labeling, which unambiguously confirms that the recovered peptide was labeled by subtiligase (SI Appendix, Fig. S2; see Materials and Methods for details).
Fig. 1.
Proteolytic substrate identification of caspase-2 and caspase-6 substrates, using subtiligase-enrichment tagging technology. Schematic of the N-terminomics methodology used for direct capture of proteolytic peptides. Jurkat cells are harvested and lysed. The α-amines of most proteins are naturally blocked by acetylation (AcNH-) unless there has been a proteolytic event (20). Endogenous proteases are preinactivated in the freshly prepared extract by addition of protease inhibitors, and excess thiol protease inhibitors are neutralized with DTT before the addition of exogenous caspase. The engineered peptide ligase, subtiligase, is added together with a biotinylated ester (SI Appendix, Fig. S2) to tag the free N-termini of endogenous proteins as well as the N-termini generated by caspase proteolysis. The biotinylated proteins are captured using avidin beads and trypsinized, and the N-terminal peptides are subsequently released using TEV protease. This leaves behind a nonnatural Abu residue that aids in identification. Substrate identification is performed using LC-MS/MS.
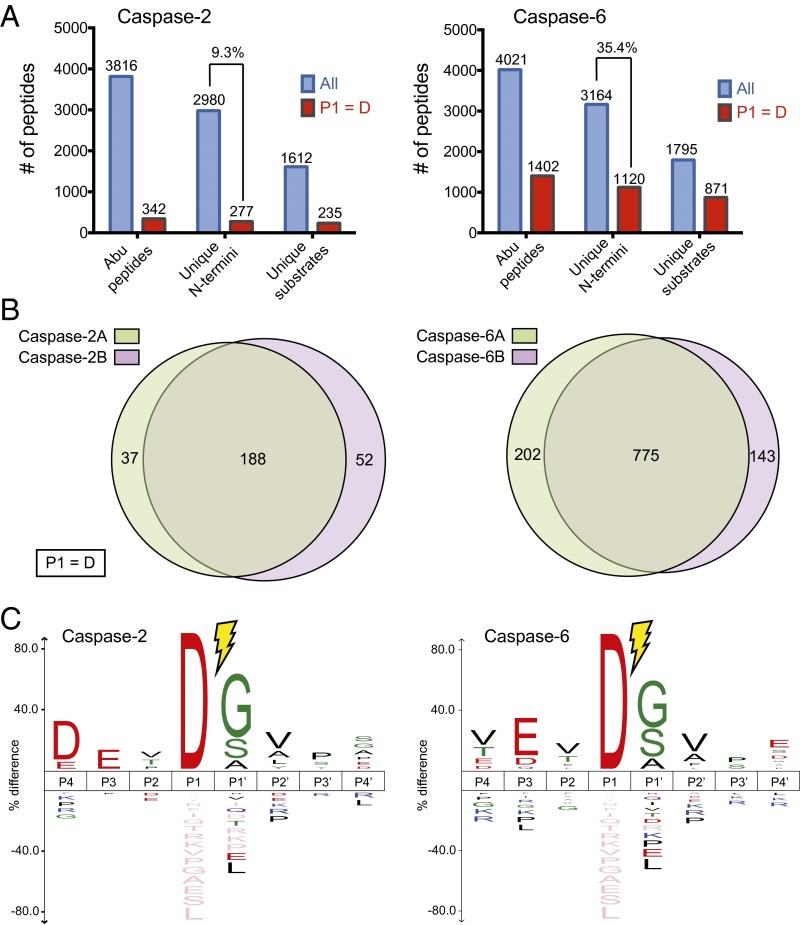
Caspases classically cleave their targets after an aspartic acid residue (P1 = D) (21). We found 277 aspartate cleavage sites in 235 proteins on addition of caspase-2 (averaging 1.2 sites per protein) and 1,120 cleavage sites in 871 putative caspase-6 substrate proteins (averaging 1.3 sites per protein) (Fig. 2A and Datasets S1 and S2). More than 70% of the cleavage sites were identified in two replicates (Fig. 2B), indicating good sampling coverage. The false discovery rates were <1.3% for both datasets at the peptide level and were similar to our previous studies on other caspases. The majority of the N termini we observed (∼4,000 Abu-tagged peptides) are derived from endogenous noncaspase endoproteolytic events, which we have previously observed in healthy cell extracts (3). In general, less than 3% of aspartic acid cleavage sites are detected in the absence of exogenous caspase, suggesting little caspase activation. Several small-scale studies have previously identified a total of 37 and 68 substrates for caspase-2 and caspase-6, respectively (11, 22, 23). More than half of the previously reported caspase-2 substrates were found in our dataset (19 of the 37), and we identified 13 of 61 caspase-6 substrates reported in the CutDB database (cutdb.burnham.org/).
Fig. 2.
Discovery of caspase-2 and caspase-6 substrates in cell lysates. (A) Shown are the total number of unique peptides (tagged with Abu), unique cleavage sites, and unique substrates at the protein level identified for caspase-2 (Left) and caspase-6 (Right). The subset of identifications featuring an Asp at P1 position is shown in red. Total peptides are colored in blue, whereas peptides originating only from a caspase cleavage event (aspartate at P1 position) are colored in red. (B) The discovery experiments show greater than 70% overlap between the two biological replicates in the cleavage sites identified for both caspase-2 and caspase-6. (C) Sequence recognition motifs of caspase cleavages for caspase-2 and caspase-6 shows highest consensus DEVD↓(G/S/A) and VEVD↓(G/S/A) motifs, respectively (P value = 0.05). Consensus sequences were generated using iceLogo (24).
We compiled all the respective aspartate P1 recognition sequences of caspase-2 and caspase-6 (Fig. 2C) (24). The most prominent sequence cleaved by caspase-2 is DEVD↓(G/S/A), which is remarkably similar to the consensus cleavage motif for the executioner caspase-3 and caspase-7 (11). Our results did not show a strong specificity at the P5 position, with the possible exception of glutamate (SI Appendix, Fig. S3). Caspase-6 showed an initiator-like VEVD↓(G/S/A) consensus motif and only a faint preference for aspartate at P4, dissimilar to caspase-2, caspase-3, and caspase-7 at the P4 position. Interestingly, the caspase-6 data also showed a small enrichment for a negatively charged residue at the P5 position. It is interesting to us that the consensus motifs for caspase-2 and caspase-6 are more similar to executioner and initiator caspases, respectively, despite their domain architectures and homologies suggesting the opposite.
Bioinformatics Analysis of Caspase-2 and Caspase-6 Substrates.
We performed bioinformatics analysis of the 276 and 871 caspase-2 and caspase-6 substrates identified in cell lysate, respectively. We observed virtually identical substrate cellular localization patterns for both datasets, with 35% of the substrates annotated as found in the cytoplasm, 37% in the nucleus, 5% in the endoplasmic reticulum, 6% at the cell membrane, 3% in the mitochondria, 2% as secreted proteins, and 12% as found in other organelles (or unknown) (SI Appendix, Fig. S4). This distribution is very similar to the distribution of substrates seen when we have induced apoptosis in intact cells (3). Moreover, the distribution is similar to the subcellular localization of all proteins found in the human proteome, with the exception of secreted and cell membrane proteins, which are typically underrepresented in proteomic experiments based on their extraction efficiency in whole-cell lysates. Thus, the distribution of substrates is not biased by subcellular location for either caspase-2 or caspase-6.
To obtain a general overview of caspase-2 and caspase-6 biology, we performed a gene ontology enrichment analysis on both datasets (SI Appendix, Fig. S4 and Datasets S3 and S4). For caspase-2, we found an enrichment for RNA processing, regulation of cell death, intracellular transport, chromosome organization, and cytoskeleton. For caspase-6, we found enrichment for regulation of transcription, cell cycle, cell death, RNA splicing, cytoskeleton, and response to DNA damage. Examples of substrates for each category are shown in SI Appendix, Fig. S4.
Quantitative Measurement of Caspase-Cleaved Substrates, Using Selected Reaction Monitoring.
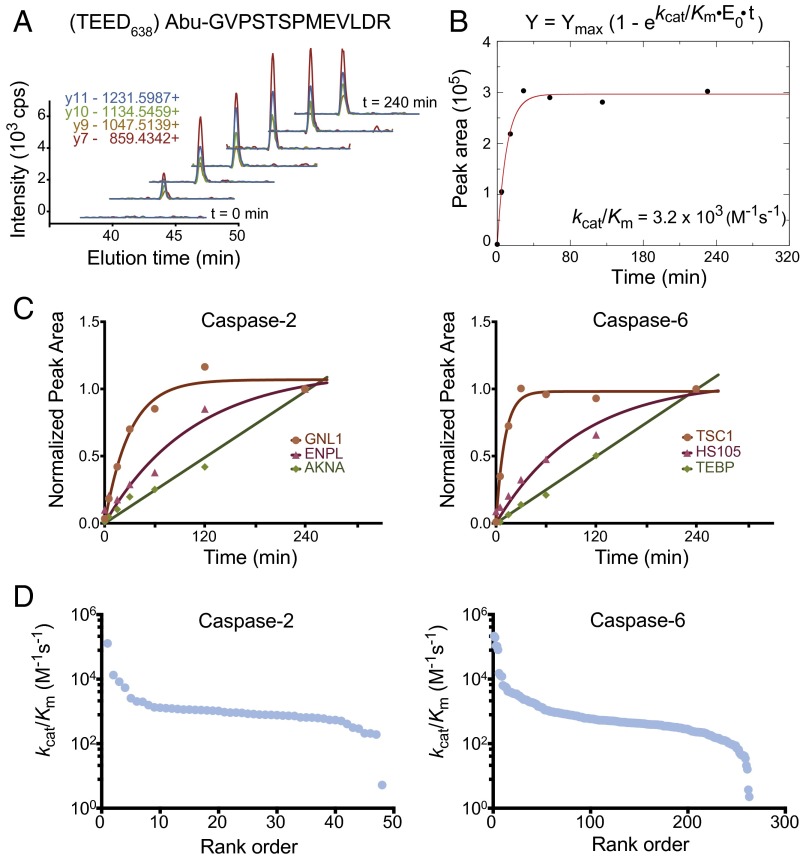
To determine the rates of proteolysis of the natural protein substrates, we developed a scheduled selected reaction monitoring (SRM) method on a triple-quadrupole mass spectrometer (SI Appendix, Fig. S5). The 342 and 1,402 aspartate-cleaved peptides identified for caspase-2 and caspase-6, respectively, were evaluated for their suitability for SRM analysis. Using the sample from the discovery experiment (Fig. 2), SRM training data were acquired on a QTRAP 5500 LC-MS/MS to identify suitable peptides. Peptide transitions, known as precursor and product ion pairs, were chosen from the top seven most intense transitions and were searched within a 15-min retention time window. We selected only peptides showing four strong and unambiguous transitions, which ultimately led us to monitor 152 and 471 peptides (plus a control) for caspase-2 and caspase-6, respectively. As earlier, we added caspase-2 and caspase-6 to preneutralized cell lysate and quenched the caspases using the pan caspase inhibitor zVAD-fmk at each of seven points (0, 5, 15, 30, 60, 120, and 240 min) before N-terminal enrichment and LC-MS/MS quantification. The appearance of each proteolytic substrate was monitored as a function of time (Fig. 3A). Using pseudofirst-order kinetics, we performed automated nonlinear fitting to determine the catalytic efficiency (kcat/KM) of each caspase substrate (Fig. 3B and Datasets S5 and S6). In total, we were able to measure 49 and 276 kcat/KM values for caspase-2 and caspase-6, respectively. These values ranged two to four log units for caspase-2 and caspase-6, respectively (Fig. 3D). Most progress curves were monotonic and parabolic, allowing calculation of kcat/KM values. The fastest substrates were cleaved at a rate of up to 105 M−1⋅s−1 (Fig. 3 C and D and SI Appendix, Fig. S6), which rivaled several optimized synthetic fluorescent substrates (25–27). However, the catalytic efficiency values for the slowest (<102 M−1⋅s−1) and fastest (>105 M−1⋅s−1) reactions are less reliable than the intermediate cleavage reactions; the slowest reactions did not go to completion within the 240-min assay time, whereas the fastest reactions were generally complete by the second time interval (15 min), so only one or two data points could be fit. In future studies, various enzyme concentrations could be used to determine more accurately the proteolysis of these few substrates.
Fig. 3.
Determination of catalytic efficiency values (kcat/KM) for caspase-2 and caspase-6 substrates, using SRM. (A) Example chromatograms of the four transitions monitored for a peptide from the TSC1 protein shows accumulation of cleavage product over time. (B) The peak integrations were summed over the four transitions monitored to estimate extent of cleavage product and plotted as a function of time to determine the progress rate curve. (C) The assays were analyzed using pseudofirst-order kinetic conditions, and kcat/KM values were calculated as described in the Materials and Methods for 49 and 276 cleavage sites for caspase-2 and caspase-6, respectively. Three examples are shown for caspase-2 (Left) and caspase-6 (Right). (D) A wide range of kcat/KM values were observed over two to four logs for caspase-2 and caspase-6 substrates, respectively. The fastest protein substrates (105 M−1⋅s−1) can only be estimated because they saturated too rapidly. Most of the substrates are in the order of 103 M−1·s−1 and fit well because multiple points were obtained before saturation.
Western Blots.
Quantitation of cleavage rates for hundreds of substrates by immunoblot is impractical, given the lack of suitable antibodies and prohibitive cost. Nonetheless, we chose to monitor the rates and extents of cleavage for two caspase-6 targets, the new substrate tuberous sclerosis 1 protein homolog (TSC1 or hamartin) and the classic substrate poly (ADP ribose) polymerase 1, for which suitable antibodies were available. We performed the same time course in extracts as done for the SRM analysis and monitored the disappearance of the full-length proteins and appearance of the cleaved products, using traditional Western blotting (SI Appendix, Fig. S7). Full-length hamartin showed a marked reduction in intensity over time with simultaneous increase of cleaved product, consistent with cleavage at position 638 identified in the discovery experiment. Full-length poly (ADP ribose) polymerase 1 showed similar results, with a decrease in the full-length protein and increase of the cleaved product, whereas the intensity of the GAPDH loading control remained unchanged. The rate constants obtained by Western blot analysis were within fivefold of those obtained by MS for these two substrates: TSC1 (Western blot : 7.1 × 102 and SRM: 3.2 × 103 M−1⋅s−1) and poly (ADP ribose) polymerase 1 (Western blot : 1.1 × 102, SRM: 1.5 × 102 M−1⋅s−1). The small discrepancies in rate constants are likely a result of inherent variability and sensitivity between the two methods.
Pairwise Comparisons of Rates for Common Substrates Cleaved by Caspase-2, Caspase-3, Caspase-6, Caspase-7, and Caspase-8 Show Virtually No Correlation and Highlight Their Unique Substrate Preferences.
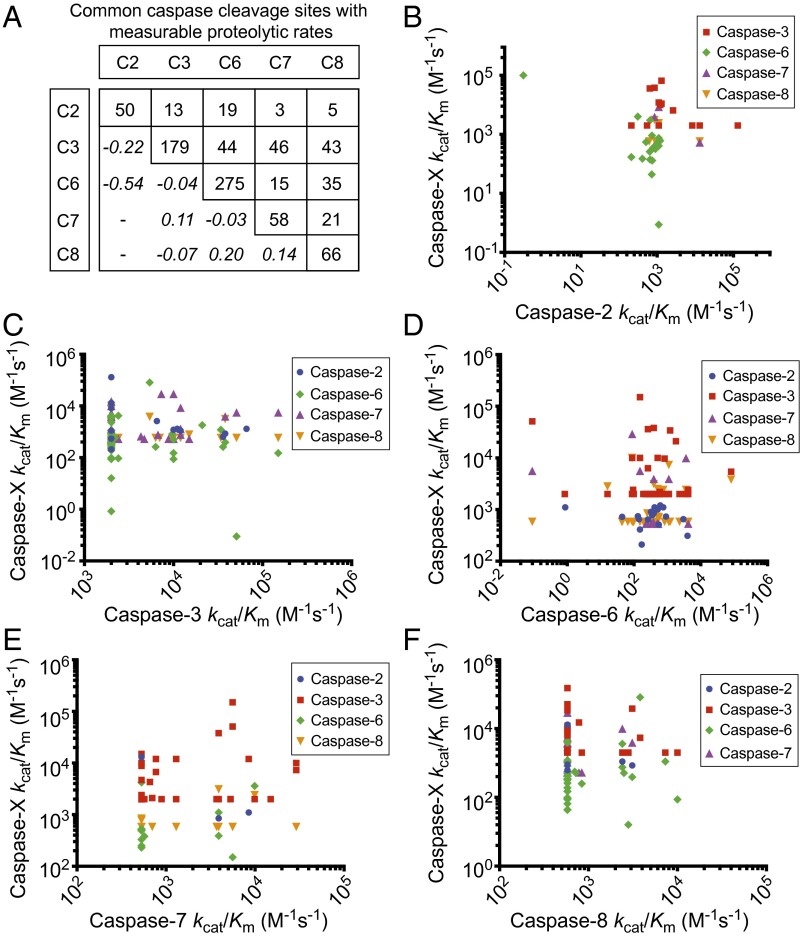
We performed a pairwise comparison of the rates of proteolysis obtained for identical cleavage sites that were shared between the caspases, comparing caspase-2 and caspase-6 SRM data obtained here with values obtained for caspase-3, caspase-7, and caspase-8 previously from our laboratory (10). The number of common substrate sites shared between the two caspases ranged from as few as three (between caspases-2 and caspase-8) to as many as 46 (between caspase-3 and caspase-7) (Fig. 4A). We found virtually no correlation among the rates at which these common substrates were cleaved (Fig. 4 B–F and Dataset S7). This is true for all 10 pairwise comparisons. The only small connection observed among the five caspases studied is a weak anticorrelation between caspase-2 and caspase-6 substrates. This suggests that individual caspases have preferred protein substrates, and thus specific roles. This quantitative difference is not evident when looking at consensus cleavage motifs. For example, caspase-2, caspase-3, and caspase-7 show a consensus DEVD sequence motif in cell lysates, but the common substrates are cleaved at very different rates (Fig. 4B, C, and E).
Fig. 4.
Pairwise comparisons of common substrates of caspase-2, caspase-3, caspase-6, caspase-7, and caspase-8 shows uncorrelated catalytic efficiency values. (A) Shows the number of common substrates with available rates of proteolysis determined among caspase-2, caspase-3, caspase-6, caspase-7, and caspase-8. (B–F) Shows the poor pairwise comparisons for the rate of proteolysis of the common substrates among the individual caspases. Pearson correlation coefficients are shown in italic in A (for n > 5). The vertical set of points for caspase-3, caspase-7, and caspase-8 (C, E, and F) reflect the lower limit of detection for slow substrates (10).
Comparisons of Sequence Cleavage Patterns for Protein and Peptide Substrates.
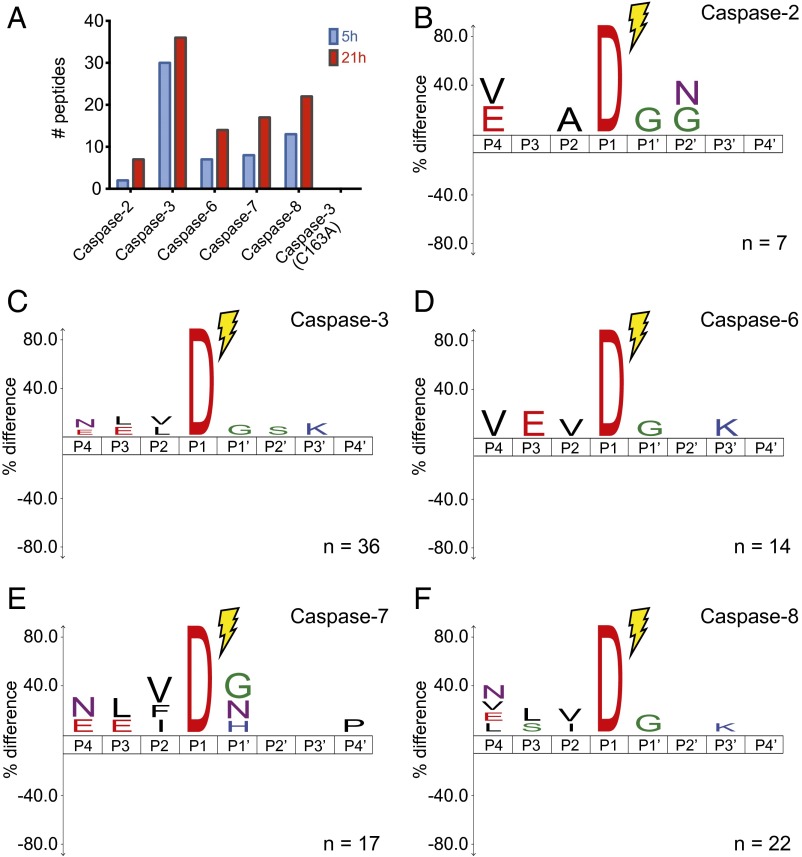
We wished to assess whether the intrinsic rate and specificity toward linear substrate motifs differs among the caspases, and thus sought a peptide-library approach that would remove the influence of protein structure. An unbiased approach for characterizing linear peptide specificity of proteases, called multiplex substrate profiling by MS (MSP-MS), has been applied to characterizing protease cleavage patterns (28). This in vitro technique uses a highly diversified tetra-decapeptide library designed to present virtually all possible pairwise amino acid combinations at a minimum twofold redundancy within a five-position linear motif to a peptidase of interest (currently designed with 228 unique peptides). We would expect to observe dyads from the P1 Asp to the other subsites at P4, P3, and P1′. To test this, equal amounts of caspase-2, caspase-3, caspase-6, caspase-7, and caspase-8 were added separately to the peptide library and were samples taken at 5 and 21 h for analysis. The number of unique Asp-cleaved peptides identified increased with time and ranged from 36 peptides cleaved for caspase-3 to as few as seven for caspase-2 (Fig. 5A and Dataset S8). The number of observed cuts roughly track previous kinetic experiments on optimized substrates, showing that caspase-3 is the most active, followed by caspase-8, caspase-7, caspase-6, and caspase-2.
Fig. 5.
Caspase-2, caspase-3, caspase-6, caspase-7, and caspase-8 sequence specificity obtained by MSP-MS. Each caspase was incubated with a 228-member degenerate peptide library with the MSP-MS method to evaluate the protease specificity in vitro (28). (A) Total number of peptides cleaved by each enzyme (presence of aspartate at C-termini) after 5 and 24 h incubation. (B–F) Sequence recognition motifs of caspase-2, caspase-3, caspase-6, caspase-7, and caspase-8, respectively (P value = 0.05).
Although the number of cleavage events was far less than those sampled in the N-terminomics experiments, recognition motifs were evident that are similar to the N-terminomics data (Fig. 4 B–F). All show an enrichment for aspartate at P1 and small amino acids at P1′. The consensus (E/V)XAD↓G for caspase-2, VEVD↓G for caspase-6, and (N/E)(L/E)VD↓G for caspase-3, caspase-7, and caspase-8 are not dissimilar from the respective consensus sequences derived from the N-terminomics data shown for capase-2 and caspase-6 in Fig. 2 C and D, and previously reported for caspase-3, caspase-7, and caspase-8 (10). The greatest divergence is at P4, between the peptide and protein data, but is most likely a result of the limited number of peptide sequences present in the peptide library. In fact, DEVD or similar motifs are not in the peptide library, as the library was designed with a focus on pairwise diversity.
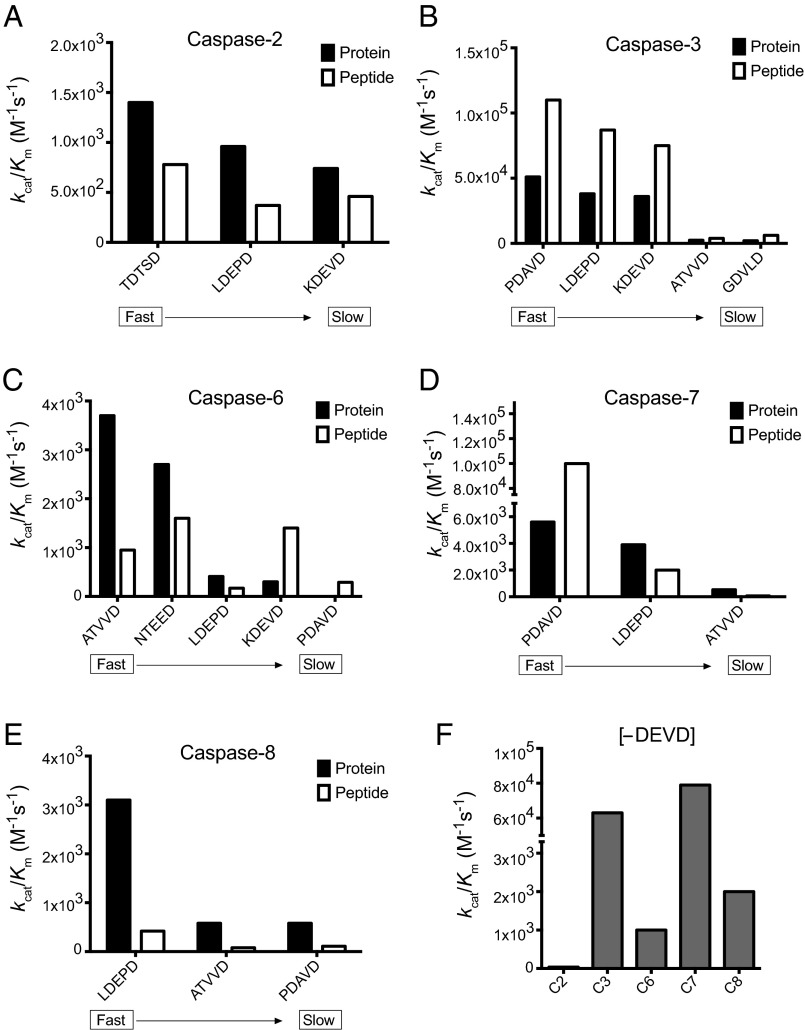
We next wished to quantitatively study the different rates of cleavage toward protein and peptide substrate sites to determine to what extent cleavage rates depended on the primary versus tertiary context. For example, it has been previously reported that a tetrapeptide substrate for caspase-1 is cleaved with similar efficiency to a protein substrate (pro-IL1β) containing the same cleavage site (29). Similarly, we determined the catalytic efficiencies for cleavage of a linear peptide from at least one representative protein substrate identified in the N-terminomics experiment with fast, medium, and slow protein substrate catalytic efficiencies observed for each of the five caspases. We designed tetra-decapeptides corresponding to the P5 to P4ʹ site residues of the protein substrate. At the C terminus of these peptides, we included a five-residue linker (GGSRR) containing the Arg–Arg sequence that would facilitate efficient ionization, thus increasing the sensitivity for the LC-MS/MS measurement. We used SRM to monitor the proteolysis of each peptide over time for each of the five caspases (see Materials and Methods for more details). As earlier, we performed automated nonlinear fitting to determine the rate constant (kcat/KM) of each caspase toward its peptide substrates (Fig. 6 A–E). Remarkably, in most cases, the kcat/KM values for the cleavages were within a factor of two between linear peptide and intact protein substrates. There were a few notable exceptions for caspase-6 and caspase-8, where the protein substrates were two- to fourfold more active. Previous structural studies on caspase-2 showed that it also can bind to the P5 residue, and our N-terminomics data support this as well (SI Appendix, Fig. S3). To test this at the primary sequence level, we made a peptide lacking a P5 residue in the consensus motif (DEVD↓GLGVAGGSRR) (Fig. 6F). Caspase-3 and caspase-7 cleaved this very rapidly (kcat/KM >104), but caspase-2 was 10-fold reduced from cleaving the KDEVD peptide, which extends to P5 (Fig. 6A) (kcat/KM <102). These data corroborate the proteomics and structural data, suggesting the substrate recognition sequence extends to the P5 position for caspase-2.
Fig. 6.
Rates of proteolysis for protein substrates compared with their corresponding peptide sequences. (A–E) Proteolysis rate constants for fast, medium, and slow substrates of caspase-2, caspase-3, caspase-6, caspase-7, and caspase-8, and corresponding tetradecapeptide sequences. P5-P1 residue sequences that match the corresponding protein cut sites are shown on the x-axis. (F) Cleavage rate constants for a DEVD↓GLGVAGGSRR peptide, lacking a P5 residue, highlighting the importance of this residue for caspase-2.
Discussion
Importance of Sequence and Structure in Guiding Specificity in Caspases.
Our data support the view that both sequence and structure determine where a protein is cut. We find caspase-2 and caspase-6 cut their substrates on average 1.2–1.3 times per protein, similar to the average number of cuts per substrate protein seen previously for caspase-3, caspase-7, and caspase-8 (10) and observed globally in apoptosis (18, 30). Less than 1/250 of aspartic acids are cleavage sites, clearly showing that extended sequence is critical (8, 31). Moreover, using structural bioinformatics for sites of known or modeled proteins, we find that caspase-2 and caspase-6 have similar secondary structural preferences for cutting loops > helices > sheets (SI Appendix, Fig. S8). The same pattern was seen globally during apoptosis (18). It is not surprising that protein structure should play a role, as substrates are seen to bind over an eight-residue stretch in the active sites of the proteases (32, 33), which requires unfolding of the cut site in the protein. In future studies, one could compare the proteolytic rates in a native cell lysate with those in a fully denatured or partially predigested cell lysate.
In contrast, we see many instances in which different caspases cut the same protein at exactly the same site, but at radically different rates. This must reflect the intrinsic specificity of each caspase, and not the structural plasticity of the substrate protein. We find the consensus substrate patterns derived from cutting a naive synthetic peptide library using MSP-MS or from natural proteins using subtiligase N-terminomics to be similar, suggesting differences in the intrinsic specificity of the caspases and not the substrate scaffold. This is remarkable, given that the methods are totally orthogonal and that neither substrate set has identical sequences from which consensus patterns are derived. Perhaps most striking is that the differences in rates of cleavage for intact protein substrates compared with their matched synthetic peptides for each of the five caspases are generally small, within a factor of two to four. Caspase-7 has been shown to have a highly charged exosite region distant from the active site that imparts a 30-fold advantage to cleaving the protein, poly (ADP ribose) polymerase 1 (34). Although our data could suggest some exosite component to account for more rapid cleavage of the protein substrate compared with the corresponding peptide substrate, the rate differences are small and could be a result of differences in the kinetic methods used for the protein and peptide substrates.
The use of structural information (secondary structure, solvent accessibility, degrees of disordered regions), in combination with the primary sequence of validated apoptotic substrates, has been previously used to predict caspase cleavage sites (31). In these studies, all possible caspase cleavage sites in the human proteome were ranked on the basis of their support vector machine (SVM) score, representing the likelihood of a given site to be a “real” caspase cleavage site. This generated 5 × 106 predictions of all octapeptides showing an aspartate at P1. We extracted the SVM score for each caspase-2 and caspase-6 cleavage site from this dataset and compared it with the overall predictions. This analysis showed an average SVM score of 0.54 for the caspase-2 cleavage sites observed in our studies, whereas the average SVM score for all predicted sites is only −1.99 (Gaussian distribution ranging from −6 to 4) (SI Appendix, Fig. S9 and Fig. S10). Similarly, we obtained an average SVM score of 0.26 for caspase-6. These data further validate the methodology used by Barkan et al. to predict caspase cleavage sites, exploiting both the primary sequence and structural features of the substrates (31). It is possible these predictive models could be improved for the specific caspases if they used training sets based on the catalytic efficiencies for individual substrates for each of the caspases generated here, instead of a more generic motif derived from apoptosis data that likely emphasizes capase-3, capase-7, and capase-8 cleavage sites.
Dissecting Functional Differences Among Caspase Family Members.
Our studies highlight tools and results that allow greater distinction among caspases in their specificity, cellular substrates, and cell biology. The N-terminomics discovery approach provided reproducible lists of hundreds of natural proteins cleaved by exogenously added caspase-2 and capase-6. Although it is possible that some of these cleavages are derived indirectly from activation of other procaspases in the neutralized extract, we believe this is unlikely for the vast majority of substrates. First, there are many unique substrates for caspase-2 and capase-6 that do not overlap, and for this massive difference to result from secondary activation would require downstream activation of different procaspases for caspase-2 and capase-6 with unique specificity. Second, there are a number of common substrates, cleaved at the same site but at very different rates, that again would require activation of a unique procaspase for each substrate. The progress curves for the SRM analysis of >300 substrates for caspase-2 and capase-6 show no lag phase, which would be expected for a cascade activation event. Furthermore, the substrate cleavage sequence patterns determined from N-terminomics for the caspase-2 and capase-6 differed from each other and were mirrored in the MSP-MS data. The rates of cleavage were similar for the N-terminomics extract and synthetic peptide data.
Gene ontology (GO) analysis provides some clues to help organize the cellular logic for cleaving these client proteins. Caspase-2 cleaves proteins involved in RNA processing, and caspase-6 cleaves proteins involved in cell cycle, DNA damage, and splicing. Both have strong components involving cytoskeleton and cell death, which we have seen in apoptotic datasets (18). Importantly, we believe that the identified substrates are a valuable resource for understanding caspase biology, and the identification of these specific targets could further our understanding of their respective biological function. We find caspase substrates identified to belong to three different categories. First, we identified many previously unidentified substrates of caspase-2 and capase-6 and report 128 (of 235) and 553 (of 871) caspase substrates that have not been previously reported in our apoptotic database of caspase substrates (of 1,268) (8). Second, we identified many known substrates, but in many instances the exact location was not known until our studies here. For example, we could confirm cleavage of BNIP-2 by caspase-6 at IDLD83GLDT, as speculated in previous studies (35). Finally, there are substrates for which the cleavage site is known, but the enzyme that catalyzes the cleavage was until now unknown, and in fact we can deorphan that cleavage. For example, we found that the previously unidentified caspase substrate tuberous sclerosis (TSC1), also called hamartin, is cleaved by caspase-6 at position 638 (TEED638). This substrate has been seen many times in our apoptotic datasets (8), confirming its biological importance, but the identity of the protease remained unknown until now. Hamartin has been shown to confer protection in CA3 hippocampal neurons from the effects of stroke (36). Also consistent with the conjecture in the literature that caspase-6 plays a role in neurons, we identified the substrate PARK7, a peptidase involved in Parkinson’s disease from which an N-terminal fragment was shown to promote apoptosis via increased reactive oxygen species (ROS) production (37). We also identified superoxide dismutase (SODC or SOD1) as a caspase-6 substrate, an important protein capable of misfolding and forming aggregates that leads to amyotrophic lateral sclerosis (also known as Lou Gehrig's disease). The use of different cell lines, such as a neuronal cell line to study the specific involvement of caspase-6 in neurodegeneration, should help clarify caspase biology in specific tissues.
Of the tools we have used to distinguish the functions of caspases, SRM analysis appears to be the most comprehensive. From a technical point of view, it provides multiple-point confirmation that a protein is cleaved. In the substrate discovery phase, biological replicates provide some confidence that the method is reproducible; however, the seven-point kinetic analysis improves the confidence level in determining which proteins are truly cleaved. SRM provides an estimation of catalytic efficiencies for natural caspase substrates (Dataset S7) and can reveal that the rates of cleavage for the same site can differ by more than 100-fold for each enzyme. This strongly supports a view that each caspase likely plays a unique and nonredundant role and opens a window into understanding the catalytic cascade orchestrated by multiple caspases.
Although it is not in the scope of this article to functionally annotate the individual roles of these cleaved substrates in driving caspase biology, we would hope these data will inspire the community to pursue detailed studies on individual substrates. For example, previous proteomic data showed gasdermin D is uniquely cut by caspase-1, but not the other inflammatory caspases, caspase-4 or caspase-5, in cell extracts (9). Recently, two laboratories have shown that gasdermin D is a key mediator in the biology of caspase-1 (38, 39). To facilitate further the analysis of the data here, we show a Venn diagram for the unique and overlapping substrates for which we have SRM data (SI Appendix, Fig. S12). The vast majorities are unique to individual caspases, but some are surprisingly promiscuous. In our SRM dataset, 11 substrates are cleaved by four or five caspases, and one is cleaved by all, DGCR8. DGCR8 complexes the Drosha enzyme and is essential for microRNA processing (40). Broadly, these studies highlight the value of determining both site specificity and kinetic activity for substrates within the context of natural proteins and linear peptides to understand how substrate sequence and structure can functionally distinguish family members involved in posttranslational modifications.
Materials and Methods
Cloning and Recombinant Protein Expression and Purification.
Caspase-2 and caspase-6 were cloned into His6-affinity tag containing vector pET23b (Novagen). The active enzymes were expressed in Escherichia coli BL21 (DE3) pLysS cells (Stratagene). Cells were grown in 2xYT media supplemented with 200 μg/mL ampicillin at 37 °C to an OD600nm at approximately ∼0.6–0.8. Caspase overexpression was induced with 0.2 mM IPTG at 16 °C overnight. Cells were harvested by centrifugation and lysed by microfluidization (Microfluidics). The cell lysates were clarified by centrifugation, and the soluble fractions were purified by Ni-NTA superflow resin (Millipore). His-tagged caspase was then loaded on mono Q ion exchange column for further purification. Peak fractions were combined, concentrated, and stored at −80 °C.
N-Terminal Labeling and Enrichment.
For each discovery replicate, 1 × 109 Jurkat cells were harvested by centrifugation and lysed (0.1% Triton x-100) in the presence of protease inhibitors [5 mM EDTA, 1 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), 1 mM phenylmethanesulfonyl fluoride (PMSF), 10 mM iodoacetamide (IAM)]. Excess cysteine-protease inhibitors were neutralized with 20 mM DTT, and caspase buffer was added [20 mM Hepes, 50 mM NaCl, 15 mM MgCl2, 1.5% (wt/vol) sucrose, 10 mM DTT] before addition of 500 nM exogenous caspase-2 or caspase-6 for up to 4 h. As a function of time, the exogenous caspase activity was then neutralized with 100 μM zVAD-fmk. N-terminal labeling was then performed with 1 μM subtiligase and 1 mM TEVTest4B (SI Appendix, Fig. S2) for 2 h. Tagged protein fragments were precipitated using acetonitrile, then denatured (8 M Gdn-HCl) and reduced (2 mM TECP), and thiols were alkylated (4 mM IAM), before ethanol precipitation. Biotinylated N-terminal peptides were then captured with NeutrAvidin agarose beads for 48 h. The beads were washed using 4 M Gdn-HCl, trypsinized, and released from the beads using TEV protease. The tryptic peptides fractionated into 15 fractions/sample, using high pH reverse phase C18 chromatography, and were then desalted with a C18 Ziptip (Millipore).
Discovery MS.
LC-MS/MS was carried out by reverse phase LC interfaced with a LTQ Orbitrap Velos (Thermo Fisher Scientific) mass spectrometer. A nanoflow HPLC (NanoAcquity UPLC system, Waters Corporation) was equipped with a trap column (180 μm × 20 mm, 5 μm SymmetryC18; Waters) and an analytical column (100 μm × 100 mm, 1.7 μm BEH130C18; Waters). Peptides were eluted over a linear gradient over the course of 60 min from 2% to 30% acetonitrile in 0.1% formic acid. MS and MS/MS spectra were acquired in a data-dependent mode with up to six higher-energy collisional dissociation (HCD) MS/MS spectra acquired for the most intense parent ions per MS. For data analysis, peptide sequences were assigned using the ProteinProspector (v5.10.15) database search engine (prospector.ucsf.edu/prospector/mshome.htm) against the Swiss-Prot human protein database (2013.6.27). Search parameters included a precursor mass tolerance of 20 ppm, fragment ion mass tolerance of 20 ppm, up to two missed trypsin cleavages, constant carbamidomethylation of Cys, variable modifications of N-terminal addition of Abu amino acid, acetylation of protein N terminus, and oxidation of methionine. The identified peptides were searched against a random decoy protein database for evaluating the false-positive rates. The false-discovery rate never exceeded 1.3% at the maximum expectation value of peptide set at 0.05.
SRM Method Development.
The caspase peptides (P1 = D) observed in the caspase-2 and caspase-6 datasets were imported into SkyLine (v2.5.0) (41) to build a scheduled SRM method. For each peptide, based on the spectral library generated from our discovery dataset presented earlier, we selected the seven most intense parent ion-fragment ion transitions (y- and b-series ions only). The retention times observed for each peptide during the discovery experiments were used for this initial method with a window of 15 min. A small portion of the discovery samples was reserved to develop the SRM method. These peptides were injected on an AB Sciex QTRAP 5,500 triple quadrupole mass spectrometer interfaced in-line with a nanoAcquity UPLC system (Waters) identical to that on the LTQ Orbitrap Velos [trapping column: Symmetry C18 Column (0.18 • 20 mm, 5 μm; Waters); analytical column: BEH130 C18 column (0.075 • 200 mm column, 1.7 μm; Waters)]. The SRM data obtained were imported into SkyLine for analysis. Positive peptide identification by SRM was based on detection of at least four coeluting targeted transitions above baseline noise, retention time within ±5 min of that obtained in the discovery dataset, and relative intensity of fragment ions similar to that found in discovery spectral library. Each peptide detected from the caspase-2 and caspase-6 datasets was manually analyzed, and four transitions (of seven) were selected for further method refinement. The preferred transitions were defined as the most intense peaks, least noise or background interference, and higher m/z within the monitored window between 300 and 1,250 Da. Ultimately, our final SRM method included 152 and 471 peptides (plus two controls each) for caspase-2 and caspase-6, respectively. This scheduled SRM method was developed with a retention time window of ±5 min and was used for each point sampled here.
SRM Acquisition, Time Course, and Data Analysis.
The subtiligase N-terminomics enrichment method described earlier was used to monitor the appearance of cleaved products as a function of time, with 2 × 109 and 4 × 109 Jurkat cells for caspase-2 and caspase-6, respectively. For each sample, an aliquot was taken out after 0, 5, 15, 30, 60, 120, and 240 min incubation with 500 nM exogenous protease and quenched with 100 µM zVAD-fmk. Samples were processed for data acquisition on the QTrap 5500 with one and two injections per sample for caspse-2 and caspase-6, respectively. The SRM data for each sample were imported into Skyline for quantification. The sum of the peak area of the four transitions for each peptide was measured as a function of time. The global mean of the control peptides for each point was used to normalize each summation. Nonlinear fitting was performed using Grace (plasma-gate.weizmann.ac.il/Grace/), using pseudofirst-order kinetics: Y = Ymax (1 – ek*t), where Y is the sum of the peak area, Ymax is the peak area at maximal value, t is the time of incubation, and k is the observed rate of proteolysis (kcat/KM), multiplied by the enzyme concentration (E0).
Peptide Cleavage Site Identification by MSP-MS.
Intrinsic substrate specificity was determined with the MSP-MS method (28), with modifications as reported (42). Briefly, 228 tetra-decapeptides were split in two pools of 114 peptides each (500 nM) in assay buffer (20 mM Hepes, 50 mM NaCl, 15 mM Mg2Cl, 1.5% sucrose, 10 mM DTT), to which was added caspase-2 or caspase-6 (both at 200 nM final concentration), or an equal volume of buffer as a no-enzyme control in a final reaction volume of 150 μL for each reaction. Samples were incubated at room temperature (19–22 °C), 30 μL aliquots were removed at 15, 60, 300, and 1,260 min, and quenched with formic acid to a final concentration of 0.4% vol/vol. Samples were desalted by C18 ZipTip (Millipore), and then lyophilized and stored at –80 °C. Peptides were sequenced by LC-MS/MS on an LTQ-OrbitrapXL (Thermo) mass spectrometer equipped with a nanoAcquity UPLC system (Waters), using identical methods to those reported (42). Data were processed and searched as reported (42).
Rate of Proteolysis of Tetra-Decapeptides Peptides Determined by Quantitative MS.
To compare the catalytic efficiency of the caspases against synthetic peptides versus intact protein measurements, a peptide library was designed by selecting a set of fast, medium, and slow substrates for each studied caspase to manage the number of peptides required. Peptides were obtained from GenScript Inc. and Shengnuo Inc. (Dataset S8). The peptide library (4 µM) was added to caspase buffer (20 mM Hepes, 50 mM NaCl, 15 mM MgCl2, 1.5% sucrose, 10 mM DTT) before addition of a given caspase at 500 nM. For each sample, an aliquot was taken out after 0, 5, 15, 30, 60, 120, 240, and 360 min and 24 h incubation and quenched with 100 µM zVAD-fmk. Samples were processed as mentioned earlier, and data were acquired on a QExactive Plus (Thermo), using parallel reaction monitoring mode. The data were imported into Skyline for quantification, as described earlier for protein substrates.
Supplementary Material
Acknowledgments
We thank Justin Rettenmaier, Zachary Hill, Amy Weeks, Matthew Ravalin, Rebecca Levin, Julia Seaman, and Nicholas Agard for helpful discussions and technical help. We are grateful to Julie Zorn, Yinyan Tang, and Dongju Wang for providing caspases. MS was performed at the Bio-Organic Biomedical Mass Spectrometry Resource at the University of California at San Francisco (Alma Burlingame, director), supported by Biomedical Research Technology Program of the National Institutes of Health National Center for Research Resources (Grants P41RR001614 and 1S10RR026662) and the Howard Hughes Medical Institute. A.P.W. is supported by the Damon Runyon Cancer Research Foundation (DRG 11-112 and DFS 16-15). This project was supported in part by grants from the National Institutes of Health (R21CA186007 to C.S.C.; and R01GM081051, R01GM097316, and R01CA154802 to J.A.W.). O.J. is the recipient of a Banting Postdoctoral Fellowship funded by the Canadian Institutes of Health Research and the Government of Canada and a fellowship from the University of California at San Francisco Program for Breakthrough Biomedical Research, which is funded in part by the Sandler Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524900113/-/DCSupplemental.
References
- 1.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281(5381):1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 2.Yuan J, Kroemer G. Alternative cell death mechanisms in development and beyond. Genes Dev. 2010;24(23):2592–2602. doi: 10.1101/gad.1984410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford ED, Wells JA. Caspase substrates and cellular remodeling. Annu Rev Biochem. 2011;80:1055–1087. doi: 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- 4.Lamkanfi M, Declercq W, Kalai M, Saelens X, Vandenabeele P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 2002;9(4):358–361. doi: 10.1038/sj.cdd.4400989. [DOI] [PubMed] [Google Scholar]
- 5.Salvesen GS, Ashkenazi A. Snapshot: caspases. Cell. 2011;147(2):476–476.e1. doi: 10.1016/j.cell.2011.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Dix MM, Simon GM, Cravatt BF. Global identification of caspase substrates using PROTOMAP (protein topography and migration analysis platform) Methods Mol Biol. 2014;1133:61–70. doi: 10.1007/978-1-4939-0357-3_3. [DOI] [PubMed] [Google Scholar]
- 7.Staes A, et al. Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat Protoc. 2011;6(8):1130–1141. doi: 10.1038/nprot.2011.355. [DOI] [PubMed] [Google Scholar]
- 8.Crawford ED, et al. The DegraBase: a database of proteolysis in healthy and apoptotic human cells. Mol Cell Proteomics. 2013;12(3):813–824. doi: 10.1074/mcp.O112.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agard NJ, Maltby D, Wells JA. Inflammatory stimuli regulate caspase substrate profiles. Mol Cell Proteomics. 2010;9(5):880–893. doi: 10.1074/mcp.M900528-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agard NJ, et al. Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proc Natl Acad Sci USA. 2012;109(6):1913–1918. doi: 10.1073/pnas.1117158109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wejda M, et al. Degradomics reveals that cleavage specificity profiles of caspase-2 and effector caspases are alike. J Biol Chem. 2012;287(41):33983–33995. doi: 10.1074/jbc.M112.384552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh JG, et al. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci USA. 2008;105(35):12815–12819. doi: 10.1073/pnas.0707715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timmer JC, et al. Structural and kinetic determinants of protease substrates. Nat Struct Mol Biol. 2009;16(10):1101–1108. doi: 10.1038/nsmb.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talanian RV, et al. Substrate specificities of caspase family proteases. J Biol Chem. 1997;272(15):9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 15.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 16.Graham RK, Ehrnhoefer DE, Hayden MR. Caspase-6 and neurodegeneration. Trends Neurosci. 2011;34(12):646–656. doi: 10.1016/j.tins.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Flygare JA, Arkin MR. Inhibiting caspase-6 activation and catalytic activity for neurodegenerative diseases. Curr Top Med Chem. 2014;14(3):319–325. doi: 10.2174/1568026613666131126224039. [DOI] [PubMed] [Google Scholar]
- 18.Mahrus S, et al. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134(5):866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss A, Wiskocil RL, Stobo JD. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J Immunol. 1984;133(1):123–128. [PubMed] [Google Scholar]
- 20.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc Natl Acad Sci USA. 2009;106(20):8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornberry NA, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272(29):17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 22.Klaiman G, Petzke TL, Hammond J, Leblanc AC. Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteomics. 2008;7(8):1541–1555. doi: 10.1074/mcp.M800007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igarashi Y, et al. PMAP: databases for analyzing proteolytic events and pathways. Nucleic Acids Res. 2009;37(Database issue):D611–D618. doi: 10.1093/nar/gkn683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nat Methods. 2009;6(11):786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- 25.Stennicke HR, Renatus M, Meldal M, Salvesen GS. Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem J. 2000;350:563–568. [PMC free article] [PubMed] [Google Scholar]
- 26.McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15(2):322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- 27.Zorn JA, Wolan DW, Agard NJ, Wells JA. Fibrils colocalize caspase-3 with procaspase-3 to foster maturation. J Biol Chem. 2012;287(40):33781–33795. doi: 10.1074/jbc.M112.386128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donoghue AJ, et al. Global identification of peptidase specificity by multiplex substrate profiling. Nat Methods. 2012;9(11):1095–1100. doi: 10.1038/nmeth.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rano TA, et al. A combinatorial approach for determining protease specificities: application to interleukin-1beta converting enzyme (ICE) Chem Biol. 1997;4(2):149–155. doi: 10.1016/s1074-5521(97)90258-1. [DOI] [PubMed] [Google Scholar]
- 30.Dix MM, Simon GM, Cravatt BF. Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell. 2008;134(4):679–691. doi: 10.1016/j.cell.2008.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barkan DT, et al. Prediction of protease substrates using sequence and structure features. Bioinformatics. 2010;26(14):1714–1722. doi: 10.1093/bioinformatics/btq267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chai J, et al. Crystal structure of a procaspase-7 zymogen: mechanisms of activation and substrate binding. Cell. 2001;107(3):399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 33.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384(Pt 2):201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boucher D, Blais V, Denault JB. Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis. Proc Natl Acad Sci USA. 2012;109(15):5669–5674. doi: 10.1073/pnas.1200934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valencia CA, Cotten SW, Liu R. Cleavage of BNIP-2 and BNIP-XL by caspases. Biochem Biophys Res Commun. 2007;364(3):495–501. doi: 10.1016/j.bbrc.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 36.Papadakis M, et al. Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat Med. 2013;19(3):351–357. doi: 10.1038/nm.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert G, et al. The caspase 6 derived N-terminal fragment of DJ-1 promotes apoptosis via increased ROS production. Cell Death Differ. 2012;19(11):1769–1778. doi: 10.1038/cdd.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kayagaki N, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526(7575):666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 39.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 40.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Donoghue AJ, et al. Destructin-1 is a collagen-degrading endopeptidase secreted by Pseudogymnoascus destructans, the causative agent of white-nose syndrome. Proc Natl Acad Sci USA. 2015;112(24):7478–7483. doi: 10.1073/pnas.1507082112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.