Abstract
Genetic studies of autism spectrum disorder (ASD) have established that de novo duplications and deletions contribute to risk. However, ascertainment of structural variants (SVs) has been restricted by the coarse resolution of current approaches. By applying a custom pipeline for SV discovery, genotyping, and de novo assembly to genome sequencing of 235 subjects (71 affected individuals, 26 healthy siblings, and their parents), we compiled an atlas of 29,719 SV loci (5,213/genome), comprising 11 different classes. We found a high diversity of de novo mutations, the majority of which were undetectable by previous methods. In addition, we observed complex mutation clusters where combinations of de novo SVs, nucleotide substitutions, and indels occurred as a single event. We estimate a high rate of structural mutation in humans (20%) and propose that genetic risk for ASD is attributable to an elevated frequency of gene-disrupting de novo SVs, but not an elevated rate of genome rearrangement.
Introduction
Structural variants (SVs), such as deletions and duplications, are a major source of genetic differences between humans and contribute significantly to risk of common disease.1 In particular, studies of copy-number variation (CNV) have been seminal in establishing a role for rare genetic variants in the etiology of autism spectrum disorder (ASD [MIM: 209850]).2, 3 Despite this success, characterization of SVs from individual genomes remains a major challenge. Identification of SVs in human populations and disease has been restricted by the limited sensitivity of microarray- and sequencing-based approaches.4, 5, 6
Large CNVs detectable by microarrays represent a small fraction of structural variation in the genome. Recent methodological advances have enabled the discovery of a wide variety of SV classes from whole-genome sequencing (WGS) datasets, including small deletions and duplications down to 50 bp in length, inversions, translocations, mobile-element insertions (MEIs), and more-complex rearrangements. By applying a combination of specialized methods, each tailored to specific classes of variation, the 1000 Genomes (1000G) Project has produced the most complete catalog of SVs to date by creating an integrated call set of eight classes of SV by using low-coverage (7.4×) WGS in 2,504 human genomes.7 In a study of 250 population-control families, analysis of low-coverage (13×) WGS data allowed for detection of de novo deletions, tandem duplications, and MEIs.8 However, advanced analytical methods for SV discovery and genotyping have not been applied in genetic studies of ASD. Initial forays into the application of WGS to the detection of SVs in neurodevelopmental disorders have been restricted to CNVs larger than 1 kb,9 focused on a subset of variant calls prioritized by putative clinical relevance,9, 10 or limited to the characterization of CNVs previously detected by microarrays.6
More comprehensive ascertainment of SV is needed for elucidating the genetic mechanisms that underlie ASD risk. In this study, we applied a suite of complementary SV-discovery methods, coupled with custom methods for SV genotyping and detection of de novo mutations, to assess global patterns and rates of structural mutation in ASD.
Material and Methods
Recruitment
Individuals were primarily referred from clinical departments at Rady Children’s Hospital, including the Autism Discovery Institute, the Departments of Psychiatry, Neurology, and Speech and Occupational Therapy, and the Developmental Evaluation Clinic. Further referrals came directly through our project website. The Autism Center of Excellence at the University of California, San Diego (UCSD), contributed a further 11 trios. Each child included in the study has an existing ASD diagnosis and received a diagnosis of ASD on the basis of an evaluation by a licensed clinician.11 Prior to appointments, families were provided with institutional-review-board-approved consent forms and Health Insurance Portability and Accountability consent forms. DNA was obtained from 5 ml blood draws. We recalled a subset of individuals with specific genetic findings to confirm the original ASD diagnoses. These included individuals with SVs in TMEM185A (MIM: 300031), TESC (MIM: 611585), NRXN1 (neurexin 1 [MIM: 600565]), and CACNG2 (MIM: 602911). A diagnosis of ASD was confirmed in all affected individuals.
WGS
WGS was performed on 246 samples, which included 11 monozygotic twin pairs. One sibling from each twin pair was excluded from the dataset, which brought the final sample size to 235. WGS of 206 samples was performed with an Illumina HiSeq at the Illumina Fast Track service laboratory in San Diego. For 161 samples, preparations consisted of 313 bp libraries and 100 bp paired-end reads. For the remaining 45 samples, library size and read length were 493 and 125 bp, respectively. In addition, a subset of our data consisted of 40 samples sequenced with an Illumina HiSeq at the Beijing Genomics Institute as described previously (SVs were not reported in this publication),12 and genomes were realigned to the human reference genome (UCSC Genome Browser build hg19) with the Burrows-Wheeler Aligner (BWA-mem version 0.7.12).13
To generate sequence alignment and variant calls on families, we implemented our WGS analysis pipeline on the Comet compute cluster at UCSD. Short reads were mapped to the hg19 reference genome by BWA-mem v.0.7.12.13 Subsequent processing was carried out with SAMtools v.1.2,14 Genome Analysis Toolkit (GATK) v.3.3,15 and Picard Tools v.1.129, which consisted of the following steps: sorting and merging of the BAM files, indel realignment, removal of duplicate reads, and recalibration of base quality scores for each individual.16
SV Detection
We utilized three complementary algorithms to detect SVs. ForestSV is a statistical-learning approach that integrates a wide variety of features, including signal from read depth and discordant paired-ends, from WGS data to identify deletions and duplications.17 Lumpy uses signal from discordant paired ends and split reads to identify breakpoints for deletions, duplications, inversions, translocations, and complex SVs.18, 19 Finally, Mobster uses signal from discordant paired ends and split reads in combination with consensus sequences of known active transposable elements to identify MEIs.20
SV Post-processing
We assembled call sets of deletions, duplications, inversions, complex SVs, and MEIs detected in 246 individuals. For monozygotic twins, we generated a consensus call set for each twin pair from the raw SV calls as an initial processing step.
SV Filtering
SVs were filtered if they overlapped centromeres, segmental duplications (genomicSuperDups), regions with low mappability and 100 bp reads (wgEncodeCrgMapabilityAlign100-mer), and regions subject to somatic V(D)J recombination (parts of antibodies and T cell receptor genes) by 50%. Genome annotations used for filtering were downloaded from UCSC Genome Browser build hg19. Filtered regions are provided in the Web Resources.
ForestSV
ForestSV was run with default parameters. Large SVs that were fragmented into multiple calls by ForestSV were stitched together as a function of their separation distance and divided by the total length of the individual calls. SVs between individuals were collapsed on the basis of >50% reciprocal overlap, and the same median start and end coordinates were assigned to each call.
Lumpy
SVs were called within families according to the default parameters of the SpeedSeq SV pipeline (v.0.0.3a), which uses Lumpy (v.0.2.9) to process samples and SVtyper (v.0.0.2) to genotype variants.18, 19 The pipeline outputs deletions, duplications, inversions, and breakpoints that cannot be assigned to one of the three classes. To detect complex SVs, we wrote a custom algorithm to cluster overlapping pairs of breakpoints and resolve the patterns and ordering of breakpoint alignments to the reference genome. We detected five classes of complex SVs, both intra- and interchromosomal. For intrachromosomal events, if two or more sets of breakpoints overlapped within an individual, we considered them to be part of the same SV event, and then on the basis of the patterns and orientations of discordant paired ends, we determined the SV type (as shown in Figure S3). For interchromosomal duplications, we required that two or more sets of breakpoints map to the same two chromosomes and that at least one breakpoint from each set map within one read length of each other (which restricted the size of target-site duplications and deletions that we could detect). Calls between individuals were considered to be the same SV if they shared the same start and end coordinates within a margin of error defined by the read length (100 bp for most samples).
Mobster
Mobile elements were called by Mobster v.0.1.6 within families and were included in the call set if they had at least five reads, including discordant paired ends at both the 3′ and 5′ sides of the insert point,20 supporting the call in one individual in the family. The parameters used in the Mobster properties file are available for download in the Web Resources. Calls between individuals were considered to be the same SV if they shared the same insertion coordinates within a margin of error defined by the read length (100 bp for most samples).
SV Genotyping and Calling of De Novo Mutations
We utilized gtCNV and SVtyper,18 two complementary methods for assigning genotype likelihoods to SVs. Specifically, gtCNV integrates signal from depth of coverage, paired ends, and split reads and is most suitable for CNVs (i.e., deletions or duplications). SVtyper does not use coverage signal and therefore is more suitable for genotyping balanced SVs and the smallest (<500 bp) CNVs. We used estimates of genotype likelihood to derive a quality score for each SV site, defined as the median genotype likelihood for individuals genotyped as non-reference. We systematically genotyped a merged set of CNV calls (biallelic deletions and duplications) from ForestSV and Lumpy. CNVs that were called by both methods (i.e., overlapping CNVs in the same individual) and had different breakpoints were both genotyped, and the coordinates with the best genotype likelihoods were retained in the SV call set (Data S1). All 246 individuals were genotyped with gtCNV for all CNVs called by the two algorithms.
We filtered the finalized call sets solely on quality scores by using thresholds of 12 for deletions genotyped with gtCNV, 8 for duplications genotyped with gtCNV, and 100 for SV breakpoints genotyped with SVtyper. The false-discovery rate (FDR) of the combined call set was estimated from Illumina 2.5M SNP array data with the intensity-rank-sum (IRS) test implemented in the Structural Variation Toolkit (see Web Resources).
From the finalized call set, we extracted de novo mutations that had a non-reference genotype in the child, reference genotypes in both parents, and a parent allele frequency of 0 in the cohort.
gtCNV
To classify CNV genotypes, we developed gtCNV, a likelihood-based support-vector-machine (SVM) approach that genotypes deletions and duplications. The classifier was trained on high-coverage CNV data from 27 individuals sequenced as part of the 1000G Project.21
When training the SVM, we selected read depth, discordant paired ends, and split reads as features. We extracted features for all deletion and duplication calls made by ForestSV and Lumpy. When determining coverage, we masked regions overlapping segmental duplications. For each SV, we calculated mean coverage, which we then normalized to the mean chromosomal coverage for each sample. We also extracted all discordant paired ends and split reads (mapping quality > 20) by implementing the SAMtools application programming interface for Python in pysam.14 Discordant paired ends were defined as reads with insert sizes more than 5 SDs from the mean.
The SVM training utilized a radial-basis-function (RBF) kernel, which we implemented in Python by using scikit-learn.22, 23 In order to determine the optimal parameters of the RBF kernel, we used the IRS test to estimate the FDR for deletions and duplications in the call set. Optimal parameters were C = 1 and gamma = 0.005, which had an FDR of 7.0% for deletions at a quality score of ≥12. The optimal parameters for duplications were C = 1 and gamma = 0.01, which had an FDR of 9.2% at a quality score of ≥8. The gtCNV software can be found on GitHub (see Web Resources), and the method will be further detailed in a companion paper in the near future.
SVtyper
Genotyping of Lumpy calls was performed with SVtyper as part of the SpeedSeq SV pipeline.18 SVtyper is a Bayesian SV-breakpoint-genotyping algorithm that estimates the likelihood that a genotype is non-reference on the basis of allele counts at each junction. A quality score for each individual SV locus was derived on the basis of the median genotype likelihood for individuals genotyped as non-reference. An optimal quality-score threshold for Lumpy was determined as described in the section above. We performed family-based calling and genotyping for Lumpy calls and kept variants that had a median quality score ≥ 100 across the cohort. For complex variants with multiple overlapping breakpoints, we kept variants that had a median quality score ≥ 100 for at least one breakpoint.
We assessed the performance of SVtyper by using the IRS test described above. The FDR of CNV sites was 3.3% for deletions, 9.5% for tandem duplications, 0% for deletions in complex events, and 11.5% for duplications in complex events (complex combined FDR = 7.5%).
Sensitivity Analysis of CNV Detection and Genotyping Pipeline
To assess the sensitivity of our CNV-calling pipeline, we applied it to 27 samples sequenced with a high-coverage PCR-free protocol in phase 3 of the 1000G Project. Raw CNV calls from ForestSV and Lumpy were merged, genotyped, and then filtered as detailed above. Because our genotyping method, gtCNV, was originally trained on these data, we used a leave-one-out strategy to generate genotype likelihoods for calls in each sample (we excluded the test sample from the training set before genotyping SV calls in the sample). We then intersected our call set to the non-reference deletions and biallelic duplications found in the 1000G phase 3 SV call set for these 27 samples. Calls that had 50% reciprocal overlap with phase 3 CNVs were counted as overlapping within each sample. Sensitivity values were then calculated and binned according to CNV size (<100 bp, 100 bp to 1 kb, and >1 kb).
Parent of Origin of De Novo SVs
For deletions, we extracted from the VCF file generated by GATK HaplotypeCaller all SNPs that mapped within the deletion breakpoints and that were homozygous alternate (alt) in the proband, heterozygous in one parent, and homozygous reference in the other parent. The parental origin was then inferred to be on the haplotype of the parent who had homozygous reference alleles for informative SNP markers. For duplication CNVs, we extracted all SNPs that mapped within the breakpoints and that were heterozygous in the proband, had a ∼2:1 ratio of reference to alt alleles (or vice versa), and were heterozygous in one parent and homozygous reference in the other parent. The allele with double the expected number of reads indicates which parental haplotype the duplication originated on.
In the case of the MEI in C3orf35 (chromosome 3 open reading frame 35 [MIM: 611429]), we validated the MEI (and flanking 3′ UTR sequence) by cloning it into a vector and sequencing it. The paternal origin was determined from an informative variant within the cloned locus (rs35484794).
From the exonic NRXN1 deletion, which is de novo in the mother, we selected three SNPs (rs2042471, rs12468395, and rs13031783) that were hemizygous in the mother. SNPs were PCR amplified and Sanger sequenced from the mother and grandparents. We further performed paternity testing (DNA Solutions) of saliva and confirmed that both grandparents are the biological parents of the mother.
CNV Validation by SNP Microarray
We performed genome-wide assessment of CNVs in the majority of individuals (n = 205) in this study via Illumina 2.5M SNP microarrays. CNVs were detected by trio-based calling implemented in the PennCNV algorithm24 and were retained if they had at least eight supporting probes. For de novo CNVs with fewer than eight probes, we assessed the median log R ratio (LRR) of the probes within the CNV locus for all individuals in the study and considered the CNV validated if the child’s median LRR was more than 2 SDs below (for deletions) or above (for duplications) the mean in the cohort.
PCR Validation of SVs
We designed PCR primers flanking breakpoints for small CNVs, complex SVs, and balanced SVs. We attempted validation of nine putative de novo MEIs, six Alu insertions, and three L1 insertions. For Alu elements, primers were designed to flank the insertion point, and for L1 elements, one primer was designed to flank the insertion point, and two were designed within the element (both sense and antisense because the orientations of the insertions were unknown). Primers for SV validation are listed Table S5. PCR amplification validated three de novo Alu elements when it was run on an agarose gel; the remaining putative de novo variants were false positives. PCR products were cloned with TOPO-TA vectors. Resulting clones were screened and sequenced with M13 primers from both ends of the vector insert. We assigned the subfamily by using BLAST to compare the sequence results with the consensus Alu sequences.
Assembly of Breakpoints
For deletion and duplication SVs, we used Velvet25 to perform de novo assembly of clipped reads and determined the precise breakpoint down to a single-base-pair resolution for 60.8% of deletions (n = 11,168). We observed that 17.9% of deletions had an inserted sequence at the breakpoint. For duplications, we determined the breakpoints for 31% (n = 733). Breakpoint positions were assigned to SV coordinates where applicable in Data S1.
SV Burden
We assessed the burden of de novo SVs between ASD individuals in this study and the combined control individuals from this study and a study from the Genome of the Netherlands (GoNL) Consortium by using a case-control permutation test implemented in PLINK.26
MEI Permutations
To permute the enrichment and/or depletion of MEI insertions in genomic features, we used BedTools27 to shuffle the position of the observed MEI sites across the genome while maintaining the orientation of the MEI (sense or antisense) but excluding any overlap with the filtered regions above. We counted the number of times that a shuffled MEI overlapped the following genomic features: exons, introns (sense and antisense orientations separately), promoters, 5′ UTRs, and 3′ UTRs. We performed 10,000 permutations and compared the observed overlap to the expected overlap.
Overlap between SVs and Known Polymorphic SV Events
Deletions, duplications, and inversions were intersected with the 1000G SV call set with BedTools and were considered part of the same polymorphic or recurrent SV event if they had >50% reciprocal overlap. MEIs were considered to overlap if their insertion point was located within 100 bp of an MEI event of the same class from the 1000G integrated SV set or the database of retrotransposon insertion polymorphisms.
Overlap between SVs and Published CNV Data
We permuted the expected overlap between SVs and CNV regions (CNVRs) previously associated with ASD, intellectual disability (ID), and developmental delay (derived from two large-scale microarray CNV studies28, 29). These CNVRs are significantly more abundant in affected individuals than in control individuals and are either hotspots flanked by segmental duplications or enrichment peaks derived from the intersection of multiple breakpoints.
Using BedTools, we randomly shuffled the position of the observed rare SVs in children (including SVs that are de novo or have a frequency < 1% in parents) while maintaining the size of the CNVs and the chromosome but excluding any overlap with sequencing gaps. We counted the number of times that at least 90% of a shuffled CNV overlapped a CNVR. When a single gene was implicated by a CNVR, we stipulated that the CNV had to overlap only one exon of the gene to be counted. This method is conservative because it allows small CNVs overlapping only a small proportion of larger CNVRs to be counted, i.e., the overlap is not required to be reciprocal.
When performing gene-set enrichment analysis with published exome sequencing data, we determined the number of SVs overlapping genes affected by one or more loss-of-function SNVs and indels in studies of ASD and ID, and we then permuted the SV positions while maintaining the total number of genes disrupted.
Detection of De Novo SNVs and Indels
We called putative de novo SNVs by using ForestDNM, a custom machine-learning pipeline that uses a random forest classifier to predict the validation status of putative de novo SNVs identified by the GATK UnifiedGenotyper.12 Putative de novo indels were called with three different algorithms: GATK, Platypus, and Scalpel.15, 30, 31 First, we called variants genome-wide by using GATK and Platypus. Then, we used Scalpel for targeted de novo assembly of the locus around this set of putative de novo indels. We kept de novo indels called by at least two out of three algorithms. We then excluded (1) any indels observed more than once in the GATK or Platypus VCF files of the entire cohort and (2) common indels in the population from 1000G data. The genome-wide burden of de novo SNVs in case and control individuals was 66.9 and 63.3, respectively; for indels, it was 6.67 and 6.11 for case and control individuals, respectively. Analysis of de novo SNVs and indels will feature in a future publication.
Mutational Clustering
To assess whether de novo SVs cluster with nucleotide substitutions or indels, we used a window-based permutation approach. We took windows of 100 bp, 1 kb, 10 kb, 100 kb, 1 Mb, and 10 Mb around the breakpoints of de novo SVs and intersected the windows with de novo SNVs and indels in the same individuals. We then used BedTools to shuffle the position of these windows in the genome either randomly (excluding regions that were filtered during SV calling) or across detected inherited SV breakpoints and calculated the expected number of windows overlapping DNMs by performing 100,000 permutations.
Transmission-Disequilibrium Test
Using a haplotype-based group-wise transmission-disequilibrium test32 and assuming an additive model, we tested whether variants private to families in our study and not present in the 1000G call set were transmitted to affected children more than expected by chance.
Results
Genome Sequencing Uncovers a Diverse Landscape of Structural Variation
We recruited ASD-affected individuals and their families (235 subjects, including 71 affected individuals and 26 typically developing siblings) from Rady Children’s Hospital, San Diego, and local pediatric clinics. WGS of blood-derived genomic DNA was performed at a mean coverage of 40.6× (Table S1 and Material and Methods).
We developed a SV-discovery pipeline that utilizes a combination of three specialized methods each optimized to capture a specific subtype of variation (Figure S1 and Material and Methods): (1) ForestSV17 is a statistical-learning approach that we developed to integrate a variety of features from WGS data into a random-forest classifier and is optimized to detect deletions and duplications; (2) Lumpy19 utilizes information from discordant paired ends and split reads and is optimal for the detection of balanced rearrangements, such as inversions and translocations; and (3) Mobster20 utilizes discordant paired ends and split reads to detect MEIs. As we have shown here, this combination of methods is highly efficient and provides accurate detection of most known classes of SV. For each subject, unfiltered SV calls from the three methods were merged into a set of consensus calls (see Material and Methods).
The final call set from our 235 study subjects included 1,225,067 SVs (5,213 SVs/genome) from 29,719 sites (Figure 1). The primary variant calls comprised seven major classes, including deletions (3,383 alleles/individual; 18,359 sites), duplications (423 alleles/individual; 2,360 sites), inversions (51 alleles/individual; 211 sites), and four classes of MEIs (1,105 alleles/individual; 7,915 sites) (Figure 1, Data S1, and Table S2). FDRs for deletion and duplication calls were estimated from Illumina 2.5M SNP array data (with the Structural Variation Toolkit; see Material and Methods), which were collected on a majority of samples (n = 205) in our study. The FDR was determined to be 7.0% for deletions and 9.2% for duplications (Figure S2). We assessed our sensitivity for detecting deletions and biallelic duplications by applying our methods to 27 individuals sequenced at high coverage in the 1000G Project.21 We captured a majority (59%) of SVs in the phase 3 call set. In addition, 40% of deletions and 99% of duplications were unique to our call set (Figure S2). The sensitivity for detecting 1000G phase 3 deletions was 75%, 61%, and 25% for lengths >1,000 bp, 100–1,000 bp, and <100 bp, respectively (Table S3).
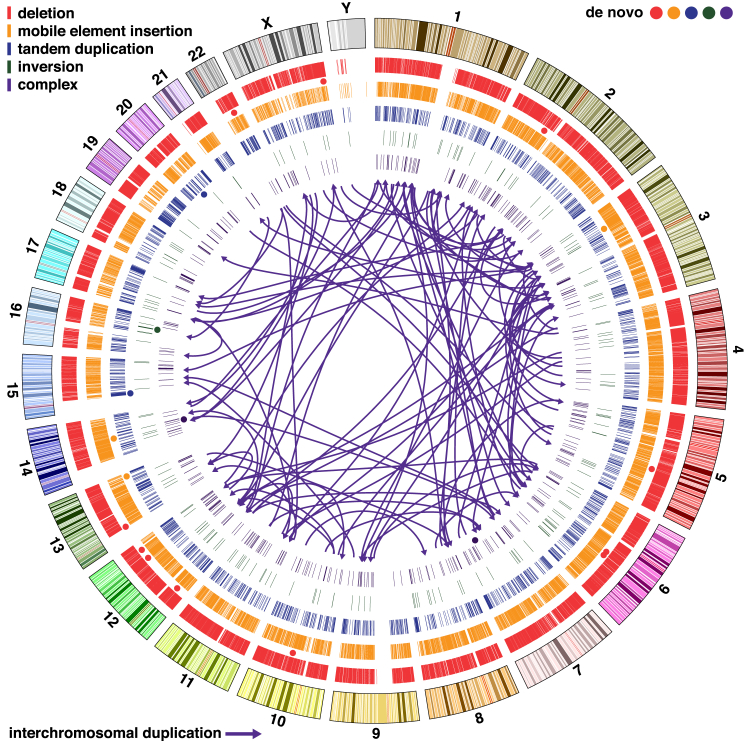
Figure 1.
Structural Variation Detected from WGS in 235 Individuals
Circos plot in which concentric circles represent the following (from outermost to inner): ideogram of the human genome with colored karyotype bands (UCSC Genome Browser build hg19), deletions, MEIs (four different classes), tandem duplications, balanced inversions, and complex SVs (four different classes). Circles indicate the location of de novo SVs, and their colors match the five SV types. Arrows represent interchromosomal duplications.
The complete call set and detailed descriptive information for all calls are provided in Data S1. A comparison of our SV call set with the phase 3 SV call set from the 1000G Project is described in Figure S2.
Detection of Complex Rearrangements
A recent study using a combination of microarrays and sequencing of large-insert (“jumping”) libraries has shown that a variety of complex SVs are observed in a subset (24%) of ASD.6 In all subjects in our study, we identified dense clusters of SVs with overlapping breakpoints. Most of such instances could be resolved into one of four “complex” SV classes: tandem duplications with nested deletions, non-tandem duplications, deletion-inversion-deletion events, and duplication-inversion-duplication events6 (Figure S3 and Table S4). Non-tandem duplications were the most common form of complex SV (Table S4), and these have not been documented in previous genome-wide studies. Insertions occurred in direct and inverted orientations with equal probability, and 22% were interchromosomal (Figure 1C [arrows] and Figure S4). The majority (73%) had target-site deletions at the insertion point. We detected an average of 251 complex SVs per individual; thus, complex SVs represent common forms of genetic variation in humans.
SV Genotyping and Detection of De Novo Mutations
Previous studies by our group and others found that de novo SVs occur at significantly higher rates in ASD-affected individuals than in typically developing offspring.2, 4 The more comprehensive SV dataset here provides an opportunity to investigate de novo structural mutation with much greater sensitivity. Identification of de novo SVs from WGS data, however, is a significant challenge. Given the expected number of false positives in our call set (>200/subject), the overwhelming majority of putative de novo mutations will be errors.8 To address this challenge, we performed joint genotyping of SVs across all samples by using gtCNV, a SVM-based algorithm we developed here to estimate genotype likelihoods for deletions and duplications on the basis of multiple features including read depth, discordant paired ends, and split reads. Breakpoints called by Lumpy were genotyped with SVtyper, which performs Bayesian likelihood estimation on the basis of the observed discordant paired ends and split reads at each junction.18 Putative de novo SVs were validated by microarray analysis or through PCR and Sanger sequencing (Table S5). We detected 31 de novo SVs and validated 19 in 97 offspring. De novo SVs consisted of a diversity of mutation classes, including deletions (n = 11), duplications (n = 2), inversions (n = 1), Alu insertions (n = 3), and complex SVs (n = 2; Table 1); their positions are indicated by circles in the Circos plot in Figure 1. The overall FDR for de novo SV calls was 39% (12/31). Compared to the 93% FDR from a recent study by the GoNL Consortium,8 this represents a substantial improvement in the accuracy of calling de novo SVs. Furthermore, 12 false-positive de novo mutations in a call set of 29,719 SV sites represents a very low error rate overall (0.04%).
Table 1.
De Novo SVs
| hg19 Coordinatesa | Type | Size (bp)b | Locus | Mechanism | Parental Origin | ID | Status | Gender |
|---|---|---|---|---|---|---|---|---|
| chr3: 37,476,966–37,476,979 | AluYb8 | 14|134 | C3orf35 (3′ UTR) | MEI | paternal | 74-0115-01 | ASD | female |
| chr13: 107,803,685–107,803,696 | AluYa5 | 12|277 | intergenic | MEI | NA | REACH000120 | ASD | male |
| chr7: 112,115,899–112,123,778 | complex | 70|454|128 | IFRD1, LSMEM1 | MMBIR | NA | REACH000141 | ASD | male |
| chr14: 61,548,613–61,552,405 | complex | 23|140 | SLC38A6 (intron 16/16) | MMBIR | paternal | REACH000182 | ASD | male |
| chr2: 74,482,718–74,511,562 | deletion | 28,844 | SLC4A5 (5/32 exons) | MMBIR | NA | REACH000239 | ASD | male |
| chr6: 93,142,763–93,142,954 | deletion | 192 | intergenic | NHEJ | NA | REACH000001 | ASD | male |
| chr10: 69,823,502–69,823,806 | deletion | 305 | HERC4 (intron 3/25) | MMBIR | NA | REACH000288 | ASD | male |
| chr12: 117,519,631–117,537,968 | deletion | 18,338 | TESC (1/8 exons) | NHEJ | paternal | REACH000163 | ASD | male |
| chr13: 21,131,642–21,135,198 | deletion | 3,557 | intergenic | NAHR | paternal | REACH000292 | ASD | male |
| chr22: 36,969,581–37,097,776 | deletion | 128,195 | CACNG2 (1/4 exons) | NHEJ | paternal | REACH000001 | ASD | male |
| chrX: 148,682,301–148,736,750 | deletion | 54,450 | TMEM185A (4/7 exons) | NAHR | maternal | REACH000145 | ASD | male |
| chr15: 22,701,351–28,574,000 | duplication | 5,872,650 | 15q11.2–13.1 (PWS/AS)c | NAHR | maternal | REACH000316 | ASD | female |
| chr20: 29,804,201–30,388,100 | duplication | 583,900 | 20q11.21 (18 genes) | NAHR | paternal | REACH000141 | ASD | male |
| chr16: 60,410,404–61,926,470 | inversion | 1,516,067 | CDH8 (9/12 exons) | NHEJ | NA | L7H6W_01 | ASD | male |
| chr14: 58,985,087–58,985,102 | AluYa5 | 16|312 | KIAA0586 (intron 30/31) | MEI | NA | REACH000176 | control | female |
| chr5: 111,391,882–111,398,120 | deletion | 6,238 | intergenic | NHEJ | maternal | REACH000300 | control | female |
| chr6: 100,911,772–100,916,248 | deletion | 4,476 | SIM1 (upstream) | NHEJ | NA | REACH000162 | control | male |
| chr12: 19,257,899-19,293,874 | deletion | 35,975 | PLEKHA5 (3/35 exons) | NHEJ | maternal | REACH000076 | control | female |
| chr12: 98,296,358–98,297,335 | deletion | 977 | intergenic | NHEJ | NA | REACH000236 | control | male |
Abbreviations are as follows: AS, Angelman syndrome; ASD, autism spectrum disorder; NA, not available; MEI, mobile-element insertion; MMBIR, microhomology-mediated break-induced replication; NAHR, non-allelic homologous recombination; NHEJ, non-homologous end joining; and PWS, Prader-Willi syndrome.
Coordinates are based on breakpoint sequence alignments; however, coordinates for three NAHR-mediated SVs (15q11.2–13.3, 20q11.21, and Xq28) are based on ForestSV boundaries.
Pipes (|) separate the sizes of individual elements within complex structural variants; further details can be found in Figure 4 and Table S5.
Critical region for PWS and AS.
High Rate of De Novo Structural Mutation in Humans
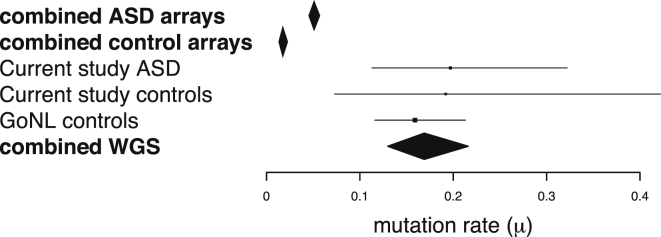
In this study, de novo SVs were observed in 19.7% of affected individuals (95% confidence interval [CI] = 11.3%–32.2%) and 19.2% of control individuals (95% CI = 7.3%–42.2%), a 3-fold and 10-fold higher rate, respectively, than what was reported in previous studies of ASD (Figure 2). The higher rate of de novo SV observed here is driven by the fact that our methods have increased sensitivity for detecting copy-neutral and smaller SVs. The majority of de novo SVs (58% [11/19]) were undetectable by a high-density (2.5M) SNP microarray (Figure S6).
Figure 2.
Frequency of De Novo SVs
A forest plot indicates the average mutation frequency per genome (μ) from published microarray studies of ASD, from the ASD-affected and control individuals in our study, and from a whole-genome study from the GoNL Consortium. Error bars represent the 95% CIs according to a Poisson distribution, and boxes are proportional to the sample sizes tested.
Unlike in previous studies, the rate of de novo SVs was not higher in affected individuals (Figure 2) than in control individuals in this study (p = 0.39) or than in a combined set of 276 control trios from this study and a study from the GoNL Consortium (p = 0.17). Although the mutation rate was not elevated in affected individuals, de novo SVs were larger (median length of 10.9 and 1.2 kb in ASD and control individuals, respectively; permutation p = 0.026), and a greater proportion of SVs intersected an exon of at least one gene (11.1% and 2.8% in case and control individuals, respectively; permutation p = 0.01).
Fine-Scale Characterization of De Novo SVs
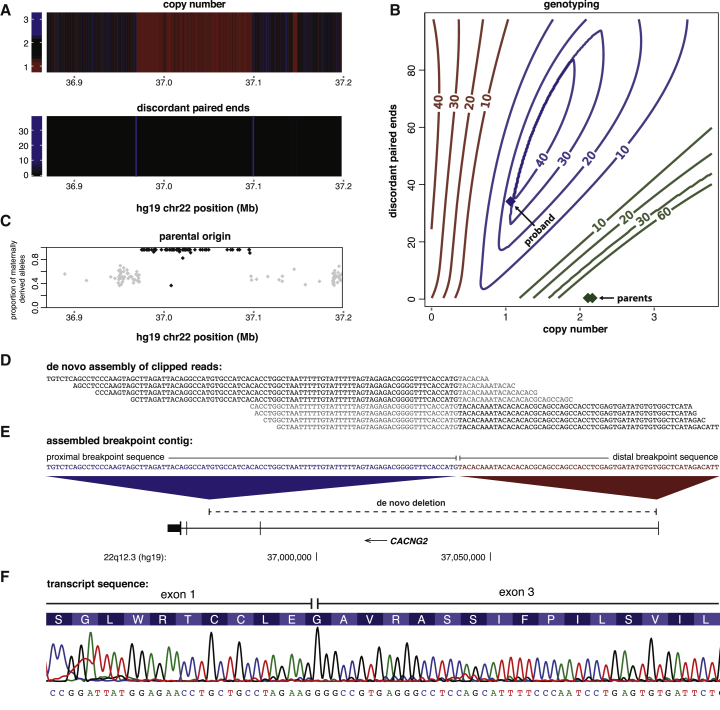
Multilayered genetic information extracted from the genome sequences of individuals can provide further insights into the origin, mutational mechanism, and functional impact of de novo SVs. For de novo events, we assessed the parent of origin and the junction sequences obtained by local de novo assembly of breakpoints. As an illustrative example, a de novo CACNG2 deletion detected in an individual is presented in Figure 3. After detection of the deletion (Figure 3A) and genotype-likelihood estimation of family members (Figure 3B), the paternal origin of the deletion could be inferred from allelic ratios of SNPs within the deleted region (Figure 3C). The complete sequence of the breakpoint junction could be assembled from reads that partially mapped near the deletion boundaries (Figure 3D). From the assembled breakpoint sequence, we inferred that the deletion eliminates exon 2 and all but 634 bp of intron 1 (Figure 3E) and that the deletion occurred by a non-homologous end-joining mechanism.33 The mutant transcript lacking exon 2 of CACNG2 was confirmed in a fibroblast line derived from the individual and is predicted to result in the in-frame deletion of 30 amino acids of its extracellular AMPA receptor-binding domain (Figure 3F).
Figure 3.
Detection, Genotyping, and Sequence Characterization of De Novo SVs
(A) Heatmaps show a deletion signal from the total sequence coverage (copy number) and the number of discordant paired ends.
(B) SVs were genotyped with gtCNV, a SVM algorithm we developed. The contour plot shows the Phred-scaled genotype likelihoods for homozygous reference (green), heterozygous (blue), and homozygous (red) genotypes (for simplicity, only read depth and discordant paired ends are plotted). The colored diamonds indicate the genotype likelihoods for the proband and the parents.
(C) A majority of SNP alleles between the deletion boundaries were derived from the mother (shown in black), confirming a deletion of the paternal haplotype.
(D) De novo assembly of clipped reads resolved the breakpoint to single-base-pair resolution. Unaligned sequences within clipped reads are highlighted in gray.
(E) Aligning the assembled contig to the genome revealed the deletion breakpoint. Unique sequence proximal and distal to the breakpoint suggests a non-homologous-end-joining (NHEJ) mechanism.
(F) The mutant transcript of CACNG2 was sequenced from a fibroblast line derived from the individual and results in an in-frame deletion of exon 2.
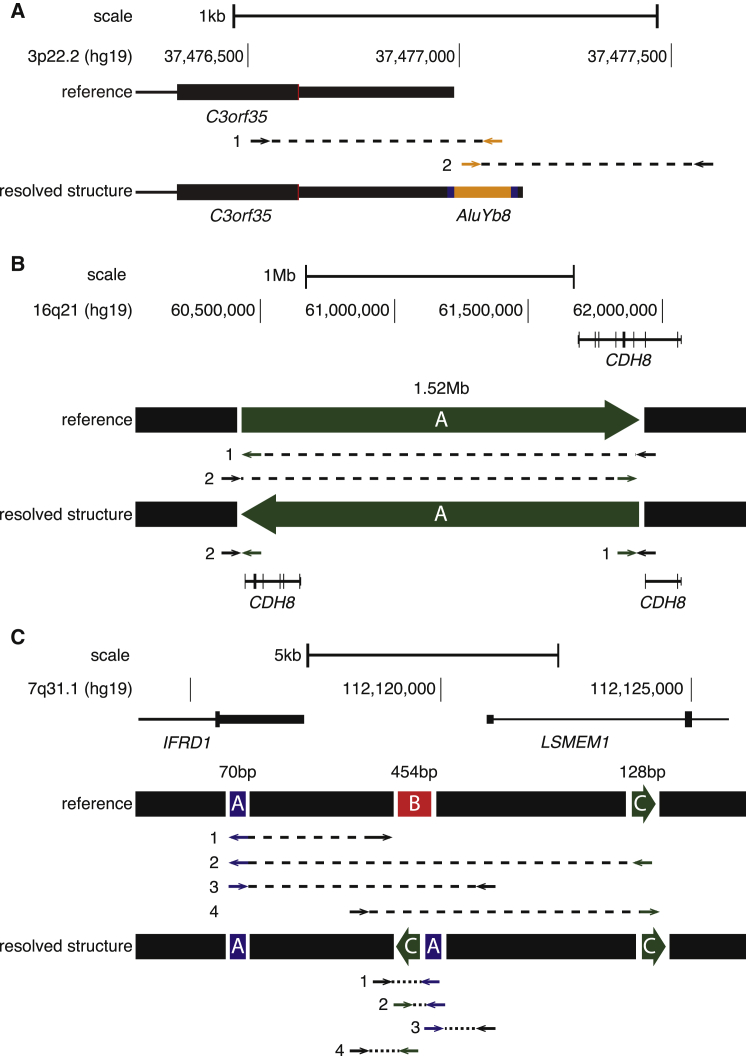
MEIs, balanced SVs, and complex rearrangements have not been systematically ascertained genome-wide in previous studies of ASD. Our results highlight how detection of these SV classes is useful for gene identification. For example, one validated MEI was a partial AluYb8 insertion in the 3′ UTR of C3orf35 (Figure 4A). This single observation was surprising given the low rate of de novo MEIs and the strong depletion of MEIs within 3′ UTRs in our call set (odds ratio [OR] = 0.44; 95% CI = 0.34–0.60; Figure S7). Additionally, we identified a de novo inversion (1.52 Mb) that disrupts cadherin-8 (CDH8 [MIM: 603008]; Figure 4B). These results strengthen the evidence from previous studies implicating C3orf354 and CDH834 in ASD.
Figure 4.
De Novo SVs of Genes Detectable through Genome Sequencing
Discordant paired-end mapping identified de novo SVs.
(A) A de novo AluYb8 element insertion into the 3′ UTR of C3orf35. Discordant paired ends and split reads mapped to both the 3′ and 5′ sides of the insert point, as well as the Alu. The partial AluYb8 (134 bp) was inserted into the positive strand with a 14 bp target-site duplication (shown in blue).
(B) A 1.52 Mb simple inversion with a distal breakpoint in intron 3 of CDH8.
(C) A non-tandem duplication and a non-tandem inverted duplication inserted into the promoter of LSMEM1 with a concomitant deletion at the insertion point (note that segments are not shown to scale). Arrows indicate the discordant orientation and location of paired-end reads in relation to the reference genome (UCSC Genome Browser build hg19) and the concordant pattern of paired-end reads in relation to the resolved structure. Black segments are unchanged in the SV events, green segments are inverted, blue segments are duplicated, and red segments are deleted.
Complex Mutation Clusters
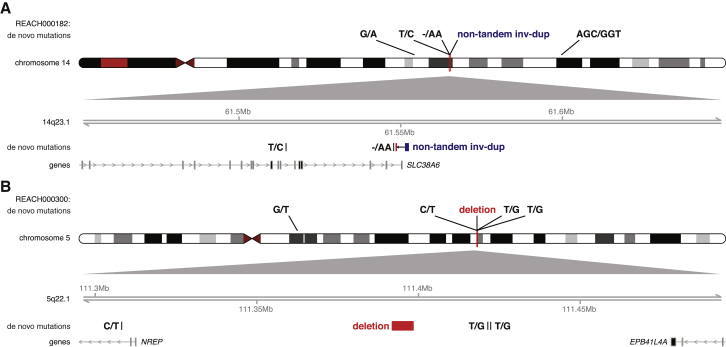
The complexity of de novo SVs consisted not only of clusters of deletions, duplications, and inversions occurring as single events (Figure 4C) but also of co-occurring de novo nucleotide substitutions and indels in the surrounding region (Figure 5). We observed greater clustering of de novo SVs and point mutations within individual genomes than would be expected by chance. In total, six de novo mutations (five SNVs and one indel) were located within 100 kb of de novo SV breakpoints, a 72-fold enrichment over random mutation (permutation p = 0.0001; Table S6). Adjacent de novo SVs and SNVs were located tens of kilobases apart; therefore, the enrichment of de novo substitutions around SV breakpoints could not be explained as an artifact because of the mismapping of reads at the junction. An alternative hypothesis for the mutational clustering is that the mutation rate of SNVs, indels, and SVs is elevated within certain mutational hotspots. If this were the case, we would expect de novo SNVs and indels to also cluster near breakpoints of SVs that are inherited. However, when we repeated the analysis, building a null model by shuffling the de novo SV breakpoints across the inherited SV breakpoints, instead of randomly, gave us similar results (Table S6). Figure 5 and Table S7 detail examples of complex mutation clusters.
Figure 5.
Mutational Clustering of SVs, Indels, and SNVs
Two examples of complex mutation clusters are shown in individuals.
(A) REACH000182, a 143 bp sequence near the 3′ UTR of SLC38A6, was duplicated and inserted into intron 16 of the gene with a concomitant deletion of 23 bp at the insertion site. Additionally, a de novo indel and SNV occurred at 211 bp and 34,611 bp proximal to the insertion site.
(B) REACH000300, a 6.2 kb de novo deletion, was detected at 5q22.2, and three de novo SNVs occurred within 100 kb of the breakpoints. The 200 kb zoomed-in locus below the ideogram shows the positions of the de novo mutations in relation to each other. Gene tracks below the mutation show the longest transcript of each gene within the locus (arrows indicate the strand, and bars indicate the exons of genes).
Pathogenic Inherited SVs
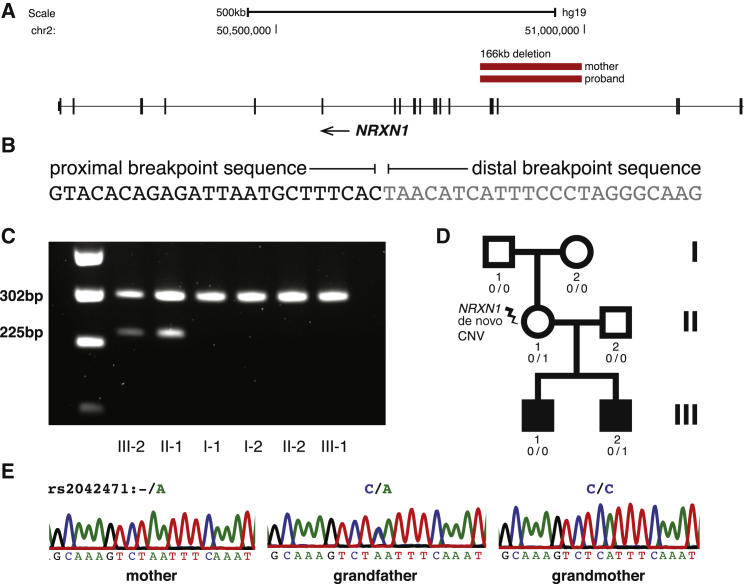
We examined the call set for known pathogenic SVs and observed five rare or de novo CNVs overlapping known ASD or ID risk variants in affected individuals (expected = 2; 95% CI = 0–5; p = 0.063; OR = 2.42). We did not observe significant overlap with genes affected by de novo loss-of-function variants identified in ASD and ID by exome sequencing (observed = 13; expected = 14.9; 95% CI = 8–23; p = 0.72; OR = 0.84). Inherited risk variants were identified in four (6%) unrelated affected individuals. One de novo SV identified in this study, a duplication of 15q11.2–13.1 (MIM: 608636; Table 1), has also been previously implicated in ASD.35 We observed two paternally inherited deletions of 15q11.2 (MIM: 615656).36 Inherited X-linked variants included a Xp21.1–21.2 duplication-inversion-duplication event that duplicates the Dp71 isoform of DMD (MIM: 300377) and disrupts TAB3 (MIM: 300480; Figure S3). Duplications and deletions of DMD are associated with Duchenne muscular dystrophy, and some alleles can predispose to ASD.36, 37, 38 Lastly, we detected a maternally inherited deletion of NRXN1.36 Follow-up genetic analysis of the extended family revealed that the deletion occurred de novo in the mother and that the mutation originated in the grandmother (Figure 6). This observation highlights the fact that although these disease-associated variants were inherited from a parent, they occur within regions that are prone to frequent recurrent rearrangements and are likely to be mutations that occurred in recent ancestry.
Figure 6.
Identification and Validation of the Pathogenic NRXN1 Deletion
(A) A 166 kb deletion disrupting three exons of NRXN1 leads to a frameshift in the longer isoform (α-NRXN1).
(B) Breakpoint mapping shows a unique sequence flanking the breakpoint, suggesting a NHEJ mechanism.
(C) A forward PCR primer was designed proximal to the breakpoint, and two reverse primers were designed (one within the deletion region produces a 302 bp product, and one spanning the breakpoints produces a 225 bp product in the presence of a deletion). We confirmed the deletion in this pedigree in the proband (III-2) and mother (II-1), but not in the father, sibling, or maternal grandparents.
(D) Pedigree of the family affected by multiplex ASD. The NRXN1 deletion occurred de novo in the mother and was passed on to her younger son. The mother is unaffected, and the older son has ASD but did not inherit the deletion, suggesting that other de novo and/or inherited variants contribute to ASD in this family.
(E) Sanger sequencing of rs2042471 within the NRXN1 locus indicated that this deletion originated on the grandmaternal haplotype.
Discussion
We have assembled what is, to our knowledge, the most complete set of SVs in ASD to date. WGS of trio families reveals a diverse landscape of structural variation throughout the genome and a higher rate and complexity of structural mutation than previously recognized. Structural mutations detected in individuals include previously undetectable events that disrupt genes and are likely to influence disease risk.
The combined frequency of de novo SVs that we observed (μ = 0.195) is more than triple the estimate from a previous WGS study of autism (μ = 0.058).9 Our estimate is also slightly higher than the rate observed in a family-based study by the GoNL Consortium (μ = 0.16).8 The mutation rate reported here will ultimately prove to be an underestimate as well given the challenges of detecting SVs by using short-read next-generation sequencing technology.
With the improved ascertainment of small deletions, inversions, and MEIs, we observed a similar overall mutation rate in case and control individuals, unlike in previous studies by our group2 and others that were based on microarray technology.29, 39 Thus, a genetic contribution of de novo SVs to ASD is evident not from an elevated frequency of genomic rearrangement but instead from the greater proportion of new mutations that disrupt genes. In this respect, the contribution of de novo structural mutation to ASD bears a similarity to that of de novo loss-of-function mutations detected by exome sequencing.40, 41
Studies of genetic diversity in populations reveal a diverse spectrum of SVs7 but do not fully illuminate the mutational process that gives rise to that diversity. Here, we have shown that one-third of de novo SVs consist of mobile elements, balanced mutations, or complex mutations, underlining the role that these mutational mechanisms play in generating genetic diversity and disease risk. Candidate loci for ASD were identified from two such de novo SVs, including a MEI in C3orf35 and an inversion disrupting CDH8. Published studies of ASD provide additional evidence implicating both genes, including a de novo deletion disrupting C3orf355 and segregating CDH8 deletions observed in ASD-affected families.34
Our results highlight how clusters of SVs arise through complex mutational events that generate combinations of deletions, duplications, insertions, and inversions (and sometimes all of the above). Adding further complexity to the mutational process, we have shown that 16% (3/19) of de novo SVs co-occur with clusters of de novo SNVs and indels. These results expand upon our previous study reporting the observation of de novo nucleotide substitution “showers.”12 Our current findings suggest that sequence variation and structural variation can arise from common mechanisms. We hypothesize that such complex mutation clusters form as a consequence of break-induced replication (BIR) during the repair of double-stranded breaks. BIR is significantly more error prone than normal DNA replication and occurs over hundreds of kilobases,42 a scale that is similar to the length of the mutation clusters observed here.
The observation of complex mutation clusters is interesting in light of a previous study from the 1000G Project, which found that SNPs and indels in the population are enriched within 400 kb of deletion breakpoints. It was further hypothesized that the observed enrichment of SNPs and indels is due to relaxed selection at these loci.43 On the basis of our results, we suggest that the observed correlation between SNPs and SVs is in part attributable to the underlying mutational processes and is not driven entirely by selection.
With our high-coverage and complementary SV-discovery methods, we were able to detect 5,213 SVs per individual, 27% more alleles per genome than we and others recently reported in the 1000G call set (n = 4,095/genome).7 However, as we demonstrated (Figure S2), our methods do not present a complete catalog of SVs. Furthermore, the short-read shotgun sequencing technology used here possesses significant technical limitations that impeded our ascertainment of SVs. Application of new long-read sequencing technologies44 will be another significant step toward uncovering the complexity of structural variation in autism.
Conflicts of Interest
J.S. declares that a patent has been issued to the Cold Spring Harbor Laboratory by the US Patent and Trademark Office on genetic methods for the diagnosis of autism (patent number 8554488).
Acknowledgments
We would like to thank the families who volunteered for the study. J.S. received grants from the NIH (MH076431 and HG007497) and the Simons foundation (SFARI 275724). L.M.I. received grants from the NIH (HD065288, MH104766, and MH105524). W.M.B. was supported by a fellowship from the Autism Science Foundation. D.A. was supported by NIH predoctoral training grant T32 GM008666. We would like to thank Wayne Pfeiffer, Mahidhar Tatineni, and the San Diego Supercomputer Center for hosting the computing infrastructure necessary for completing this project.
Published: March 24, 2016
Footnotes
Supplemental Data include seven figures, seven tables, and a supplemental data set and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2016.02.018.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes SV data, ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/phase3/integrated_sv_map/
Autism Center of Excellence, https://autism-center.ucsd.edu
BED file of filtered regions, http://bit.ly/1PDkVPQ
Mobster properties file, http://bit.ly/1PlX4IB
OMIM, http://www.omim.org
Picard Tools, http://broadinstitute.github.io/picard/
REACH Project, http://reachproject.ucsd.edu
SpeedSeq pipeline, https://github.com/hall-lab/speedseq
Structural Variation Toolkit, https://sourceforge.net/projects/svtoolkit/
UCSC Genome Browser hg19 data, http://hgdownload.soe.ucsc.edu/goldenPath/hg19/database/
Supplemental Data
References
- 1.Malhotra D., Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S., Autism Sequencing Consortium Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand H., Collins R.L., Hanscom C., Rosenfeld J.A., Pillalamarri V., Stone M.R., Kelley F., Mason T., Margolin L., Eggert S. Paired-Duplication Signatures Mark Cryptic Inversions and Other Complex Structural Variation. Am. J. Hum. Genet. 2015;97:170–176. doi: 10.1016/j.ajhg.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Hsi-Yang Fritz M., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kloosterman W.P., Francioli L.C., Hormozdiari F., Marschall T., Hehir-Kwa J.Y., Abdellaoui A., Lameijer E.W., Moed M.H., Koval V., Renkens I., Genome of Netherlands Consortium Characteristics of de novo structural changes in the human genome. Genome Res. 2015;25:792–801. doi: 10.1101/gr.185041.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuen R.K., Thiruvahindrapuram B., Merico D., Walker S., Tammimies K., Hoang N., Chrysler C., Nalpathamkalam T., Pellecchia G., Liu Y. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 10.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 11.Lord C., Risi S., Lambrecht L., Cook E.H., Jr., Leventhal B.L., DiLavore P.C., Pickles A., Rutter M. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J. Autism Dev. Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- 12.Michaelson J.J., Shi Y., Gujral M., Zheng H., Malhotra D., Jin X., Jian M., Liu G., Greer D., Bhandari A. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 2012;151:1431–1442. doi: 10.1016/j.cell.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michaelson J.J., Sebat J. forestSV: structural variant discovery through statistical learning. Nat. Methods. 2012;9:819–821. doi: 10.1038/nmeth.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang C., Layer R.M., Faust G.G., Lindberg M.R., Rose D.B., Garrison E.P., Marth G.T., Quinlan A.R., Hall I.M. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat. Methods. 2015;12:966–968. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thung D.T., de Ligt J., Vissers L.E., Steehouwer M., Kroon M., de Vries P., Slagboom E.P., Ye K., Veltman J.A., Hehir-Kwa J.Y. Mobster: accurate detection of mobile element insertions in next generation sequencing data. Genome Biol. 2014;15:488. doi: 10.1186/s13059-014-0488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudmant P.H., Mallick S., Nelson B.J., Hormozdiari F., Krumm N., Huddleston J., Coe B.P., Baker C., Nordenfelt S., Bamshad M. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abraham A., Pedregosa F., Eickenberg M., Gervais P., Mueller A., Kossaifi J., Gramfort A., Thirion B., Varoquaux G. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. 2014;8:14. doi: 10.3389/fninf.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu T.F., Lin C.J., Weng R.C. Probability estimates for multi-class classification by pairwise coupling. J. Mach. Learn. Res. 2004;5:975–1005. [Google Scholar]
- 24.Wang K., Li M., Hadley D., Liu R., Glessner J., Grant S.F., Hakonarson H., Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinto D., Delaby E., Merico D., Barbosa M., Merikangas A., Klei L., Thiruvahindrapuram B., Xu X., Ziman R., Wang Z. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narzisi G., O’Rawe J.A., Iossifov I., Fang H., Lee Y.H., Wang Z., Wu Y., Lyon G.J., Wigler M., Schatz M.C. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat. Methods. 2014;11:1033–1036. doi: 10.1038/nmeth.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S.R., Wilkie A.O., McVean G., Lunter G., WGS500 Consortium Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen R., Wei Q., Zhan X., Zhong X., Sutcliffe J.S., Cox N.J., Cook E.H., Li C., Chen W., Li B. A haplotype-based framework for group-wise transmission/disequilibrium tests for rare variant association analysis. Bioinformatics. 2015;31:1452–1459. doi: 10.1093/bioinformatics/btu860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagnamenta A.T., Khan H., Walker S., Gerrelli D., Wing K., Bonaglia M.C., Giorda R., Berney T., Mani E., Molteni M. Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J. Med. Genet. 2011;48:48–54. doi: 10.1136/jmg.2010.079426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J., Zwaigenbaum L., Szatmari P., Scherer S.W. Molecular Cytogenetics of Autism. Curr. Genomics. 2004;5:347–364. [Google Scholar]
- 36.Abrahams B.S., Arking D.E., Campbell D.B., Mefford H.C., Morrow E.M., Weiss L.A., Menashe I., Wadkins T., Banerjee-Basu S., Packer A. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol. Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pagnamenta A.T., Holt R., Yusuf M., Pinto D., Wing K., Betancur C., Scherer S.W., Volpi E.V., Monaco A.P. A family with autism and rare copy number variants disrupting the Duchenne/Becker muscular dystrophy gene DMD and TRPM3. J. Neurodev. Disord. 2011;3:124–131. doi: 10.1007/s11689-011-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Y.H., Yuen R.K., Jin X., Wang M., Chen N., Wu X., Ju J., Mei J., Shi Y., He M. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deem A., Keszthelyi A., Blackgrove T., Vayl A., Coffey B., Mathur R., Chabes A., Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abyzov A., Li S., Kim D.R., Mohiyuddin M., Stütz A.M., Parrish N.F., Mu X.J., Clark W., Chen K., Hurles M. Analysis of deletion breakpoints from 1,092 humans reveals details of mutation mechanisms. Nat. Commun. 2015;6:7256. doi: 10.1038/ncomms8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dijk E.L., Auger H., Jaszczyszyn Y., Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.