Abstract
The emergence of mosquito-borne RNA viruses, such as West Nile virus (WNV), is facilitated by genetically complex virus populations within hosts. Here, we determine whether WNV enzootic (Culex tarsalis, Cx. quinquefasciatus, and Cx. pipiens) and bridge vectors (Aedes aegypti) have differential impacts on viral mutational diversity and fitness. During systemic mosquito infection, WNV faced stochastic reductions in genetic diversity that rapidly was recovered during intra-tissue population expansions. Interestingly, this intrahost selection and diversification was mosquito-species dependent with Cx. tarsalis and Cx. quinquefasciatus exhibiting greater WNV divergence. However, recovered viral populations contained a preponderance of potentially deleterious mutations (i.e. high mutational load) and had lower relative fitness in avian cells compared to input virus. These findings demonstrate that the adaptive potential associated with mosquito transmission varies depending on the mosquito species and carries a significant fitness cost in vertebrates.
INTRODUCTION
The emergence of arthropod-borne RNA viruses (arboviruses) is an ongoing problem that imposes significant heath and economic burdens on communities worldwide. West Nile (WNV), chikungunya (CHIKV), dengue (DENV), and Zika viruses are all in various states of emergence at local or global scales (Kramer et al., 2008; Lanciotti, 2014; Musso et al., 2015). The mechanisms underlying arbovirus emergence are complex and include, for example, altered land use and increased global travel. In addition, RNA viruses have an inherent ability to rapidly mutate and thus generate opportunities for adaptation in novel environments through an error-prone polymerase (Holland et al., 1982). WNV is an excellent example of an introduced RNA virus that adapted to a new environment (i.e. new genotype linked to a shorter extrinsic incubation period in local mosquitoes), promoting its spread throughout the Americas (Moudy et al., 2007). Several studies have assessed how different host types impact WNV population structure, and have shown that WNV populations are more diverse in mosquitoes compared to birds (Grubaugh et al., 2015; Jerzak et al., 2005; Jerzak et al., 2007). In mosquitoes, purifying selection is weak and virus diversification is driven by the action of RNA interference (RNAi), which creates an intracellular milieu that favors rare genotypes (Brackney et al., 2009; Brackney et al., 2015). In contrast, purifying selection in birds is strong (Jerzak et al., 2008) and the innate antiviral responses are dominated by type I interferon. Thus, in the WNV system, mosquitoes and birds have distinct impacts on virus population biology: Mosquitoes allow for increased adaptive plasticity, while birds maintain high fitness through purifying selection (Deardorff et al., 2011).
A wide array of studies has suggested that arboviruses can adapt to microhabitat-specific conditions. CHIKV is capable of adapting to transmission by Aedes albopictus during a single round of infection (Stapleford et al., 2014). WNV evolutionary dynamics have been shown to vary in response to environmental conditions (Bertolotti et al., 2008). Indeed, different avian hosts of WNV have distinct impacts on virus population structure and fitness (Grubaugh et al., 2015). The impacts of different mosquito species on WNV population biology and fitness, however, have not been directly addressed. This is a critical shortcoming in the field because throughout its distribution, WNV is maintained in its enzootic cycle by several Culex species, including Culex tarsalis, Cx. pipiens quinquefasciatus, and Cx. pipiens pipiens (Kramer et al., 2008). In addition, mosquitoes of several divergent genera have been found infected and/or demonstrated to be competent vectors (Bernard et al., 2001; Turell et al., 2005). These include Aedes mosquitoes that may act as a “bridge” between enzootic cycles and mammals, including humans (Kilpatrick et al., 2005). Although it seems clear that infection of mosquitoes leads to genetically complex virus populations (Jerzak et al., 2008; Sim et al., 2015; Stapleford et al., 2014), the impact of any particular mosquito species on WNV population biology has not been determined. In addition, the full range of selective forces acting on WNV during systemic infection of mosquitoes is poorly understood, and important inconsistencies persist in the literature. For example, whereas some studies have documented the existence of population bottlenecks during arbovirus transmission by mosquitoes (Ciota et al., 2012a; Forrester et al., 2012), others have not (Brackney et al., 2011; Gutierrez et al., 2015). The genetic implications of high mutational diversity coupled with population bottlenecks during mosquito transmission have not been fully elaborated, and the impacts of different vector species on virus population diversity and fitness are not known.
Therefore, we determined the extent to which mosquito vectors of WNV differ in their propensity to drive virus diversification and impact fitness. In particular, using next-generation sequencing (NGS) we characterized virus populations within distinct tissue compartments including midguts, hemolymph, salivary glands, and expectorated saliva of Cx. tarsalis, Cx. quinquefasciatus, Cx. pipiens, and Ae. aegypti during a single mosquito infection. Taken together, our results 1) demonstrate that mosquito species have differential impacts on virus evolution, 2) illustrate sequential reductions and expansions in virus population size that occur during the spread of virus from one mosquito compartment to another, and 3) confirm the importance of purifying selection during vertebrate infection in maintaining WNV fitness. Ironically, our results document a profound loss in relative fitness imposed by arthropod transmission of an arbovirus.
RESULTS
Vector competence and anatomical barriers to virus transmission
Arboviruses must overcome anatomical barriers within mosquitoes for transmission to occur (Figure 1A). Three enzootic vectors, Cx. tarsalis, Cx. quinquefasciatus, and Cx. pipiens, and one potential bridge vector, Ae. aegypti, were exposed to WNV derived from an infectious clone of the NY99 genotype (WNVic) to evaluate vector competence and obtain samples for analysis. Vector competence was determined by examining the percent of midguts, legs (containing hemolymph), salivary glands, and saliva infected with WNV after 14 days extrinsic incubation. Differences in overall vector competence were mostly related to differences in the strengths of barriers to infection and escape from the midguts and salivary glands of tested mosquitoes (Figure 1B). Furthermore, the mosquitoes with the highest viral genome equivalents (GE) in their salivary glands were more likely to have virus detected in their saliva (Figures 1C, p < 0.05 Mann-Whitney test [data for test not shown]). However, viral GE in the midguts and saliva were not significantly different among mosquito species (Figure 1C, p > 0.05) despite species-specific differences in susceptibility to oral infection and transmission, respectively. Moreover, the rates of WNV replication in the midguts are not directly correlated with the midgut infection and escape barriers (Figure 1D). Importantly, these data demonstrate a context-dependent relationship between the strength of any given anatomical barrier and viral load.
Figure 1. Vector competence of mosquitoes and characterization of specimens used in this study.
(A) Overview of the anatomical barriers to virus transmission. Infected tissues indicate that WNV could overcome the barrier (e.g. infected legs indicate there was not a midgut escape barrier in that mosquito).
(B) ercent of tissues and saliva with WNV RNA determined by qRT-PCR (n = 32 mosquitoes for each species) at 14 days post exposure.
(C) WNV GE per tissue or saliva sample from only the WNV-infected tissues determined by qRT-PCR (*, p < 0.05; ns, not significant).
(D) NV replication rates determined by collecting midguts at 3-7 and 14 days post infection (n = 16).
Intrahost WNV population structure is mosquito species-dependent
Three mosquitoes from each species that had detectable WNV RNA in all four compartments were used to assemble three biological replicates of each tissue per species. WNV RNA was examined using NGS to define species- or tissue-dependent impacts on virus mutational diversity. Approximately 6% of > 22 million reads obtained from each specimen aligned to WNV, resulting in > 12,000× coverage depth across the viral genome (Figure S1, Table S1). However, the coverage depth from one biological replicate of Cx. pipiens salivary glands and all three Cx. pipiens saliva was much lower (< 100×) precluding viral population analysis. Analysis of the remaining samples was limited to the protein coding due to large variation in the sequencing coverage of the untranslated regions (presumably caused by secondary structures in these regions).
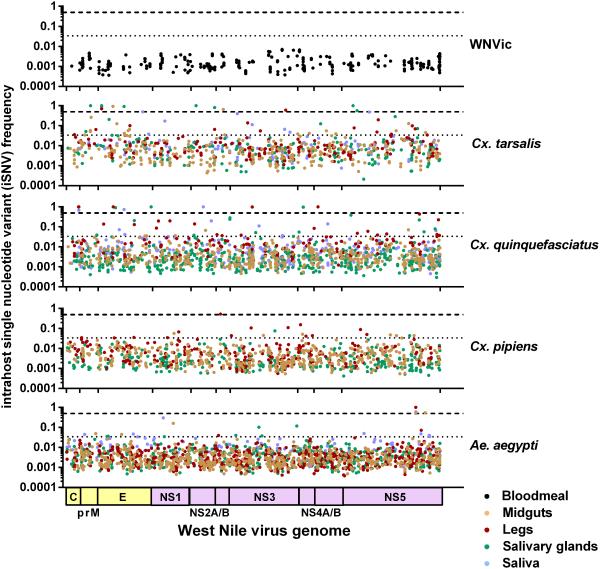
Intrahost single nucleotide variants (iSNVs) from each biological replicate were combined by species and tissue to assess their genome positions and frequencies. As expected, the relatively homogenous input WNVic population diversified within each mosquito species (i.e. iSNVs reached higher frequency, Figure 2). However, the number of iSNV sites that reached high frequency (HF, > 0.034, upper 5% of a gamma distribution), changed the consensus sequence (frequency > 0.5), and went to fixation (frequency = 1) were mosquito species- and tissue-dependent (Table S2). The most HF iSNVs, consensus changes, and fixations were found in Cx. quinquefasciatus and Cx. tarsalis tissues, however most of the consensus sequence changes detected in the salivary glands of Cx. tarsalis were not found in the saliva (Table S3). In addition, none of the observed consensus changes (Table S3) resemble known WNV lineage-defining mutations (e.g. V to A substitution at envelope codon position 159 that place it in the “WN02” lineage (Ebel et al., 2004)).
Figure 2. More high frequency single nucleotide variants are generated during virus replication in Cx. quinquefasciatus and Cx. tarsalis.
iSNVs from each biological replicate (n = 3) were plotted by their position on the WNV genome and their frequency in each mosquito species and tissue. The WNV genome consists of three structural protein coding regions (shown in yellow), capsid (C), premembrane (prM), and envelope (E), and seven nonstructural (NS) protein coding regions (shown in purple). The cut-offs for high frequency iSNVs (0.034, dotted line) and consensuses sequence changes (0.5, dashed line) are shown. iSNV sites are categorized by frequency in Table S2 and all consensus sequence changes are listed in Table S3.
Intrahost viral population structure was further assessed using several genetic diversity indices: richness (sites per million WNV reads), complexity (uncertainty associated with randomly sampling an allele), distance (iSNVs and amino acid variants per coding sequence), and divergence (genetic differences between two populations). Despite similar levels of richness and complexity (Figures 3A and 3B, p > 0.05), WNV populations in Cx. quinquefasciatus and Cx. tarsalis contained greater genetic diversity at both the iSNV (Figure 3C) and amino acid levels (Figure 3D) , and diverged further from the input WNV as compared to WNV in Cx. pipiens and Ae. aegypti (Figure 3E and Figure S2). The increased genetic diversity in Cx. quinquefasciatus, and to a lesser extent Cx. tarsalis, was largely due to increased accumulation of HF iSNVs (including consensus changes and fixations, Figure 2).
Figure 3. Diversification of WNV during systemic infection of mosquitoes.
(A-D) Intrahost genetic diversity was characterized by measuring (A) richness, (B) complexity (the proportion of different variants in a mutant spectrum), (C) iSNV distance, and (D) amino acid distance. Data shown as means with 95% confidence intervals (*, p < 0.05). (E) Mean genetic divergence (accumulation of independent mutations between two isolated populations) of each population from the input virus (y-axis), between tissues (x-axis, cumulative), and among biological replicates (circles, shown to axis scale). Individual comparisons are shown in Figure S2.
iSNVs generated during replication in all mosquito species seemed to be distributed uniformly across the viral coding sequence (Figure 2). To investigate this further, we combined the WNV populations from each tissue to determine whether particular WNV genomic regions were more diverse (Figure S3). Nucleotide and amino acid diversities were greatest in Cx. quinquefasciatus and Cx. tarsalis, and were uniformly distributed across the protein coding sequence. In Cx. quinquefasciatus, the capsid coding region was more diverse due to a relatively small number of HF iSNVs (Table S2).
Viral population declines and expansions during systemic mosquito infection
To assess population bottlenecks during mosquito infection we tracked the spread of individual unique iSNVs and the 30 most common predicted haplotypes (conservatively predicted using only HF iSNVs, Tables S4-S7) during systemic spread between mosquito tissues. Most input (e.g. < 5% in saliva, Figure S4A) and locally-derived (Figure 4A) unique iSNVs were not transferred between tissues. For example, > 90% of iSNVs detected in the saliva were not detected in other tissues. In addition, the original WNVic input haplotype was dominant in most tissues, except in Cx. quinquefasciatus (Figure 4B). In this species a new dominant haplotype was detected in the hemolymph (Table S5). These observations led us to hypothesize that bottlenecks within mosquitoes limit haplotype spread. We thus evaluated intra-tissue viral demographics using Tajima’s D (i.e. comparison of pairwise mismatches and segregating sites) and Harpending’s raggedness index (i.e. the distribution of pairwise mismatches). D values were consistently negative and not significantly different among species, tissues, or replicates (mean = −0.73, p > 0.05); and the distributions of pairwise mismatches (Figure 4C) revealed several multimodal curves. Phylogenies reconstructed from predicted haplotypes were also consistent with population declines and expansions (Figure 4D). In tissues with extensive diversification, the trees demonstrated strong spatial structure (clades corresponded to intra-tissue populations) and star-like branching topologies (see biological replicates A and B in Cx. tarsalis and A, B, and C in Cx. quiquefasciatus). Together, these results suggest periodic population declines followed by expansions and are consistent with sequential population bottlenecks and recoveries involving founder’s effects.
Figure 4. Recovery of viral genetic diversity during intra-tissue population expansions following bottlenecks.
(A) roportion of iSNVs found in the subsequent viral population (e.g. carry-through from bloodmeal to the midgut) (mean with 95% confidence interval). Carry-through of the input iSNVs to the rest of the tissues are shown in Figure S4A.
(B) Haplotypes were predicted from each viral population (See Table S4-S7) were characterized by compartment of origin (e.g. haplotype in a saliva population was originally detected in the legs). The average composition of haplotype origins is shown for each tissue.
(C) The distribution of pairwise mismatches between haplotypes. Shown are the average mismatch distributions for each tissue and the expected distribution for a constant population size (dashed line). “Ragged” lines indicate recent population expansions following declines.
(D) hylogenies of all of the predicted haplotypes from each species. The letters represent distinct clades from biological replicates (A, B, and C) and the asterisk marks the input virus branch. The proportions of predicted haplotypes from each tissue with mutations are shown in
(E) Genetic divergence of neutral alleles was calculated to determine the amount of genetic variance between populations caused by bottlenecks and drift. Larger FST values indicate a more severe bottleneck (smaller Ne). The proportion of infected midguts (triangles), legs (squares), salivary glands (diamonds), and saliva (circles) was obtained from Figure 1B.
The number of founder WNV genomes in the midgut from the bloodmeal was calculated to determine the population bottleneck size at the point of initial mosquito infection (i.e. the effective population size [Ne]). We estimated Ne by the genetic variance within and between populations caused by drift by calculating FST from neutral alleles (Table S8). Between 52 and 129 WNV genomes initiated mosquito midgut infection and were not significantly different among species (p > 0.05). Ne calculations between the other tissues produced highly variable results and could not be confidently estimated in this study (data not shown). However, we were able to determine the relative bottleneck severity between tissues and discovered a correlation with the anatomical barrier strength (Figure 4E).
The phylogenies also showed that some prevalent de novo haplotypes that were transferred between tissues acquired additional, locally derived, secondary mutations (Figure 4D, Tables S4-S7). Populations containing these haplotypes were also the most divergent (Figure S4). Haplotypes in the salivary glands and saliva with mutations shared with haplotypes arising in the midguts or legs acquired on average 2.5× more locally derived mutations than haplotypes without common mutations (2.4 compared to 0.9, p < 0.05 - Mann-Whitney test).
Intrahost purifying selection is host-dependent
Selection becomes the predominant force changing variant frequencies as Ne increases. We estimated the rates of intra-tissue Ne expansion and Ne at the time of sequencing using GE as a proxy. Viral GE increased at faster rates during replication in Cx. tarsalis and Cx. quinquefasciatus midguts (Figure 1D) even though the number of GEs in the midguts at the time of sequencing were not significantly different between species (Figure 1C, p > 0.05), and these differences may also influence selection.
Viral intrahost length variants (iLVs, including single and double nucleotide insertions and deletions) in the coding sequence are predicted to be deleterious and thus rapidly removed by selection. Therefore we first assessed the level of purifying selection by measuring the accumulation of HF iLVs per coding sequence (Figure 5A). The input WNVic did not contain any HF iLVs, therefore all HF iLVs must have accumulated during replication in mosquitoes. We did not detect species or tissue-dependent effects on viral iLVs accumulation (p > 0.05). Next we calculated the ratio of nonsynonymous (dN) to synonymous (dS) substitutions per coding sequence site (dN/dS) using both iSNVs and iLVs. Viral populations from the species in which WNV diverged the most, Cx. tarsalis and Cx. quinquefasciatus, had higher dN/dS ratios (Figure 5B) and dN rates (Figure 5C) compared to Cx. pipiens and Ae. aegypti, but similar dS rates (Figure 5D). These data suggest that slight differences in mosquito-specific selection may lead to higher population divergence. Specifically, more controlled viral replication during infection of Cx. pipiens and Ae. aegypti midguts (Figure 1D), is likely apparent in our data as stronger purifying selection (Figure 5B).
Figure 5. Host-specific strength of purifying selection in vivo.
(A-D) The strengths of purifying selection were compared between mosquito species and between mosquitoes and birds. Wild caught birds (American crows, house sparrows, and American robins) and 2 days old chicks (n = 3 for each species) were previously inoculated with the same WNVic used in this study and the serum was sequenced at 3 days post infection (Grubaugh et al., 2015). (A) Accumulation of potentially deleterious mutations was estimated by the number of high frequency iLVs. (B) The ratios of nonsynonymous (dN) to synonymous (dS) substitutions per nonsynonymous and synonymous coding sequence site, respectively (dN/dS, > 1 [dotted line] weak purifying selection, < 1 strong purifying selection), (C) the dN rates, and (D) the dS rates were used to infer the strength of purifying selection. All data were summarized using the mean (A, B) or geometric mean (C, D) and the 95% confidence interval from each tissue and biological replicate per host (*, p < 0.05; ns, not significant).
In addition, these data, combined with previously reported data on intrahost WNV populations in various avian species (Grubaugh et al., 2015), demonstrate that purifying selection is weaker in mosquitoes compared to birds.
Fitness of saliva WNV populations is lower in avian cells relative to the input virus
The fitness of WNV within mosquito saliva was estimated using two methods. First, relative fitness was measured using in vitro competition against a genetically marked reference virus during infection of chicken fibroblasts (DF-1 cells) (Figure 6A, Figure S5A). WNV in mosquito saliva was consistently displaced by the reference virus during direct competition. WNV recovered from Cx. quinquefasciatus represented only 10-20% of the total following 8 days of competition and was significantly lower than the proportion of the WNVic during competition (p > 0.05, Mann-Whitney test). WNV from Cx. tarsalis, Cx. pipiens, and Ae. aegypti saliva was undetectable by 2 days post infection. However, the range of high accuracy for the quantitative sequencing assay is 0.1 to 0.9 (Fitzpatrick et al., 2010), therefore we can only determine that the undetectable competitor WNV is < 10%. Second, we calculated the ratios of viral GE to PFUs to determine if lower relative fitnesses were due to losses in infectivity (Figure 6B). We found no significant differences among species or in comparison to the input WNV (p > 0.05).
Figure 6. Viral populations in mosquito saliva have a lower fitness in bird cells relative to the input virus.
(A) ompetitive replicative fitness in DF-1 cells of the WNVic and WNV recovered from mosquito saliva (competitors) compared to a WNV reference (WNV-REF) during co-infection. The proportion of the competitor genotypes from the DF-1 supernatants were determined by quantitative sequencing (Figure S5A) and were normalized by the fold change (log2 transformed) from the inoculum. Values below the dotted line at 0 represent samples with decreased competitive fitness compared to WNV-REF. The proportion of competitor from saliva samples that went to extinction (proportion = 0) was reset to 0.01 for fold change calculations. Cx. quinques, Culex quinquefasciatus.
(B) NV GE:PFU ratios from the bloodmeal and all saliva samples containing virus were calculated to determine if differences in relative fitness were due to differences in infectivity (mean with 95% confidence interval; ns, not significant).
(C-D) The highest nonsynonymous variant detected in each recovered saliva sample was engineered into the WNVic to determine the fitness of mosquito-derived mutations (Table S9).
(C) Competitive fitness of the mutants relative to the WNVic during DF-1 cell infection was determined as described in (A) (Figure S5B). (D) Replicative fitness of the mutants compared to the WNVic and WNV-REF during DF-1 cell infection was determined by qRT-PCR (n = 4 for each virus). Replication fitness in Ae. albopictus clone C6/36 cells is shown in Figure S5C. Cx.t, Culex tarsalis; Cx.q, Cx. quinquefasciatus; Ae.a, Aedes aegypti.
The competitive fitness data suggests that several mutations with decreased relative fitness are incorporated into the WNV populations during systemic mosquito infection and are transmitted in the expectorated saliva. Therefore we tested the relative fitness of the highest frequency nonsynonymous mutation detected in each sequenced saliva sample by engineering the mutations into the WNVic (Table S9). Five of the nine mutations lowered the fitness in DF-1 cells relative to the WNVic and the remaining four had no effect (Figure 6C, Figure S5B). A I449T mutation to the WNV nonstructural protein 3 (NS3) derived from Cx. tarsalis also had severe replication defects in DF-1 cells (Figure 6D), while mutation NS5-W808C from Ae. aegypti had diminished replication in both avian (DF-1) and mosquito cells (Ae. albopictus clone C6/36) relative to the WNVic (Figure S5C). These data are consistent with the findings that ~60% of SNVs are predicted to be deleterious or lethal (Sanjuan et al., 2004).
DISCUSSION
Species-dependent impacts on viral genetic diversity
Using NGS we characterized WNV populations during systemic mosquito infection from individuals of four important WNV vectors. Our data confirm that replication in mosquitoes promotes WNV diversification (Brackney et al., 2009; Jerzak et al., 2005; Jerzak et al., 2008). Importantly, however, our data also show that the degree of viral divergence is species-dependent. Our first observation was that a much greater number of WNV consensus mutations occurred during replication in the tissues of Cx. quinquefasciatus and Cx. tarsalis as compared to Cx. pipiens and Ae aegypti. Measures of genetic distance from the input virus also demonstrated that WNV replication in Cx. quinquefasciatus and Cx. tarsalis produced twice as many iSNVs compared to Cx. pipiens and Ae. aegypti. The nucleotide diversity from Cx. pipiens naturally infected with WNV (Jerzak et al., 2005) and field-derived Ae. aegypti experimentally infected with DENV (Sim et al., 2015) were similar to our intrahost WNV data from the same species (~1 per genome). However our calculations of WNV nucleotide diversity during replication in Ae. aegypti were about 4× lower than what was reported during CHIKV infection (Stapleford et al., 2014). These results suggest that the evolutionary outcomes of RNA virus transmission by an arthropod are the product of specific virus-vector interactions that influence genetic drift and selection.
Repeated stochastic reductions in genetic diversity
Vector competence is largely determined by barriers to infection and escape from key mosquito tissues, principally the midgut and salivary glands. These anatomical barriers impose bottlenecks as arboviruses spread in mosquito tissues (Ciota et al., 2012a; Forrester et al., 2012). Several aspects of our data suggest that RNA viruses undergo stochastic reductions in genetic diversity in mosquitoes. First, the majority of the iSNVs and novel haplotypes generated within one tissue were not detected in the subsequent tissue (e.g. iSNVs generated in the midguts were rarely detected in the legs). Second, Tajima’s D (negative values), Harpending’s raggedness (multimodal mismatch distributions), and the phylogenies (spatial structure and star-like topologies) suggest that genetic bottlenecks arise when new tissues are colonized (Harpending et al., 1993; Rogers and Harpending, 1992; Slatkin and Hudson, 1991; Tajima, 1989).
The magnitude of these bottlenecks is however dependent upon the virus diversity, and amount of virus in the bloodmeal, and virus-vector pairing. We exposed mosquitoes to a high dose (~2 × 108 PFUs) of virus containing one dominant haplotype and several very low frequency variants. Ciota et al. found that high frequency haplotypes (≥ 0.15) are more likely to survive the bottlenecks within Cx. pipiens (Ciota et al., 2012a). The same was true of WNV infection in Cx. tarsalis and Ae. aegypti wherein the input haplotype was dominant in all tissues, but not in Cx. quinquefasciatus in which the input haplotype went to extinction in the legs. Forrester et al. determined the bottleneck severity is inversely proportional to amount of virus in the bloodmeal (Forrester et al., 2012). Therefore transmission cycles involving vertebrate hosts with high peak viremia, such as birds infected with WNV (Komar et al., 2003), may have less severe midgut bottlenecks compared to cycles involving hosts that develop lower viremias, such as rodents infected with some subtypes of Venezuelan equine encephalitis virus (Carrara et al., 2005). The high dose that we used for this experiment is representative of the dose present in natural avian bloodmeals; therefore the observation that midgut infection has the weakest bottleneck (52-129 founder genomes) among the barriers tested likely reflects natural WNV transmission. Moreover, the midgut infection bottleneck size is also dependent upon the virus strain and mosquito species pair. For example, Gutierrez et al. calculated about a 6× greater number of founder genomes with enzootic VEEV strain paired with Cx. taeniopus (Ne ~ 520) than with an epizootic strain paired with Ae. taeniorhynchus (Ne ~ 83) (Gutierrez et al., 2015). Our data demonstrate that population bottlenecks occur across a range of mosquito vectors of WNV and that bottleneck severity (small Ne) may be related to the strength of the anatomical barrier (small proportion of infected tissues). Specifically, within Cx. quinquefasciatus we predict that the least severe bottlenecks occur during midgut infection (~100% infection rate) and the strongest during egress into the saliva (~45%).
Recovery of genetic diversity during population expansions
Following stochastic reductions in genetic diversity, we observed rapid recovery in virus population size and diversity during expansions in the next tissue/compartment that was likely promoted by RNAi (Brackney et al., 2009; Brackney et al., 2015). However, our data does not support that differences in RNAi targeting were responsible for the differences in intra-tissue divergence detected among species. RNAi promotes diversification by selecting for rare haplotypes until they are no longer rare. This diversifying selection is best measured by Shannon entropy where genetic complexity is the greatest when two alleles at a locus occur at equal frequencies (0.5) and complexity decreases as one allele becomes more dominant. Therefore diversifying selection as imposed by RNAi will act to increase genetic complexity more so than divergence. During mosquito infection, WNV complexity was not significantly different among species while more variants trended towards fixation during replication in Cx. quinquefascaitus and Cx. tarsalis. This suggests that bottlenecks and selection are more likely responsible for the species differences in viral divergence than RNAi.
Stochastic forces alter the genetic composition of the viral populations as they pass through the mosquito anatomical barriers, but as the population expands, even weak selection may play a role in WNV replication. All viral populations studied accumulated large numbers iLVs, but our measures of dN/dS suggest that selection may affect intra-tissue divergence. Specifically, the mosquito species with the most intra-tissue divergence, Cx. quinquefasciatus and Cx. tarsalis, also had the highest nonsynonymous mutation rates (dN) despite Cx. pipiens and Ae. aegytpi having similar dS values. These variations appear to be due to differences in the strength of purifying selection and may be mediated by mosquito’s ability to control WNV replication. Pressure against viral replication removes the least fit genomes first (e.g. nonsynonymous mutations with lowered fitness), and control of replication, such as in Cx. pipiens and Ae. aegypti midguts, lowers dN. Thus, the comparatively rapid rates of WNV population expansion within Cx. tarsalis and Cx. quinquefasciatus may facilitate higher genetic divergence because there is less pressure from purifying selection.
Haplotypes arising de novo in mosquitoes were more likely to accumulate further mutations than the input haplotype (~2.5×). In fact, the intra-tissue populations with the most genetic diversity (especially within Cx. quinquefasciatus) appear to have been seeded by a haplotype that arose within mosquitoes (rather than the input WNV haplotype). We hypothesized that the mutations arising on de novo haplotypes could have 1) decreased the replication fidelity or 2) helped the virus to explore an adaptive landscape. Increased viral genetic diversity has been shown to be beneficial in mosquitoes (Fitzpatrick et al., 2010), possibly by providing a mechanism for escaping RNAi (Brackney et al., 2009; Brackney et al., 2015) or by cooperative interactions between haplotypes and viral proteins (Ciota et al., 2012b). However, decreased fidelity haplotypes often have lower relative fitness through accumulation of deleterious mutations (Coffey et al., 2011; Van Slyke et al., 2015). De novo haplotypes increased in frequency and persisted in multiple tissues but did not cause an accumulation of iLVs compared to the other species; therefore it is not likely that these mutations caused changes in replication fidelity. Multi-step pathways that cause accumulations of viral mutations have been discovered in other arbovirus systems (Stapleford et al., 2014; Tsetsarkin et al., 2014). The principle of a multi-step pathway is that even weak selection for a beneficial mutation allows a network of secondary adaptive mutations. Overall, our results suggest that multi-step adaptive pathways may arise during a single systemic mosquito infection.
Lower relative fitness in avian cells
Repeated reductions in genetic diversity may lead to the accumulation of mutations that confer low fitness (Chao, 1990; Duarte et al., 1992). As has been suggested for DENV (Sim et al., 2015), the immediate deleterious effects of bottlenecks and high mutation rates appeared to be avoided by WNV through the rapid recovery of viral genetic diversity during intra-tissue replication. However, rapidly expanding populations and high multiplicities of cellular infection (MOIs) may allow mosquito tissues to tolerate new mutations, which could be either costly or beneficial in a new environment (Stern et al., 2014). Surprisingly, we detected a severely lower competitive fitness of the saliva derived WNV populations relative to the input virus in avian cells. Several changes to the viral population structure may account for the relative fitness declines in the absence of notable consensus changes in the recovered saliva populations. All viral populations in our studies accumulated abundant iLVs (> 0.1 per coding sequence, all frame-shifting) and nonsynonymous mutations (> 0.5 per amino acid sequence), most of which are predicted to be lethal or deleterious (Sanjuan et al., 2004). In fact, five of most frequent nonsynonymous mutations detected from the nine recovered saliva populations engineered into the WNVic decreased the relative fitness of the virus. Furthermore, limiting our analysis to the protein coding sequence likely missed potentially import mutations to the untranslated regions that may have negatively influence RNA structure and fitness. However, the viral genetic diversity detected in mosquito saliva may have benefits not measured in our experiments that could facilitate rapid adaptation in new environments. In addition, the mechanisms of fitness recovery in a highly purifying avian environment require further study.
Arbovirus transmission cycles lead to slow rates of evolution (Woelk and Holmes, 2002). A common explanation for this is that mutations occurring in mosquitoes have a fitness trade-off in vertebrates (and vice versa). However, this hypothesis has been debated due to conflicting results (Ciota et al., 2008; Coffey et al., 2008; Vasilakis et al., 2009). These data, combined with our previous studies (Deardorff et al., 2011; Grubaugh et al., 2015), support a fitness trade-off in birds but not in mosquitoes. The difference between studies may be partially attributable to methods for measuring relative fitness (competitive vs replicative), MOI differences (low MOIs to allow for variants to reach their true fitness levels (de la Torre and Holland, 1990)), and the replication environment (in vivo vs in vitro). These differing results may also represent the complex nature of virus-host interactions. For example, flavivirus-Culex-bird and alphavirus-Aedes-rodent cycles may fundamentally differ in their evolutionary dynamics.
Conclusions
In addition to RNAi, genetic drift and weak purifying selection can significantly alter viral populations. Our data demonstrate that Cx. quinquefasciatus may be significant drivers of WNV divergence and are more likely to transmit virus with consensus sequence changes compared to other mosquito species examined. We previously demonstrated in wild birds that WNV disease-susceptibility was negatively associated with maintaining viral fitness (Grubaugh et al., 2015). Taken together, we hypothesize that transmission cycles involving Cx. quinquefasciatus and American robins (disease resistant) would be more likely to produce novel WNV genotypes while maintaining high viral fitness than transmission cycles involving Cx. pipiens and American crows (disease susceptible). In addition, we have outlined the stochastic and deterministic forces that continuously shape viral populations (Figure 7). At anatomical barriers, viral populations undergo population bottlenecks that greatly reduced genetic diversity through drift and founder’s effects. A small virus population seeds subsequent tissues and then rapidly expands. Population fluctuations and genetic diversity led to tissue-specific viral haplotypes distinct from the input virus population. The impacts of repeated bottlenecks on the virus populations are important for two main reasons. 1) The high variance in variant frequencies detected among the mosquito-borne viruses should allow the populations to explore very different adaptive landscapes (Fisher, 1930; Wright, 1977), such as would be expected between mosquitoes and birds. 2) However, genetic drift coupled with weak purifying selection in mosquitoes may also lead to the accumulation of deleterious mutations (i.e. mutational load). Therefore, the collective fitness of the expectorated WNV from mosquitoes started at a point of lower relative fitness (i.e. in a fitness landscape valley) than the input viruses during avian cell infection. Thus, most of the mosquito-derived viruses were rapidly removed by strong purifying selection and/or were displaced by the more fit input viruses during mass selection. The observed lower fitness relative to the input virus is reminiscent of that predicted to occur as a result of Muller’s ratchet (Muller, 1964), which has been observed to result in virus fitness declines in vitro (Chao, 1990; Duarte et al., 1992), but not previously in vivo. Taken together, our results illustrate the irony of arthropod transmission, and may explain why arboviruses have low long-term rates of amino acid substitution compared to other host-specific RNA viruses (Woelk and Holmes, 2002).
Figure 7. Impacts of anatomical barriers and mosquito species on viral population structure.
(A) osquitoes feed upon a bloodmeal containing a relatively homogenous WNV population than seeded infection in the midgut epithelial cells. Within the tissue, the viruses rapidly diversified during a phase of population expansion and weak purifying selection. Only a few viruses escaped and seeded infection in the next set of cells, reducing genetic diversity. The cycling of stochastic reductions and rapid diversification led to unique subpopulations in each tissue and transmitted in the saliva.
(B) The genetic diversity of the transmitted viral populations is dependent upon the vector species, but all accumulate potentially deleterious mutations such as frame-shifting insertions and deletions and low fitness amino acid substitutions (mutations per genome represent both iSNVs and HF iLVs). The virus and mutation colors represent tissue of origin (bloodmeal = black, midgut = orange, hemolymph (legs) = red, salivary glands = green, saliva = blue) and “X” represents predicted deleterious mutations.
EXPERIMENTAL PROCEDURES
For detailed procedures, see the Supplemental Experimental Procedures.
Mosquito infections
Midguts, legs, salivary glands, and saliva were collected from Cx. quinquefasciatus, Cx. pipiens, Cx. tarsalis, and Ae. aegypti mosquitoes after ingesting bloodmeals containing ~2 × 108 PFU/mL of WNVic (strain NY99 collected during the 2000 outbreak in New York City).
Production of mutant viruses
Mutations were engineered into the WNVic using mutagenic primers and fragments containing overlapping sequences, amplified by rolling circle amplification, linearized, capped, and rescued in BHK-21 cells. The presence of desired mutations was confirmed by Sanger sequencing (Table S9).
Phenotypic assessment
Viral GE was determined by qRT-PCR and viral PFUs were determined by standard plaque assay in Vero cells. Replication fitness of the reconstructed WNV mutants was compared to the WNVic during infection of chicken DF-1 and Ae. albopicutus clone C6/36 cells at MOIs of 0.01. Competitive fitness of saliva WNV populations and the reconstructed WNV mutants (competitors) was determined by directly comparing their replication to a reference WNV during co-infection in DF-1 cells at MOIs of ~ 0.0008 and 0.01, respectively (Fitzpatrick et al., 2010). Quantitative Sanger sequencing was used to determine the proportion of competitor to reference WNV genotypes.
Sequencing
Total RNA from mosquito tissues and saliva after 14 days extrinsic incubation were amplified and prepared for NGS (100 nt paired-end reads) on the Illumina HiSeq 2500 platform at Beckman Coulter Genomics. All NGS data can be accessed from the NCBI BioProject PRJNA311123.
Intrahost variants
The reads aligned to the WNVic sequence, duplicate reads were removed, and variants were called using VPhaser2 (Yang et al., 2013). Variants with significant strand bias were removed. Analysis was limited to the protein coding sequences sequence (nucleotide positions 97-10,395) and was done separately for iSNVs and iLVs.
Genetic diversity
Genetic diversity was calculated using four separate methods: richness (sum of the iSNV sites detected in each population and normalized by the number of WNV reads [i.e. sites per million WNV reads]), distance (sum of the iSNV and amino acid substitution frequencies from each population [i.e. iSNVs and amino acid variants per coding sequence]), complexity (Shannon entropy [i.e. uncertainty associated with randomly sampling an allele]), and divergence (FST [i.e. genetic differences between two populations]).
Viral demographics
Haplotypes were reconstructed using QuasiRecomb 1.2 (Topfer et al., 2013) and manually edited to only include HF iSNVs (frequencies > 0.034). The thirty most common predicted haplotypes from each population (Tables S4-S7) were used to calculate Tajima’s D (Tajima, 1989) and Harpending’s raggedness index (Harpending et al., 1993; Rogers and Harpending, 1992). Haplotype phylogenies were constructed using BEAST (v1.8) (Drummond et al., 2012). Ne was determined by the genetic variance within and between populations using FST with only third codon synonymous iSNVs not predicted to co-occur on haplotypes with nonsynonymous mutations (Monsion et al., 2008).
Selection
The strength of purifying selection was measured by the accumulation of iLVs (all predicted to be deleterious) and the ratios of nonsynonymous (dN) to synonymous (dS) substitutions per nonsynonymous and synonymous coding sequence site.
Statistics
All tests were done using Kruskal-Wallis with Dunn’s corrections unless otherwise specified.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank A. Gendernalik, C. Rueckert, C. Nguyen, Z. Desmond, E. Petrilli, P. Buxton, S. Taylor, N. Duggal, A. Sherman, and A. Brault for laboratory support, providing mosquitoes, and/or useful discussions. G.D.E. and his lab are supported by the NIH under grant number AI067380.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures, nine tables, and experimental procedures.
AUTHOR CONTRIBUTIONS
Conceptualization, G.D.E. and N.D.G.; Methodology, G.D.E., N.D.G., W.C.B., J.W.L., and R.A.M.; Investigation, N.D.G., J.W.L., J.R.F., S.M.G.L., and A.N.P.; Writing – Original Draft, N.D.G. and G.D.E.; Writing – Reviewing & Editing, G.D.E., N.D.G., W.C.B., J.W.L., R.A.M., A.N.P., S.M.G.L., and J.R.F.
REFERENCES
- Bernard KA, Maffei JG, Jones SA, Kauffman EB, Ebel G, Dupuis AP, 2nd, Ngo KA, Nicholas DC, Young DM, Shi PY, et al. West Nile virus infection in birds and mosquitoes, New York State, 2000. Emerging infectious diseases. 2001;7:679–685. doi: 10.3201/eid0704.010415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti L, Kitron UD, Walker ED, Ruiz MO, Brawn JD, Loss SR, Hamer GL, Goldberg TL. Fine-scale genetic variation and evolution of West Nile Virus in a transmission "hot spot" in suburban Chicago, USA. Virology. 2008;374:381–389. doi: 10.1016/j.virol.2007.12.040. [DOI] [PubMed] [Google Scholar]
- Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS pathogens. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney DE, Pesko KN, Brown IK, Deardorff ER, Kawatachi J, Ebel GD. West Nile virus genetic diversity is maintained during transmission by Culex pipiens quinquefasciatus mosquitoes. PloS one. 2011;6:e24466. doi: 10.1371/journal.pone.0024466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney DE, Schirtzinger EE, Harrison TD, Ebel GD, Hanley KA. Modulation of Flavivirus Population Diversity by RNA Interference. Journal of virology. 2015;89:4035–4039. doi: 10.1128/JVI.02612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara AS, Gonzales G, Ferro C, Tamayo M, Aronson J, Paessler S, Anishchenko M, Boshell J, Weaver SC. Venezuelan equine encephalitis virus infection of spiny rats. Emerging infectious diseases. 2005;11:663–669. doi: 10.3201/eid1105.041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L. Fitness of RNA virus decreased by Muller's ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- Ciota AT, Ehrbar DJ, Van Slyke GA, Payne AF, Willsey GG, Viscio RE, Kramer LD. Quantification of intrahost bottlenecks of West Nile virus in Culex pipiens mosquitoes using an artificial mutant swarm. Infection Genetics and Evolution. 2012a;12:557–564. doi: 10.1016/j.meegid.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Ehrbar DJ, Van Slyke GA, Willsey GG, Kramer LD. Cooperative interactions in the West Nile virus mutant swarm. BMC evolutionary biology. 2012b;12:58. doi: 10.1186/1471-2148-12-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciota AT, Lovelace AO, Jia Y, Davis LJ, Young DS, Kramer LD. Characterization of mosquito-adapted West Nile virus. The Journal of general virology. 2008;89:1633–1642. doi: 10.1099/vir.0.2008/000893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey LL, Beeharry Y, Borderia AV, Blanc H, Vignuzzi M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey LL, Vasilakis N, Brault AC, Powers AM, Tripet F, Weaver SC. Arbovirus evolution in vivo is constrained by host alternation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6970–6975. doi: 10.1073/pnas.0712130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre JC, Holland JJ. RNA Virus Quasispecies Populations Can Suppress Vastly Superior Mutant Progeny. Journal of virology. 1990;64:6278–6281. doi: 10.1128/jvi.64.12.6278-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff ER, Fitzpatrick KA, Jerzak GV, Shi PY, Kramer LD, Ebel GD. West Nile virus experimental evolution in vivo and the trade-off hypothesis. PLoS pathogens. 2011;7:e1002335. doi: 10.1371/journal.ppat.1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte E, Clarke D, Moya A, Domingo E, Holland J. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel GD, Carricaburu J, Young D, Bernard KA, Kramer LD. Genetic and phenotypic variation of West Nile virus in New York, 2000-2003. The American journal of tropical medicine and hygiene. 2004;71:493–500. [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. The Clarendon press; Oxford: 1930. [Google Scholar]
- Fitzpatrick KA, Deardorff ER, Pesko K, Brackney DE, Zhang B, Bedrick E, Shi PY, Ebel GD. Population variation of West Nile virus confers a host-specific fitness benefit in mosquitoes. Virology. 2010;404:89–95. doi: 10.1016/j.virol.2010.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester NL, Guerbois M, Seymour RL, Spratt H, Weaver SC. Vector-Borne Transmission Imposes a Severe Bottleneck on an RNA Virus Population. PLoS pathogens. 2012 doi: 10.1371/journal.ppat.1002897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh ND, Smith DR, Brackney DE, Bosco-Lauth AM, Fauver JR, Campbell CL, Felix TA, Romo H, Duggal NK, Dietrich EA, et al. Experimental evolution of an RNA virus in wild birds: evidence for host-dependent impacts on population structure and competitive fitness. PLoS pathogens. 2015;11:e1004874. doi: 10.1371/journal.ppat.1004874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez S, Thebaud G, Smith DR, Kenney JL, Weaver SC. Demographics of Natural Oral Infection of Mosquitos by Venezuelan Equine Encephalitis Virus. Journal of virology. 2015;89:4020–4022. doi: 10.1128/JVI.03265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpending HC, Sherry ST, Rogers AR, Stoneking M. The Genetic-Structure of Ancient Human-Populations. Curr Anthropol. 1993;34:483–496. [Google Scholar]
- Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, Vandepol S. Rapid Evolution of Rna Genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. The Journal of general virology. 2005;86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak GV, Bernard K, Kramer LD, Shi PY, Ebel GD. The West Nile virus mutant spectrum is host-dependant and a determinant of mortality in mice. Virology. 2007;360:469–476. doi: 10.1016/j.virol.2006.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerzak GV, Brown I, Shi PY, Kramer LD, Ebel GD. Genetic diversity and purifying selection in West Nile virus populations are maintained during host switching. Virology. 2008;374:256–260. doi: 10.1016/j.virol.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Kramer LD, Campbell SR, Alleyne EO, Dobson AP, Daszak P. West Nile virus risk assessment and the bridge vector paradigm. Emerging infectious diseases. 2005;11:425–429. doi: 10.3201/eid1103.040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N, Langevin S, Hinten S, Nemeth N, Edwards E, Hettler D, Davis B, Bowen R, Bunning M. Experimental infection of North American birds with the New York 1999 strain of West Nile virus. Emerging infectious diseases. 2003;9:311–322. doi: 10.3201/eid0903.020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer LD, Styer LM, Ebel GD. A global perspective on the epidemiology of West Nile virus. Annual review of entomology. 2008;53:61–81. doi: 10.1146/annurev.ento.53.103106.093258. [DOI] [PubMed] [Google Scholar]
- Lanciotti RSV. Transcontinental movement of Asian genotype chikungunya virus [letter] Emerging infectious diseases. 2014 doi: 10.3201/eid2008.140268. A.M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsion B, Froissart R, Michalakis Y, Blanc S. Large Bottleneck Size in Cauliflower Mosaic Virus Populations during Host Plant Colonization. PLoS pathogens. 2008:4. doi: 10.1371/journal.ppat.1000174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudy RM, Meola MA, Morin LL, Ebel GD, Kramer LD. A newly emergent genotype of West Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. The American journal of tropical medicine and hygiene. 2007;77:365–370. [PubMed] [Google Scholar]
- Muller HJ. The Relation of Recombination to Mutational Advance. Mutation research. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Musso D, Cao-Lormeau VM, Gubler DJ. Zika virus: following the path of dengue and chikungunya? Lancet. 2015;386:243–244. doi: 10.1016/S0140-6736(15)61273-9. [DOI] [PubMed] [Google Scholar]
- Rogers AR, Harpending H. Population-Growth Makes Waves in the Distribution of Pairwise Genetic-Differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Sanjuan R, Moya A, Elena SF. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8396–8401. doi: 10.1073/pnas.0400146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Aw PP, Wilm A, Teoh G, Hue KD, Nguyen NM, Nagarajan N, Simmons CP, Hibberd ML. Tracking Dengue Virus Intra-host Genetic Diversity during Human-to-Mosquito Transmission. PLoS neglected tropical diseases. 2015;9:e0004052. doi: 10.1371/journal.pntd.0004052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR. Pairwise Comparisons of Mitochondrial-DNA Sequences in Stable and Exponentially Growing Populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleford KA, Coffey LL, Lay S, Borderia AV, Duong V, Isakov O, Rozen-Gagnon K, Arias-Goeta C, Blanc H, Beaucourt S, et al. Emergence and Transmission of Arbovirus Evolutionary Intermediates with Epidemic Potential. Cell host & microbe. 2014;15:706–716. doi: 10.1016/j.chom.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Stern A, Bianco S, Yeh MT, Wright C, Butcher K, Tang C, Nielsen R, Andino R. Costs and benefits of mutational robustness in RNA viruses. Cell reports. 2014;8:1026–1036. doi: 10.1016/j.celrep.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical-Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topfer A, Zagordi O, Prabhakaran S, Roth V, Halperin E, Beerenwinkel N. Probabilistic inference of viral quasispecies subject to recombination. Journal of computational biology : a journal of computational molecular cell biology. 2013;20:113–123. doi: 10.1089/cmb.2012.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin KA, Chen RB, Yun RM, Rossi SL, Plante KS, Guerbois M, Forrester N, Perng GC, Sreekumar E, Leal G, et al. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014 doi: 10.1038/ncomms5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turell MJ, Dohm DJ, Sardelis MR, Oguinn ML, Andreadis TG, Blow JA. An update on the potential of north American mosquitoes (Diptera: Culicidae) to transmit West Nile Virus. Journal of medical entomology. 2005;42:57–62. doi: 10.1093/jmedent/42.1.57. [DOI] [PubMed] [Google Scholar]
- Van Slyke GA, Arnold JJ, Lugo AJ, Griesemer SB, Moustafa IM, Kramer LD, Cameron CE, Ciota AT. Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes. PLoS pathogens. 2015;11:e1005009. doi: 10.1371/journal.ppat.1005009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakis N, Deardorff ER, Kenney JL, Rossi SL, Hanley KA, Weaver SC. Mosquitoes Put the Brake on Arbovirus Evolution: Experimental Evolution Reveals Slower Mutation Accumulation in Mosquito Than Vertebrate Cells. PLoS pathogens. 2009:5. doi: 10.1371/journal.ppat.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelk CH, Holmes EC. Reduced positive selection in vector-borne RNA viruses. Mol Biol Evol. 2002;19:2333–2336. doi: 10.1093/oxfordjournals.molbev.a004059. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution and the Genetics of Populations. Vol. 3. University of Chicago Press; Chicago: 1977. [Google Scholar]
- Yang X, Charlebois P, Macalalad A, Henn MR, Zody MC. V-Phaser 2: variant inference for viral populations. BMC genomics. 2013;14:674. doi: 10.1186/1471-2164-14-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.