Abstract
The ability to accurately and systematically evaluate the cellular mechanisms underlying human neurodegenerative disorders such as Alzheimer's disease (AD) should lead to advancements in therapeutics. Recent developments in human induced pluripotent stem cells (iPSCs) have afforded the opportunity to use human neurons and glia to study cellular changes involved in neurological diseases. iPSCs have the potential to be differentiated into AD-relevant cell types, including forebrain neurons, astrocytes, and microglia. This permits the evaluation of individual cell types in isolation or in concert, thus modeling the interdependence of cell types within the brain. When discussing the potential of modeling AD with iPSCs, it is important to remember that the umbrella diagnosis of “Alzheimer's disease” represents a disease that is heterogeneous in terms of age of onset, underlying causes, and at times precise pathology. The ability of iPSCs to be derived from an array of AD patients allows for a closer examination of the mechanism of disease progression in particular subsets of subjects, who may have different mutations and allelic variants affecting their risk for disease. Disease mechanisms can be probed both by the genetic manipulation of iPSCs and by modifications to the cellular environment by chemical treatment. These studies may lead not only to the refinement of known pathways implicated in AD, but also to the identification of novel pathways heretofore unaffiliated with disease pathology. In this review, we describe the potential of iPSC models to transform our understanding of AD and to lead to valuable advancements in therapeutics.
Keywords: Stem cell, iPS, Alzheimer's disease, Aβ, Tau, Induced pluripotent stem cell
1. Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative disorder characterized by dysfunction and deterioration of neurons within the cerebral cortex resulting in loss of memory and progressive cognitive decline. According to the Alzheimer's Association, AD is the 6th leading cause of death in the United States and the number one cause of dementia (Thies and Bleiler, 2013). The earliest symptoms of AD are subtle difficulties with memory storage and recall. Episodic memory impairment becomes more striking as the disease progresses and, in the final stages of disease; AD patients suffer from amnesia and an acute decline in cognitive faculties, including executive function and language.
An overall reduction in brain volume and degeneration of the hippocampus, a key brain region for memory consolidation and spatial orientation, indicate AD pathology. AD is hallmarked by the presence of intracellular neurofibrillary tangles and the deposition of extracellular neuritic plaques. Hyperphosphorylation of the microtubule-associated protein tau leads to a conformational change that causes tau to dissociate from microtubules and aggregate into paired helical filaments (reviewed in Ballatore et al. (2007)). Neuritic plaques arise from the aggregation of amyloid-β (Aβ) peptides. While mutation in the gene encoding tau (MAPT) results in other forms of dementia including frontotemporal dementia, mutation of Presenilins (PSEN1, PSEN2) or amyloid precursor protein (APP) cause early-onset, familial forms of AD (EOAD, fAD). APP encodes the precursor protein to Aβ, while PSEN encodes the catalytic site of an enzyme that cleaves APP to generate Aβ. These genetic and pathological findings place Aβ in a central role in AD pathogenesis. In addition to these findings in human subjects, decades of research have provided evidence that high concentrations of Aβ are neurotoxic and lead to inflammation, synaptotoxic effects, and neuronal loss (reviewed in Hardy and Selkoe (2002) and Walsh et al. (2003)).
An estimated 5 million Americans are suffering from AD. As the “baby boomer” generation reaches a vulnerable age, that number is expected to nearly triple by 2050. Between 2000 and 2010, the occurrence of AD was increased by 68% (Thies and Bleiler, 2013). During that same period, the incidence of other major causes of death including heart disease, stroke and cancer were down an average of 15% (Thies and Bleiler, 2013). Thus there is an immediate need to develop efficacious therapies, a goal which in part has been stymied due to lack of faithful cellular and animal models.
Current treatment regimes are purely palliative and do not modify the disease process. The two classes of drugs approved for AD treatment, acetylcholinesterase inhibitors and N-methyl-d-aspartate (NMDA) receptor modulators, are thought to work by maximizing productive neurotransmission and are only mildly and transiently beneficial. Despite strong supporting results of novel therapeutic strategies in animal models, recent drug trials have not yielded safe and effective disease-modifying treatments.
One possible explanation for this failure to translate laboratory results into clinical success may be the insufficiency of our disease models (reviewed in Franco and Cedazo-Minguez (2014) and Wojda and Kuznicki (2013)). To date, mechanistic work has relied heavily on non-neuronal human cells and non-human animal models. Non-neuronal cells lack the unique cellular structures and pathways of neurons and therefore may fail to capture a complete picture of disease-relevant cell biological changes. Mice are attractive disease models as transgenic technology allows for genetic manipulation, and assessments of cognitive function have been well established in these animals. However, in order to reproduce AD neuropathological hallmarks, mice must be made to overexpress one or more mutant human genes. This allows for the analysis of only a small number of contributing factors to what is widely accepted as a multifactorial disease. The brain anatomy and genetics of non-human animals are less complex and therefore key effectors may be lacking in these models. Notably, the majority of murine models of AD fail to recapitulate the widespread AD-related neuronal loss, which is itself a critical feature to disease progression. The reliance on known FAD mutations to model disease and the requirement for overexpression of these mutant human genes hinders study of sporadic AD, which is significantly more prevalent in the population. Consequently, it is important to develop discovery and pre-clinical neuronal and glial systems to complement animal models. Human induced pluripotent stem cells (iPSCs) offer an attractive model that should recapitulate critical disease-relevant cell biological changes. However, it is clear that we cannot gain significant insights from the study of iPSC lines from only a few late-onset AD subjects. Genetic studies and clinical observations suggest that AD is heterogeneous in terms of age-of-onset and genetic contributors (Fig. 1). The ability to generate and study cells from an array of subjects with sporadic, late-onset AD allows us to probe the mechanisms of disease progression in different subsets of subjects.
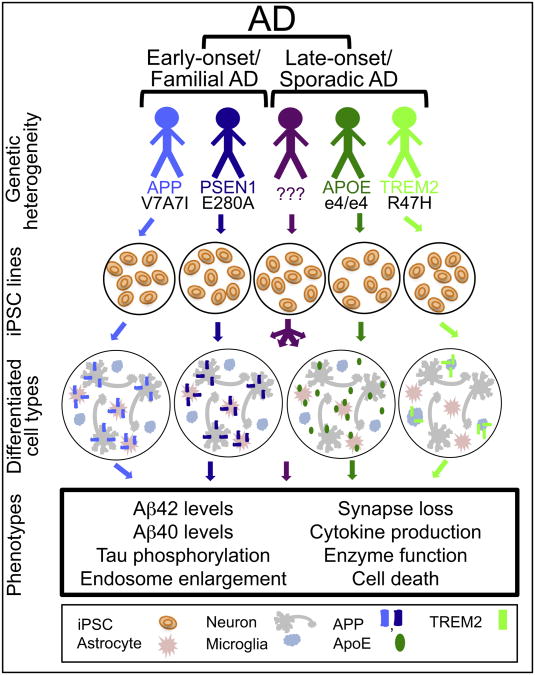
Fig. 1.
Considerations for modeling AD-relevant processes using iPSCs. Alzheimer's disease modeling in a dish requires the consideration of several factors. Firstly, AD is not a homogeneous disease, with varying age-of-onset and underlying genetic influences. Multiple cell types are involved in AD pathogenesis, and the specific question being addressed will determine which cell type(s) should be interrogated. A host of phenotypes can be investigated in iPSC-derived neurons and glia that recapitulate various aspects of the AD. Genetic studies suggest that different mechanisms may underlie AD pathogenesis in different individuals. The identification of phenotypic differences between lines from different individuals with AD has the potential to provide a molecular basis for subdividing the disease, which ultimately could have implications for patient treatment.
iPSCs are derived from somatic cells and retain their potential to give rise to all cell types. This technology allows for the modeling of mature cell types that are directly affected by disease. In the case of AD, neurons and glia can be derived from iPSCs of known genetic backgrounds and used to probe mechanisms of disease and test therapeutic interventions. The choice of which cell type(s) to interrogate, and the cell and molecular phenotypes studied are two important considerations when modeling various aspects of the disease (Fig. 1). In this review we will explore the use of iPSCs as a model of AD and discuss their utility as a tool for mechanism and therapeutic discovery.
2. Human iPSC differentiation into neural subtypes relevant to AD
The discovery of methods to generate iPSCs (Takahashi et al., 2007) was particularly important for diseases of the nervous system, since it allows for the potential to provide a virtually limitless supply of cell types (human brain cells) which were previously unattainable. The field of embryonic stem (ES) cell biology had established numerous methods for differentiation of ES cells to neural fates, and the iPSC field rapidly adapted and optimized these methods for use in iPSC differentiation. While much progress has been made in generating a number of cell types found in the brain, further work is required to truly recapitulate the full repertoire of cells found in the mature brain.
2.1. Forebrain neurons
Alzheimer's disease typically begins with loss of episodic memory, but ultimately leads to additional cortical symptoms related to language, attention, and visuospatial orientation. Amyloid plaque deposition, neuritic plaques, neurofibrillary tangles and neuronal loss are extensive in the cerebral cortex. Therefore, studies modeling AD with iPSCs often begin with the generation of forebrain neurons. Differentiation to forebrain neuronal fates is referred to as the “default” for neuronal differentiation strategies, as differentiation to the neural lineage in the absence of exogenous patterning factors typically leads to neurons expressing markers of cortical neuronal fates. The base protocols used for these differentiations include dual-SMAD inhibition in a monolayer culture (Chambers et al., 2009), an embryoid body or aggregate method (Eiraku et al., 2008) or more recently, the formation of cerebral organoids (Lancaster et al., 2013).
Both glutamatergic and GABAergic neuronal fates have been implicated in different aspects of AD pathogenesis. Through modulation of patterning pathways, differentiation can be directed to generate cultures of more dorsal, glutamatergic fates (Boisvert et al., 2013; Vazin et al., 2014; Zeng et al., 2010) or else of more ventral, GABAergic fates (Cunningham et al., 2014; Nicholas et al., 2013; Vazin et al., 2014; Yan et al., 2013). It was recently shown that iPSC-derived GABAergic neurons are less susceptible to Aβ induced cell death (Vazin et al., 2014), thus demonstrating the differential phenotypes that can be understood by specifying cell fates.
The loss of basal forebrain cholinergic neurons is associated with deficits in spatial learning and memory in AD. Since cholinergic neurons are among the first to degenerate in AD, there has been much interest in directing differentiation to this fate. Cholinergic neurons can be generated from human ES cells by sequential treatment with growth factors that are found in the forebrain during different developmental stages (Bissonnette et al., 2011). Alternatively, cholinergic neurons can be generated by the overexpression of transcription factors. Expression of Lhx8 and Gbx1 is necessary and sufficient to generate basal forebrain cholinergic neurons (Bissonnette et al., 2011). Production of cholinergic neurons by nucleofection with Lhx8/Gbx1-IRES-GFP allows for additional purification of cell populations by FAC-sorting. iPSC-derived basal forebrain cholinergic neurons derived from sporadic AD patients of the ApoE3/E4 background using this method displayed typical AD biochemical features including an increase in the Aβ42 to Aβ40 ratio and an increased vulnerability to glutamate-mediated cell death (Duan et al., 2014).
2.2. Astrocytes
In addition to neurons, astrocytes play an important role in AD pathogenesis. Astrogliosis is a prominent hallmark of the disease, and astrocytes are important for neural homeostasis. In response to insult, astrocytes act to prevent further damage, although aberrant astrocyte activity can contribute to pathology. Derivation and culture of astrocytes from iPSCs will allow research into the mechanisms by which astrocytes lead to, as well as protect from, pathology.
Astrocytes and neurons share a common progenitor, and astrocytes arise spontaneously from neuronal differentiation schemes. As such, astrocytes can be isolated and purified from neuron populations using cell specific markers. Yuan et al. (2011) identified the cell surface signatures for astrocytes as CD184 and CD44. Neural stem cell populations sorted on the basis of being CD184+/CD44+ were enriched in cells that would mature into GFAP positive astrocytes (Yuan et al., 2011).
Differential culture conditions favor neuronal growth versus astrocyte proliferation. Alternatively, culture conditions can be optimized to produce astrocytes (Emdad et al., 2012; Krencik and Zhang, 2011; Yuan et al., 2011; Zhang et al., 2013). Krenicik and Zhang (2011) demonstrated that regular dissociation of neuroepithelial clusters in the presence of mitogens leads to the generation of astroglial cells. Subsequent withdrawal of mitogen and addition of ciliary neurotrophic factor (CNTF) will allow differentiation into functional astrocytes (Krencik and Zhang, 2011). Mature, functional astrocytes also have been obtained by treatment of iPSC-derived neuroepithelial cells with FGF2 and CNTF followed by CNTF alone (Emdad et al., 2012). Recently, direct reprogramming of mouse fibroblasts with NFIA, NFIB, and SOX9 has resulted in astrocyte generation (Caiazzo et al., 2015). Introduction of these same transcription factors to human cells was able to generate a small number GFAP and S100B positive cells (Caiazzo et al., 2015), indicating that a similar protocol may soon be available for the robust generation of human astrocytes.
2.3. Microglia
Accumulating evidence from genetic and animal models suggests that microglia-mediated neuroinflammatory processes are a significant contributor to AD pathogenesis. Unlike neurons, astrocytes, and oligodendrocytes, which arise from an ectodermal lineage, microglia arise from myeloid precursor cells generated in the yolk sac, which then migrate in to the brain at embryonic time points. Numerous labs are developing protocols for iPSC differentiation to microglial fate based upon the fields' collective knowledge of how these cells develop endogenously. To date, an efficient protocol for directing hiPSCs to microglial fate through the myeloid lineage has not been published to date. Differentiation of mouse embryonic stem cells through a modified neural differentiation to yield microglia capable of chemokine-directed migration and phagocytosis has been reported (Beutner et al., 2010). The behavior of microglia in vitro is quite different from their behavior in vivo, both in terms of gene expression profiles and morphology (reviewed in Ransohoff and Cardona (2010)), complicating the assessment of success in their derivation from stem cells.
3. Human iPSC patient derived lines
Two major contributors to AD pathology are age and genetics. The average age of onset for the more pervasive, age-related form of AD, sporadic AD, is 65 years with diagnosis increasingly more likely each subsequent year (Selkoe, 2001). Heri-table AD, termed familial AD (FAD) or early onset AD (EOAD), emerges decades sooner than sporadic AD, with diagnosis generally occurring between 30 and 60 years of age. Although FAD is estimated to represent less than 5% of all AD cases (Bekris et al., 2010), the insights gained from the analysis of the genetics of fAD have proven invaluable. FAD results from mutations in the γ-secretase complex members presenilin 1 (PSEN1) and presenilin 2 (PSEN2) or in the precursor protein to Aβ, APP. These mutations are inherited in an autosomal-dominant fashion and are fully penetrant, and result in an accelerated onset of symptoms. Importantly, these mutations are sufficient to reproduce certain disease phenotypes in culture without additional genetic or environmental factors. Therefore, FAD mutations have been the favored laboratory model for AD. In contrast, the etiology of sporadic AD is nebulous and likely more heterogeneous in nature.
Both genetic and extrinsic influences contribute to pathology, contributing to the challenge of modeling sporadic AD in vitro or in animal models. Mitigating factors such as incomplete reprogramming and variable efficiency in cell differentiation can each lead to phenotypic variability within culture models (reviewed in Soldner and Jaenisch (2012)). Added to this, the diversity of patient genetic backgrounds as well as other extenuating factors, such as disease progression, can make properly controlling experiments difficult and therefore confound the interpretation of results. The use of genome-editing to correct or manufacture mutations allows for “wild type” controls to be generated from patient cells and “disease” models from non-affected donors. To further enhance cell models, the use of reporters, chemical selection, and FACs purification allow incompletely differentiating or confounding cell types to be removed from cultures, thus enhancing the homogeneity of the culture (for example, see Yuan et al. (2011) and Zhang et al. (2013)).
3.1. fAD lines
Mutations in APP represent a subset of known FAD mutations. Pathogenic mutations can result in preferential cleavage of APP by β-secretase, thus leading to elevated Aβ production. APP mutations also can facilitate the production of the 42 amino acid Aβ species, Aβ42, which has an increased propensity for aggregation (Hardy and Selkoe, 2002). Mutations within the Aβ sequence may result in AD phenotypes through other means. The “Arctic” mutation (E693G), for example, leads to lower Aβ40 and Aβ42 plasma levels, but results in production of Aβ with a higher propensity for protofibril formation (Nilsberth et al., 2001).
To assess the ability of iPSC-derived neurons to replicate known phenotypes, multiple studies have now been published where iPSC lines were generated from fAD subjects with duplication or mutation of APP. In the first of these, lines from two FAD patients possessing duplications of the APP gene were generated (Israel et al., 2012). Both of these lines produced neurons that, relative to neurons from age-matched, non-demented controls, had elevated levels of Aβ40, active GSK3β, and phosphorylated tau and total tau. Interestingly, these neurons were responsive to β- and γ-secretase inhibitors to lower Aβ levels, but only β-secretase inhibitors rescued the observed tau phenotypes, suggesting that APP cleavage products other than Aβ play a role in induction of GSK3β activity and phospho-tau (Israel et al., 2012). These neurons, when co-cultured with astrocytes, produced enlarge, Rab5 positive early endosomes (Israel et al., 2012). The presence of enlarged endosomes within the AD patient brain is an established phenotype and coincides with elevated Aβ40 and 42 prior to plaque deposition (Cataldo et al., 2000; Cataldo et al., 2004).
In another study, an iPSC line was derived from patients carrying a rare autosomal-recessive mutation in the APP gene that produces Aβ lacking the Glu22, AD (APP-E693Δ). This mutation leads to early-onset AD symptoms and only low levels of PIB-PET detectable amyloid in (Kondo et al., 2013). An abundance of intracellular Aβ oligomers was noted in neuronal cells derived from this lineage, while extracellular plaque formation was absent. Further, the intracellular Aβ oligomers provoked both endoplasmic reticulum and oxidative stress responses in AD (APP-E693Δ)-derived neural cells. Interestingly, an additional FAD-related APP mutant line, APP-V717L, was tested in parallel and produced large quantities of extracellular Aβ42 and lacked intracellular accumulation and the accompanying stress response hallmarks (Kondo et al., 2013) in contrast to the results in the APP-E693Δ mutant derived neurons. This analysis demonstrates the ability of APP mutations to produce drastically different phenotypes while still producing AD symptoms in patients.
In work from our lab, iPSC lines from two subjects containing the London mutation (APPV717I) were generated. When directed to forebrain neuronal fates, this mutation increased β-secretase cleavage of APP, and altered γ-secretase cleavage such that Aβ42 and Aβ38 levels were increased. At early neuronal differentiation time points, total tau levels were elevated, and in mature neurons, the relative level of phospho- to total-tau also was increased. Tau levels were reversed with early Aβ antibody intervention (Muratore et al., 2014), demonstrating that the tau phenotype was dependent in part upon altered Aβ levels induced by the mutation. Recent studies of forebrain neurons from novel fAD lines harboring the London mutation, as well as APP duplication lines, confirmed the elevation of intracellular tau protein levels (Moore et al., 2015). Interestingly, in the same study it was shown that neurons from lines with PSEN1 fAD mutations did not show the elevated tau phenotype. Further, inhibition of gamma-secretase activity did not rescue elevations in tau, while inhibition of beta-secretase did rescue the tau pheno-type (Moore et al., 2015). Together, these studies potentially point to a role for Aβ, as well as other APP cleavage products, in downstream effects on tau (Moore et al., 2015; Muratore et al., 2014).
In addition to APP, mutations in the catalytic subunit of the enzyme that cleaves APP also lead to FAD. Approximately 90% of mutations linked to FAD are in PSEN1 (reviewed in Wolfe (2010)). In iPSC-derived neurons, the AD-associated PSEN1L166P mutation produced an increase in the Aβ42/40 ratio owing to a selective decrease in Aβ40 production (Koch et al., 2012). Additional PSEN1 mutations, PSEN1A246E and PSEN1M146L, also elevate the Aβ42/40 ratio, thus mimicking an AD-like amyloidogenic process (Mahairaki et al., 2014; Sproul et al., 2014; Yagi et al., 2011). Neuroprogenitor cells from the PSEN1A246E and PSEN1M146L were shown to possess 14 genes that were differentially regulated in FAD lines compared to controls (Sproul et al., 2014). Five of these genes were previously demonstrated to be differentially expressed in the brains of late and intermediate AD patients, thus demonstrating the power of iPSC to replicate additional phenotypic features of disease.
Mutations in PSEN2 also exhibit γ-secretase elicited changes in APP processing. Like PSEN1 mutant cells, iPSC-derived neurons carrying PSEN2N141I had an increase in Aβ42 secretion (Yagi et al., 2011). PSEN2N141I and PSEN1A246E iPSC-derived neurons lacked both neurofibrillary tangles and the presence of hyperphosphorylated tau. Further study is necessary to determine if the failure to display this AD attribute was due to the short experimental window in which this study was conducted or an inability of tau to be affected in these cells. Each line also responded to γ-secretase inhibition as predicted, demonstrating the ability of iPSC-derived neurons to replicate known drug actions (Yagi et al., 2011).
3.2. Late-onset, “sporadic” AD lines
The majority of AD patients suffer from late-onset, sporadic AD. Sporadic AD is defined by a lack of autosomal-dominant inheritance. While lumped together into one category, the underlying causes of sporadic AD are multifactorial, and likely do not represent a single disease. This is evident from several studies that compared iPSC lines derived from different sporadic AD patients, each of which displayed distinct cellular phenotypes. In the study by Israel et al. (2012), neurons derived from one of two sporadic AD patient iPSC lines exhibited similar phenotypes (elevated Aβ40 and phospho-tau levels) to those of FAD patient-derived iPSC neurons, whereas the other sporadic AD line did not.
The heterogeneous nature of the phenotypes found in sporadic lines was further demonstrated in another study by evaluation of two different sAD iPSC lines, AD3E211 and AD8K213. Intracellular production and accumulation of Aβ oligomers were elevated in AD8K213 neural cells compared to AD3E211 and control neuronal cells, suggesting that these neurons retain a cellular environment capable of producing Aβ aggregates (Kondo et al., 2013). The Aβ accumulation in AD8K213 neurons induced endoplasmic reticulum and oxidative stress without caspase-4 activation, which is dissimilar to the stress produced in an FAD line assessed concurrently (Kondo et al., 2013).
Additional recent studies have demonstrated the utility of sAD iPSCs in investigating the molecular mechanisms behind disease phenotypes. In an elegant study interrogating genetic risk variants in sAD, 13 hiPSC lines (6 control and 7 sAD) were analyzed that were genotyped for risk (R) or protective (P) variants of the SORL1 gene (Young et al., 2015). At the basal level, no significant effect on SORL1 expression was observed of diagnosis or SORL1 haplotype. However, interestingly, BDNF treatment induced upregulation of SORL1 only in the lines with the P haplotype. Further, the fold reduction in Aβ40 secretion following BDNF treatment was significantly reduced in cells with the R haplotype, suggesting a mechanism behind the increased risk. The authors went on to show that the effect of BDNF on Aβ secretion was SORL1-dependent through targeted knock down of SORL1 with shRNA constructs (Young et al., 2015). This study suggests that iPSC technology may be a valuable tool in exploring the heterogeneous nature of the etiology of sAD through the interrogation of functional effects of genetic variants linked to risk and protection from AD.
4. Modeling late-stage AD pathology in vitro
As summarized above, phenotypes such as altered Aβ levels, altered tau levels and phosphorylation state, and cellular stress have been widely observed and reported in iPSC-derived neurons from AD backgrounds. However, Aβ rich extracellular plaques and hyperphosphorylated tau tangles mark late-stage AD, and these are challenging to model in vitro. With few exceptions (Choi et al., 2014), plaques and tangles have not been reported in in vitro cell models. The inability to recapitulate late-stage AD phenotypes in culture may be due in part to the maturity level of in vitro neurons or to culture procedures that require frequent media changes causing extracellular species to be cleared from cultures. We outline impediments and possible solutions to modeling late-stage processes of AD in culture in a later section of this review.
4.1. Aβ pathology
As outlined above, several studies have shown altered Aβ phenotypes in iPSC models of AD. In Down's syndrome iPSC models, plaque-like structures were identified by staining neural cultures with BTA1, a fluorescent thioflavin T analog with a high affinity for amyloid deposits, two months from the start of differentiation (Shi et al., 2012). Additional antibody staining revealed extracellular Aβ42-positive aggregates localized to neurites (Shi et al., 2012).
While not in iPSC-derived neural cultures, a recent paper by Choi et al. (2014) presented the formation of both plaques and tangles in an in vitro cellular model. Two modifications were likely contributing to these findings: 1) both mutant human APP and PSEN1 were overexpressed well above endogenous levels and 2) the cells were embedded in matrigel, which likely limited the diffusion of toxic Aβ species, thus maintaining a high local Aβ concentration necessary for fibril and subsequent plaque formation. It remains to be seen whether endogenous levels of fAD- or sAD-derived Aβ can induce plaque formation in vitro if cells are plated in a 3-dimensional matrix.
While overexpression of mutant proteins can be a valuable tool for accelerating pathology in cellular and animal systems, it may potentially mask certain processes relevant to the mechanisms of disease progression. Proteolytic processing of overexpressed APP has been shown to be dramatically different than that of endogenous APP. For example, in HEK293T cells, the APPsα:APPsβ ratio in media is 10-fold higher in cells overexpressing human APP relative to cells expressing endogenous APP (Rice et al., 2013). This remarkable difference highlights the non-physiological nature of the processing of overexpressed APP. A recent study in hiPSCs has highlighted the importance of interrogating drug responsiveness in the cell types of interest that express endogenous levels of relevant proteins (Liu et al., 2014). In this study, Liu et al. generated iPSCs from multiple PSEN1 mutation carriers, differentiated these cells to neural fates, and examined their responsiveness to gamma-secretase modulators (GSMs). This study revealed differences in the types of Aβ generated in response to a GSM in neurons derived from human subjects relative to several previous reports, all of which had tested the effects of the modulator in APP-overexpressing cell lines.
4.2. Tau pathology
Increases in phosphorylated tau at multiple residues have been reported in iPSC models of fAD and sAD (Israel et al., 2012; Muratore et al., 2014). However, the formation of tangles has been limited to the in vitro model of combined APP/PSEN1 overexpression in a support matrix (Choi et al., 2014). A major challenge to in vitro recapitulation of tau pathology has been the relative immaturity of the neurons generated from standard differentiation protocols. While electrically active, the RNA expression profile for iPSC-derived neurons are more similar to those of late fetal neurons (Brennand et al., 2015). One particularly relevant constraint of immaturity is in the limited tau isoforms expressed in iPSC-derived neurons (reviewed in Spillantini and Goedert (2013)). Tau is alternatively spliced at exon 10 to make isoforms with (4R) and without (3R) the exon. Developing neurons express 3R, while the adult brain has equal expression of 3R and 4R. In many tauopathies, 4R predominates, indicating that 4R is an important form of neurotoxic tau and its elevated presence over 3R in culture is critical to fully modeling the role of tau in pathology. To overcome this, current optimization of differentiation protocols aims to accelerate the maturation of neurons. For example, exogenous neurogenin 2 is sufficient to convert iPSCs into induced neuronal cells in less than 2 weeks (Zhang et al., 2013). Expression of progerin in iPSC-derived fibroblasts and neurons is sufficient to induce age-related markers such as dendrite degeneration, loss of tyrosine-hydroxylase expression, and mitrochondrial enlargement (Miller et al., 2013). The use of cellular stressors including reactive oxygen species and adenosine also has been suggested to induce an “aging” phenotype (Campos et al., 2014). Optimization and refinement of these and other techniques may allow for more complete modeling of late stage disease processes.
4.3. Cell loss
In all of the AD iPSC models studied to date, little overt neuronal loss has been noted despite cell loss being a prominent feature of AD. This is perhaps owing to factors discussed above including clearance of neurotoxic species, immaturity of neurons, or lack of aging in culture compared to decades of pathology in vivo. While it may not be necessary to recapitulate this late phenotype when studying initiating factors in the disease process, one approach to attempt to circumvent this issue is the application of exogenous neurotoxic species to iPSC-derived neurons. For example, differentiation of human ES cells treated with oligomeric Aβ40 and Aβ42 yielded opposing effects, with oligomeric Aβ40 increasing and Aβ42 decreasing the number of functional neurons without affecting the total number of neurons derived (Wicklund et al., 2010), suggesting that Aβ42 oligomers may impair neuron function and contribute to disease. Treatment with fibrillar Aβ40 or Aβ42 induced gliogenesis, further defining the pathogenic differences between oligomeric and fibrillar Aβ species. An additional study demonstrated that oligomeric prefibrillar Aβ led to an increase in cell death within the glutamatergic neuron population (Vazin et al., 2014). By examining AD-related insults to wild-type human neurons, studies such as these will lead to a better understanding of the mechanisms through which neurotoxic species lead to neurodegeneration.
5. Novel pathway discovery
An important utility of iPSCs is the discovery and refinement of novel pathways involved in AD pathology. Genome wide association screens have identified a number of genes predicted to be linked to AD. Genetically altering these target genes will enable validation of their involvement in AD and uncover additional members of their pathogenic pathways. There are multiple tools available to perturb gene expression, outlined below.
5.1. Genetic techniques
iPSCs are amenable to manipulation and genome editing while maintaining genome stability. As such, a variety of gene-editing techniques have been developed to facilitate investigation of specific genes and their functions (reviewed in Li et al. (2014)). Genomic engineering by zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENS), and the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 RNA-guided nuclease system are used to permanently alter target genome sequences. For example, TALENS have been used to generate isogenic cells to examine a series of allelic PSEN1 mutations and determine their effects on disease-related phenotypes (Woodruff et al., 2013). Importantly, this study interrogated the effects of PSEN1 mutations in cells expressing endogenous levels of the gene. Through these studies the authors conclude that PSEN1 mutations dominantly gain a toxic activity, rather than simply acting in a loss-of-function manner.
In addition to genome-editing strategies, RNA manipulation can be accomplished by use of viral vectors such as lentivirus or adeno-associated virus to modulate gene expression. One valuable feature of adeno-associated viruses is the variable efficiency of serotypes for different cell types. For example, serotypes 9, 6, and 2 are best for use in iPSC-derived neurons, while serotype 9 is less efficient in human astrocytes (Sullivan and Young-Pearse, unpublished observation). Should these techniques be unavailable or inefficient, electroporation-based transfection methods also may be used to introduce nucleic acids that are difficult to transfect into cells by other methods.
5.2. APOE and related genes
Apolipoprotein E (ApoE) is the strongest risk locus for late-onset AD. Of the 3 common alleles, ε2, ε3 and ε4, ApoEε4 has been associated with increased risk and decreased age of AD onset. ApoE plays an important role in lipid transport but has been implicated in other process, including Aβ homeostasis, synaptic function, immune regulation, and intracellular signaling (reviewed in Schellenberg and Montine (2012)). With this wide range of functional duties, there are many potential impacts of ApoE allele variants on neurodegeneration. Defining the precise function or functions of ApoE that lead to AD pathology is likely to allow for more specific therapeutic targeting.
Additional AD-associated genes have been proposed to act in concert with APOE to lead to disease phenotypes. For example, other putative regulators of lipid metabolism (SORL1, Clu, and ABCA7) have been associated with AD and may act in concert with APOE (reviewed in Schellenberg and Montine (2012)). SORL1 encodes a lipoprotein receptor that can bind ApoE and may therefore be acting in the same pathways. Clusterin is a lipoprotein in the periphery and brain and is involved with lipid transport. ABCA7 is involved in efflux of phospholipids and possible cholesterol transport in phagocytosis. While these genes all have functions in lipid metabolism, how they act and interact to lead to AD remains unknown. By genetically modifying both astrocytes and/or neurons derived from iPSCs, researchers may be able to identify their specific interactions and pathogenic functions in the cell types of interest.
5.3. Inflammation genes
As discussed above, microglial biology has been implicated in AD pathology. Aβ can initiate the inflammatory response including release of cytokines, chemokines, reactive oxygen species and neurotoxic products. This leads to neuron and synapse damage, further implicating inflammation in AD pathology (reviewed in Birch et al. (2014)). Genetic linkage studies have implicated several microglial-specific genes to AD including TREM2 and CD33 (Bertram et al., 2008; Hollingworth et al., 2012).
The strength of the correlation between TREM2 and AD is comparable to that of inherited APOE ε4, but mutations in TREM2 are more rare (Jiang et al., 2013). TREM2 encodes the triggering receptor expressed on myeloid cells 2 protein and is upregulated in plaque-associated microglia. Together with the transmembrane adapter protein TYROBP, TREM2 mediates phagocytosis of bacteria and apoptotic cells (Mosher and Wyss-Coray, 2014). The TREM2 risk variant impairs protein function, limiting its ability to clear amyloid and cellular debris (Guerreiro et al., 2013). It appears that the link between these genes and pathology is imbalance in cellular protection mechanisms.
Expressed on the surface of myeloid progenitor cells, mature monocytes, and mactrophages, CD33 functions as a carbohydrate binding protein and can repress monocyte-derived pro-inflammatory cytokines. Elevated expression of CD33 is correlated with severity of cognitive decline in AD patients (Griciuc et al., 2013). The CD33 risk allele rs3865444C is associated with increased activated microglia, diminished Aβ42 internalization, and accumulation of neuritic and fibrillar amyloid (Bradshaw et al., 2013). The importance of these pathways in AD pathogenesis is clear, making the establishment and publication of efficient derivation protocols for microglia from iPSCs an urgent need in the field.
5.4. Endocytosis
The endocytosis of surface APP is a critical step in Aβ generation because the acidic pH found within endocytic compartments facilitates β- and γ-secretase cleavage whereas α-secretase cleavage occurs in the more neutral pH at the plasma membrane. BIN1, PICALM, and CD2AP have been linked to AD through GWAS studies and have known functions in the endocytosis pathway. BIN1 is a membrane adapter protein that can complex and act in clathrin-mediated endocytosis. PICALM recruits clathrin and adapter proteins to sites of vesicle assembly. CD2AP is involved in dynamin actin remodeling in endocytosis. The study of endocytic pathways is highly amenable to in vitro cellular modeling. Therefore, the ability to manipulate cultured iPSCs and their derivatives will permit the delineation of their roles in AD pathology.
5.5. Genes of unknown function
Some of the genes that have been identified to be associated with AD have less obvious roles in known disease pathways. EphA1, for example, has been found to be associated with AD, but has yet to be extensively studied in the context of neurode-generation. EphA1 is a member of the ephrin family of receptor protein tyrosine kinsases. Ephrin receptors have been implicated in developmental pathways, but little is known about the role of EphA1 gene product in degeneration. EphA1 is expressed in CD4 and T lymphocytes and monocytes, suggesting a possible role in immune-mediated phenotypes.
Similarly, MS4A4A, MS4A4E, and MS4A6E have all been linked to AD and their role in disease remains unclear. These genes are members of the 15-gene MS4A family that encodes proteins containing multiple membrane spanning domains. MS4A4A was initially identified by CD20 homology and is expressed in myeloid cells and monocytes, suggesting a possible immune-related function for the resulting gene product and its other protein family members (reviewed in Schellenberg and Montine (2012)). Manipulations of these genes in iPSC derived neural cultures may aid in elucidating their role in degenerative processes.
6. Potential and challenges for future discovery and screening
The ability to generate human neurons and glia from both disease and wild-type backgrounds has the possibility to increase our understanding of the underlying biology of disease. As more genetic information about AD is obtained, it becomes an increasing priority to translate those genotypes into their functional biological outcomes. Drug discovery also will be facilitated by permitting high through-put screening in relevant cell types. iPSCs will allow screening to occur in a disease-relevant context with the potential to enable a more rapid transition from the bench to the clinic. The ability to culture cells from sAD and fAD backgrounds will further permit more tailored treatment strategies by delineating the differences between and within these disease sub-types. However, multiple technical hurdles must be overcome in order to fully realize the potential of iPSCs.
6.1. Potential for using iPSC to translate genetics into biology
The ability to translate genetic information into functional biological phenotypes is a critical step in decoding the mechanisms of disease action. For example, known disease relevant mutations can be studied in culture and their specific contributions to disease phenotypes can be determined. This will help improve our understanding of disease pathogenesis and progression by defining the contributions of each AD-related gene and variant known. The capacity to generate neuron subtypes will allow interrogation of variants within discrete cell populations and will define specific genetic contributions to disease within each cell class. This may add to our appreciation of cell type involvement in disease and could help to identify new drug targets.
The use of gene manipulation by techniques described above will allow disease relevant cell types to be probed to identify biological signatures and to provide novel mechanistic insights. By modulating a specific gene expression profile, the effect on downstream pathway members may be monitored, and allow for refinement of proposed disease relevant pathways. Measuring the phenotypic outcomes of these alterations will reveal the biological effects of gene expression and solidify or contradict their classification as a disease-relevant gene.
6.2. Potential for using iPSCs for high through-put screening
Chemical library screens conducted on iPSC derived neurons offer a powerful opportunity to uncover drugs that are effective in patient cells without the added concern of translating rodent results to human applications. In one such screen, several hundred compounds were tested on commercially available wild-type iPSC-derived neurons to identify drugs that could ameliorate the effects of Aβ42 induced toxicity. Several small molecule inhibitors were subsequently identified that can block toxic effects, including Cdk2 inhibitors (Xu et al., 2013). This study suggests that insights into disease can be derived from screens using iPSC-derived neurons to yield novel mechanistic insights while identifying putative candidates for intervention.
One caveat to the use of iPSC cells for drug screening is the variation between treatment efficiency at different differentiation time points. For example, susceptibilities to γ-secretase inhibitors (Yahata et al., 2011) and to Aβ immunotherapy (Muratore et al., 2014) were shown to differ between early and late differentiation stages. It therefore becomes important to tightly temporally control neuron differentiation consistently between all cells involved in a screen.
A primary difficulty for studying neurodegenerative disease in culture is that disease manifests itself within the three dimensional, interconnected structure of the aged human brain. Within this complex structure, neurodegeneration takes decades to manifest, a significant constraint on culture models. Because of these caveats, interpretations of results will have to be tempered to reflect the capacities of the model in use, and results confirmed using complementary in vivo models.
6.3. Existing barriers to optimal iPSC modeling of AD
Several barriers to optimal AD modeling with iPSCs have been discussed above, and include: 1) the level of maturity of the neurons arising from our current neuronal differentiation protocols, which results in the lack of proper adult tau splicing, 2) the lack of efficient protocols for generating microglia, a cell type with a central role in AD pathogenesis, and 3) the limited number of differentiation protocols that establish a densely packed, three dimensional structure that more properly recapitulates the in vivo environment of the brain. In addition to the lack of proper circuitry formation, monolayer cultures also allow for a much greater range of diffusion of neurotoxic species (and thus lower local concentrations) than would be available in vivo in the brain. To this final point, recent advancements in cerebral organoid protocols (Lancaster et al., 2013) have allowed researchers to recapitulate certain aspects of cortical development in a dish using hiPSCs. In the next few years, advancements on these and other protocols for directing iPSCs to neural and micro-glial fates have the potential to greatly increase the utility of iPSCs for pathway discovery.
7. Conclusions/summary
The advent of IPSC technology has provided the field of neurodegeneration with an exciting new apparatus for research. Characterizing the mechanisms underlying AD is essential to advancing our understanding of disease with the continued goal of identifying effective therapies to restrain or impede disease progression. iPSCs allow for enhanced disease modeling as they can be generated from disease or non-diseased human patients, thus eliminating the confounding species-specific effects that may exist in murine models. In vitro analysis of varying disease-relevant cell types will allow experiments to be better tailored to the specific functional properties of the cells being interrogated and will isolate the pathological involvement of each cell type. Derivation of neurons from iPSCs generated from sporadic and familial AD patients permits the analysis of genetic background on downstream gene expression, and the phenotypic consequences of these genetic backgrounds. The understanding of known genetic mutations may shed light onto the mechanism of pathogenesis and lead to the uncovering of novel drug targets. In this way, iPSCs may transform our current understanding of AD and could lead to therapeutic advancements that may change the way we treat AD.
Acknowledgments
We thank Drs. Matthew LaVoie, Dominic Walsh, and members of the Young-Pearse lab for helpful comments on this manuscript. This work was supported by grants from the National Institute of Health (U01 AG046152, R01 MH101148, and R33 AG049864), and the Harvard NeuroDiscovery Center. Acknowledgment is made to the donors of the ADR, a program of the BrightFocus Foundation, for support of this research (TLY-P).
References
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23:213–227. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, Abdelrahim M, Matikainen-Ankney B, Chao SH, MrSich M, Rakic P, Fang G, Zhang B, Yates JR, 3rd, Gage FH. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;3:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, HuLince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE. Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutner C, Roy K, Linnartz B, Napoli I, Neumann H. Generation of microglial cells from mouse embryonic stem cells. Nat Protoc. 2010;5:1481–1494. doi: 10.1038/nprot.2010.90. [DOI] [PubMed] [Google Scholar]
- Birch AM, Katsouri L, Sastre M. Modulation of inflammation in transgenic models of Alzheimer's disease. J Neuroinflamm. 2014;11:25. doi: 10.1186/1742-2094-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette CJ, Lyass L, Bhattacharyya BJ, Belmadani A, Miller RJ, Kessler JA. The controlled generation of functional basal forebrain cholinergic neurons from human embryonic stem cells. Stem Cells. 2011;29:802–811. doi: 10.1002/stem.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert EM, Denton K, Lei L, Li XJ. The specification of telencephalic glutamatergic neurons from human pluripotent stem cells. J Vis Exp. 2013;74 doi: 10.3791/50321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, Morris MC, Evans DA, Johnson K, Sperling RA, Schneider JA, Bennett DA, De Jager PL. CD33 Alzheimer's disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16:848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, Sessa A, Colasante G, Bartolomeo R, Massimino L, Ferroni S, Settembre C, Benfenati F, Broccoli V. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Rep. 2015;4:25–36. doi: 10.1016/j.stemcr.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos PB, Paulsen BS, Rehen SK. Accelerating neuronal aging in in vitro model brain disorders: a focus on reactive oxygen species. Front Aging Neurosci. 2014;6:292. doi: 10.3389/fnagi.2014.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA. Endocytic pathway abnormalities precede amyloid beta deposition in sporadic Alzheimer's disease and Down syndrome: differential effects of APOE genotype and presenilin mutations. Am J Pathol. 2000;157:277–286. doi: 10.1016/s0002-9440(10)64538-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Petanceska S, Terio NB, Peterhoff CM, Durham R, Mercken M, Mehta PD, Buxbaum J, Haroutunian V, Nixon RA. Abeta localization in abnormal endosomes: association with earliest Abeta elevations in AD and Down syndrome. Neurobiol Aging. 2004;25:1263–1272. doi: 10.1016/j.neurobiolaging.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D'Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham M, Cho JH, Leung A, Savvidis G, Ahn S, Moon M, Lee PK, Han JJ, Azimi N, Kim KS, Bolshakov VY, Chung S. hPSC-derived maturing GABAergic inter-neurons ameliorate seizures and abnormal behavior in epileptic mice. Cell Stem Cell. 2014;15:559–573. doi: 10.1016/j.stem.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Bhattacharyya BJ, Belmadani A, Pan L, Miller RJ, Kessler JA. Stem cell derived basal forebrain cholinergic neurons from Alzheimer's disease patients are more susceptible to cell death. Mol Neurodegener. 2014;9:3. doi: 10.1186/1750-1326-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Emdad L, D'Souza SL, Kothari HP, Qadeer ZA, Germano IM. Efficient differentiation of human embryonic and induced pluripotent stem cells into functional astrocytes. Stem Cells Dev. 2012;21:404–410. doi: 10.1089/scd.2010.0560. [DOI] [PubMed] [Google Scholar]
- Franco R, Cedazo-Minguez A. Successful therapies for Alzheimer's disease: why so many in animal models and none in humans? Front Pharmacol. 2014;5:146. doi: 10.3389/fphar.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE. Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78:631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer's disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Sweet R, Sims R, Harold D, Russo G, Abraham R, Stretton A, Jones N, Gerrish A, Chapman J, Ivanov D, Moskvina V, Lovestone S, Priotsi P, Lupton M, Brayne C, Gill M, Lawlor B, Lynch A, Craig D, McGuinness B, Johnston J, Holmes C, Livingston G, Bass NJ, Gurling H, McQuillin A, Holmans P, Jones L, Devlin B, Klei L, Barmada MM, Demirci FY, DeKosky ST, Lopez OL, Passmore P, Owen MJ, O'Donovan MC, Mayeux R, Kamboh MI, Williams J. Genome-wide association study of Alzheimer's disease with psychotic symptoms. Mol Psychiatry. 2012;17:1316–1327. doi: 10.1038/mp.2011.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, Goldstein LS. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Yu JT, Zhu XC, Tan L. TREM2 in Alzheimer's disease. Mol Neurobiol. 2013;48:180–185. doi: 10.1007/s12035-013-8424-8. [DOI] [PubMed] [Google Scholar]
- Koch P, Tamboli IY, Mertens J, Wunderlich P, Ladewig J, Stuber K, Esselmann H, Wiltfang J, Brustle O, Walter J. Presenilin-1 L166P mutant human pluripotent stem cell-derived neurons exhibit partial loss of gamma-secretase activity in endogenous amyloid-beta generation. Am J Pathol. 2012;180:2404–2416. doi: 10.1016/j.ajpath.2012.02.012. [DOI] [PubMed] [Google Scholar]
- Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, Takahashi K, Asaka I, Aoi T, Watanabe A, Watanabe K, Kadoya C, Nakano R, Watanabe D, Maruyama K, Hori O, Hibino S, Choshi T, Nakahata T, Hioki H, Kaneko T, Naitoh M, Yoshikawa K, Yamawaki S, Suzuki S, Hata R, Ueno S, Seki T, Kobayashi K, Toda T, Murakami K, Irie K, Klein WL, Mori H, Asada T, Takahashi R, Iwata N, Yamanaka S, Inoue H. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Krencik R, Zhang SC. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat Protoc. 2011;6:1710–1717. doi: 10.1038/nprot.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Nakano T, Hotta A. Genetic correction using engineered nucleases for gene therapy applications. Dev Growth Differ. 2014;56:63–77. doi: 10.1111/dgd.12107. [DOI] [PubMed] [Google Scholar]
- Liu Q, Waltz S, Woodruff G, Ouyang J, Israel MA, Herrera C, Sarsoza F, Tanzi RE, Koo EH, Ringman JM, Goldstein LS, Wagner SL, Yuan SH. Effect of potent gamma-secretase modulator in human neurons derived from multiple presenilin 1-induced pluripotent stem cell mutant carriers. JAMA Neurol. 2014;71:1481–1489. doi: 10.1001/jamaneurol.2014.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahairaki V, Ryu J, Peters A, Chang Q, Li T, Park TS, Burridge PW, Talbot CC, Jr, Asnaghi L, Martin LJ, Zambidis ET, Koliatsos VE. Induced pluripotent stem cells from familial Alzheimer's disease patients differentiate into mature neurons with amyloidogenic properties. Stem Cells Dev. 2014;23:2996–3010. doi: 10.1089/scd.2013.0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, Studer L. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S, Evans LD, Andersson T, Portelius E, Smith J, Dias TB, Saurat N, McGlade A, Kirwan P, Blennow K, Hardy J, Zetterberg H, Livesey FJ. APP metabolism regulates tau proteostasis in human cerebral cortex neurons. Cell Rep. 2015;11:689–696. doi: 10.1016/j.celrep.2015.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher KI, Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer's disease. Biochem Pharmacol. 2014;88:594–604. doi: 10.1016/j.bcp.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, Walsh DM, Selkoe DJ, Young-Pearse TL. The familial Alzheimer's disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, Sasai Y, Alvarez-Buylla A, Rubenstein JL, Kriegstein AR. Functional maturation of hPSC-derived forebrain inter-neurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, Stenh C, Luthman J, Teplow DB, Younkin SG, Naslund J, Lannfelt L. The ‘Arctic’ APP mutation (E693G) causes Alzheimer's disease by enhanced Abeta protofibril formation. Nat Neurosci. 2001;4:887–893. doi: 10.1038/nn0901-887. [DOI] [PubMed] [Google Scholar]
- Ransohoff RM, Cardona AE. The myeloid cells of the central nervous system parenchyma. Nature. 2010;468:253–262. doi: 10.1038/nature09615. [DOI] [PubMed] [Google Scholar]
- Rice HC, Young-Pearse TL, Selkoe DJ. Systematic evaluation of candidate ligands regulating ectodomain shedding of amyloid precursor protein. Biochemistry. 2013;52:3264–3277. doi: 10.1021/bi400165f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg GD, Montine TJ. The genetics and neuropathology of Alzheimer's disease. Acta Neuropathol. 2012;124:305–323. doi: 10.1007/s00401-012-0996-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, MacLean G, Orkin SH, Livesey FJ. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Sci Transl Med. 2012;4:124ra29. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Jaenisch R. Medicine: iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/S1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- Sproul AA, Jacob S, Pre D, Kim SH, Nestor MW, Navarro-Sobrino M, Santa-Maria I, Zimmer M, Aubry S, Steele JW, Kahler DJ, Dranovsky A, Arancio O, Crary JF, Gandy S, Noggle SA. Characterization and molecular profiling of PSEN1 familial Alzheimer's disease iPSC-derived neural progenitors. PLoS One. 2014;9:e84547. doi: 10.1371/journal.pone.0084547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thies W, Bleiler L. 2013 Alzheimer's disease facts and figures. Alzheimers Dement. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Vazin T, Ball KA, Lu H, Park H, Ataeijannati Y, Head-Gordon T, Poo MM, Schaffer DV. Efficient derivation of cortical glutamatergic neurons from human pluripotent stem cells: a model system to study neurotoxicity in Alzheimer's disease. Neurobiol Dis. 2014;62:62–72. doi: 10.1016/j.nbd.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Hartley DM, Selkoe DJ. The many faces of Aβ: strucures and activity. Curr Med Chem – Immunol Endocr Metab Agents. 2003;3:277–291. [Google Scholar]
- Wicklund L, Leao RN, Stromberg AM, Mousavi M, Hovatta O, Nordberg A, Marutle A. Beta-amyloid 1-42 oligomers impair function of human embryonic stem cell-derived forebrain cholinergic neurons. PLoS One. 2010;5:e15600. doi: 10.1371/journal.pone.0015600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojda U, Kuznicki J. Alzheimer's disease modeling: ups, downs, and perspectives for human induced pluripotent stem cells. J Alzheimers Dis. 2013;34:563–588. doi: 10.3233/JAD-121984. [DOI] [PubMed] [Google Scholar]
- Wolfe MS. Structure, mechanism and inhibition of gamma-secretase and presenilin-like proteases. Biol Chem. 2010;391:839–847. doi: 10.1515/BC.2010.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff G, Young JE, Martinez FJ, Buen F, Gore A, Kinaga J, Li Z, Yuan SH, Zhang K, Goldstein LS. The presenilin-1 DeltaE9 mutation results in reduced gamma-secretase activity, but not total loss of PS1 function, in isogenic human stem cells. Cell Rep. 2013;5:974–985. doi: 10.1016/j.celrep.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lei Y, Luo J, Wang J, Zhang S, Yang XJ, Sun M, Nuwaysir E, Fan G, Zhao J, Lei L, Zhong Z. Prevention of beta-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of cyclin-dependent kinases and associated cell cycle events. Stem Cell Res. 2013;10:213–227. doi: 10.1016/j.scr.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- Yahata N, Asai M, Kitaoka S, Takahashi K, Asaka I, Hioki H, Kaneko T, Maruyama K, Saido TC, Nakahata T, Asada T, Yamanaka S, Iwata N, Inoue H. Anti-Abeta drug screening platform using human iPS cell-derived neurons for the treatment of Alzheimer's disease. PLoS One. 2011;6:e25788. doi: 10.1371/journal.pone.0025788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Shin S, Jha BS, Liu Q, Sheng J, Li F, Zhan M, Davis J, Bharti K, Zeng X, Rao M, Malik N, Vemuri MC. Efficient and rapid derivation of primitive neural stem cells and generation of brain subtype neurons from human pluripotent stem cells. Stem Cells Transl Med. 2013;2:862–870. doi: 10.5966/sctm.2013-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JE, Boulanger-Weill J, Williams DA, Woodruff G, Buen F, Revilla AC, Herrera C, Israel MA, Yuan SH, Edland SD, Goldstein LS. Elucidating molecular phenotypes caused by the SORL1 Alzheimer's disease genetic risk factor using human induced pluripotent stem cells. Cell Stem Cell. 2015;16:373–385. doi: 10.1016/j.stem.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan SH, Martin J, Elia J, Flippin J, Paramban RI, Hefferan MP, Vidal JG, Mu Y, Killian RL, Israel MA, Emre N, Marsala S, Marsala M, Gage FH, Goldstein LS, Carson CT. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Guo M, Martins-Taylor K, Wang X, Zhang Z, Park JW, Zhan S, Kronenberg MS, Lichtler A, Liu HX, Chen FP, Yue L, Li XJ, Xu RH. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS One. 2010;5:e11853. doi: 10.1371/journal.pone.0011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Sudhof TC. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]