Abstract
We used high hydrostatic pressure as a tool for exploring the conformational landscape of human cytochrome P450 3A4 (CYP3A4) by electron paramagnetic resonance and fluorescence spectroscopy. Site-directed incorporation of a luminescence resonance energy transfer donor-acceptor pair allowed us to identify a pressure-dependent equilibrium between two states of the enzyme, where an increase in pressure increased the spatial separation between the two distantly located fluorophores. This transition is characterized by volume change (ΔV°) and P1/2 values of −36.8 ± 5.0 mL/mol and 1.45 ± 0.33 kbar, respectively, which corresponds to a Keq° of 0.13 ± 0.06, so that only 15% of the enzyme adopts the pressure-promoted conformation at ambient pressure. This pressure-promoted displacement of the equilibrium is eliminated by the addition of testosterone, an allosteric activator. Using site-directed spin labeling, we demonstrated that the pressure- and testosterone-sensitive transition is also revealed by pressure-induced changes in the electron paramagnetic resonance spectra of a nitroxide side chain placed at position 85 or 409 of the enzyme. Furthermore, we observed a pressure-induced displacement of the emission maxima of a solvatochromic fluorophore (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin) placed at the same positions, which suggests a relocation to a more polar environment. Taken together, the results reveal an effector-dependent conformational equilibrium between open and closed states of CYP3A4 that involves a pronounced change at the interface between the region of α-helices A/A′ and the meander loop of the enzyme, where residues 85 and 409 are located. Our study demonstrates the high potential of pressure-perturbation strategies for studying protein conformational landscapes.
Introduction
Cytochromes P450 constitute a superfamily of heme-thiolate enzymes found in all domains of life. These enzymes serve as terminal monooxygenases in multienzyme systems that catalyze oxidation of a wide variety of endogenous and exogenous lipophilic compounds. P450s play diverse physiological functions ranging from oxidation of exogenous compounds to the synthesis of steroid hormones, prostaglandins, and second messengers. In particular, the membrane-bound P450 enzymes that are abundant in liver play a key role in the metabolism and subsequent disposal of drugs, toxins, and other xenobiotics from the body. The conformational flexibility of cytochromes P450 continues to be demonstrated by an increasing number of resolved structures of these enzymes and their complexes with various ligands (1, 2, 3, 4). The conformational changes that accompany enzyme-substrate interactions are now recognized as a common feature that contributes to the ability of drug-metabolizing cytochromes P450 to metabolize a vast range of substrates (5, 6).
In addition to its the role in adaptation of the P450 active site to substrates of various shapes and sizes, conformational mobility plays an important role in the P450 catalytic mechanisms. Modulation of solvent accessibility and the degree of hydration of the heme pocket during the catalytic cycle is believed to be of vital importance for the catalytic efficiency of these enzymes (7, 8, 9, 10, 11). The exclusion of solvent molecules from the active site upon substrate binding, as well as the proton shuttling through a protein-solvent network of water molecules, requires a direct contact of the heme pocket of P450 with the bulk water. However, the active-site cavity of most bacterial and some eukaryotic P450s is essentially closed, and the pathway that is followed by water from the active site to the bulk solvent is not obvious (12, 13, 14). In contrast, in several other P450 structures (15, 16, 17, 18, 19, 20, 21, 22, 23) the active-site cavity is fairly open, thus providing no clue as to possible mechanisms underlying the precise control of water accessibility. Despite numerous studies on this subject (13, 14, 24), the general mechanisms of water access and its modulation in catalytic turnover remain obscure.
The last but not least important role of the conformational mobility of cytochromes P450 is related to the allosteric mechanisms that reveal themselves in multiple instances of heterotropic cooperativity (i.e., the activation of metabolism of one substrate by another one) observed with some drug-metabolizing P450s, of which human P450 3A4 (CYP3A4) is the most prominent. Important support for allosteric modulation of CYP3A4 is provided by recent observations of a peripheral ligand-binding site in the enzyme (25, 26, 27, 28). Interactions of this site, which is located at the distal surface of the enzyme and surrounded by the F/F′ and G/G′ loops (25, 27), were hypothesized to be involved in CYP3A4 activation by steroids, α-naphthoflavone (ANF), and other heterotropic activators (26, 27, 29).
This mechanism suggests that allosteric effectors induce a conformational rearrangement that facilitates the formation of a catalytically competent enzyme-substrate complex (27, 30, 31, 32, 33). Our recent design of a P450-compatible luminescence resonance energy transfer (LRET) donor-acceptor pair (34) allowed us to introduce a new, to our knowledge, method for detecting conformational mobility in CYP3A4. For this purpose, we used a set of double-cysteine mutants of the enzyme labeled with erythrosine iodoacetamide (ERIA, phosphorescent donor) and DY-731 maleimide (DYM, fluorescent acceptor). We found that the interactions of the protein with allosteric activators, such as testosterone (TST) and ANF, resulted in a large-scale conformational transition that increased the spatial separation between the region of α-helices A, B, and K and the β-bundle (which is sometimes designated the β-domain (35)) and the heme-containing core (α-domain) of the enzyme (34). In contrast, no considerable changes in the interprobe distances were elicited by such substrates as bromocriptine (BCT) and 1-pyrenebutanol. Besides providing strong support for the involvement of large-scale conformational mobility in the mechanisms of heterotropic cooperativity in CYP3A4 these results provide important information regarding the structural nature of effector-induced conformational changes.
Here, we complement the above approach for detecting conformational transitions in P450 with site-directed incorporation of paramagnetic and fluorescent probes and pressure-perturbation electron paramagnetic resonance (EPR) and fluorescence spectroscopy. We use hydrostatic pressure as a variable parameter that affects the protein conformational landscape. Along with the effects of temperature, varying pressure is indispensable for understanding the mechanisms of protein conformational transitions in detail. The basis of pressure effects is the change in system volume that accompanies biochemical processes (36, 37, 38, 39). According to Le Chatelier’s principle, increased pressure enhances processes that are accompanied by a decrease in system volume and, conversely, inhibits processes that occur with a volume increase. An important part of the volume changes that occur during protein transitions stems from the changes in interactions with solvent (40, 41, 42, 43, 44, 45). These include both water penetration into the cavities and water constriction around solvent-exposed polar groups (38, 45, 46, 47, 48, 49, 50). Thus, the volume change that results from penetration of one water molecule into a protein cavity is equal to −18 mL/mol, whereas the solvation of a singly charged ion in water is characterized by ΔV values of the order of −10 mL/mol (38). Generally speaking, a pressure increase results in enhancement of protein hydration, which therefore constitutes the core of pressure-induced protein transitions (40, 42, 44, 50, 51, 52, 53). The pivotal role of protein-solvent interactions in catalytic mechanisms of cytochromes P450 (7, 8, 9, 11, 22) makes pressure perturbation the method of choice for studies of conformational dynamics (54, 55, 56).
To detect conformational rearrangements, we used three complementary methods. First, we used LRET-based monitoring of the distance between donor and acceptor fluorophores (ERIA and DYM, respectively) placed at distant loci of CYP3A4 (C64 and C468 of the respective double-cysteine mutant), in a manner similar to that described in our recent study of ligand-induced conformational changes (34). Monitoring pressure-dependent changes of interprobe distances by LRET enables detection of large-scale transitions in proteins and hence provides a general picture of protein conformational dynamics. However, this approach gives only limited information as to the location of flexible regions and the nature of local conformational rearrangements involved in these large-scale changes. Thus, as a complementary second approach, we probed local conformational rearrangements using site-directed spin labeling (SDSL) and pressure-perturbation EPR spectroscopy. This approach is based on monitoring pressure-dependent changes in the environment of a nitroxide side chain (R1) introduced using a sulfhydryl (SH)-reactive methanethiosulfonate reagent ((1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate (MTSSL)) (57, 58, 59). Third, we monitored the changes caused by pressure in the fluorescence of the environment-sensitive, coumarin-based, SH-reactive fluorescent probe (7-diethylamino-3-((((2-maleimidyl)ethyl)amino)carbonyl) coumarin (MDCC)) introduced at the same sites as the spin label.
A combination of the above techniques allowed us to demonstrate an effector-dependent conformational equilibrium between the open and closed states of CYP3A4 that involves a pronounced change in protein conformation in the region of the meander loop. To our knowledge, this is the first study to combine a pressure-perturbation approach with EPR and fluorescence spectroscopy to explore the conformational landscapes of proteins.
Materials and Methods
Materials
Glucose oxidase, catalase, D,L-glucose, D,L-dithiothreitol (DTT), Igepal CO-630, and TST were obtained from Sigma-Aldrich (St. Louis, MO). ERIA was obtained from AnaSpec (San Jose, CA). Tris(2-carboxyethyl)phosphine (TCEP) and MDCC were obtained from Life Technologies/Molecular Probes (Eugene, OR). DYM was obtained from Dyomics (MoBiTec; Göttingen, Germany). MTSSL was obtained from Toronto Research Chemicals (Toronto, Canada). All other chemicals were of ACS grade and were used without further purification.
Site-directed mutagenesis and protein purification
Cysteine-depleted mutants of CYP3A4 were generated using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) and a template consisting of the cDNA of the N-terminally truncated (Δ3-12) CYP3A4 S18F variant with a tetrahistidine tag attached at the C-terminus (33). Proteins were expressed in Escherichia coli TOPP3 cells as described previously (60). Purification of the proteins via a two-column procedure (61) allowed us to obtain electrophoretically homogeneous preparations with >90% P450 holo-protein (heme-containing protein) in most cases. It should be noted, however, that the low yield of expression of single-cysteine mutants such as C121, C495, and C85 (<50 nmol/L) did not allow us to achieve a holo-protein content higher than 80% in the purified preparations. The preparations of these mutants used in this study contained up to 30% of P450 apo-protein. Control of the holo-protein content was based on approximation of the UV-VIS (250–700 nm) spectra of ferric heme protein, with a combination of prototypical spectra of absorbance of the three states of the holo-protein (high-spin, low-spin, and P420) and the prototypical spectrum of absorbance of apo-CYP3A4. The latter spectral standard was obtained by peroxide-dependent heme depletion and standardized with the use of the extinction coefficient at 278 nm calculated from the amino acid sequence of CYP3A4.
Modification of proteins with thiol-reactive reagents
Modification of the double-cysteine mutant CYP3A4(C64,C468) with ERIA and DYM yielding the double-labeled protein (C64/C468-ER/DY) was performed as previously described (34). The procedure used for modification of single-cysteine mutants of CYP3A4 with MDCC was similar to that described previously for P450eryF, with the exception that the use of DTT was avoided, and 3 mM TCEP was used as an SH protector instead. Therefore, the procedure of DTT removal was omitted. The incubation time required for 95% modification was in the range of 1–3 h depending on the position of modification. To modify single-cysteine CYP3A4 mutants with MTSSL, 50–150 μM solutions of the proteins in storage buffer (0.1 M Na-Hepes buffer, pH 7.4, containing 10% glycerol, 3 mM TCP, 150 mM KCl, and 0.5 mM EDTA) were passed through a Bio-Spin-6 spin column (Bio-Rad, Hercules, CA) equilibrated with 0.1 M Na-Hepes buffer, pH 7.4, containing 50 mM KCl. A 3–5 mM solution of MTSSL in methanol was then added to reach a 1.5:1 molar ratio of MTSSL to protein. After 1 h of incubation, excess label was removed by passing the solution through a BioSpin-6 spin column. The resulting 40–120 μM solution of the labeled protein was directly transferred to the EPR tube for pressure-perturbation EPR studies. Modification of the single-cysteine mutants with MDCC or MTSSL did not result in any significant decrease in the heme-protein content, and the yield of the MDCC and R1 adducts was as high as 80–90% relative to the initial amount of protein taken for modification.
Pressure-perturbation LRET experiments were done using a custom-built, high-pressure optical cell (62) connected to a manual pressure generator (High Pressure Equipment, Erie, PA) capable of generating a pressure of up to 6000 bar. The excitation light was provided with a pulsing M505F1 light-emitting diode (Thorlabs, Newton, NJ) emitting at 505 nm. Spectra of delayed emissions were recorded with an FLS920 spectrofluorometer (Edinburgh Instruments Ltd., Livingston, UK) equipped with custom-designed software for delayed fluorescence spectroscopy. The instrument was connected to the high-pressure cell with a VIS-NIR liquid light guide (Oriel Instruments/Newport, Irvine, CA). Excitation was performed with a square-wave modulation with a 3.5 ms interval between the pulses. Emission was recorded with a delay of 500 μs after the falling edge of the pulse. The integration period after each pulse was equal to 1.25 ms and the results of 75 integration cycles were accumulated per each data point. The spectra were recorded in the region of 660–860 nm with a step of 2 nm and an emission slit width of 18 nm. The experiments were performed with an 8–15 μM protein concentration at 4°C in 0.1 M Na-Hepes buffer under anaerobiosis, which was achieved by using an oxygen-scavenging system consisting of glucose oxidase (30 units/mL), 60 mM glucose, and 2000 units/mL catalase.
Pressure-perturbation fluorescence spectroscopy experiments with MDCC-labeled proteins were performed using a custom-built, high-pressure optical cell (see above) connected to an FLS920 spectrofluorometer (Edinburgh Instruments) as described above. Excitation was performed at 405 nm with a LDH405 pulsed diode laser head equipped with a PDL 800-D diode laser driver (Picoquant, Berlin, Germany). The experiments were performed at 25°C with a protein concentration of 3–5 μM in 0.1 M Na-Hepes buffer, pH 7.4. The spectra were recorded in the wavelength range of 430–700 nm with a step of 2 nm and an emission slit width of 2 nm.
Pressure-perturbation EPR spectroscopy
The computer-controlled pressure intensifier (model HUB440, rated at 3.79 kbar) and ceramic sample cells (rated to 3 kbar) used in these experiments were jointly developed with Pressure BioSciences (Easton, MA) and are now commercially available. The intensifier is feedback regulated to maintain constant pressure. The ceramic cells (HUB440-Cer, also now commercially available) were fabricated for use in a five-loop, four-gap resonator operating at X-band microwave frequency (59). The ceramic cell had a pressure-independent EPR signal that was subtracted from all spectra after data acquisition. The pressurization fluid was water, and pressure was monitored with two separate transducers connected in-line with the sample cell as described earlier (59). EPR spectra were acquired on a Varian E-109 spectrometer (Palo Alto, CA) fitted with the above-mentioned five-loop, four-gap resonator operating with 2 mW incident power at room temperature. The sweep width for all spectra was 100 G. The spectra were collected at room temperature (21–23°C) in 100 mM Na-Hepes buffer, pH 7.4, containing 50 mM KCl at a protein concentration of 40–60 μM.
To analyze the series of spectra obtained in our fluorescence and EPR spectroscopy experiments, we applied principal component analysis (PCA). In the experiments with pressure-induced displacement of the emission band of MDCC, this technique was combined with a linear least-squares approximation of the principal vectors by a combination of prototypical spectra of MDCC emission obtained at ambient and high hydrostatic pressures. The spectral standards and the procedure used for their generation are described in the Supporting Material.
The effect of pressure on protein equilibria was interpreted based on the equation for the pressure dependence of the equilibrium constant (54):
| (1) |
or in integral form (63):
| (2) |
where Keq is the equilibrium constant of the reaction at pressure P, P1/2 is the pressure at which Keq = 1 (half pressure of the conversion), ΔV° is the standard molar reaction volume, and K°eq is the equilibrium constant extrapolated to zero pressure, K°eq = eP1/2ΔV°/RT. For equilibrium A B and K°eq = [B]/[A], Eq. 2 may be transformed into the following relationship:
| (3) |
where C0 = [A] + [B]. To determine the ΔV° and P1/2 parameters from the experimental data sets describing pressure-induced changes in the amplitude of a signal derived from the fluorescence or EPR spectra (Ap), this equation was complemented with the offset (A0) and scaling factor (Amax) parameters and used in the following form:
| (4) |
Fitting of the experimental data to this equation with a combination of Neilder-Mead and Marquardt nonlinear regression algorithms resulted in a set of A0, Amax, ΔV°, and P1/2 parameters. We performed all data treatment and fitting, as well as the data acquisition in our fluorimetric experiments, using our custom-designed SpectraLab software (64) rewritten for the Windows XP environment using Borland Delphi 7 from Borland (Scotts Valley, CA) and the Win32Forth Public Domain Forth interpreter (www.win32forth.org). The software package is available from the corresponding author upon request.
Results and Discussion
Pressure-perturbation LRET spectroscopy
In a recent study (34), we demonstrated that binding of ANF or TST to CYP3A4 resulted in a considerable decrease in efficiency of LRET between ERIA and DYM labels placed at three pairs of residues (64/468, 377/468, and 64/121). These results were interpreted as evidence of a ligand-induced allosteric transition that increases the spatial separation between two domains of the enzyme. One is the β-domain, which bears residues 64 and 377 and includes the sheets β1 and β2 along with the helices A, B, and K′ and the connecting loops (35). The second is the remainder of the protein, the so-called α-domain, where residues 121 and 468 are located. Such a concerted movement of the β-domain may be interpreted as a ligand-induced transition between closed and open conformations (34) that affects the openness of most of the known substrate access channels (65) located at the interface between the two domains. This transition is anticipated to alter protein-solvent interactions and thus evoke an important volume change. We therefore initiated a study of the effect of pressure on LRET efficiency in double-labeled CYP3A4(C64/C468)ER/DY protein in the absence and presence of TST.
We carried out pressure-perturbation LRET experiments in solution and in the absence of detergents and phospholipids. Similarly to other membranous P450 proteins, CYP3A4 in solution is known to be represented by large oligomers ranging from 215 to 450 kDa (66, 67). Dissociation of these aggregates by pressure is extremely unlikely, as no traces of monomers were detectable in either concentrated or diluted solutions of CYP3A4 by analytical ultracentrifugation (66, 67), which is known to generate hydrostatic pressures up to 500 bar (68). Thus, the pressure-induced changes described in this study are thought to reflect the conformational mobility in the protein oligomers rather than to result from any pressure-induced change in the state of aggregation.
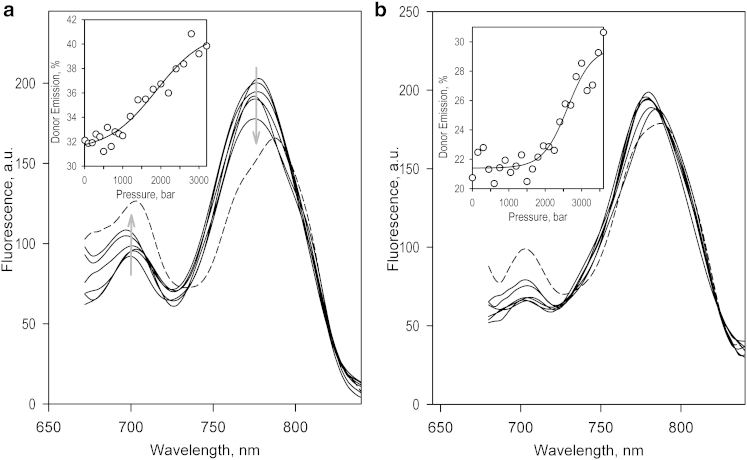
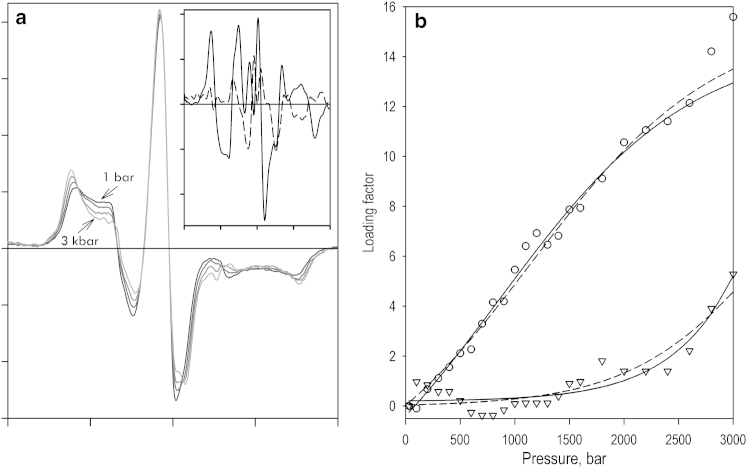
Fig. 1 illustrates the effect of pressure on the normalized spectra of phosphorescence of CYP3A4(C64,C468)-ERIA/DYM with no ligand added (Fig. 1 a) and in the presence of TST (Fig. 1 b). Application of elevated pressure to the ligand-free enzyme resulted in a considerable decrease in the efficiency of LRET, which was completely reversible upon decompression from pressures up to 2.5 kbar. At higher pressures, we observed a gradual denaturation of the protein, which was revealed in a stepwise irreversible decrease in the LRET efficiency, a red shift of the maximum of DYM emission (Fig. 1 a, dashed line), and some precipitation of the protein. The values of ΔV° and P1/2 of this irreversible transition derived from the fitting of the pressure dependencies of LRET efficiency to Eq. 4 were equal to −70.0 ± 3.7 mL/mol and 2.73 ± 0.76 kbar, respectively. These values, which represent the averages of the results of three individual experiments, are in a reasonable agreement with the parameters of the pressure-induced P450→P420 transition (inactivation) of CYP3A4 derived in our studies using pressure-perturbation absorbance spectroscopy (61, 69).
Figure 1.
(a and b) Effects of pressure on LRET in CYP3A4(C64,C468)-ERDY with no ligand added (a) and in the presence of 200 μM TST (b). The spectra shown in solid lines were recorded at 1, 600, 1200, 2500, and 3200 bar. The spectra shown in dashed lines correspond to a pressure of 4900 (a) or 3600 (b) bar. The insets show the pressure dependencies of the relative intensity of donor emission. The solid lines represent the fitting of the data sets to Eq. 4. Conditions: 10 μM CYP3A4(C64,C468)-ER/DY in 0.1 M Na-Hepes buffer, pH 7.4, oxygen-scavenging system with glucose oxidase and catalase, 4°C.
In this study, we focus on the reversible changes in LRET efficiency observed below 2.5 kbar, which we interpret as a pressure-dependent displacement of a preexisting conformational equilibrium. As shown in Fig. 1 b, addition of TST results in an almost complete elimination of spectral changes observed at pressures below 2.5 kbar. These results are consistent with a pressure-sensitive equilibrium between the two protein conformations, which is displaced toward the pressure-promoted state in the presence of TST. Averaging the values derived from six individual experiments by fitting the pressure dependencies of LRET efficiency to Eq. 4 results in ΔV° and P1/2 values of −36.8 ± 5.0 mL/mol and 1.45 ± 0.33 kbar, respectively (ΔG° = −5.3 ± 1.1 kJ/mol, Keq° = 0.13 ± 0.06), so that only 15% of the enzyme adopts the pressure-promoted conformation at ambient pressure.
Our interpretation that the observed pressure-induced changes reflect changes in LRET efficiency is confirmed by control experiments in which we studied the selective effect of pressure on the fluorescence of protein-incorporated DY-731. In these experiments, we used direct excitation of the above fluorophore in double-labeled CYP3A4(C64/C468)-ER/DY at 650 nm. The results of these studies, which are illustrated in Fig. S2, show no effect of pressure on the fluorescence of protein-incorporated DY-731 up to a pressure of 2.5 kbar. Importantly, the pressure dependencies of the irreversible decrease of fluorescence intensity observed at higher pressures are very similar to the pressure profile of the changes in delayed emission observed in the presence of TST (Fig. 1 b, inset).
The fact that the pressure-induced reversible changes in LRET efficiency were essentially eliminated by the addition of TST indicates that this ligand causes a profound displacement of a pressure-sensitive conformational equilibrium between the open and closed enzyme conformers toward the pressure-promoted (open) state. This parallelism between the effects of pressure and interactions with TST on protein conformation suggests that the TST-induced allosteric transition in CYP3A4 is associated with a decrease in the system volume, possibly due to enhanced protein hydration.
SDSL and pressure-perturbation EPR spectroscopy
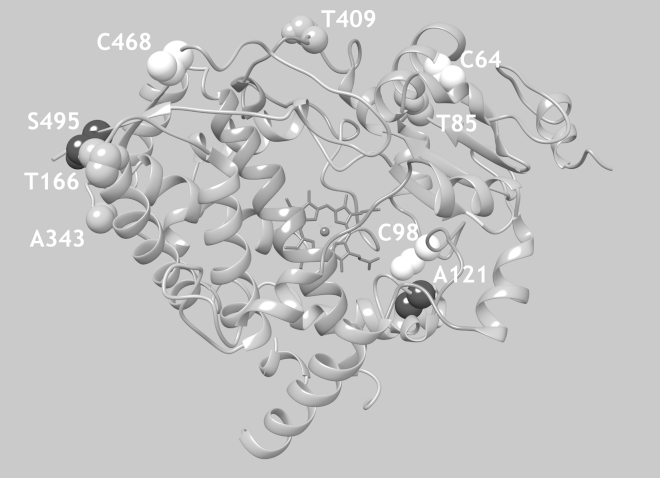
The above observation prompted us to use SDSL and pressure-perturbation EPR spectroscopy (58, 59) to study the underlying mechanisms. The sites used to introduce the R1 nitroxide side chain in CYP3A4 are shown in Fig. 2. Along with the use of the naturally present C64, C98, and C468 SH groups, we also incorporated new points for R1 attachment by making T85C, A121C, T166C, A343C, T409C, and S495C replacements on the background of the CYP3A4 construct depleted of the native cysteine residues (except for the heme ligating C443). The selection of these sites was prompted by our recent study of high-pressure adaptation in cytochromes P450 from deep-sea bacteria, in which we demonstrated a pressure-actuated conformational switch in the high-pressure-adapted (piezophilic) cytochromes P450 261C1 and 261D1 (70). A comparison of the primary structures of these proteins with those of their mezophilic (not pressure-adapted) progenitors CYP261C2 and CYP261D2 allowed us to localize two regions affected by piezophilic adaptation: 1) the interface between the α-helices A, B′, K, and the meander loop; and 2) the interface between the C/D, J/J′, and the C-terminal loops of the protein (Fig. 2). On the assumption that the substitutions evolved in piezophiles point to the sites of pressure-sensitive conformational mobility common to cytochromes P450, we elected to place R1 at sites 85 and 409 located in region 1, and at sites 166, 343, and 495 located in region 2 in CYP3A4. In addition, we also placed R1 at site 121 in the B/C loop of the enzyme, which is a region of high conformational mobility associated with enzyme-substrate interactions in cytochromes P450.
Figure 2.
Points of incorporation of the environment-sensitive EPR label (MTSSL) and fluorescent probe (MDCC). The cartoon representation of the structure of CYP3A4 is based on the coordinates of the enzyme complex with peripherally bound progesterone (PDB ID: 1W0F). Side chains of the naturally present cysteine residues C64, C98, and C468 are shown in white spheres. Gray spheres show the residues in the regions of the meander and C-terminal loops, respectively, which were chosen for label incorporation on the basis of our studies of high-pressure adaptation in CYP261 enzymes. Residues A121 and S495 are shown in black spheres.
All of the single-cysteine constructs used in this study produced heme-proteins with absorbance spectra typical of cytochromes P450. As shown in Table S1, some of these mutants exhibited a modest displacement of the spin equilibrium in the ligand-free enzyme toward the high-spin state as compared with the wild-type enzyme. This displacement was most pronounced in the mutants C409, C468, and C495 (Table S1). All proteins retained a low-to-high spin shift (type I spectral changes) upon TST binding, although the parameters of the interactions reveal some variations (Table S1). Modification of these mutants with MTSSL caused no denaturation of the proteins, although in some cases this modification increased the content of the P420 state of the protein (Table S1).
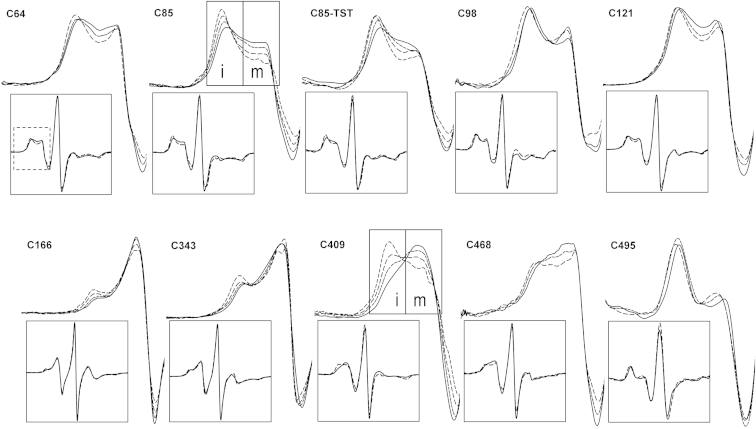
Pressure-induced changes in the EPR spectra of R1 in spin-labeled derivatives of CYP3A4 mutants are illustrated in Fig. 3. The most significant spectral changes were found when R1 was introduced at residue C85 in the loop between β1.2 strand and α-helix B, and residue C409 in the “meander” loop. In the panels of Fig. 3 corresponding to these positions, we illustrate the most important spectral features used in our interpretation. Outlined areas indicate the portions of the spectra that display intensity corresponding to mobile (m) and immobile (i) motional states of the nitroxide. In response to an increase in pressure, the spectra of R1 at C85 and C409 show a strong and reversible redistribution of the two populations toward the more immobilized state, which implies a divergence of the protein between two conformational states existing in a pressure-sensitive equilibrium. Importantly, addition of TST considerably decreases the amplitude of the pressure-induced changes observed at both positions. This effect is illustrated in Fig. 3 for C85-R1. Low-amplitude changes of the same type were also observed for R1 at C166 and C343, although in the latter these changes were seen only at pressures of >2 kbar. The changes observed with R1 placed at C468 may be indicative of a low-amplitude change in the opposite direction (toward a more mobile state of the nitroxide side chain).
Figure 3.
EPR spectra of R1 incorporated into various locations of CYP3A4 as a function of pressure. The data shown were obtained with no ligand added for all positions of the label except for C85, in which case the spectra recorded in the presence of TST (C85+TST) are also shown for comparison. The main panels show the low-field regions of the spectra, and the complete spectra are shown in the insets. The dashed box in the inset of the top leftmost panel indicates the zoomed-in region in the main panels. EPR spectra are shown for the pressures of 30 (solid line), 1000 (long dash), 2000 (medium dash), and 3000 bar (short dash). Outlined areas in the plots for C85-R1 and C409-R1 indicate the portions of the spectra that display intensity corresponding to mobile (m) and immobile (i) motional states of the nitroxide. In response to an increase in pressure, the spectra of residues C85-R1 and C409-R1 exhibit a shift in population toward the component corresponding to an immobilized state. Subtle changes of the same kind are also detectable for residues C166-R1 and C343-R1.
In contrast, no redistribution of the populations of the two conformers is seen with the label placed at any other position. The only effect of pressure on the spectra of R1 in C64, C121, C468, and to a lesser extent C98 and C495 was a change in the lineshape of the low-field portion of the spectra, likely due to compressibility of the protein.
These results are consistent with our LRET studies. Together, they indicate that the protein is subject to a pressure-dependent equilibrium between two conformers, which is displaced toward the pressure-promoted state in the presence of TST. The effect of this transition on the environment of the spin labels is most pronounced in the responses of R1 at positions 409 and 85, which are located in proximity to each other. These probes are placed at the conceptual interface between the α- and β-domains, which is formed by the meander loop in the α-domain and the A/A′ loop in the β-domain (Fig. 2).
Site-directed labeling of CYP3A4 with environment-sensitive fluorescent probes and pressure-perturbation fluorescence spectroscopy
To further explore the nature of the pressure-sensitive conformational equilibrium, we used the environment-sensitive, thiol-reactive fluorescent probe MDCC attached to the same positions as used in our SDSL studies. This coumarin-based fluorophore exhibits a pronounced red shift of the emission band upon transition to more polar environment. As shown in Table S1, modification of our single-cysteine mutant did not cause any significant changes in the spin state of the mutants or parameters of their interactions with TST.
Control experiments with the reduced adduct of MDCC with reduced glutathione (GSH) demonstrated that even in the absence of the protein matrix, the probe exhibited some pressure-induced displacement of the emission band toward a longer wavelength. Application of PCA to the series of spectra obtained in these experiments yielded the first principal component (PC), revealing a pressure-induced shift of the fluorescence band from 478 to 490 nm with a weak, close-to-linear pressure dependence of the loading factor, suggesting a transition with ΔV° of −9.2 ± 3.9 mL/mol and P1/2 equal to 1.25 ± 0.25 kbar. A detailed analysis of this transition in the protein-free label can be found in the Supporting Material.
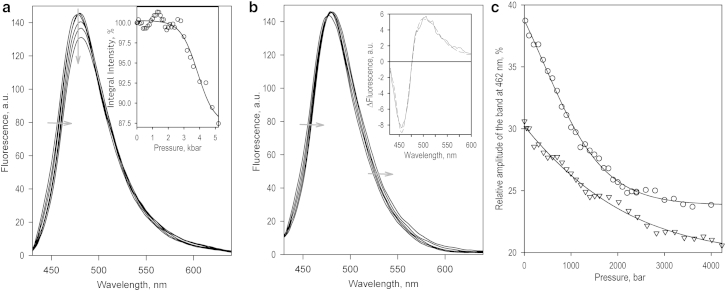
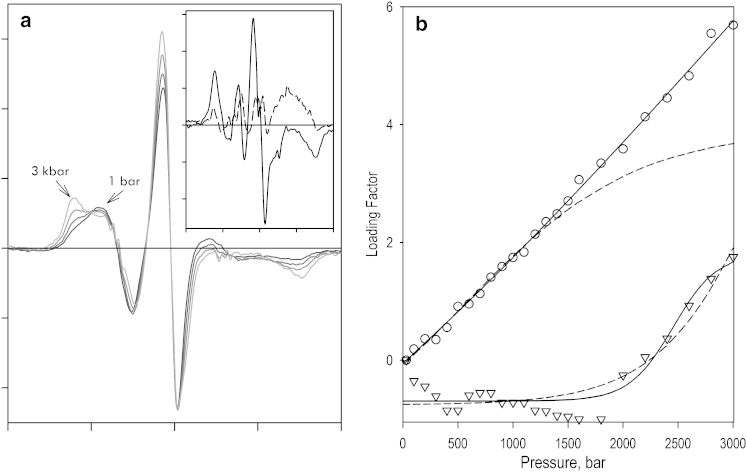
Incorporation of MDCC into the single-cysteine CYP3A4 (C409) changes the pressure dependence of the emission of the fluorophore considerably compared with the protein-free label. As shown in Fig. 4, application of increasing hydrostatic pressure to CYP3A4(C409)MDCC results in a decrease in the intensity of fluorescence along with a red shift in its position (Fig. 4 a). The dependence of the integral intensity of fluorescence on pressure (Fig. 4 a, inset) suggests a transition with ΔV° of −50 ± 20 mL/mol and P1/2 of 3.4 ± 0.5 kbar. The parameters of this apparent pressure-induced inactivation are close to those revealed for irreversible inactivation transitions observed by either LRET or absorbance spectroscopy (see above).
Figure 4.
Effects of pressure on the fluorescence of MDCC incorporated into CYP3A4(C409). (a) Series of emission spectra recorded at 20, 800, 1600, 3200, 4000, and 4800 bar. Gray arrows show the direction of the pressure-induced changes in the line shape. The inset illustrates the pressure dependence of the integral intensity of fluorescence, and the solid line shows the results of its fitting to Eq. 4, which resulted in estimates of ΔV° and P1/2 equal to −45.8 ± 7.7 mL/mol and 3.87 ± 0.11 kbar, respectively (ρ2 = 0.949). (b) The same series of spectra after their normalization based on the intensity of fluorescence. The direction of pressure-induced displacement of the fluorescence band is indicated by gray arrows. The inset shows the spectrum of the first PCs obtained in the analysis of this data set by PCA. (c) Pressure-induced changes in the relative amplitude of the protein-specific, blue-shifted (462 nm) band of MDCC emission with no ligand added (circles) and in the presence of 200 mM TST (triangles). The solid line shows the fitting of the data sets with the equation for pressure dependence of equilibrium (Eq. 4).
In contrast to the changes in the fluorescence intensity of CYP3A4(C409)MDCC, the pressure-induced displacement of the fluorescence band was completely reversible upon decompression. Alongside the reversibility in the changes in LRET efficiency and EPR spectra discussed above, this reversibility suggests that these changes reflect a displacement of a physiologically relevant equilibrium between different states of the enzyme, rather than pressure-induced inactivation. Application of PCA to the series of spectra normalized based on the integral intensity of fluorescence (Fig. 4 b) yields the first PC, revealing a pressure-induced transition between states with different positions of the MDCC fluorescence band (Fig. 4 b, inset).
It should be noted that the band of fluorescence of the protein-bound MDCC at low pressures is shifted to a shorter wavelength compared with the protein-free label and could not be adequately approximated with the spectra of the glutathione (GSH)-MDCC adduct. Reconstruction of the spectra corresponding to the end points of the pressure-induced transition in CYP3A4(C409)MDCC suggests that the protein-bound label, in addition to the states with λmax at 478 and 491 nm seen in MDCC-GSH, also adopts a state with λmax of 462 nm that is indicative of its relocation to a less polar environment. In contrast, the spectra of fluorescence of CYP3A4(C409)MDCC at high (>2000 bar) pressure do not show any considerable deviations from their fitting with the GSH-MDCC-derived standards. This observation is also consistent with a pressure-induced relocation of the probe to a more solvent-exposed position, where its environment becomes similar to that characteristic of the protein-free label.
A combination of PCA with an approach based on approximation of the PCs with linear combinations of the prototypical spectra of MDCC fluorescence (Fig. S2) allowed us to interpret the pressure-induced changes reflected in the first PC in terms of the changes in the fraction of the protein-specific 462 nm band (Fig. 4 c). Approximation of the dependence obtained in the absence of added ligand (Fig. 4 c, circles) yields ΔV° and P1/2 values of −32.0 ± 4.6 mL/mol and 0.62 ± 0.14 kbar, respectively. The difference between these values and those observed with MDCC-GSH in solution (Table 1) suggests that the observed transition reflects a pressure-induced displacement in the protein conformational equilibrium.
Table 1.
Parameters of the Pressure-Induced Transition in Single-Cysteine CYP3A4 Mutants Labeled with MDCC Observed by Pressure-Perturbation Fluorescence Spectroscopy
| Ligand | Position of the Label | P1/2 (Bar) | ΔV (mL/mol)a | ΔG (kJ/mol) | K°eqb |
|---|---|---|---|---|---|
| None | C85 | 1163 ± 315 | −29.4 ± 3.8 (<0.01)∗ | −3.42 ± 1.02 | 0.26 ± 0.12 |
| C409 | 617 ± 144 | −32.0 ± 4.6 (<0.01)∗ | −2.02 ± 0.83 | 0.46 ± 0.16 | |
| C64 | 134 ± 731 | −13.1 ± 4.4 (0.16) | −0.26 ± 1.01 | 0.92 ± 0.37 | |
| C166 | −802 ± 447 | −11.8 ± 3.2 (0.23) | 0.90 ± 0.27 | 1.44 ± 0.15 | |
| C468 | −73 ± 1327 | −11.5 ± 0.6 (0.19) | 0.10 ± 1.53 | 1.10 ± 0.65 | |
| C495 | 878 ± 873 | −13.7 ± 0.6 (0.06) | −1.22 ± 1.25 | 0.63 ± 0.31 | |
| BCT, 15 μM | C85 | 1066 ± 789 | −31.0 ± 14.0 (0.03)∗ | −3.01 ± 0.95 | 0.30 ± 0.11 (0.69) |
| C409 | 935 ± 855 | −30.3 ± 10.2 (<0.01)∗ | −3.18 ± 3.72 | 0.43 ± 0.44 (0.80) | |
| C64 | −344 ± 404 | −11.2 ± 4.4 (0.36) | 0.34 ± 0.30 | 1.15 ± 0.14 (0.38) | |
| C166 | −1399 ± 1875 | −11.3 ± 2.0 (0.24) | 1.48 ± 1.84 | 1.95 ± 1.39 (0.34) | |
| C468 | 202 ± 369 | −12.7 ± 0.2 (0.09) | −0.26 ± 0.47 | 0.91 ± 0.17 (0.64) | |
| C495 | 103 ± 1672 | −13.6 ± 2.5 (0.08) | −0.25 ± 2.29 | 1.01 ± 0.87 (0.51) | |
| TST, 200 μM | C85 | 179 ± 81 | −10.2 ± 5.6 (0.61) | −0.19 ± 0.18 | 0.93 ± 0.07 (<0.01)∗ |
| C409 | −713 ± 291 | −15.6 ± 2.3 (0.01)∗ | 1.20 ± 0.68 | 1.65 ± 0.47 (0.01)∗ | |
| C64 | −525 ± 323 | −12.4 ± 2.8 (0.15) | 0.67 ± 0.55 | 1.32 ± 0.29 (0.24) | |
| C166 | −488 ± 434 | −11.0 ± 0.8 (0.26) | 0.53 ± 0.44 | 1.24 ± 0.22 (0.69) | |
| C468 | −1009 ± 1395 | −10.8 ± 0.1 (0.28) | 1.09 ± 1.50 | 1.63 ± 0.95 (0.46) | |
| C495 | 412 ± 620 | −11.5 ± 1.9 (0.22) | −0.50 ± 0.79 | 0.83 ± 0.26 (0.45) | |
| None | GSH | 1245 ± 258 | −9.2 ± 3.9 | −1.17 ± 0.72 | 0.63 ± 0.18 |
The values given in the table represent the averages of the results of three to six individual experiments. The individual estimates of ΔV° and P1/2 were obtained from fitting of the pressure dependencies of the loading factor of the first PC found with PCA with Eq. 4. These estimated were used to calculate the values of ΔG and K°eq given in the table. The “±” values show the confidence interval calculated for p = 0.05.
Values in parentheses represent the p-values from Student’s t-test for the hypothesis of equality of the respective values of ΔV° with that determined for MDCC-GSH adduct in solution; ∗p ≤ 0.05 emphasizes the cases with a statistically significant difference from the case of protein-free label in solution.
Values in parentheses represent the p-values from Student’s t-test for the hypothesis of equality of the values of K°eq determined in the presence of BCT or TST with those observed in the absence of ligand; ∗∗p ≤ 0.05 emphasizes the cases with a statistically significant effect of ligands on the position of equilibrium.
As shown in Fig. 4 c (triangles), interactions of the enzyme with TST cause a decrease in the amplitude of the pressure-induced effect together with a decrease in the molar volume change and displacement of the P1/2 to negative pressures. The respective values of the equilibrium constant for the pressure-promoted and “relaxed” (low-pressure) conformers at ambient pressure are equal to 0.46 ± 0.16 and 1.65 ± 0.47, and indicate a TST-induced displacement of the equilibrium toward the pressure-promoted state.
Table 1 compares the parameters of the pressure-induced red shift obtained with a series of CYP3A4 mutants labeled with MDCC. The mutants C85 and C409 exhibited similar behavior, whereas all others were considerably different. The parameters of the pressure-induced transition observed with the labels placed at positions 64, 121, 166, and 468 were essentially similar to those observed with MDCC-GSH in solution, and no effect of TST was evident, in contrast to its pronounced effect in C409 and C85. These data are in good agreement with the results of pressure-perturbation EPR spectroscopy. Taken together, our findings suggest that only the probes located at residues 409 and 85 reveal a pressure-sensitive and TST-dependent conformational equilibrium associated with a pronounced change in the environment of the spin label or fluorescent probe and an effect of TST.
To probe whether the effect of TST on the pressure-related behavior of CYP3A4 is specific to this allosteric modulator rather than common to interactions of CYP3A4 with different substrates, we also probed pressure-induced changes in the fluorescence of MDCC-labeled CYP3A4 mutants in the presence of BCT. This dopamine agonist and anticonvulsant drug is a high-affinity (Kd ≤ 1 μM) CYP3A4 substrate, and its interaction with the enzyme results in a pronounced low-to-high spin shift but lacks any known activating effect (69, 71, 72, 73). As can be seen in Table 1, in contrast to TST, addition of a saturating concentration of BCT had no considerable effect on pressure-induced changes in any of the studied MDCC-labeled proteins.
The values of ΔV° obtained with MDCC-labeled C85 and C409 (Table 1) are in good agreement with the value determined in our LRET experiments (−36.8 ± 5.0 mL/mol). At the same time, our results are indicative of decreased values of P1/2 in C85-MDCC and C409-MDCC as compared with that derived from the LRET studies (1.45 ± 0.33 kbar). This difference was most pronounced for C409-MDCC (see Table 1), where it resulted in an increase in the value of Keq° from 0.13 ± 0.06 (as in the CYP3A4(C64/C468)ER/DY construct) to 0.46 ± 0.16. Despite these variations, which may reveal the effects of labeling and/or mutagenesis on the conformational landscape, the barotropic behavior of C85-MDCC and C409-MDCC is generally similar to that revealed in the LRET studies. This congruence suggests that in both cases the observed changes reflect a displacement of conformational equilibrium between two states with different spatial separation between the α- and β-domains. This equilibrium is therefore associated with considerable structural changes at the interface between the α-helix A and the meander loop, where residues 85 and 409 are located.
Analysis of the results of pressure-perturbation EPR by PCA
The protein loci involved in the pressure-sensitive changes identified by SDSL-EPR experiments coincide with those suggested on the basis of the studies with MDCC-labeled enzyme. However, definitive conclusions about the interrelationship between the conformational equilibria revealed by the two techniques require a comparison of their thermodynamic parameters (ΔV° and Keq°). Determination of these parameters from the results of pressure-perturbation EPR is complicated by the fact that changes in the EPR spectra caused by physical factors (e.g., pressure) usually represent a superposition of several distinct transitions. This complexity renders inadequate a model with only two end states that can be analyzed with existing analytical methods (74, 75). Instead, PCA, which is commonly employed in the analysis of spectral changes in complex systems (76, 77, 78), becomes the method of choice.
Although the literature provides several examples of the use of PCA and singular value decomposition, a closely related technique, to analyze the EPR spectra of paramagnetic probes introduced into proteins (79, 80, 81), the full potential of these methods remains to be realized. A major difficulty of interpreting the results of PCA is that the set of differential spectra (eigenvectors) and the dependencies of their amplitudes (eigenvalues or loading factors) on a variable parameter Z (e.g., pressure) have no straightforward concordance with real, physically relevant processes. Instead, these processes may be construed as linear combinations of individual PCs found by PCA (76, 78). This complexity is addressed in the approach known as target factor analysis (TFA) (78), which is briefly discussed in the Supporting Material. As shown below, the application of the PCA-TFA technique combined with evaluation of thermodynamic and/or kinetic parameters of the underlying processes with complementary techniques provides a promising approach that allows the changes in EPR spectra to be resolved into distinct, physically relevant components.
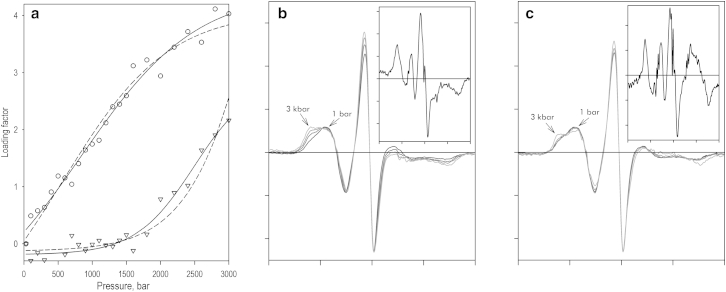
The results of applying PCA to a series of EPR spectra obtained in a pressure-perturbation experiment with CYP3A4(C85)R1 are illustrated in Fig. 5. More than 97% of the observed changes were covered by the first two PCs, and the spectrum of the first PC (Fig. 5 a, inset) represents the predominant part of the changes in the low-field part of the spectra. As discussed above, these changes are indicative of the displacement of equilibrium between two protein conformers with different mobility of the nitroxide side chain. Importantly, the pressure dependence of the loading factor of the first PC (Fig. 5 b, circles) may be adequately approximated with Eq. 4 and ΔV° and P1/2, values determined in our studies of solvatochromic shift in CYP3A4(C85)MDCC (Fig. 5 b, circles and dashed line). Optimization of these parameters for the best fitting of this data set (ρ2 = 0.992) results in values of −28.0 ± 1.3 mL/mol and 0.86 ± 0.03 kbar (Fig. 5 b, circles and solid line).
Figure 5.
Analysis of pressure-induced changes in the EPR spectra of CYP3A4(C85)R1 by PCA. (a) Normalized EPR spectra were obtained at pressures of 1 bar and 1, 2, and 3 kbar. The inset shows the spectra of the first (solid line) and second (dashed line) PCs of the observed changes. (b) Pressure dependencies of the loading factors for the first (circles) and second (triangles) PCs. Dashed lines show the approximations of the data sets with Eq. 4 using the values of ΔV° and P1/2 determined in the studies with CYP3A4(C85)MDCC. In the case of the first PC, these parameters represent a pressure-induced solvatochromic shift, whereas in the case of the second PC, we used values characterizing the pressure dependence of fluorescence intensity. Solid lines show the results obtained by optimizing these parameters for the best fitting of the data sets.
The pressure dependence of the loading factor for the second principal vector (Fig. 5 b, triangles) may be satisfactorily approximated (ρ2 = 0.856; Fig. 5 b, triangles and dashed line) with Eq. 4 and the values of ΔV° and P1/2, derived from the pressure-induced decrease in fluorescence of CYP3A4(C85)MDCC (−38.7 ml/mol and 3.38 kbar, respectively). Optimization of these values for the best approximation of the data set increases the square correlation coefficient to 0.883 (Fig. 5 b, triangles and solid line) and results in values of −45.6 ± 1.8 mL/mol and 4.65 ± 0.07 kbar for ΔV° and P1/2, respectively. Attempts to improve the approximation of the two theoretical target functions (Fig. 5 b, dashed lines) using the TFA approach, by combining the pressure dependencies of the loading factors of the first two or three PCs, did not improve the quality of the approximation. Therefore, the two abstract transitions found in the PCA are very close to those expected based on the results of our fluorometric studies with MDCC-labeled proteins.
The results of applying PCA to a series of EPR spectra obtained in pressure-perturbation experiments with CYP3A4(C409)R1 are shown in Fig. 6. More than 99% of the observed changes were covered by the first two PCs, and the pressure dependence of the loading factor for the second PC (Fig. 6 b, triangles) closely resembles the plot expected for the pressure-induced inactivation with the parameters found in our studies with MDCC-labeled proteins (dashed line over triangles). Fitting of this dependence to Eq. 4 resulted in a satisfactory approximation (Fig. 6 b, solid line over triangles), with values of ΔV° and P1/2 equal to −123 ± 68 mL/mol and 2.46 ± 0.12 kbar, respectively (ρ2 = 0.860). In contrast, the plot of the loading factor for the first PC versus pressure (Fig. 6 b, circles) noticeably deviates from the theoretical curve built for the pressure-induced solvatochromic shift in CYP3A4(C409)MDCC (Fig. 6 b, dashed line over circles). We hypothesize, therefore, that the application of PCA to this data set results in PCs that reflect linear combinations of the underlying processes rather than providing straightforward representations, as observed with CYP3A4(C85).
Figure 6.
Analysis of pressure-induced changes in the EPR spectra of CYP3A4(C409)R1 by PCA. (a) Normalized EPR spectra obtained at pressures of 1 bar and 1, 2, and 3 kbar. The inset shows the spectra of the first two PCs of the observed changes. (b) Pressure dependencies of the loading factors for the first (circles) and second (triangles) PCs. Dashed lines show the approximations of the whole data set (for the case of triangles) or its initial part, including the points corresponding to pressures up to 1200 bar (for the case of circles) with Eq. 4, using the values of ΔV° and P1/2 determined in the studies with CYP3A4(C409)MDCC. In the case of the first PC, these parameters represent a pressure-induced solvatochromic shift, whereas in the case of the second PC, we used values characterizing the pressure dependence of fluorescence intensity. The solid line for the second PC (triangles) shows the results obtained by optimizing these parameters for the best fitting of the data sets. In the case of the first PC (circles), the solid line corresponds to a linear approximation of the data.
Our application of TFA-PCA to the spectra of CYP3A4(C409)R1 illustrated in Fig. 7 was based on the assumption that the observed spectral changes represent an overlap of two independent, physically relevant processes (reversible conformational shift and pressure-induced inactivation). The predominant majority of the respective changes are thus captured by the first two PCs. Consistent with these inferences, the theoretical plots calculated for the two pressure-dependent transitions (conformational shift and inactivation) observed in CYP3A4(C409)MDCC may be satisfactorily approximated with linear combinations of the vectors of loading factors for the first two PCs (Fig. 7 a). This allowed us to transform the matrices of PCs to correspond to the above approximation. The details of this procedure are described in the Supporting Material. The resulting differential spectra of the two hypothetical processes are shown in the insets to Fig. 7, b and c. The main plots in these panels show the reconstitution of the series of EPR spectra representing each of the two target processes, namely, the conformational shift (Fig. 7 b) and inactivation (Fig. 7 c).
Figure 7.
Analysis of pressure-induced changes in the EPR spectra of CYP3A4(C409)R1 by TFA-PCA. (a) Approximation of the two pressure-induced transitions observed in fluorescence spectroscopy experiments (dashed lines), with linear combinations of the sets of loading factors for the first two PCs obtained in a pressure-perturbation experiment with CYP3A4(C409)R1. The main (reversible) transition is shown in circles, and the transition corresponding to apparent inactivation is shown in triangles. The solid lines represent the fitting of the data sets with Eq. 4. (b and c) Sets of EPR spectra obtained from the original data by sorting out the changes corresponding to reversible (b) and irreversible (c) components. The spectra shown in (b) represent pressures of 1, 1000, 2000, and 3000 bar, and those in (c) correspond to 1, 2000, 2400, and 3000 bar. The insets show the respective spectra of the first PCs of the pressure-induced changes.
Fitting of the combined pressure dependence obtained for the reversible TST-dependent conformational shift (Fig. 7 a, circles) resulted in ΔV° and P1/2 values of −29.2 ± 2.3 mL/mol and 0.84 ± 0.05 kbar, respectively (ρ2 = 0.981; circles, solid line). The respective fitting for the inactivation transition yielded ΔV° and P1/2 values equal to −57.9 ± 8.5 mL/mol and 2.49 ± 0.07 kbar, respectively (ρ2 = 0.961; triangles, solid line). In both cases, the obtained parameters were in agreement with those obtained for these two processes in our measurements of the solvatochromic shift.
Therefore, application of the PCA-TFA approach allowed us to resolve the pressure-induced changes in the EPR spectra obtained with CYP3A4(C85)R1 and CYP3A4(C409)R1 into two superimposed transitions that correspond to the pressure-induced and TST-sensitive conformational shift and pressure-induced inactivation detected in our studies using pressure-perturbation fluorescence spectroscopy and LRET.
Conclusions
Taken together, our results reveal an effector-dependent conformational equilibrium between the open and closed states of CYP3A4 that involves a pronounced conformational change at the conceptual interface between the α- and β-domains of CYP3A4 and in the region of the meander loop of the enzyme, where residues 85 and 409 are located. We can conclude, therefore, that the pressure-induced changes in the environment of the probes attached to residues 85 and 409 are the most affected by the TST-induced conformational transition.
It is important to note that the location of this apparent hinge region was predicted on the basis of our studies of the evolutionary adaptation of cytochromes P450 of piezophilic bacteria (CYP261 proteins) that enabled them to function at high hydrostatic pressure. This suggests that the large-scale conformational dynamics demonstrated in this study is common to various evolutionarily distant cytochromes P450. The pressure-sensitive equilibrium between the open and closed conformers may play an important role in P450 catalytic mechanisms by modulating solvent and substrate accessibility during the catalytic cycle. These results provide a striking demonstration of the exceptional value of exploring enzyme conformational dynamics based on studies of the conformational adaptation that evolved in pressure-tolerant enzymes of piezophilic (deep-sea) species (39).
The modulation of conformational equilibrium by TST observed here may provide evidence of the involvement of this transition in allosteric mechanisms that regulate the function of the human drug-metabolizing ensemble. Our recent demonstration of a causal relationship between oligomerization of CYP3A4 and its ability to be activated by allosteric effectors (82) suggests that modulation of the CYP3A4 conformation by TST may involve its binding at the peripheral ligand-binding site located at the interface between the interacting subunits (82, 83). Further exploration of the molecular mechanisms of this modulation may provide an important clue to the mechanisms of P450 allostery.
Our study demonstrates the unique power of pressure-perturbation techniques, and pressure-perturbation EPR spectroscopy in particular, for studying protein conformational dynamics. The combination of pressure-perturbation EPR spectroscopy with LRET and solvatochromic shift techniques, supplemented with global data analysis using PCA-TFA, is a promising approach for exploring protein conformational landscapes.
Author Contributions
D.R.D. and W.L.H. designed the study. D.R.D., Z.Y., and N.Y.D. performed research. D.R.D., Z.Y., and W.L.H. analyzed the data. D.R.D., W.L.H., and J.R.H. wrote the manuscript.
Acknowledgments
This research was supported by National Institutes of Health grants GM054995 (to J.R.H) and EY005216 (to W.L.H.), and a Jules Stein Professorship (to W.L.H.).
Editor: Elizabeth Rhoades.
Footnotes
James Halpert’s present address is School of Pharmacy, University of Connecticut, Storrs, Connecticut.
Four figures, one table and a textual addendum are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00214-9.
Supporting Material
References
- 1.Wilderman P.R., Halpert J.R. Plasticity of CYP2B enzymes: structural and solution biophysical methods. Curr. Drug Metab. 2012;13:167–176. doi: 10.2174/138920012798918417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pochapsky T.C., Kazanis S., Dang M. Conformational plasticity and structure/function relationships in cytochromes P450. Antioxid. Redox Signal. 2010;13:1273–1296. doi: 10.1089/ars.2010.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gay S.C., Roberts A.G., Halpert J.R. Structural features of cytochromes P450 and ligands that affect drug metabolism as revealed by X-ray crystallography and NMR. Future Med. Chem. 2010;2:1451–1468. doi: 10.4155/fmc.10.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otyepka M., Berka K., Anzenbacher P. Is there a relationship between the substrate preferences and structural flexibility of cytochromes P450? Curr. Drug Metab. 2012;13:130–142. doi: 10.2174/138920012798918372. [DOI] [PubMed] [Google Scholar]
- 5.Ma Q., Lu A.Y.H. The challenges of dealing with promiscuous drug-metabolizing enzymes, receptors and transporters. Curr. Drug Metab. 2008;9:374–383. doi: 10.2174/138920008784746337. [DOI] [PubMed] [Google Scholar]
- 6.Atkins W.M., Qian H. Stochastic ensembles, conformationally adaptive teamwork, and enzymatic detoxification. Biochemistry. 2011;50:3866–3872. doi: 10.1021/bi200275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haines D.C., Tomchick D.R., Peterson J.A. Pivotal role of water in the mechanism of P450BM-3. Biochemistry. 2001;40:13456–13465. doi: 10.1021/bi011197q. [DOI] [PubMed] [Google Scholar]
- 8.Fishelovitch D., Shaik S., Nussinov R. How does the reductase help to regulate the catalytic cycle of cytochrome P450 3A4 using the conserved water channel? J. Phys. Chem. B. 2010;114:5964–5970. doi: 10.1021/jp101894k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar D., Altun A., Thiel W. Water as biocatalyst in cytochrome P450. Faraday Discuss. 2011;148:373–383. doi: 10.1039/c004950f. discussion 421–441. [DOI] [PubMed] [Google Scholar]
- 10.Sineva E.V., Davydov D.R. Cytochrome P450 from Photobacterium profundum SS9, a piezophilic bacterium, exhibits a tightened control of water access to the active site. Biochemistry. 2010;49:10636–10646. doi: 10.1021/bi101466y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madrona Y., Hollingsworth S.A., Poulos T.L. P450cin active site water: implications for substrate binding and solvent accessibility. Biochemistry. 2013;52:5039–5050. doi: 10.1021/bi4006946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oprea T.I., Hummer G., Garcia A.E. Identification of a functional water channel in cytochrome P450 enzymes. Proc. Natl. Acad. Sci. USA. 1997;94:2133–2138. doi: 10.1073/pnas.94.6.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wade R.C., Winn P.J., Sudarko A survey of active site access channels in cytochromes P450. J. Inorg. Biochem. 2004;98:1175–1182. doi: 10.1016/j.jinorgbio.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Rydberg P., Rod T.H., Ryde U. Dynamics of water molecules in the active-site cavity of human cytochromes P450. J. Phys. Chem. B. 2007;111:5445–5457. doi: 10.1021/jp070390c. [DOI] [PubMed] [Google Scholar]
- 15.Williams P.A., Cosme J., McRee D.E. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 16.Williams P.A., Cosme J., Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 17.Scott E.E., He Y.A., Stout C.D. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc. Natl. Acad. Sci. USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoch G.A., Yano J.K., Johnson E.F. Structure of human microsomal cytochrome P450 2C8. Evidence for a peripheral fatty acid binding site. J. Biol. Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 19.Gay S.C., Sun L., Stout C.D. Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole: ligand-induced structural response through alpha-helical repositioning. Biochemistry. 2009;48:4762–4771. doi: 10.1021/bi9003765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowland P., Blaney F.E., Bridges A.M. Crystal structure of human cytochrome P450 2D6. J. Biol. Chem. 2006;281:7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- 21.Podust L.M., Kim Y., Waterman M.R. The 1.92-A structure of Streptomyces coelicolor A3(2) CYP154C1. A new monooxygenase that functionalizes macrolide ring systems. J. Biol. Chem. 2003;278:12214–12221. doi: 10.1074/jbc.M212210200. [DOI] [PubMed] [Google Scholar]
- 22.Zhao B., Guengerich F.P., Waterman M.R. Role of active site water molecules and substrate hydroxyl groups in oxygen activation by cytochrome P450 158A2: a new mechanism of proton transfer. J. Biol. Chem. 2005;280:42188–42197. doi: 10.1074/jbc.M509220200. [DOI] [PubMed] [Google Scholar]
- 23.Lee D.S., Yamada A., Shiro Y. Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis. Crystallographic, spectroscopic, and mutational studies. J. Biol. Chem. 2003;278:9761–9767. doi: 10.1074/jbc.M211575200. [DOI] [PubMed] [Google Scholar]
- 24.Cojocaru V., Winn P.J., Wade R.C. The ins and outs of cytochrome P450s. Biochim. Biophys. Acta. 2007;1770:390–401. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Williams P.A., Cosme J., Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 26.Roberts A.G., Atkins W.M. Energetics of heterotropic cooperativity between alpha-naphthoflavone and testosterone binding to CYP3A4. Arch. Biochem. Biophys. 2007;463:89–101. doi: 10.1016/j.abb.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davydov D.R., Halpert J.R. Allosteric P450 mechanisms: multiple binding sites, multiple conformers or both? Expert Opin. Drug Metab. Toxicol. 2008;4:1523–1535. doi: 10.1517/17425250802500028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sevrioukova I.F., Poulos T.L. Anion-dependent stimulation of CYP3A4 monooxygenase. Biochemistry. 2015;54:4083–4096. doi: 10.1021/acs.biochem.5b00510. [DOI] [PubMed] [Google Scholar]
- 29.Davydov D.R., Rumfeldt J.A.O., Halpert J.R. Peripheral ligand-binding site in cytochrome P450 3A4 located with fluorescence resonance energy transfer (FRET) J. Biol. Chem. 2012;287:6797–6809. doi: 10.1074/jbc.M111.325654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atkins W.M., Wang R.W., Lu A.Y.H. Allosteric behavior in cytochrome p450-dependent in vitro drug-drug interactions: a prospective based on conformational dynamics. Chem. Res. Toxicol. 2001;14:338–347. doi: 10.1021/tx0002132. [DOI] [PubMed] [Google Scholar]
- 31.Isin E.M., Guengerich F.P. Substrate binding to cytochromes P450. Anal. Bioanal. Chem. 2008;392:1019–1030. doi: 10.1007/s00216-008-2244-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isin E.M., Guengerich F.P. Multiple sequential steps involved in the binding of inhibitors to cytochrome P450 3A4. J. Biol. Chem. 2007;282:6863–6874. doi: 10.1074/jbc.M610346200. [DOI] [PubMed] [Google Scholar]
- 33.Tsalkova T.N., Davydova N.Y., Davydov D.R. Mechanism of interactions of alpha-naphthoflavone with cytochrome P450 3A4 explored with an engineered enzyme bearing a fluorescent probe. Biochemistry. 2007;46:106–119. doi: 10.1021/bi061944p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sineva E.V., Rumfeldt J.A.O., Davydov D.R. A large-scale allosteric transition in cytochrome P450 3A4 revealed by luminescence resonance energy transfer (LRET) PLoS One. 2013;8:e83898. doi: 10.1371/journal.pone.0083898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graham-Lorence S., Peterson J.A. P450s: structural similarities and functional differences. FASEB J. 1996;10:206–214. doi: 10.1096/fasebj.10.2.8641554. [DOI] [PubMed] [Google Scholar]
- 36.Heremans K. High pressure effects on proteins and other biomolecules. Annu. Rev. Biophys. Bioeng. 1982;11:1–21. doi: 10.1146/annurev.bb.11.060182.000245. [DOI] [PubMed] [Google Scholar]
- 37.Somero G.N. Adaptations to high hydrostatic pressure. Annu. Rev. Physiol. 1992;54:557–577. doi: 10.1146/annurev.ph.54.030192.003013. [DOI] [PubMed] [Google Scholar]
- 38.Mozhaev V.V., Heremans K., Balny C. High pressure effects on protein structure and function. Proteins. 1996;24:81–91. doi: 10.1002/(SICI)1097-0134(199601)24:1<81::AID-PROT6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 39.Davydov D.R. Merging thermodynamics and evolution: how the studies of high-pressure adaptation may help to understand enzymatic mechanisms. J. Thermodyn. Catal. 2012;3:1000e1110. [Google Scholar]
- 40.Masson P., Reybaud J. Hydrophobic interaction electrophoresis under high hydrostatic pressure: study of the effects of pressure upon the interaction of serum albumin with a long-chain aliphatic ligand. Electrophoresis. 1988;9:157–161. doi: 10.1002/elps.1150090402. [DOI] [PubMed] [Google Scholar]
- 41.Low P.S., Somero G.N. Protein hydration changes during catalysis: a new mechanism of enzymic rate-enhancement and ion activation/inhibition of catalysis. Proc. Natl. Acad. Sci. USA. 1975;72:3305–3309. doi: 10.1073/pnas.72.9.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Low P.S., Somero G.N. Activation volumes in enzymic catalysis: their sources and modification by low-molecular-weight solutes. Proc. Natl. Acad. Sci. USA. 1975;72:3014–3018. doi: 10.1073/pnas.72.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber G., Drickamer H.G. The effect of high pressure upon proteins and other biomolecules. Q. Rev. Biophys. 1983;16:89–112. doi: 10.1017/s0033583500004935. [DOI] [PubMed] [Google Scholar]
- 44.Shimizu S. Estimating hydration changes upon biomolecular reactions from osmotic stress, high pressure, and preferential hydration experiments. Proc. Natl. Acad. Sci. USA. 2004;101:1195–1199. doi: 10.1073/pnas.0305836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boonyaratanakornkit B.B., Park C.B., Clark D.S. Pressure effects on intra- and intermolecular interactions within proteins. Biochim. Biophys. Acta. 2002;1595:235–249. doi: 10.1016/s0167-4838(01)00347-8. [DOI] [PubMed] [Google Scholar]
- 46.Royer C.A. Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta. 2002;1595:201–209. doi: 10.1016/s0167-4838(01)00344-2. [DOI] [PubMed] [Google Scholar]
- 47.Imai T., Hirata F. Hydrophobic effects on partial molar volume. J. Chem. Phys. 2005;122:094509. doi: 10.1063/1.1854626. [DOI] [PubMed] [Google Scholar]
- 48.Mozhaev V.V., Heremans K., Balny C. Exploiting the effects of high hydrostatic-pressure in biotechnological applications. Trends Biotechnol. 1994;12:493–501. [Google Scholar]
- 49.Chalikian T.V., Filfil R. How large are the volume changes accompanying protein transitions and binding? Biophys. Chem. 2003;104:489–499. doi: 10.1016/s0301-4622(03)00037-1. [DOI] [PubMed] [Google Scholar]
- 50.Helms V. Protein dynamics tightly connected to the dynamics of surrounding and internal water molecules. ChemPhysChem. 2007;8:23–33. doi: 10.1002/cphc.200600298. [DOI] [PubMed] [Google Scholar]
- 51.Mentré P., Hui Bon Hoa G. Effects of high hydrostatic pressures on living cells: a consequence of the properties of macromolecules and macromolecule-associated water. Int. Rev. Cytol. 2001;201:1–84. doi: 10.1016/s0074-7696(01)01001-4. [DOI] [PubMed] [Google Scholar]
- 52.Kharakoz D.P. Partial volumes and compressibilities of extended polypeptide chains in aqueous solution: additivity scheme and implication of protein unfolding at normal and high pressure. Biochemistry. 1997;36:10276–10285. doi: 10.1021/bi961781c. [DOI] [PubMed] [Google Scholar]
- 53.Kornblatt J.A., Kornblatt M.J. The effects of osmotic and hydrostatic pressures on macromolecular systems. Biochim. Biophys. Acta. 2002;1595:30–47. doi: 10.1016/s0167-4838(01)00333-8. [DOI] [PubMed] [Google Scholar]
- 54.Hui Bon Hoa G., McLean M.A., Sligar S.G. High pressure, a tool for exploring heme protein active sites. Biochim. Biophys. Acta. 2002;1595:297–308. doi: 10.1016/s0167-4838(01)00352-1. [DOI] [PubMed] [Google Scholar]
- 55.Bancel F., Hoa G.H., Lange R. High pressure: a new tool to study P450 structure and function. Methods Enzymol. 2002;357:145–157. doi: 10.1016/s0076-6879(02)57674-6. [DOI] [PubMed] [Google Scholar]
- 56.Davydov D.R., Hui Bon Hoa G., Peterson J.A. Dynamics of protein-bound water in the heme domain of P450BM3 studied by high-pressure spectroscopy: comparison with P450cam and P450 2B4. Biochemistry. 1999;38:751–761. doi: 10.1021/bi981397a. [DOI] [PubMed] [Google Scholar]
- 57.Hubbell W.L., Cafiso D.S., Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 58.McCoy J., Hubbell W.L. High-pressure EPR reveals conformational equilibria and volumetric properties of spin-labeled proteins. Proc. Natl. Acad. Sci. USA. 2011;108:1331–1336. doi: 10.1073/pnas.1017877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lerch M.T., Horwitz J., Hubbell W.L. Circular dichroism and site-directed spin labeling reveal structural and dynamical features of high-pressure states of myoglobin. Proc. Natl. Acad. Sci. USA. 2013;110:E4714–E4722. doi: 10.1073/pnas.1320124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harlow G.R., Halpert J.R. Analysis of human cytochrome P450 3A4 cooperativity: construction and characterization of a site-directed mutant that displays hyperbolic steroid hydroxylation kinetics. Proc. Natl. Acad. Sci. USA. 1998;95:6636–6641. doi: 10.1073/pnas.95.12.6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davydov D.R., Baas B.J., Halpert J.R. Allosteric mechanisms in cytochrome P450 3A4 studied by high-pressure spectroscopy: pivotal role of substrate-induced changes in the accessibility and degree of hydration of the heme pocket. Biochemistry. 2007;46:7852–7864. doi: 10.1021/bi602400y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hui Bon Hoa G., Marden M.C. The pressure dependence of the spin equilibrium in camphor-bound ferric cytochrome P-450. Eur. J. Biochem. 1982;124:311–315. doi: 10.1111/j.1432-1033.1982.tb06593.x. [DOI] [PubMed] [Google Scholar]
- 63.Weber G. Chapman and Hall; New York: 1991. Protein Interactions. [Google Scholar]
- 64.Davydov D.R., Deprez E., Archakov A.I. High-pressure-induced transitions in microsomal cytochrome P450 2B4 in solution: evidence for conformational inhomogeneity in the oligomers. Arch. Biochem. Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 65.Yu X., Cojocaru V., Wade R.C. Conformational diversity and ligand tunnels of mammalian cytochrome P450s. Biotechnol. Appl. Biochem. 2013;60:134–145. doi: 10.1002/bab.1074. [DOI] [PubMed] [Google Scholar]
- 66.Fernando H., Davydov D.R., Halpert J.R. Role of subunit interactions in P450 oligomers in the loss of homotropic cooperativity in the cytochrome P450 3A4 mutant L211F/D214E/F304W. Arch. Biochem. Biophys. 2007;460:129–140. doi: 10.1016/j.abb.2006.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davydov D.R., Fernando H., Halpert J.R. Kinetics of dithionite-dependent reduction of cytochrome P450 3A4: heterogeneity of the enzyme caused by its oligomerization. Biochemistry. 2005;44:13902–13913. doi: 10.1021/bi0509346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Molina-García A.D. Hydrostatic pressure in ultracentrifugation (revisited) Prog. Colloid Polym. Sci. 1999;113:57–61. [Google Scholar]
- 69.Davydov D.R., Halpert J.R., Hui Bon Hoa G. Conformational heterogeneity of cytochrome P450 3A4 revealed by high pressure spectroscopy. Biochem. Biophys. Res. Commun. 2003;312:121–130. doi: 10.1016/j.bbrc.2003.09.247. [DOI] [PubMed] [Google Scholar]
- 70.Davydov D.R., Sineva E.V., Halpert J.R. CYP261 enzymes from deep sea bacteria: a clue to conformational heterogeneity in cytochromes P450. Biotechnol. Appl. Biochem. 2013;60:30–40. doi: 10.1002/bab.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peyronneau M.A., Delaforge M., Mansuy D. High affinity of ergopeptides for cytochromes P450 3A. Importance of their peptide moiety for P450 recognition and hydroxylation of bromocriptine. Eur. J. Biochem. 1994;223:947–956. doi: 10.1111/j.1432-1033.1994.tb19072.x. [DOI] [PubMed] [Google Scholar]
- 72.Isin E.M., Guengerich F.P. Kinetics and thermodynamics of ligand binding by cytochrome P450 3A4. J. Biol. Chem. 2006;281:9127–9136. doi: 10.1074/jbc.M511375200. [DOI] [PubMed] [Google Scholar]
- 73.Sevrioukova I.F., Poulos T.L. Structural and mechanistic insights into the interaction of cytochrome P4503A4 with bromoergocryptine, a type I ligand. J. Biol. Chem. 2012;287:3510–3517. doi: 10.1074/jbc.M111.317081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oganesyan V.S. Computational approaches for simulating motional EPR spectra. In: Gilbert B.C., Chechik V., Murphy D.M., editors. Vol. 24. Royal Society of Chemistry; Cambridge: 2015. pp. 32–61. (Electron Paramagnetic Resonance). [Google Scholar]
- 75.Pedone A., Biczysko M., Barone V. Environmental effects in computational spectroscopy: accuracy and interpretation. ChemPhysChem. 2010;11:1812–1832. doi: 10.1002/cphc.200900976. [DOI] [PubMed] [Google Scholar]
- 76.Ross R.T., Leurgans S. Component resolution using multilinear models. Methods Enzymol. 1995;246:679–700. doi: 10.1016/0076-6879(95)46029-2. [DOI] [PubMed] [Google Scholar]
- 77.Hendler R.W., Shrager R.I. Deconvolutions based on singular value decomposition and the pseudoinverse: a guide for beginners. J. Biochem. Biophys. Methods. 1994;28:1–33. doi: 10.1016/0165-022x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 78.Tam K.Y., Chau F.T. Multivariate study of kinetic data for a 2-step consecutive reaction using target factor-analysis. Chemom. Intell. Lab. Syst. 1994;25:25–42. [Google Scholar]
- 79.Landahl E.C., Rice S.E. Model-independent decomposition of two-state data. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2013;88:062713. doi: 10.1103/PhysRevE.88.062713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zapata A.L., Kumar M.R., Farmer P.J. A singular value decomposition approach for kinetic analysis of reactions of HNO with myoglobin. J. Inorg. Biochem. 2013;118:171–178. doi: 10.1016/j.jinorgbio.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 81.Keszler A., Kalyanaraman B., Hogg N. Comparative investigation of superoxide trapping by cyclic nitrone spin traps: the use of singular value decomposition and multiple linear regression analysis. Free Radic. Biol. Med. 2003;35:1149–1157. doi: 10.1016/s0891-5849(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 82.Davydov D.R., Davydova N.Y., Halpert J.R. Pivotal role of P450-P450 interactions in CYP3A4 allostery: the case of α-naphthoflavone. Biochem. J. 2013;453:219–230. doi: 10.1042/BJ20130398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davydov D.R., Davydova N.Y., Halpert J.R. Interactions among cytochromes P450 in microsomal membranes: oligomerization of cytochromes P450 3A4, 3A5, and 2E1 and its functional consequences. J. Biol. Chem. 2015;290:3850–3864. doi: 10.1074/jbc.M114.615443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.