Abstract
We have developed retroviral replicating vectors (RRV) derived from Moloney murine gammaretrovirus with an amphotropic envelope and an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES)-transgene cassette downstream of the env gene. During long-term (180 days) replication of the vector in animals, a bulge of 7 adenosine residues (A's) in the J-K bifurcation domain sometimes serially added A's. Therefore, vectors with 4–12 A's in the A bulge in the J-K bifurcation domain were generated, and the impact of the variants on transgene protein expression, vector stability, and IRES sequence upon multiple infection cycles was assessed in RRV encoding yeast-derived cytosine deaminase and green fluorescent protein in vitro. For transgene protein expression, after multiple infection cycles, RRV-IRES with 5–7 A's gave roughly comparable levels, 4 and 8 A's were within about 4–5-fold of the 6 A's, whereas 10 and 12 A's were marked lower. In terms of stability, after 10 infection cycles, expansion of A's appeared to be a more frequent event affecting transgene protein expression than viral genome deletions or rearrangement: 4 and 5 A's appeared completely stable; 6, 7, and particularly 8 A's showed some level of expansion in the A bulge; 10 and 12 A's underwent both expansion and transgene deletion. The strong relative translational activity of the 5 A's in the EMCV IRES has not been reported previously. The 5A RRV-IRES may have utility for preclinical and clinical applications where extended replication is required.
Introduction
We have developed retroviral replicating vectors (RRV) derived from Moloney murine leukemia virus (MLV), a gammaretrovirus, with an amphotropic envelope and an encephalomyocarditis virus (EMCV) internal ribosomal entry site-transgene cassette downstream of the env gene.1 A version of this RRV, Toca 511 (vocimagene amiretrorepvec), encodes a modified yeast cytosine deaminase transgene (yCD2) and, with subsequent 5-fluorocytosine (5-FC) administration, is in clinical trials for treatment of recurrent high-grade glioma (NCT02414165, NCT01156584, NCT01470794 & NCT01985256).
The internal ribosome entry site (IRES) allows translation of RNAs in a cap-independent manner. The IRES from EMCV has been studied extensively and is widely used in retroviral and other mammalian expression vectors.2,3 The proper folding and secondary structure of the IRES dictate its translation efficiency, and sequence changes may or may not affect this. Palmenberg and coworkers showed that, independent of the 5′-IRES region, the J and K hairpin structures in the 3′ half of the IRES play a critical role in translation initiation4–6 (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/hgtb). Between these hairpins is a single-stranded region consisting of a stretch of consecutive adenosine residues, termed the “A bulge” or “bifurcation loop,” which has been shown to directly bind eIF4G, a critical mediator of the recruitment ribosomes to mRNA.7
The sequence of the IRES in Toca 511 was derived from pEMCF,2 which carries a bulge of seven adenosine residues (A's) instead of the six A's originally reported.4,8 The IRES with 7 A's is more widely used in bicistronic expression vectors than the IRES with 6 A's and has shown comparable expression level to the native IRES containing 6 A's. Because no apparent reduction in transgene protein expression was observed in pACE-CD and pACE-GFP originally described by Wang et al. and Logg et al.,9,10 we retained the IRES with 7 A's in Toca 511. Although the negative impact on initiation of protein translation of modifications in the J and K hairpin structure and specifically the oligo-adenosine loop has been reported,11–13 the impact of changes in the oligo-adenosine loop has not been investigated fully in plasmid-derived, bicistronic expression vectors and certainly not in the RRV context. In the present study, we noted some variation in the size of this loop in RRV sequences from infected tissue in permissive BALB/c mice 180 days after infection and extensive viral replication, in the absence of 5-FC treatment. To investigate this phenomenon further, we generated a series of vectors bearing 4–12 A's in the A bulge in the J-K bifurcation domain and evaluated the impact of the variants on transgene expression, the rate of mutation contributed by reverse transcriptase during viral replication, as well as vector stability. We show here that neither deletion nor insertion of A's in the A bulge affects RRV replication in this series of vectors, that 6 A's provide maximal yCD2 and green fluorescent protein (GFP) expression, and that <5 and >8 A's have a large effect on transgene protein expression without a significant effect on RNA expression. In the context of RRV, longer A's (>8) appear to be more susceptible to further lengthening and risk of gene deletions. Depending on the situation, it may be appropriate in the future to use a vector with an IRES with 5, 6, or 7 A's in the A bulge, preferably with 5 or 6 A's to allow more viral replication cycles before reaching A's >8, where translation is dramatically reduced. Currently, the only direct clinical experience is with RRV carrying the 7 A's IRES, and IRES carrying 5 or 6 A's may require further evaluation before clinical use.
Materials and Methods
Intravenous injection of Toca 511 into BALB/c mice
An amount of 2.35 × 106 or 2.35 × 105 transduction units (TU) of Toca 511 was intravenously administered to 8-week-old female BALB/c mice. Approximately 180 days postinfection, genomic DNA from various tissues was harvested for biolocalization study. Genomic DNA from abnormal tissues such as thymus or lymph node was extracted for sequence analysis of the envelope and IRES-yCD2 cassette.
Construction of RRV containing various numbers of A's in the A bulge of the J-K bifurcation domain
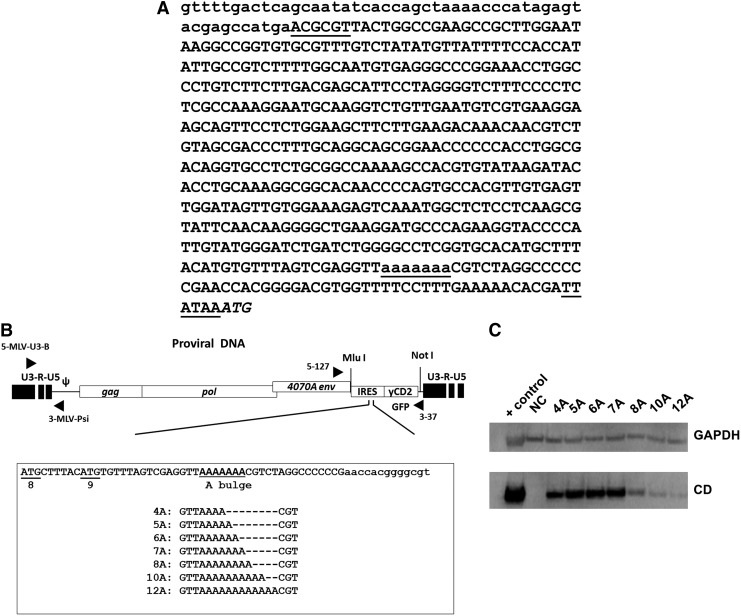
RRV containing an EMCV IRES and encoding yCD2 or GFP1,14 were generated to have 4, 5, 6, 7, 8, 10, or 12 A's in the A bulge in the J–K bifurcation domain. Each construct was generated by DNA synthesis (BioBasics Inc.) of the entire IRES cassette with an MluI at the 5′ end and a PsiI at the 3′ end, respectively, for direct replacement of the equivalent cassette in the RRV backbone (Fig. 1A and B). All DNA fragments were confirmed by sequencing before and after cloning into the RRV backbone. The RRV constructs containing the yCD2 transgene were designated using the name of the transgene followed by the number of A's in the A bulge (i.e., yCD2-4A contains yCD2 transgene and 4 A's in the A bulge in the IRES).
Figure 1.
Replicating retroviral vectors containing IRES with various numbers of A's in the A bulge. (A) DNA sequence spanning the 3′ of MLV env and EMCV IRES in Toca 511 that contains 7 A's in the A bulge. Lower-case letters indicate the 3′ of the env sequence; underlined capital letters indicate MluI (ACGCGT) and PsiI (TTATAA) restriction enzyme sites flanking the IRES sequence; underlined lower-case letters indicate the 7 A's in the A bulge; italicized capital letters indicate the start codon of a transgene. (B) Diagram of the A bulge in the J-K bifurcation domain in EMCV IRES incorporated into RRV expressing yCD2 or GFP. The native ATG8 (AUG in RNA) and ATG9 are underlined; enlarged and underlined sequence represents the A bulge in the J-K bifurcation domain; lower-case letters indicate the 5′ sequences in the pyrimidine-rich region in the 3′ IRES. (C) yCD2 protein expression from transiently transfected 293T cells. GAPDH detection was included as a loading control. Positive control (+) is lysate from U87-MG cells infected with RRV-yCD2 vector. EMCV, encephalomyocarditis virus; IRES, internal ribosome entry site; MLV, murine leukemia virus; NC, negative control from naïve cell lysates.
Cell culture
293T cells were obtained through a materials transfer agreement with the Indiana University Vector Production Facility and Stanford University deposited with ATCC (SD-3515; lot No. 2634366). Human glioblastoma cells U87-MG (ATCC; HTB-14), human prostate tumor cells PC-3 (ATCC; CRL-1435), human fibrosarcoma cells HT-1080 (ATCC; CCL-121), and 293T cells were cultured in complete DMEM containing 10% FBS (Hyclone), sodium pyruvate, GlutaMAX (Thermo Fisher Scientific), and antibiotics penicillin 100 IU/ml and streptomycin 100 IU/ml (Corning).
Virus production, infection, and titer
Virus stock was first produced by transient transfection of 293T cells using calcium phosphate precipitation method. Cells were seeded at 2 × 106 cells per 10 cm petri dish the day before transfection. Cells were transfected with 20 μg of designated plasmid DNA the next day. Eighteen hours posttransfection, cells were washed with PBS twice and incubated with fresh complete culture medium. Viral supernatant was collected approximately 42 hr posttransfection and filtered through a 0.45 μm syringe filter unit. Viral supernatants were stored in aliquots at −80°C. RRV-producer cells were established by infection of HT-1080 cells at multiplicity of infection (MOI) of 0.1. Viral titer from transiently transfected 293T cells as well as from RRV-producer cells was performed by quantitative polymerase chain reaction as described.1 The viral titers obtained from infected RRV-producer HT-1080 cells were measured before infecting naïve U87-MG cells in order to calculate MOI.
Quantification of cellular viral RNA by quantitative real-time polymerase chain reaction
Total RNA was extracted from naïve and maximally RRV-infected U87-MG cells (infected cells reach maximum infectivity approximately day 6 to day 8 postinfection at MOI of 0.01 and remain persistent thereafter) using the RNeasy Kit (Qiagen). Reverse transcription was carried out with 100 ng total RNA using High Capacity cDNA Reverse Transcription Kit (ABI). Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed to measure the mRNA expression level of unspliced and spliced cellular viral RNA with the following parameters: 95°C 10 min, and 40 cycles of 95°C 15 sec and 60°C 30 sec. The cellular viral RNA expression levels were measured using two primer sets, located in the env (5′Env2: 5′-ACCCTCAACCTCCCCTACAAGT-3′, 3′Env2: 5′-GTTAAGCGC CTGATAGGCTC-3′, probe: 5′FAM-AGCCACCCCCAGGAACTGGAGATAGA-3′BHQ) and in yCD2 region (5′yCD2: 5′-ATCATCATGTACGGCATCCCTAG-3′, 3′yCD2: 5′-TGAACTGCTTCATCAGCTTCTTAC-3′, probe: 5′FAM-TCATCGTCAACAACCACCAC CTCGT-3′BHQ), respectively. The relative level of viral RNA from each vector was calculated using 2−ΔΔ(Ct) method with respect to the vector containing the 6 A's.
Immunoblot
Transiently transfected 293T cells or maximally infected U87-MG cells were harvested and lysed for immunoblotting. Equal amount of proteins from lysates were resolved on Criterion XT Precast Gel 4–12% Bis-Tris gels (Bio-Rad; cat No. 345-0124). Mouse antihuman GAPDH (Millipore; cat No. MAB374) antibody at 1: 500 dilution was used to detect the expression of GAPDH, and mouse anti-yCD2 (Tocagen; clone 9A11) antibody at 1:1,000 dilution was used to detect the expression of yCD2 protein. The specificity of anti-yCD2 antibody is shown in Supplementary Fig. S2. Detection of protein expression was visualized using Clarity Western ECL Substrate (Bio-Rad; cat No. 170-5060). Quantity One software (Bio-Rad) was used to quantify the signal of yCD2 and GAPDH detected on the immunoblots. Quantification of yCD2 protein expression level was obtained using the Quantity One Image Sofware version 4.6.7 (BioRad). Ratios of measured GAPDH pixel density relative to U87-MG cell infected with yCD2-6A were used to normalize yCD2 expression levels.
Cell-based yCD2 enzymatic assay
A cell-based enzymatic activity of yCD2 was performed to measure the conversion of 5-FC to 5-FU by high-performance liquid chromatography (HPLC). Briefly, 4 × 105 U87-MG cells were seeded in T75 flask and then infected with IRES-yCD2 variants at MOI of 0.05. Cells were passaged at 1:5 on day 5 and day 7 postinfection and harvested at day 10 postinfection. Approximately 1.6 × 105 cells in 200 μl media were subsequently transferred to 4 wells in 96-well plate and 5-FC was added to the wells to achieve final concentration of 3.57 mg/ml. Two hours postadministration of 5-FC, 28 μl of 10% trichloroacetic acid (Sigma-Aldrich; cat. No. T9159-500G) was added to the wells to terminate the reaction. Supernatants were collected and filtered through a glass fiber filter (Millipore; cat. No. MSFBN6B10) followed by a 0.45 μm filter (Millipore; cat. No. MSHVN4510). Filtrates were then analyzed for 5-FU content on HPLC. The chromatography was conducted on a reverse-phase C18 column with an isocratic run using a low-pH phosphate buffer and methanol as previously described.14
Flow cytometry
Cells harvested for flow cytometric analysis were washed with PBS and centrifuged at 1000 rpm for 5 min. Cell pellets were resuspended in PBS containing 1% paraformaldehyde. The percentage of GFP-positive cells was determined by flow cytometry using proper gating to exclude GFP-negative cells. Percentage of GFP-positive cells was measured by FACSCanto II using FL1 channel (BD Biosciences). GFP protein expression levels were quantified by using mean fluorescence intensity (MFI).
Vector copy number of proviral DNA
Proviral vector copy numbers from infected U87-MG cells (MOI 0.01) during the course of infection were determined by qPCR as previously described.1 The average vector copy number per diploid genome/cell was determined from the standards of known vector copy numbers and the amount of genomic DNA input (assuming 7 pg genomic DNA per diploid cell) in the qPCR. The average vector copy number per cell was determined from the amount of genomic DNA input and the number of transduction events (derived from the standards and Ct values) in the qPCR.
Vector stability assay
Vector stability was measured from genomic DNA of maximally infected U87-MG cells after 1 round of infection or from genomic DNA of maximally infected U87-MG cells at each round of infection from a serial infection of U87-MG cells as described previously.1 PCR was performed using the following primers: 5–127 (forward), 5′-CTGATCTTACTCTTTGGACCTTG-3′; 3–37 (reverse), 5′-CCCCTTTTT CTGGAGACTAAATAA-3′, which resulted in an ∼1.2 kb fragment spanning the IRES-yCD2 cassette. SuperTaq Plus polymerase (Ambion; cat No. AM2056) was used for PCR.
PCR and TA cloning for sequence analysis
PCR fragments using the primers and SuperTaq Plus polymerase described were isolated from 0.8% agarose gel and sublconed into TOPO vector provided in the TOPO TA Cloning Kit for Sequencing (Invitrogen; cat No. K4530-20). After selection of bacterial colonies and extraction of plasmid DNA, samples were sequenced using the 5–127 and 3–37 primers. Minimum of 10 colonies of each variants were selected for plasmid DNA extraction and sequencing analysis.
Results
RRV can undergo changes in the length of the oligo adenosine in the A bulge of the EMCV IRES in vivo
The expression of yCD2 and the conversion of 5-FC to 5-FU by yCD2 have been demonstrated to be efficient and stable both in vitro and in vivo when cells are infected with an RRV with a 7A IRES (Toca 511).1,14 In a vector biolocalization study conducted as part of a preclinical package to support initiation of clinical trials, Toca 511 was injected intravenously into a permissive mouse strain (BALB/c mice) to evaluate long-term vector bio-localization. 5-FC was not administered to the animals as the goal was to observe biodistribution of the virus. As expected 10–20% of the mice (depending on the cohort) at the higher doses (see Materials and Methods) displayed histologically confirmed abnormalities in lymphoid tissues associated with viremia and lymphoma at 180 days postinfection (data not shown). Genomic DNA from the abnormal thymus or lymph nodes of three mice was harvested for molecular PCR cloning of proviral sequences, followed by sequencing analysis. There were multiple copies of the virus including recombinants with endogenous mouse MCF envelope sequences present, as occurs with ecotropic MLV infection in mice.15 Further analyses are planned for a future publication. However, one additional feature revealed by the sequence analysis of the envelope-IRES-yCD2 transgene cassettes that we have investigated in the work described here was an expansion or contraction of oligo A sequences in the A bulge of the J-K domain of the IRES in some vector sequences after extensive viral replication. As shown in Table 1, tissues from three mice contained vectors with heterogeneous expansions of 7–8, 9, 10, and 11 A's and a contraction of 7–6 A's. It appears that the oligo A number drifts preferentially toward longer oligo A in the A bulge. However, the nature of this preference was undefined in the in vivo study.
Table 1.
Changes in the bulge A region of internal ribosome entry site in tissues harvested from Toca 511–treated BALB/c mice
| Animal | Treatment | Source of gDNA | Changes in bulge A (excluding other mutations such as recombination) |
|---|---|---|---|
| 1 | i.v. Toca 511 | Thymus | Contraction of 7(A) to 6(A) in IRES accompanied by deletion in CD |
| Expansion of 7(A) to 8(A) in IRES | |||
| Expansion of 7(A) to 9(A) in IRES | |||
| 2 | i.v. Toca 511 | Lymph node | Expansion of 7(A) to 10(A) in IRES |
| Expansion of 7(A) to 11(A) in IRES | |||
| 3 | i.v. Toca 511 | Thymus | Expansion of 7(A) to 8(A) in IRES |
IRES, internal ribosome entry site; gDNA, genomic DNA.
Transgene expression in RRV containing various numbers of A's in the A bulge varies, but produces similar titers in RRV-producer cells
It has been demonstrated that the J-K domain is important for translational initiation.4,7 The observation made from our in vivo study and the 7 A's in pEMCF as opposed to the 6 A's in ECMV IRES originally described led us to investigate the impact on yCD2 expression of IRES with various numbers of A's in the A bulge, and, in particular, the impact on transgene protein expression in the context of RRV. Therefore, we generated a series of deletion and insertion mutants specifically in the A bulge to mimic sequences observed from the in vivo study. RRV containing an EMCV IRES and encoding yCD2 or GFP were generated to have 4, 5, 6, 7, 8, 10, or 12 A's in the A bulge in the J-K bifurcation domain (Fig. 1A and B). As we are interested in using such vectors to treat human tumors in patients, we did not further investigate the mouse models at this time, but rather looked at the effect, in a human tumor cell line, of alterations in the A bulge of the EMCV IRES on RRV carrying the IRES and a transgene. We first examined whether the alteration in the A bulge would affect viral titer. Virus stocks were initially produced by transient transfection in 293T cells, followed by infection of HT-1080 cells at MOI of 0.1 to generate RRV-producer cells. Table 2 shows that RRV containing various number of A's from transiently transfected 293T cells produced similar titers. The viral titer of each vector produced by the RRV-producer HT-1080 cells in the subsequent infection was also determined. Similar to viral titer data obtained from transiently transfected 293T cells, RRV-producer cells containing various numbers of A's also produced comparable titers, suggesting that the number of the A's in the A bulge does not affect viral production.
Table 2.
Viral titers of retroviral replicating vectors containing various numbers of A's in the A bulge produced from transiently transfected 293T cells and retroviral replicating vectors-producer HT-1080 cells
| Titer (TU/ml) | ||
|---|---|---|
| Vector | 293T | HT1080 |
| yCD2-4A | 6.4 ± 0.7E6 | 1.8 ± 0.6E7 |
| yCD2-5A | 4.3 ± 0.4E6 | 2.2 ± 0.5E7 |
| yCD2-6A | 3.4 ± 0.4E6 | 1.2 ± 0.1E7 |
| yCD2-7A | 5.6 ± 0.4E6 | 1.5 ± 0.6E7 |
| yCD2-8A | 4.0 ± 0.2E6 | 1.8 ± 0.3E7 |
| yCD2-10A | 5.7 ± 0.8E6 | 1.4 ± 0.3E7 |
| yCD2-12A | 6.8 ± 0.5E6 | 2.7 ± 0.5E7 |
Titers were performed in triplicate, and the titer values represent means ± SD from one of two independent experiments.
We next checked yCD2 protein expression mediated by IRES variants in transiently transfected 293T cells. The data showed that yCD2 protein expression levels mediated by RRV variants containing 5 and 6 A's were comparable to that of the 7A. In contrast, yCD2 protein expression levels mediated by RRV variants containing 4, 8, 10, and 12 A's were substantially reduced (Fig. 1C).
RRV containing various numbers of A's in the A bulge replicate at similar rates
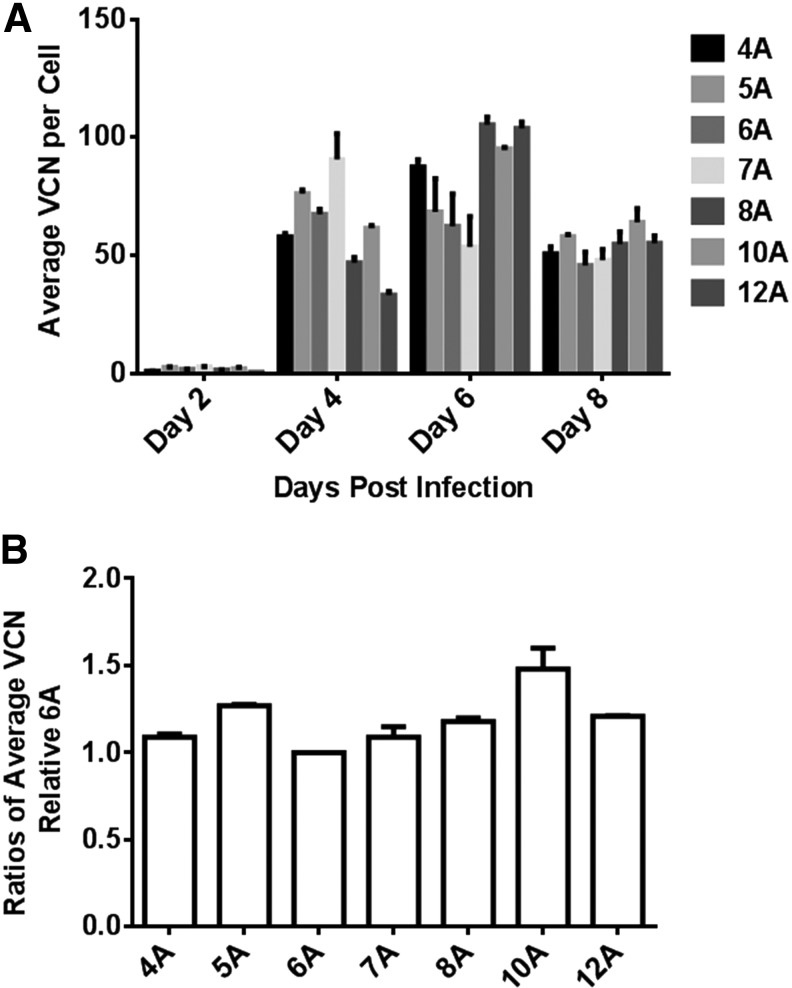
Given that the number of the A's in the A bulge does not affect viral titer produced from cells initially infected with low MOI, it is likely that these vectors also replicate at similar rates. We examined the replication kinetics of these vectors by measuring the average vector copy number during the course of infection. Viral supernatants from RRV-producer cells were used to infect naïve U87-MG cells at MOI of 0.01. At each passage a portion of cells were harvested for genomic DNA extraction for qPCR analysis. Replication kinetics measured by vector copy number per cell from genomic DNA was used instead of viral particle number from supernatant partly because of moderate correlation between viral particle number and viral infectivity. In addition, the vector copy number can later be used for normalization of intracellular transgene protein expression. Figure 2A shows that the vector copy number varied among vectors at day 4 and day 6 postinfection and stabilized by day 8 postinfection with comparable average vector copy numbers (Fig. 2B).
Figure 2.
Replication kinetics of RRV-yCD2 variants in U87-MG cells. (A) Replication kinetics of RRV-yCD2 carrying various lengths of A's in the A bulge was measured by the average VCN per cell in infected U87-MG cells (MOI of 0.01) at indicated time points during the course of infection. (B) Ratios of the average vector copy number relative to the yCD2-6A vector in maximally infected U87-MG cells (day 8 postinfection). The data shown represent mean ± SD (n = 3) of triplicates performed from one of two independent experiments. MOI, multiplicity of infection; VCN, vector copy number.
RRV containing various numbers of A's express similar levels of transcripts but different levels of transgene protein
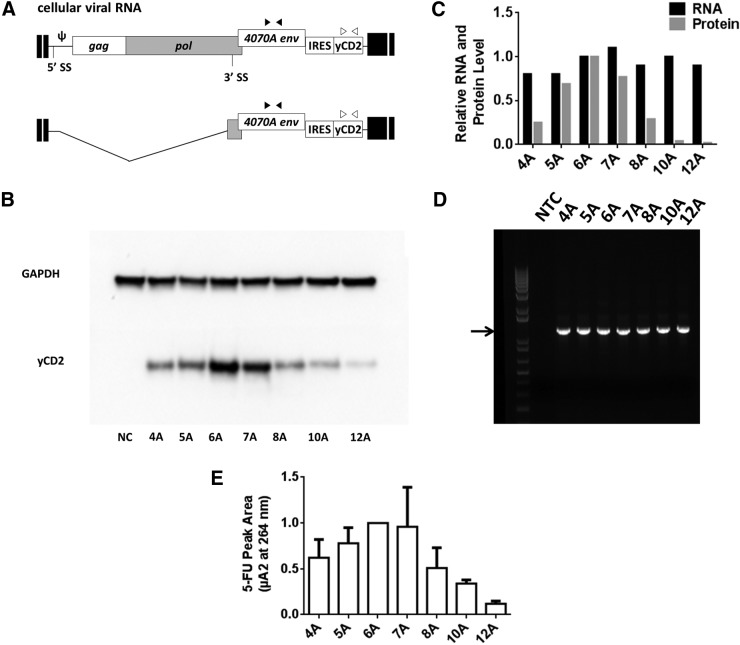
The yCD2 protein expression from transiently transfected 293T cells was substantially less from vectors carrying the 8, 10, or 12 A's than those carrying the 5, 6, and 7 A's, whereas the 4A version was intermediate level for expression (Fig. 1C). However, we cannot rule out the possibility that the difference in yCD2 protein expression could be because of the transfection efficiency rather than the length of the A's. In order to demonstrate that the decrease in transgene expression mediated by IRES variants is regulated at the translational level, cellular viral RNA of maximally infected U87-MG cells was harvested and levels measured by qRT-PCR, and yCD2 protein levels were examined by immunoblotting. The cellular viral RNA levels were measured using two different primer sets, located in the env and in yCD2 region, respectively (Fig. 3A). The relative level of RNA from each vector was calculated using the 2−ΔΔ(Ct) method with respect to the vector containing 6 A's. The cellular viral RNA level ratios ranged from 0.9 to 1.2, and the values of ratios from each primer set were comparable (Table 3). Together, the data suggest that there is no significant difference in viral RNA expression levels because of the modifications in the IRES. In examining the yCD2 protein expression level of these vectors by immunoblotting, we found that the yCD2 protein expression levels of the vectors containing the 5 and 7 A's after a single round of infection cycle were 69% and 77% that of the yCD2-6A vector. In contrast, a marked reduction of yCD2 protein expression was observed in the vectors containing the 4 and 8 A's. Similar to data observed in 293T transfection, a substantial reduction of yCD2 protein expression was observed in the vectors containing the 10 and 12 A's. The yCD2 protein expression levels of these vectors range from 4% to 25% that of the yCD2-6A vector (Fig. 3B). The drastic reduction of the yCD2 protein expression with similar expression levels of the cellular viral RNA (Fig. 3C) suggests that the length of oligo A in the bulge A of the IRES can have a large effect on protein expression at the posttranscriptional level.
Figure 3.
RNA and protein expression from U87-MG cells infected with RRV containing various numbers of A's in the A bulge. (A) Schematic diagram of cellular viral RNA isoforms. The env2 and yCD2 primer sets, which recognize both unspliced and spliced viral RNA in the env and the yCD2 region, respectively, were used to measure the level of cellular viral RNA by qRT-PCR. Filled triangles, env2 primer set; open triangles, yCD2 primer set. (B) Immunoblot of yCD2 and the GAPDH protein. Twenty micrograms of cell lysate of maximally infected U87-MG cells from one round of infection was loaded to each lane, and equivalent loading and blotting efficiency controlled for by detection of the ubiquitous marker GAPDH. NC, negative control from naïve cell lysates. (C) Histograms represent the RNA and protein expression levels relative to the yCD2-6A vector. RNA expression was performed in triplicates and the relative level of viral RNA from each vector was calculated using 2−ΔΔ(Ct) method with respect to the vector containing the 6 A's. Protein expression levels of yCD2 were calculated relative to vector containing the 6 A's. (D) Stability of proviral DNA of IRES-yCD2 cassette in RRV-IRES-yCD2 variants from one round of infection showed no detection of deletion mutants. Arrow indicates the expected 1.2 kb PCR product containing the IRES-yCD2 cassette. (E) Cell-based enzymatic activity of yCD2 in infected U87-MG cells was measured by HPLC to detect the amount of 5-FU. The 5-FU peak area of each vector is plotted relative to yCD2-6A vector that is set to 1. Data shown in (B–D) are from representative examples of two or three independent experiments. Data shown in (E) represent mean ± SD relative to 6A from three independent experiments.
Table 3.
Cellular viral RNA expression levels relative to yCD2-6A
| Env | yCD2 | |
|---|---|---|
| 4A | 1.16 ± 0.29 | 0.98 ± 0.17 |
| 5A | 0.90 ± 0.12 | 0.83 ± 0.05 |
| 6A | 1.00 ± 0.00 | 1.00 ± 0.00 |
| 7A | 1.07 ± 0.04 | 0.96 ± 0.20 |
| 8A | 0.98 ± 0.11 | 1.10 ± 0.29 |
| 10A | 0.90 ± 0.18 | 0.86 ± 0.12 |
| 12A | 1.01 ± 0.12 | 0.98 ± 0.06 |
Values represent means ± SD from three independent experiments.
To ensure that the reduction in yCD2 protein expression in RRV with 4, 10, and 12 A's is not because of deletion in the IRES-yCD2 cassette outside the region of which the yCD2 primer set binds in qRT-PCR, we next examined their vector stability from genomic DNA of infected U87-MG cells. Genomic DNA of infected U87-MG cells was harvested at day 10 postinfection to match the time point of the samples harvested for qRT-PCR and immunoblotting. A 1.2 kb PCR product of the proviral DNA was amplified using a primer set outside (Fig. 1B) of the IRES-yCD2 cassette to assess the integrity of the integrated viral genome.1,9 No detection of deletion mutants (PCR products <1.2 kb represent partial or complete deletion of viral genome in the IRES-yCD2 region) was observed (Fig. 3D). Together, the data indicate that the vectors are stable in such a short-term replication setting and the reduction of yCD2 protein expression is not because of deletion in the IRES-yCD2 cassette.
We also measured relative intracellular yCD2 enzymatic activity employing a cell-based assay by adding 5-FC to the cultures and measuring 5-FU after 2 hr by HPLC. The differences in activity were ranked similarly to the immunoblot data (Fig. 3E), and a correlation (R2 = 0.8995; Supplementary Fig. S3) was observed between yCD2 expression and enzymatic activity, suggesting expression of similar ratios of active enzyme regardless of the amount of protein expressed. To confirm the generality of the observations with the yCD2 gene, we measured the effect of the number of A's in the A bulge with another transgene for which the protein expression assay was well defined.
Therefore, we generated an equivalent set of RRV encoding GFP. Because the GFP expression level can be quantified by flow cytometry, we examined the replication kinetics of RRV-GFP variants by measuring percentage of GFP-positive cells during the course of infection. Despite their marked difference in GFP protein expression, all RRV-GFP variants, except RRV-GFP with 12 A's, showed similar replication kinetics (Supplementary Fig. S4A). The apparent lag in RRV-GFP viral replication for the 12A construct is likely because of the overall low expression level of GFP excluded from the GFP-positive gating, as the average vector copy number in maximally infected U87-MG cells is similar to that of RRV-GFP with 6 A's (Supplementary Fig. S4B). Consistent with the data observed in yCD2 vectors, a reduction of GFP protein expression was observed in vectors containing the 4, 8, 10, and 12 A's with minimal change at the viral RNA level (Supplementary Fig. S4C). When the GFP protein expression in maximally infected U87-MG cells is normalized to vector copy number per cell and cellular viral RNA expression level, a substantial reduction of GFP protein expression was observed in vectors containing the 4, 8, 10, and 12 A's (Supplementary Fig. S4D). Furthermore, because of the sensitivity of the detection method, a remarkable difference in GFP expression level was revealed, showing approximately 96% and 99% decrease in GFP expressed by the vectors containing the 10 and 12 A's, respectively. As seen with yCD2, vectors containing 5 and 7 A's showed comparable GFP expression levels although at a reduced level compared with vector containing the 6 A's. Again, the PCR result confirmed that the reduction of GFP protein expression is not because of deletion of the IRES-GFP cassette in proviral DNA (Supplementary Fig. S4E). Overall, the results from IRES-GFP vectors were consistent with those observed with IRES-yCD2 vectors. In both sets of vectors, vectors containing the 5 and 7 A's express comparable level of transgene protein, with 5 A's slightly less than 7 A's, and vectors containing the 6 A's express the highest level of transgene. The reduced protein translation efficiency in RRV with 4 A's compared with RRV with 6 A's is also consistent with findings of the mutant 768Δ4 reported by Hoffman and Palmenberg.5
In vitro viral replication and analysis of mutations in the A bulge of RRV carrying various numbers of A's
In order to mimic the in vivo study in which extensive rounds of viral replication occurred and length variation in the A bulge was observed, we performed in vitro replication experiments to examine the viral genomic stability of these vectors particularly in the A bulge. It has been reported that repeat of A's in DNA template can produce artifacts in PCR when using Taq DNA polymerase.16 To ensure that expansion of oligo A in the A bulge observed in vivo previously and in vitro replication described below is not contributed by such an event, we performed the PCR using plasmid DNA as the template. Sequence analysis from PCR cloning using plasmid DNA with 4, 5, 6, 7, and 8 A's as template did not produce any mutation. In contrast, plasmid DNA carrying the 10A variant resulted in 1 clone that showed contraction to 9 A's. Likewise, the 12A variant gave rise to 1 clone that showed contraction to 8 A's. The data indicate that the Taq polymerase effect is minimal and appears to favor contraction; they are consistent with the study of Shinde et al., in which they reported no mutations observed even after 60 PCR cycles for (A)n with 8 or less repeat units.16
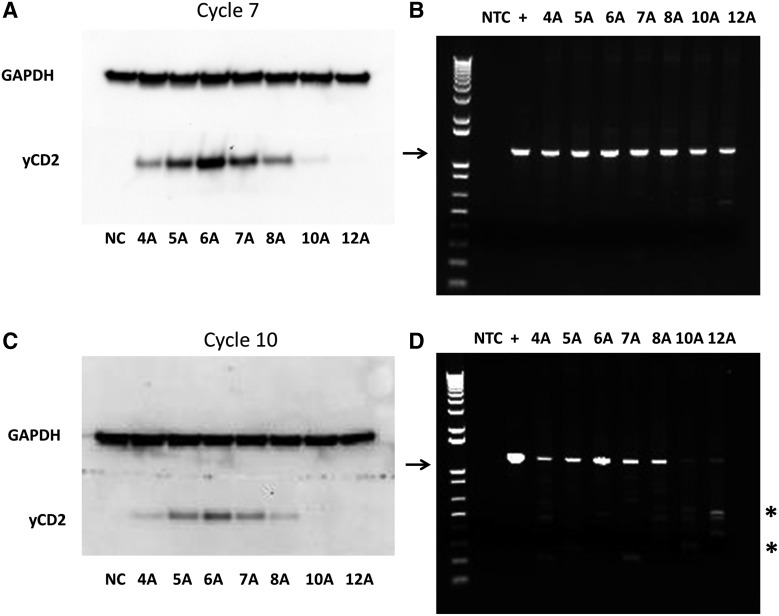
After confirming that PCR artifact is minimal, serial infection cycles were performed. In this experiment, the viral supernatant from RRV-producer cells was used to infect naïve U87-MG cells at MOI of 0.01 to allow time for the virus to replicate to day 8 to reach maximal infectivity. The same procedure was repeated over serial infection cycles by collecting viral supernatant from maximally infected U87-MG cells, and infecting fresh U87-MG cells for 12 cycles. Cells at indicated infection cycles were harvested to examine the changes in the yCD2 protein expression from cell lysates and the length of A's in the A bulge in proviral DNA by immunoblotting and by TA cloning of the 1.2 kb PCR product, respectively. We first compared the expression of yCD2 between infection cycle 1 and 7. The expression levels of yCD2 among the RRV carrying various numbers of A's from infection cycle 1 were consistent with data shown previously in Fig. 3B. After 7 cycles of infection in vitro, the yCD2 expression in RRV carrying the 10 and 12 A's was substantially reduced (Fig. 4A). Notably, the reduction in yCD2 expression observed in 10A and 12A variants is not predominantly because of deletion in the IRES-yCD2 cassette as evident by the PCR result (Fig. 4B).
Figure 4.
Protein expression level of yCD2 in RRV-IRES-yCD2 variants decreases because of expansion of the oligo A length in the A bulge. Protein expression of yCD2 in RRV-IRES-yCD2 variants was evaluated at infection cycle 7 (A) and cycle 10 (C) to correlate with expansion of the oligo A length in A bulge A observed in sequencing results. Vector stability analyzed by PCR from maximally infected cells at infection cycle 7 (B) and cycle 10 (D) is shown to detect deletion of IRES-yCD2 cassette and noted as an additional factor in some variants contributing to the reduction of yCD2 protein expression. DNA molecular marker (1 kb plus marker; Invitrogen) is included in the first lane of each gel. Arrow indicates the expected 1.2 kb PCR product containing the IRES-yCD2 cassette. Asterisk indicates a deletion of the IRES-yCD2 cassette. +, positive control using RRV-IRES-6A plasmid DNA as a template in PCR; NTC, no template control.
In parallel, we also performed sequence analysis to examine changes that might have occurred in the A bulge after 7 cycles of infection (Table 4). Sequence analysis revealed that the length of variants carrying the 4 and 5 A's remained unchanged. Variants carrying the 6 and 7 A's remained relatively stable. Eight out of 10 clones from the 6A variant remained the same length; 2 out of 10 clones showed expansion to 7 A's. For the variant carrying 7 A's, 7/10 clones remained the same length, whereas others expanded to 8 A's and 10 A's. For the variant carrying the 8 A's, 3/10 clones remained the same length. In addition, a range of expansion from 9 A's up to 22 A's was observed. Interestingly, 1/10 clones showed a contraction to 7 A's (Table 4). However, the length of expansion observed in the variant carrying 8 A's does not grossly affect the overall yCD2 protein expression (Fig. 4A; compare to Fig. 3B). In contrast, variants originally carrying the 10 and 12 A's had extensively expanded A's ranging from 12 A's to 54 A's and these expansions correlate with a substantial reduction in yCD2 expression (Table 4 and Fig. 4A; compare to Fig. 3B). Furthermore, data from infection cycle 10 indicate that variants with 10 and 12 A's had mostly deleted sequences in the IRES-yCD2 cassette and resulted in undetectable yCD2 protein expression (Fig. 4C and D). However, the length of oligo A in the A bulge in variants with 4–8 A's remained roughly stable after infection cycle 10; the variants carrying the 4 and 5 A's continue to remain stable over time; the variant carrying the 6 A's showed 2/12 clones expanded to 7 A's; and the variant carrying the 7 A's was also relatively stable and did not show further expansion of the proportion of mutations from that observed in infection cycle 7. While the 8A variant also did not change the proportion of mutants from 7–10 infection cycles, it had already incorporated more mutations at cycle 7. In addition, the expansion of oligo A in variants carrying the 10 and 12 A's appears to have compromised the viral genome stability as indicated by the deletion of the IRES-yCD2 cassette in PCR (Fig. 4D). In contrast to data from infection cycle 7 for the 10A and 12A variants, in which the reduction of yCD2 expression appears to associated with expansion of the oligo A in the A bulge, the reduction of yCD2 expression in cycle 10 is presumably due mainly to the emergence of deletion mutants, on top of the oligo A expansion.
Table 4.
Mutation rate in the A bulge from different infection cycles in vitro
| Plasmid DNA | |
|---|---|
| 4A | 4A (11/11) |
| 5A | 5A (12/12) |
| 6A | 6A (11/11) |
| 7A | 7A (12/12) |
| 8A | 8A (12/12) |
| 10A | 9A (1/11); 10A (10/11) |
| 12A | 8A (1/11); 12A (10/11) |
| Proviral DNA from infection cycle 7 | |
|---|---|
| 4A | 4A (10/10) |
| 5A | 5A (10/10) |
| 6A | 6A (8/10); 7A (2/10) |
| 7A | 7A (7/10); 8A (2/10); 10A (1/10) |
| 8A | 7A (1/10); 8A (3/10); 9A (1/10); 10A (1/10); 11A (2/10); 18A (1/10)l 22A (1/10) |
| 10A | 12A (1/10); 13A (2/10); 14A (1/10); 15A (1/10); 18A (2/10); 20A (1/10); 22A (1/10); 25A (1/10) |
| 12A | 13A (1/10); 18A (2/10); 20A (1/10); 27A (1/10); 28A (1/10); 31A (2/10); 39A (1/10); 54A (1/10) |
| Proviral DNA from infection cycle 10 | |
|---|---|
| 4A | 4A (10/10) |
| 5A | 5A (10/10) |
| 6A | 6A (10/12); 7A (2/12) |
| 7A | 7A (10/15); 8A (5/15) |
| 8A | 7A (2/13); 8A(4/13); 9A (3/13); 10A (1/3); 11A (1/13); 12A (1/13); 12A (2/13) |
| 10A | 11A (3/10); 20A (1/10); 30A (2/10); 40A (1/10); 53A (2/10); 54A (1/10) |
| 12A | 26A (4/10); 27A (1/10); 29A (2/10); 32A(1/10); 36A (1/10); 55A (1/10) |
All together, the data indicate that the A bulge in IRES in the context of RRV is susceptible to mutation, particularly expansion, during reverse transcription, and the longer the length of oligo A, the less stable they are. Our data also indicate that RRV with 5 and 6 A's are more stable than RRV with 7 A's.
Discussion
We have demonstrated in a human tumor cell line that the length of the A bulge in the J-K bifurcation domain affects expression of the transgene downstream of the IRES, presumably through effects on translation efficiency. Previous findings imply that the context around nucleotide triplet AUG11 in the IRES, the spacing between the pyrimidine-rich region located in the 3′ IRES, and the first AUG in the cistron as well as the arrangement of cistron on the mRNA all play a role in modulating protein translation.11–13
Our data, in RRV, show that the presence of 6 A's provides the highest level of transgene protein expression and that alteration of the numbers of A's in the A bulge by contraction or expansion of 2–4 nucleotides can significantly affect the protein expression level of the transgene downstream of the IRES. The protein expression results suggest that the optimum IRES configuration in general is with 6 A's in the A bulge, while 7 A's are acceptable, due, in part, to the rescue by polypyrimidine tract binding (PTB) protein previously described by Kaminski and Jackson, showing that lengthening the A bulge from 6 A's to 7 A's rendered IRES function dependent on PTB.17 It is therefore reasonable to speculate that the vector variants with 4, 5, 8, 10, and 12 A's also require binding of PBT to the polypyrimidine tract in combination with other IRES-trans acting factors for efficient protein translation. Recent data reported by Chamond et al. revealed no apparent difference in ribosome 40S binding efficiency to 5A IRES-luc and 6A IRES-luc RNA. Their data further refined the previously proposed functional role of IRES trans-acting factors, eIF4G/A, in binding to the J-K domain in IRES.18 Although the variants in our studies may allow efficient binding of the 40S ribosomal subunit to the initiation site, distortion of the secondary structure of the J-K bifurcation domain of the IRES may interfere with the stabilizing function of eIF4G/A factors for efficient translation initiation.5,18 Interestingly, our attempt in examining whether the length of A's in the A bulge affects RNA folding of the IRES in general using mFold program revealed no difference in their secondary structure or minimum free energy under physiological condition.
The expansion of oligo A has also been described in an artificial bicistronic flavivirus genome consisting of the EMCV IRES in the 3′ UTR to mediate protein expression of viral surface glycoproteins prM and E required for viral replication in vitro.19 Nevertheless, despite enhanced viral replication, the expansion of the oligo A also led to decrease in protein expression mediated by IRES as seen in our study. Other than the EMCV IRES synthetic constructs made for bicistronic expression vectors, the mutations in the number of A's in the A bulge have not been described in EMCV.
The alterations in the number of oligo A residues we observed in the RRV after extensive replication in vivo and in vitro settings are likely not driven by any kind of selective pressure, as these experiments were performed in the absence of 5-FC, but rather happen because of mutation-prone reverse transcriptase activity that could be influenced by the secondary structure of the RNA.20 The expansion of A's in the A bulge could be attributable to MLV reverse transcriptase, an RNA-dependent DNA polymerase, which is known to perform template switching during reverse transcription upon entry into host cell. Template switching during reverse transcription leading to high frequency of direct repeat deletion mutants has been reported in retroviral-based vectors containing heterologous sequence.2,20–23 Although duplication of direct repeat sequence occurs at much lower frequency, duplication mutants has been described in retroviral-based vector containing the herpes simplex virus thymidine kinase gene.24 Another possible mechanism could be the slipped-strand mispairing of the DNA polymerase during reverse transcription, PCR amplification, or sequencing. However, a preference for contraction over expansion is observed in general.16,25–27 By both mechanisms, these mutants can generate stutter products in sequence chromatograms and thus confound the true number of mononucleotides that have gone through expansion during in vitro selection. Nevertheless, plasmid controls indicate that the Taq polymerase bias during PCR amplification is minimal and appears, if anything, to favor contraction as reported.16 In addition, successful DNA sequencing reads of plasmid DNA containing A/T repeat up to 50 bp have been reported.28 Because a trend of expansion from early to late infection cycle during the course of in vitro selection was observed among variants with >7 A's in our experiments, we favor the mechanism of misalignment during template switching of reverse transcriptase. Shinde et al have proposed that when the polymerase active site is occupied by mononucleotides, the polymerase complex has higher frequency of dissociation from the template and this creates an opportunity for misalignment.16 The data suggest that the MLV reverse transcriptase during in vitro selection is biased toward expansion, and that expansion of oligo A may partly contribute to instability of vector over time. Although the exact molecular mechanism is not clear, the data provide insight into a possible mechanism of transgene inactivation in addition to physical deletion of transgene sequences.
In conclusion, in RRV including the ECMV IRES, although the 7 A's version has been used extensively, successfully, and without unexpected toxicities in clinical trials, the 5 or 6 A's version of the IRES may have further advantages, not only because of somewhat enhanced genome stability and transgene protein expression from the 5 A's and 6 A's IRES, respectively, but also because the more frequent direction of oligo A number drift seems to be preferentially toward longer oligo A in the A bulge in RRV context.
Supplementary Material
Acknowledgments
We thank Nicholas A. Boyle, PhD, MBA, and Alessandro Lobbia, PhD, for critical reading of the manuscript. The original RRV system was developed with support from NIH Grants U01-NS59821 and R01-CA105171 to N.K. The work was also supported by Accelerate Brain Cancer Cure, American Brain Tumor Association, Musella Foundation, National Brain Tumor Society, Voices Against Brain Cancer, and the Department of Health and Human Services US.
Author Disclosure
Tocagen has licensed intellectual property for which C.R.L. and N.K. are inventors. A.H.L., C.B., K.C., H.G., C.I., J.M.R., and D.J.J. are fulltime employees of Tocagen and hold stock or stock options in Tocagen. Y.L., C.R.L., and N.K. were/are paid consultants to Tocagen. C.R.L. and N.K. hold stock or stock options in Tocagen.
References
- 1.Perez OD, Logg CR, Hiraoka K, et al. . Design and selection of Toca 511 for clinical use: modified retroviral replicating vector with improved stability and gene expression. Mol Ther 2012;20:1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logg CR, Logg A, Tai CK, et al. . Genomic stability of murine leukemia viruses containing insertions at the Env-3′ untranslated region boundary. J Virol 2001;75:6989–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y, Aran J, Gottesman MM, Pastan I. Co-expression of human adenosine deaminase and multidrug resistance using a bicistronic retroviral vector. Hum Gene Ther 1998;9:287–293 [DOI] [PubMed] [Google Scholar]
- 4.Duke GM, Hoffman MA, Palmenberg AC. Sequence and structural elements that contribute to efficient encephalomyocarditis virus RNA translation. J Virol 1992;66:1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman MA, Palmenberg AC. Mutational analysis of the J-K stem-loop region of the encephalomyocarditis virus IRES. J Virol 1995;69:4399–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman MA, Palmenberg AC. Revertant analysis of J-K mutations in the encephalomyocarditis virus internal ribosomal entry site detects an altered leader protein. J Virol 1996;70:6425–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolupaeva VG, Pestova TV, Hellen CU, Shatsky IN. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem 1998;273:18599–18604 [DOI] [PubMed] [Google Scholar]
- 8.Palmenberg AC, Kirby EM, Janda MR, et al. . The nucleotide and deduced amino acid sequences of the encephalomyocarditis viral polyprotein coding region. Nucleic Acids Res 1984;12:2969–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logg CR, Logg A, Matusik RJ, et al. . Tissue-specific transcriptional targeting of a replication-competent retroviral vector. J Virol 2002;76:12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang WJ, Tai CK, Kasahara N, Chen TC. Highly efficient and tumor-restricted gene transfer to malignant gliomas by replication-competent retroviral vectors. Hum Gene Ther 2003;14:117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochkov YA, Palmenberg AC. Translational efficiency of EMCV IRES in bicistronic vectors is dependent upon IRES sequence and gene location. BioTechniques 2006;41:283–284, 286, 288 passim [DOI] [PubMed] [Google Scholar]
- 12.Davies MV, Kaufman RJ. The sequence context of the initiation codon in the encephalomyocarditis virus leader modulates efficiency of internal translation initiation. J Virol 1992;66:1924–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hennecke M, Kwissa M, Metzger K, et al. . Composition and arrangement of genes define the strength of IRES-driven translation in bicistronic mRNAs. Nucleic Acids Res 2001;29:3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostertag D, Amundson KK, Lopez Espinoza F, et al. . Brain tumor eradication and prolonged survival from intratumoral conversion of 5-fluorocytosine to 5-fluorouracil using a nonlytic retroviral replicating vector. Neuro Oncol 2012;14:145–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan H. Leukemogenesis by Moloney murine leukemia virus: A multistep process. Trends Microbiol 1997;5:74–82 [DOI] [PubMed] [Google Scholar]
- 16.Shinde D, Lai Y, Sun F, Arnheim N. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res 2003;31:974–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski A, Jackson RJ. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 1998;4:626–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chamond N, Deforges J, Ulryck N, Sargueil B. 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of Type II IRESs. Nucleic Acids Res 2014;42:10373–10384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlinger KK, Kofler RM, Heinz FX, et al. . Selection and analysis of mutations in an encephalomyocarditis virus internal ribosome entry site that improve the efficiency of a bicistronic flavivirus construct. J Virol 2007;81:12619–12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duch M, Carrasco ML, Jespersen T, et al. . An RNA secondary structure bias for non-homologous reverse transcriptase-mediated deletions in vivo. Nucleic Acids Res 2004;32:2039–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu WS, Bowman EH, Delviks KA, Pathak VK. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J Virol 1997;71:6028–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathak VK, Temin HM. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: Deletions and deletions with insertions. Proc Natl Acad Sci U S A 1990;87:6024–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stuhlmann H, Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol 1992;66:2378–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parthasarathi S, Varela-Echavarria A, Ron Y, et al. . Genetic rearrangements occurring during a single cycle of murine leukemia virus vector replication: Characterization and implications. J Virol 1995;69:7991–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel TA. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem 1986;261:13581–13587 [PubMed] [Google Scholar]
- 26.Levinson G, Gutman GA. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol Biol Evol 1987;4:203–221 [DOI] [PubMed] [Google Scholar]
- 27.Seki M, Akiyama M, Sugaya Y, et al. . Strand asymmetry of +1 frameshift mutagenesis at a homopolymeric run by DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem 1999;274:33313–33319 [DOI] [PubMed] [Google Scholar]
- 28.Kieleczawa J. Fundamentals of sequencing of difficult templates—an overview. J Biomol Tech 2006;17:207–217 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.