Abstract
Genes for the plastid-encoded RNA polymerase (PEP) persist in the plastid genomes of all photosynthetic angiosperms. However, three unrelated lineages (Annonaceae, Passifloraceae and Geraniaceae) have been identified with unusually divergent open reading frames (ORFs) in the conserved region of rpoA, the gene encoding the PEP α subunit. We used sequence-based approaches to evaluate whether these genes retain function. Both gene sequences and complete plastid genome sequences were assembled and analyzed from each of the three angiosperm families. Multiple lines of evidence indicated that the rpoA sequences are likely functional despite retaining as low as 30% nucleotide sequence identity with rpoA genes from outgroups in the same angiosperm order. The ratio of non-synonymous to synonymous substitutions indicated that these genes are under purifying selection, and bioinformatic prediction of conserved domains indicated that functional domains are preserved. One of the lineages (Pelargonium, Geraniaceae) contains species with multiple rpoA-like ORFs that show evidence of ongoing inter-paralog gene conversion. The plastid genomes containing these divergent rpoA genes have experienced extensive structural rearrangement, including large expansions of the inverted repeat. We propose that illegitimate recombination, not positive selection, has driven the divergence of rpoA.
Before inexpensive DNA sequencing, the plastid genomes (plastomes) of flowering plants (angiosperms) were surveyed for gene content using Southern hybridization1,2,3. These surveys revealed remarkably conserved gene order and content across almost all angiosperms, yet also discovered a few isolated lineages with highly divergent, rearranged plastomes lacking genes and introns. The subsequent publication of more than 800 complete plastomes has confirmed most of these early results. Plastomes typically contain 79 protein-coding genes, 30 tRNA and 4 rRNA genes4. Plastid encoded genes are often categorized as either photosynthesis related or housekeeping, and the latter are generally found to have been lost from plastomes and either functionally replaced or transferred to the nucleus5,6,7. Among the housekeeping genes encoded by the plastome are the four subunits of the eubacterial-like RNA polymerase (PEP) that is responsible for most photosynthetic gene expression8.
The genes encoding the three largest PEP subunits, β, β′ and β″, are cotranscribed from rpoB, rpoC1 and rpoC2, respectively. The α subunit is encoded by rpoA, the last gene in the conserved rpl23 transcriptional unit consisting mostly of ribosomal protein genes8. The three large subunit genes have only been found missing from a few parasitic and mycoheterotrophic plant plastomes9,10. In these cases it appears that the single-subunit nuclear-encoded RNA polymerase (NEP) has taken over transcription of the residual plastome, which no longer encodes a functional photosynthetic apparatus7. All deletions of individual PEP subunits from Nicotiana tabacum (tobacco) produced photosynthetically defective transformants, demonstrating that each of the four subunits is an essential gene11. PEP and NEP transcribe many of the same genes using distinct promoters; species lacking PEP have also lost PEP-specific promoters and nuclear-encoded σ factors12,13.
Southern hybridization surveys identified three unrelated lineages, Pelargonium (Geraniaceae), Annona (Annonaceae) and Passiflora (Passifloraceae), which appeared to lack the plastid rpoA gene1,14. Subsequent work6 identified highly divergent rpoA sequences encoded in the plastomes of P. x hortorum and Passiflora biflora, however no further data are available for Annona.
The P. x hortorum plastome is the largest and most complex angiosperm plastome yet discovered and houses three distinct, divergent rpoA-like ORFs15. No other plastome is known to harbor multiple paralogs of this gene, and it is difficult to judge which, if any, of these divergent genes are functional. Moreover, it is unclear whether they have diverged due to positive or relaxed selection or by some unusual, locus-specific neutral process.
Determining the functionality of rpoA poses several difficulties. Due to its location at the end of a conserved transcriptional unit, mRNA expression data are uninformative, as it has been shown that the entire plastome can be transcribed via read-through16. There is no published nuclear genome data for Pelargonium, Passiflora or Annona. It is possible that rpoA has been transferred to the nucleus and that the divergence of the gene reflects relaxed selection on the plastid copy in the wake of its functional replacement by a nuclear paralog. Although the transfer of rpoA has not been demonstrated in angiosperms, it was detected in the moss Physcomitrella patens17, and it was inferred that rpoA has been transferred to the nucleus twice in the bryophytes18. Following functional transfer to the nucleus, the original plastome gene copy may degrade slowly, making it difficult to judge the functionality of an ORF if the gene has been transferred relatively recently19.
Due to the intractability of reverse genetics in most plastomes, we have adopted a sequence-based approach to address whether Pelargonium, Passiflora and Annona plastomes still encode a functional PEP α subunit. We conducted substitution rate analyses to explore selective forces acting on the rpoA sequences in these plastomes. The results of our in silico analyses suggest that these rpoA-like sequences are functional genes, some of which have been evolving in manners unlike that of other plastid genes due to illegitimate recombination. Furthermore, illegitimate recombination is also evident in the large changes in the inverted repeat (IR) boundaries in all three lineages.
Results
Plastome sequence of Annona cherimola
The plastome of Annona cherimola is 201,723 bp with a 69,771 bp large single copy (LSC) region, a 64,493 bp IR and a small single copy (SSC) region of only 2,966 bp (Fig. S1). The IR has greatly expanded at both the IRB/SSC and IRB/LSC boundaries. Expansions at the IRB/LSC boundary duplicated 24 genes, from rps19 through most of psbA. The IRB/SSC expansion included 11 genes from ycf1 through trnL-UAG. This resulted in a very small SSC containing a single complete gene (rpl32) and a nearly complete copy of ndhF. The Annona plastome comprises 165 genes: 113 unique genes and 52 duplicated genes in the expanded IR. Gene order is highly conserved compared to the ancestral plastid genome organization for angiosperms7 with a single inversion involving six genes (ycf3 - atpE) in the LSC (Fig. S1). Gene content is also highly conserved with no apparent gene loss, however, rpoA is highly divergent with a nucleotide sequence identity of 57% compared to Chloranthus, which is sister to the magnoliid clade (Table 1).
Table 1. Summary of conserved domain database (CDD) search results for Annonaceae and Passifloraceae data sets.
| Annonaceae Comparison | N-terminal | dimer | β | β′ | nt identity (%) | aa identity (%) | ORF length (bp) |
|---|---|---|---|---|---|---|---|
| Annona | Y | Y | Y | Y | 57.0 | 40.7 | 1,035 |
| Asimina | Y | Y | Y | Y | 74.5 | 63.8 | 1,020 |
| Calycanthus | Y | Y | Y | Y | 89.0 | 86.6 | 1,020 |
| Cananga | Y | Y | Y | Y | 55.9 | 38.5 | 1,143 |
| Chloranthus | Y | Y | Y | Y | 100.0 | 100.0 | 1,002 |
| Drimys | Y | Y | Y | Y | 90.2 | 85.4 | 1,017 |
| Liriodendron | Y | Y | Y | Y | 91.3 | 89.8 | 1,014 |
| Magnolia | Y | Y | Y | Y | 91.8 | 89.4 | 1,014 |
| Piper | Y | Y | Y | Y | 85.7 | 80.2 | 1,017 |
| Passifloraceae Comparison | nt identity | aa identity | ORF length | ||||
| Hevea | Y | Y | Y | Y | 92.0 | 88.9 | 1,023 |
| Jatropha | Y | Y | Y | Y | 91.8 | 87.9 | 1,017 |
| Linum | Y | Y | Y | Y | 86.9 | 80.8 | 1,011 |
| Manihot | Y | Y | Y | Y | 91.9 | 87.8 | 1,029 |
| Oxalis | Y | Y | Y | Y | 89.8 | 85.3 | 1,017 |
| P. biflora | Y | Y | Y | Y | 53.6 | 37.4 | 1,071 |
| P. ciliata | Y | Y | Y | Y | 93.4 | 88.8 | 1,005 |
| P. cirrhiflora | Y | Y | Y | Y | 93.8 | 90.6 | 1,017 |
| P.quadrangularis | Y | Y | Y | Y | 93.3 | 89.1 | 1,017 |
| Populus | Y | Y | Y | Y | 100.0 | 100.0 | 1,017 |
| Ricinus | Y | Y | Y | Y | 91.9 | 88.0 | 996 |
| Turnera | Y | Y | Y | Y | 80.7 | 71.0 | 891 |
Predictions of the PEP α subunit N-terminus, homodimer interface, beta and beta prime interfaces are indicated (Y = Yes, N = No). The pairwise identity of each sequence with the outgroups Populus or Chloranthus is given for nucleotide (nt) and amino acid (aa) alignments. Generic names in Annonaceae comparison are in bold; other genera represent related families of magnoliids or the outgroup Chloranthus. P. = Passiflora; species in bold in Passifloraceae comparison are members of the genus Passiflora; other genera are related familes of rosids.
High levels of rpoA sequence divergence in three unrelated angiosperm lineages
Comparison of both nucleotide and amino acid sequence divergence of rpoA for members of the three unrelated lineages of angiosperms, Annonaceae, Passiflora (Passifloraceae) and Pelargonium (Geraniaceae) were performed (Tables 1, 2). For Annonaceae the three genera examined (Annona, Asimina and Cananga) have nucleotide and amino acid sequence identities ranging from 56–75% and 39–64%, respectively, in comparison to Chloranthus (Table 1). This is in contrast to the 86–92% and 80–90% nucleotide and amino acid sequence identities, respectively, for the five other magnoliids examined. Sequence identities within Passiflora were variable for one species, P. biflora, which showed high levels of rpoA divergence, 54% and 37% nucleotide and amino acid sequence identities, respectively (Table 1). Levels of sequence identity of the other three species of Passiflora and eight species from other families of rosids were substantially higher (81–94% and 71–91%, respectively). Within Pelargonium, levels of sequence identity of rpoA were among the lowest, with nucleotide and amino acid identities ranging from 30–49% and 15–34%, respectively (Table 2). The levels of sequence divergence are much lower in related rosids (78–92% nucleotide and 65–86% amino acid identity), including four other genera of Geraniales, one of which is a member of the Geraniaceae (i.e., Hypseocharis).
Table 2. Summary of conserved domain database (CDD) search results for Pelargonium data set.
| Outgroup and Geraniales | N-terminal | dimer | β | β′ | nt identity (%) | aa identity (%) | ORF length (bp) |
|---|---|---|---|---|---|---|---|
| Eucalyptus | Y | Y | Y | Y | 100.0 | 100.0 | 1,014 |
| Francoa | Y | Y | Y | Y | 92.2 | 85.8 | 1,020 |
| Melianthus | Y | Y | Y | Y | 91.3 | 84.4 | 1,020 |
| Viviana | Y | Y | Y | Y | 84.0 | 73.2 | 1,014 |
| Hypseocharis | Y | Y | Y | Y | 77.8 | 65.4 | 1,089 |
| Pelargonium Clade A1 | |||||||
| P. citronellum | Y | Y | N | N | 46.0 | 31.6 | 885 |
| P. cucullatum | Y | Y | N | N | 46.0 | 31.6 | 885 |
| P. nanum | Y | Y | Y | Y | 46.2 | 31.9 | 885 |
| P. quercifolium | Y | Y | Y | Y | 46.0 | 31.6 | 885 |
| Clade A2 | |||||||
| P. alternans | Y | Y | Y | Y | 46.3 | 31.6 | 885 |
| P. echinatum | Y | Y | Y | Y | 45.1 | 32.0 | 858 |
| P. fulgidum | Y | Y | Y | Y | 44.9 | 31.4 | 855 |
| P. incrassatum | Y | Y | Y | Y | 46.2 | 32.2 | 885 |
| P. luridum | Y | Y | Y | Y | 46.2 | 31.6 | 885 |
| Clade B | |||||||
| P. australe | Y | Y | Y | Y | 44.0 | 34.1 | 828 |
| P. cotyledonis | Y | Y | Y | Y | 48.8 | 33.8 | 945 |
| P. exstipulatum | Y | Y | Y | Y | 46.0 | 33.0 | 879 |
| P. grossularioides | Y | Y | Y | Y | 45.1 | 31.0 | 864 |
| P. reniforme | Y | Y | Y | Y | 46.2 | 32.7 | 885 |
| Clade C1 | |||||||
| P. dolomiticum | Y | Y | N | N | 34.1 | 24.4 | 750 |
| P. trifidum | Y | Y | N | N | 32.6 | 23.5 | 708 |
| P. myrrhifolium | Y | Y | N | N | 35.1 | 25.2 | 714 |
| P. tetragonum | Y | Y | Y | Y | 46.0 | 29.8 | 912 |
| P. worcesterae | Y | Y | Y | Y | 46.2 | 30.1 | 912 |
| Clade C2 | |||||||
| P. endlicherrianum_578 | Y | Y | N | N | 32.3 | 25.0 | 1,701 |
| P. endlicherrianum_597 | Y | Y | N | N | 31.6 | 25.1 | 1,737 |
| P. spinosum_578 | Y | Y | N | N | 30.1 | 19.9 | 1,773 |
| P. spinosum_597 | Y | Y | N | N | 35.0 | 23.0 | 1,788 |
| P. transvaalense_597-1 | Y | Y | N | N | 35.3 | 21.1 | 1,788 |
| P. transvaalense_597-2 | Y | Y | Y | Y | 36.0 | 20.5 | 1,788 |
| P. transvaalense_597-3 | Y | Y | N | N | 35.4 | 21.1 | 1,788 |
| P. transvaalense_597-4 | Y | Y | N | N | 35.5 | 21.8 | 1,782 |
| P. transvaalense_597-5 | Y | Y | N | N | 35.5 | 21.4 | 1,866 |
| P. transvaalense_597-6 | Y | Y | N | N | 33.7 | 19.3 | 1,788 |
| Clade C2, sect. Ciconium | |||||||
| P. alchemilloides_521 | Y | Y | N | N | 34.8 | 33.5 | 702 |
| P. alchemilloides_578 | Y | Y | Y | Y | 31.8 | 15.9 | 1,737 |
| P. alchemilloides_597 | Y | Y | N | N | 30.7 | 14.8 | 1,794 |
| P. quinquelobatum_521 | Y | Y | N | N | 33.6 | 20.6 | 1,560 |
| P. quinquelobatum_578 | Y | Y | Y | Y | 32.0 | 15.9 | 1,737 |
| P. quinquelobatum_597 | Y | Y | N | N | 30.7 | 15.3 | 1,794 |
| P. tongaense_521 | Y | Y | Y | Y | 33.0 | 20.6 | 1,554 |
| P. tongaense_578 | Y | Y | Y | Y | 31.9 | 15.7 | 1,737 |
| P. tongaense_597 | Y | Y | N | N | 30.3 | 14.9 | 1,815 |
| P. xhortorum_521 | Y | Y | Y | Y | 33.4 | 21.1 | 1,566 |
| P. xhortorum_578 | Y | Y | Y | Y | 31.9 | 15.9 | 1,737 |
| P. xhortorum_597 | Y | Y | N | N | 30.6 | 14.9 | 1,794 |
Predictions of the PEP α subunit N-terminus, homodimer interface, beta and beta prime interfaces are indicated (Y = Yes, N = No). The pairwise identity of each sequence with outgroup Eucalyptus is given for nucleotide (nt) and amino acid (aa) alignments.
Detection of plastid rpoA transcripts by RT-PCR
Transcripts were confirmed for the two longer rpoA-like ORFs of P. x hortorum, ORF578 and ORF597 (Fig. S2); there are at least dicistronic transcripts for both of these ORFs. The result does not preclude ORF transcripts being present as monocistrons or as polycistrons, including genes further upstream.
Conservation of PEP promoters and sigma factors
A database comprising contigs from the published20 high-coverage nuclear transcriptome assembly of P. x hortorum was queried with rpoA nucleotide and amino acid sequences from A. thaliana. No nuclear-encoded rpoA paralog transcript was detected in either the nucleotide or the translated database by BLAST search. Other nuclear-encoded components of the PEP holoenzyme, e.g. sigma factors, were found using the same BLAST parameters and were recently reported in Zhang et al.21.
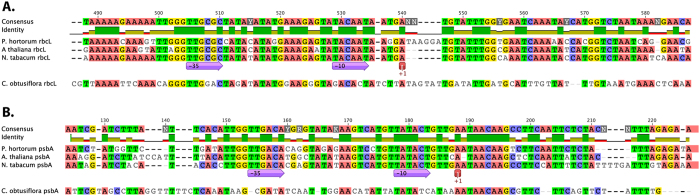
In silico examination of PEP promoters upstream of the rbcL and psbA coding regions revealed that P. x hortorum sequences closely resembled those of A. thaliana and N. tabacum. The -35 and -10 elements, as well as the transcription start sites, were 100% identical across all three species, unlike in Cuscuta obtusiflora, a parasitic plant lacking PEP (Fig. 1A,B).
Figure 1. Alignment of PEP promoter regions.
(A) Alignment of promoter region for rbcL in three species with functional PEP (Nicotiana tabacum, Arabidopsis thaliana, Pelargonium x hortorum) and one lacking PEP (Cuscuta obtusiflora). (B) Alignment of promoter region for psbA in three species with functional PEP (N. tabacum, A. thaliana, P. x hortorum) and one lacking PEP (C. obtusiflora). The conserved −10 and −35 elements are indicated by block arrows and the transcription start site is indicated by a red box (+1).
Analysis of signals of selection
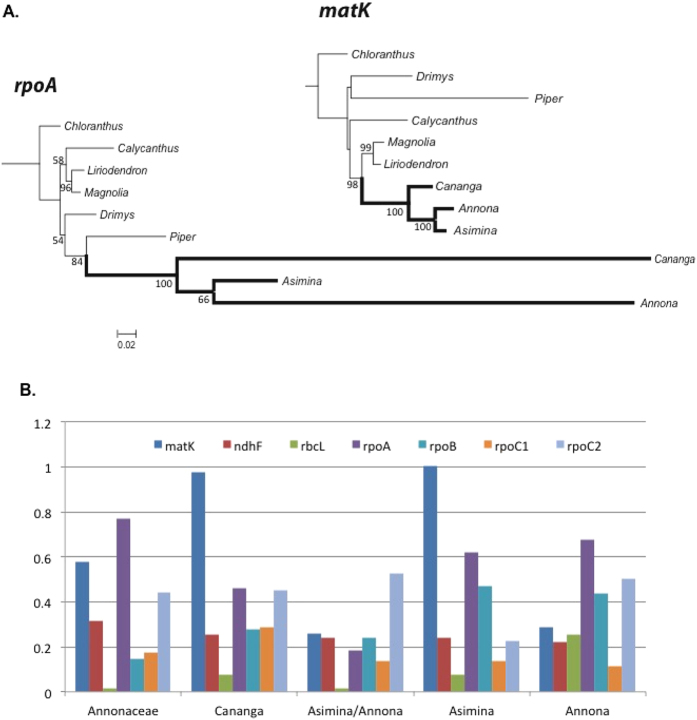
The dN/dS ratio was calculated for the three different lineages of angiosperms. Seven plastid genes (rpoA, rpoB, rpoC1, rpoC2, ndhF, matK and rbcL) were analyzed in PAML for three datasets to compare the dN/dS ratio of rpoA to the other rpo genes as well as to other non-rpo plastid genes. These same seven genes were used to generate constraint trees for each dataset. Constraint tree topologies were identical to the matK trees for Annonaceae (Fig. 2A) and Passifloraceae (Fig. 3A). The seven gene constraint tree for Geraniaceae is shown as an inset in Fig. 4.
Figure 2. Representative maximum likelihood trees and dN/dS values for the nine taxa of Annonaceae.
(A) Likelihood scores for the matK and rpoA trees were −5638.2661 lnL and −5506.0125 lnL, respectively. Branches in bold are members of Annonaceae. Bootstrap values greater than 50 are shown at the nodes. Scale bar indicates non-synonymous substitutions per codon. (B) Histogram of dN/dS values for seven genes for the Annonaceae. For each gene, dN/dS values (y axis) are given for all branches of interest: the branch leading to the family, the internal branch to Annona/Asimina, and the terminal branches to Annona, Asimina, and Cananga. Only one ratio was marginally >1, the terminal branch to Asimina for matK (dN/dS = 1.0069).
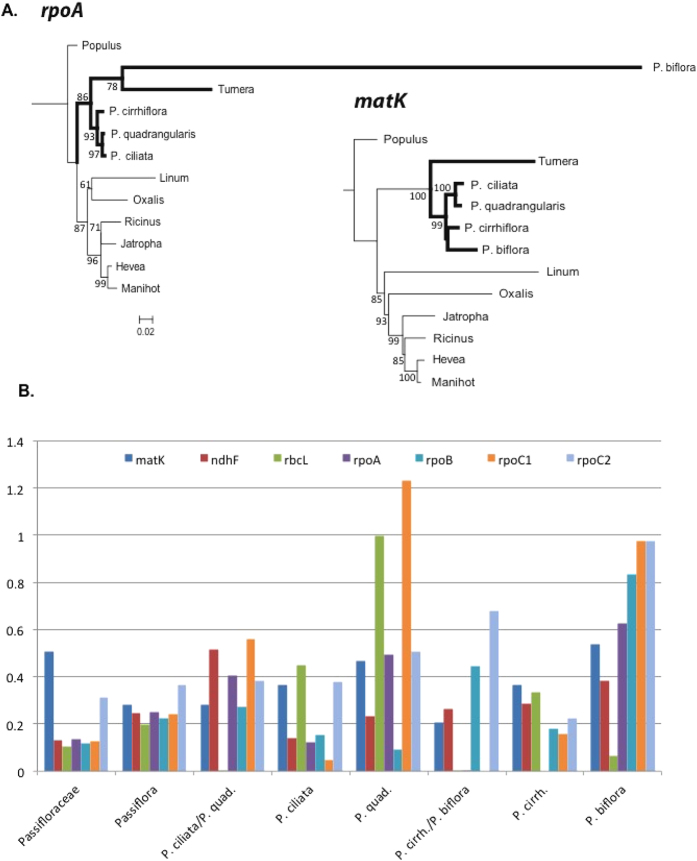
Figure 3. Representative maximum likelihood trees and dN/dS values for 12 taxa of Passifloraceae.
(A) Likelihood scores for the matK and rpoA trees were lnL −7243.3426 and −4810.9045 lnL, respectively. Branches in bold are members of Passifloraceae. Bootstrap values greater than 50 are shown at the nodes. Scale bar indicates non-synonymous substitutions per codon. (B) Histogram of dN/dS ratios for seven genes for the Passifloraceae. For each gene, dN/dS values (y axis) are given for all branches of interest: the branch leading to the family as well as all internal and terminal branches. The primary branch of interest is the terminal branch to P. biflora, the only species with a divergent rpoA gene. The terminal branch to P. quadrangularis for rpoC1 has a dN/dS value >1, but this is likely an artifact, as the branch length is extremely short. The lack of a bar for rbcL is due to a dS value of 0.
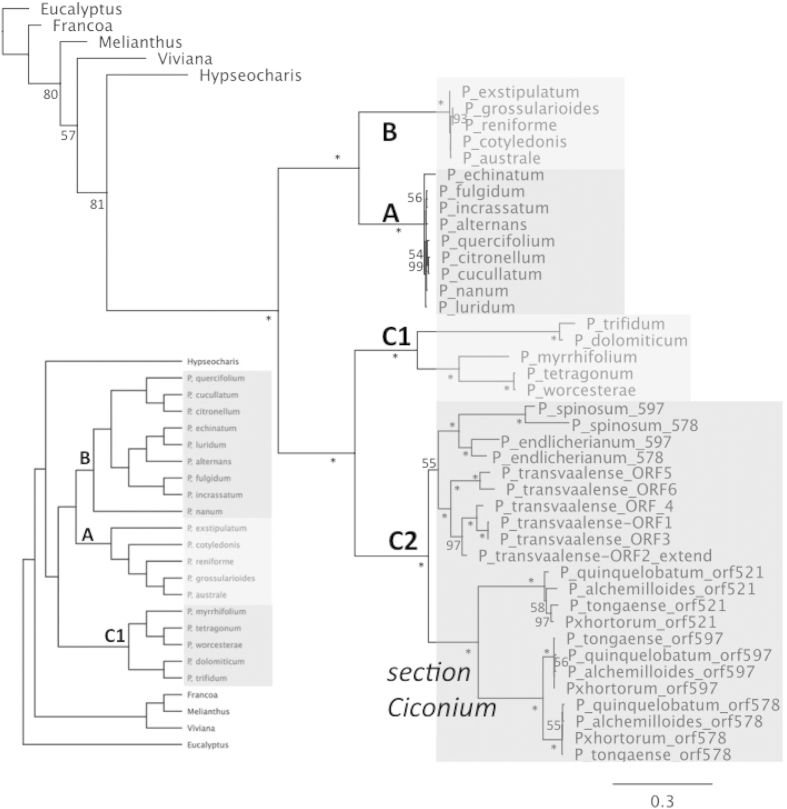
Figure 4. Maximum likelihood tree generated for all 46 rpoA ORFs from 26 Pelargonium species with likelihood score −21428.281249 lnL.
Species in clade C2 contain two (P. spinosum and P. endlicherianum), three (four species from section Ciconium) or six (P. transvaalense) rpoA paralogs. Bootstrap values greater than 50 are shown at the nodes; values of 100 are indicated by asterisks. Scale bar indicates non-synonymous substitutions per codon. The constraint tree (inset) does not contain clade C2 taxa as these species all contain multiple rpoA sequences.
Annonaceae
Maximum likelihood trees for matK and rpoA (Fig. 2A) were generated from the Annonaceae dataset, which comprised eight magnoliids including three genera in the Annonaceae, and Chloranthus of the Chloranthales (Table 3). The matK tree had the same topology as most other individual plastid genes (not shown) where the branch leading to Piper was long but branches within Annonaceae relatively short. However, in the rpoA tree branch lengths within Annonaceae were sufficiently long to produce an incorrect topology through long-branch attraction to Piper. The five branches of interest are highlighted in Fig. 2B. The terminal branch leading to Asimina for matK had the only dN/dS value >1 (1.0069). All rpo genes showed dN/dS values consistent with purifying selection in Annonaceae.
Table 3. Taxon sampling by data set.
| Magnoliids/Chloranthales | Accession numbers |
|---|---|
| Annona cherimola | KU563738 |
| Asimina incana | KU645794, KU645799, KU645804, KU645810, KU645815, KU645820, KU645825 |
| Calycanthus floridus | NC_004993 |
| Cananga odorata | KU645791, KU645796, KU645801, KU645806, KU645812, KU645817, KU645822 |
| Chloranthus spicatus | NC_009598 |
| Drimys granadensis | NC_008456 |
| Liriodendron tulipifera | NC_008326 |
| Magnolia kwangsiensis | NC_015892 |
| Piper cenocladum | NC_008457 |
| Malpighiales | |
| Hevea brasiliensis | NC_015308 |
| Jatropha curcas | NC_012224 |
| Linum usitatissimum | KU645792, KU645797, KU645802, KU645808, KU645813, KU645818, KU645823 |
| Manihot esculenta | NC_010433 |
| Oxalis latifolia | EU002528, KF224983, HM850223, EU002248, GQ998560, GQ998561, GQ998562 |
| Passiflora biflora | EU017067, KU645807, EU017069, EU017092, EU017096, EU017121, EU017122 |
| Passiflora ciliata | JX661956, JX662765, JX664062, JX662034, JX663490, JX664953, JX662679 |
| Passiflora cirrhiflora | KU645790, KU645795, KU645800, KU645805, KU645811, KU645816, KU645821 |
| Passiflora quadrangularis | KU645791, KU645796, KU645801, KU645806, KU645812, KU645817, KU645822 |
| Populus trichocarpa | NC_009143 |
| Ricinus communis | NC_016736 |
| Turnera ulmifolia | JX664965, JX664074, JX663502, JX662777, JX662690, JX662046, JX661965 |
| Myrtales/Geraniales | |
| Eucalyptus globulus | NC_008115 |
| Francoa sonchifolia | NC_021101 |
| Melianthus villosus | NC_023256 |
| Viviana marifolia | NC_007957 |
| Hypseocharis bilobata | NC_023260 |
| Clade A1 | |
| Pelargonium citronellum | KM527888 |
| Pelargonium cucullatum | KM527887 |
| Pelargonium nanum | KM527896 |
| Pelargonium quercifolium | KM527897 |
| Clade A2 | |
| Pelargonium alternans | NC_023261 |
| Pelargonium echinatum | KM527891 |
| Pelargonium fulgidum | KM527893 |
| Pelargonium incrassatum | KM527894 |
| Pelargonium luridum | KU535486-KU535492 |
| Clade B | |
| Pelargonium australe | KM459517 |
| Pelargonium cotyledonis | KM459516 |
| Pelargonium exstipulatum | KM527892 |
| Pelargonium grossularioides | KU535493-KU535499 |
| Pelargonium reniforme | KU535500-KU535506 |
| Clade C1 | |
| Pelargonium dolomiticum | KM527889 |
| Pelargonium trifidum | KM527898 |
| Pelargonium myrrhifolium | KM527895 |
| Pelargonium tetragonum | KM527899 |
| Pelargonium worcesterae | KU535507-KU535513 |
| Clade C2 | |
| Pelargonium endlicherrianum | KU535514-KU535522 |
| Pelargonium spinosum | KU535523-KU535530 |
| Pelargonium transvaalense | KM527900 |
| Clade C2, sect. Ciconium | |
| Pelargonium alchemilloides | KU535531-KU535539 |
| Pelargonium quinquelobatum | KU535540-KU535548 |
| Pelargonium tongaense | KU535549-KU535557 |
| Pelargonium x hortorum | NC_008454 |
Single accession numbers represent complete plastomes and seven accession numbers are for taxa with individual sequences for each gene.
Passiflora
Maximum likelihood trees for matK and rpoA were constructed from the Passiflora dataset, consisting of 12 taxa from the Malpighiales, including four Passiflora species (Fig. 3A, Table 3). The matK tree has the same topology as most other individual plastid genes (not shown), with a long branch leading to Turnera (Passifloraceae) but relatively short branches within Passiflora. In the rpoA tree, however, the long terminal branch leading to P. biflora resulted in long-branch attraction to Turnera. For dN/dS ratios, the branches of interest are highlighted in Fig. 3B. The principal branch of interest was the terminal branch leading to P. biflora, the only species with a divergent rpoA. The only gene for which a branch had a dN/dS value >1 (1.2312) was rpoC1, on the terminal branch leading to P. quadrangularis.
Pelargonium
The Pelargonium dataset consisted of 26 species representing all major clades (Table 3). Pelargonium rpoA genes showed a complex pattern of divergence by clade that confounded the analysis of evolutionary rates. A maximum likelihood tree of all rpoA genes/ORFs from the Pelargonium dataset was generated (Fig. 4). To overcome the potential for error due to the difficulties in aligning rpoA sequences across clades and with outgroups four different alignment algorithms were utilized in Pelargonium rate comparisons (Table S2).
The rpoA genes in clades A and B were somewhat divergent between the two clades, sharing only 66–71% nucleotide sequence identity, but showed high identity within each clade. The five rpoA genes representing clade B shared 94% sequence identity. However, this percentage was lowered by indels associated with tandem repeats at the 3′ end of the gene immediately preceding the predicted stop codon (Fig. S3). When this repeat-rich region was excluded from the alignment the remaining sequences share over 98% identity. In fact, four of the five genes were 100% identical when the 3′ end was excluded, and the fifth, P. exstipulatum, differed by only two nucleotides, both of which were nested in tandem repeats and caused non-synonymous substitutions.
Nine rpoA genes representing clade A shared 92% identical sites, or 95% identical sites if the 3′ end was excluded. Similar to clade B, different numbers of tandem repeats towards the 3′ end caused length differences in clade A rpoA (Fig. S4). Although indels associated with tandem repeats underlie the length differences between rpoA genes of clades A and B, the repeats were nonhomologous sequences. In clade B there were two different tandem repeat units that underlie the length differences: a 6 bp motif of GCGAGG was present in all the ORFs, ranging from two repeat units in P. australe to eight in the same region of P. grossularioides. In P. cotyledonis, two copies of this 6 bp tandem repeat were nested inside a unique 39 bp repeat, which expanded to four tandem copies, the last base pair of which was the first base pair of the predicted TAA stop codon (Fig. S3). The 6 bp repeat from clade B was not found in any clade A rpoA sequence, instead, a 9 bp repeat unit, present as both tandem and dispersed repeats at the 3′ end of the gene in all clade A species, appeared to have caused a deletion of 30 bp between two direct, dispersed 9 bp repeat units in P. echinatum and P. fulgidum. These two taxa are not sister species, thus it appeared that this deletion occurred twice independently in clade A.
The C1 and C2 clades were highly divergent both within and between clades, and the C2 clade contained species with multiple (2, 3 or 6) rpoA-like ORFs (Fig. 4). For clade C2 species it was not clear which of the paralogous ORFs might be functional. ORFs from clade C2 were excluded from dN/dS analysis (see Gene Conversion below).
Clade C1 was represented by five species whose ORFs fell into two groups of more closely related sequences. Pelargonium dolomiticum and P. trifidum shared 96% nucleotide sequence identity. Pelargonium tetragonum and P. worcesterae had 99% identity and were identical in length at 912 bp; P. myrrhifolium was more closely related to this second pair but shared only 61% identity with P. tetragonum. Between the groups, P. dolomiticum and P. tetragonum had only 64% identity.
The branches of interest for the Pelargonium rates analyses were different from those in the previous two data sets: the terminal branches were excluded as intra-clade divergence among species was extremely low due to dense taxon sampling in this dataset. Low sequence divergence between closely related taxa caused error values to be returned in the calculation of dN/dS where either or both of the parameters were calculated to be zero or close to zero (not shown). Therefore the branches of interest were chosen as those where the greatest divergence in rpoA has occurred and are highlighted in Fig. 5.
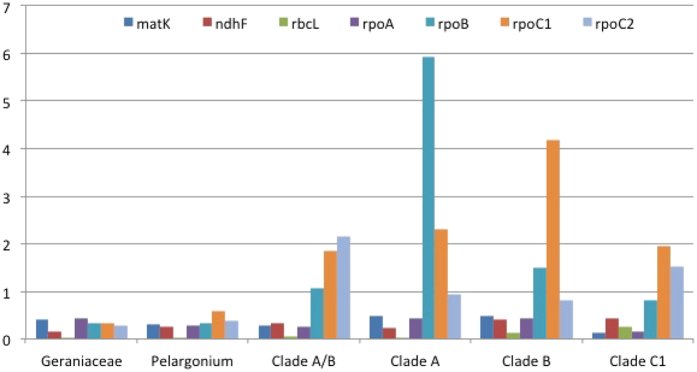
Figure 5. Histogram of dN/dS ratios for seven genes for Geraniaceae.
In addition to MAFFT results presented here, three other alignment algorithms were used (See Table S2). For each gene, dN/dS values are given for all branches of interest, the branch leading to the family (Geraniaceae), to Pelargonium, to the branch to clades A/B, to clade A, to clade B, and to clade C1.
Rates analyses of matK, ndhF and rbcL for Pelargonium detected low dN/dS values consistent with purifying selection across all alignments for all branches of interest (Fig. 5; Table S2). For the rpo genes, a pattern emerged that was consistent across all alignment methods used: dN/dS values for rpoA were uniformly low (<1), consistent with purifying selection on all branches of interest (Fig. 5; Table S2). However, dN/dS values for the other rpo genes were elevated along several branches of interest (Fig. 5; Table S2). On the branch leading to clades A and B, rpoB, rpoC1 and rpoC2 all showed dN/dS values >1. The same was seen for the branches leading to each clade (A and B) except for rpoC2 on the clade A branch, where dN/dS values were near or >1 depending on the alignment method used. On the branch leading to the C1 clade, rpoC1 and rpoC2 but not rpoB showed dN/dS values >1.
Detection of conserved domains
For each of the three datasets, rpoA genes from the outgroup taxa were queried against the Conserved Domain Database (CDD) for detection of functional domains that lie in the N-terminal region of the α-subunit. In each case the three functional domains, involved in the interaction of the α-subunit with itself and the β and β′ subuints, were predicted as present (Tables 1 and 2). Having verified the predictive capability of the CDD in these conserved plastid genes, all the other rpoA genes were queried against the database to predict the presence of the three interaction domains.
In Annonaceae, all rpoA ORFs were predicted to encode all three interaction domains including those from Annona, Asimina and Cananga, despite their substantial sequence divergence from the outgroup Chloranthus (Table 1). Likewise, in Passiflora, all rpoA ORFs were predicted to encode the three conserved domains (Table 1). In Passiflora the divergence was restricted to a single species surveyed, P. biflora (Fig. 3).
In Pelargonium, all ORFs were predicted to encode the N-terminal region of the α-subunit as well as the homodimer interface. However, the conservation of functional domains showed a more complex pattern that differed by clade (Table 2). Clade B was the simplest as all five rpoA sequences were predicted to contain all three functional domains despite retaining just 44%–49% sequence identity with outgroup Eucalyptus.
In Pelargonium clade A all nine rpoA genes were predicted to encode the N-terminus containing the homodimer interface (Table 2), but the CDD search did not predict the other functional domains for two of the four species in clade A1 (P. citronellum and P. cucullatum). All five species from clade A2 were predicted to contain all three functional domains. Divergence from Eucalyptus in clade A is similar to that in clade B, ranging from 45%–46% sequence identity.
The Pelargonium C clade contained the most divergent and puzzling rpoA-like ORFs with respect to the prediction of conserved functional domains (Table 2). All five taxa representing clade C1 were predicted to encode the homodimer interface, which spans the beginning and end of the α-subunit N-terminus (Fig. S5), but only P. tetragonum and P. worcesterae were predicted to contain the other two functional domains (Fig. S5). These two species had the highest sequence identity to the outgroup and at 912 bp were closest in length to rpoA in most angiosperms (versus 1014 bp in Eucalyptus), whereas the other three C1 taxa had shorter genes of 708 bp–750 bp.
Likewise, CDD analyses identified the α-subunit N-terminal region and homodimer domain in all clade C2 taxa rpoA-like ORFs. Using high-coverage Illumina sequence data we found two sequencing errors in the rpoA-like ORFs of the published P. x hortorum plastome annotation15. Both errors were single base pairs missing from ORFs, leading to a premature stop codon (ORF578) and to the division of one long ORF into two shorter ORFs (ORF521, formerly ORF221 and ORF332). The re-annotation of these ORFs was confirmed by comparison with those from the three closely related taxa in section Ciconium. After correction the plastomes each contained three long rpoA-like ORFs of similar length (1566 bp, 1737 bp, and 1794 bp in P. x hortorum; Table 4, Fig. S6). These ORF names were used for the homologous ORFs in the other clade C2 species, even though some differ slightly in length; homology was inferred from synteny.
Table 4. Revised naming system and basic statistics for P. x hortorum and other sect. Ciconium rpoA ORFs.
| Old P. x hortorum ORF name | New ORF name | Length in bp/aa | P. tongaense (bp/aa) | P. alchemill. (bp/aa) | P. quinque. (bp/aa) | pairwise identity (%) | identical sites (%) |
|---|---|---|---|---|---|---|---|
| ORF574 | ORF597 | 1794/597 | 1815/604 | 1794/597 | 1794/597 | 99.40 | 98.80 |
| ORF365 | ORF578 | 1737/578 | 1737/578 | 1737/578 | 1737/578 | 99.50 | 99 |
| ORFs332+221 | ORF521 | 1566/521 | 1554/517 | 702/233* | 1560/519 | 92.70 | 87 |
*P. alchemilloides ORF521 homolog ends after 702 bp but is otherwise in frame through conserved stop codon after 1470 bp/490aa.
In the two species containing two rpoA-like ORFs, P. endlicherianum and P spinosum, all ORFs were predicted to encode the homodimer interface, yet neither contained the other two functional domains (Table 2). Pelargonium transvaalense contained six rpoA-like ORFs predicted to encode the N-terminal domain of the α-subunit and the homodimer interface, however only ORF597–2 contained the other two functional domains. In the four section Ciconium taxa, at least one of the ORFs in each species was predicted to encode all three functional domains. One homolog, ORF578, was predicted to encode all domains in all four taxa. Although the length of the other two ORFs varied between species, ORF578 was identical in length at 1737 bp in all four taxa and also displayed the highest percentage (99%) of identical sites across the four species.
Detection of gene conversion among rpoA paralogs
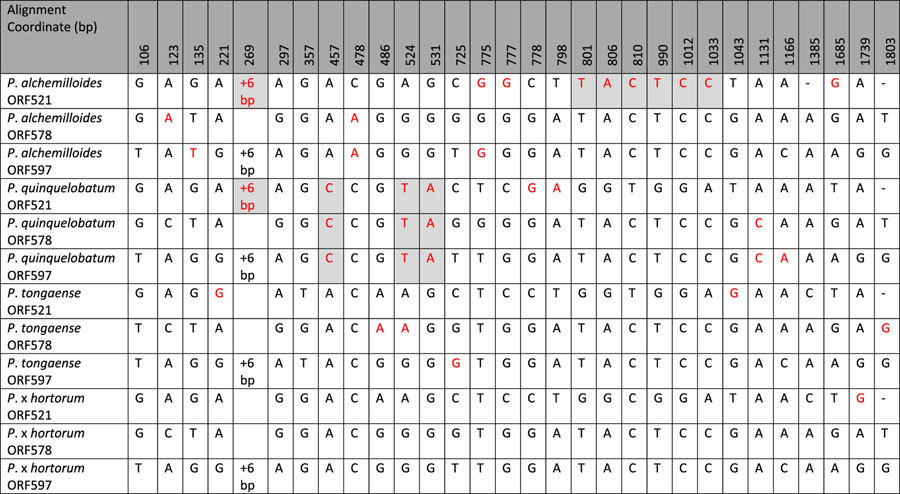
The likelihood tree generated from clade C2 rpoA-like ORFs showed a pattern suggesting that gene conversion was an important phenomenon underlying the evolution of these unusual ORFs (Fig. 4). First, ORFs from the two taxa containing only two ORFs grouped together by species and not by ORF, suggesting that these ORFs have not been evolving independently since their duplication in the ancestor of C2 taxa. For example, the two ORFs in P. endlicherianum shared only 63–69% sequence identity with those from P. spinosum, whereas the ORFs in each species shared 86% and 72% identity with its paralog, respectively. The six ORFs in P. transvaalense grouped together as well, despite their apparent common ancestry with the ORFs in section Ciconium. For the four section Ciconium taxa (Fig. 4), the ORFs grouped by ORF in the likelihood tree rather than by species, despite showing evidence of gene conversion among ORFs, likely reflecting the relatively recent divergence of these taxa.
ORGCONV22 found evidence of recombination among ORFs in all four species of section Ciconium (Table 5), predicting that gene conversion took place in all species in a region from approximately the 120th to 720th (600 bp) position in alignment of the three ORFs. This was the region predicted by the CDD to encode the N-terminus of the α-subunit containing the functional domains. Visual inspection of alignments for mutations potentially resulting from gene conversion was conducted. A parsimony criterion was used: substitutions common to multiple ORFs within a species but not among homologous ORFs across species were scored as putative gene conversion events (Table 6). Both ORGCONV and manual assessment indicated that gene conversion occurred among paralogs in all four section Ciconium species.
Table 5. Gene conversion events detected by ORGCONV.
| Converted Sequence | Donor | Start | End | P-value (L/N) | P-value (L-N) |
|---|---|---|---|---|---|
| P_alchemilloides_ORF597 | P_alchemilloides_ORF521 | 130 | 713 | 1.13E-07 | 5.14E-06 |
| P_quinquelobatum_ORF597 | P_quinquelobatum_ORF521 | 119 | 713 | 2.30E-10 | 1.09E-08 |
| P_tongaense_ORF597 | P_tongaense_ORF521 | 124 | 713 | 5.52E-10 | 2.74E-08 |
| Pxhortorum_ORF578 | Pxhortorum_ORF521 | 223 | 300 | 1.31E-03 | 5.20E-03 |
| Pxhortorum_ORF597 | Pxhortorum_ORF521 | 669 | 713 | 1.03E-03 | 1.77E-02 |
The donor and acceptor of each putative gene conversion event are given along with the coordinates of the converted region and the p-value of the conversion event.
Table 6. Gene conversion events detected by manual count from an alignment of all 12 ORFs from the four Pelargonium section Ciconium species.
Discussion
Of the PEP subunits, α is the least conserved23, so its degree of divergence may not be useful in determining functionality. Likelihood-based calculation of dN/dS ratios to detect selection may be inappropriate for some of these ORFs, as some appear to be evolving in ways not anticipated by standard evolutionary models. For example, gene conversion, which is known to occur between paralogs, can produce spurious signals of selection under likelihood-based models24. Furthermore, alignment error could lead to spurious signals of selection25, as some of the divergent rpoA-like ORFs share less than 40% amino acid sequence identity with outgroup sequences within the same angiosperm order15. At this level of divergence, different alignment methods can produce different estimates of evolutionary rates, none of which is obviously superior to the others. For this investigation we employed a multifaceted, in silico approach to study the evolution of divergent rpoA sequences in three unrelated lineages.
For the Annonaceae and Passiflora, both the CDD predictions and dN/dS values for rpoA strongly suggest that the divergent genes are functional. Members of both lineages for which plastome sequences are available and which have highly divergent rpoA sequences show evidence of substantial and repeated expansions and contractions of the inverted repeat (IR), including genomic rearrangement in the vicinity of rpoA. Illegitimate recombination is a logical cause of the divergence of rpoA in Passiflora and Annona. For Annonaceae, more plastomes (e.g. Asimina and Cananga) will be needed to determine whether divergence of rpoA is consistently associated with large shifts in the IR boundaries.
The Berberis bealei plastome shows a similar pattern with a 12 kb expansion of the IR that duplicates 15 genes, including the region where rpoA resides26. This expansion was noted previously in 26 species of Berberis using comparative restriction site and gene mapping27. Although Ma et al.26 reported that rpoA was absent from the B. bealei plastome, given the similaries between it and the species studied here, we searched for a divergent rpoA that could have been overlooked in the original analyses. Indeed, we identified a copy (coordinates 78645–79644, NC_022457) of rpoA with 67% nucleotide sequence identity to another member of the same family, Nandina domestica, which retained all three functional domains according to a CDD search.
Shifts in IR boundaries in Pelargonium have been even more extreme28. In Pelargonium, dN/dS values for rpoA indicated that this gene is under purifying selection and therefore likely functional. Furthermore the persistence of PEP promoters and the identification of all six PEP sigma factor sequences, but no rpoA homolog in the nuclear transcriptome of Pelargonium x hortorum20 corroborate functionality. The CDD results are less definitive, with all three functional domains predicted for most but not all species. This complex pattern of functional domain conservation is inconsistent with a single loss of rpoA function in Pelargonium. If indeed failure to predict all three functional domains indicates a lack of function, then multiple independent losses of rpoA would be required to achieve the pattern represented in Table 2. In addition to being unparsimonious, this scenario does nothing to explain how rpoA may have retained functionality in some clades despite an unparalleled degree of divergence from the outgroup Eucalyptus.
In the species with mulitple paralogs, represented by P. x hortorum, the IR region has expanded to three times the normal angiosperm size (75,741 bp)15. It is possible that once fixed inside the IR these peculiar rpoA paralogs become more difficult to purge from the plastome, as the rate of sequence evolution in the IRs is slower than in single copy regions29.
Passiflora biflora, Annona cherimola, Berberis bealei and especially Geraniaceae display myriad plastome abnormalities including structural rearrangement, loss of genes and introns, and the divergence of genes that are conserved in almost all other photosynthetic angiosperms6,7. Illegitimate recombination during plastid DNA repair explains the seemingly opposite nature of the genomic divergence between Geraniaceae genera. For example, in Erodium illegitimate recombination led to the deletion of one copy of the IR30, whereas in Pelargonium it led to an expansion and rearrangement of the IR15. In both cases, illegitimate repair of plastid DNA may have caused structural changes that did not delete any genes or their regulatory elements and thus the mutant plastomes were able to reach fixation.
In view of the high levels of sequence divergence of rpoA in these four unrelated lineages of angiosperms and the much lower levels of divergence in related species, the question as to why this gene has diverged so significantly remains. We propose that the divergence is a result of two factors, the inherently labile nature of the gene product, which is known from bacteria to be the least conserved of the polymerase subunits23, and the high degree of genomic rearrangement by illegitimate recombination in the rearranged plastomes. The dN/dS values <1 for the Annonaceae, Passiflora and Pelargonium species included in these analyses also suggest that the divergence of these genes has resulted from a neutral process and is not the result of positive selection. Unlike another gene found to be divergent or missing in Geraniaceae, accD, the conserved domains of rpoA consist of amino acids dispersed across the ORF rather than a single block of contiguous, conserved amino acids that form the catalytic domain of accD. The dispersed nature of the functional domains in rpoA may permit substantial divergence of much of the gene, as long as a number of individual non-contiguous, conserved amino acids are undisturbed.
The especially high level divergence in Pelargonium clade C2 rpoA (Table 2) may be due to gene conversion among paralogs, which is simply a special case of illegitimate recombination. The frequency of gene conversion events is difficult to estimate, but it is sufficiently frequent in section Ciconium rpoA sequences to cause the genes to group together by species, rather than by gene, in a phylogenetic reconstruction. The effect of gene conversion overrides the phylogenetic signal one would expect if these genes were evolving independently. The presence of multiple shared pseudogenes of petD and rps11 upstream from the ORFs (Fig. S6) suggests that gene conversion has taken place not only in coding sequences but in intergenic regions as well. We propose that the same error-prone recombination-based DNA repair mechanism likely underlies the divergence of rpoA in all four lineages examined, and that this mechanism is likely also responsible for the abnormal fluidity of the IR boundary in Annona, Berberis, Passiflora and Pelargonium.
Previous studies have hypothesized that aberrant DNA repair was responsible for accelerated rates of nucleotide substitution, gene and intron loss, and genomic rearrangement of plastid genomes in Geraniaceae28,30,31 and Campanulaceae32. With our present findings we propose a more specific hypothesis: These unusual phenomena, including the divergence of rpoA and movement of the IR boundaries, are likely due to the failure to suppress illegitimate recombination during replication or repair of plastid DNA, both of which are dependent on recombination33.
The Whirly genes encode single stranded DNA binding proteins that suppress illegitimate recombination in Arabidopsis and maize34. We envision a scenario in which these or other proteins that normally suppress illegitimate recombination in plastids are either insufficiently expressed or compromised in their function. As a result of increased illegitimate recombination, the repeat content of affected plastomes increases, which in turn provides an increasing number of substrates for further illegitimate recombination. The process is brought to an end by increased expression or the spread of alleles that more effectively suppress illegitimate recombination.
As long as illegitimate recombination occurs, nothing precludes it occurring within protein-coding genes and affecting their evolution. As with point mutations, most illegitimate recombination events within protein-coding genes are likely to be deleterious and are subject to purifying selection. However, in the less constrained subset of protein-coding genes that includes rpoA, the outcomes of some of these events are more likely to be neutral and arrive at fixation. The unparalleled divergence of the rpoA genes in the four lineages discussed here suggests that they evolved not simply through an accumulation of single nucleotide substitutions but also through at least one mechanism capable of causing multiple coincident substitutions and indels. Short homology-dependent illegitimate recombination, as seen in Whirly mutants, induces these types of mutations34.
Material and Methods
Taxon sampling
Taxon sampling included representatives of the Annonaceae, Geraniaceae, Passifloraceae and associated outgroups (Table 3). For some species of Geraniaceae plastomes have already been completed and published15,21,28,31,35 and gene sequences were extracted from Genbank. For other Geraniaceae and for Passifloraceae, genes were extracted from draft plastomes and individual gene sequences have been submitted to GenBank (Table 3).
DNA isolation
Total genomic DNA used for all newly generated sequences was extracted by a modified version (including the use of 2% PVP in the extraction buffer) of the hexadecyltrimethylammonium bromide protocol from Doyle & Doyle36.
Plastome sequencing, assembly and annotation
Sequencing of Passiflora cirrhiflora (454), P. quadrangularis and P. biflora (Sanger) was carried out using products of rolling circle amplification of purified plastomes as described in Jansen et al.37. Sanger sequence reads were assembled using consed38 and 454 reads utilized Newbler39 and MIRA40 as described in Chumley et al.15 and Blazier et al.35. For Annona and Geraniaceae, total genomic DNA was sequenced on the Illumina HiSeq 2000 at the Genome Sequence and Analysis Facility (GSAF) at the University of Texas at Austin. Approximately 60 million 100 bp paired-end reads were generated from a sequencing library with ~750 bp inserts. Subsequent to filtering, raw reads were assembled de novo with Velvet v. 1.2.0741 using a range of kmer sizes from 71 to 93, with and without scaffolding enabled. Plastid contigs were identified by BLAST searches against a database of angiosperm plastid protein-coding genes using custom Python scripts. Nuclear and mitochondrial contigs containing plastid DNA insertions were excluded using 1000x coverage cutoff. Assembly and filtering were performed on the Lonestar Linux Cluster at the Texas Advanced Computing Center (TACC). For all genomes, initial annotation was performed with Dogma42 and annotations were checked by comparisons to other annotated plastid genes in Genbank using Geneious 7.0.4 (www.biomatters.com).
Reverse transcription PCR
Total RNA isolated from P. x hortorum was used for RT-PCR to detect transcription of the rpoA ORFs. Newly emergent leaves of Pelargonium x hortorum cv ‘Ringo White’ were collected from live plants grown in the University of Texas at Austin (UT) greenhouse and frozen in liquid nitrogen. Total RNA was isolated by the same protocol used in Zhang et al.20. Approximately 1 μg of P. x hortorum DNase-free RNA was thawed on ice and used as the template for reverse transcription PCR (RT-PCR). The RT reactions utilized ImProm-II™ Reverse Transcriptase (Promega, Madison WI) following the manufacturer’s protocol. For each reaction a control reaction was performed where no enzyme was added. rpoA mRNA sequences were reverse transcribed from within the rpoA ORFs. Products were amplified from the RT template with the forward primers located in the upstream genes, petD and rps11 (Fig. S2). Reverse transcription products, 3 μL each, were used as templates for PCR reactions using the Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Pittsburgh PA) according to the manufacturer’s protocol and MgCl2-free buffer. Magnesium chloride concentration was adjusted to 2 mM. Primers were designed manually to amplify transcripts of the two largest rpoA-like ORFs in P. x hortorum. All primer sequences were selected by visual inspection of the P. hortorum plastome sequence and are given in Table S1. Amplification products were Sanger sequenced at the Institute of Cellular and Molecular Biology core facility at the University of Texas at Austin.
Sequence alignment and rates analyses
Gene sequences were extracted from draft or complete plastomes using the default settings for plastid genes in DOGMA42, for rpoA sequences, the identity setting was lowered to 25%. All sequence editing and alignment was conducted in Geneious 7.0.4 (www.biomatters.com). Alignment of rpo genes was conducted using the L-INS-i algorithm in MAFFT as implemented in Geneious, as a single locally alignable block flanked by long terminal gaps was expected43. For other plastid genes, the MAFFT G-INS-i algorithm was used, as a global alignment without large terminal gaps was expected. Individual gene trees were constructed by the same methods as the seven-gene constraint trees described below.
Constraint trees for the three datasets (Annonaceae, Geraniaceae and Passiflora) were created using a concatenated nucleotide alignment of seven plastid genes (rpoA, rpoB, rpoC1, rpoC2, ndhF, matK and rbcL). For Geraniaceae, Clade C2 species were omitted due to the presence of multiple rpoA paralogs. Constraint trees were generated by Garli44 using the GTR model in Geneious. Codon alignments were created using MAFFT in Geneious. For the Pelargonium data set, three additional alignment algorithms (CLUSTALW, MUSCLE and the Geneious aligner) were used in order to control for alignment error with difficult sequences45,46. All dN/dS ratios were calculated using the lineage specific seven-gene constraint tree.
Plastid genes were analyzed with codon-based models to quantify the rates of synonymous (dS) and nonsynonymous (dN) substitution. Analyses were conducted in PAML47 4.7 on the Lonestar Linux Cluster at TACC using custom Python scripts. Codon frequencies were calculated by the F3×4 model, and a free-ratio model was used to compute dN/dS values. Transition/transversion and dN/dS ratios were estimated with the initial values of 2 and 0.4, respectively48,49. A dN/dS ratio of 50 was selected as an arbitrary cutoff over which a value was assumed to be an artifact.
Promoter analysis
The upstream regions of psbA and rbcL were aligned by MAFFT in Geneious and conserved PEP promoter elements were annotated in accordance with Gruissem and Zurawski50. Upstream regions of Cuscuta obtusiflora, a parasitic plant lacking PEP, were included for comparison.
Conserved domain prediction
Conserved domains in rpoA-like ORFs were predicted by the Conserved Domain Database at NCBI (CDD v.3.10) at an E-value of 0.01 and low-complexity filters applied51.
Detection of gene conversion
Gene conversion among Pelargonium rpoA-like ORFs was investigated both manually and using the ORGCONV algorithm22. For manual detection, the alignment was inspected for SNPs shared by two or three rpoA paralogs in a single species that were not shared across paralogs in multiple species.
Additional Information
How to cite this article: Blazier, J. C. et al. Divergence of RNA polymerase α subunits in angiosperm plastid genomes is mediated by genomic rearrangement. Sci. Rep. 6, 24595; doi: 10.1038/srep24595 (2016).
Supplementary Material
Acknowledgments
Support was provided by the National Science Foundation (IOS-1027259 to RJK and TAR and a NSF GRF predoctoral fellowship to JCB) and from Vice President for Educational Affairs Abdulrahman O. Alyoubi at King Abdulaziz University, Jeddah, Saudi Arabia, to JCB, RKJ and JSMS. The authors thank Mike Moore, the Soltis Lab, Anne K. Hansen and Joshua McDill for unpublished sequence data, Jin Zhang for custom Python scripts and Robin Parer at Geraniaceae.com for providing living material of Geraniaceae species. We also thank the Genome Sequencing and Analysis Facility at the University of Texas at Austin for performing the Illumina sequencing, the Texas Advanced Computing Center at the University of Texas at Austin for access to supercomputers and TEX for serving as a repository for voucher specimens.
Footnotes
Author Contributions J.C.B. sequenced and annotated the Annona plastid genome, designed and performed analyses, co-wrote the manscript and produced figures; T.A.R. isolated DNAs from Annona and Pelargonium, performed RT-PCR experiment, performed bioinformatic analyses of PEP promotors, assisted in the design of analyses, co-wrote the manuscript and produced figures; M.-L.W. provided Pelargonium sequences and assisted in rate analyses; S.K.R. assisted in the collection of Passiflora plastid genome and gene sequence data; J.S.M.S. assisted in the design of analyses and edited the manuscript; R.K.J. assisted in the design of analyses and edited the manuscript and figures. All authors read and approved the final manuscript.
References
- Downie S. R. & Palmer J. D. in Molecular Systematics of Plants (eds Soltis P. S., Soltis D. E. & Doyle J. J.) Ch. 2, 14–35 (Springer, 1992). [Google Scholar]
- Doyle J. J., Doyle J. L. & Palmer J. D. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot. 20, 272–294 (1995). [Google Scholar]
- Gantt J. S., Baldauf S. L., Calie P. J., Weeden N. F. & Palmer J. D. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10, 3073–3078 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. In Cell and Molecular Biology of Plastids (ed Bock R.) Vol. 19, 29–63 (Springer, 2007). [Google Scholar]
- Millen R. S. et al. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 13, 645–658 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K. et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104, 19369–19374 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T. A. & Jansen R. K. In Chloroplast Biotechnology: Methods and Protocols (ed Maliga P.) Ch 1, 3–38 (Humana Press, 2014). [Google Scholar]
- Liere K. & Borner T. In Cell and Molecular Biology of Plastids (ed Bock R.) Vol. 19, 121–174 (Springer, 2007). [Google Scholar]
- Delannoy. E., Fujii S., Colas des Francs-Small C., Brundrett M. & Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 28, 2077–2086 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S. et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 25, 3711–3725 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serino G. & Maliga P. RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 117, 1165–1170 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K., Berg S. & Krupinska K. Plastid transcription in the holoparasitic plant genus Cuscuta: Parallel loss of the rrn16 PEP-promoter and of the rpoA and rpoB genes coding for the plastid-encoded RNA polymerase. Planta. 216, 815–823 (2003). [DOI] [PubMed] [Google Scholar]
- Wickett N. J. et al. Transcriptomes of the parasitic plant family Orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr Biol. 21, 2098–2104 (2011). [DOI] [PubMed] [Google Scholar]
- Stefanovic S., Kuhlman P., Calie P. & Palmer J. D. Rapid evolution of plastid RNA polymerases in three unrelated flowering plant lineages. Botany and Plant Biology Joint Congress, Chicago, IL USA http://2007.botanyconference.org/engine/search/ index.php?func=detail&aid=1823, date of access 06/01/2015 (2007).
- Chumley T. W. et al. The complete chloroplast genome sequence of Pelargonium x hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 23, 2175–2190 (2006). [DOI] [PubMed] [Google Scholar]
- Shi C. et al. Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: An exemplary study of ycf15 function and evolution in angiosperms. PLos ONE. 8, e59620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura C., Kobayashi Y., Aoki S., Sugita C. & Sugita M. Complete chloroplast DNA sequence of the moss Physcomitrella patens: Evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucl Acids Res. 31, 5324–5331 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet B., Wickett N. J., Shaw A. J. & Cox C. J. Phylogenetic significance of the rpoA loss in the chloroplast genome of mosses. Taxon. 54, 353–360 (2005). [Google Scholar]
- Jansen R. K., Saski C., Lee S.-B., Hansen A. K. & Daniell H. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28, 835–847 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ruhlman T. A., Mower J. P. & Jansen R. K. Comparative analyses of two Geraniaceae transcriptomes using next-generation sequencing. BMC Plant Biol. 13, 228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ruhlman T. A., Sabir J. S. M., Blazier J. C. & Jansen R. K. Coordinated rates of evolution between interacting plastid and nuclear genes in Geraniaceae. Plant Cell. 27, 563–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W. OrgConv: detection of gene conversion using consensus sequences and its application in plant mitochondrial and chloroplast homologs. BMC Bioinformatics. 11, 114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M. C. & Hallick R. B. Chloroplast rpoA, rpoB, and rpoC genes specify at least three components of a chloroplast DNA-dependent RNA polymerase active in tRNA and mRNA transcription. J Biol Chem. 263, 14302–14307 (1988). [PubMed] [Google Scholar]
- Casola C. & Hahn M. W. Gene conversion among paralogs results in moderate false detection of positive selection using likelihood methods. J Mol Evol. 68, 679–687 (2009). [DOI] [PubMed] [Google Scholar]
- Schneider A. et al. Estimates of positive Darwinian selection are inflated by errors in sequencing, annotation, and alignment. Gen Biol Evol. 1, 114–118 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. The complete chloroplast genome sequence of Mahonia bealei (Berberidaceae) reveals a significant expansion of the inverted repeat and phylogenetic relationship with other angiosperms. Gene. 528, 120–131 (2013). [DOI] [PubMed] [Google Scholar]
- Kim. Y.-D. & Jansen R. K. Characterization and phylogenetic distribution of a chloroplast DNA rearrangement in the Berberidaceae. Plant Syst Evol. 190, 157–185 (1994). [Google Scholar]
- Weng M.-L., Blazier J. C., Govindu M. & Jansen R. K. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats and nucleotide substitution rates. Mol Biol Evol. 31, 645–659 (2014). [DOI] [PubMed] [Google Scholar]
- Perry A. S. & Wolfe K. H. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J Mol Evol. 55, 501–508 (2002). [DOI] [PubMed] [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L. & Jansen R. K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol Biol Evol. 28, 583–600 (2011). [DOI] [PubMed] [Google Scholar]
- Guisinger M. M., Kuehl J. V., Boore J. L. & Jansen R. K. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci USA. 105, 18424–18429 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard-Kubow K., Sloan D. B. & Galloway L. F. Correlation between sequence divergence and polymorphism reveals similar evolutionary mechanisms acting across multiple timescales in a rapidly evolving plastid genome. BMC Evol Biol. 14, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A. & Madesis P. In Cell and Molecular Biology of Plastids (ed Bock R.) Vol. 19, 65–119 (Springer, 2007). [Google Scholar]
- Maréchal A. et al. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc Natl Acad Sci USA. 106, 14693–14698 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazier J. C., Guisinger M. M. & Jansen R. K. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol Biol. 76, 263–272 (2011). [DOI] [PubMed] [Google Scholar]
- Doyle J. J. & Doyle J. L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19, 11–15 (1987). [Google Scholar]
- Jansen R. K. et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Method Enzymol. 395, 348–384 (2005). [DOI] [PubMed] [Google Scholar]
- Gordon D., Abajian C. & Green P. Consed: A graphical tool for sequence finishing. Genome Res. 8, 195–202 (1998). [DOI] [PubMed] [Google Scholar]
- Margulies M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 437, 376–380 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B., Wetter T. & Suhai S. Genome sequence assembly using trace signals and additional sequence information. Proc German Conf Bioinformatics. 99, 45–56 (1999). [Google Scholar]
- Zerbino D. R. & Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S. K., Jansen R. K. & Boore J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20, 3252–3255 (2004). [DOI] [PubMed] [Google Scholar]
- Katoh K., Asimenos G. & Toh H. In Bioinformatics for DNA Sequence Analysis (ed Posada D.) Vol 537, 39–64 (Humana Press, 2009). [Google Scholar]
- Zwickl D. J. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. (2008) Available at: http://repositories.lib.utexas.edu/handle/2152/2666, date of access 12/10/2014.
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- Sloan D. B., Oxelman B., Rautenberg A. & Taylor D. R. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol. 9, 260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M.-L., Ruhlman T. A., Gibby M. & Jansen R. K. Phylogeny, rate variation, and genome size evolution of Pelargonium (Geraniaceae). Mol Phylogenet Evol. 64, 654–670 (2012). [DOI] [PubMed] [Google Scholar]
- Gruissem W. & Zurawski G. Analysis of promoter regions for the spinach chloroplast rbcL, atpB and psbA genes. EMBO J. 4, 3375 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.