Abstract
We recently demonstrated that acute myeloid leukemia (AML) cell lines and patient-derived blasts release exosomes that carry RNA and protein; following an in vitro transfer, AML exosomes produce proangiogenic changes in bystander cells. We reasoned that paracrine exosome trafficking may have a broader role in shaping the leukemic niche. In a series of in vitro studies and murine xenografts, we demonstrate that AML exosomes downregulate critical retention factors (Scf, Cxcl12) in stromal cells, leading to hematopoietic stem and progenitor cell (HSPC) mobilization from the bone marrow. Exosome trafficking also regulates HSPC directly, and we demonstrate declining clonogenicity, loss of CXCR4 and c-Kit expression, and the consistent repression of several hematopoietic transcription factors, including c-Myb, Cebp-β and Hoxa-9. Additional experiments using a model of extramedullary AML or direct intrafemoral injection of purified exosomes reveal that the erosion of HSPC function can occur independent of direct cell–cell contact with leukemia cells. Finally, using a novel multiplex proteomics technique, we identified candidate pathways involved in the direct exosome-mediated modulation of HSPC function. In aggregate, this work suggests that AML exosomes participate in the suppression of residual hematopoietic function that precedes widespread leukemic invasion of the bone marrow directly and indirectly via stromal components.
INTRODUCTION
The bone marrow (BM) is a complex tissue, uniquely adapted to regulate the function of hematopoietic stem and progenitor cells (HSPC) during homeostasis, disease and in response to injury.1 Many patients with acute myeloid leukemia (AML) present with cytopenias at diagnosis or during relapse, often without significant marrow effacement.2,3 Recent data from a xenograft model suggests that invasion of the BM by AML cells actively suppresses hematopoietic progenitor differentiation, but a mechanism for an altered HSPC function in the leukemic niche remains elusive.4,5
Exosomes are secreted vesicles (30–110 nm) of endocytic origin that traffic RNA and protein between cells.6 We previously characterized exosome biogenesis in AML blasts (primary cells, Molm-14 and HL-60 cell lines) and identified a range of coding and noncoding RNAs relevant to disease pathogenesis.7 We hypothesized that AML exosomes may contribute to the functional suppression of HSPC in the leukemic BM microenvironment. To examine the potential contributions of exosome trafficking to hematopoietic suppression in the BM, we studied the engraftment of Molm-14 cells, a highly penetrant model of high-risk FLT3-ITD+ AML8 and HL-60 cells, a predominantly extramedullary model of promyelocytic leukemia, in NOD/SCID/IL-2rγnull (NSG) mice. In comparison with AML patient samples, this provided a more cohesive platform for adaptation to the tissue culture environment, which was necessary in order to experimentally distinguish the exosome impact on different niche cell types from conventional juxtacrine or paracrine signaling mechanisms.9
A critical feature for in vitro modeling of HSPC behavior in the BM is the compartmental oxygen (O2) concentration (1–6%), hypoxic compared with the bloodstream or the tissue culture environment (20%).10 Because hypoxia not only exerts a significant influence on HSPC function and leukemogenesis but also on exosome biogenesis,11,12 we performed in vitro experiments at physiologically appropriate oxygen conditions.13
Our Molm-14 xenograft studies show systematic functional alterations in murine stromal and hematopoietic cell populations, with evidence for in vivo transfer of human RNA transcripts. We replicated these results using an extramedullary HL-60 model of AML and direct intrafemoral injection of purified exosomes. The involvement of exosomes in the suppression of canonical hematopoietic cell function is further supported by extensive in vitro experiments and proteomics data that identify several putative targets mediating these changes in HSPC function. AML exosomes appear to dysregulate HSPC both directly and indirectly via stromal components.
MATERIALS AND METHODS
Cells, cell lines and low-oxygen cell culture
Molm-14, HL-60 and OP9 cells were previously described.7 For low-O2 culture, cells were cultured in RPMI (Life Technologies, Grand Island, NY, USA) with 10% vesicle-free (VF) fetal bovine serum (FBS) using a G-Rex gas-permeable flask (Wilson-Wolf Corp, St Paul, MN, USA) in a BioSpherix chamber (Lacona, NY, USA) at 1–3% O2 or a standard incubator at 20% O2 and at 5% CO2. VF FBS was produced by centrifugation (Gemini Bio-Products, West Sacramento, CA, USA) at 100 000 g for 6 h. Primary AML cells were maintained in EGM-2 media (Lonza, Allendale, NJ, USA) with OHSU IRB-approved protocols. Human CD34+ cord-blood progenitors (New York Blood Center) were enriched using MACS cell separation (Miltenyi Biotec, San Diego, CA, USA) and cultured in serum-free media (StemCell Technologies, Vancouver, BC, Canada) supplemented with 100 U/ml penicillin/streptomycin, 40 ng/ml FLT3L, 25 ng/ml stem cell factor (SCF) and 50 ng/ml thrombopoietin (Miltenyi Biotec).
Exosome preparation and RNA extraction
As described,7 AML cells were cultured for 48 h, media spun at 300 g for 10 min, supernatant at 2000 g for 20 min and 10 000 g for 20 min followed by supernatant centrifugation at 100 000 g for 2 h. Exosome pellets were resuspended in 10% VF-FBS/RPMI used in all in vitro experiments or used for RNA extraction. In xenograft and IF experiments, exosomes were resuspended in Hank's balanced salt solution media (Life Technologies). Media from exosome preparations after spinning at 10 000g is defined as exosome-containing media (ECM). An amount of 2 ml of ECM was cultured with 3 × 104 OP9 per well in a six-well plate (4.8 × 109 Molm-14 exosomes/well per nanoparticle tracking analysis (NTA) analysis). Concentrated exosomes were resuspended in 2 ml of 10% VF-FBS RPMI.
Murine xenograft studies
NSG xenograft recipients (6–8-week old) were used with IACUC approval. Conditioned Molm-14 cells (1 × 105), cord-blood CD34+ cells or 5 × 106 HL-60 cells were resuspended in Hank's balanced salt solution media and injected via tail vein. Hank's balanced salt solution medium was used as vehicle control in all xenograft experiments. Human CD45 chimerism (BioLegend, HI30, San Diego, CA, USA) was monitored by flow cytometry. Animals were killed at 3–5-weeks post engraftment, and peripheral blood (PB) and BM were collected. Adherent BM stromal cells were propagated in Iscove's MDM (Life Technologies) with 10% VF FBS (detailed description in Supplementary Materials and Methods).
Intrafemoral injection (IF)
For a modified IF procedure,14,15 AML exosomes (5.8–6.8 × 1011 Molm-14 exosomes or 5.2–6.0 × 1011 HL-60 exosomes per NTA quantification) were injected into one femur of isoflurane-anesthetized animals; Hank's balanced salt solution vehicle control was injected in the contralateral femur. Animals were killed 48 h later for BM collection and c-Kit+ progenitor cell enrichment (detailed description in Supplementary Materials and Methods).
RNA analysis and qRT-PCR
RNA was extracted using miRNeasy or RNeasy (Qiagen, Valencia, CA, USA) and quantified using a Nanodrop 2000c (Thermo Scientific, Grand Island, NY, USA) and Agilent Bioanalyzer (Agilent, Santa Clara, CA, USA). cDNA was synthesized using a SuperScript III First Strand Synthesis kit (Invitrogen, Grand Island, NY, USA) with oligo-dT priming, followed by PCR. SYBR Green PCR (Applied Biosystems, Grand Island, NY, USA) was used for quantitative PCR with reverse transcription (qRT-PCR) analysis. The ΔΔCT method was used for quantification. Species-specific primers are listed at: http://www.ohsu.edu/xd/health/services/doernbecher/research-education/research/research-labs/kurre-lab-protocols.cfm.
Nanoparticle tracking analysis
Exosome samples were resuspended and serial dilutions were prepared in nanofiltered (Whatman Anotop 25, Piscataway, NJ, USA, 0.02 μm) molecular-grade water (Thermo Scientific) using low-adhesion 1.7-ml tubes (Genemate, Kaysville, UT, USA). Diluted samples (1 × 108–1 × 109 particles/ml) were loaded into the NanoSight LM10 chamber, the laser engaged and microparticles visualized. Sixty second videos were acquired with a Hamamatsu (Bridgewater, NJ, USA) C11440 ORCA-Flash 2.8 camera and analyzed by Nanosight NTA (Malvern, UK) 2.3 software.
In vitro co-culture and colony-forming unit assay
BM and PB from NSG or C57BL/6 mice were harvested, resuspended in hemolytic buffer and peripheral blood mononuclear cells (PBMCs) collected by centrifugation.16 For enrichment, c-Kit-PE antibody (BD Biosciences, San Jose, CA, USA, clone 2B8) and EasySep Mouse PE Selection kits (StemCell Technologies) were used (90–93% mCD45/c-Kit purity confirmed). Murine c-Kit-enriched cells were cultured in Iscove's Modified Dulbecco's Medium/10% VF FBS per 50 ng/ml mIL-3 and mSCF (R&D Systems, Minneapolis, MN, USA). c-Kit+ cells (1 × 106) were incubated with exosomes from 6–7 × 107 Molm-14 or HL-60 cells (at 5.8–6.8 × 1011 exosomes per well from Molm-14 cells or 5.2–6.0 × 1011 exosomes per well from HL-60 per NTA analysis), 2.5–4 × 107 primary AML cells or 3–3.5 × 107 human CD34+ cells for 48 h with VF media controls. Mouse methylcellulose complete media (R&D Systems) was used for colony-forming unit, CFU-C, assays in triplicate with 3 × 103 c-Kit+ cells or 2 × 105 PBMCs per 35-mm dish (37 °C, 5% CO2, ≥ 95% humidity for 7 days). Human CD34+ cells were cultured in serum-free media, with cytokines as above. Human methylcellulose media (StemCell Technologies) was used for CFU-C assays with 100 CD34+ cells/35-mm dish cultured for 14 days).
Cell migration assay
Lineage-depleted (Lin-) BM (Progenitor enrichment kit, StemCell Technologies) was cultured in triplicate at 5 × 105 cells in 2 ml Iscove's Modified Dulbecco's Medium, 10% VF-free FBS, 50 ng/ml SCF and IL-3 for 24 h ± 1 ml of ECM (at 2.4 × 109 Molm-14 exosomes/well per NTA analysis). Washed cells were placed on 8-μm transwell inserts (Corning, Corning, NY, USA) in plates containing media ± CXCL12 (50 ng/ml). After 2 h, transwells were removed and migrated cells were counted using a Guava PCA (Millipore, Billerica, MA, USA).
Immunohistochemistry
Femurs were fixed in 10% formalin for 24 h at 4 °C, transferred to cold 70% ethanol, embedded in paraffin and sectioned at 5 μm thickness. Slides were rehydrated, high-pressure treated, blocked and stained with mouse anti-human CD45 (Clone HI30, BioLegend) at 1:150 dilution and biotinylated anti-mouse IgG (mouse IgG1 isotype control). Images were taken with a Leica ICC50 microscope (Buffalo Grove, IL, USA) with high-definitixon camera (×20 lens) and processed with Adobe Photoshop (San Jose, CA, USA).
Flow cytometry analysis
CD45 chimerism was analyzed on Calibur/ CANTO/ LSR II cytometers (BD Biosciences) using anti-human or anti-mouse CD45 (BioLegend), murine c-Kit+ (BioLegend) or CXCR4 (Clone 2811, eBioscience, San Diego, CA, USA) antibodies, and analyzed with FlowJo (Tree Star, Ashland, OR, USA).
Fluorescence microscopy
Lin- cells were exposed to exosomes from N-Rh-PE-labeled Molm-14, washed twice in phosphate-buffered saline and labeled with APC-anti-Sca-1 (Clone D7, BioLegend) and PacBlue-anti-c-Kit (Clone 2B8, BioLegend). Fluorescence microscopy was performed on an Olympus IX71 (Olympus, Waltham, MA, USA) with a × 60 1.4 NA oil lens; Z-stacks were acquired every 0.2 μm with a Z-stack center bright-field reference, and images processed with DeltaVision SoftWoRx Explorer (Piscataway, NY, USA).
Tandem Mass Tag spectrometry analysis
NSG mouse c-Kit+ cells (1×106) were co-cultured with AML exosomes from 6–7 × 107 Molm-14 cells (~5.8–6.8 × 1011 Molm-14 exosomes per well) in the presence of cytokines (SCF, IL-3, 50 ng/ml) at 3% O2 for 48 h. Cells were washed, pelleted and snap-frozen. Proteins were digested with trypsin, peptides solid-phase extracted17 and digests labeled with tandem mass tag (TMT) 10-plex reagents18 (Thermo Scientific). Followed by mixing, labeled digests were separated into multiple fractions by high pH reverse-phase chromatography followed by nano-reverse-phase chromatography at low pH. Peptides were analyzed during elution using an Orbitrap Fusion mass spectrometer (Thermo Scientific) to measure TMT reporter ion intensities. Results were processed with Protein Discoverer 1.419 (Thermo Scientific) to identify peptides (default TMT template using 16 604 mouse Sprot v2013.04 sequences) and export reporter ion intensities. Summed reporter ion intensities from unique peptides per protein were analyzed using edgeR (Bioconductor, Seattle, WA, USA) with multiple test correction to calculate false-discovery rates for differential candidates.20 Further details of the TMT analysis and results are provided in Supplementary Material and Methods.
Statistical analysis
Continuous variables are summarized as mean ± s.d. A two-sample t-test was employed for comparison between samples derived from the same source, but different conditions. For comparisons between cells and exosomes, or left versus right femur IF injection, a paired t-test was used. With the exception of TMT analysis, statistical significance was set at P ≤ 0.05.
RESULTS
AML xenografts and exosomes modulate BM compartmental signaling
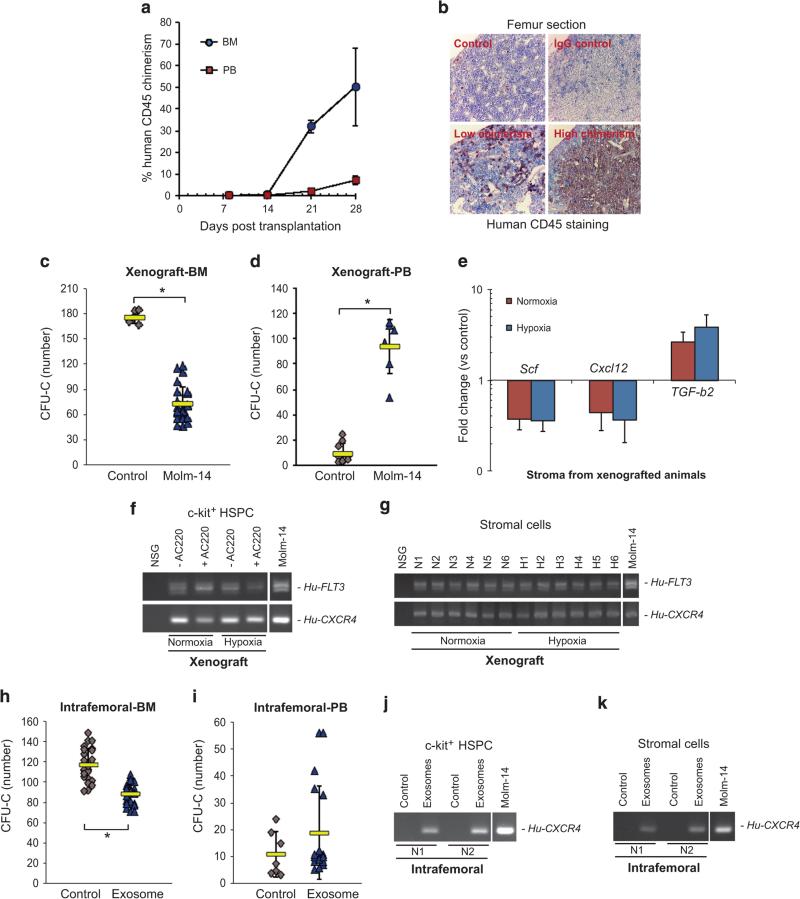
In a series of studies, we established xenografts of Molm-14 AML as a highly penetrant model, observing consistent kinetics across multiple cohorts, critical for downstream correlative in vitro experiments8 (Figure 1a). Animals killed at day 28 showed highly reproducible metaphyseal effacement of the femur cavity by Molm-14 cells (chimerism >50%), unlike animals studied at day 21 (chimerism < 5%) (Figure 1b). In all subsequent analyses we therefore focused on the earlier (day 21) time point. Here, we found a significant reduction (P < 0.01) in methylcellulose colonies (CFU-C) (Figure 1c) with a concomitant increase in PB CFU-C formation (P < 0.01) (Figure 1d). To demonstrate exosome trafficking to HSPC and stromal cells, we harvested c-Kit+ cells and stroma from the BM of animals with low xenograft burden, and controls. We treated the cells with a potent FLT3 inhibitor, AC220 (10 nm) for 96 h to eliminate potential Molm-14 cell contamination and confirmed the elimination of human cells by flow cytometry (Supplementary Figures S1A and B). We then isolated RNA from both c-Kit+ and BM stromal cells, and using RT-PCR, detected human leukemia cell-derived transcripts in c-Kit+ progenitors (Figure 1f) and BM stroma (Figure 1g) of engrafted animals, but not controls. Having demonstrated transfer of RNA between AML blasts and cells of the hematopoietic compartment, we explored potential mechanisms for the loss of progenitors from the marrow. qRT-PCR analysis of xenograft recipient BM stromal cells revealed that expression of the critical HSPC retention factors Scf and Cxcl12 was downregulated (Figure 1e). Next, to examine the direct involvement of leukemia-derived exosomes in HSPC mobilization, we injected purified Molm-14 exosomes into the femoral cavity with contralateral injection of vehicle control in the same animal. We observed a significant reduction in colony-forming capacity in the exosome-injected BM (paired t-test, P < 0.01) (Figure 1h). We replicated these findings with IF injection of exosomes from primary patient samples (n = 3) and similarly observed a consistent impairment in progenitor cell clonogenicity (P < 0.05) (Supplementary Fig. S2A). Remarkably, for a one-time injection in only one femur, we found a correlation with increased progenitor frequency in the peripheral blood (Figure 1i), along with human transcripts in both c-Kit+ cells and stroma isolated from IF-injected animals (Figures 1j and k). These results demonstrate that a minority of leukemia cells present in the BM suppress critical stromal growth and retention factors, causing progenitor redistribution to the PB. Several of our observations implicate AML exosomes.
Figure 1.
AML xenografts and exosomes modulate BM compartmental signaling. (a) BM and PB chimerism were measured by flow cytometry for human CD45+ cells. Data from 6 independent experiments with total 67 animals engrafted with either hypoxia- or normoxia-conditioned Molm-14. The results are presented as mean ± s.e.m. (b) Femur sections of engrafted mice were examined by IHC. Low chimerism: < 5% huCD45+; high chimerism: > 50% huCD45+ in the BM. (C-G) NSG mice were engrafted with Molm-14 (Xenograft) (n = 10) or (H-K) exposed to Molm-14-derived exosomes through IF injection (Intrafemoral) (n = 8). (c and d) CFU-C assays were performed on c-Kit+ progenitor cells and PBMC isolated from xenografted animals. (e) The stromal regulatory gene profile in xenografted recipient BM stroma was evaluated by qRT-PCR. (f and g) Transfers of human FLT3 and CXCR4 in AC220-treated c-Kit+ cells and recipient BM stroma were determined by RT-PCR. N1 to N6 and H1 to H6 represent the individual animal. (H-I) CFU-C assays were performed on c-Kit+ progenitor cells and PBMC from IF-injected animals. (j and k) The presence of human transcripts (FLT3 and CXCR4) in c-Kit+ progenitor and BM stromal cells from IF-injected animals was examined by RT-PCR. Vehicle medium was used as the control. Data are representative of 6 independent xenograft experiments and four independent IF experiments. The results are presented as mean ± s.d. *P < 0.01.
AML cells increase exosome production under physiological oxygen conditions attenuating stromal cell expression of HSC maintenance factors
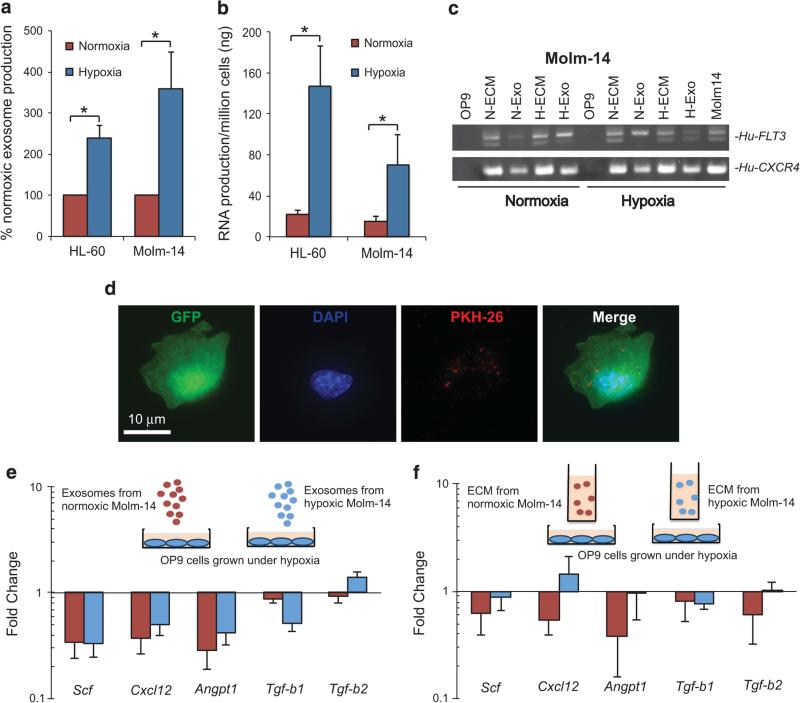
Low oxygen concentrations are central to both HSPC function and leukemogenesis.1,10 Here, we examined the release of exosomes by AML cells under controlled low-oxygen conditions (1% O2) while maintaining optimum growth rates and viability (Supplementary Figure S3A).21 Exosomes released from both Molm-14 and HL-60 cells were characterized by NTA (Supplementary Figures S4A and B), with exosome release and average RNA concentration per cell increasing 2–3-fold compared with conventional tissue culture O2 levels (Figures 2a and b). Under these compartmentally appropriate O2 conditions we studied AML exosome transfer to OP9 murine BM stromal cells, thereby enabling us to distinguish AML exosome- induced signaling from contact-dependent and other paracrine effects. we cultured OP9 with ECM, containing both exosomes and soluble media factors, or purified exosomes collected from Molm-14 cells grown under normoxia or hypoxia, respectively. Consistent with our xenograft studies, RT-PCR analysis of OP9 RNA demonstrated the transfer of human FLT3 and CXCR4 transcripts in vitro (Figure 2c). We further corroborated AML exosome entry into OP9 cells in vitro using fluorescently (PKH26) labeled exosomes (Figure 2d).7 Next, we investigated whether exosomal RNA transfer affects the expression of genes important in the stromal regulation of hematopoiesis (Scf, Cxcl12, Angpt1, Tgfb1 and Tgfb2).22 Our data demonstrate a significant decrease in the expression of these regulatory genes except Tgfb2 (Figures 2e and f). We further studied and confirmed loss of Scf and Cxcl12 expression in primary murine BM stromal cells (Supplementary Figure S5A). Our data support the hypothesis that leukemic exosomes dysregulate stromal HSPC maintenance factors. Importantly, the data indicate that additional paracrine factors in ECM, predicted to be present in tissue compartments, modulate signaling outcomes, providing context for the differences seen between xenograft and IF injection studies.
Figure 2.
AML cells release more exosomes under physiological oxygen condition and AML exosomes attenuate stromal cell expression of HSC maintenance factors. (a) Exosome production from Molm-14 and HL60 under normoxic and hypoxic conditions was compared by nanoparticle tracking analysis. Data represents 6 independent experiments. *P < 0.01. (b) The exosome RNA concentration was determined by spectrophotometry. Data represents nine independent experiments *P < 0.05. (c) Transfer of transcripts from (human) Molm- 14 exosomes to (murine) OP9 cells was detected by qRT-PCR. N-ECM, H-ECM, N-Exo, H-Exo represent exosome-containing media (ECM) or exosomes (Exo) produced under normoxic (N) or hypoxic (H) conditions. (d) GFP-expressing OP-9 stromal cells were exposed to PKH-26- labeled Molm-14 exosomes for 3 h. (e and f) The stromal regulatory gene profile in OP9 under normoxia (red bar) or hypoxia (blue bar) after exposure to exosomes or ECM from Molm-14 cultured under hypoxia was evaluated by qRT-PCR normalized to GAPDH. Data represent three independent experiments.
AML exosomes modulate HSPC function directly
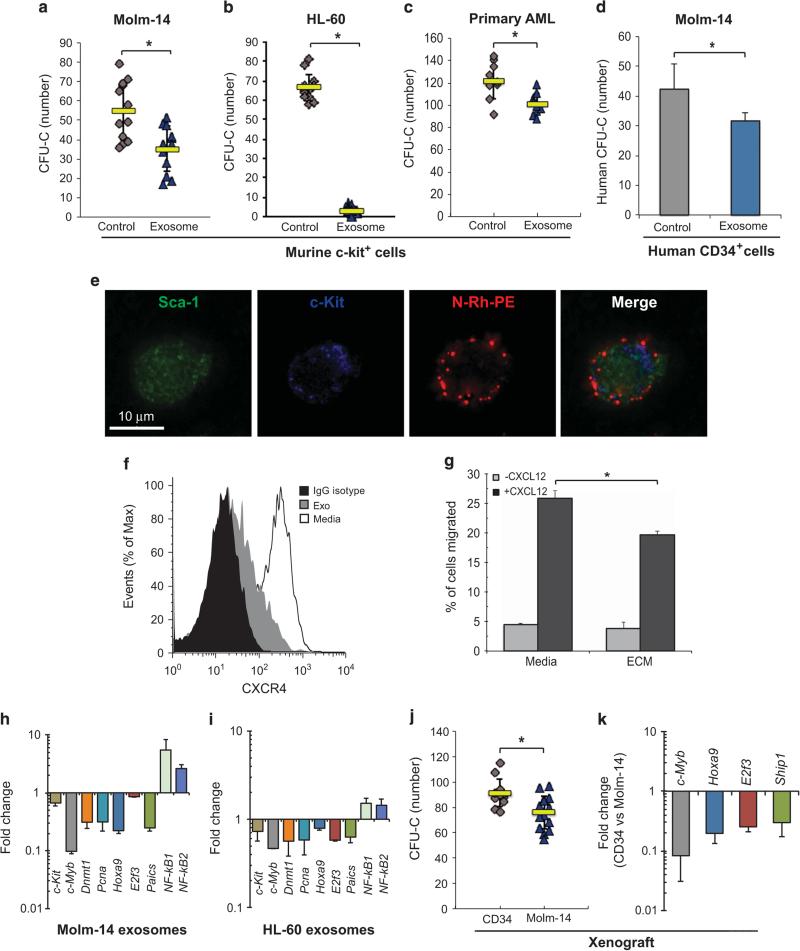
Xenograft and IF studies suggest that AML acts on the stroma to mobilize colony-forming progenitors from the BM. Here, we explored the direct impact of exosomes on HSPC in vitro and observed that the progenitor frequency was reduced by 36% after exposure to Molm-14 exosomes (Figure 3a), and even more strikingly upon exposure to HL-60 exosomes (Figure 3b), analogous to our in vivo observations. We also performed CFU assays with exosomes isolated from primary blasts of three AML patients, confirming a significant decrease (P < 0.01) in the colony-forming capacity of murine c-Kit+ cells (Figure 3c). These observations were not limited to murine progenitors, as human cord-blood CD34+ cells incubated with Molm-14 exosomes similarly showed a significant reduction in CFU-C (P = 0.02) (Figure 3d).
Figure 3.
AML exosomes modulate the function of HSPC. CFU-C assay for murine c-Kit+ progenitor cells after in vitro exposure to exosomes from Molm-14 (a) or HL-60 cells (b) cultured under hypoxia for 48 h. 10% VF-FBS medium was used as the control. (c) CFU-C for murine c-Kit+ progenitor cells after 48-hr in vitro exposure to exosomes from AML primary cells. (d) CFU-C for human CD34+ cells after 48 h in vitro exposure to exosomes from Molm-14. (e) Murine lin- cells were exposed to N-Rh-PE labeled Molm-14 exosomes overnight, labeled for Sca-1 and c-Kit, and imaged using deconvolution microscopy. (f) Expression of CXCR4 on lineage-depleted BM cells after 24-h exposure to Molm-14 exosomes. (g) Migration of lineage-depleted bone marrow cells along a CXCL12 gradient after 24-h culture with Molm-14 ECM or media alone. (h and i) Hematopoietic gene profile of c-Kit+ cells was examined by qRT-PCR after exposure to exosomes from Molm-14 or HL-60. (j) CFU-C for murine c-Kit+ progenitor cells collected from animals engrafted with Molm-14 (n = 6) or human CD34+ cells (n = 4). (k) Gene expression in c-Kit+ progenitor cells from xenografted animals, normalized to Gapdh. Data are representative of three independent experiments. *P < 0.01.
Given the broad reproducibility, we next validated the in vitro uptake of Molm-14 exosomes (N- Rh-PE-labeled) by murine Lin-/Sca-1+/c-Kit+ cells using deconvolution microscopy (Figure 3e) and tested key regulatory elements involved in HSPC localization. We observed that co-culture with Molm-14 exosomes caused a significant downregulation of CXCR4 in murine lineage-depleted BM cells (Figure 3f), resulting in a compromise of their ability to migrate along a CXCL12 gradient (Figure 3g). As further corollary, we observed changes in expression of a set of transcription factors with involvement in HSPC function (c-Kit, c-Myb, Dnmt1, Pcna, Hoxa9, E2f3, Paics, NF-kB1 and NF-kB2)23–29 following 48 h of culture with exosomes from Molm-14 or HL-60 cells (Figures 3h and i). To determine whether these changes occur in vivo, we returned to our xenograft model, using mice engrafted with human CD34+ (cord blood) cells as controls. After collecting c-Kit+ marrow progenitors, we confirmed significantly reduced colony formation in Molm-14 xenograft recipients (Po0.01) (Figure 3j). Finally, we tested the expression of hematopoietic transcription factors in c-Kit+ cells from the same cohorts and found that c-Myb, Hoxa9, E2f3 and Ship1 were also consistently downregulated in AML xenograft versus human CD34+ control animals (Figure 3k). Both in vitro and in vivo data show that AML exosomes not only downregulate the expression of stromal regulators of HSPC but also suppress hematopoietic function directly.
AML exosomes from extramedullary myeloid tumors traffic to the BM niche and dysregulate niche signaling
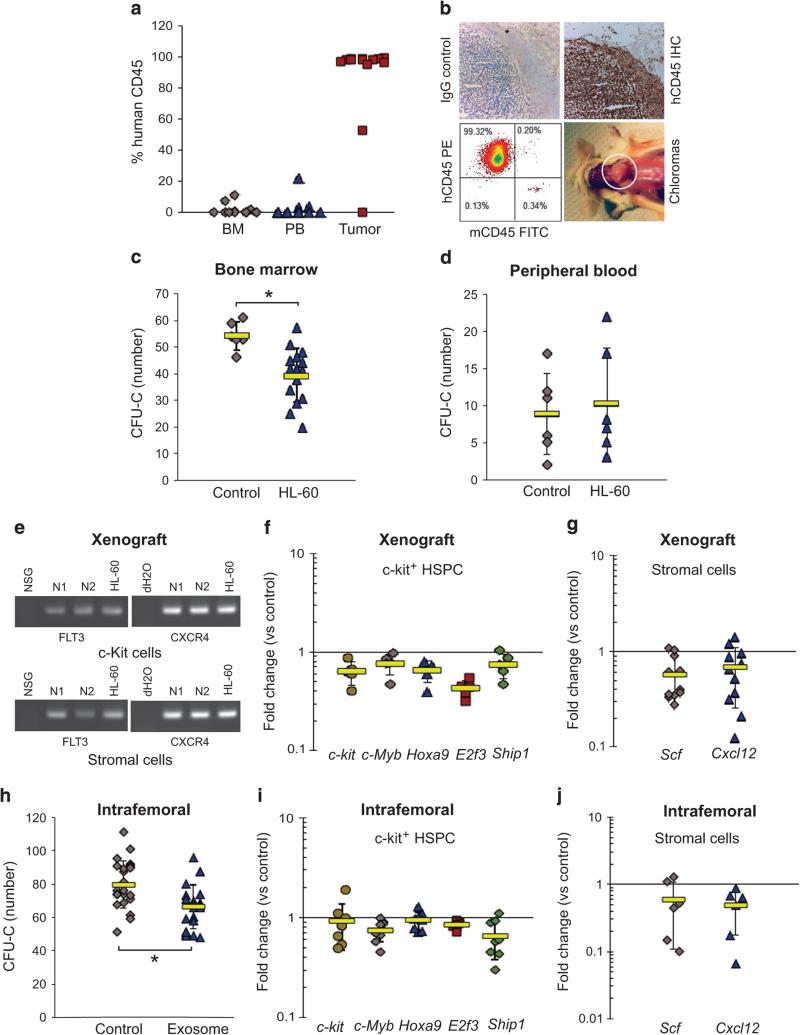
Next, we used xenografts of HL-60 leukemia cells that form predominantly extramedullary chloromas to broaden the context of our observations. Animals showed essentially no BM invasion as determined by immunohistochemistry and flow cytometry for human CD45 (Figures 4a and b), allowing a unique opportunity to rule out cell–cell contact contributions to the observed effects on the hematopoietic compartment. When we examined the function of murine c-Kit+ hematopoietic progenitor cells from the BM of HL-60 xenografted mice, we found a significant reduction (P < 0.01) in colony formation, with no detectable leukemia invasion of the BM (Figure 4c). The increase in PB progenitors in this model was more modest, perhaps suggesting that mobilization requires greater aggregate exposures to leukemia (that is, dose) (Figure 4d). Yet, we detected leukemia transcripts in recipient c-Kit+ progenitor cells and stroma, again consistent with trafficking of leukemia exosomes to BM (Figure 4e). Moreover, qRT-PCR analysis of c-Kit+ cells and BM stroma from xenograft animals revealed a 20–40% loss in expression of the hematopoietic transcription factor genes c-Kit, c-Myb, Hoxa9, E2f3 and Ship1 in HSPCs (Figure 4f), and the HSPC retention factor genes Scf and Cxcl12 (20–30%) in BM stroma were consistently downregulated (Figure 4g). When we injected purified HL-60 exosomes or vehicle control into recipient animal femurs, the paired statistical analysis of media- versus exosome- injected femur again showed significant CFU-C deficits in the exosome-injected BM (paired t-test, P < 0.01) (Figure 4h). qRT-PCR analysis of transcripts from these animals confirmed decreased expression of c-Kit, c-Myb, Hoxa9, E2f3 and Ship1 in c-Kit+ progenitor cells (Figure 4i) and Scf and Cxcl12 in BM stroma (Figure 4j). Results show that extramedullary myeloid tumors traffic human transcripts to BM cells, in turn dysregulating hematopoietic function.
Figure 4.
AML exosomes from extramedullary myeloid tumors dysregulate BM niche signaling. HL-60 cells were conditioned at 1% O2 before xenografting. (a) Bone marrow, peripheral blood and extramedullary tumor chimerism were measured by flow cytometry for human CD45. Data are representative of three independent experiments (n = 23). (b) Chloroma sections were examined by IHC and flow cytometry for human CD45. (c and d) CFU-C for murine c-Kit+ progenitor cells and PBMC after HL-60 xenograft. (e) Transfer of human FLT3 and CXCR4 into NSG c-Kit+ progenitor cells and stroma was detected by RT-PCR. Gene expression profiles in c-Kit+ progenitor cells (f) and stroma (g) from xenografted animals were examined by qRT-PCR. Data are representative of three independent experiments. (h) CFU-C on c-Kit+ progenitor cells from animals exposed to HL-60-derived exosomes through intrafemoral (IF) injection (n = 8). Vehicle medium was used as the control. (i and j) Expression of hematopoietic and stromal regulatory genes was quantified by qRT-PCR.
AML exosomes regulate HSPC via signaling networks
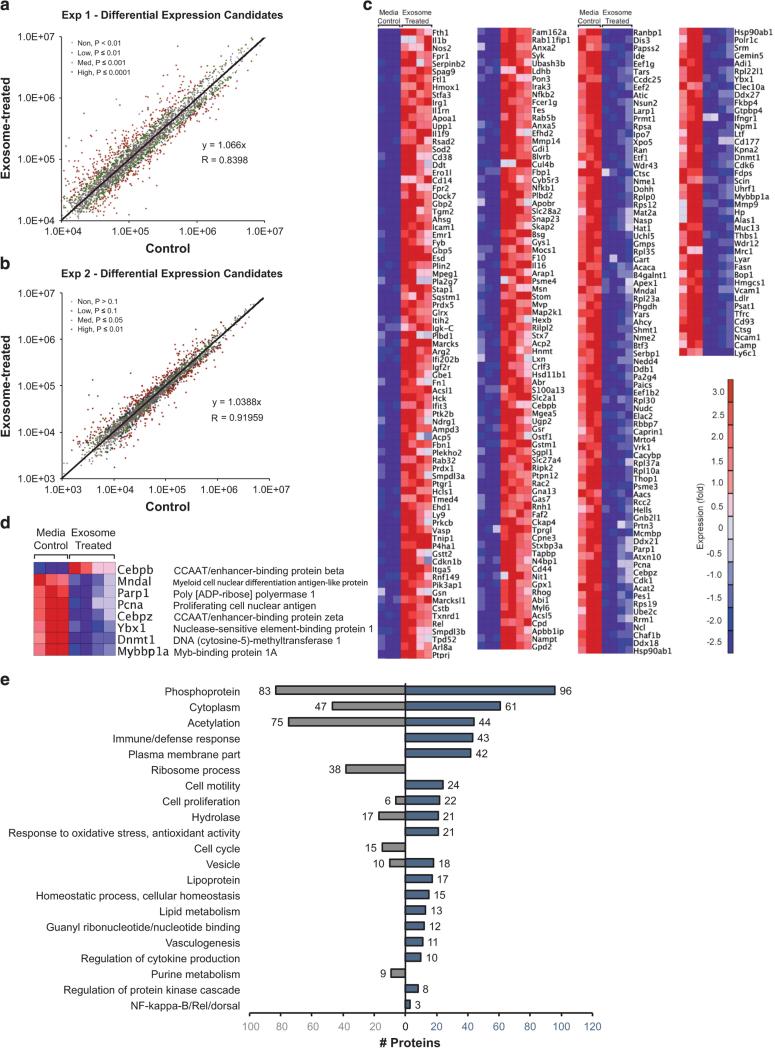
To better understand the cellular targets for AML exosomes that traffic to HSPC, we performed unbiased, high-throughput quantitative proteomic analysis. In two independent experiments, we compared relative protein abundances between c-Kit-enriched HSPC after exposure to Molm-14 exosomes versus media and identified 2837 and 2476 quantifiable proteins, respectively (Figures 5a and b). Across both experiments, 282 proteins were differentially expressed between exosome-treated and control cells, of which 161 were upregulated and 121 were down-regulated (Figure 5c). Because of their substantial effect on cells, we examined each data set for differential expression of transcription factors and found a total of seven downregulated and one upregulated (CEBPB) protein (Figure 5d). DAVID functional annotation analysis30,31 revealed that 27% (43/161) of the upregulated proteins are involved in immune processes (Figure 5e). Other prominent categories included acetylation, cell proliferation and antioxidant activity, all central to HSPC integrity (Figure 5e). A remarkable 31% (38/121) of the downregulated targets are involved in protein synthesis (Figure 5e). In the upregulated protein set, 15% are involved in cell motility, and 6% in cytokine production (Figure 5e).
Figure 5.
Differentially expressed proteins and gene ontology. We performed two TMT experiments to compare proteins in exosome-treated HSPC, compared with controls. From five control and eight exosome-treated samples across 2 experiments, we obtained 282 overlapping proteins differentially expressed (P ≤ 0.0001 for Exp. 1, P ≤ 0.10 for Exp. 2) between exosome- treated and control samples. Because of higher sampling of peptides, Exp. 1 yielded the highest quality data and was thus subjected to more stringent statistical criteria. (a and b) Averaged summed reporter ion intensities for proteins in exosome-treated versus control samples used for a normalization check of data from Exp. 1 and Exp. 2, respectively, showing differentially abundant proteins in color with indicated P-values. (c) Heat map of differentially expressed proteins in Molm-14 exosome-treated versus control HSPC from Exp. 1. (d) Relative protein expression of differentially expressed transcription factors. (e) Up- and downregulated gene ontology processes from DAVID (P < 0.001; see Supplementary File 1) for differently expressed proteins in exosome-treated versus control HSPC.
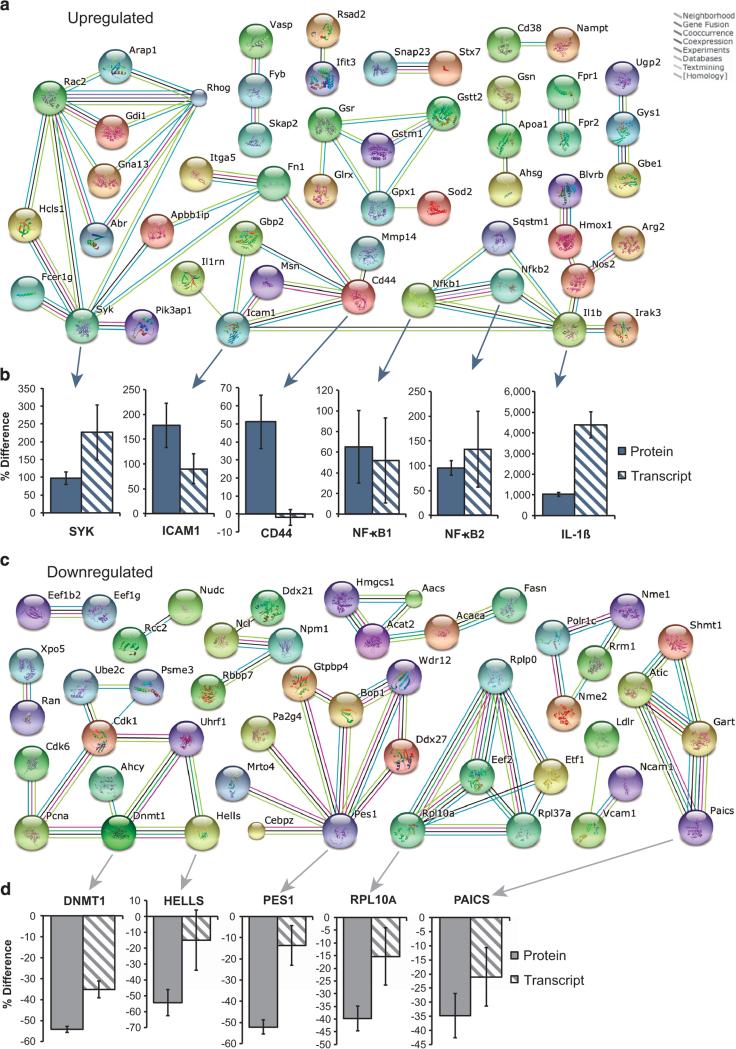
Finally, we used the String database32 to examine functional interactions between differentially expressed proteins, and transcriptionally validated several select candidates with qRT-PCR. Of these, CD44 and two NF-κB proteins have known links to compartmental function in the AML BM33,34 (Figures 6a and b). Intriguingly, in the downregulated protein set, we saw decreased expression for several known epigenetic regulators of hematopoiesis (Dnmt1, Hells and Pcna)35 (Figures 6c and d). Together, the data provide a link to the phenotypes of HSPC mobilization and constrained hematopoietic function we observed following exposure to AML exosomes in vivo.
Figure 6.
Network interactions for differentially expressed proteins in exosome-treated HSPC. (a) Upregulated protein interaction network (generated on String DB 9.1, with highest confidence setting). (b) Fold change (exosome-treated vs control) in upregulated candidate proteins by TMT and verification by qRT-PCR on cDNA from exosome-treated HSPC. (c) Downregulated protein interaction network. (d) Fold change (exosome-treated vs the control) in downregulated candidate proteins by TMT and verification by qRT-PCR on cDNA from exosome-treated HSPC. Graphs reflect means ± s.e.m.
DISCUSSION
There is considerable interest in the dysregulation of hematopoiesis during conversion from a homeostatic microenvironment to a leukemic niche.3–5,22 One recent xenograft study illustrated that AML blasts alter the differentiation of residual murine recipient progenitor cell pool in NSG mice.4 We now provide in vitro and in vivo evidence that AML exosomes account, at least in part, for modulation of residual compartmental function in the BM, indirectly via dysregulation of stromal cell supportive factors and chemokines, and directly via functional decline in clonogenic progenitor activity and reduction of CXCR4 expression.
We previously characterized AML exosome biogenesis and validated key attributes of their release in primary AML patient samples.7 Here, we chose AML cell lines for their predictable xenograft kinetics (Molm-14) or extramedullary chloroma formation (HL-60), which allowed in vivo measurements at low/no leukemic BM occupancy to minimize bias due to competitive HSPC displacement. Additionally, the reduced sample-to-sample variability for both Molm-14 and HL-60 cell lines was critical to dissect the regulation of stromal and hematopoietic cell populations.
We found that the presence of either Molm-14 or HL-60 leukemia cells in animals significantly compromised residual progenitor colony frequency in the BM, similar to a study of primary AML grafts, but at much lower leukemic burden.4 The HSPC redistribution to the peripheral blood we observed in Molm-14 xenografts echoes murine models of CML, where cytokines and retention factors, including Scf and Cxcl12, mediate hematopoietic suppression.3,22 Here, we demonstrate not only a decline in expression of CXCR4-CXCL12 and c-KIT-SCF but also the consistent presence of human transcripts in murine stromal and hematopoietic cells harvested from xenografted animals.36,37 The extra-medullary HL-60 xenograft model is particularly instructive in that the impact of xenografting is robustly reproduced, even though leukemia cells are not present in the BM. This line of evidence receives additional support from paired CFU-C analysis after IF injection of Molm-14 and HL-60 exosomes versus contralateral controls. Together, the data suggests that paracrine exosome trafficking from AML blasts to HSPC and BM stromal cells contributes to the suppression of hematopoiesis.
Unlike specific unidirectional ligand-receptor activities seen with cytokines or chemokines, exosomes have no known cell- or tissue-specific entry restriction, implying that all BM cells are permissive. Thus, multiple cellular components can be simultaneously regulated and signaling is ultimately determined by exosome cargo (protein and RNA), integrated at the level of gene expression in each ‘recipient’ cell type.9 These complex events are difficult to isolate in the in vivo environment. However, in carefully calibrated in vitro experiments at physiological oxygen concentrations, we demonstrate that AML exosomes directly promote the loss of HSPC clonogenicity, CXCR4 expression and CXCL12-directed migration in conjunction with downregulation of Scf and Cxcl12 in stromal cells. Thus, our data consistently show that exosomes cooperatively erode critical aspects of BM function and redistribute HSPC to the peripheral circulation.36–38
There are likely many cellular targets to account for exosome action in HSPC. We utilized a highly sensitive proteomics technique that identified proteins important in hematopoietic function and delineated regulatory networks for select candidates. For example, we find upregulation of SYK, a kinase expressed in hematopoietic cells39 which prevents differentiation of FLT3-ITD AML.40,41 Interleukin-1ß (IL1-ß), a cytokine dependent on SYK that reduces hematopoietic progenitor cell frequency,42 was upregulated 10-fold in exosome-exposed cells.43 It induces the secretion of G-CSF and other cytokines that prompt egress of HSPC from the leukemic niche.3,44 Similarly, the loss of expression of the adhesion protein VCAM1 in exosome-treated cells is consistent with exosome dysregulation of HSPC retention.
The downregulation of proteins involved in methylation maintenance was particularly intriguing. Two key epigenetic regulators, DNMT1 and HELLS (along with the binding partner PCNA), were all downregulated at the level of both transcript and protein in AML exosome-treated cells and cells from Molm-14-xenografted animals, suggesting that more long-lived epigenetic changes are provoked in HSPC. Similarly, the downregulation of 39 ribosomal or ribosomal biogenesis related proteins in exosome-treated BM cells is entirely consistent with the critical role protein synthesis has for HSPC function.45
Accumulated studies suggest that extracellular vesicles, including exosomes shed from tumor cells, play a critical role in remodeling the tumor microenvironment, in turn promoting the survival of tumor stem cells resistant to the treatment.3–5,22,46,47 Our leukemia focused studies (BM-myeloid tumor (Molm-14) vs extramedullary myeloid tumor (HL-60)) suggest that leukemia-derived exosomes exert target-cell-specific effects that drive coordinate compartmental remodeling and successive loss of hematopoietic activity during leukemic invasion of the BM. Our studies reinforce the importance of more thoroughly understanding exosome biogenesis, especially the selective incorporation of RNAs and proteins. Our work implies that leukemia exosome cargo dysregulates factors important to hematopoiesis. It is conceivable that those may present therapeutic targets to relieve HSPC loss and stem cell niche-perturbing effects of leukemogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
This manuscript is dedicated to the late Dr Zili Zhang, a thoughtful scientist, inspired colleague and, above all, a good friend. We acknowledge the assistance of Dr Muneesh Tewari, Dr Ashok Reddy, John Klimek, Rajani Kaimal, Aurelie Snyder and Jennifer Wherley. Studies were supported in part by the St. Baldrick's Foundation (PK), the Hyundai Hope on Wheels Foundation (PK), NIH/NIAID T32 grant (5T32AI78903-5) (NH), NIH core grants P30EY10572 and P30CA069533 and a Canary Foundation/American Cancer Society (ACS) Postdoctoral Fellowship (PFTED-09-249-01-SEID) with support by the Hillcrest Committee of Southern Oregon and the ACS Great West Division (JRC).
Footnotes
AUTHOR CONTRIBUTIONS
JH designed and performed experiments, interpreted data and prepared the manuscript; NH designed and performed experiments and interpreted data; NG performed experiments; ANK interpreted data and edited the manuscript; LLD interpreted data and edited the manuscript; PAW performed experiments and interpreted data; TM interpreted data and edited the manuscript; JRC performed experiments; AN performed experiments and interpreted data; CR interpreted data and edited the manuscript; ML interpreted data and edited the manuscript; BHC interpreted data and edited the manuscript; PK designed experiments, interpreted data and edited the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Ayala F, Dewar R, Kieran M, Kalluri R. Contribution of bone microenvironment to leukemogenesis and leukemia progression. Leukemia. 2009;23:2233–2241. doi: 10.1038/leu.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, et al. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer Cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang B, Ho YW, Huang Q, Maeda T, Lin A, Lee SU, et al. Altered microenviron-mental regulation of leukemic and normal stem cells in chronic myelogenous leukemia. Cancer Cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miraki-Moud F, Anjos-Afonso F, Hodby KA, Griessinger E, Rosignoli G, Lillington D, et al. Acute myeloid leukemia does not deplete normal hematopoietic stem cells but induces cytopenias by impeding their differentiation. Proc Natl Acad Sci USA. 2013;110:13576–13581. doi: 10.1073/pnas.1301891110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colmone A, Amorim M, Pontier AL, Wang S, Jablonski E, Sipkins DA. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science. 2008;322:1861–1865. doi: 10.1126/science.1164390. [DOI] [PubMed] [Google Scholar]
- 6.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 7.Huan J, Hornick NI, Shurtleff MJ, Skinner AM, Goloviznina NA, Roberts CT, Jr, et al. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 2013;73:918–929. doi: 10.1158/0008-5472.CAN-12-2184. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Jacamo R, Konopleva M, Garzon R, Croce C, Andreeff M. CXCR4 down-regulation of let-7a drives chemoresistance in acute myeloid leukemia. J Clin Invest. 2013;123:2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts CT, Jr, Kurre P. Vesicle trafficking and RNA transfer add complexity and connectivity to cell-cell communication. Cancer Res. 2013;73:3200–3205. doi: 10.1158/0008-5472.CAN-13-0265. [DOI] [PubMed] [Google Scholar]
- 10.Harrison JS, Rameshwar P, Chang V, Bandari P. Oxygen saturation in the bone marrow of healthy volunteers. Blood. 2002;99:394. doi: 10.1182/blood.v99.1.394. [DOI] [PubMed] [Google Scholar]
- 11.Hatfield KJ, Bedringsaas SL, Ryningen A, Gjertsen BT, Bruserud O. Hypoxia increases HIF- 1alpha expression and constitutive cytokine release by primary human acute myeloid leukaemia cells. Eur Cyt Netw. 2010;21:154–164. doi: 10.1684/ecn.2010.0204. [DOI] [PubMed] [Google Scholar]
- 12.Lam BS, Adams GB. Blocking HIF1alpha activity eliminates hematological cancer stem cells. Cell Stem Cell. 2011;8:354–356. doi: 10.1016/j.stem.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Jacamo R, Shi YX, Wang RY, Battula VL, Konoplev S, et al. Human extra-medullary bone marrow in mice: a novel in vivo model of genetically controlled hematopoietic microenvironment. Blood. 2012;119:4971–4980. doi: 10.1182/blood-2011-11-389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan D. In situ (in vivo) gene transfer into murine bone marrow stem cells. Methods Mol Biol. 2009;506:159–169. doi: 10.1007/978-1-59745-409-4_12. [DOI] [PubMed] [Google Scholar]
- 15.Mazurier F, Doedens M, Gan OI, Dick JE. Rapid myeloerythroid repopulation after intrafemoral transplantation of NOD-SCID mice reveals a new class of human stem cells. Nat Med. 2003;9:959–963. doi: 10.1038/nm886. [DOI] [PubMed] [Google Scholar]
- 16.Skinner AM, Grompe M, Kurre P. Intra-hematopoietic cell fusion as a source of somatic variation in the hematopoietic system. J Cell Sci. 2012;125:2837–2843. doi: 10.1242/jcs.100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilmarth PA, Tanner S, Dasari S, Nagalla SR, Riviere MA, Bafna V, et al. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility? J Proteome Res. 2006;5:2554–2566. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAlister GC, Huttlin EL, Haas W, Ting L, Jedrychowski MP, Rogers JC, et al. Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 20.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringner M, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci USA. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schepers K, Pietras EM, Reynaud D, Flach J, Binnewies M, Garg T, et al. Myeloproliferative neoplasia remodels the endosteal bone marrow niche into a self-reinforcing leukemic niche. Cell Stem Cell. 2013;13:285–299. doi: 10.1016/j.stem.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh AK, Shanafelt TD, Cimmino A, Taccioli C, Volinia S, Liu CG, et al. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113:5568–5574. doi: 10.1182/blood-2008-10-185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno Y, Yasunaga S, Janmohamed S, Ohtsubo M, Saeki K, Kurogi T, et al. Hoxa9 transduction induces hematopoietic stem and progenitor cell activity through direct down-regulation of geminin protein. PloS One. 2013;8:e53161. doi: 10.1371/journal.pone.0053161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chong JL, Tsai SY, Sharma N, Opavsky R, Price R, Wu L, et al. E2f3a and E2f3b contribute to the control of cell proliferation and mouse development. Mol Cell Biol. 2009;29:414–424. doi: 10.1128/MCB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sattler M, Verma S, Pride YB, Salgia R, Rohrschneider LR, Griffin JD. SHIP1 an SH2 domain containing polyinositol-5-phosphatase, regulates migration through two critical tyrosine residues and forms a novel signaling complex with DOK1 and CRKL. J Biol Chem. 2001;276:2451–2458. doi: 10.1074/jbc.M006250200. [DOI] [PubMed] [Google Scholar]
- 27.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, et al. Hypoxiainducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee DW, Futami M, Carroll M, Feng Y, Wang Z, Fernandez M, et al. Loss of SHIP-1 protein expression in high-risk myelodysplastic syndromes is associated with miR-210 and miR-155. Oncogene. 2012;31:4085–4094. doi: 10.1038/onc.2011.579. [DOI] [PubMed] [Google Scholar]
- 29.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]
- 34.Jacamo R, Chen Y, Wang Z, Ma W, Zhang M, Spaeth EL, et al. Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-kappaB mediates chemoresistance. Blood. 2014;123:2691–2702. doi: 10.1182/blood-2013-06-511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizuno S, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, et al. Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood. 2001;97:1172–1179. doi: 10.1182/blood.v97.5.1172. [DOI] [PubMed] [Google Scholar]
- 36.Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Cosgun KN, Rahmig S, Mende N, Reinke S, Hauber I, Schafer C, et al. Kit regulates HSC engraftment across the human-mouse species barrier. Cell Stem Cell. 2014;15:227–238. doi: 10.1016/j.stem.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 38.Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, et al. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mocsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat Rev Immunol. 2010;10:387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puissant A, Fenouille N, Alexe G, Pikman Y, Bassil CF, Mehta S, et al. SYK is a critical regulator of FLT3 in acute myeloid leukemia. Cancer Cell. 2014;25:226–242. doi: 10.1016/j.ccr.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn CK, Berchuck JE, Ross KN, Kakoza RM, Clauser K, Schinzel AC, et al. Proteomic and genetic approaches identify Syk as an AML target. Cancer Cell. 2009;16:281–294. doi: 10.1016/j.ccr.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross O, Poeck H, Bscheider M, Dostert C, Hannesschlager N, Endres S, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–436. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 43.Yonemura Y, Ku H, Hirayama F, Souza LM, Ogawa M. Interleukin 3 or interleukin 1 abrogates the reconstituting ability of hematopoietic stem cells. Proc Natl Acad Sci USA. 1996;93:4040–4044. doi: 10.1073/pnas.93.9.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartwig UF, Keller U, Huber C, Peschel C. Regulation of hematopoietic growth factor production by genetically modified human bone marrow stromal cells expressing interleukin-1beta antisense RNA. J Interferon Cytokine Res. 2001;21:851–860. doi: 10.1089/107999001753238105. [DOI] [PubMed] [Google Scholar]
- 45.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a prometastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barcellos-Hoff MH, Lyden D, Wang TC. The evolution of the cancer niche during multistage carcinogenesis. Nat Rev Cancer. 2013;13:511–518. doi: 10.1038/nrc3536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.