Abstract
Verticillium wilt is a disastrous vascular disease in plants caused by Verticillium dahliae. Verticillium pathogens secrete various disease-causing effectors in cotton. This study identified a subtilase gene GbSBT1 from Gossypium babardense and investigated the roles against V. dahliae infection. GbSBT1 gene expression is responsive to V. dahliae defense signals, jasmonic acid, and ethylene treatments. Moreover, the GbSBT1 protein is mainly localized in the cell membrane and moves into the cytoplasm following jasmonic acid and ethylene treatments. Silencing GbSBT1 gene expression through virus-induced GbSBT1 gene silencing reduced the tolerance of Pima-90 (resistant genotype), but not facilitated the infection process of V. dahliae in Coker-312 (sensitive genotype). Moreover, the ectopically expressed GbSBT1 gene enhanced the resistance of Arabidopsis to Fusarium oxysporum and V. dahliae infection and activated the expression levels of defense-related genes. Furthermore, pull-down, yeast two-hybrid assay, and BiFC analysis revealed that GbSBT1 interacts with a prohibitin (PHB)-like protein expressed in V. dahliae pathogens during infection. In summary, GbSBT1 recognizes the effector PHB protein secreted from V. dahliae and is involved in Verticillium-induced resistance in cotton.
Introduction
Verticillium wilt, a devastating disease of more than 200 crops worldwide, is typically caused by the soil-borne fungus Verticillium dahliae [1]. V. dahliae secretes various effectors to evade the guard system or destroy the innate immune system of host plants [2,3,4,5]. The V. dahliae genome encodes approximately 780 secreted proteins containing signal peptides as candidate effectors [6]. These secreted proteins, including diverse polysaccharide lyases, could cleave different forms of pectins in host plants to help Verticillium wilt-causing pathogens invade xylem vessels [3,7]. V. dahliae also secretes isochorismatases (without signal peptide) that suppress salicylate-mediated innate immunity in host plants [4]. In the absence of its corresponding R protein (Ve1), the Verticillium effector Ave1 functions in the apoplast of host plants to promote pathogenicity [3]. Nevertheless, the mechanism by which these effectors from Verticillium pathogens are recognized or primed by host plants remains largely unknown.
Subtilisin-like proteases (subtilase) are extracellular and broad-spectrum serine proteases containing a catalytic triad motif that consists of aspartate, histidine, and serine [8]. The subtilase gene family in Arabidopsis is composed of 56 members classified into six distinct subfamilies [9,10]. Recent studies have revealed that subtilase genes are specifically induced following pathogenic infection and are hypothesized to be involved in pathogen recognition and immune priming. SBT3.3 rapidly responds to pathogenic infection and activates innate immunity preceding the activation of SA responsive genes [11]. P69, the first identified plant subtilase to be identified located in the vacuole and intercellular space, is specifically responsible for the in vivo pathogenesis-associated processing of LRP (a leucine-rich repeat protein) [12,13]. Furthermore, AtSBT1.1 specifically cleaves proAtPSK4 into the mature peptide growth factor AtPSK4 to promote callus formation in culture and fungal resistance in Arabidopsis [14]. Combined data indicate that subtilases function as catalytic proteases to recognize pathogenic attacks [10].
This study characterized an extracellular subtilase gene (GbSBT1) from the Gossypium barbadense variety Pima-90. GbSBT1 knockdown reduced the defenses of G. barbadense against V. dahliae attack, and the cotton plants exhibited a more severe wilting phenotype than the control plants. Ectopically expressed GbSBT1 gene enhanced the disease tolerance of Arabidopsis against F. oxysporum and V. dahliae. Importantly, GbSBT1 interacted with the protein prohibitin (PHB) secreted by V. dahliae. Our results proved that GbSBT1 functions as a sensing protein during V. dahliae infection and activates downstream resistance response in cotton.
Materials and Methods
Plant materials and pathogen culture
Cotton seeds (G. barbadense variety Pima-90 and Gossypium hirsutum variety Coker-312) were soaked in corrosive sublimate (1/1000, v/v) for 5 min and then washed three times with sterile water. The aseptic seeds were grown in MS medium, and 1-week-old cotton seedlings were used in subsequent experiments. Wild-type (WT; ecotype Columbia, Col-0) and transgenic Arabidopsis plants were grown in a greenhouse under long-day conditions (22°C, 16/8 h light/dark).
The defoliating isolate V991 of V. dahliae was grown on Czapek’s medium agar medium (NaNO3, 0.3% w/v; MgSO4, 0.1% w/v; KH2PO4, 0.1% w/v; FeSO4, 0.0002% w/v; KCl, 0.1% w/v; sucrose, 3% w/v; and agar 3% w/v; pH 6.0) for 7 days [15]. Conidial spores were harvested and adjusted to 1 × 106 spores mL−1 with sterile distilled water. For F. oxysporum, the isolate was inoculated on Bilai’s medium plates. After 4 days, conidial spores were harvested and adjusted to 1 × 106 spores mL−1 with sterile distilled water. V. dahliae and F. oxysporum spore suspensions were then used in the inoculation experiments in cotton and Arabidopsis, respectively.
GbSBT1 gene isolation from sea-island cotton
To clone the subtilase gene from sea-island cotton, first-strand cDNA was synthesized from 1 μg of total RNA by using PrimeScript RT reagents and a gDNA Eraser kit (TaKaRa, Japan). The synthesized first-strand cDNA served as the template in reverse transcript polymerase chain reaction (RT-PCR). Gene-specific primers (S1 Table) were used to amplify the full-length subtilase gene. The PCR reactions (25 μL) contained 10 ng first-strand cDNA, 1 U ExTaq, 10 pM dNTPs, 5 pM MgCl2, and 10 pM primers. RT-PCR was performed under the following conditions: initial reaction at 94°C for 5 min, followed by 30 cycles at 94°C for 30 s, 54°C for 30 s, 72°C for 2 min and 30 s, and 72°C for 10 min. The amplified gene was cloned into the pGEM-T vector (Clontech, USA) and then confirmed by DNA sequencing. The cloned subtilase gene was named GbSBT1.
The conservative domains of GbSBT1 (the signal peptide, the putative inhibitor domain, and the putative catalytic sites) were searched online through the domain Blast (www.ncbi.nlm.nih.gov). We aligned GbSBT1 with the subtilase proteins of three organisms by using Bioedit to analyze the relationship of GbSBT1 with other subtilases. A neighbor-joining tree of different subtilases was constructed using MEGA 3.0 [16].
Expression pattern analysis of the GbSBT1 gene
Cotton seeds were soaked in corrosive sublimate (1/1000, v/v) for 5 min and then washed three times with sterile water. The aseptic seeds were grown in MS medium at the appropriate condition (28±2°C, 14/10 h light/dark), and 2-week-old cotton seedlings were used in subsequent experiments. Sterilized young cotton seedlings were placed in V991 suspension liquid (1 × 106 conidia mL−1). Cotton roots were harvested at 2, 4, 6, 8, 12, 24, 36, and 48 h after inoculation. Cotton roots inoculated with 1/2MS medium without mycelia served as the control. Total RNA was extracted using RNA prep Pure Plant Kit (DP441; Tiangen, China) in accordance with the manufacturer’s instructions.
The expression pattern of the GbSBT1 gene was analyzed using qRT-PCR in accordance with the manual of SYBR premix Ex-Taq (Takara, Japan) in a DNA Engine Option 3 System (MJ Research, USA). The ubiquitin gene was amplified using the primers as endogenous controls. Changes in gene expression were calculated using the comparative ΔCT method. Each sample was repeated at least thrice, and the amplification results were analyzed using the Option 3 software. The primers used in qRT-PCR are listed in S1 Table.
Subcellular localization of GbSBT1 protein
To investigate the subcellular localization of the GbSBT1 protein, we cloned the coding region of the GbSBT1 gene and its ORF without signal peptide-encoding nucleotides into the pEG101-YFP vector to generate pEG101-(CaMV35S::GbSBT1 or GbSBT1△SP-YFP::NOS) constructs. The recombinant plasmids were transformed into the Agrobacterium strain EHA105. Nearly fully expanded 3-week-old tobacco (Nicotiana benthamiana) leaves were infiltrated with the Agrobacterium that was diluted into OD600 of 0.6–0.8 with the solution (10 mM MES, pH 5.6; 10 mM MgCl2, and 150 mM acetosyringone). After 2–4 days, the fluorescence signals in infiltrated leaves were analyzed through confocal microscopy (Leica TCS SP5).
To determine whether or not GbSBT1 is localized on the cell membrane, we co-expressed the GbSBT1-GFP fusion protein with plasma membrane integral protein PIP1-mCherry (cell membrane-localized) in tobacco leaf epidermal cells according to the above-mentioned method. Three days after infiltration, the tobacco leaves were cut into strips and then digested into the protoplasts according to the reported method [17]. Confocal images of tobacco protoplasts expressing both GbSBT1-GFP and plasma membrane integral protein PIP1-mCherry proteins were observed.
Virus-induced GbSBT1 gene silencing (VIGS) in cotton and V. dahliae resistance analysis
VIGS was used to investigate the function of the SBT1 gene in resistant (Pima-90) and sensitive genotypes (Coker-312) as previously described method [18,19]. GbSBT1 and GhSBT1 gene-specific fragments (corresponding to the 225th-448th amino acids (aa) in the GbSBT1 protein and not detected in other genes at the genomic level) were amplified using gene-specific primers (S1 Table) and then inserted into the pDONR201 vector. The GbSBT1-specific fragment was subsequently recombined with the pYL156 vector. Plasmids containing the binary TRV vectors pTRV-RNA1, pTRV-RNA2, and pYL156 derivatives were transformed into Agrobacterium strain GV3101. Agrobacterium strains with different derivatives were grown in LB medium containing 50 μg mL−1 kanamycin, 50 μg mL−1 rifampicin, 10 mM MES, and 200 μM acetosyringone. The Agrobacterium cultures were harvested by centrifugation, resuspended in a solution (10 mM MES and 200 μM MgCl2), and then incubated for at least 3 h at room temperature.
The Agrobacterium harboring pTRV-RNA1 was mixed with the Agrobacterium harboring pTRV-RNA2 and pTRV-GbSBT1 or pTRV-GhSBT1 at a 1:1 ratio. A needle-free injection syringe was used to inject the mixed Agrobacterium cultures into the cotyledons of 2-week-old cotton plants. The injected cotton plants were grown in a greenhouse for 2 weeks to repress GbSBT1 or GhSBT1 gene expression.
Conidial suspensions of V. dahliae isolate V991 (1 × 106 spores mL−1) were stem-inoculated into both control and VIGS plants by using a syringe needle approximately 1 cm below the cotyledons [19]. After 7 days, the disease symptom, which was divided into 5 levels (level 0–4) on the basis of severity, was investigated in the cotton seedlings. The plant disease indexes (DI) were then calculated as follows: DI = (∑(n × number of seedlings at level n))/(4 × number of total seedlings × 100), where n denotes the severity of the disease level of the cotton seedlings [19]. VIGS experiments were repeated at least thrice using more than 10 cotton plants for each constructs.
To analyze V. dahliae infection in the control and VIGS, diaminobenzidine (DAB) staining of cotton leaves was performed as previously described [19]. The DAB staining experiments were repeated at least thrice.
Generation of transgenic GbSBT1 Arabidopsis plants and pathogen resistance analysis
Wild-type Arabidopsis thaliana plants (ecotype Columbia, Col-0) were grown in the greenhouse under long-day conditions (22°C, 16/8 h light/dark) according to Huang et al. [20]. We cloned the coding region of the GbSBT1 gene into the pDONR201 vector to generate the pDONR-GbSBT1 construct. The GbSBT1 gene was then recombined into the pEG101 vector via the Gateway LR recombination reaction (Invitrogen, CA, USA) to generate the pEG101-35S::GbSBT1::NOS expression cassette. The construct was transferred into Agrobacterium tumefaciens GV3101 and then introduced into Arabidopsis (ecotype Columbia) plants using the floral dip method [21]. Fully mature seeds were collected and screened by 20 mg L−1 BSATA. Herbicide-resistant plants were confirmed via PCR and self-crossed to generate homogenous T3 lines.
The WT and transgenic lines were grown in a greenhouse for 2 weeks and then used for F. oxysporum and V. dahliae resistance analysis. The leaves of the WT and transgenic lines were inoculated with 10 μL conidial suspensions (1 × 107 conidia ml−1) of F. oxysporum and V. dahliae. Distilled water with 0.2% Tween-20 without conidia served as the control. The severity of the disease in the WT and transgenic GbSBT1 lines was assessed after a week. At least three biological replicates were performed in the F. oxysporum and V. dahliae resistance analysis.
Expression analysis of defense-related genes and trypan blue staining of Arabidopsis leaves followed by pathogen inoculation
qRT-PCR was used to analyze the expression patterns of defense-related genes in WT and transgenic Arabidopsis plants after pathogen inoculation. Leaves of the WT and transgenic Arabidopsis plants were collected for RNA isolation in different intervals after F. oxysporum inoculation. qRT-PCR analysis was performed as described above [20]. The primers used in qRT-PCR are listed in S1 Table, and in Arabidopsis, Actin 2 was chosen as the control. Each sample was repeated at least thrice, and the amplification results were analyzed using the Option 3 software.
Conidial suspensions of F. oxysporum were inoculated on leaves of 4-week-old WT and transgenic GbSBT1 Arabidopsis lines for 2 days. To determine pathogen colonization, the inoculated leaves were stained with trypan blue [19]. The leaves were boiled in a lactophenol-trypan blue solution (10 mL lactic acid, 10 mL glycerol, 10 mg phenol, and 10 mg trypan blue dissolved in 10 mL distilled water) for 2 min and then cleared with chloral hydrate (2.5 g mL−1) overnight. The trypan blue staining experiments were repeated at least thrice.
Identification of GbSBT1-interacting proteins through pull-down assay
The Agrobacterium strain GV3101 containing the pEG101-35S::GbSBT1-GFP::Nos plasmid and the control plasmid were transformed into cotton cotyledons for the GbSBT1-GFP fusion protein or GFP expression (control). After 12 h, V. dahliae V991 was inoculated into the cotyledons of Coker-312. The total proteins of the cotton cotyledons expressing the GbSBT1-GFP fusion protein or the GFP protein were extracted after 24 h for pull-down assay. This assay was performed in accordance with the protocol of FOCUS Plant Proteome (Sangon, China), which is briefly described below.
The GFP monoclonal antibody was covalently linked to Sepharose beads and washed twice with washing buffer. The extracted total protein of the cotton cotyledons inoculated with V991 was loaded into the column and then incubated with the GFP antibody beads. After 30 min of incubation, the Sepharose beads combined with GFP antibody-cotton proteins were washed thrice. Finally, GbSBT1-interacting candidates were eluted with the lysis buffer and then collected in the loading buffer.
The eluted proteins were confirmed via SDS-PAGE. The GbSBT1-interacting proteins and the control (GFP-binding) were identified using Nano-Liquid Chromatography (UltiMate3000 RSLCnano Liquid Chromatography, Bruker Daltonics) and Quadrupole-Time-of-Flight Mass Spectrometer (maXis impact UHR-TOF MS, Thermofisher). The sequenced peptides were mapped to the V. dahliae genome and annotated by the hits. The GbSBT1-interacting proteins without background noise were then analyzed further.
Yeast two-hybrid assay and BiFC confirmation in vivo
To test protein interaction in vitro, we cloned the genes encoding the benzodiazepine receptor family protein (Accession NO: XP_009649206), peptidyl-tRNA hydrolase (Accession NO: XP_009656275), PHB (Accession NO: XP_009649890), and GbSBT1 into both pGBKT7 and pGADT7 vectors. The yeast two-hybrid assay was performed using a Yeast Transformation System kit (Clontech, CA, USA) in accordance with the manufacturer’s protocol. The transformed AH109 yeast cells were grown on SD/-T-L and then incubated at 28°C for 3 days. The positive colonies were subsequently transferred into the selective and stringent SD/-T-L-H medium supplemented with 2 mM 3-AT medium.
For BiFC analysis, the coding region of GbSBT1 was cloned into pEarleyGate202 vector and PHB was cloned into pEarley-Gate201 vector. These vectors were transformed into the Agrobacterium strains EHA105. Equal volume suspensions of different Agrobacterium strains carrying GbSBT1-pEarleyGate202, PHB- pEarley-Gate201, p19 protein were mixed prior to infiltration. The re-suspended cells were infiltrated into leaves of tobacco plants as described previously. The fluorescence signals in infiltrated leaves were analyzed through confocal microscopy (Leica TCS SP5).
Results
Identification of the GbSBT1 gene and phylogenetic analysis
Our previous study revealed more than 100 differentially expressed genes from the Verticillium-resistant cotton variety 7124 by using subtractive suppression hybridization [15]. Among these differentially expressed genes, the cDNA upregulated by V. dahliae attack was further studied in this work. The full-length cDNA was cloned by RT-PCR and was confirmed by DNA sequencing. This gene encoded a protein containing an evolutionary conserved structure that is highly similar to the subtilase proteins of other plants; thus, this gene was named GbSBT1 and was deposited in GenBank (Accession NO: KT336228).
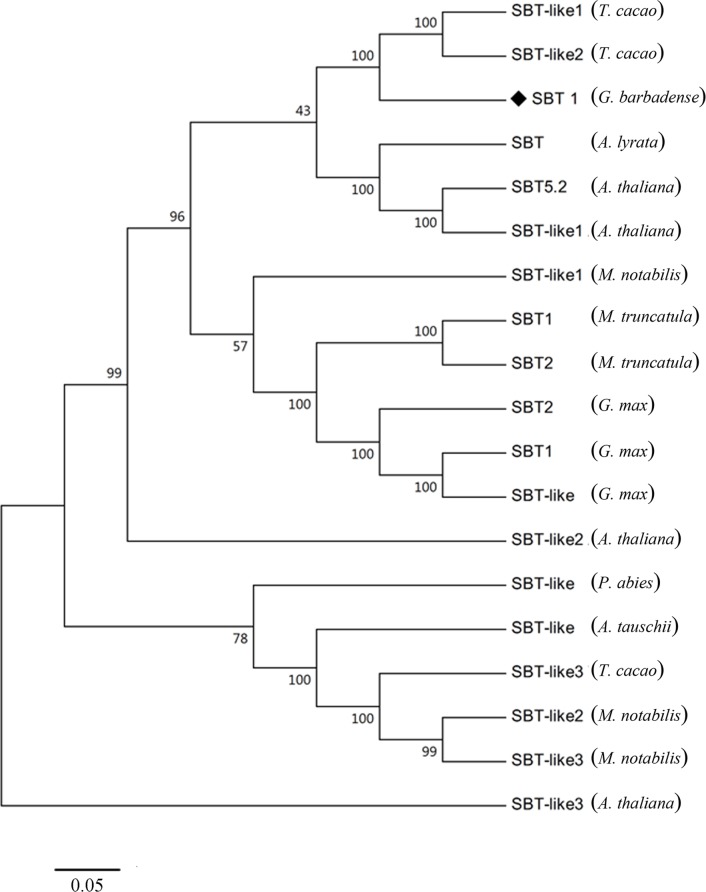
The GbSBT1 gene encodes a protein containing 769 aa, corresponding to a molecular mass of 81.78 kDa and a pI of 6.34. The GbSBT1 protein contains a putative signal peptide, an inhibitor domain, five conserved active sites (Ser-544, Asp-145, and His-223), and two catalytic triads of serine protease (Fig 1). A neighbor-joining phylogenetic tree of 19 subtilases indicated that that GbSBT1 is closely related to AtSBT5.2 (Fig 2). In Arabidopsis, AtSBT5.2 exhibits oxido-reductase activity and responds to an external biotic stimulus [10], indicating that the GbSBT1 gene may function in biotic stress response.
Fig 1. Multiple sequence alignment analysis of GbSBT1 and other plant subtilases.
GbSBT1 (KT336228, Gossypium barbadense), subtilase-like (XP_007017870, Theobroma cacao), SBT5.2 (NP_564107, Arabidopsis thaliana), SBT-like (AAM65424, A. thaliana). SP: signal peptide; Inhibitor I9: serine protease inhibitor domain; black triangles indicate conserved active sites; blank triangles indicate catalytic triads of serine protease; and blank box A indicates PA/protease or protease-like domain interface.
Fig 2. Phylogenetic analysis of GbSBT1 including the subtilases of different plant species.
Nineteen of the complete amino acid sequences of subtilases were used to generate the neighbor–joining tree, and the numbers next to each node provide bootstrap values from 1000 replicates. The GbSBT1 gene is marked by “◆.” GbSBT1 (KT336228, Gossypium barbadense), SBT-like Picea abies (BAA13135, Picea sitchensis), AtSBT5.2 (NP_564107, Arabidopsis thaliana), SBT-like1 Theobroma cacao (XP_007017870, T. cacao), SBT-like2 T. cacao (XP_007017871, T. cacao), SBT Arabidopsis lyrata (XP_002893091, A. lyrata), SBT-like1 A. thaliana (AAM65424, A. thaliana), SBT-like1 Morus notabilis (EXB60669, M. notabilis), SBT1 Medicago truncatula (XP_003603196, M. truncatula), SBT2 M. truncatula (XP_003603807, M. truncatula), SBT1 Glycine max (NP_001236511, G. max), SBT2 G. max (NP_001238252, G. max), SBT-like G. max (AAK53589, G. max), SBT-like2 A. thaliana (NP_564106, A. thaliana), SBT-like Aegilops tauschii (EMT32761, A. tauschii), SBT-like3 T. cacao (XP_007028364, T. cacao), SBT-like2 M. notabilis (EXB44295, M. notabilis), SBT-like3 M. notabilis (EXB44294, M. notabilis), SBT-like3 A. thaliana (NP_565309, A. thaliana).
Expression pattern of GbSBT1 gene following V. dahliae inoculation
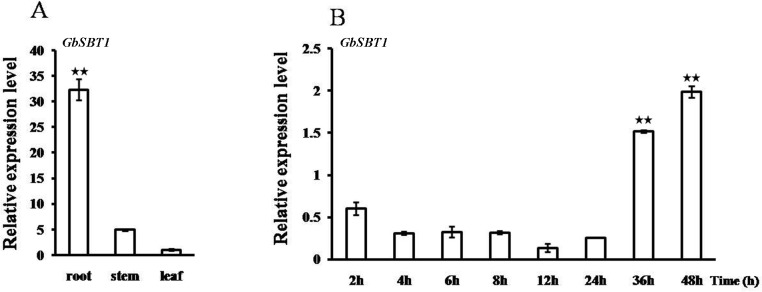
qPCR was used to analyze the expression pattern of GbSBT1 in various tissues of sea-island cotton. Fig 3A shows that the GbSBT1 gene was ubiquitously expressed in all of the tested tissues. Among the tested tissues, GbSBT1 expressed much higher in root where the V. dahliae enters through the roots more often than other organs.
Fig 3. Induced GbSBT1 expression following V. dahliae V991 infection.
(A) Expression analysis of the GbSBT1 gene in vegetative tissues (roots, stems, and leaves) via qPCR. The Ubiquitin gene is used as an internal control. At least three biological replicates were performed. (B) Expression analysis of GbSBT1 in roots following V. dahliae strain V991 infection. The comparative CT method is adopted, and the expression is normalized. Each sample was repeated at least thrice. Error bars represent SE. Double asterisks represent a significant expression change of GbSBT1 compared to uninfected condition (P < 0.01) in t-test.
In Arabidopsis, the induction of the SBT3.3 gene was transient; that is, its expression peaked at 48 h post-inoculation (hpi) of the bacterial pathogen Pseudomonas syringae DC3000 and then abruptly decayed thereafter [11]. When cotton plants were inoculated with the V991 strain, GbSBT1 expression in the roots decreased within 24 h, significantly increased thereafter, and then peaked at 48 hpi (Fig 3B). The rapid increase in GbSBT1 expression suggests that GbSBT1 is associated with cotton resistance to Verticillium.
GbSBT1 protein is localized in the plasma membrane and is affected by jasmonic acid (JA) and ethylene
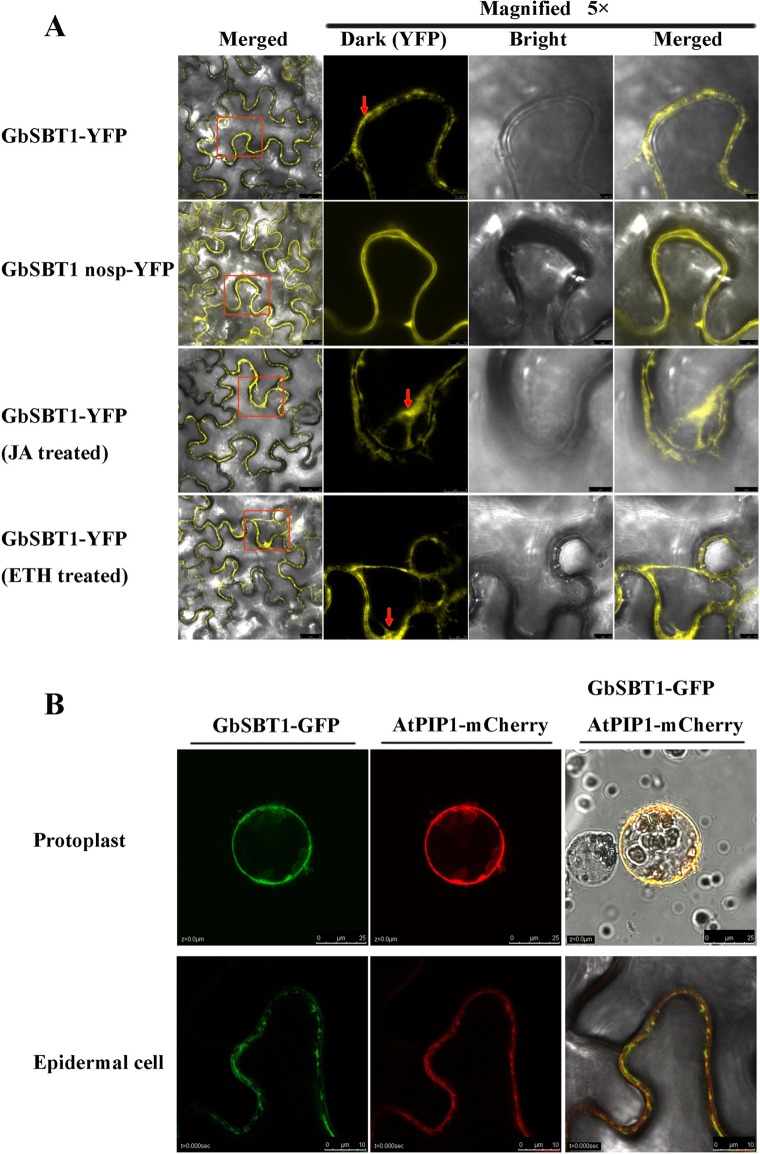
To analyze GbSBT1 subcellular localization, we expressed the ORF of the GbSBT1 gene fused with eYFP in tobacco leaves. As shown in Fig 4A, GbSBT1-eYFP fluorescence signals were detected in the plasma membrane. When magnifying the accumulation area, GbSBT1-eYFP signals were unevenly distributed in the cell membrane (Fig 4A), which provides the conduits for the exchange of informational molecules that are central to cell growth and defense response in plants. When GbSBT1 without the signal peptide was expressed in tobacco leaves, GbSBT1 (no SP)-YFP signals were uniformly distributed in the plasma membrane. The co-localization signals with plasma membrane integral protein PIP1-mcherry further demonstrate that GbSBT1 mostly targets the cell membrane (Fig 4B). Overall, GbSBT1 is mainly localized in the cell membrane. GbSBT1 extracellular localization may be linked to the acceptance of external signals.
Fig 4. Extracellular localization of the GbSBT1 protein in tobacco leaves determined through confocal microscopy.
(A) Localization of GbSBT1-YFP in tobacco leaves; localization of GbSBT1 (without signal peptide)-YFP in tobacco leaves; localization of GbSBT1-YFP in tobacco leaves under JA treatment; localization of GbSBT1-YFP in tobacco leaves under ethylene treatment. Arrows indicate the difference in protein localization between normal and phytohormone treatments; (B) GbSBT1 protein co-localized with plasma membrane integral protein PIP1 on the cell membrane using protoplast and tobacco lead epidermal cells as the protein expressing materials. Upper panels: protoplast; lower panels: tobacco leaf epidermal cells.
Verticillium wilt Ve1-mediated resistance is compromised in the Arabidopsis JA response mutant jar1-1 [22], and the JA signaling pathway can be activated in cotton by inoculation with V. dahliae [19,23], suggesting that cotton Verticillium resistance is related to JA signal transduction. To elucidate whether or not phytohormones affect GbSBT1 localization, we treated the cotton seedlings with methyl jasmonate and ethylene. Similar to the normal condition, GbSBT1 was unevenly localized in the cell membrane prior to the treatments with ethephon (1 mM) and JA (10 ppm). GbSBT1-YFP signal was induced and moved into cytoplasm after spraying these phytohormones (Fig 4A). Tracking the movement of the GbSBT1-YFP complex in vivo revealed that GbSBT1 particles rapidly moved along microfilaments in the cytoplasm (S1 Video). The movement of the GbSBT1 protein from the cell membrane into the cytoplasm suggests that GbSBT1 senses the combined signals of ethylene and JA treatments.
Knockdown of GbSBT1 gene expression results in lost resistance of cotton plants to V. dahliae infection
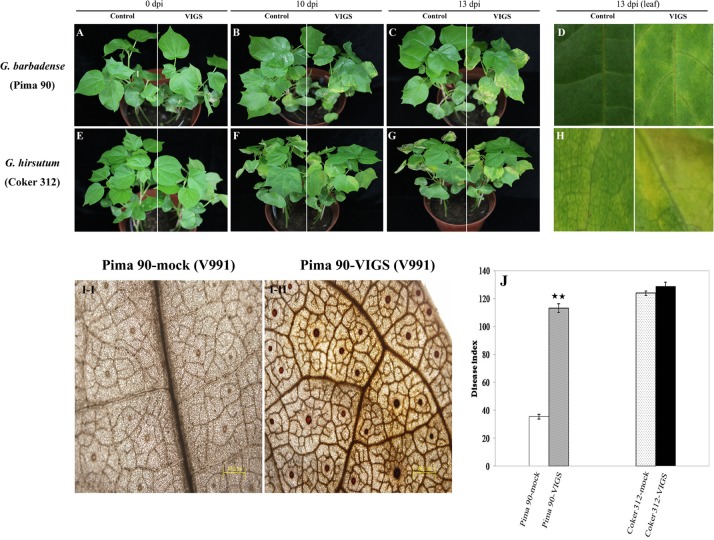
We generated GbSBT1 gene-silenced cotton plants through VIGS to study the function of the GbSBT1 gene during V. dahliae infection. After growing in a greenhouse for 7 days, the V. dahliae resistance of the control (Pima-90) and VIGS plant lines was subsequently examined and quantified. qPCR results showed that GbSBT1 was successfully repressed in VIGS plants. Results indicated that GbSBT1 expression in the VIGS plants was repressed significantly (about 76% lower than control) compared with control plants (S1 Fig). The GbSBT1 gene-silenced and control plants were inoculated with V. dahliae strain V991. As shown in Fig 5A, GbSBT1-silenced plants grew as normal as the control plants before fungal inoculation. Ten days after inoculation, the silenced plants began to display disease symptoms in the form of margin-wilted leaves; by contrast, the control cotton plants grew normally (Fig 5B). Thirteen days after inoculation, the GbSBT1-silenced plants were severely affected and the majority of leaves turned yellow and tended to wilt (Fig 5C, right panel). Moreover, the rate of diseased plants and the disease index were significantly higher in the GbSBT1-silenced plants than in the control plants (Fig 5J). This result suggests that GbSBT1 knockdown in cotton plants increases plant susceptibility to V. dahliae infection.
Fig 5. Reduced resistance of cotton to Verticillium dahliae after GbSBT1 gene silencing.
(A–C) Disease symptoms in Gossypium barbadense Pima-90 and the VIGS plants inoculated with V. dahliae strain V991; (D) Enlarged region of disease symptoms on leaf of Pima-90 inoculated with V. dahliae strain V991. Left panels: Pima-90; right panels: Pima-90 containing silenced GbSBT1 gene; dpi: days post-inoculation. (E–G) Disease symptoms in Gossypium hirsutum Coker-312 and the VIGS plants inoculated with V. dahliae strain V991; (H) Enlarged region of disease symptoms on leaf of Coker-312 inoculated with V. dahliae strain V991. Left panels: Coker-312; right panels: Coker-312 containing silenced GhSBT1 gene; dpi: days post-inoculation. (I–I/II) DAB staining in the control and VIGS cotton leaves of G. barbadense after V. dahliae V991 inoculation. (J) Disease index measurement in Pima-90 and Coker-312 cotton plants after inoculation with V991. VIGS experiments were repeated at least three times with more than 10 cotton plants for each construct. Double asterisks represent significant difference between VIGS plants and wild-type plants (P < 0.01) in t-test.
The subtilase 1 gene in Coker-312 was further silenced and the disease symptom was investigated to validate the effect of subtilase 1 gene expression on V. dahliae resistance in different genotypes (resistant: Pima-90; sensitive: Coker-312). Ten days after inoculation, the GhSBT1-silenced and control plants were both severely affected, and the bottom leaves began to defoliate (right panel, Fig 5E). As the infection progressed, the control and silenced plants still showed no obvious difference in symptoms. The disease index showed no significant difference between the control and VIGS plants (Fig 5J). These results demonstrated that subtilase1 suppression in the sensitive genotype failed to facilitate V. dahliae infection and that the GbSBT1 gene is involved in R-gene resistance.
Reactive oxygen accumulation is related to hypersensitive cell death, which could lead to the spontaneous formation of lesions in plants. In addition, H2O2 accumulation could impede V. dahliae infection in cotton [5]. The changes in H2O2 levels in the control and VIGS cotton plants were observed through DAB staining to investigate whether or not high levels of H2O2 are produced during V. dahliae infection. As shown in Fig 5I, control Pima-90 plants have no DAB staining. Conversely, the VIGS plants showed obvious DAB staining in the leaf vessels, indicating that V. dahliae pathogens ingress into cotton plants and induce H2O2 accumulation.
Ectopically expressed GbSBT1 gene in Arabidopsis confers disease resistance to F. oxysporum and V. dahliae
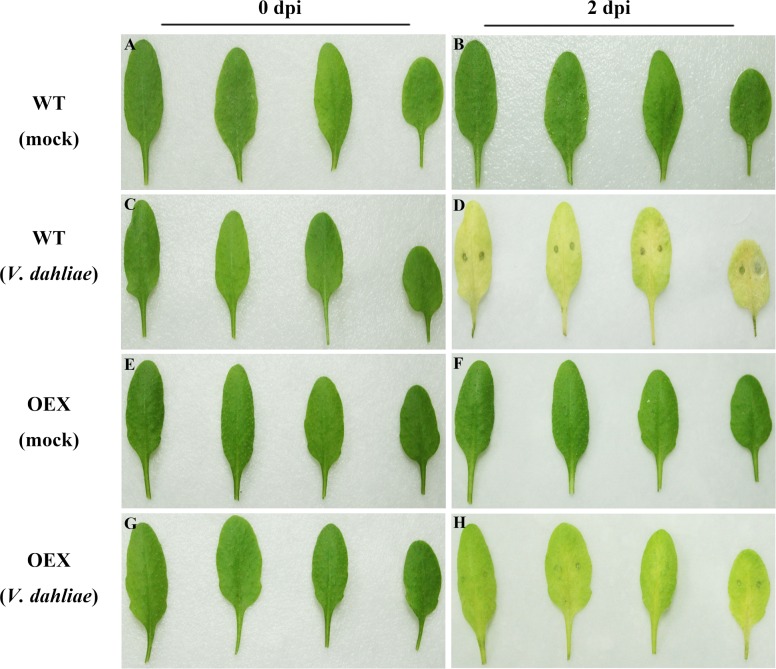
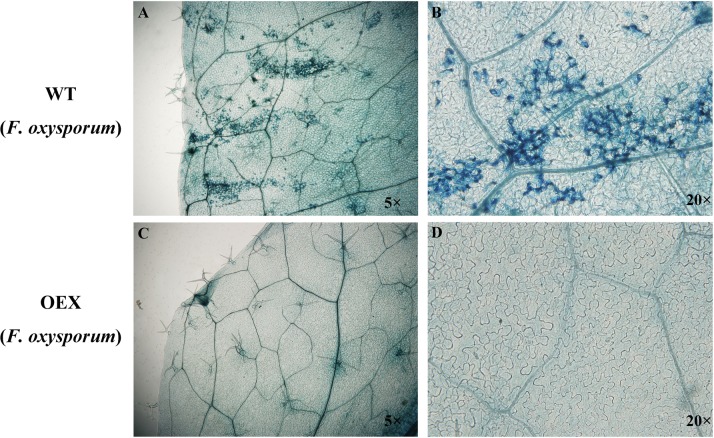
To better understand the molecular mechanisms of GbSBT1 in the immune response, we generated and self-crossed transgenic GbSBT1 Arabidopsis plants. After three rounds of self-crossing, two homologous lines showing different expression levels of the GbSBT1 gene were randomly selected and subjected to F. oxysporum and V. dahliae challenge. qPCR analysis of GbSBT1 expression levels in transgenic Arabidopsis lines OEX1 and OEX2 were carried out (S2 Fig). Obvious differences in disease symptoms between the WT and transgenic plants were observed at 2 days post-inoculation (dpi). The necrotic lesions were larger in the WT leaves than in the transgenic leaves (Fig 6). The disease spot diameter was also significantly smaller in the transgenic plants (0.36 ± 0.31 mm) than in the control plants (2.52 ± 0.66 mm). Similarly, necrotic lesions of V. dahliae on Arabidopsis leaves were smaller in the transgenic plants than in the WT plants (Fig 7). This result suggests that GbSBT1 overexpression inhibits the spread of necrotic lesions in transgenic leaves. Moreover, the control plants showed more pathogen aggregates near their leaf vessels, whereas the transgenic plants showed a negative result for trypan blue staining (Fig 8). As the infection progressed, the control plants wilted, whereas the transgenic GbSBT1 plants grew normally. These results demonstrate that the ectopic expression of GbSBT1 confers the increased tolerance of Arabidopsis against F. oxysporum infection.
Fig 6. Enhanced disease resistance of Arabidopsis against Fusarium oxysporum after GbSBT1 overexpression.
Wild-type (WT): Arabidopsis Columbia type Col-0; OEX: transgenic GbSBT1 lines. Left panels: un-inoculated plants; right panels: plants 2 days after F. oxysporum inoculation. (A–B) WT plants 0 and 2 days after ddH2O inoculation; (C–D) WT plants 0 and 2 days after F. oxysporum inoculation; (E–F) transgenic GbSBT1 plants 0 and 2 days after inoculation with ddH2O containing 0.2% Tween-20; (G–H) transgenic GbSBT1 plants 0 and 2 days after F. oxysporum inoculation. At least three biological replicates were performed in the F. oxysporum resistance analysis.
Fig 7. Enhanced disease resistance of Arabidopsis against V. dahliae after GbSBT1 overexpression.
Wild-type (WT): Arabidopsis Columbia type Col-0; OEX: transgenic GbSBT1 lines. Left panels: un-inoculated plants; right panels: plants 2 days after V. dahliae inoculation. (A–B) WT plants 0 and 2 days after ddH2O inoculation; (C–D) WT plants 0 and 2 days after V. dahliae inoculation; (E–F) transgenic GbSBT1 plants 0 and 2 days after inoculation with ddH2O containing 0.2% Tween-20; (G–H) transgenic GbSBT1 plants 0 and 2 days after V. dahliae inoculation. At least three biological replicates were performed in the V. dahliae resistance analysis.
Fig 8. Trypan blue staining of Fusarium oxysporum-inoculated leaves.
(A) Wild-type (WT) plants inoculated with F. oxysporum; (B) 20×magnification of the infected area in WT; (C) GbSBT1 overexpression in F. oxysporum-inoculated Arabidopsis leaves; (D) 20×magnification of the infected area in GbSBT1-overexpressing Arabidopsis. The trypan blue staining experiments were repeated at least three times.
Overexpression of GbSBT1 gene in Arabidopsis enhances the expression of defense-related genes
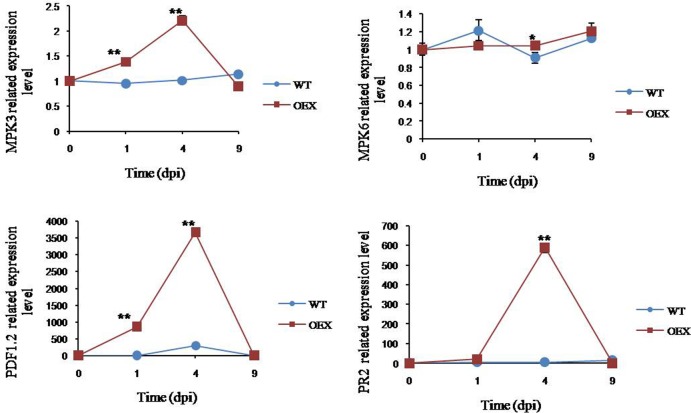
Given their consistent resistance effects on GbSBT1-overexpressing Arabidopsis plants and VIGS cotton plants, the transcripts of the genes involved in JA and immune transduction were analyzed in Arabidopsis. The expression levels of PR-2 and PDF1.2 were upregulated in the overexpressing lines following F. oxysporum inoculation (Fig 9). PR-2 was induced, peaked at 4 dpi, and then abruptly decreased. The highest level of PR-2 expression was nearly 10 times that of the WT plants at 4 dpi. PDF1.2 induction was peaked at 4 dpi; the highest expression of this gene was approximately 98.8 times as that of the control plants. The increased expression of PRs in GbSBT1 OEX lines contributed to the resistant phenotype.
Fig 9. qPCR analysis of gene expression in WT and transgenic GbSBT1 lines after F. oxysporum infection.
Gene expression levels were normalized to the constitutive ACT2 gene expression. Data represent the mean of gene expression levels with SD at different infection stages; n = 3 replicates. dpi: days post-inoculation. Error bars represent the standard deviation for three biological experiments, and three technical replicates were analyzed. Statistical significance was determined using two-tailed unpaired Student’s t-tests, and P values <0.05 were considered statistically significant (** for P values <0.01).
The activation of mitogen-activated protein kinases (MAPKs) is linked to innate immune responses [24,25]. The expression levels of MPK6 and MPK3 were assessed in the WT plants and GbSBT1 OEX lines to demonstrate whether or not the enhanced resistance phenotype is mediated by GbSBT1 expression (Fig 9). After F. oxysporum inoculation, the expression levels of MPK6 and MPK3 were higher in the GbSBT1 OEX lines than in the WT plants. MAPK activation in the GbSBT1 OEX lines indicates that GbSBT1 is a positive regulator of induced resistance response.
GbSBT1 interacts with PHB protein secreted by V. dahliae strain V991
The GbSBT1-interacting proteins were identified using GFP monoclonal antibody by extracting the total proteins of the V. dahliae-inoculated cotton leaves to characterize whether or not GbSBT1 senses the infection signals from pathogens, the total proteins of cotton leaves inoculated with V. dahliae were extracted to identify the GbSBT1 interacting proteins using GFP monoclonal antibody. The pull-down results indicated that 128 proteins were characterized after NLM analysis including 15 secreted proteins and 113 unsecreted proteins (S2 Table). Gene ontology showed that these secreted proteins were assigned to biological processes, including metabolic process, pigmentation, and response to stimulus. After searching the functions of the homologes online, 15 secreted proteins containing a 15–30 aa signal peptide were found to be involved in protein translocation across organelles or cell membrane (Table 1). Gene enrichment analysis also revealed that most of the secreted proteins participated in signal transduction and protein transport. The Het-C protein (gi|697081603), similar to the sugar ester transfer protein, drastically impairs ascospore production [26]. In summary, most of the secreted proteins are involved in cell wall biogenesis and hyphal growth during pathogenic ingression into host plants.
Table 1. GbST1-interacting proteins secreted from V. dahliae V991.
| Protein accession | Protein putative identity | Signal peptide | Peptide | Score |
|---|---|---|---|---|
| gi|697086726 | peptidyl-tRNA hydrolase | MSDTLHLILTTSAVTFLSGFALGVFAI | 13 | 378 |
| gi|697069413 | prohibitin | MAAALNFISKAAVPAFFGASLLSTAI | 3 | 95 |
| gi|697065929 | acetyl-CoA acetyltransferase | MANLPSVYIVSAARTPVGSFLGSLSSLSAV | 3 | 75 |
| gi|697067287 | oligosaccharyltransferase alpha subunit | MRAFAIATGLLSLVSAAVASSD | 4 | 60 |
| gi|697067481 | carbamoyl-phosphate synthase arginine-specific large chain | MPSAMACSLAGRAPAVLRHGRRMALPVRSFTSLTAASKSFTPAQSQ | 3 | 59 |
| gi|697088233 | zinc transporter SLC39A9 | MGGLFLLLVLCAVMAVASFLAG | 4 | 59 |
| gi|697067381 | uridylate kinase | MHATPRTLWRSLPTACTASRIASST | 4 | 54 |
| gi|697086240 | phosphoribosylaminoimidazole-succinocarboxamide synthase | MSSSAPLTTLSLPSLERIASGK | 2 | 44 |
| gi|697066901 | disulfide-isomerase erp38 | MVLLKSFVLGALAATAAAK | 3 | 36 |
| gi|697068043 | benzodiazepine receptor family protein | MTTFIPALTLPRQVFDHPATSILLPIALGTAVGYTSSR | 3 | 29 |
| gi|697089965 | CTP synthase | MKYLLVSGGVISGPLRRAFCS | 2 | 28 |
| gi|697081603 | Het-C protein | MASFHFGKGSWFLVFCIVLVLLPGRAAAF | 2 | 25 |
| gi|697068103 | helicase SWR1 | MAGLAAGFSSSPPGRGF | 2 | 20 |
| gi|697071463 | short-chain dehydrogenase/reductase SDR | MAAHRSATRALARAFTARTAAA | 2 | 18 |
| gi|697077881 | hypothetical protein VDAG_09776 | MSSNTLDNTALLATGICMVILTSAVFGF | 2 | 18 |
The proteins listed here were all detected at least thrice. The signal peptides were predicted by SignalP 4.1 online. “Peptide” indicates the number of detected peptide in the putative protein. Putative proteins were removed if number of detected peptide less than two (Peptide<2).The score indicates the total number of detection in the pellet pulled down by the antibody after digestion.
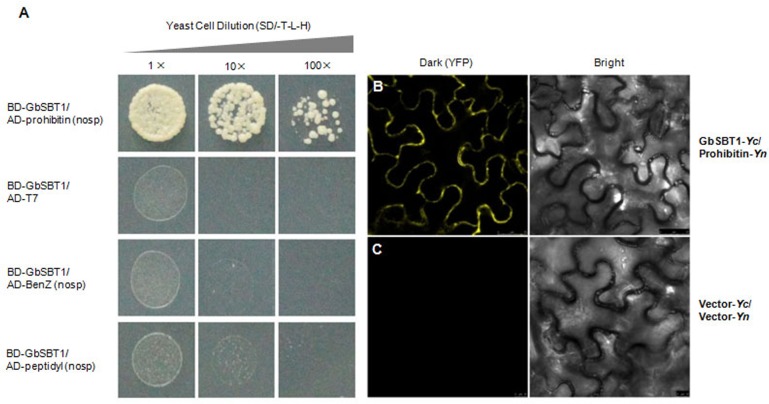
To further confirm which protein directly interacts with GbSBT1 during V. dahliae ingression into cotton, three proteins (based on the number of detected peptides in pull-down assay) were tested their interactions with GbSBT1 in yeast-two-hybridization. As shown in Fig 10A, GbSBT1 did not interact with the benzodiazepine receptor family protein and peptidyl-tRNA hydrolase, whereas the yeast cells co-transformed with GbSBT1 and PHB grew on the selective medium SD/-T-L-H. This result indicates that GbSBT1 can directly bind to PHB.
Fig 10. Interaction between GbSBT1 and prohibitin secreted by Verticillium dahliae.
(A) The yeast cells co-expressing GbSBT1 and benzodiazepine receptor family protein or peptidyl-tRNA hydrolase could not grow on the selective medium SD/-T-L-H, whereas the yeast cells co-transformed with GbSBT1 and prohibitin could grow on the selective medium SD/-T-L-H; (B-C) BiFC of epidermal cells co-expressing split YFP fusions of GbSBT1 and prohibitin or empty vector controls. Combinations of N- and C-terminal YFP fragments (Yn and Yc, respectively) were infiltrated as vector controls or fused to the N terminus of GbSBT1 and prohibitin as follows: B: GbSBT1-Yc and prohibitin -Yn, Scale bar: 25 μm; C: vector-Yc and vector-Yn, Scale bar: 10 μm. The interaction and co-localization were observed at the plasma membrane. Right is the corresponding bright-field
Bimolecular fluorescence complementation (BiFC) analysis was used to confirm the interaction between GbSBT1 and PHB in vivo. Fluorescence formed through formation of a fluorescent complex comprising of two fragments of YFP could be observed in the plasma membrane of tobacco leaf cells, indicating that PHB and GbSBT1 actually interacted in plants and as predicted (Fig 10B and 10C).
Discussion
V. dahliae secretes various effectors for its invasion and colonization in cotton. For instance, V. dahliae secretes pectinase or xylanase to degrade the cell wall of the elements of the vascular system and disrupt water transport in cotton [7]. The isochorismatase VdIsc1 is also secreted by V. dahliae to suppress the salicylate-mediated innate immunity of host cells [4]. Although the effector candidates have been identified in the V. dahliae genome and the functions of several effectors have been recently characterized, little is known about how the cotton plant interferes with the infection and colonization of the filamentous pathogen V. dahliae. In this study, we analysis the function of GbSBT1 and provide the evidences that GbSBT1 is involved in Verticillium-induced resistance in cotton.
Host proteases are important defense components against pathogens [27]. Host plants use proteases to monitor or facilitate effector-mediated degradation and subsequently activate effector-triggered immunity [28,29,30]. The extracellular subtilase P69C specifically processes an extracellular leucine-rich repeat LRP protein in tomato to initiate various signaling processes [31]. In Arabidopsis, SBT3.3 expression initiates a durable auto-induction mechanism that activates the salicylate-dependent expression of defense genes for amplified immune response [11]. Whether or not the GbSBT1 protein activates immune response remains unknown. Similar to the reported subtilases, GbSBT1 is localized in the cell membrane and in the extracellular apoplast, where V. dahliae ingression into cotton plant occurs and which serves as the recognition site between cotton plant and V. dahliae. In contrast to the SBT3.3 gene in Arabidopsis, the GbSBT1 gene is activated by JA and ethylene treatments instead of SA stimulus, demonstrating that JA signaling is required for the resistance of cotton plant against V. dahliae [23]. Moreover, the GbSBT1 protein moves from the cell membrane into the cytoplasm after V. dahliae attack and JA treatment, indicating that GbSBT1 is related to JA-induced resistance against V. dahliae [23].
The JA-responsive gene PDF1.2 encodes a protein that exhibits antimicrobial activity, which is dependent on COI1 and EIN2, and is correlated with resistance to necrotrophic pathogens [32,33]. Increased PDF1.2 expression indicated that GbSBT1 overexpression in Arabidopsis can induce the JA signaling pathway when responding to F. oxysporum attack. GbSBT1 overexpression in Arabidopsis also enhanced resistance against F. oxysporum and upregulated the expression of the PR1 and PR2 genes after fungal inoculation. Correspondingly, GbSBT1 knockdown in Pima-90 resulted in lost resistance to V. dahliae and reduced expression of the PR genes (S3 Fig). The changes in the expression of PR genes in both Arabidopsis and cotton plants indicate that GbSBT1 gene expression activates induced resistance. Furthermore, the MPK6 and MPK3 genes that are associated with innate immune response were activated by GbSBT1 gene expression in Arabidopsis, demonstrating that GbSBT1 plays important roles in innate immune responses. Considering that the SBT1 protein has no amino acid difference between from Coker-312 (sensitive to V. dahliae) and Pima-90 (resistant to V. dahliae), so we silenced the SBT1 gene in two genetic backgrounds and investigated their disease symptoms. The lost resistance to V. dahliae in Pima-90 and the absence of severe susceptibility of Coker-312 shows that the GbSBT1 gene is involved in R-gene resistance.
In tomato, cysteine protease is required for Cf2-dependent disease resistance and auto-necrosis suppression. During incompatible interaction, Avr2 physically interacts with and inhibits the extracellular Cys-protease Rcr3 in resistant tomato varieties [34]. The localization of GbSBT1 protein made it possible for its recognition with protein secreted by V. dahliae. Therefore, we employed the pull-down assay to identify GbSBT1-interacting proteins and thus analyze further the role of GbSBT1 in Verticillium resistance. A total of 15 secreted proteins in V991 pathogen were identified as candidates. Searching for the functions of the homologs of these proteins revealed that the secreted proteins are likely related to the pathogenicity of V. dahliae during the invasion of host plants. The first type of secreted proteins, such as Het-C protein (gi|697081603), is required for hyphal and spore growth, and deletion of these genes results in the loss of cell wall integrity. The second type of secreted proteins facilitates pathogen evasion from host surveillance. All of the pull-down proteins of V. dahliae are involved in pathogenicity, indicating that GbSBT1 may effectively recognize the effectors of V. dahliae.
To confirm further which specific effector interacts with GbSBT1, we determined the interaction of three proteins in the yeast two-hybrid system and confirmed that PHB is the GbSBT1-interactor. PHB, which is localized in the cell membrane, is a highly conserved protein exhibiting a common structure and a domain that is similar to that of other organisms. In human, the PHB1 of the host binds to RtxA1-D2, and downregulation of PHB1 genes by small interfering RNAs reduces RtxA1-D2 cytotoxicity against HeLa cells. PHB1 is directly involved in RtxA1 cytotoxicity when Vibrio cholerae infects HeLa cells [35]. Furthermore, the Vi capsular polysaccharide of Salmonella typhi targets the PHB family in intestinal epithelial cells and suppresses early inflammatory responses [36]. These findings reveal that PHB modulates early inflammatory responses during infection. In plants, PHB3 affects NO accumulation and NO-mediated processes [37]. Moreover, a PHB3 mutant exhibits an extreme constitutive ethylene response in etiolated seedlings and reduces ethylene-inducible genes, such as AtEBP and PDF1.2, suggesting that AtPHB3 plays dual roles in ethylene signaling [38]. These findings indicate that PHB is associated with inflammatory response in human, and ethylene or NO mediated cellular responses in plants. In this study, we found that a PHB interacts with GbSBT1 by using pull-down and yeast two-hybrid assays. Based on the function of GbSBT1 during Verticillium ingression, we hypothesized that the PHB from V. dahliae interacts with GbSBT1 and then activates the transcription of genes related to Verticillium resistance signaling in cotton. However, the mechanism underlying GbSBT1-PHB interaction and the roles of PHB in Verticillium resistance warrant further investigation.
Supporting Information
VIGS experiments were repeated at least three times with more than 10 cotton plants for each construct. Double asterisks represent significant difference between VIGS plants and wild-type plants (P < 0.01) in t-test.
(PDF)
The comparative CT method is adopted, and the expression is normalized. Each sample was repeated at least thrice. Error bars represent SE. Double asterisks represent a significant expression change of GbSBT1 compared to uninfected condition (P < 0.01) in t-test.
(PDF)
Total RNA was isolated from leaves of the control and inoculated VIGS cotton plants. Changes in the expression of defense-related genes, including PR1 and PR2, were analyzed via qRT-PCR. The ubiquitin7 gene (DQ116441) of cotton plant served as an internal control. The primers (GbPR1 and GbPR2) used in qRT-PCR analysis are listed in S1 Table. Gene expression levels were normalized to the constitutive Ubiquitin gene expression. The expression levels are the means with SD; n = 3 replicates. dpi: days post-inoculation. Double asterisks represent a very significant difference between VIGS plants and wild-type plants (P < 0.01) in t-test.
(PDF)
(DOCX)
(DOCX)
(MP4)
Acknowledgments
We thank Dr. Guiliang Jian for providing the defoliating isolate V991 of V. dahliae.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Basic Research Project of MOST in China (973 Project) (2013CB733903), the National High-Tech (863) Program of China (No. 2012AA02A703), the Natural Science Foundation of China (31071458) and China Transgenic Program (2013AA08005-003). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gao X, Wheeler T, Li Z, Kenerley CM, He P, Shan LB (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J 66: 293–305. 10.1111/j.1365-313X.2011.04491.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Bebany AF, Rampitsch C, Daayf F (2010) Proteomic analysis of the phytopathogenic soilborne fungus Verticillium dahliae reveals differential protein expression in isolates that differ in aggressiveness. Proteomics 10: 289–303. 10.1002/pmic.200900426 [DOI] [PubMed] [Google Scholar]
- 3.de Jonge R, van Esse HP, Maruthachalam K, Bolton MD, Santhanam P, Saber MK, et al. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Natl Acad Sci U S A 109: 5110–5115. 10.1073/pnas.1119623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T, Song T, Zhang X, Yuan H, Su L, Li W, et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun 5: 4686 10.1038/ncomms5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie C, Wang C, Wang X, Yang X (2013) Proteomics-based analysis reveals that Verticillium dahliae toxin induces cell death by modifying the synthesis of host proteins. J Gen Plant Pathol 79: 335–345. [Google Scholar]
- 6.Klosterman SJ, Subbarao KV, Kang S, Veronese P, Gold SE, Thomma BP, et al. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog 7: e1002137 10.1371/journal.ppat.1002137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Z, Liu H, Wang C, Xu JR (2013) Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14: 274 10.1186/1471-2164-14-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dodson G, Wlodawer A (1998) Catalytic triads and their relatives. Trends Biochem Sci 23: 347–352. [DOI] [PubMed] [Google Scholar]
- 9.Che P, Bussell JD, Zhou W, Estavillo GM, Pogson BJ, Pogson BJ, et al. (2010) Signaling from the Endoplasmic Reticulum Activates Brassinosteroid Signaling and Promotes Acclimation to Stress in Arabidopsis. Sci Signal 3: ra69–ra69. 10.1126/scisignal.2001140 [DOI] [PubMed] [Google Scholar]
- 10.Figueiredo A, Monteiro F, Sebastiana M (2014) Subtilisin-like proteases in plant-pathogen recognition and immune priming: a perspective. Front Plant Sci 5: 739 10.3389/fpls.2014.00739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez V, Lopez A, Mauch-Mani B, Gil MJ, Vera P (2013) An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathog 9: e1003445 10.1371/journal.ppat.1003445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorda L, Coego A, Conejero V, Vera P (1999) A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J Biol Chem 274: 2360–2365. [DOI] [PubMed] [Google Scholar]
- 13.Tornero P, Conejero V, Vera P (1997) Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem 272: 14412–14419. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava R, Liu JX, Howell SH (2008) Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J 56: 219–227. 10.1111/j.1365-313X.2008.03598.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuo K, Wang J, Wu W, Chai Y, Sun X, Tang K (2005) Identification and characterization of differentially expressed ESTs of Gossypium barbadense infected by Verticillium dahliae with suppression subtractive hybridization. Mol Biol 39: 191–199. [PubMed] [Google Scholar]
- 16.Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5: 150–163. [DOI] [PubMed] [Google Scholar]
- 17.Sheen J (2002) A transient expression assay using Arabidopsis mesophyll protoplasts.
- 18.Fradin EF, Zhang Z, Juarez Ayala JC, Castroverde CD, Nazar RN, Robb J, et al. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol 150: 320–332. 10.1104/pp.109.136762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao W, Long L, Zhu L-F, Xu L, Gao W-H, Sun LQ, et al. (2013) Proteomic and Virus-induced Gene Silencing (VIGS) Analyses Reveal That Gossypol, Brassinosteroids, and Jasmonic acid Contribute to the Resistance of Cotton to Verticillium dahliae. Mol Cell Proteomics 12: 3690–3703. 10.1074/mcp.M113.031013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Wang J, Zhang L, Zuo K (2013) A cotton annexin protein AnxGb6 regulates fiber elongation through its interaction with actin 1. PLoS One 8: e66160 10.1371/journal.pone.0066160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang X, Henriques R, Lin S- S, Niu Q- W, Chua N- H (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protocols 1: 641–646. [DOI] [PubMed] [Google Scholar]
- 22.Fradin EF, Abd-El-Haliem A, Masini L, van den Berg GC, Joosten MH, Thomma BP (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol 156: 2255–2265. 10.1104/pp.111.180067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, He X, Luo X, Xu L, Liu L, Min L, et al. (2014) Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN1 expression. Plant Physiol 166: 2179–2194. 10.1104/pp.114.246694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beckers GJ, Jaskiewicz M, Liu Y, Underwood WR, He SY, Zhang S, et al. (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21: 944–953. 10.1105/tpc.108.062158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Xie J, Yan C, Zou X, Ren D, Zhang S (2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase‐mediated oxidative burst are two independent signaling events in plant immunity. Plant J 77: 222–234. 10.1111/tpj.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saupe S, Descamps C, Turcq B, Begueret J (1994) Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc Natl Acad Sci U S A 91: 5927–5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Hoorn RA (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223. 10.1146/annurev.arplant.59.032607.092835 [DOI] [PubMed] [Google Scholar]
- 28.Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S (2004) A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem 279: 26370–26377. [DOI] [PubMed] [Google Scholar]
- 29.Tian M, Benedetti B, Kamoun S (2005) A Second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol 138: 1785–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian M, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S (2007) A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 143: 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tornero P, Mayda E, Gómez MD, Cañas L, Conejero V, Vera P (1996) Characterization of LRP, a leucine-rich repeat (LRR) protein from tomato plants that is processed during pathogenesis. Plant J 10: 315–330. [DOI] [PubMed] [Google Scholar]
- 32.Penninckx IA, Thomma BP, Buchala A, Metraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10: 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, et al. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci U S A 95: 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, et al. (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747. [DOI] [PubMed] [Google Scholar]
- 35.Kim BA, Lim JY, Rhee JH, Kim YR (2016) Characterization of prohibitin 1 as a host partner of Vibrio vulnificus RtxA1 toxin. J Infec Dis 213: 131–138. [DOI] [PubMed] [Google Scholar]
- 36.Sharma A, Qadri A (2004) Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc Natl Acad Sci U S A 101: 17492–17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Ries A, Wu K, Yang A, Crawford NM (2010) The Arabidopsis Prohibitin Gene PHB3 Functions in Nitric Oxide-Mediated Responses and in Hydrogen Peroxide-Induced Nitric Oxide Accumulation. Plant Cell 22: 249–259. 10.1105/tpc.109.072066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christians MJ, Larsen PB (2007) Mutational loss of the prohibitin AtPHB3 results in an extreme constitutive ethylene response phenotype coupled with partial loss of ethylene-inducible gene expression in Arabidopsis seedlings. J Exp Bot 58: 2237–2248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VIGS experiments were repeated at least three times with more than 10 cotton plants for each construct. Double asterisks represent significant difference between VIGS plants and wild-type plants (P < 0.01) in t-test.
(PDF)
The comparative CT method is adopted, and the expression is normalized. Each sample was repeated at least thrice. Error bars represent SE. Double asterisks represent a significant expression change of GbSBT1 compared to uninfected condition (P < 0.01) in t-test.
(PDF)
Total RNA was isolated from leaves of the control and inoculated VIGS cotton plants. Changes in the expression of defense-related genes, including PR1 and PR2, were analyzed via qRT-PCR. The ubiquitin7 gene (DQ116441) of cotton plant served as an internal control. The primers (GbPR1 and GbPR2) used in qRT-PCR analysis are listed in S1 Table. Gene expression levels were normalized to the constitutive Ubiquitin gene expression. The expression levels are the means with SD; n = 3 replicates. dpi: days post-inoculation. Double asterisks represent a very significant difference between VIGS plants and wild-type plants (P < 0.01) in t-test.
(PDF)
(DOCX)
(DOCX)
(MP4)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.