ABSTRACT
Autophagy and inflammation are 2 fundamental biological processes involved in both physiological and pathological conditions. Through its crucial role in maintaining cellular homeostasis, autophagy is involved in modulation of cell metabolism, cell survival, and host defense. Defective autophagy is associated with pathological conditions such as cancer, autoimmune disease, neurodegenerative disease, and senescence. Inflammation represents a crucial line of defense against microorganisms and other pathogens, and there is increasing evidence that autophagy has important effects on the induction and modulation of the inflammatory reaction; understanding the balance between these 2 processes may point to important possibilities for therapeutic targeting. This review focuses on the crosstalk between autophagy and inflammation as an emerging field with major implications for understanding the host defense on the one hand, and for the pathogenesis and treatment of immune-mediated diseases on the other hand.
KEYWORDS: autoimmune diseases, autoinflammatory diseases, autophagy, carcinogenesis, infectious diseases, inflammation
Introduction
Autophagy is the physiological cellular process through which intracellular components undergo lysosome-mediated self-digestion and recycling.1 In conditions of cellular stress such as starvation, hypoxia, or exposure to toxic molecules, the autophagy machinery is activated in order to maintain cell homeostasis. An increasing body of evidence has emerged supporting the view that autophagy is involved in several physiological processes including cell metabolism, cell survival, and host defense.2 It can be argued that autophagy is a primordial form of eukaryotic innate immunity against invading microorganisms. Research performed in the past decade has also proposed that, in mammals, these primordial functions of autophagy have evolved and now encompass multiple innate and adaptive immune mechanisms.3 Moreover, defective autophagy is associated with several pathological conditions such as cancer, autoimmune disease, neurodegenerative disease, and senescence.4 These new insights strongly suggest that besides its classical role as a “housekeeping” mechanism, autophagy can also be considered as crucial for host defense in general, and regulation of inflammation in particular. In this review we present the newest concepts that link these 2 fundamental biological processes, autophagy, and inflammation.
Autophagy and host defense
Several types of autophagy have been recognized, including macroautophagy, microautophagy, chaperone-mediated autophagy, and noncanonical autophagy.5 In this review we will focus on macroautophagy, which will be henceforth referred to as autophagy. This process is activated in response to a multitude of stress factors including starvation, hypoxia, and other toxins that result in accumulation of damaged organelles and protein aggregates. As a consequence, a double-layer membrane, the phagophore, is formed to engulf the damaged cytosolic components (Fig. 1). After complete sequestration of these components, the resulting autophagosome fuses with the lysosome, which enables the breakdown of the autophagosome inner membrane and exposes the cargo to the lysosomal hydrolases. The resulting breakdown products are subsequently released in the cytosol and are available to be reused for the synthesis of required macromolecules.5 Several autophagy-related (ATG) genes and proteins are involved in the induction of autophagy (Fig. 1), while a subset of them also have autophagy-independent functions.6
Figure 1.
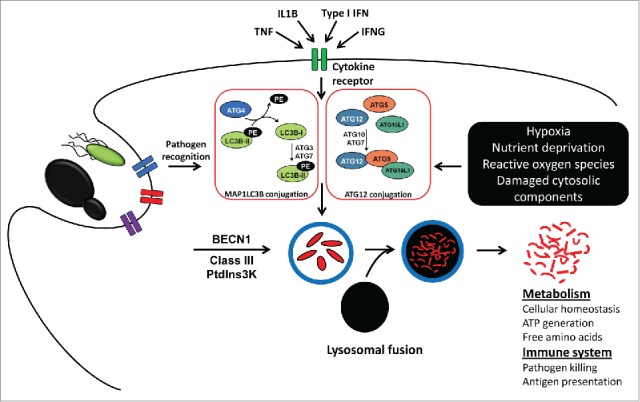
The process of autophagy in eukaryotic cells and its involvement in cellular homeostasis, metabolism, and the immune system. Autophagy is induced both by metabolic and immune signals comprising pathogen recognition and stimulation by proinflammatory cytokines. Upon autophagy triggering through any of these pathways, the MAP1LC3B (depicted as unconjugated LC3B-I and PE-conjugated LC3B-II) and ATG12 conjugation systems, as well as BECN1 and class III PtdIns3K, are activated that are critical for autophagosome formation. After engulfment of the autophagic cargo, autophagosomes proceed to lysosomal fusion, leading to degradation of the sequestered content. In case the autophagic cargo originates from intracellular organelles or protein complexes, the degraded fragments are utilized for metabolic purposes to generate ATP and provide the cell with free amino acids. In contrast, autophagy-mediated degradation of extracellular material, especially microbes, enables pathogen killing and activation of adaptive immunity by MHC-dependent antigen presentation.
Among the most important functions of autophagy in addition to maintaining cell homeostasis is its role in host defense. Autophagy improves host defense mechanisms through several biological functions: the direct elimination of the invading pathogens (involving also xenophagy and MAP1LC3/LC3 [microtubule-associated protein 1 light chain 3]-associated phagocytosis-LAP),7 control of adaptive immunity through regulation of antigen handling and presentation,8,9 induction of innate immune memory or trained immunity,10 and modulation of inflammation. Indeed, autophagy has been suggested to represent an ancient form of innate immune response to an infection.3 Engagement of various families of pattern-recognition receptors (PRRs) has been reported to induce autophagy through pathways that are similar to those activated in response to nutrient deprivation, and mediated via MTOR (mechanistic target of rapamycin [serine/threonine kinase]) and adenosine monophosphate (AMP)-activated protein kinase (AMPK). In this respect, engagement of TLR4 (toll-like receptor 4) by lipopolysaccharide (LPS) of Gram-negative bacteria results in the ubiquitination of BECN1/beclin-1 by TRAF6 (TNF receptor-associated factor 6, E3 ubiquitin protein ligase).11 TRAF6 also activates the serine/threonine protein kinase ULK1 (unc-51 like autophagy activating kinase 1), both pathways leading to the activation of autophagy.12 Interestingly, the LPS-induced autophagy is dependent on MAPK/p38 and TICAM1/TRIF (toll-like receptor adaptor molecule 1), but not on MYD88 (myeloid differentiation primary response 88).13 Similarly, the engagement of several other TLRs also induces autophagy.14 Not only TLRs, but nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) as well have been demonstrated to induce autophagy: NOD2 induces autophagy in dendritic cells and directs antigen handling and presentation,15 a process that involves targeting of ATG16L1 to the plasma membrane at the site of bacterial entry.16
In addition to PRRs, several proinflammatory cytokines including TNF (tumor necrosis factor)17 and IL1B (interleukin 1, β)18 induce autophagy and thus improve the control of infection. Furthermore, both type I (IFNA/IFNα-IFNB1/IFNβ-IL6) and type II (IFNG/IFNγ) interferons are major factors in activating autophagy.19 For instance, activation of macrophages by IFNG is crucial for intracellular killing of Mycobacterium tuberculosis,20,21 and type I IFN for host defense against viruses.22 In mice, IFNG induces the expression of autophagic p47 immunity-related guanosine-5′-triphosphate (GTP)ases (IRG) proteins,21 with IRGM1 (immunity-related GTPase family M member 1) being an important factor for inducing antimycobacterial autophagy. These findings are corroborated by the genetic association of IRGM polymorphisms with tuberculosis in West Africans, and increased Bacillus Calmette-Guérin (BCG) vaccination efficacy through enhanced antigen presentation.23-25 In contrast to the proinflammatory cytokines, T helper 2 (Th2) type responses and secretion of anti-inflammatory cytokines such as IL4 and IL13 antagonize autophagy induction through activation of MTOR.26 Altogether, the T helper 1/T helper 2 (Th1/Th2) responses seem to be correlated to the activation of autophagy, with autophagy being induced in Th1 type responses and inhibited when Th2 responses are elicited.
In addition, it must be underlined that some infectious agents are able to manipulate and inhibit autophagy, a de facto mechanism of evasion from the host defense. In line with this, human immunodeficiency virus (HIV) infection of CD4 lymphocytes reduces their content of BECN1 and MAP1LC3B-II, 2 crucial components for the induction of autophagy.27 Moreover, M. tuberculosis is also able to inhibit autophagy and phagolysosome biogenesis, a process that greatly contributes to the escape from the bactericidal mechanisms of the host.28 The fact that autophagy was targeted by microorganisms during evolution, with inhibitory effects increasing survival of the pathogens, demonstrates its importance for host defense.
In this review we will further focus on the modulatory effects of autophagy on inflammation, a broad concept with important consequences for the pathogenesis not only of infections, but of autoinflammatory diseases and malignancies as well.3
The impact of autophagy on inflammation
Innate immunity represents the first line of defense against pathogens. It requires adequate pathogen recognition, which triggers activation of inflammation resulting in recruitment of immune cells, such as phagocytes, secretion of cytokines and chemokines, which in turn help recruit other immune cells and prime dendritic cells (DCs) for activation of adaptive immune responses. An increasing number of studies have recently shown that autophagy has important effects on the induction of the inflammatory reaction. However, when the inflammatory process is not properly controlled, autophagy can be detrimental for the host. In atherosclerosis, excessively stimulated autophagy can lead to endothelial cell death that can contribute to plaque destabilization, maintaining the inflammatory status of the plaque.29 In chronic obstructive pulmonary disease (COPD) cigarette smoking triggers a form of autophagy, namely mitophagy (autophagy-dependent selective elimination of mitochondria), through stabilization of the mitophagy regulator PINK1 (PTEN-induced putative kinase 1). This eventually leads to epithelial cell death, and thus can contribute to persisting inflammatory responses in COPD.30
Autophagy effects on inflammasome activation and IL1B production
Production of cytokines from the IL1 family, and especially IL1B, is one of the main mechanisms through which innate and adaptive immune responses are induced by an infection. The regulation of the production of this cytokine is thus crucial for understanding inflammation and host defense. Probably the best studied aspect of the interaction between autophagy and inflammation is represented by the effects of autophagy on inflammasome activation and IL1B release. An overview of the interplay between autophagy and IL1B-inflammasome activation is depicted in Figure 2. The inflammasome consists of protein complexes formed by one or more members of the NLR family of receptors together with CASP1/caspase-1, leading to the activation of this cysteine protease, and processing of the inactive pro-IL1B and pro-IL18 into active cytokines.31 The first observation regarding the effect of autophagy on inflammasome activation was that of Saitoh et al. who reported that macrophages of Atg16l1 knockout mice respond with increased production of IL1B after stimulation with LPS. This was due to an exaggerated activation of CASP1 in the Atg16l1-deficient mice, mediated by the adapter molecule TICAM.32 Additional studies support the concept that autophagy regulates inflammasome activation.33-36
Figure 2.
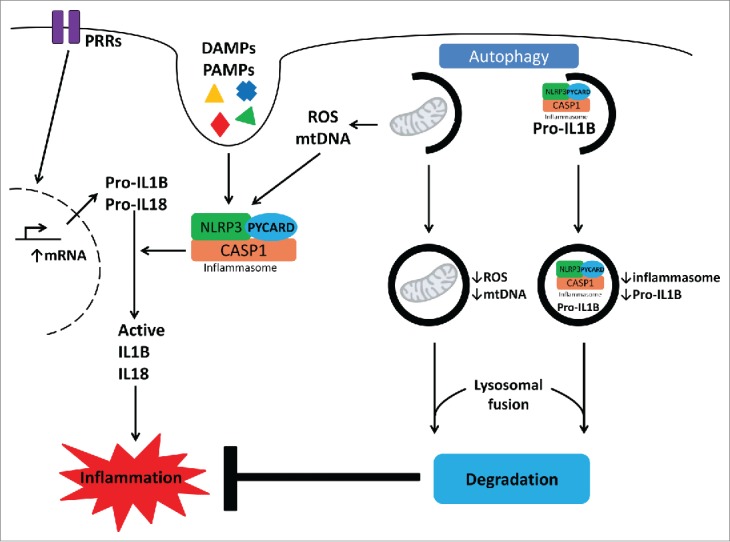
The interplay between autophagy and IL1B-inflammasome activation. Production of the pro-inflammatory cytokines IL1B and IL18 is pivotal in antimicrobial host defense, since this leads to activation of both innate and adaptive immune responses including Th1 and Th17. Autophagy potently regulates these immune responses by several means. First, autophagy inhibits IL1B and IL18 production through digestion of dysfunctional mitochondria and thereby prevention of mitochondrial ROS release that is known to activate the inflammasome. Second, autophagy is capable of targeting inflammasome complexes for degradation, which prevents cleavage of pro-IL1B and pro-IL18 into their biologically active counterparts. Finally, the autophagy machinery engulfs and eradicates pro-IL1B proteins, representing another level of IL1B regulation. DAMP, danger-associated molecular pattern; PAMP, pathogen-associated molecular pattern; PRR, pattern recognition receptor; ROS, reactive oxygen species.
Several mechanisms have been proposed to mediate these anti-inflammatory effects of autophagy. During homeostatic conditions autophagy is responsible for the clearing of the cytoplasm of nonfunctional mitochondria and other organelles. If autophagy is defective this leads to an accumulation of depolarized mitochondria, that release inflammasome activators such as reactive oxygen species (ROS) or mitochondrial DNA (mtDNA).33,34,37 Moreover, autophagy may also remove aggregated inflammasome structures, thus contributing to dampening proinflammatory responses.36 These processes might play an important role during infections. This is underscored by the observation that influenza virus triggers RIPK2 (receptor-interacting serine-threonine kinase 2) to activate ULK1, to enhance mitophagy and control inflammasome activation.38 These studies in mice point toward a regulatory effect of autophagy on CASP1 activation through modulation of the NLRP3 (NLR family, pyrin domain containing 3) inflammasome.
The important role of autophagy for IL1B secretion was also confirmed in human primary cells, in which inhibition of autophagy leads to increased production of IL1B.39 Interestingly, the same study in humans demonstrated that TNF production is decreased by autophagy inhibition, suggesting divergent effects of autophagy on the production of these cytokines.39 However, important differences may exist between mice and humans regarding the effect of autophagy on IL1B production. In mice, this effect is ascribed to inhibition of inflammasome activation by autophagy, whereas in humans IL1B mRNA transcription is elevated when autophagy is inhibited, whereas no effects are observed on CASP1 activation.39,40 One potential mechanism for this observation was proposed to be due to targeting pro-IL1B for degradation,40 while a recent study also reported autophagy-dependent degradation of the PELI (pellino 3 ubiquitin protein ligase) with subsequent inhibition of IL1B expression during TLR4 signaling.41 These discrepancies between mice and humans emphasize that caution should be exercised when extrapolating studies from mice to humans. Moreover, it has recently been demonstrated that a noncanonical secretion pathway controlled by autophagy plays an important role in the secretion of proteins that lack a signal sequence, such as IL1B. In contrast to basal autophagy, which inhibits IL1B secretion,33 induced autophagy augments IL1B secretion via an unconventional secretion mechanism that utilizes the autophagic machinery.35
The effect of autophagy on other inflammatory pathways
Autophagy does not only have inhibitory effects on inflammasome activation, but also on inflammatory mediators that are independent of CASP1 activation. In line with this, autophagy suppresses CAPN/calpain-dependent IL1B activation, and this effect is linked to the release of ROS as well.42 In addition, autophagy reduces NFKB (nuclear factor of kappa light polypeptide gene enhancer in B-cells) activation by selective degradation of BCL10 complexes.43 This process is mediated through NSFL1C (NSFL1 [p97] cofactor [p47]), a negative regulator of IKBKB/IKK acting via degradation of the polyubiquitinated NFKB modulator IKBKG/NEMO.44
In addition to these effects on proinflammatory cytokines, autophagy also amplifies TLR signaling to improve the delivery of microbial ligands to cytoplasmic receptors such as TLR7 that promote the synthesis of type I IFN.45 This could also lead, however, to inappropriate immune activation, as shown by the increased trafficking of DNA-containing antigens and increased IFNA secretion.7,46 However, the effects of autophagy on the stimulation of type I IFN is complex, with the ATG12–ATG5 complex inhibiting the signaling induced by RIG-I-helicase like receptors (RLRs) by direct binding to the caspase-recruitment domain of the RLR adaptor molecule MAVS.47
These data suggest therefore that inflammation and autophagy are intertwined processes during host defense, and derangements in the crosstalk between these 2 processes can have important consequences for the pathogenesis and treatment of several diseases.
Autophagy in the pathogenesis and treatment of immune-mediated disease
Due to the mechanisms described above, autophagy has emerged as a central player in the pathogenesis of immune-mediated diseases, including autoinflammatory and autoimmune diseases, infections, and cancer.
The impact of autophagy on autoinflammatory and autoimmune diseases
The first major autoinflammatory disease in which autophagy has been suggested to play a significant role is Crohn disease (CD). CD is a chronic inflammatory disorder of the intestine caused by a combination of multiple factors including defective host response, altered mucosal barrier function, and exaggerated cytokine production. Genetic variants of important autophagy genes such as ATG16L1, IRGM, and ULK1, have been reproducibly associated with susceptibility and clinical outcome of the disease, demonstrating the important role of autophagy in CD.48-51 Functional studies have revealed that autophagy and subsequently inflammatory pathways are heavily influenced by genetic variation in autophagy genes, the ATG16L1 variant in particular, as has been demonstrated for defective Paneth cell function,52 impaired bacterial defense,53,54 aberrant antigen presentation,55,56 and increased production of proinflammatory cytokines including IL1B and IL18.57-59 The IRGM promoter polymorphism risk allele influences IRGM expression, at least in part through posttranscriptional regulation by miRNAs.60,61 Furthermore, a well-established genetic risk factor with the highest predictive value for CD is for the gene encoding NOD2, a PRR that recognizes peptidoglycans and activates immune pathways and autophagy.62-65 Another indication that autophagy is a central process in intestinal homeostasis is the recent finding of CALCOCO2/NDP52 mutations in CD patients, a gene that codes for an important factor in phagophore targeting of intracellular bacteria (Fig. 3).66,67
Figure 3.
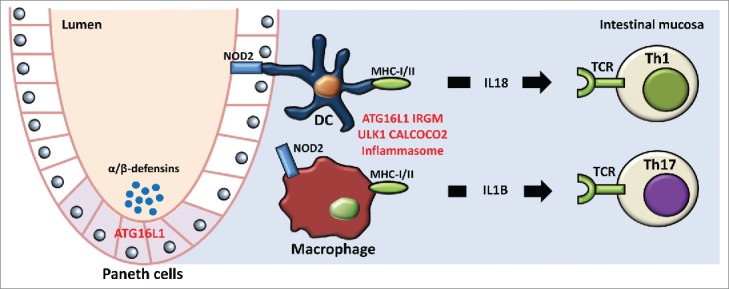
Interplay between autophagy and inflammation in the pathogenesis of Crohn disease. Multiple facets of inflammatory pathways involved in CD pathogenesis are influenced by autophagy, ranging on the one hand from defective microbial recognition and phagocytosis in dendritic cells (DCs) through the PRR NOD2, diminished antigen processing and presentation on MHC class I/II caused by genetic variants in ATG16L1, IRGM, and CALCOCO2, and on the other hand to a loss of inhibition of IL1B and IL18 production, important pro-inflammatory mediators leading to generation of Th1 and Th17 responses. Furthermore, genetic variation in ATG16L1 impairs the capacity of Paneth cells to excrete antimicrobial peptides, including DEFA/α- and DEFB/β-defensins. All together, these defects allow for microbial overgrowth and lead to hyperinflammation. DC, dendritic cell; TCR, T cell receptor.
In addition to CD, important insights have also been provided by the assessment of autophagy induction and inflammasome activation in patients with chronic granulomatous diseases (CGD). CGD is a primary immunodeficiency characterized by defective ROS production due to mutations in the proteins forming the NOX (NADPH oxidase) complex.68 In addition to the defects in host defense, CGD is characterized by the presence of hyperinflammatory features, such as colitis that is indistinguishable from CD in 30% of the patients.69 Earlier studies have demonstrated that IL1B production capacity of monocytes and macrophages isolated from CGD patients is increased compared to controls,70,71 bringing into question the importance of NADPH-dependent ROS production for inflammasome activation. Since NADPH-oxidase dependent ROS was shown to be important for the induction of LAP,72 this led to the hypothesis that defective LAP in CGD may lead to hyperinflammation. Indeed, a recent study showed defective LAP in both patients with CGD and mice with mutations in genes encoding the NOX complex, which contribute to the exaggerated IL1B production.73 More importantly, treatment of CGD patients having colitis with recombinant IL1RN/IL1RA (interleukin 1 receptor antagonist) significantly improve the clinical picture.73,74 IL1RN not only reduces excessive IL1-driven inflammation, but also restores deficient LAP in NOX-deficient cells. IL1RN also has intracellular effects, and cellular uptake of IL1RN has been reported.75 Therefore, IL1RN might influence LAP through several possible mechanisms: i) direct inhibition of IL1 bioactivity, ii) intracellular interaction with the LAP machinery or iii) indirectly via effects on other processes such as inflammasome activation which is responsible for pro-IL1B processing.
Epidemiological studies assessing genetic variants in autophagy genes demonstrated their association with susceptibility to autoimmune disease as well, such as variants in ATG5 associated with systemic lupus erythematosus (SLE)76-78 and systemic sclerosis.79 Furthermore, ATG5 expression is elevated in both blood and brain-residing T-cells in multiple sclerosis patients and is associated with the relapsing-remitting disease phenotype.80 These ATG5 genetic variants are functionally linked to a differential ATG5 expression and exhibit gene-gene interactions with ATG7 and IRGM,81 thereby likely influencing the degree of activation of the autophagy process. Interestingly, ATG5 also plays an important role in clearing apoptotic cells,82 and a hallmark of SLE is the deficiency to clear apoptotic cells. Similar to CD, IRGM has also been implicated in multiple sclerosis based on the observation that IRGM expression is strongly induced in affected lesions of multiple sclerosis patients. Consistently, IRGM1-deficient mice are resistant to the experimental model of multiple sclerosis, antigen-induced experimental autoimmune encephalomyelitis (EAE), by promoting blood-brain barrier integrity and preventing T cell infiltration into the central nervous system.83,84
SLE is characterized by deregulation of autoantibody-producing B cells, infiltration of target organs by inflammatory T cells and aberrant immune cell activation. The latter is caused by abnormal activation of myeloid cells triggered by the presence of immune complexes composed of self-DNA from apoptotic cells and antibodies generated by autoreactive B cells.85,86 While T cells are considered as a major cell type driving initiation and progression of SLE,87,88 innate immune cells are also implicated in the disease process. In this respect, inappropriate handling of apoptotic bodies by phagocytes leads to accumulation of autoantigens that could promote loss of tolerance, predisposing the organism to autoimmune responses.89,90 Autophagy is likely involved in these processes, since it influences T cell development and activation, but also because it executes clearance of apoptotic cells and contributes to the elimination of their immunogenic potential.91
In rheumatoid arthritis (RA), a chronic inflammatory disorder affecting the joints, autophagy has a critical role in TNF-induced joint destruction (in murine experimental arthritis); TNF activates autophagy by induction of BECN1 and ATG7 and the conversion from MAP1LC3B-I to MAP1LC3B-II in osteoclasts, providing a mechanism for TNF-associated synovial inflammation and bone resorption.92 These findings suggest that TNF promotes autophagy-dependent osteoclast differentiation and survival, which is confirmed in human synovial fibroblasts.93-95
The role of autophagy in infectious diseases
In the fight against pathogenic microorganisms, autophagy (this form of autophagy is called xenophagy) contributes to both innate immune defense and to development of adaptive immune memory, and excellent recent reviews have described these mechanisms in great detail.96,97 The role of autophagy in infectious diseases is apparent at the level of microbial handling by the innate immune system, as well as antigen processing and presentation of a wide variety of microorganisms ranging from viruses, to bacteria, fungi, and protozoa.
Within the context of innate host defense, autophagy provides the machinery for intracellular killing of microbes by means of phagophore engulfment and lysosomal fusion. Proper designation of microorganisms for autophagy processing is facilitated by protein adaptors such as SQSTM1/p62, NBR1 and CALCOCO2, that are activated downstream of microbial recognition by pattern-recognition receptors (TLR, NLR, RLR), providing anchors for phagophore assembly around the selected cargo.67,98,99 Several intracellular signaling pathways have been described to induce autophagy by PRRs. Activation of a TLR4-TICAM1 pathway has been reported to induce autophagy by Gram-negative bacteria such as Pseudomonas aeruginosa, and cleavage of TICAM1 by CASP1 inhibits autophagy.100 In addition, another important factor for TLR-induced autophagy is activation of the IRF8 transcription factor, which in turn activates genes involved in autophagosome formation.101 Induction of autophagy by NLR receptors is induced by the interaction with the peptidoglycans that activate a RIPK2/RIP2 (receptor-interacting serine-threonine kinase 2)-dependent signaling pathway.102,103 Furthermore, a very important process during the induction of autophagy is represented by the interaction with molecular sensors of metabolites. In this respect, during antimycobacterial responses induction of autophagy necessitates activation of AMPK that induces PPARGC1A (peroxisome proliferator-activated receptor gamma, coactivator 1 α), leading to activation of oxidative phosphorylation, inhibition of MTOR, and induction of autophagy.104 A similar dependency on MTOR inhibition and induction of ROS is required for induction of autophagy by S. pneumonia.105
Autophagy is also important for the induction of T cell responses, and recent studies have underlined its crucial role for lymphocyte survival and memory. During infection with lymphocytic choriomeningitis virus autophagy is not required for proliferation and CD8+ T-effector function, but is crucial for long-term cell survival and memory.106 A similar role of autophagy for CD8+ memory was reported during influenza and MCMV infections.107 During influenza infection, Atg5 knockout mice display a defective lymphocyte survival and proliferation.108 Interestingly, chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation.109
At the level of autophagy-inflammation interaction, host defense mechanisms are directly influenced by the modulation of proinflammatory cytokines, as described above in detail. However, the host defense against various classes of microorganisms is not influenced to the same extent. One of the most important infections for which autophagy makes a major impact is tuberculosis. Several studies have demonstrated the crucial role of autophagy for elimination of mycobacteria.20,110 M. tuberculosis induces ubiquitin-mediated targeting in macrophages, resulting in delivery of the pathogen to autolysosomes. This response requires the autophagy receptors SQSTM1, CALCOCO2 and the DNA-responsive kinase TBK1 (TANK-binding kinase 1),110 while inhibition of MTOR through AMPK activation is a necessary change in metabolic status of the cell activating autophagy.104 In addition, antimycobacterial peptides are produced in a SQSTM1-dependent manner.111,112
Autophagy is also important in host defense against other intracellular pathogens such as Toxoplasma gondii,113,114 Salmonella spp.115,116 and Listeria monocytogenes.117 The role of autophagy for host defense against viruses strongly depends on the type of infection, and involves several mechanisms, especially those involved in autophagosome generation and maturation (as detailed in ref. 96), rather than inflammation. Autophagy is important for removal of herpes simplex virus-1118 and Rift valley virus,119 and restricts HIV infection by degrading the HIV1 transactivator Tat in CD4+ lymphocytes.120 In addition, autophagy not only contributes to the elimination of the virus, but also controls IL1B induction and aberrant induction of IL17-dependent lung pathology in respiratory syncytial virus infection.121 Autophagy also proved important for the host defense against the fungal pathogen Cryptococcus neoformans, through mechanisms involving the nonlytic exocytosis of the fungus, but also interfering with production of the inflammatory mediators IL6 and CXCL10/IP-10 (chemokine [C-X-C motif] ligand 10).122 In contrast, no effect of autophagy for host defense against Candida albicans has been demonstrated,123,124 despite the demonstration of CLEC7A/dectin-1-dependent accumulation of MAP1LC3B to phagosomes that increases in vitro killing of the fungus.125
Notably, pathogens have also evolved strategies to evade the immune response by taking advantage of the autophagy machinery. For instance, M. tuberculosis latently infects human immune cells by concealed residence in the autophagosome and by preventing lysosomal fusion.20 In addition, active mycobacterial infection counteracts autophagy through the induction of MIR30A that inhibits BECN1.126 Salmonella in turn attempts to inhibit autophagy and increase its survival by activating PTK2/focal adhesion kinase and MTOR.127 Furthermore, Listeria monocytogenes and Shigella, as well as viral pathogens such as HIV, have evolved different strategies to evade the autophagy machinery by inhibition of autophagosome formation or by modulation of positive (IFNA and IFNB-IL6 signaling) and negative upstream regulators (MTOR kinase signaling pathway) of autophagy.128-130
Genetic variants of autophagy genes have been investigated for their effects on antimicrobial immune responses, which revealed that the CD-associated ATG16L1 polymorphism impairs defense mechanisms against Salmonella typhimurium and increases susceptibility toward Helicobacter pylori infection.53,131 In addition, genetic variants of both ATG16L1 and IRGM are associated with sepsis, potentially through modulation of immunoparalysis.132,133 However, in a large cohort no significant predisposition was observed for polymorphisms in autophagy genes to develop M. tuberculosis infections, although autophagy profoundly influences the host immune response against this pathogen.134,135 Similar findings have been reported for mucosal and systemic infections caused by the fungus Candida spp, indicating the redundant role of autophagy in anticandidal host defense.136,137
The impact of autophagy-inflammation interplay on carcinogenesis
Both autophagy and inflammation are involved in several steps of carcinogenesis and cancer progression. Autophagy has been reported to have both antitumoral effects and tumor-promoting effects in cancer. The generally accepted explanation for this apparent ambiguous role of autophagy in cancer is that its role might differ in the different stages of carcinogenesis and in different tumor types and is related to changes in the tumor microenvironment and treatment. In addition, the dual role of autophagy in cancer might reflect the interplay between autophagy and other fundamental biological processes such as apoptosis, senescence, cell metabolism, and the immune system.138,139
In the early stages of cancer development, defects in autophagy machinery or inhibition of autophagy in premalignant cells might result in impaired degradation and removal of damaged organelles, unfolded proteins and ROS. These cytosolic components, which might accumulate in the cells, produced as a result of activation of oncogenic pathways result in DNA damage and maintain the carcinogenic phenotype.140,141 In mice, monoallelic deletion of haplo-insufficient Becn1, results in autophagy deficiency, leads to spontaneous development of malignant tumors, and stimulates development of hepatitis-B-induced premalignant liver lesions.142 Other studies indicate that BECN1 represents an important tumor suppressor and an increased BECN1 expression is associated with a favorable prognosis in several cancers.143-146 Moreover somatic mutations in ATG genes have been described in different malignant tumors.147,148 In addition, somatic mutations and/or genetic amplifications in AKT1 and class I phosphoinositide 3-kinase (PI3K) and somatic or germ-line mutations leading to inactivation of the tumor suppressor gene PTEN are often found in malignant tumors.149-153As a result, signaling molecules upstream of MTOR such as AKT1 and class I PI3K are activated and exert inhibitory effects on autophagy in malignant transformed cells. AKT1 can inhibit autophagy either through activation of MTOR or through MTOR-independent mechanisms such as BECN1 phosphorylation and formation of an autophagy-inhibitory BECN1-YWHA/14–3–3-VIM/vimentin intermediate filament complex.154 The class III phosphatidylinositol 3-kinase (PtdIns3K) is an intracellular lipid kinase that has crucial functions in autophagosome formation, vesicular trafficking and endocytosis.155 In addition these lipid kinases represent key signaling molecules that play important roles in carcinogenesis. Three classes of phosphoinositide and phosphatidylinositol 3-kinases have been described according to their structure and substrate specificity. Of these, the functions of class I and of class III enzymes are best understood. Class III PtdIns3k phosphorylates phosphatidylinositol to form phosphatidylinositol-3-phosphate. The catalytic subunit of class III PtdIns3k, PIK3C3/Vps34, binds to other regulatory proteins, including ATG14, and contributes to the induction of autophagosome formation. In cancer, however, class I PI3Ks are often activated and therefore their role has been better explored as potential targets for therapy in malignant processes. In contrast to class III PtdIns3K, class I PI3K is activated downstream of receptor tyrosine kinases and G-protein coupled receptors and upon activation phosphorylates phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol 3,4,5-trisphosphate. Through this, PI3K activates AKT and MTOR, which in turn promotes cell survival and proliferation and suppresses autophagy. 156-158 Altogether, these studies suggest that autophagy genes can act as tumor suppressor genes in several tumors, and suppressed or defective autophagy may play an important role in tumor initiation.
Conversely, in the more advanced stages of tumor development and progression, once the carcinogenic phenotype has been established, the cancer cells rely on autophagy to support the requirements for energy and provide substrates necessary for proliferation. This is especially important for surviving in an unfavorable microenvironment characterized by severe hypoxia and nutrient-deficient conditions. Therefore activation of autophagy in this context could be regarded as a survival factor, promoting cell proliferation and tumor progression. This is particularly relevant in the context of therapeutic response to cytotoxic agents and ionizing radiation. The strong induction of autophagy observed as a result of treatment with ionizing radiation or chemotherapy may contribute to cell survival and the acquired resistance to therapy in many cancer cells including breast cancer, pancreatic cancer, and malignant glioma.159-162 For this reason, inhibition of autophagy with chloroquine or hydroxychloroquine in combination with radiotherapy or chemotherapy has been investigated as a strategy to prevent therapy resistance and sensitize tumors to therapy.163-167 However, in the context of excessive activation autophagy may lead to cell death in cells lacking apoptotic machinery, in which the programmed cell death takes place preferentially through autophagic cell death.168-172 Several mechanisms have been postulated to explain these apparently conflicting roles of autophagy in tumorigenesis. These include involvement of autophagy in cell death and cell survival through an intricate interplay with apoptosis and necrosis by shaping the metabolic and oxidative stress environment of the cell, its role in quality control for the homeostasis of proteins and organelles, and its function in the modulation of tumor-targeted immune responses.138,173 However the exact regulatory mechanisms of this dynamic process are not completely understood and due to the length and the focus of this review this will not be further discussed here. We will focus instead on the role and the consequences of the interplay between autophagy and inflammation in cancer pathogenesis and treatment.
The relationship between inflammation and cancer has long been recognized. Many chronic inflammatory conditions, such as inflammatory bowel disease, chronic hepatitis, or pancreatitis are clearly recognized as precursors of malignancies in the respective organs.174-177 Moreover, the immune system is the most important defense mechanism that identifies and eliminates malignant transformed cells and prevents tumor progression and metastasis. Autophagy has a modulatory role that shapes the interface between cancer and immune response, including effects on both tumor cells and immune cells, which subsequently influence the survival and function of, and interplay between, these cells and ultimately results in either tumor-promoting or tumor-suppressing effects (Fig. 4).
Figure 4.
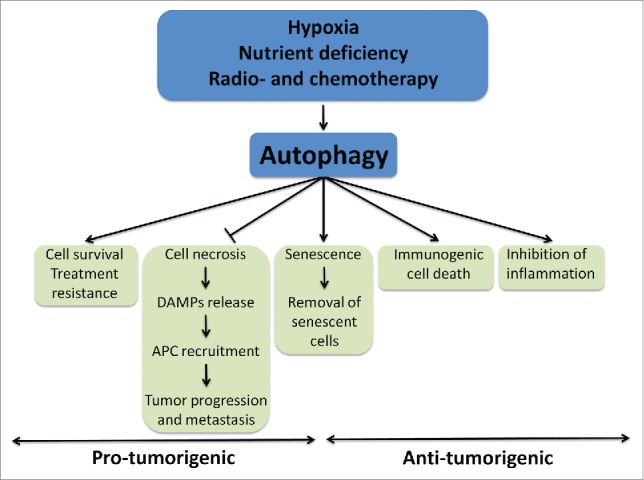
The role of crosstalk between autophagy and inflammation in cancer. Induction of autophagy is a double-edged sword with both protumorigenic and antitumorigenic effects. Processes downstream of autophagy that influence tumor progression and treatment resistance are comprised of cell survival pathways and recruitment of innate immune cells following release of DAMPs by necrotic tumor cells. Conversely, antitumorigenic signaling is evoked by autophagy-mediated immunogenic cell death and inhibition of tumor-associated inflammation. An outstanding example of the diverse and apparently opposite effects of autophagy on cancer initiation and progression is the induction of cellular senescence and subsequent removal of senescent cells; depending on the cellular context this is either pro- or anti-tumorigenic. APC, antigen-presenting cell.
It is well known that tumor cells rely on aerobic glycolysis to support their energy requirements and to enable increased protein production (historically known as the Warburg effect).178,179 This mode of growth, however, requires an increased influx of glucose, which is often not available in parts of the tumor that have no or only limited access to (neo)vascularization. As described above, in conditions of glucose or amino acid deprivation and hypoxia autophagy is activated as an important survival mechanism. However, oncogenic events, such as mutations in the PI3K-AKT-MTOR signaling pathway, have anti-apoptotic effects and inhibit autophagy simultaneously. This impairs the activation of autophagy as a compensatory survival mechanism and renders the tumor cells prone to necrosis.180 In cancer, necrosis is seen as an unfavorable event because it results in release of highly inflammatory molecules, such as uric acid, ATP/UTP, HMGB1 (high mobility group box 1), and other damage-associated molecular patterns (DAMPs).181-183 All together, these inflammatory mediators and DAMPs promote and maintain malignant features such as cell survival and proliferation and contribute to recruitment of macrophages and other specialized antigen-presenting cells such as DCs.184 These cells in turn stimulate inflammation in the peritumoral stroma and induce a pro-tumorigenic environment that promotes angiogenesis, tumor progression, and metastasis.180
Although not an inflammatory mechanism itself, it is also important to underline that antigen recognition and presentation is the cornerstone of the specific immune responses against malignant cells. Hence, proper functioning of these processes heavily depends on the integrity of the autophagy machinery. It has for instance been shown that autophagy facilitates antigen presentation by tumor cells, such as human embryonic kidney cells (HEK293T) and melanoma cells, thereby facilitating the recognition of these cells by immune cells and their subsequent removal.185
In addition, autophagy induces cellular senescence, a physiological state of sustained cell cycle arrest characterized by a senescence-associated secretory phenotype. This consists of increased expression and secretion of several cytokines, chemokines, growth factors, and proteases in the senescent cells,186 which stimulate on the one hand the recruitment and activation of phagocytes responsible for the removal of these cells, and on the other hand contribute to maintenance of the senescent phenotype. Subsequently, failure to induce autophagic flux and immune-mediated senescence increases the susceptibility to hepatocellular carcinoma in a mouse model.187
The interplay between autophagy and inflammation is particularly interesting in the context of therapeutic implications, and the concerted targeting of these 2 processes can be explored for novel strategies. TNFSF10/TRAIL (tumor necrosis factor [ligand] superfamily, member 10) induces cell death in different cancer cells and therefore contributes to immune cell-mediated cytotoxicity.188-190 However, pharmacological treatment with recombinant ligands has been hampered by the occurrence of therapy resistance.191 Nevertheless, blocking autophagy in the FRO anaplastic thyroid cancer cell line with ATG7 siRNA sensitizes the cells to TNFSF10-induced apoptosis, indicating that modulation of autophagy holds important therapeutic potential for anticancer treatment.192
Moreover, autophagy plays an important role in the response to chemotherapy of some tumors, in particular through influencing the immune response in the tumor microenvironment. Treatment with chemotherapy can induce immunogenic cell death.193 In this process, macrophages and DCs are attracted and recruited in the peritumoral stroma by molecules exposed at the cell surface or released in the tumor microenvironment as a result of apoptosis and necrosis following treatment with chemotherapeutic agents. DCs engulf the damaged tumor cells (remnants) and activate T-cells through antigen-presentation. These cells exert an antitumoral effect on the remaining unaffected cancer cells, thus enhancing the antitumoral effect of the chemotherapeutic agent. Michaud et al. have shown in cell lines and mouse models that autophagy is crucial for ATP release by the chemotherapy-induced dying tumor cells and for the induction of the antitumoral immunogenic response.194 In this study, therapy with methotrexate results in production of significantly lower amounts of ATP by autophagy-deficient tumors and a lack of antitumoral immunogenic responses. The immunogenicity of the autophagy-deficient cells could be restored after treatment with ARL67156, an inhibitor of ecto-ATPases that increase the extracellular ATP concentration, and after treatment with recombinant human IL1B, the production of which depends on the availability of ATP.194 Building upon these findings, Ko et al. showed that autophagy-dependent ATP release from stressed or dying tumors after radiotherapy contributes to lymphocyte infiltration in the tumor microenvironment and to the efficacy of radiotherapy in immunocompetent mice.195 Furthermore, Rao et al. report that non-small lung carcinoma tumors developing in autophagy-deficient KRAS; atg5fl/fl mice have an accelerated growth compared to tumors developing in autophagy-competent KRAS; Atg5fl/+ animals. In this model, the autophagy deficient tumors also show increased infiltration with FOXP3+ regulatory T cells and increased expression of ENTPD1/CD39 ecto-ATPase. These effects can be reversed by pharmacological inhibition of ENTPD1, thus supporting the concept that autophagy deficiency contributes to carcinogenesis through inhibition of anticancer immunosurveillance.196
Therefore, these data indicate that the immunogenic responses and immunosurveillance against cancer are dependent on the interplay between autophagy and inflammation. In addition the autophagy status contributes significantly to tumorigenesis and to the efficacy of chemo- and radiotherapy.
Conclusions and future perspectives
Autophagy influences several key components of the immune response, and thus plays a critical role in regulating inflammatory responses. It has even been suggested that autophagy evolved as a primordial host defense mechanism of eukaryotes.3 As evolution progressed, autophagy has intimately integrated with the other components of host defense, including the inflammatory reaction.
Despite the increasing knowledge about the role of autophagy in modulation of inflammation, this field is still only in its beginnings. Little is known about the involvement of the autophagy-related SQSTM1-like receptors for the modulation of inflammation, although some initial data have suggested important effects.197,198 The pathophysiological relevance of autophagy-inflammation interplay has not been studied yet in the context of colonization of mucosal membranes, and detection of tissue invasion. The influence of autophagy for directing polarization of adaptive immune responses (e.g., Th1 vs. Th2 vs. Th17) also warrants additional study in the coming years. In addition, the study of the balance between induction of autophagy and inhibition of inflammation is only in its beginning in the setting of understanding the pathophysiology of different types of cancer, and remains nearly untouched in autoimmune and autoinflammatory diseases. Understanding its role in tissue repair and remodeling is also an important challenge for the future.
Finally and most importantly, modulation of autophagy activity may represent a promising therapeutic approach for a wide range of inflammatory conditions. In this respect, a recent study reported the development of an autophagy-inducing peptide, Tat-BECN1, which decreased the replication of several human pathogens in vitro, and reduced mortality in mice infected with chikungunya or West Nile virus.199 However, at the same time, autophagy is a fundamental evolutionarily conserved process, and detailed knowledge about autophagy is necessary regarding its role in different diseases, cell types, and timelines, in order to avoid improper (or even dangerous) manipulation of a process fundamental for cell integrity. Nevertheless, future investigations are warranted in order to pursue novel autophagy-directed therapeutic approaches to effectively treat the ever-growing number of patients suffering from immune-mediated disorders.
Abbreviations
- AMP
adenosine monophosphate
- AMPK
5′ AMP-activated protein kinase
- ATG
autophagy related
- ATP
adenosine triphosphate
- BCG
Bacillus Calmette-Guérin
- BECN1
Beclin 1, autophagy related
- CD
Crohn disease
- CGD
chronic granulomatous disease
- CXCL10/IP-10
chemokine (C-X-C motif) ligand 10
- DAMP
danger-associated molecular pattern
- DC
dendritic cell
- EAE
experimental autoimmune encephalomyelitis
- GTP
guanosine-5′-triphosphate
- IFN
interferon
- IKBKB/IκB kinase
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
- IL
interleukin
- IL1RN/ILR1A
interleukin 1 receptor antagonist
- IRGM
immunity-related GTPase family, M
- MAP1LC3B
microtubule-associated protein 1 light chain 3 beta
- LAP
LC3-associated phagocytosis
- LPS
lipopolysaccharide
- mtDNA
mitochondrial DNA
- MAPK
mitogen-activated protein kinase
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- MYD88
myeloid differentiation primary response 88
- NFKB
nuclear factor of kappa light polypeptide gene enhancer in B-cells
- NLR
NOD-like receptor
- NLRP3
NLR family, pyrin domain containing 3
- NADPH
nicotinamide adenine dinucleotide hydrogen phosphate
- NOD
nucleotide-binding oligomerization domain
- PAMP
pathogen-associated molecular pattern
- PRR
pattern-recognition receptor
- PTEN
phosphatase and tensin homolog
- PINK1
PTEN-induced putative kinase 1
- RIPK2
receptor-interacting serine-threonine kinase 2
- RA
rheumatoid arthritis
- RLR
RIG-I-like receptor
- ROS
reactive oxygen species
- SLE
systemic lupus erythematosus
- Th
T helper
- TLR
toll-like receptor
- TNF
tumor necrosis factor
- TRAF
TNF receptor-associated factor
- TNFSF10/TRAIL
tumor necrosis factor (ligand) superfamily, member 10
- TICAM1/TRIF
toll-like receptor adaptor molecule 1
- ULK1
unc-51 like autophagy activating kinase 1.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
MGN was supported by a Vici grant of the Netherlands Organization for Scientific Research and by an ERC Consolidator Grant (#310372). TSP was supported by a Veni grant of the Netherlands Organization for Scientific Research and by the Alpe d'HuZes fund of the Dutch Cancer Society (KUN2014–6728). FV was supported by a Veni grant of the Netherlands Organization for Scientific Research.
References
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; http://dx.doi.org/ 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol 2012; 30:611-46; PMID:22449030; http://dx.doi.org/ 10.1146/annurev-immunol-020711-074948 [DOI] [PubMed] [Google Scholar]
- 3.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13:722-37; PMID:24064518; http://dx.doi.org/ 10.1038/nri3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heath RJ, Xavier RJ. Autophagy, immunity and human disease. Curr Opin Gastroenterol 2009; 25:512-20; PMID:19826372; http://dx.doi.org/ 10.1097/MOG.0b013e32833104f1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 2011; 27:107-32; PMID:21801009; http://dx.doi.org/ 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 6.Virgin HW, Levine B. Autophagy genes in immunity. Nat Immunol 2009; 10:461-70; PMID:19381141; http://dx.doi.org/ 10.1038/ni.1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, Sasai M, Latz E, Brinkmann MM, Iwasaki A, et al.. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity 2012; 37:986-97; PMID:23219390; http://dx.doi.org/ 10.1016/j.immuni.2012.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005; 307:593-6; PMID:15591165; http://dx.doi.org/ 10.1126/science.1104904 [DOI] [PubMed] [Google Scholar]
- 9.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature 2008; 455:396-400; PMID:18701890; http://dx.doi.org/ 10.1038/nature07208 [DOI] [PubMed] [Google Scholar]
- 10.Buffen K, Oosting M, Quintin J, Ng A, Kleinnijenhuis J, Kumar V, van de Vosse E, Wijmenga C, van Crevel R, Oosterwijk E, et al.. Autophagy controls BCG-induced trained immunity and the response to intravesical BCG therapy for bladder cancer. PLoS pathogens 2014; 10:e1004485; PMID:25356988; http://dx.doi.org/ 10.1371/journal.ppat.1004485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 2010; 3:ra42; PMID:20501938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al.. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406-16; PMID:23524951; http://dx.doi.org/ 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Liu XD, Gong X, Eissa NT. Signaling pathway of autophagy associated with innate immunity. Autophagy 2008; 4:110-2; PMID:18059159; http://dx.doi.org/ 10.4161/auto.5225 [DOI] [PubMed] [Google Scholar]
- 14.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J 2008; 27:1110-21; PMID:18337753; http://dx.doi.org/ 10.1038/emboj.2008.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med 2010; 16:90-7; PMID:19966812; http://dx.doi.org/ 10.1038/nm.2069 [DOI] [PubMed] [Google Scholar]
- 16.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al.. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11:55-62; PMID:19898471; http://dx.doi.org/ 10.1038/ni.1823 [DOI] [PubMed] [Google Scholar]
- 17.Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem 2011; 286:26987-95; PMID:21646350; http://dx.doi.org/ 10.1074/jbc.M111.223610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartman ML, Kornfeld H. Interactions between naive and infected macrophages reduce Mycobacterium tuberculosis viability. PloS One 2011; 6:e27972; PMID:22125644; http://dx.doi.org/ 10.1371/journal.pone.0027972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, et al.. Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol 2010; 12:1154-65; PMID:21102437; http://dx.doi.org/ 10.1038/ncb2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004; 119:753-66; PMID:15607973; http://dx.doi.org/ 10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- 21.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 2006; 313:1438-41; PMID:16888103; http://dx.doi.org/ 10.1126/science.1129577 [DOI] [PubMed] [Google Scholar]
- 22.Talloczy Z, Jiang W, Virgin HWt, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S Am 2002; 99:190-5; PMID:11756670; http://dx.doi.org/ 10.1073/pnas.012485299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng CG, Zheng L, Jankovic D, Báfica A, Cannons JL, Watford WT, Chaussabel D, Hieny S, Caspar P, Schwartzberg PL, et al.. The immunity-related GTPase Irgm1 promotes the expansion of activated CD4+ T cell populations by preventing interferon-gamma-induced cell death. Nat Immunol 2008; 9:1279-87; PMID:18806793; http://dx.doi.org/ 10.1038/ni.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Intemann CD, Thye T, Niemann S, Browne EN, Amanua Chinbuah M, Enimil A, Gyapong J, Osei I, Owusu-Dabo E, Helm S, et al.. Autophagy gene variant IRGM-261T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathogens 2009; 5:e1000577; PMID:19750224; http://dx.doi.org/ 10.1371/journal.ppat.1000577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med 2009; 15:267-76; PMID:19252503; http://dx.doi.org/ 10.1038/nm.1928 [DOI] [PubMed] [Google Scholar]
- 26.Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity 2007; 27(3):505-17; PMID:17892853; http://dx.doi.org/ 10.1016/j.immuni.2007.07.022 [DOI] [PubMed] [Google Scholar]
- 27.Zhou D, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS 2008; 22:695-9; PMID:18356598; http://dx.doi.org/ 10.1097/QAD.0b013e3282f4a836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deretic V, Singh S, Master S, Harris J, Roberts E, Kyei G, Davis A, de Haro S, Naylor J, Lee HH, et al.. Mycobacterium tuberculosis inhibition of phagolysosome biogenesis and autophagy as a host defence mechanism. Cell Microbiol 2006; 8:719-27; PMID:16611222; http://dx.doi.org/ 10.1111/j.1462-5822.2006.00705.x [DOI] [PubMed] [Google Scholar]
- 29.Martinet W, De Meyer GR. Autophagy in atherosclerosis: a cell survival and death phenomenon with therapeutic potential. Circ Res 2009; 104:304-17; PMID:19213965; http://dx.doi.org/ 10.1161/CIRCRESAHA.108.188318 [DOI] [PubMed] [Google Scholar]
- 30.Mizumura K, Choi AM, Ryter SW. Emerging role of selective autophagy in human diseases. Front Pharmacol 2014; 5:244; PMID:25414669; http://dx.doi.org/ 10.3389/fphar.2014.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: an integrated view. Immunol Rev 2011; 243:136-51; PMID:21884173; http://dx.doi.org/ 10.1111/j.1600-065X.2011.01046.x [DOI] [PubMed] [Google Scholar]
- 32.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al.. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008; 456:264-8; PMID:18849965; http://dx.doi.org/ 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- 33.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011; 469:221-5; PMID:21124315; http://dx.doi.org/ 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- 34.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al.. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011; 12:222-30; PMID:21151103; http://dx.doi.org/ 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J 2011; 30:4701-11; PMID:22068051; http://dx.doi.org/ 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13:255-63; PMID:22286270; http://dx.doi.org/ 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol 2010; 11:136-40; PMID:20023662; http://dx.doi.org/ 10.1038/ni.1831 [DOI] [PubMed] [Google Scholar]
- 38.Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, Huang G, Green M, Kundu M, Chi H, et al.. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol 2013; 14:480-8; PMID:23525089; http://dx.doi.org/ 10.1038/ni.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crisan TO, Plantinga TS, van de Veerdonk FL, Farcaş MF, Stoffels M, Kullberg BJ, van der Meer JW, Joosten LA, Netea MG. Inflammasome-independent modulation of cytokine response by autophagy in human cells. PloS One 2011; 6:e18666; PMID:21490934; http://dx.doi.org/ 10.1371/journal.pone.0018666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, et al.. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem 2011; 286:9587-97; PMID:21228274; http://dx.doi.org/ 10.1074/jbc.M110.202911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giegerich AK, Kuchler L, Sha LK, Knape T, Heide H, Wittig I, Behrends C, Brüne B, von Knethen A. Autophagy-dependent PELI3 degradation inhibits proinflammatory IL1B expression. Autophagy 2014; 10:1937-52; PMID:25483963; http://dx.doi.org/ 10.4161/auto.32178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS, Bhattacharya D, Yang H, et al.. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S Am 2012; 109:E3168-76; PMID:23093667; http://dx.doi.org/ 10.1073/pnas.1210500109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity 2012; 36:947-58; PMID:22658522; http://dx.doi.org/ 10.1016/j.immuni.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata Y, Oyama M, Kozuka-Hata H, Han X, Tanaka Y, Gohda J, Inoue J. p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO. Nat Commun 2012; 3:1061; PMID:22990857; http://dx.doi.org/ 10.1038/ncomms2068 [DOI] [PubMed] [Google Scholar]
- 45.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007; 315:1398-401; PMID:17272685; http://dx.doi.org/ 10.1126/science.1136880 [DOI] [PubMed] [Google Scholar]
- 46.Chaturvedi A, Dorward D, Pierce SK. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity 2008; 28:799-809; PMID:18513998; http://dx.doi.org/ 10.1016/j.immuni.2008.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, et al.. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S Am 2007; 104:14050-5; PMID:17709747; http://dx.doi.org/ 10.1073/pnas.0704014104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al.. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet 2007; 39:207-11; PMID:17200669; http://dx.doi.org/ 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- 49.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al.. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 2007; 39:596-604; PMID:17435756; http://dx.doi.org/ 10.1038/ng2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henckaerts L, Cleynen I, Brinar M, John JM, Van Steen K, Rutgeerts P, Vermeire S. Genetic variation in the autophagy gene ULK1 and risk of Crohn's disease. Inflamm Bowel Dis 2011; 17:1392-7; PMID:21560199; http://dx.doi.org/ 10.1002/ibd.21486 [DOI] [PubMed] [Google Scholar]
- 51.Morgan AR, Lam W-J, Han D-Y, Fraser AG, Ferguson LR. Association Analysis of ULK1 with Crohn's Disease in a New Zealand Population. Gastroenterol Res Pract 2012; 2012:715309; PMID:22536218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al.. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008; 456:259-63; PMID:18849966; http://dx.doi.org/ 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, et al.. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A 2014; 111:7741-6; PMID:24821797; http://dx.doi.org/ 10.1073/pnas.1407001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, et al.. A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 2014; 506:456-62; PMID:24553140; http://dx.doi.org/ 10.1038/nature13044 [DOI] [PubMed] [Google Scholar]
- 55.Wildenberg ME, Vos ACW, Wolfkamp SCS, Duijvestein M, Verhaar AP, Te Velde AA, van den Brink GR, Hommes DW. Autophagy attenuates the adaptive immune response by destabilizing the immunologic synapse. Gastroenterology 2012; 142:1493-503.e6; PMID:22370477; http://dx.doi.org/ 10.1053/j.gastro.2012.02.034 [DOI] [PubMed] [Google Scholar]
- 56.Strisciuglio C, Miele E, Wildenberg ME, Giugliano FP, Andreozzi M, Vitale A, Capasso F, Camarca A, Barone MV, Staiano A, et al.. T300A variant of autophagy ATG16L1 gene is associated with decreased antigen sampling and processing by dendritic cells in pediatric Crohn's disease. Inflamm Bowel Dis 2013; 19:2339-48; PMID:24022642; http://dx.doi.org/ 10.1097/MIB.0b013e3182a6a11c [DOI] [PubMed] [Google Scholar]
- 57.Plantinga TS, Crisan TO, Oosting M, van de Veerdonk FL, de Jong DJ, Philpott DJ, van der Meer JW, Girardin SE, Joosten LA, Netea MG. Crohn's disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut 2011; 60:1229-35; PMID:21406388; http://dx.doi.org/ 10.1136/gut.2010.228908 [DOI] [PubMed] [Google Scholar]
- 58.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al.. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008; 456:264-8; PMID:18849965 [DOI] [PubMed] [Google Scholar]
- 59.Crisan TO, Plantinga TS, van de Veerdonk FL, Farcaş MF, Stoffels M, Kullberg BJ, van der Meer JW, Joosten LA, Netea MG. Inflammasome-independent modulation of cytokine response by autophagy in human cells. PLoS One 2011; 6:e18666; PMID:21490934; http://dx.doi.org/ 10.1371/journal.pone.0018666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hébuterne X, et al.. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat Genet 2011; 43:242-5; PMID:21278745; http://dx.doi.org/ 10.1038/ng.762 [DOI] [PubMed] [Google Scholar]
- 61.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH, et al.. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet 2008; 40:1107-12; PMID:19165925; http://dx.doi.org/ 10.1038/ng.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al.. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 2001; 411:599-603; PMID:11385576; http://dx.doi.org/ 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- 63.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al.. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001; 411:603-6; PMID:11385577; http://dx.doi.org/ 10.1038/35079114 [DOI] [PubMed] [Google Scholar]
- 64.Travassos LH, Carneiro LAM, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al.. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol 2010; 11:55-62; PMID:19898471; http://dx.doi.org/ 10.1038/ni.1823 [DOI] [PubMed] [Google Scholar]
- 65.Sorbara MT, Ellison LK, Ramjeet M, Travassos LH, Jones NL, Girardin SE, Philpott DJ. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 2013; 39:858-73; PMID:24238340; http://dx.doi.org/ 10.1016/j.immuni.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 66.Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, Jiang T, Stade B, Bromberg Y, Ellinghaus E, Keller A, et al.. Association between variants of PRDM1 and NDP52 and Crohn's disease, based on exome sequencing and functional studies. Gastroenterology 2013; 145:339-47; PMID:23624108; http://dx.doi.org/ 10.1053/j.gastro.2013.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215-21; PMID:19820708; http://dx.doi.org/ 10.1038/ni.1800 [DOI] [PubMed] [Google Scholar]
- 68.Holmes B, Page AR, Good RA. Studies of the metabolic activity of leukocytes from patients with a genetic abnormality of phagocytic function. J Clin Investigat 1967; 46:1422-32; PMID:6036538; http://dx.doi.org/ 10.1172/JCI105634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Genetic, biochemical, and clinical features of chronic granulomatous disease. Med (Baltimore) 2000; 79:170-200; PMID:10844936; http://dx.doi.org/ 10.1097/00005792-200005000-00004 [DOI] [PubMed] [Google Scholar]
- 70.van de Veerdonk FL, Smeekens SP, Joosten LA, Kullberg BJ, Dinarello CA, van der Meer JW, Netea MG. Reactive oxygen species-independent activation of the IL-1beta inflammasome in cells from patients with chronic granulomatous disease. Proc Natl Acad Sci U S A 2010; 107:3030-3; PMID:20133696; http://dx.doi.org/ 10.1073/pnas.0914795107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meissner F, Seger RA, Moshous D, Fischer A, Reichenbach J, Zychlinsky A. Inflammasome activation in NADPH oxidase defective mononuclear phagocytes from patients with chronic granulomatous disease. Blood 2010; 116:1570-3; PMID:20495074; http://dx.doi.org/ 10.1182/blood-2010-01-264218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang J, Canadien V, Lam GY, Steinberg BE, Dinauer MC, Magalhaes MA, Glogauer M, Grinstein S, Brumell JH. Activation of antibacterial autophagy by NADPH oxidases. Proc Natl Acad Sci U S A 2009; 106:6226-31; PMID:19339495; http://dx.doi.org/ 10.1073/pnas.0811045106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Luca A, Smeekens SP, Casagrande A, Iannitti R, Conway KL, Gresnigt MS, Begun J, Plantinga TS, Joosten LA, van der Meer JW, et al.. IL-1 receptor blockade restores autophagy and reduces inflammation in chronic granulomatous disease in mice and in humans. Proc Natl Acad Sci U S A 2014; 111:3526-31; PMID:24550444; http://dx.doi.org/ 10.1073/pnas.1322831111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van de Veerdonk FL, Netea MG, Dinarello CA, van der Meer JW. Anakinra for the inflammatory complications of chronic granulomatous disease. Neth J Med 2011; 69:95; PMID:21411850 [PubMed] [Google Scholar]
- 75.Abbate A, Salloum FN, Vecile E, Das A, Hoke NN, Straino S, Biondi-Zoccai GG, Houser JE, Qureshi IZ, Ownby ED, et al.. Anakinra, a recombinant human interleukin-1 receptor antagonist, inhibits apoptosis in experimental acute myocardial infarction. Circulation 2008; 117:2670-83; PMID:18474815; http://dx.doi.org/ 10.1161/CIRCULATIONAHA.107.740233 [DOI] [PubMed] [Google Scholar]
- 76.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, et al.. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 2009; 41:1228-33; PMID:19838195; http://dx.doi.org/ 10.1038/ng.468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.International Consortium for Systemic Lupus Erythematosus G, Harley JB, Alarcon-Riquelme ME, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, et al.. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008; 40:204-10; PMID:18204446; http://dx.doi.org/ 10.1038/ng.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou X-j, Lu X-l, Lv J-c, Yang HZ, Qin LX, Zhao MH, Su Y, Li ZG, Zhang H. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 2011; 70:1330-7; PMID:21622776; http://dx.doi.org/ 10.1136/ard.2010.140111 [DOI] [PubMed] [Google Scholar]
- 79.Mayes MD, Bossini-Castillo L, Gorlova O, Martin JE, Zhou X, Chen WV, Assassi S, Ying J, Tan FK, Arnett FC, et al.. Immunochip analysis identifies multiple susceptibility loci for systemic sclerosis. Am J Hum Genet 2014; 94:47-61; PMID:24387989; http://dx.doi.org/ 10.1016/j.ajhg.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb AL, Frausto RF, Bhan V, Kiosses WB, Whitton JL, Robertson GS, et al.. Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy 2009; 5:152-8; PMID:19066443; http://dx.doi.org/ 10.4161/auto.5.2.7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou XJ, Lu XL, Lv JC, Yang HZ, Qin LX, Zhao MH, Su Y, Li ZG, Zhang H. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 2011; 70:1330-7; PMID:21622776; http://dx.doi.org/ 10.1136/ard.2010.140111 [DOI] [PubMed] [Google Scholar]
- 82.Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, Hengartner MO, Green DR. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A 2011; 108:17396-401; PMID:21969579; http://dx.doi.org/ 10.1073/pnas.1113421108 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Wang C, Wang C, Dong H, Wu XM, Wang C, Xia F, Li G, Jia X, He S, Jiang X, et al.. Immune-related GTPase Irgm1 exacerbates experimental auto-immune encephalomyelitis by promoting the disruption of blood-brain barrier and blood-cerebrospinal fluid barrier. Mol Immunol 2013; 53:43-51; PMID:22796503; http://dx.doi.org/ 10.1016/j.molimm.2012.06.009 [DOI] [PubMed] [Google Scholar]
- 84.Xu H, Wu Z-Y, Fang F, Guo L, Chen D, Chen JX, Stern D, Taylor GA, Jiang H, Yan SS. Genetic deficiency of Irgm1 (LRG-47) suppresses induction of experimental autoimmune encephalomyelitis by promoting apoptosis of activated CD4+ T cells. Faseb J 2010; 24:1583-92; PMID:20056715; http://dx.doi.org/ 10.1096/fj.09-137323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gualtierotti R, Biggioggero M, Penatti AE, Meroni PL. Updating on the pathogenesis of systemic lupus erythematosus. Autoimmun Rev 2010; 10:3-7; PMID:20863908; http://dx.doi.org/ 10.1016/j.autrev.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 86.Orme J, Mohan C. Macrophages and neutrophils in SLE-An online molecular catalog. Autoimmun Rev 2012; 11:365-72; PMID:22036828; http://dx.doi.org/ 10.1016/j.autrev.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 87.Fernandez D, Perl A. Metabolic control of T cell activation and death in SLE. Autoimmun Rev 2009; 8:184-9; PMID:18722557; http://dx.doi.org/ 10.1016/j.autrev.2008.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Giovannetti A, Pierdominici M, Di Iorio A, Cianci R, Murdaca G, Puppo F, Pandolfi F, Paganelli R. Apoptosis in the homeostasis of the immune system and in human immune mediated diseases. Curr Pharm Des 2008; 14:253-68; PMID:18220836; http://dx.doi.org/ 10.2174/138161208783413310 [DOI] [PubMed] [Google Scholar]
- 89.Baumann I, Kolowos W, Voll RE, Manger B, Gaipl U, Neuhuber WL, Kirchner T, Kalden JR, Herrmann M. Impaired uptake of apoptotic cells into tingible body macrophages in germinal centers of patients with systemic lupus erythematosus. Arthritis Rheum 2002; 46:191-201; PMID:11817590; http://dx.doi.org/ 10.1002/1529-0131(200201)46:1%3c191::AID-ART10027%3e3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- 90.Bijl M, Reefman E, Horst G, Limburg PC, Kallenberg CGM. Reduced uptake of apoptotic cells by macrophages in systemic lupus erythematosus: correlates with decreased serum levels of complement. Ann Rheum Dis 2006; 65:57-63; PMID:15919679; http://dx.doi.org/ 10.1136/ard.2005.035733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 2007; 128:931-46; PMID:17350577; http://dx.doi.org/ 10.1016/j.cell.2006.12.044 [DOI] [PubMed] [Google Scholar]
- 92.Lin N-Y, Beyer C, Giessl A, Kireva T, Scholtysek C, Uderhardt S, Munoz LE, Dees C, Distler A, Wirtz S, et al.. Autophagy regulates TNFalpha-mediated joint destruction in experimental arthritis. Ann Rheum Dis 2013; 72:761-8; PMID:22975756; http://dx.doi.org/ 10.1136/annrheumdis-2012-201671 [DOI] [PubMed] [Google Scholar]
- 93.Connor AM, Mahomed N, Gandhi R, Keystone EC, Berger SA. TNFalpha modulates protein degradation pathways in rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther 2012; 14:R62; PMID:22417670; http://dx.doi.org/ 10.1186/ar3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shin Y-J, Han S-H, Kim D-S, Lee GH, Yoo WH, Kang YM, Choi JY, Lee YC, Park SJ, Jeong SK, et al.. Autophagy induction and CHOP under-xpression promotes survival of fibroblasts from rheumatoid arthritis patients under endoplasmic reticulum stress. Arthritis Res Ther 2010; 12:R19; PMID:20122151; http://dx.doi.org/ 10.1186/ar2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu K, Xu P, Yao J-F, Zhang Y-G, Hou W-K, Lu S-M. Reduced apoptosis correlates with enhanced autophagy in synovial tissues of rheumatoid arthritis. Inflamm Res 2013; 62:229-37; PMID:23178792; http://dx.doi.org/ 10.1007/s00011-012-0572-1 [DOI] [PubMed] [Google Scholar]
- 96.Arroyo DS, Gaviglio EA, Peralta Ramos JM, Bussi C, Rodriguez-Galan MC, Iribarren P. Autophagy in inflammation, infection, neurodegeneration and cancer. Int Immunopharmacol 2014; 18:55-65; PMID:24262302; http://dx.doi.org/ 10.1016/j.intimp.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vural A, Kehrl JH. Autophagy in macrophages: impacting inflammation and bacterial infection. Scientifica (Cairo) 2014; 2014:825463; PMID:24818040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kirkin V, Lamark T, Sou Y-S, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al.. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505-16; PMID:19250911; http://dx.doi.org/ 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- 99.Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem 2011; 286:26987-95; PMID:21646350; http://dx.doi.org/ 10.1074/jbc.M111.223610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jabir MS, Ritchie ND, Li D, Bayes HK, Tourlomousis P, Puleston D, Lupton A, Hopkins L, Simon AK, Bryant C, et al.. Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and beta-interferon production during Pseudomonas aeruginosa infection. Cell Host & Microbe 2014; 15:214-27; PMID:24528867; http://dx.doi.org/ 10.1016/j.chom.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gupta M, Shin DM, Ramakrishna L, Goussetis DJ, Platanias LC, Xiong H, Morse HC 3rd, Ozato K. IRF8 directs stress-induced autophagy in macrophages and promotes clearance of Listeria monocytogenes. Nature communications 2015; 6:6379; PMID:25775030; http://dx.doi.org/ 10.1038/ncomms7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, Turner LJ, Thomas BJ, Malosse C, Gantier MP, Casillas LN, et al.. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host & Microbe 2014; 15:623-35; PMID:24746552; http://dx.doi.org/ 10.1016/j.chom.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 103.Juarez E, Carranza C, Hernandez-Sanchez F, Loyola E, Escobedo D, León-Contreras JC, Hernández-Pando R, Torres M, Sada E. Nucleotide-oligomerizing domain-1 (NOD1) receptor activation induces pro-inflammatory responses and autophagy in human alveolar macrophages. BMC Pulmonary Med 2014; 14:152; PMID:25253572; http://dx.doi.org/ 10.1186/1471-2466-14-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang CS, Kim JJ, Lee HM, Jin HS, Lee SH, Park JH, Kim SJ, Kim JM, Han YM, Lee MS, et al.. The AMPK-PPARGC1A pathway is required for antimicrobial host defense through activation of autophagy. Autophagy 2014; 10:785-802; PMID:24598403; http://dx.doi.org/ 10.4161/auto.28072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li P, Shi J, He Q, Hu Q, Wang YY, Zhang LJ, Chan WT, Chen WX. Streptococcus pneumoniae Induces Autophagy through the Inhibition of the PI3K-I/Akt/mTOR Pathway and ROS Hypergeneration in A549 Cells. PloS One 2015; 10:e0122753; PMID:AMBIGUOUS; http://dx.doi.org/ 10.1371/journal.pone.0122753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu X, Araki K, Li S, Han JH, Ye L, Tan WG, Konieczny BT, Bruinsma MW, Martinez J, Pearce EL, et al.. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol 2014; 15:1152-61; PMID:25362489; http://dx.doi.org/ 10.1038/ni.3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, Watson AS, Cerundolo V, Townsend AR, Klenerman P, et al.. Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife 2014; 3; PMID:25385531; http://dx.doi.org/ 10.7554/eLife.03706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schlie K, Westerback A, DeVorkin L, Hughson LR, Brandon JM, MacPherson S, Gadawski I, Townsend KN, Poon VI, Elrick MA, et al.. Survival of effector CD8+ T cells during influenza infection is dependent on autophagy. J Immunol 2015; 194:4277-86; PMID:25833396[http://dx.doi.org/ 10.4049/jimmunol.1402571 [DOI] [PubMed] [Google Scholar]
- 109.Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H, Gravekamp C, Cuervo AM, Macian F. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol 2014; 15:1046-54; PMID:25263126; http://dx.doi.org/ 10.1038/ni.3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell 2012; 150:803-15; PMID:22901810; http://dx.doi.org/ 10.1016/j.cell.2012.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HW 4th, Kyei GB, Johansen T, Vergne I, et al.. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 2010; 32:329-41; PMID:20206555; http://dx.doi.org/ 10.1016/j.immuni.2010.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A 2007; 104:6031-6; PMID:17389386; http://dx.doi.org/ 10.1073/pnas.0700036104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, et al.. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 2008; 4:458-69; PMID:18996346; http://dx.doi.org/ 10.1016/j.chom.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liesenfeld O, Parvanova I, Zerrahn J, Han SJ, Heinrich F, Muñoz M, Kaiser F, Aebischer T, Buch T, Waisman A, et al.. The IFN-gamma-inducible GTPase, Irga6, protects mice against Toxoplasma gondii but not against Plasmodium berghei and some other intracellular pathogens. PloS One 2011; 6:e20568; PMID:21698150; http://dx.doi.org/ 10.1371/journal.pone.0020568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al.. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011; 333:228-33; PMID:21617041; http://dx.doi.org/ 10.1126/science.1205405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304[http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 117.Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al.. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol 2009; 11:1233-40; PMID:19749745; http://dx.doi.org/ 10.1038/ncb1967 [DOI] [PubMed] [Google Scholar]
- 118.Yakoub AM, Shukla D. Autophagy stimulation abrogates herpes simplex virus-1 infection. Sci Rep 2015; 5:9730; PMID:25856282; http://dx.doi.org/ 10.1038/srep09730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moy RH, Gold B, Molleston JM, Moy RH, Gold B, Molleston JM, Schad V, Yanger K, Salzano MV, Yagi Y, et al.. Antiviral autophagy restrictsRift Valley fever virus infection and is conserved from flies to mammals. Immunity 2014; 40:51-65; PMID:24374193; http://dx.doi.org/ 10.1016/j.immuni.2013.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sagnier S, Daussy CF, Borel S, Robert-Hebmann V, Faure M, Blanchet FP, Beaumelle B, Biard-Piechaczyk M, Espert L. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol 2015; 89:615-25; PMID:25339774; http://dx.doi.org/ 10.1128/JVI.02174-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reed M, Morris SH, Owczarczyk AB, Lukacs NW. Deficiency of autophagy protein Map1-LC3b mediates IL-17-dependent lung pathology during respiratory viral infection via ER stress-associated IL-1. Mucosal Immunol 2015; PMID:25669150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nicola AM, Albuquerque P, Martinez LR, Dal-Rosso RA, Saylor C, De Jesus M, Nosanchuk JD, Casadevall A. Macrophage autophagy in immunity to Cryptococcus neoformans and Candida albicans. Infect Immun 2012; 80:3065-76; PMID:22710871; http://dx.doi.org/ 10.1128/IAI.00358-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smeekens SP, Malireddi RK, Plantinga TS, Buffen K, Oosting M, Joosten LA, Kullberg BJ, Perfect JR, Scott WK, van de Veerdonk FL, et al.. Autophagy is redundant for the host defense against systemic Candida albicans infections. Eur J Clin Microbiol Infect Dis 2014; 33:711-22; PMID:24202731; http://dx.doi.org/ 10.1007/s10096-013-2002-x [DOI] [PubMed] [Google Scholar]
- 124.Rosentul DC, Plantinga TS, Farcas M, Oosting M, Hamza OJ, Scott WK, Alexander BD, Yang JC, Laird GM, Joosten LA, et al.. Role of autophagy genetic variants for the risk of Candida infections. Med Mycol 2014; 52:333-41; PMID:24713404; http://dx.doi.org/ 10.1093/mmy/myt035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tam JM, Mansour MK, Khan NS, Seward M, Puranam S, Tanne A, Sokolovska A, Becker CE, Acharya M, Baird MA, et al.. Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J Infect Dis 2014; 210:1844-54; PMID:24842831; http://dx.doi.org/ 10.1093/infdis/jiu290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen Z, Wang T, Liu Z, Zhang G, Wang J, Feng S, Liang J. Mycobacteria tuberculosis induced miR-30A inhibit autophagy process as a possible mechanism of immune escape in human macrophage. Jpn J Inf Dis 2015; PMID:25866116 [DOI] [PubMed] [Google Scholar]
- 127.Owen KA, Meyer CB, Bouton AH, Casanova JE. Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLoS Pathogens 2014; 10:e1004159; PMID:24901456; http://dx.doi.org/ 10.1371/journal.ppat.1004159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S, et al.. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe 2011; 9:376-89; PMID:21575909; http://dx.doi.org/ 10.1016/j.chom.2011.04.010 [DOI] [PubMed] [Google Scholar]
- 129.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science 2005; 307:727-31; PMID:15576571; http://dx.doi.org/ 10.1126/science.1106036 [DOI] [PubMed] [Google Scholar]
- 130.Dreux M, Chisari FV. Viruses and the autophagy machinery. Cell Cycle 2010; 9:1295-307; PMID:20305376; http://dx.doi.org/ 10.4161/cc.9.7.11109 [DOI] [PubMed] [Google Scholar]
- 131.Raju D, Hussey S, Jones NL. Crohn disease ATG16L1 polymorphism increases susceptibility to infection with Helicobacter pylori in humans. Autophagy 2012; 8:1387-8; PMID:22885761; http://dx.doi.org/ 10.4161/auto.21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kimura T, Watanabe E, Sakamoto T, Takasu O, Ikeda T, Ikeda K, Kotani J, Kitamura N, Sadahiro T, Tateishi Y, et al.. Autophagy-related IRGM polymorphism is associated with mortality of patients with severe sepsis. PLoS One 2014; 9:e91522; PMID:24626347; http://dx.doi.org/ 10.1371/journal.pone.0091522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Savva A, Plantinga TS, Kotanidou A, Farcas M, Baziaka F, Raftogiannis M, Orfanos SE, Dimopoulos G, Netea MG, Giamarellos-Bourboulis EJ. Association of autophagy-related 16-like 1 (ATG16L1) gene polymorphism with sepsis severity in patients with sepsis and ventilator-associated pneumonia. Eur J Clin Microbiol Infect Dis 2014; 33:1609-14; PMID:24791954; http://dx.doi.org/ 10.1007/s10096-014-2118-7 [DOI] [PubMed] [Google Scholar]
- 134.Kleinnijenhuis J, Oosting M, Plantinga TS, van der Meer JW, Joosten LA, Crevel RV, Netea MG. Autophagy modulates the Mycobacterium tuberculosis-induced cytokine response. Immunology 2011; 134:341-8; PMID:21978003; http://dx.doi.org/ 10.1111/j.1365-2567.2011.03494.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Songane M, Kleinnijenhuis J, Alisjahbana B, Sahiratmadja E, Parwati I, Oosting M, Plantinga TS, Joosten LA, Netea MG, Ottenhoff TH, et al.. Polymorphisms in autophagy genes and susceptibility to tuberculosis. PLoS One 2012; 7:e41618; PMID:22879892; http://dx.doi.org/ 10.1371/journal.pone.0041618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rosentul DC, Plantinga TS, Farcas M, Oosting M, Hamza OJ, Scott WK, Alexander BD, Yang JC, Laird GM, Joosten LA, et al.. Role of autophagy genetic variants for the risk of Candida infections. Med Mycol 2014; 52:333-41; PMID:24713404; http://dx.doi.org/ 10.1093/mmy/myt035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Smeekens SP, Malireddi RK, Plantinga TS, Buffen K, Oosting M, Joosten LA, Kullberg BJ, Perfect JR, Scott WK, van de Veerdonk FL, et al.. Autophagy is redundant for the host defense against systemic Candida albicans infections. Eur J Clin Microbiol Infect Dis 2014; 33:711-22; PMID:24202731; http://dx.doi.org/ 10.1007/s10096-013-2002-x [DOI] [PubMed] [Google Scholar]
- 138.Rosenfeldt MT, Ryan KM. The multiple roles of autophagy in cancer. Carcinogenesis 2011; 32:955-63; PMID:21317301[http://dx.doi.org/ 10.1093/carcin/bgr031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mah LY, Ryan KM. Autophagy and cancer. Cold Spring Harb Perspect Biol 2012; 4:a008821; PMID:22166310[http://dx.doi.org/ 10.1101/cshperspect.a008821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy 2010; 6:838-54; PMID:20505317; http://dx.doi.org/ 10.4161/auto.6.7.12113 [DOI] [PubMed] [Google Scholar]
- 141.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al.. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137:1062-75; PMID:19524509; http://dx.doi.org/ 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al.. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809-20; PMID:14638851; http://dx.doi.org/ 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miracco C, Cevenini G, Franchi A, Luzi P, Cosci E, Mourmouras V, Monciatti I, Mannucci S, Biagioli M, Toscano M, et al.. Beclin 1 and LC3 autophagic gene expression in cutaneous melanocytic lesions. Hum Pathol 2010; 41:503-12; PMID:20004946 [DOI] [PubMed] [Google Scholar]
- 144.Cicchini M, Chakrabarti R, Kongara S, Price S, Nahar R, Lozy F, Zhong H, Vazquez A, Kang Y, Karantza V. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy 2014; 10:2036-52; PMID:25483966; http://dx.doi.org/ 10.4161/auto.34398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672-6; PMID:10604474; http://dx.doi.org/ 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- 146.Zhao Y, Chen S, Gou WF, Xiao LJ, Takano Y, Zheng HC. Aberrant Beclin 1 expression is closely linked to carcinogenesis, differentiation, progression, and prognosis of ovarian epithelial carcinoma. Tumour Biol 2014; 35:1955-64; PMID:24132590; http://dx.doi.org/ 10.1007/s13277-013-1261-6 [DOI] [PubMed] [Google Scholar]
- 147.Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Kim SS, Ahn CH, Yoo NJ, Lee SH. Frameshift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with microsatellite instability. J Pathol 2009; 217:702-6; PMID:19197948; http://dx.doi.org/ 10.1002/path.2509 [DOI] [PubMed] [Google Scholar]
- 148.Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol 2008; 39:1059-63; PMID:18495205; http://dx.doi.org/ 10.1016/j.humpath.2007.11.013 [DOI] [PubMed] [Google Scholar]
- 149.Hou P, Liu D, Shan Y, Hu S, Studeman K, Condouris S, Wang Y, Trink A, El-Naggar AK, Tallini G, et al.. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res 2007; 13:1161-70; PMID:17317825; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-1125 [DOI] [PubMed] [Google Scholar]
- 150.Xing M. Genetic alterations in the phosphatidylinositol-3 kinase/Akt pathway in thyroid cancer. Thyroid: Off J Am Thyroid Assoc 2010; 20:697-706; PMID:20578891; http://dx.doi.org/ 10.1089/thy.2010.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al.. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 1997; 16:64-7; PMID:9140396; http://dx.doi.org/ 10.1038/ng0597-64 [DOI] [PubMed] [Google Scholar]
- 152.Wu G, Mambo E, Guo Z, Hu S, Huang X, Gollin SM, Trink B, Ladenson PW, Sidransky D, Xing M. Uncommon mutation, but common amplifications, of the PIK3CA gene in thyroid tumors. J Clin EndocrinolMetab 2005; 90:4688-93; PMID:15928251; http://dx.doi.org/ 10.1210/jc.2004-2281 [DOI] [PubMed] [Google Scholar]
- 153.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2002; 2:489-501; PMID:12094235; http://dx.doi.org/ 10.1038/nrc839 [DOI] [PubMed] [Google Scholar]
- 154.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012; 338:956-9; PMID:23112296; http://dx.doi.org/ 10.1126/science.1225967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 2010; 11:329-41; PMID:20379207; http://dx.doi.org/ 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- 156.Hawkins PT, Stephens LR. PI3K signalling in inflammation. Biochimica et Biophysica acta 2015; 1851:882-97; PMID:25514767; http://dx.doi.org/ 10.1016/j.bbalip.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 157.Reidick C, El Magraoui F, Meyer HE, Stenmark H, Platta HW. Regulation of the tumor-suppressor function of the class III phosphatidylinositol 3-kinase complex by ubiquitin and SUMO. Cancers 2014; 7:1-29; PMID:25545884; http://dx.doi.org/ 10.3390/cancers7010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.F OF, Rusten TE, Stenmark H. Phosphoinositide 3-kinases as accelerators and brakes of autophagy. FEBS J 2013; 280:6322-37; PMID:23953235; http://dx.doi.org/ 10.1111/febs.12486 [DOI] [PubMed] [Google Scholar]
- 159.Chaachouay H, Ohneseit P, Toulany M, Kehlbach R, Multhoff G, Rodemann HP. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol 2011; 99:287-92; PMID:21722986; http://dx.doi.org/ 10.1016/j.radonc.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 160.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, Zhuang L, Luo J, Chen H, Liu L, et al.. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology 2013; 145:1133-43e12; PMID:23916944; http://dx.doi.org/ 10.1053/j.gastro.2013.07.048 [DOI] [PubMed] [Google Scholar]
- 161.Wang P, Zhang L, Chen Z, Meng Z. MicroRNA targets autophagy in pancreatic cancer cells during cancer therapy. Autophagy 2013; 9:2171-2; PMID:24145177; http://dx.doi.org/ 10.4161/auto.26463 [DOI] [PubMed] [Google Scholar]
- 162.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer 2009; 125:717-22; PMID:19431142; http://dx.doi.org/ 10.1002/ijc.24402 [DOI] [PubMed] [Google Scholar]
- 163.Zanotto-Filho A, Braganhol E, Klafke K, Figueiró F, Terra SR, Paludo FJ, Morrone M, Bristot IJ, Battastini AM, Forcelini CM, et al.. Autophagy inhibition improves the efficacy of curcumin/temozolomide combination therapy in glioblastomas. Cancer Lett 2015; 358:220-31; PMID:25542083; http://dx.doi.org/ 10.1016/j.canlet.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 164.Rosenfeld MR, Ye X, Supko JG, Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, et al.. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 2014; 10:1359-68; PMID:24991840; http://dx.doi.org/ 10.4161/auto.28984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M, et al.. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 2014; 10:1369-79; PMID:24991839; http://dx.doi.org/ 10.4161/auto.29118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, et al.. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 2014; 10:1391-402; PMID:24991838; http://dx.doi.org/ 10.4161/auto.29119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Poklepovic A, Gewirtz DA. Outcome of early clinical trials of the combination of hydroxychloroquine with chemotherapy in cancer. Autophagy 2014; 10:1478-80; PMID:24991829; http://dx.doi.org/ 10.4161/auto.29428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927-39; PMID:16179260; http://dx.doi.org/ 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 169.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 2007; 17:1-11; PMID:17208179; http://dx.doi.org/ 10.1016/j.cub.2006.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bursch W, Karwan A, Mayer M, Dornetshuber J, Fröhwein U, Schulte-Hermann R, Fazi B, Di Sano F, Piredda L, Piacentini M, et al.. Cell death and autophagy: cytokines, drugs, and nutritional factors. Toxicology 2008; 254:147-57; PMID:18694801; http://dx.doi.org/ 10.1016/j.tox.2008.07.048 [DOI] [PubMed] [Google Scholar]
- 171.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science 2014; 345:1250256; PMID:25237106; http://dx.doi.org/ 10.1126/science.1250256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu Y, Levine B. Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ 2015; 22:367-76; PMID:25257169[http://dx.doi.org/ 10.1038/cdd.2014.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15:81-94; PMID:24401948; http://dx.doi.org/ 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kuper H, Ye W, Broome U, Romelsjö A, Mucci LA, Ekbom A, Adami HO, Trichopoulos D, Nyrén O. The risk of liver and bile duct cancer in patients with chronic viral hepatitis, alcoholism, or cirrhosis. Hepatology 2001; 34:714-8; PMID:11584367; http://dx.doi.org/ 10.1053/jhep.2001.28233 [DOI] [PubMed] [Google Scholar]
- 175.Xing S, Li ZW, Tian YF, Zhang LM, Li MQ, Zhou P. Chronic hepatitis virus infection increases the risk of pancreatic cancer: a meta-analysis. Hepatobiliary Pancreat Dis Int 2013; 12:575-83; PMID:24322741; http://dx.doi.org/ 10.1016/S1499-3872(13)60091-0 [DOI] [PubMed] [Google Scholar]
- 176.Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andrén-Sandberg A, Domellöf L. Pancreatitis and the risk of pancreatic cancer. International pancreatitis study group. New Engl J Med 1993; 328:1433-7; PMID:8479461; http://dx.doi.org/ 10.1056/NEJM199305203282001 [DOI] [PubMed] [Google Scholar]
- 177.Kim ER, Chang DK. Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol 2014; 20:9872-81; PMID:25110418; http://dx.doi.org/ 10.3748/wjg.v20.i29.9872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Warburg O. On the origin of cancer cells. Science 1956; 123:309-14; PMID:13298683; http://dx.doi.org/ 10.1126/science.123.3191.309 [DOI] [PubMed] [Google Scholar]
- 179.Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011; 11:325-37; PMID:21508971; http://dx.doi.org/ 10.1038/nrc3038 [DOI] [PubMed] [Google Scholar]
- 180.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y, et al.. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 2006; 10:51-64; PMID:16843265; http://dx.doi.org/ 10.1016/j.ccr.2006.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002; 418:191-5; PMID:12110890; http://dx.doi.org/ 10.1038/nature00858 [DOI] [PubMed] [Google Scholar]
- 182.Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 2013; 38:209-23; PMID:23438821; http://dx.doi.org/ 10.1016/j.immuni.2013.02.003 [DOI] [PubMed] [Google Scholar]
- 183.Krysko O, Love Aaes T, Bachert C, Vandenabeele P, Krysko DV. Many faces of DAMPs in cancer therapy. Cell Death Dis 2013; 4:e631; PMID:23681226; http://dx.doi.org/ 10.1038/cddis.2013.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hou W, Zhang Q, Yan Z, Chen R, Zeh Iii HJ, Kang R, Lotze MT, Tang D. Strange attractors: DAMPs and autophagy link tumor cell death and immunity. Cell Death Dis 2013; 4:e966; PMID:24336086; http://dx.doi.org/ 10.1038/cddis.2013.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res 2008; 68:6889-95; PMID:18757401; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metast Rev 2010; 29:273-83; PMID:20390322; http://dx.doi.org/ 10.1007/s10555-010-9220-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lin H, Yan J, Wang Z, Hua F, Yu J, Sun W, Li K, Liu H, Yang H, Lv Q, et al.. Loss of immunity-supported senescence enhances susceptibility to hepatocellular carcinogenesis and progression in Toll-like receptor 2-deficient mice. Hepatology 2013; 57:171-82; PMID:22859216; http://dx.doi.org/ 10.1002/hep.25991 [DOI] [PubMed] [Google Scholar]
- 188.Bretz JD, Mezosi E, Giordano TJ, Gauger PG, Thompson NW, Baker JR Jr. Inflammatory cytokine regulation of TRAIL-mediated apoptosis in thyroid epithelial cells. Cell Death Differ 2002; 9:274-86; PMID:11859410; http://dx.doi.org/ 10.1038/sj.cdd.4400965 [DOI] [PubMed] [Google Scholar]
- 189.Uno K, Inukai T, Kayagaki N, Goi K, Sato H, Nemoto A, Takahashi K, Kagami K, Yamaguchi N, Yagita H, et al.. TNF-related apoptosis-inducing ligand (TRAIL) frequently induces apoptosis in Philadelphia chromosome-positive leukemia cells. Blood 2003; 101:3658-67; PMID:12506034; http://dx.doi.org/ 10.1182/blood-2002-06-1770 [DOI] [PubMed] [Google Scholar]
- 190.Song JH, Song DK, Pyrzynska B, Petruk KC, Van Meir EG, Hao C. TRAIL triggers apoptosis in human malignant glioma cells through extrinsic and intrinsic pathways. Brain Pathol 2003; 13:539-53; PMID:14655759; http://dx.doi.org/ 10.1111/j.1750-3639.2003.tb00484.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhang L, Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther 2005; 12:228-37; PMID:15550937; http://dx.doi.org/ 10.1038/sj.cgt.7700792 [DOI] [PubMed] [Google Scholar]
- 192.Jin SM, Jang HW, Sohn SY, Kim NK, Joung JY, Cho YY, Kim SW, Chung JH. Role of autophagy in the resistance to tumour necrosis factor-related apoptosis-inducing ligand-induced apoptosis in papillary and anaplastic thyroid cancer cells. Endocrine 2014; 45:256-62; PMID:23821365; http://dx.doi.org/ 10.1007/s12020-013-9997-8 [DOI] [PubMed] [Google Scholar]
- 193.Kepp O, Tesniere A, Schlemmer F, Michaud M, Senovilla L, Zitvogel L, Kroemer G. Immunogenic cell death modalities and their impact on cancer treatment. Apoptosis 2009; 14:364-75; PMID:19145485; http://dx.doi.org/ 10.1007/s10495-008-0303-9 [DOI] [PubMed] [Google Scholar]
- 194.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, et al.. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011; 334:1573-7; PMID:22174255; http://dx.doi.org/ 10.1126/science.1208347 [DOI] [PubMed] [Google Scholar]
- 195.Ko A, Kanehisa A, Martins I, Senovilla L, Chargari C, Dugue D, Mariño G, Kepp O, Michaud M, Perfettini JL, et al.. Autophagy inhibition radiosensitizes in vitro, yet reduces radioresponses in vivo due to deficient immunogenic signalling. Cell Death Differ 2014; 21:92-9; PMID:24037090; http://dx.doi.org/ 10.1038/cdd.2013.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, Sykacek P, Frank L, Schramek D, Komnenovic V, et al.. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014; 5:3056; PMID:24445999; http://dx.doi.org/ 10.1038/ncomms4056 [DOI] [PubMed] [Google Scholar]
- 197.Dupont N, Lacas-Gervais S, Bertout J, Paz I, Freche B, Van Nhieu GT, van der Goot FG, Sansonetti PJ, Lafont F. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe 2009; 6:137-49; PMID:19683680; http://dx.doi.org/ 10.1016/j.chom.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 198.Yang JQ, Liu H, Diaz-Meco MT, Moscat J. NBR1 is a new PB1 signalling adapter in Th2 differentiation and allergic airway inflammation in vivo. EMBO J 2010; 29:3421-33; PMID:20808283; http://dx.doi.org/ 10.1038/emboj.2010.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, et al.. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013; 494:201-6; PMID:23364696; http://dx.doi.org/ 10.1038/nature11866 [DOI] [PMC free article] [PubMed] [Google Scholar]


