Abstract
The NF-κB family plays key roles in immune and stress responses, and its deregulation contributes to several diseases. Therefore its modulation has become an important therapeutic target. Here, we used a high-throughput screen for small molecules that directly inhibit dimerization of the NF-κB protein p65. One of the identified inhibitors is withaferin A (WFA), a documented anticancer and anti-inflammatory compound. Computational modeling suggests that WFA contacts the dimerization interface on one subunit and surface residues E285 and Q287 on the other. Despite their locations far from the dimerization site, E285 and Q287 substitutions diminished both dimerization and the WFA effect. Further investigation revealed that their effects on dimerization are associated with their proximity to a conserved hydrophobic core domain (HCD) that is crucial for dimerization and DNA binding. Our findings established NF-κB dimerization as a drug target and uncovered an allosteric domain as a target of WFA action.
INTRODUCTION
The NF-κB family, which consists of p65/RelA, cRel, RelB, p50, and p52, is responsible for transcription activation of a large number of inflammatory genes, immune response genes, and genes promoting the survival of normal and cancer cells (1, 2). These proteins share a highly conserved DNA-binding and dimerization domain called the Rel homology region (RHR). NF-κB proteins can form homodimers and heterodimers, and this combinatorial diversity contributes to the regulation of distinct but overlapping sets of genes (3–6). The activity of NF-κB is modulated by many extracellular signals, including cytokines, tumor promoters, and chemotherapeutic agents. In unstimulated cells, NF-κB is retained in the cytoplasm in an inactive form by IκB proteins. Signals that activate NF-κB trigger ubiquitination and degradation of IκB by the proteasome, resulting in transport of NF-κB into the nucleus and activation of responsive genes (7, 8). Deregulation of NF-κB is tightly linked to chronic inflammation and cancer (9). In normal cells NF-κB activity is transient; however, in many lymphoid malignancies, certain solid tumors, and chronic inflammation, NF-κB activity becomes persistent and contributes to or causes disease (10–13). Therefore, inhibition of the NF-κB pathway has become an important target for drug development related to inflammation and cancer. Thus far, most of the efforts to modulate NF-κB have been directed toward the signaling pathway, while few attempts have been made to target NF-κB proteins.
In the present study, we conducted a screen based on a split-Renilla luciferase (RL) complementation assay for small molecules that can directly disrupt p65 dimerization. Of the 46,000 small molecules analyzed, the natural product withaferin A (WFA), a known anti-inflammatory and anticancer compound, was among the best inhibitors. We confirmed direct inhibition of p65 dimerization by WFA. Computational modeling of a WFA complex with p65-p65 and p65-p50 predicted contact with dimerization interface residues (E211 and E267 in p65 and p50, respectively) in one subunit and with surface residues E285 and Q287 in the p65 subunit. Although located far from the dimerization site, both E285 and Q287 appear to be important for dimerization and WFA sensitivity. Further investigation revealed that these residues are adjacent to a highly conserved hydrophobic core domain (HCD) that is also essential for dimerization and DNA binding, serving as a scaffold for the dimerization site. Our findings identified p65/RelA as a direct target of WFA that interferes with dimerization directly and allosterically. Furthermore, the data revealed the conserved HCD, shared by the NF-κB and nuclear factor of activated T cells (NFAT) families, as an allosteric modulator of dimerization and DNA binding.
MATERIALS AND METHODS
High-throughput drug screening.
The high-throughput drug-screening assay was performed using the GNF (San Diego, CA) liquid-handling system. The chemical compounds were added with an Echo 550 liquid handler (Labcyte Inc., Sunnyvale, CA). Luminescence signal was detected with the luminescence module of a PheraStar FS plate reader (BMG Labtech, Ortenberg, Germany). For the primary screen, 10 nl each of ∼46,000 bioactive compounds from the Grand Israel National Center for Personalized Medicine (G-INCPM) (Weizmann Institute of Science) chemical libraries was transferred into 1,536-well plates (264712; Nunc) and kept frozen at −30°C before the screen. p65–split-RL-expressing bacterial cells were lysed in 20 mM Tris, pH 8, 100 mM NaCl, 10% glycerol, 2 mM EDTA, 0.5% NP-40, 1 mM dithiothreitol (DTT), 1% protease inhibitor cocktail, and 5 μl of p65-split RL was dispensed into the assay plates. Full-length RL and lysis buffer without RL served as positive and negative controls, respectively. For inhibitory control, p65-split RL was incubated with p65 (competitor) for 25 min prior to the screen at room temperature, and 5 μl of the solution was added to the assay plates, as well. The plates were incubated for 15 min at room temperature, and 5 μl of 5 μg/ml CTZ reagent (Gold Biotechnology, Olivette, MO) in 80 mM K2HPO4, 20 mM KH2PO4 was added to each well. The signal was detected 10 min after incubation at room temperature in the dark, and 380 hits selected from the primary screen were further analyzed in a dose-response assay (0.3, 1, 3, 10, and 30 μM) in duplicate. The compounds that inhibited full-length Renilla luciferase signal were considered false positives.
Computational modeling.
Molecular docking was performed using different modules of Schrödinger Maestro Suite 2015 (Schrödinger, LLC, New York, NY). The three-dimensional crystal structure of the NF-κB protein was retrieved (Protein Data Bank [PDB] identifier [ID] 1MY5; resolution, 1.80 Å) from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (14). The structure was optimized prior to docking using the Protein Preparation Wizard in Schrödinger Maestro Suite 2015. Inconsistencies in the structure, such as missing hydrogen, incorrect bond order, and orientations of the different functional groups of the amino acids, were rectified during the optimization process (15). The prepared protein was then used for induced-fit docking (IFD). Withaferin A was prepared prior to docking using the LigPrep application in Schrödinger Maestro Suite 2015 (LigPrep, version 3.4; Schrödinger, LLC, New York, NY). The IFD was performed using the IFD application of Schrödinger Suite 2015. The IFD application in Schrödinger Suite 2015 combines grid-based ligand docking with energetics (GLIDE) and prime refinement modules (16–18). During IFD, we set the application to retain the default 20 poses for the initial GLIDE docking stage. Standard precision (SP) was selected for the GLIDE redocking stage, with the default cutoff redocking poses within 30 kcal/mol of the best-docked conformation. The poses were ordered based on the GLIDE score (GScore) (kcal/mol) and IFD score (kcal/mol). The GScore is calculated by the software as follows: GScore = 0.065 × vdW + 0.130 × Coul + Lipo + Hbond + metal + BuryP + RotB + site, where vdW is the van derWaals energy, Coul is the Coulomb energy, Lipo is the lipophilic term, Hbond is hydrogen bonding, metal is the metal-binding term, BuryP is the buried polar groups' penalty, RotB is the penalty for rotatable bonds that have been frozen, and site is the active-site polar interactions. The IFD score is calculated by the software as follows: IFD score = 1.0 × GLIDE score + 0.05 × prime_energy (the total of electrostatic, solvation, van der Waals, covalent, H bond lipophilic, pi-pi packing, and self-contact energy). Emodel (model energy score) has a more significant weighting of the force field components (electrostatic and van der Waals energies), which makes it well suited for comparing conformers but much less so for comparing chemically distinct species. We screened and selected the best-scoring conformation of the ligand by reranking according to the Emodel GLIDE score.
Plasmid construction.
A20-luciferase, Rouse sarcoma virus (RSV)-Renilla luciferase, and microRNA 22 (miR-22)-luciferase were previously described (19, 20). Interleukin 2 (IL-2)-luciferase (NFAT/AP-1 3xluc) was a gift from Orly Avni (Bar Ilan University). The p65 expression plasmid pCDNA3 was previously described (21). pCMV-SPORT6-mNFAT2 was a gift from Nir Friedman (Weizmann Institute). Green fluorescent protein (GFP)-RelA (22) was obtained from Addgene (23255). H2B-red fluorescent protein (RFP) was a kind gift from Yosef Shaul (Weizmann Institute). Point mutations were generated by site-directed mutagenesis using the transfer PCR (tPCR) method (23, 24), and all the mutants were verified by sequencing. The split-Renilla luciferase fusion plasmids were constructed by two-step PCR using the RSV-Renilla luciferase (pRL-null) plasmid and pCDNA3-p65 as backbones. As proposed by Jiang et al. (25), the N terminus of the Renilla luciferase contains positions 1 to 229, and the C terminus contains positions 230 to 311. Each fragment was followed by a linker, GGTGGCGGAGGGAGC, corresponding to amino acids GGGGS, and then positions 1 to 298 of p65. Mutations were introduced by site-directed mutagenesis as described above. Primer sequences will be provided upon request.
Cells, transfections, extract preparations, Western blotting, and antibodies.
HEK293T and HeLa cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. Transfection into HEK293T cells and A20-luciferase reporter gene analysis were carried out as described previously (19). Whole-cell extract was prepared by lysing cells in 50 mM Tris, pH 8, 250 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 1% protease inhibitor cocktail or with commercial lysis buffer (Promega). Samples were then separated by SDS-PAGE and subjected to Western blotting as described previously (26). Antibodies for p65 were purchased from Santa Cruz Biotechnology, IκBα antibody was from BD Transductions, A20 antibody was from eBioscience, and the GFP monoclonal antibody was from Abcam.
Expression and purification of His-p65 and microscale thermophoresis (MST) analyses.
p65 wild type (WT) and the E285A Q287A mutant were cloned into pRSFDuet with an N-terminal 6×His tag. Transformed BL21(DE3) bacteria were grown at 20°C following induction with 200 μM isopropyl-1-thio-β-d-galactopyranoside (IPTG) overnight. The bacteria were lysed with a cooled cell disrupter in lysis buffer containing 50 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Calbiochem) in the presence of DNase (1 μg/ml) and lysosyme (40 U/ml culture). Soluble protein was captured on a HiTrap-Chelating_HP_5 ml (GE Healthcare) and eluted with the same buffer containing 0.5 M imidazole. Fractions containing p65 were applied to a size exclusion column (HiLoad 16/60 Superdex 75; GE Healthcare) equilibrated with phosphate-buffered saline (PBS). Fractions containing p65 were loaded onto a Tricorn Q 10/100 GL column (GE Healthcare) equilibrated with 20 mM sodium phosphate, pH 7.2, 1 mM DTT. p65 was eluted from the column with a linear gradient of the same buffer containing 1 M NaCl and 1 mM DTT. Fractions containing pure p65 were pooled and frozen with liquid nitrogen.
For the nonlabeled microscale thermophoresis assays, purified WT or mutant p65 protein (3.375 μM) in 20 mM HEPES, pH 7.8, 140 mM NaCl, 0.1% pluronic was centrifuged at 10,000 × g for 10 min and then incubated with increasing concentrations of WFA (97.65 nM to 200 μM) for 5 min at room temperature. Hydrophilic capillaries were loaded with the samples, and measurements were done in a Monolith NT.LabelFree instrument (NanoTemper) using laser intensities of 20% and 40%. Data evaluation was performed with the Monolith NT.Analysis software (Nano Temper).
RNA analysis.
Total RNA was extracted from HEK293T cells using Tri-reagent (Biosolve Chemicals). cDNA was synthesized from 1 μg of total RNA using an ABI reverse-transcription kit. cDNA samples were analyzed by qPCR in an ABI 7300 real-time PCR system using Power SYBR PCR mix (ABI) as previously described (27).
Chromatin immunoprecipitation (ChIP).
HEK293T cells were transfected with either WT p65, the Y288A mutant, or mock constructs (0.5 μg) or treated with WFA (10 μM; 60 min), followed by tumor necrosis factor alpha (TNF-α) induction (20 ng/ml; 30 min) in 100-mm plates. The cells were then cross-linked with 1% formaldehyde for 10 min at room temperature, and fixing was terminated by adding 1/20 volume of 2.5 M glycine. Chromatin extraction and immunoprecipitations were carried out as previously described (27). For immunoprecipitation, 6 μl of anti-p65 or anti-Oct4 (Santa Cruz) was added to 1 ml of the soluble chromatin DNA. Samples were analyzed by quantitative PCR (qPCR) in an ABI 7300 Real Time PCR system using Power SYBR PCR mix (ABI).
Electrophoresis mobility shift assay (EMSA).
WT and mutant p65 were transcribed and translated in vitro using T7 polymerase and a rabbit reticulocyte lysate kit (Promega). Binding reaction mixtures were assembled on ice with 3 μl translated lysate in a buffer containing 10 mM Tris, pH 8, 100 mM KCl, 0.5 mM EDTA, 0.1% Triton X-100, 12.5% glycerol, 1 mM DTT, 1 μg poly(dI-dC), and 300 fmol of double-stranded NF-κB binding site probe, which was fluorescently labeled with 6-carboxyfluorescein (FAM). Samples were transferred to room temperature for 30 min and then loaded onto 6% nondenaturing Tris-borate-EDTA (TBE)-polyacrylamide gels.
GFP-p65 localization.
Subconfluent HEK293T cells were transfected with 25 ng/ml WT or mutant GFP-p65 and 40 ng/ml H2B-RFP. The cells were visualized 48 h after transfection with a fluorescence microscope (Nikon Eclipse TiS).
Software.
Computational modeling was carried out using Schrödinger Maestro Suite 2015. Structural illustrations were done using the PyMOL Molecular Graphics System, version 1.5.0.4 (Schrödinger, LLC). Amino acid sequence alignment was done using Praline (http://www.ibi.vu.nl/programs/pralinewww/). Microscopy photographs were processed with Nikon NIS-Element F 3.0 and ImageJ version 1.45 k software.
RESULTS
Design of a high-throughput screen for direct inhibitors of p65/RelA dimerization.
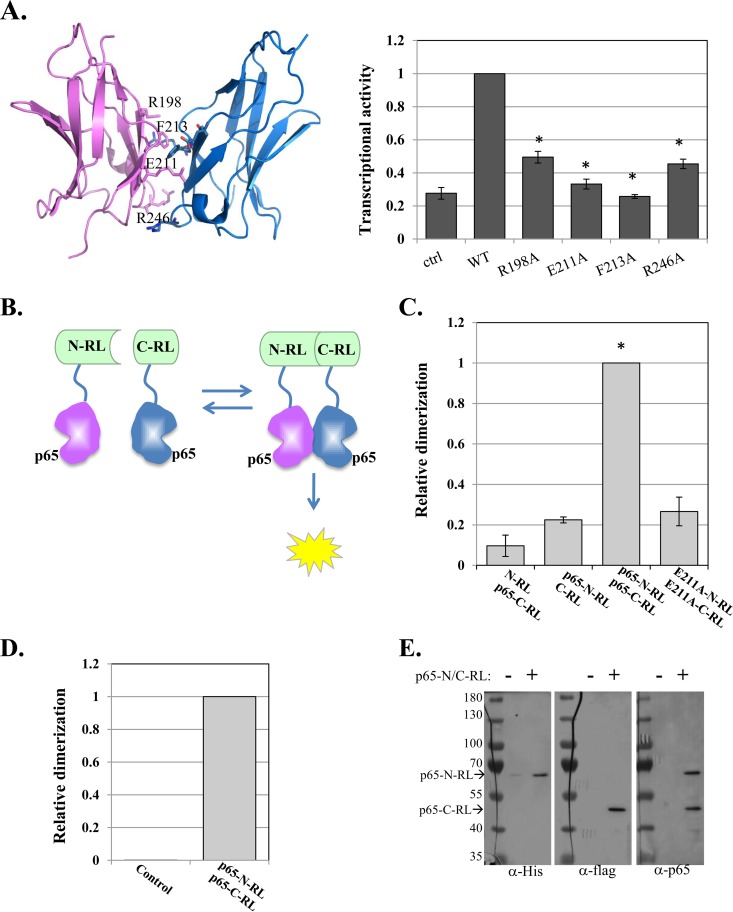
Dimerization of NF-κB is crucial for its ability to bind DNA (3). Consistent with that, mutation of four dimerization interface residues of p65—R198A, E211A, F213A, and R246A—dramatically diminished its transcriptional activity (Fig. 1A). We therefore explored the potential of NF-κB dimerization activity as a drug target. To this end, we applied a protein-protein interaction approach that is based on the split-RL complementation assay (28). In this assay, RL is split into two inactive N- and C-terminal fragments and fused to target proteins. Interaction of the target proteins brings the RL N and C termini into close proximity, restoring enzymatic activity (Fig. 1B). p65 (amino acids [aa] 1 to 298, without the nuclear localization signal [NLS] and the activation domain) was fused to the RL N and C termini and then cotransfected into cells together with a firefly luciferase (FL) reporter as a normalizing control. The p65-RL fusion proteins were also transfected with their empty counterparts to determine the background levels of RL activity. As shown in Fig. 1C, the strongest activity was conferred by the p65-RL N and C pair. To validate that this activity is dependent on dimerization, we constructed p65-split RL with an E211A dimerization site mutation. With this mutation, the RL activity is diminished (Fig. 1C), indicating that the assay is a faithful readout of dimerization.
FIG 1.
Design and application of a high-throughput screen for direct inhibitors of p65 dimerization. (A) (Left) p65-p65 homodimer structure (aa 191 to 304) (PDB ID 1MY5) (14) with dimerization interface residues labeled. (Right) WT p65 and dimerization interface mutants were transfected into cells, together with the NF-κB target reporter A20-luciferase and RSV-Renilla luciferase, which served as a control for transfection efficiency. Twenty-four hours after transfection, the cells were harvested and luciferase activities were measured. The bars represent the means and standard errors (SE) of the results of 3 independent experiments. (B) Schematic illustration of the p65–split-Renilla luciferase principle. (C) The N and C termini of the RL fused to p65 were transfected into cells, together with miR-22–FL, which served as an internal control. RL and FL activities were measured after 24 h. The bars represent the means ± standard errors of the mean (SEM) of the results of 3 independent experiments. The asterisks denote statistically significance differences (P < 0.05). The activity of the WT p65-C/N-RL pair was set to 1. (D) E. coli was transformed either with an empty plasmid or with a plasmid directing expression of p65-N/C-RL fusion proteins. Following induction, the cell lysates were analyzed for RL enzymatic activity. (E) The p65-N/C-RL proteins were analyzed by Western blotting with the antibodies indicated at the bottom.
Next, we established the p65–split-RL luminescence assay in a cell-free system to avoid indirect effects on dimerization. We engineered a plasmid that directs the expression of the two parts of p65-split RL in bacteria, each with a different tag, His or Flag. Bacterial cells transformed with this plasmid and induced to express the two parts of p65-RL were found to have strong RL enzymatic activity compared to the control bacteria (Fig. 1D). Expression of the two fusion proteins was validated by Western blotting with antibodies against the His and Flag tags (Fig. 1E). Western blotting with p65 antibody indicated that the levels of expression of the p65 fusion proteins were comparable (Fig. 1E).
The natural compound withaferin A is a direct inhibitor of p65 dimerization.
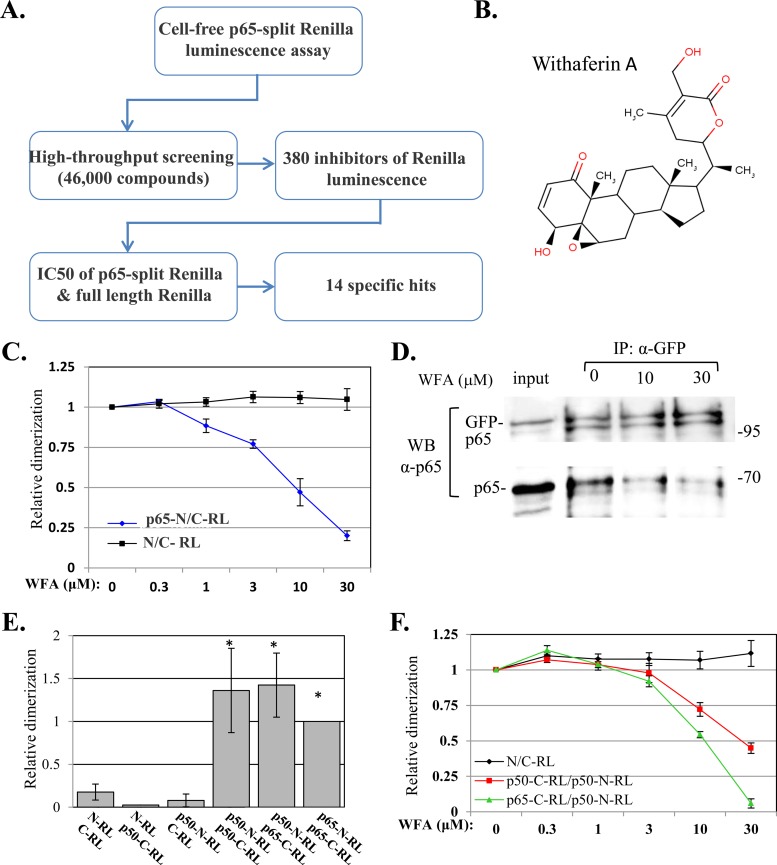
The luminescence assay was then used in a 1,536-well plate format to screen a library of ∼46,000 compounds with diverse chemical natures for inhibitors of the p65–split-RL activity (Fig. 2A). We identified 380 inhibitors (0.8%), and these were further validated in a dose-response assay using the p65-split RL and the full-length RL, which served as a control for inhibitors of the enzymatic activity rather than dimerization. The vast majority of the compounds inhibited both reporters, leaving 14 compounds that specifically inhibited the activity of p65-split RL.
FIG 2.
(A) Flow chart summarizing the high-throughput screen for direct inhibitors of p65 dimerization. (B) Chemical structure of WFA. (C) Dose-response to WFA of the empty plasmid and p65-N/C-RL. (D) Cells were cotransfected with p65 and GFP-p65 and harvested 48 h later. The cell lysates were immunoprecipitated with anti-GFP and treated with 10 μM or with 30 μM WFA for 1 h. The immune complexes were washed and then analyzed by Western blotting using anti-p65 antibody. (E) The N and C termini of the RL fused to p65 or to p50 were transfected into cells, and RL activity was measured after 24 h. The bars represent the means ± SEM of the results of 3 independent experiments. The activity of the WT p65-C/N-RL pair was set to 1. (F) Dose-response to WFA of the empty plasmid, a homodimer of p50-N/C-RL, and a heterodimer of p65-C/p50-N-RL. The asterisks denote statistically significance differences (P < 0.05).
One of the identified compounds, WFA (Fig. 2B), was of particular interest, since it was previously characterized as having anti-inflammatory and anticancer activities and was postulated to interfere with the signaling pathway of NF-κB (29). However, its ability to act as a direct inhibitor of NF-κB proteins was not documented. As shown in Fig. 2C, WFA inhibited p65-split RL effectively, with a 50% inhibitory concentration (IC50) of ∼10 μM, whereas the RL activity directed by the empty RL pair was unaffected by the drug.
To gain further support for the effect of WFA on dimerization, we performed coimmunoprecipitation assays. Cells were cotransfected with plasmids directing the expression of p65 and the GFP-p65 fusion and then subjected to immunoprecipitation using GFP antibodies. The immune complexes were then treated either with vehicle or with 10 and 30 μM WFA. Western blotting with p65 antibodies showed that p65 coprecipitated with GFP-p65 and that the addition of WFA released the p65 from its GFP-p65 counterpart, confirming its direct effect on p65 dimerization (Fig. 2D).
One of the major functional NF-κB complexes consists of a heterodimer of p65-p50. To determine the effect of WFA on this complex, we constructed a p50 (aa 1 to 361) fusion with the N- and C-terminal RL domains. These plasmids were transfected into cells and analyzed for p50-p50 homodimer and p65-p50 heterodimer formation by promoting RL enzymatic activity. Both the p50-p50 and p65-p50 pairs directed RL activity that was significantly above background (Fig. 2E). Next, we determined the effects of WFA on the RL activities directed by p50-p50 and p65-p50 and found that both were inhibited with IC50s of ∼30 and ∼10 μM, respectively (Fig. 2F). Thus, the extent of WFA inhibition of p65-p50 is comparable to that of p65-p65, while the inhibition of p50-p50 is less.
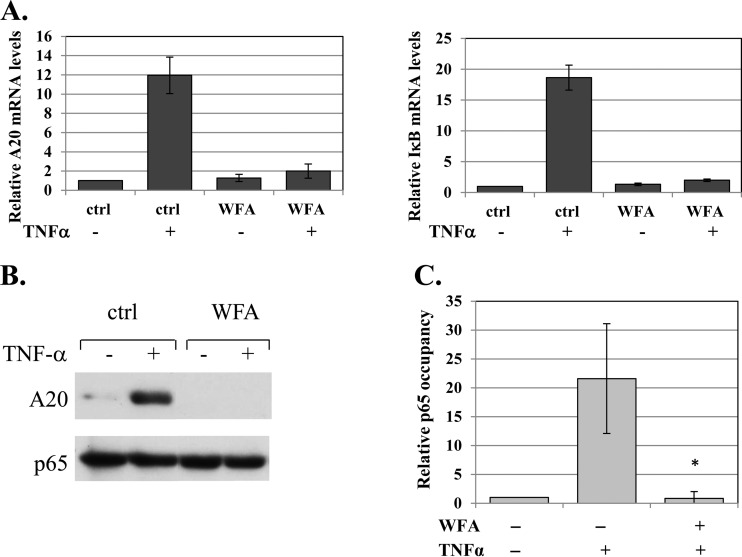
Next, we examined the effect of WFA on the activity of the endogenous NF-κB and found that the drug diminished the mRNA induction of the NF-κB A20 and IκBα target genes by TNF-α (Fig. 3A). Likewise, the induced A20 protein levels were severely diminished (Fig. 3B). We also analyzed the occupancy of the A20 promoter by p65 following TNF-α treatment in the presence or absence of WFA and found that WFA abolished the strong induction of p65 binding to the A20 promoter (Fig. 3C), consistent with the importance of dimerization for DNA binding.
FIG 3.
WFA inhibits NF-κB activity in cells. (A) Untreated or WFA-treated (10 μM; 1 h) cells were induced by TNF-α for 1 h, and levels of A20, IκBα, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNAs were determined by reverse transcription (RT)-qPCR using gene-specific primers. The bars represent the means and standard deviations (SD) of A20 and IκBα levels, normalized to GAPDH, of 2 independent experiments. (B) Untreated or WFA-treated cells (10 μM; 1 h) were induced by TNF-α for 2 h, and the level of A20 protein was determined by Western blotting. The WFA lanes were spliced to bring them close to the control lanes. The original gel is shown in Fig. S4 in the supplemental material. (C) Cells, pretreated with WFA for 60 min and then treated with TNF-α for 30 min or left untreated, were subjected to ChIP using anti-p65 or control (for background levels) antibodies. Analysis was performed by qPCR. The graphs show occupancy levels normalized to the input levels. The uninduced sample was set to 1. The results represent the averages ± SEM of the results of 3 independent experiments. The asterisk denotes a statistically significance difference (P < 0.05).
WFA-bound p65 revealed an allosteric mechanism facilitating NF-κB dimerization.
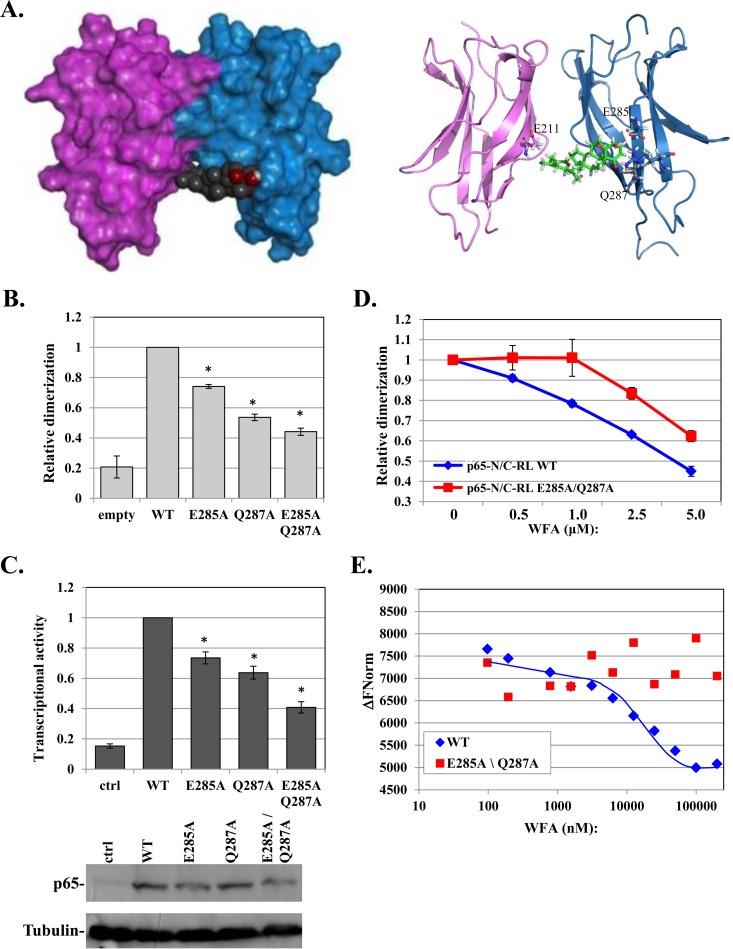
To explore the mechanism by which WFA inhibits NF-κB dimerization, we applied computational modeling for molecular docking of WFA on the p65 dimerization domain (see Materials and Methods). The model with the best docking score suggests one primary binding site spanning both subunits (Fig. 4A, left). WFA interacts with dimerization site residues (e.g., E211) in one monomer and with surface residues E285 and Q287 in the other (Fig. 4A, right). Modeling of WFA interaction with the p65-p50 heterodimer suggested a highly similar docking site with a comparable score that involves the E267 dimerization residue of p50 and E285 and Q287 of p65 (see Fig. S1 in the supplemental material). The predicted contacts of WFA with the dimerization residues, E211 in p65 and E267 in p50, are consistent with its effect on dimerization. E285 and Q287, on the other hand, are distant from the dimerization interface, raising the question of whether they contribute to the effect of WFA on dimerization. To test this possibility, these amino acids were replaced with alanine either individually or together (E285A, Q287A, and E285A Q287A) in the context of p65-split RL and the full-length protein. Surprisingly, each of these mutants decreased p65 dimerization activity, and the double mutant reduced it even further (Fig. 4B), suggesting that these residues indeed contribute to dimerization. The mutants also diminished transcriptional activity to a similar extent (Fig. 4C). Western blot analysis confirmed equivalent expression levels of the WT and mutants (Fig. 4C).
FIG 4.
WFA-bound p65 revealed E285 and Q287 as allosteric modulators of p65 dimerization. (A) Computational models of a p65-p65 homodimer (PDB ID 1MY5) (14) in a complex with WFA using the Schrödinger program. Surface and cartoon models are shown on the left and right, respectively. Contacting residues are labeled on the right. (B) Split-Renilla luciferase dimerization assay of WT p65 and the indicated mutants. (C) The effects of mutations in E285 and Q287 on p65 on transcriptional activity were determined as described for Fig. 1A. The bars represent the means and SE of the results of 3 independent experiments. Expression of the WT and mutant proteins is shown at the bottom. (D) Dose response to WFA of the WT p65 and E285A Q287A mutant N/C-RL pairs. The results represent the means ± SE of 3 independent protein preparations. (E) Purified p65 WT or E285A Q287A (3.4 μM) was incubated with increasing concentrations of WFA, and the intrinsic fluorescence levels were measured using a Monolith NT.LabelFree instrument (NanoTemper). The graph shows the changes in fluorescence intensities in response to the indicated concentrations of WFA. The MST traces are shown in Fig. S2 in the supplemental material. The asterisks denote statistically significance differences (P < 0.05).
If the effect of WFA on p65 dimerization is indeed mediated in part by E285 and Q287, we envisaged that its inhibitory effect would decrease in their absence. To test this idea, cells were transfected with p65-split RL, either WT or E285A Q287A mutant pairs. The cell lysates were then incubated with increasing amounts of WFA. Importantly, the dimerization activity of the WT p65 is clearly more sensitive than that of the E285A Q287A mutant to WFA inhibition (Fig. 4D).
To further investigate WFA interaction with the WT and the E285A Q287A p65 proteins, we used the MST method, which allows precise analysis of binding by monitoring the directed movement of the intrinsic fluorescence of p65 through tiny temperature gradients (30, 31). The WT and E285A Q287A mutant (aa 1 to 298) were expressed in Escherichia coli and purified to homogeneity. Both the WT and the E285A Q287A mutant displayed significant intrinsic fluorescence, directed by a single tryptophan (W233) (Fig. 4E). WFA decreased the intrinsic fluorescence of the WT p65 in a dose-dependent manner, while it had no significant effect on the E285A Q287A mutant (Fig. 4E). The corresponding MST traces are shown in Fig. S2 in the supplemental material. Together, these findings provide strong support to the computational-docking model, in which WFA disrupts dimerization by contacting not only the dimerization site but also E285 and Q287. Moreover, the E285 and Q287 residues emerge as allosteric modulators of p65 dimerization.
Characterization of a highly conserved HCD as a dimerization scaffold in NF-κB and NFAT.
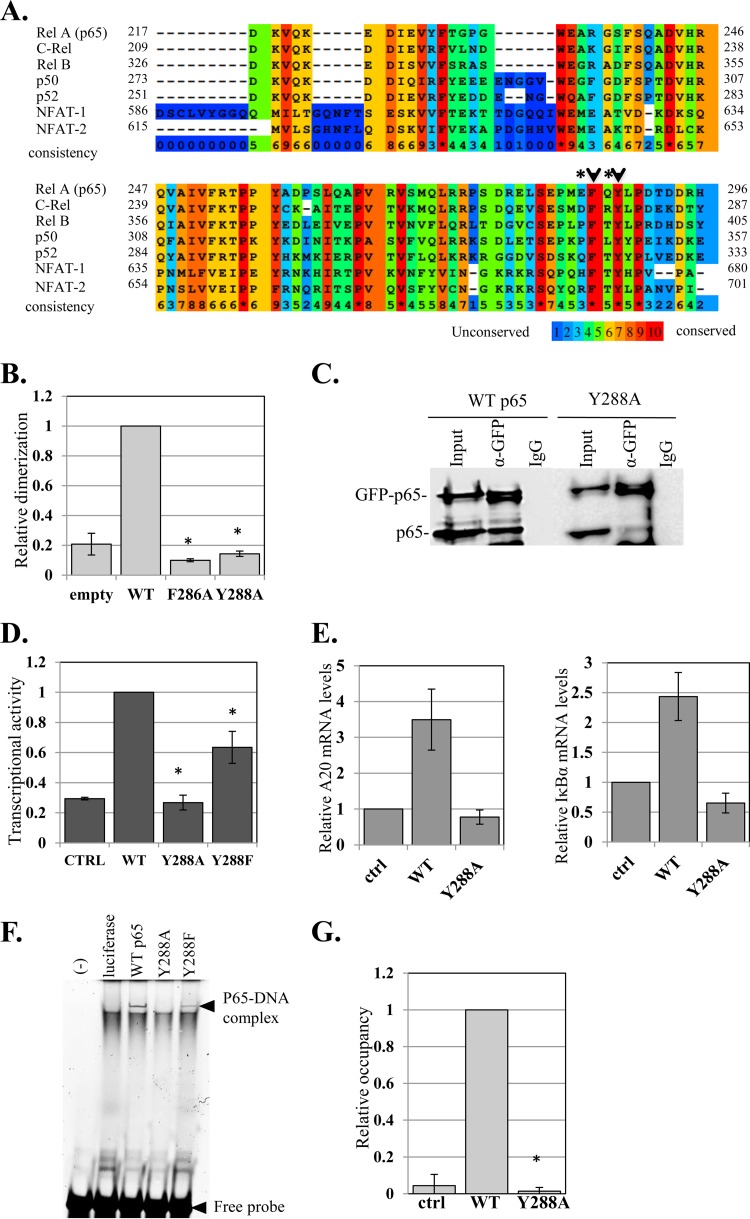
To elucidate the underlying basis for the unexpected roles of E285 and Q287 in dimerization, we examined their conservation among RHR proteins and found them to be nonconserved (Fig. 5A, asterisks). These residues, however, are adjacent to the highly conserved F286 and Y288 amino acids (Fig. 5A, arrowheads). We therefore tested whether F286 and Y288 are also involved in dimerization using the split-RL assay. Remarkably, replacing F286 or Y288 with alanine (F286A and Y288A) caused complete loss of dimerization (Fig. 5B). To gain further support for the effect of Y288 on dimerization, we applied coimmunoprecipitation assays using p65 and GFP-p65 fusion proteins (WT and mutant), as described above. The results clearly show that WT p65 was coprecipitated with WT GFP-p65, whereas GFP-p65-Y288A failed to efficiently coprecipitate p65-Y288A, confirming the dimerization defect associated with Y288A (Fig. 5C). We then analyzed the transcriptional activity of Y288A and found that the mutant is inactive, while a mutation to phenylalanine (Y288F) retained partial activity (Fig. 5D). Consistently, the Y288A mutant failed to activate the mRNA levels of the NF-κB A20 and IκBα target genes (Fig. 5E). To test whether the Y288A mutation affects nuclear localization, we used the GFP-p65 fusion proteins and the nuclear marker H2B-RFP to trace subcellular localization and found that GFP-p65-Y288A can translocate into the nucleus similarly to WT GFP-p65 and the partially active GFP-p65-Y288F (see Fig. S3 in the supplemental material).
FIG 5.
The effects of the nonconserved E285 and Q287 on dimerization are linked to the highly conserved F286 and Y288. (A) Amino acid sequence alignment of five NF-κB and two NFAT proteins. Surface residues E285 and Q287 are marked by asterisks and adjacent HCD residues with arrowheads. (B) F286 and Y288, which are adjacent to E285 and Q287, are essential for dimerization. Shown is a split-RL dimerization assay using WT p65 and F286A and Y288A mutants. (C) p65-Y288A mutation impairs dimerization. Cells were cotransfected with p65 and GFP-p65 (both WT and the Y288A mutant) and harvested 48 h later. The lysates were immunoprecipitated either with anti-GFP antibody or with a control antibody. The input (10%) and the immune complexes were analyzed by Western blotting using anti-p65 antibody. The lanes of the Y288A immunoprecipitates (IP) were spliced to bring them close to the WT lanes. The original gel is shown in Fig. S4 in the supplemental material. (D) Activation of the A20 promoter-luciferase by WT, Y288A, and Y288F p65 variants. The bars represent the means and SE of the results of 3 independent experiments. (E) HEK293T cells were transfected with p65 WT and Y288A, and 24 h later, total RNA was extracted. The levels of A20, IκBα, and GAPDH mRNAs were determined by RT-qPCR. The bars represent the means ± SD of A20 and IκBα levels normalized to GAPDH of 2 independent experiments. (F) Y288 mutations impair DNA binding. The WT and Y288A and Y288F p65 variants were synthesized in vitro in rabbit reticulocyte lysate and then subjected to EMSA using a fluorescently labeled DNA probe. The positions of the p65-DNA complex and the free DNA are indicated. (G) HEK293T cells were transfected with p65 WT and Y288A and 24 h later subjected to ChIP using anti-p65 or control (for background levels) antibodies. Analysis was performed by qPCR. The graph shows occupancy levels normalized to input levels. The uninduced sample was set to 1. The results represent the averages ± SEM of the results of at least 3 independent experiments. The asterisks denote statistically significance differences (P < 0.05).
We examined the DNA-binding activities of Y288 mutants using in vitro-synthesized p65 variants that were subjected to EMSA with a fluorescently labeled DNA probe. While the WT and Y288F proteins displayed high and moderate DNA-binding activity, respectively, the Y288A mutant lacked any detectable DNA binding (Fig. 5F). Likewise, chromatin immunoprecipitation assays showed that WT p65 was efficiently associated with the A20 gene promoter, but Y288A binding was undetected (Fig. 5G).
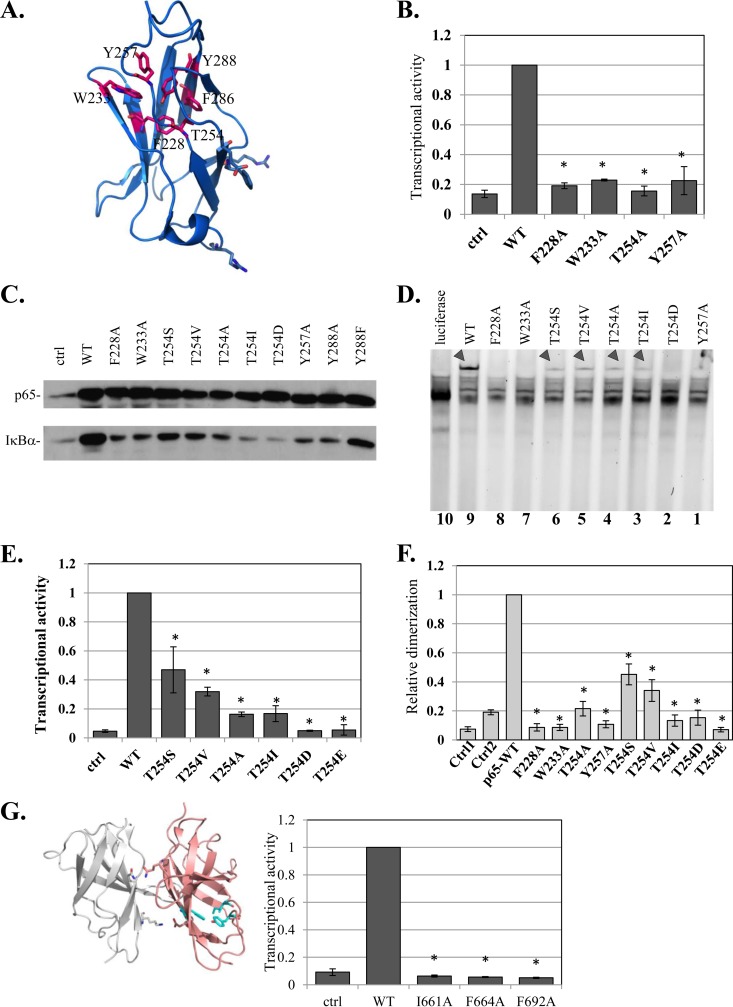
According to the various three-dimensional structures of NF-κB, it appears that F286 and Y288 are parts of a structural domain consisting of an HCD (Fig. 6A). To examine whether this HCD is linked to the function of Y288, 4 additional residues in this core—F228, W233, T254, and Y257—were replaced by alanine, and their functions were analyzed. Each of these HCD mutations caused loss of function toward the A20 reporter gene (Fig. 6B). Analysis of the expression of the endogenous IκBα target gene showed that it was stimulated by the WT p65, but its activation by all HCD mutants was severely impaired (Fig. 6C). Similar to the Y288A mutant, these HCD mutants are also defective in DNA-binding activity (Fig. 6D, lanes 3, 4, 7, and 10). These findings revealed the importance of the HCD structural arrangement for p65 activities.
FIG 6.
Characterization of a highly conserved HCD as a dimerization scaffold in NF-κB and NFAT. (A) Structural illustration of a single central domain of p65 (aa 191 to 304) (PDB ID 1MY5) (14), with HCD residues marked. (B) Analysis of transcriptional activities of F228A, W233A, T254, Y257A, and Y288A HCD mutants as described for Fig. 1A. (C) Effects of HCD mutants on the expression of the IκBα gene, an endogenous target gene. HEK293T cells were transfected with the indicated HCD mutants and harvested 24 h posttransfection. The levels of endogenous IκBα in mock- and p65 variant-transfected cells were monitored by Western blotting. (D) The effects of HCD and T254 mutants on DNA binding were analyzed as described for Fig. 3D. The arrowheads point to the p65-DNA complexes. (E) The effects of T254 mutants on transcriptional activity were determined as described for Fig. 1A. The bars represent the means ± SE of the results of 4 independent experiments. (F) Analysis of dimerization activities of F228A, W233A, T254, Y257A, and Y288A HCD mutants using the split-RL assay as described for Fig. 1C. (G) (Left) Structural illustration of NFAT1 dimer (PDB ID 2O93) (34). Residues of the HCD that correspond to those mutated in NFAT2 are shown in cyan. (Right) WT NFAT2 and the HCD mutants were cotransfected into cells with a luciferase reporter gene driven by the IL-2 promoter and analyzed as described above. The results represent the means ± SE of the results of 4 independent experiments. The asterisks denote statistically significance differences (P < 0.05).
Among the HCD residues examined above, T254 has been previously reported to induce NF-κB activity through phosphorylation, and replacement of this residue with alanine diminished p65 activity (32). As T254 is buried inside the HCD (Fig. 6A), its accessibility to protein kinases is expected to be limited. Accordingly, the effect of a previously reported T254A substitution (32) might have been the consequence of HCD rearrangement, similar to the other alanine substitutions described above. To address this issue further, T254 was replaced either with serine (S) and valine (V), which are similar to threonine; with isoleucine (I); or with the phosphomimetic residues aspartate and glutamate (D and E, respectively). The results revealed that the phosphomimetic replacement of T254 caused complete loss of transcriptional activity, whereas the T254S and T254V substitutions retained partial activity (Fig. 6E). In addition, the I substitution, which has a relatively big side chain, is also inactive (Fig. 6E). The induction levels of the endogenous IκBα recapitulate the effects of the various T254 mutants on the A20 reporter gene (Fig. 6C). Likewise, DNA-binding assays supported the importance of T254 for this activity, as any replacement of the residues diminished DNA binding (Fig. 6D).
To validate the role of HCD residues in dimerization, we employed the split-RL assay. The F228A, W233A, T254A, and Y257A HCD mutants failed to interact and restore RL enzymatic activity (Fig. 6F). T254S and T254V displayed partial dimerization activity, but the phosphomimetic T254D and T254E mutants, as well as T254I, had no significant dimerization activity (Fig. 6F). These results confirm that the HCD is a scaffold for p65 dimerization. Moreover, these findings strongly argue that an active conformation of p65 requires T254 to be within the HCD to support dimerization and transcriptional activity.
Comparing the HCD amino acid sequences in the NF-κB and NFAT families, it is clear that the residues that constitute the hydrophobic core are among the most conserved (Fig. 5A), suggesting that the NFAT HCD is also essential for function. To test this, we introduced mutations in several homologous conserved HCD residues in NFAT2 and analyzed their abilities to induce a reporter gene driven by the NFAT IL-2 target promoter. Mutations in NFAT2 HCD residues I661A, F664A, and F692A (corresponding to I642, Y645, and F673 in NFAT1) abolished its transcriptional activity (Fig. 6G), reminiscent of the effect of HCD mutation on p65 activity.
DISCUSSION
In the present study we established NF-κB dimerization activity as a drug target and identified the anticancer and anti-inflammatory compound WFA as a direct inhibitor of p65 dimerization. By combining computer-assisted molecular docking with mutagenesis of specific residues, we revealed that the mechanism by which WFA inhibits p65 dimerization is both direct and allosteric. Furthermore, the WFA-p65 complex led to the discovery of a region consisting of the HCD and adjacent surface residues as an allosteric modulator of dimerization and DNA binding. The critical residues of this domain are the most highly conserved among RHR proteins, suggesting that these residues were optimized during evolution to maintain the proper structure. This is in line with the observation that even conservative replacements of HCD residues, such as Y288F or T254S/V, resulted in significant loss of activity. While this domain is not part of the interfaces of dimerization and DNA binding, its integrity is nevertheless critical for these activities. We therefore conclude that the allosteric modulator serves as a critical structural scaffold that supports the dimerization and DNA-binding surfaces.
The DNA-binding activity of NF-κB is substantially stabilized by dimerization (3). It is possible that the effect of the HCD on NF-κB DNA binding is a consequence of its primary effect on dimerization. Interestingly, NFAT DNA-binding and transcriptional activities are not strictly dependent on its ability to dimerize (33–35), yet mutating HCD in NFAT2 completely abolished its activity, suggesting that the effect of the HCD is not limited to dimerization.
Our findings clearly show that p65/RelA T254 is an integral part of the HCD, in accordance with the various three-dimensional structures of NF-κB complexes. Furthermore, we suggest that phosphorylation of this residue, if it occurs, requires a significant structural rearrangement, as T254 is buried under a loop constituting residues 206 to 210 that is adjacent to dimerization residues 211 to 213 (Fig. 6A) (14, 36–38). Therefore, the phosphorylation of this residue, which requires its exposure on the surface, would shift the spatial location of the dimerization interface, and this is expected to inhibit rather than activate NF-κB, as previously suggested (32, 39). This is supported by our biochemical and functional evidence. For example, T254V, which has a similar chemical nature, cannot be phosphorylated and resulted in only a partial loss of activity. On the other hand, replacing T254 with phosphomimetic negatively charged D and E residues resulted in a complete loss of DNA binding, dimerization, and transactivation. Nonetheless, we do not exclude the possibility of the occurrence of phosphorylated T254 (p-T254) but suggest that in the analyzed cell types this modification is rare and acts to diminish NF-κB activity.
In summary, our findings, along with the WFA effect, suggest that targeting NF-κB dimerization with drugs has an excellent potential to modulate the activities of RHR proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Haim M. Barr, Galit Cohen, and Haleli Sharir from the HTS Unit of the G-INCPM (Weizmann Institute of Science) for their contribution to the drug screen and helpful suggestions and Shira Albeck, Tamar Unger, and Netta Gilboa from the Israel Structural Proteomic Center for purification of p65 proteins.
R.D. is the incumbent of the Ruth and Leonard Simon Chair of Cancer Research.
R.D. and S.A. conceived and designed the study, analyzed the data, and wrote the paper. S.A. carried out most of the experiments. A.P. conducted the high-throughput drug screen. A.B. performed the chromatin immunoprecipitation assays. E.B.-Z. performed the structural modeling. S.W. provided structural insights on the NF-κB HCD.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00895-15.
REFERENCES
- 1.Gilmore TD. 2003. The Re1/NF-kappa B/I kappa B signal transduction pathway and cancer. Cancer Treat Res 115:241–265. [PubMed] [Google Scholar]
- 2.DiDonato JA, Mercurio F, Karin M. 2012. NF-kappaB and the link between inflammation and cancer. Immunol Rev 246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 3.Smale ST. 2012. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol Rev 246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang VY, Huang W, Asagiri M, Spann N, Hoffmann A, Glass C, Ghosh G. 2012. The transcriptional specificity of NF-kappaB dimers is coded within the kappaB DNA response elements. Cell Rep 2:824–839. doi: 10.1016/j.celrep.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. 2012. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-kappaB family DNA binding. Nat Immunol 13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsui R, Kearns JD, Lynch C, Vu D, Ngo KA, Basak S, Ghosh G, Hoffmann A. 2015. IkappaBbeta enhances the generation of the low-affinity NFkappaB/RelA homodimer. Nat Commun 6:7068. doi: 10.1038/ncomms8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayden MS, Ghosh S. 2004. Signaling to NF-kappaB. Genes Dev 18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 8.Karin M, Lin A. 2002. NF-kappaB at the crossroads of life and death. Nat Immunol 3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Neriah Y, Karin M. 2011. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat Immunol 12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 10.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. 2009. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani A, Allavena P, Sica A, Balkwill F. 2008. Cancer-related inflammation. Nature 454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 13.Nagel D, Vincendeau M, Eitelhuber AC, Krappmann D. 2014. Mechanisms and consequences of constitutive NF-kappaB activation in B-cell lymphoid malignancies. Oncogene 33:5655–5665. doi: 10.1038/onc.2013.565. [DOI] [PubMed] [Google Scholar]
- 14.Huxford T, Mishler D, Phelps CB, Huang DB, Sengchanthalangsy LL, Reeves R, Hughes CA, Komives EA, Ghosh G. 2002. Solvent exposed non-contacting amino acids play a critical role in NF-kappaB/IkappaBalpha complex formation. J Mol Biol 324:587–597. doi: 10.1016/S0022-2836(02)01149-X. [DOI] [PubMed] [Google Scholar]
- 15.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. 2013. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 16.Farid R, Day T, Friesner RA, Pearlstein RA. 2006. New insights about HERG blockade obtained from protein modeling, potential energy mapping, and docking studies. Bioorg Med Chem 14:3160–3173. doi: 10.1016/j.bmc.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 17.Sherman W, Beard HS, Farid R. 2006. Use of an induced fit receptor structure in virtual screening. Chem Biol Drug Des 67:83–84. doi: 10.1111/j.1747-0285.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 18.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. 2006. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem 49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 19.Amir-Zilberstein L, Ainbinder E, Toube L, Yamaguchi Y, Handa H, Dikstein R. 2007. Differential regulation of NF-kappaB by elongation factors is determined by core promoter type. Mol Cell Biol 27:5246–5259. doi: 10.1128/MCB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marbach-Bar N, Ben-Noon A, Ashkenazi S, Harush AT, Avnit-Sagi T, Walker MD, Dikstein R. 2013. Disparity between microRNA levels and promoter strength is associated with initiation rate and Pol II pausing. Nat Commun 4:2118. doi: 10.1038/ncomms3118. [DOI] [PubMed] [Google Scholar]
- 21.Yamit-Hezi A, Nir S, Wolstein O, Dikstein R. 2000. Interaction of TAFII105 with selected p65/RelA dimers is associated with activation of subset of NF-kappa B genes. J Biol Chem 275:18180–18187. doi: 10.1074/jbc.275.24.18180. [DOI] [PubMed] [Google Scholar]
- 22.Chen L, Fischle W, Verdin E, Greene WC. 2001. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 23.Erijman A, Dantes A, Bernheim R, Shifman JM, Peleg Y. 2011. Transfer-PCR (TPCR): a highway for DNA cloning and protein engineering. J Struct Biol 175:171–177. doi: 10.1016/j.jsb.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Unger T, Jacobovitch Y, Dantes A, Bernheim R, Peleg Y. 2010. Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J Struct Biol 172:34–44. doi: 10.1016/j.jsb.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, Bernard D, Yu Y, Xie Y, Zhang T, Li Y, Burnett JP, Fu X, Wang S, Sun D. 2010. Split Renilla luciferase protein fragment-assisted complementation (SRL-PFAC) to characterize Hsp90-Cdc37 complex and identify critical residues in protein/protein interactions. J Biol Chem 285:21023–21036. doi: 10.1074/jbc.M110.103390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elfakess R, Dikstein R. 2008. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS One 3:e3094. doi: 10.1371/journal.pone.0003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diamant G, Amir-Zilberstein L, Yamaguchi Y, Handa H, Dikstein R. 2012. DSIF restricts NF-kappaB signaling by coordinating elongation with mRNA processing of negative feedback genes. Cell Rep 2:722–731. doi: 10.1016/j.celrep.2012.08.041. [DOI] [PubMed] [Google Scholar]
- 28.Paulmurugan R, Gambhir SS. 2003. Monitoring protein-protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal Chem 75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rai M, Jogee PS, Agarkar G, Santos CA. 2016. Anticancer activities of Withania somnifera: current research, formulations, and future perspectives. Pharm Biol 54:189–197. doi: 10.3109/13880209.2015.1027778. [DOI] [PubMed] [Google Scholar]
- 30.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. 2011. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol 9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seidel SA, Dijkman PM, Lea WA, van den Bogaart G, Jerabek-Willemsen M, Lazic A, Joseph JS, Srinivasan P, Baaske P, Simeonov A, Katritch I, Melo FA, Ladbury JE, Schreiber G, Watts A, Braun D, Duhr S. 2013. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP. 2003. Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell 12:1413–1426. doi: 10.1016/S1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 33.Stroud JC, Chen L. 2003. Structure of NFAT bound to DNA as a monomer. J Mol Biol 334:1009–1022. doi: 10.1016/j.jmb.2003.09.065. [DOI] [PubMed] [Google Scholar]
- 34.Bates DL, Barthel KK, Wu Y, Kalhor R, Stroud JC, Giffin MJ, Chen L. 2008. Crystal structure of NFAT bound to the HIV-1 LTR tandem kappaB enhancer element. Structure 16:684–694. doi: 10.1016/j.str.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hogan PG, Chen L, Nardone J, Rao A. 2003. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 36.Chen FE, Huang DB, Chen YQ, Ghosh G. 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 37.Huxford T, Huang DB, Malek S, Ghosh G. 1998. The crystal structure of the IkappaBalpha/NF-kappaB complex reveals mechanisms of NF-kappaB inactivation. Cell 95:759–770. doi: 10.1016/S0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs MD, Harrison SC. 1998. Structure of an IkappaBalpha/NF-kappaB complex. Cell 95:749–758. doi: 10.1016/S0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 39.Itoh S, Saito T, Hirata M, Ushita M, Ikeda T, Woodgett JR, Algul H, Schmid RM, Chung UI, Kawaguchi H. 2012. GSK-3alpha and GSK-3beta proteins are involved in early stages of chondrocyte differentiation with functional redundancy through RelA protein phosphorylation. J Biol Chem 287:29227–29236. doi: 10.1074/jbc.M112.372086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.