ABSTRACT
We previously found that natural single-nucleotide variations located within a proximal region of splicing acceptor 1 (SA1prox) in the HIV-1 genome could alter the viral replication potential and mRNA expression pattern, especially the vif mRNA level. Here, we studied the virological and molecular basis of nucleotide sequence variations in SA1prox for alterations of viral replication ability. Consistent with our previous findings, variant clones indeed expressed Vif at different levels and grew distinctively in cells with various APOBEC3G expression levels. Similar effects were observed for natural variations found in HIV-2 SA1prox, suggesting the importance of the SA1prox sequence. To define nucleotides critical for the regulation of HIV-1 Vif expression, effects of natural SA1prox variations newly found in the HIV Sequence Compendium database on vif mRNA/Vif protein levels were examined. Seven out of nine variations were found to produce Vif at lower, higher, or more excessive levels than wild-type NL4-3. Combination experiments of variations giving distinct Vif levels suggested that the variations mutually affected vif transcript production. While low and high producers of Vif grew in an APOBEC3G-dependent manner, excessive expressers always showed an impeded growth phenotype due to defects in single-cycle infectivity and/or virion production levels. The phenotype of excessive expressers was not due primarily to inadequate expression of Tat or Rev, although SA1prox variations altered the overall HIV-1 mRNA expression pattern. Collectively, our results demonstrate that HIV SA1prox regulates Vif expression levels and suggest a relationship between SA1prox and viral adaptation/evolution given that variations occurred naturally.
IMPORTANCE While human cells possess restriction factors to inhibit HIV-1 replication, HIV-1 encodes antagonists to overcome these barriers. Conflicts between host restriction factors and viral counterparts are critical driving forces behind mutual evolution. The interplay of cellular APOBEC3G and viral Vif proteins is a typical example. Here, we demonstrate that naturally occurring single-nucleotide variations in the proximal region of splicing acceptor 1 (SA1prox) of the HIV-1 genome frequently alter Vif expression levels, thereby modulating viral replication potential in cells with various ABOBEC3G levels. The results of the present study reveal a previously unidentified and important way for HIV-1 to compete with APOBEC3G restriction by regulating its Vif expression levels. We propose that SA1prox plays a regulatory role in Vif counteraction against APOBEC3G in order to contribute to HIV-1 replication and evolution, and this may be applicable to other primate lentiviruses.
INTRODUCTION
Following its entry into target cells, human immunodeficiency virus type 1 (HIV-1) encounters intrinsic resistance factors that provide the first line of host defense in the suppression of infection. HIV-1 encodes proteins to evade this restriction and maintain efficient replication. Antagonism between host restriction factors and viral counterparts drives them to coevolve under mutual selective pressure (1–4). We previously performed HIV-1 adaptation experiments and showed that growth-enhancing mutations were clustered in a narrow region of the C-terminal domain of Pol-integrase (IN) (5). Virological analysis of these mutations and comparative analysis of HIV-1 sequences revealed that naturally occurring single-nucleotide variations in a proximal region of splicing acceptor 1 (SA1prox) (Fig. 1A) modulated viral replication ability. Furthermore, we found that these variations in SA1prox affected HIV-1 mRNA expression patterns, especially its vif mRNA levels, thereby suggesting a role of this region in viral gene expression (6).
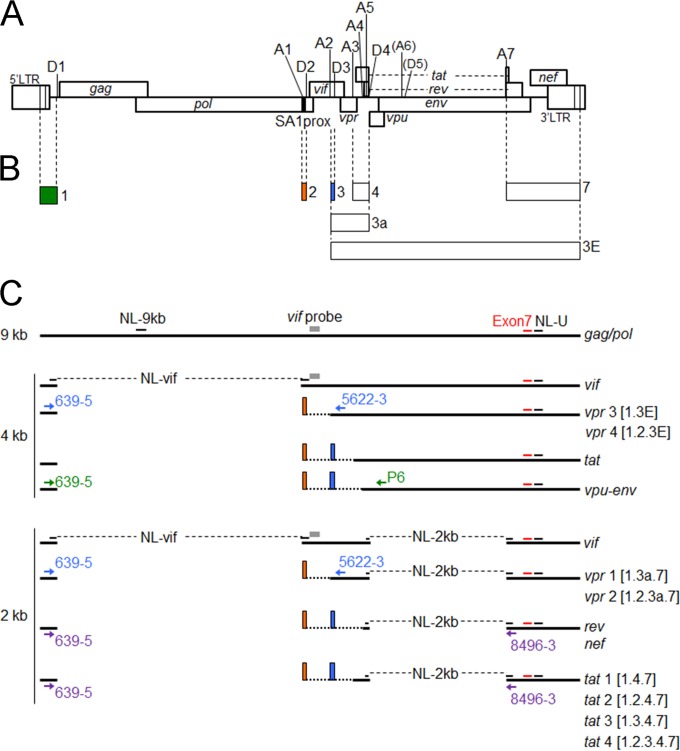
FIG 1.
Schematic representation of the HIV-1 genome and various mRNAs. (A) Organization of the HIV-1NL4-3 genome. Conserved splice donor sites (D1 to D5) and splice acceptor sites (A1 to A7) located within the HIV-1NL4-3 genome (43) (GenBank accession no. AF324493) are indicated (7–10). A6 and D5 have been reported not to be functional in the HIV-1NL4-3 genome used in this study (14). The black area in the HIV-1 genome represents SA1prox (HIV-1 NL4-3 nucleotides 4899 to 4943), as indicated. (B) Exons that form spliced HIV-1 mRNAs. Exons for vpr and tat mRNAs are indicated by boxes (exons 1, 2, 3, 4, 7, 3a, and 3E). Green, orange, and blue boxes represent noncoding exons 1, 2, and 3, respectively. (C) HIV-1 mRNAs. Unspliced (9-kb), incompletely spliced (4-kb), and completely spliced (2-kb) forms of HIV-1 mRNAs are shown. Gray boxes represent a vif probe (6) used for the Northern blot analysis. The amplified regions of the gag/pol transcript (NL-9 kb), the vif transcript (NL-vif), 2-kb transcripts (NL-2 kb), and all HIV-1 transcripts (NL-U) analyzed by qRT-PCR are indicated. The positions of primers used for the semiquantitative RT-PCR analysis of vpr mRNA species, vpu-env mRNA species, 2-kb mRNA species, and all HIV-1 mRNAs (exon 7) are shown by light blue, green, purple, and red letters/arrows/bars, respectively. Orange and blue boxes indicate noncoding exons 2 and 3, respectively. Numbers in brackets represent exon contents of vpr and tat mRNA species.
HIV-1 gene expression (transcription, capping, polyadenylation, splicing, nuclear export, and translation) is a highly controlled process. Since HIV-1 must produce several encoding proteins from the single genome, >40 mRNA species with nine viral genes are generated by alternative splicing, which utilizes four different splicing donor sites (SD1 to SD4) and seven different splicing acceptor sites (SA1 to SA7) in its genome (Fig. 1A) (7–10). Modifications in splicing efficiency cause changes in the gene expression patterns of HIV-1 and have profound effects on viral replication. Splicing regulation relies not only on the recognition of splicing sites by the cellular splicing machinery but also on a number of cis-regulatory elements located around the splicing sites (11, 12). Among various viral mRNAs, vif mRNA is known to result from the splicing of HIV-1 full-length RNA between SD1 and SA1 (Fig. 1B and C). Previous studies reported that several splicing regulatory elements are located around SA1 and that mutations to modify splicing efficiency within these elements influenced vif mRNA production and/or the expression pattern of HIV-1 mRNA species (13–15). However, the regulation of vif mRNA production and splicing at SA1 is complex, and the nucleotide sequence/mutations that control vif mRNA expression levels remain elusive.
HIV Vif (virion infectivity factor) is essential for viral replication in target cells, including CD4+ T cells and macrophages. Vif counteracts the host restriction factor APOBEC3G (A3G) (1–4, 16, 17). In the absence of Vif, A3G is encapsidated into progeny virions in producer cells. A3G exhibits cytidine deaminase activity and induces a G-to-A hypermutation in the viral genome during the reverse transcription process in target cells (18–20). Deaminase-independent inhibition of HIV-1 replication by A3G has also been reported (21–24). The primary mechanism for Vif antagonism against A3G restriction is to induce the proteasomal degradation of A3G. Vif works as an adaptor molecule that recruits A3G to an E3 ligase complex and mediates A3G polyubiquitination and subsequent proteasomal degradation (25–29). Therefore, evasion of A3G restriction by Vif is critical for HIV-1 replication. The power balance between Vif and A3G may be influenced by changes in their functions and/or expression levels and thus affect viral growth properties. Previous studies reported that elevated A3G mRNA levels and A3G-induced hypermutation rates were associated with the downregulation of plasma viral loads and disease progression in HIV-1-infected individuals (30–34). However, there have been debates on this issue (35–38). Moreover, natural variations in Vif and A3G have been reported to affect their functions (39–42). It remains unclear whether there are natural variations that alter vif mRNA levels in the HIV-1 genome.
In the present study, we focused on our previous finding that natural variations in SA1prox altered viral replication potential by affecting viral gene expression, especially vif mRNA expression levels. In order to clarify the role of SA1prox in viral replication, we attempted to identify new natural variations around SA1prox (HIV-1NL4-3 nucleotides 4899 to 4943) in the HIV Sequence Compendium and analyzed the effects of previously and newly found variations on vif mRNA and Vif protein levels and virus growth ability in cell lines expressing different levels of A3G. Our results showed that nucleotide sequence variations in SA1prox affected viral replication potential by regulating Vif expression levels through altering overall gene expression. The frequent existence of these natural variations in SA1prox within the HIV population suggests that this region is linked to viral adaptation and/or evolution.
MATERIALS AND METHODS
Proviral clones.
The proviral clones pNL4-3 (43) and pGL-AN (44) were used as parental clones in the present study. Single-nucleotide variations in HIV-1 and HIV-2 strains were searched for by using HIV-1/simian immunodeficiency virus SIVcpz complete genomes and HIV-2/SIVsmm complete genomes, respectively, in the 2014 HIV Sequence Compendium (Los Alamos National Laboratory [see http://www.hiv.lanl.gov/]). Proviral clones carrying each natural single-nucleotide mutation were generated by using the QuikChange site-directed mutagenesis kit (Agilent Technologies Inc., Santa Clara, CA).
Cells.
A reporter cell line, TZM-bl, carrying a luciferase gene driven by the viral long terminal repeat (LTR) (45, 46) and a human embryonic kidney cell line, HEK 293T (47), were cultured in minimal essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (hiFBS). The human lymphocyte cell lines H9, CEM-SS, CEM, and M8166 were cultured in RPMI 1640 supplemented with 10% hiFBS. The human lymphocyte cell line MT4/CCR5 (MT4 stably expressing CCR5) was maintained in RPMI 1640 containing 10% hiFBS and 200 μg/ml of hygromycin B (Sigma-Aldrich Co., St. Louis, MO).
Analysis of viral growth kinetics.
Virus stocks were prepared from HEK 293T cells transfected with proviral clones by the calcium phosphate coprecipitation method (43, 48) or by using Lipofectamine 2000 (Life Technologies Corporation, Carlsbad, CA). Virion-associated reverse transcriptase (RT) activity was measured as previously described (49, 50). Equal RT units were inoculated into H9, CEM-SS, and CEM cells for the analysis of HIV-1NL4-3, HIV-2GL-AN, and their variant clones. Virus replication was monitored by the activity of RT released into the culture supernatants.
Western blot analysis.
The expression levels of viral and cellular A3G proteins were determined as described previously (6). Briefly, HEK 293T cells were cotransfected with proviral clones (3.5 μg) and the pGL3 luciferase reporter vector (0.5 μg) (Promega Corporation, Madison, WI) by using Lipofectamine 2000 (Life Technologies Corporation). On day 1 posttransfection, cells were lysed in 1× TNE buffer (50 mM Tris-HCl [pH 8.0], 1% Nonidet P-40, 150 mM NaCl, 1 mM EDTA [pH 8.0], and 1% protease inhibitor cocktail [Sigma-Aldrich Co.]), and equal amounts of protein in cell lysates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblot analysis was performed by using anti-HIV-1 HXB2 Vif (catalog no. 2221; NIH Research and References Reagent Program), anti-HIV-1 Gag-p24 (183-H12-5C) (catalog no. 3537; NIH Research and References Reagent Program), anti-HIV-1 gp160 (ADP409; Immuno Ltd./MRC AIDS Directed Programme Reagent Project), anti-Rev (ab25871; Abcam, Cambridge, England), anti-HIV-1 Nef (Advanced Biotechnologies Inc., Columbia, MD), anti-HIV-1 Vpr (catalog no. 3951; NIH Research and References Reagent Program), and anti-β-actin clone AC-15 (Sigma-Aldrich Co.) antibodies. In order to determine the expression levels of A3G in cell lines, equal protein amounts (5 μg) of cell lysates prepared in 1× TNE buffer were analyzed as described above, using anti-APOBEC3G antibody (catalog no. 9968; NIH Research and References Reagent Program) and anti-α-tubulin polyclonal antibody (pAb) (code no. PM054; Medical & Biological Laboratories Co. Ltd., Nagoya, Japan).
qRT-PCR analysis of viral transcripts.
For construction of plasmid DNAs as standards for quantitative real-time reverse transcription-PCR (qRT-PCR) analysis, HIV-1NL4-3 vif and nef and HIV-2GL-AN vif sequences were amplified by PCR using cDNAs from HEK 293T cells transfected with pNL4-3 and pGL-AN, respectively. Amplified products were cloned into the pGEM-T Easy vector (Promega Corporation) to generate pNL-vif+T, pNL-2kb+T carrying the nef gene, and pGL-vif+T. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) sequence amplified by PCR using cDNA from HEK 293T cells was cloned into the pGEM-T Easy vector (Promega Corporation) to obtain pGAPDH+T. For qRT-PCR analysis, HEK 293T cells were transfected with proviral clones (2.5 μg for pNL4-3 and its variants and 3.5 μg for pGL-AN and its variants), and total RNA was extracted at 20 h posttransfection by using the RNeasy Plus minikit (Qiagen GmbH, Hilden, Germany). RNA samples were then treated with DNase I and subjected to qRT-PCR analysis by using the QuantiTect Probe RT-PCR kit (Qiagen GmbH), using the Applied Biosystems 7500 real-time PCR system (Life Technologies Corporation). The primer pairs and dually labeled probes carrying 5′ 6-carboxyfluorescein (FAM) and 3′ 6-carboxytetramethylrhodamine (TAMRA) used for qRT-PCR analysis were as follows: P1 (forward primer), P2 (reverse primer) (51), and NL-9kb-P (probe) (ACAACAACTCCCTCTCAGAAGCA) (NL-9kb in Fig. 1C) for the HIV-1NL4-3 gag/pol transcript; D1-A1 (forward primer), Vif body (reverse primer) (13), and vif mRNA specific-P (probe) (CCCTTCACCTTTCCAGAGGAGCTTTGCT) (NL-vif in Fig. 1C) for the HIV-1NL4-3 vif transcript; NL-D4A7-5 (forward primer) (CTCTATCAAAGCAACCCACCTCCC), NL8496-3 (reverse primer) (CGCAGATCGTCCCAGATAAGTG), and NL-2k-P (probe) (TCTGTCTCTGTCTCTCTCTCCACCT) (NL-2kb in Fig. 1C) for 2-kb transcripts; NL-U-F (forward primer) (CATGGAGCAATCACAAGTAGCAA), NL-U-R (reverse primer) (CTGGAAAACCCACCTCTTCCTC), and NL-U-P (probe) (AGCTAACAATGCTGCTTGTGCCTGGCT) (NL-U in Fig. 1C) for all HIV-1NL4-3 transcripts; GL-vif D1A1-re (forward primer) (CTCCGGGTGAAGGTCTATTACAGAG), GL-vif body (reverse primer) (CCAGGTGGGAACTACTATCCAG), and GL-vifmRNA-P (probe) (TCTTGCCTTCCTCCATAGTCT) for the HIV-2GL-AN vif transcript; and Hs_GAPDH-F (forward primer) (TCCTCTGACTTCAACAGCGAC), Hs_GAPDH-R (reverse primer) (CCAAATTCGTTGTCATACCAGGA), and Hs_GAPDH-P (probe) (TCCTCCACCTTTGACGCTGGGGC) for the GAPDH transcript. As standards, 103 to 107 copies of linearized pNL4-3, pNL-vif+T, pNL-2kb+T, pGL-vif+T, and pGAPDH+T were amplified in parallel with experimental samples for the HIV-1NL4-3 gag/pol transcript and all viral transcripts, including the HIV-1NL4-3 vif transcript, HIV-1NL4-3 2-kb transcripts, the HIV-2GL-AN vif transcript, and the GAPDH transcript, respectively. The expression levels of total viral transcripts in transfected HEK 293T cells were found to be similar among the viral clones tested here and were used for normalization of the transfection efficiency.
Northern blot analysis.
Northern blot analysis was performed to determine vif mRNA expression levels as previously described (6). Briefly, HEK 293T cells were transfected with proviral clones (2.5 μg), and total RNA was extracted at 20 h posttransfection by using the RNeasy Plus minikit (Qiagen GmbH). Poly(A)+ RNA was isolated with the Oligotex-dT30 Super mRNA purification kit (TaKaRa Bio Inc., Otsu, Japan) and then treated with DNase I. Samples were subjected to glyoxal-denatured 1% agarose gel electrophoresis and blotted onto a positively charged nylon membrane. vif and GAPDH mRNAs were detected by probes labeled with the PCR digoxigenin (DIG) probe synthesis kit (Fig. 1C) (Roche Diagnostics GmbH, Mannheim, Germany) and visualized with DIG High Prime DNA labeling and detection starter kit II (Roche Diagnostics GmbH).
Analysis of single-cycle infectivity.
Viruses were prepared from HEK 293T cells transfected with proviral clones by the calcium phosphate coprecipitation method (43, 48) or by using Lipofectamine 2000 (Life Technologies Corporation). The virus amount was measured by RT assays as described above, and equal numbers of RT units (2 × 103 or 1 × 104 RT units) were inoculated into TZM-bl cells (4 × 103 cells). On day 1 postinfection, cell lysates were prepared with 1× cell culture lysis reagent (Promega Corporation) for luciferase assays (Promega Corporation).
Analysis of virion production.
H9 cells (106) were cotransfected with proviral clones (2 μg) and the pGL3 luciferase reporter vector (Promega Corporation) (2 μg) by using Amaxa Cell Line Nucleofector kit V (Lonza Ltd., Basel, Switzerland), using Nucleofector II (Lonza Ltd.). The CXCR4 antagonist AMD3100 (0.5 μM) was added to the cultures throughout the experiment. Culture supernatants and cells collected on day 2 posttransfection were examined for virion production and luciferase activity, respectively. Virion production was monitored by using the HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (ZeptoMetrix Corporation, Buffalo, NY). Cell lysates were prepared with 1× cell culture lysis reagent (Promega Corporation) for luciferase assays (Promega Corporation). Luciferase activity in cell lysates was used to normalize transfection efficiency.
Analysis of HIV-1 gene expression.
For semiquantitative RT-PCR analysis, total RNA was prepared from HEK 293T cells transfected with proviral clones as described above. After DNase I treatment, samples were reverse transcribed by using the SuperScript III first-strand synthesis system (Life Technologies Corporation). To gain PCR products amplified within a linear range, conditions for semiquantitative RT-PCR were determined similarly as described previously (51). The primer pairs used for semiquantitative RT-PCR analysis were as follows (Fig. 1C): NL639-5 (forward) (GCGCCCGAACAGGGACTTGAA) and NL8496-3 (reverse) for the 2-kb mRNA species, NL639-5 (forward) and P6 (reverse) (51) for the vpu-env mRNAs, NL639-5 (forward) and NL5622-3 (reverse) (GCTCTAGTGTCCATTCATTG) for the vpr mRNAs, NL8686-5 (forward) (AGGGGACAGATAGGGTTATAG) and NL8878-3 (reverse) (TAGGTCTCGAGATACTGCTC) for all HIV-1NL4-3 mRNAs (exon 7 in Fig. 1C), and GAPDH-S (forward) (CGAGATCCCTCCAAAATCAA) and GAPDH-AS (reverse) (GTCTTCTGGGTGGCAGTGAT) for the GAPDH mRNA. PCR amplicons were separated on a 2% Metaphor agarose (Lonza Ltd.) gel in 0.5× Tris-borate-EDTA buffer by using the Mupid exU submarine electrophoresis system (Advance Co. Ltd., Tokyo, Japan) and stained with ethidium bromide. The Amersham Imager 600 instrument (GE Healthcare UK Ltd., Buckinghamshire, England) was used to visualize PCR products and to quantify signal intensities of the products. For qRT-PCR analysis, samples used for semiquantitative RT-PCR analysis were subjected to qRT-PCR analysis by using the QuantiTect Probe PCR kit (Qiagen GmbH), using the primer pairs for qRT-PCR analysis of viral transcripts described above. Tat trans-activation assays were performed as described previously (6). Briefly, Proviral clones and an LTR-driven luciferase reporter clone, 5RLTR-Luc (6), were cotransfected into HEK 293T cells with Lipofectamine 2000 (Life Technologies Corporation). Cells were lysed on day 1 posttransfection and subjected to luciferase assays (Promega Corporation).
RNA secondary structure prediction.
The mfold program, version 4.6 (52), was used to predict secondary RNA structure formed by the 46-nucleotide sequence (at HIV-1 NL4-3 nucleotides 4899 to 4943) of HIV-1NL4-3 and its variants. When multiple structures were predicted from each sequence, the structure with the lowest energy (ΔG) was selected.
RESULTS
Natural single-nucleotide variations in SA1prox (Y226tac, D229gat, P233cct, and V234gtg) alter viral replication ability in an A3G-dependent manner by changing Vif expression levels.
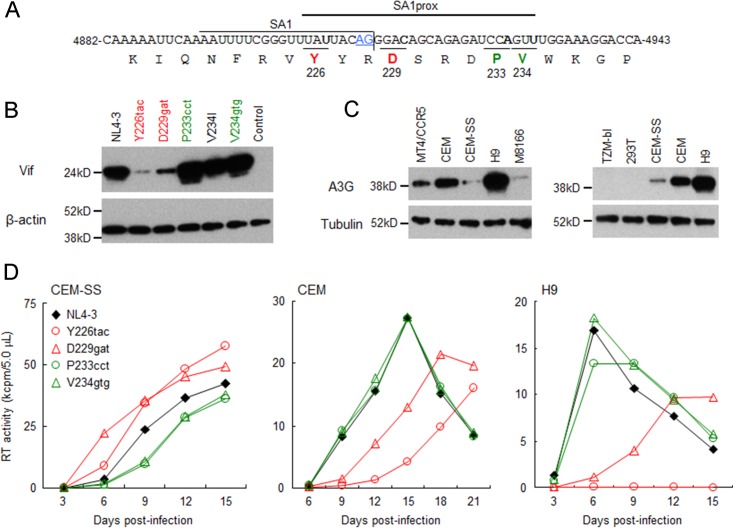
Our previously reported adaptation experiments using prototype macaque cell-tropic HIV-1s and macaque cell lines revealed that viruses frequently and reproducibly acquired growth-enhancing mutations in the C-terminal domain of Pol-IN (5). Subsequent virological studies of these mutations showed that in human lymphocyte cell lines, growth phenotypes of not only prototype macaque cell-tropic HIV-1s but also human cell-tropic HIV-1 (NL4-3 clone) were markedly altered by the introduction of naturally occurring single-nucleotide variations (Y226tac, D229gat, P233cct, and V234gtg) into SA1prox (Fig. 2A) (6). Since the common and unique characteristics of these variants relative to the parental clones were their altered vif mRNA levels (6), we determined the expression levels of the Vif protein for each NL4-3 derivative. The parental NL4-3 clone and its variants carrying Y226tac, D229gat, P233cct, V234I, or V234gtg were transfected into HEK 293T cells, and cell lysates were prepared on day 1 posttransfection for Western blot analysis. Clone V234I, carrying a slightly growth-enhancing mutation (6), was used as a control. As clearly observed in Fig. 2B, Vif expression levels for Y226tac and D229gat were remarkably reduced, whereas those for P233cct and V234gtg were higher than the NL4-3 Vif level.
FIG 2.
Effects of natural single-nucleotide variations within SA1prox on Vif protein expression levels and viral replication ability in various cell lines. Red and green letters/symbols/lines represent low and high expressers of the Vif protein, respectively. (A) Nucleotide and amino acid sequences in the region around SA1. NL4-3 sequences (nucleotides 4882 to 4943 and Pol-IN amino acids 219 to 238) are shown. SA1 (7–10) and SA1prox (6) are indicated. The AG dinucleotide of the Vif splice site is highlighted by blue letters/underlining. Letters in boldface type in the sequences represent the sites at which natural variations that alter virus replication ability (6) are located. (B) Vif expression levels in transfected cells. The proviral clones indicated were transfected into HEK 293T cells, and cell lysates were prepared on day 1 posttransfection for Western blot analysis using anti-Vif and anti-β-actin antibodies. The migration positions of mass standards are on the left. Representative data from three independent experiments are shown. Control, pUC19. (C) A3G expression levels in various cell lines. Cell lysates were prepared from the indicated cell lines and subjected to the Western blot analysis using anti-A3G and anti-tubulin antibodies. The migration positions of mass standards are on the left. Representative data from two independent experiments are shown. (D) Viral replication kinetics in various cell lines. Viruses were prepared from HEK 293T cells transfected with the proviral clones indicated and inoculated into CEM-SS, CEM, and H9 cells. Infection conditions were as follows: 104 RT units/105 CEM-SS cells and H9 cells and 103 RT units/105 CEM cells. Virus replication was monitored by the activity of RT released into the culture supernatants. Representative data from at least two independent experiments are shown.
We then investigated the effects of different Vif expression levels for these variants on antagonistic activity against A3G in multicycle viral replication. We first determined A3G expression levels in permissive (Vif-independent) and nonpermissive (Vif-dependent) cell lines (Fig. 2C). Consistent with previously reported findings (13–15, 20), the expression levels of A3G were low in permissive T-cell lines (CEM-SS, M8166, and MT4/CCR5) but high in nonpermissive T-cell lines (CEM and H9). H9 cells, which have been reported to express A3G at levels similar to those of peripheral blood mononuclear cells (53), showed the highest levels among these cell lines. We also confirmed that A3G expression was undetectable in HEK 293T and HeLa-derived TZM-bl cell lines (Fig. 2C) (20).
We then monitored the viral replication abilities of NL4-3 and its variant clones (Y226tac, D229gat, P233cct, and V234gtg) in cell lines expressing different levels of A3G (Fig. 2D). While the low-Vif producers Y226tac and D229gat grew better than NL4-3 in permissive CEM-SS cells, the replication abilities of P233cct and V234gtg with high expression levels of Vif were lower than that of NL4-3. In contrast, Y226tac and D229gat replicated very poorly in nonpermissive CEM and H9 cells, whereas P233cct and V234gtg exhibited growth potentials similar to that of NL4-3. These results show that the four natural variations in SA1prox increase or decrease Vif expression levels, thereby altering viral replication kinetics in an A3G-dependent manner.
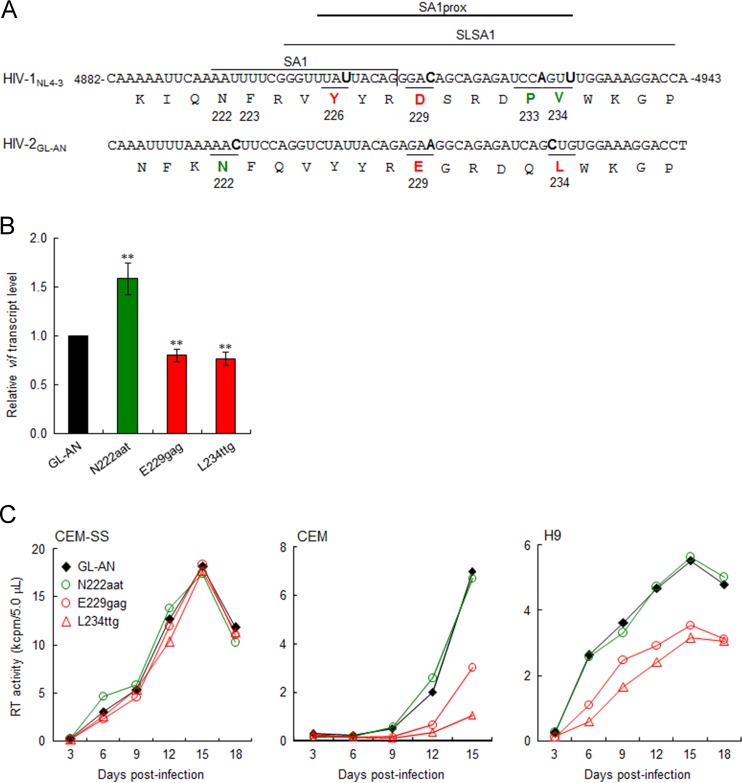
Natural variations in HIV-2 SA1prox also affect viral growth potential by modulating vif transcript levels.
A previous study reported that the nucleotide sequence around SA1 within the HIV-1 genome forms a stem-loop structure, designated SLSA1, and also that the SLSA1 structure is conserved between HIV-1NL4-3 and SIVmac239 (54). Natural variations in SA1prox that change Vif expression levels and viral growth potential were clustered within SLSA1 (Fig. 3A) (6). Since nucleotide sequences around SA1 and the genome organization of HIV-2 are similar to those of SIVmac239, we assumed that natural variations in HIV-2 SA1prox, like those in HIV-1 SA1prox, can affect vif expression levels/viral replication ability due to the conserved features of nucleotide sequences and SLSA1 structures. Using the HIV-2/SIVsmm complete genomes in the 2014 HIV Sequence Compendium (96 strains), we searched for natural variations in the HIV-2 genome (amino acid positions N222, F223, Y226, E229, Q233, and L234 in HIV-2GL-AN Pol-IN) (Fig. 3A), corresponding to the HIV-1 genomic region in which growth-altering natural variations were identified (6). As shown in Table 1, natural variations within HIV-2GL-AN SA1prox were readily found in these sites, except for the Y226 site in HIV-2GL-AN Pol-IN. Based on codon frequency differences in GL-AN and natural variants within the HIV-2/SIVsmm population, we selected N222aat, E229gag, and L234ttg for further analyses and constructed proviral clones carrying each variation. In order to evaluate the effects of these variations on vif transcript levels, parental GL-AN and its variant (N222aat, E229gag, and L234ttg) clones were transfected into HEK 293T cells, and DNase I-treated total RNA samples prepared at 20 h posttransfection were examined by qRT-PCR analysis. As shown in Fig. 3B, the vif transcript level for N222aat was higher than that for GL-AN (1.58-fold on average), but E229gag and L234ttg had lower vif transcript levels than did GL-AN (0.80- and 0.76-fold on average, respectively). Expression of the HIV-2GL-AN Vif protein could not be examined due to the unavailability of the antibody. We next compared the viral replication abilities of GL-AN and its variants in permissive CEM-SS cells and nonpermissive CEM and H9 cells (Fig. 3C). While all viral clones grew comparably well in CEM-SS cells, E229gag and L234ttg showed reduced growth potentials compared to that of GL-AN in CEM and H9 cells, despite a slight difference in vif transcript levels between GL-AN and E229gag/L234ttg. For HIV-2GL-AN, the threshold of the Vif expression level required for optimal viral replication under A3G restriction may be in a narrow range. These results show that natural variations in HIV-2 SA1prox as well as those in HIV-1 SA1prox affect the vif transcript level and viral replication ability.
FIG 3.
Effects of natural single-nucleotide variations in HIV-2 SA1prox on vif transcript levels and viral replication ability. Red and green represent low and high expressers of the vif transcript, respectively. (A) Sequences of HIV-1NL4-3 (nucleotides 4882 to 4943 and Pol-IN amino acids 219 to 238) and of the corresponding region of HIV-2GL-AN. SA1 (7–10), SLSA1 (54), and SA1prox (6) in the NL4-3 sequences are shown. Letters in boldface type in the sequences represent the sites at which natural variations that alter virus replication ability (NL4-3 [6] and GL-AN [this study]) are located. (B) vif transcript levels for GL-AN and its natural variants. The proviral clones indicated were transfected into HEK 293T cells, and at 20 h posttransfection, total RNAs were prepared for qRT-PCR analysis using a primer pair specific for the GL-AN vif transcript. vif transcript levels relative to those of GL-AN are presented. Mean values ± standard deviations from three independent experiments are shown. Significance relative to GL-AN was determined by the Student t test (**, P < 0.05). (C) Viral growth kinetics of GL-AN and its natural variants in CEM-SS, CEM, and H9 cells. Viruses were prepared from transfected HEK 293T cells, and 105 cells were infected with viruses (2 × 105 RT units for CEM-SS and CEM cells and 5 × 104 RT units for H9 cells). Virus replication was monitored by the activity of RT released into the culture supernatants. Representative data from at least two independent experiments are shown.
TABLE 1.
Codon frequencies in SA1prox of HIV-2/SIVsmma
| Amino acid position in GL-AN Pol-IN | Amino acid | Codonb | No. of sequences | % of sequences |
|---|---|---|---|---|
| 222 | N | AAC | 1 | 1.0 |
| AAT | 90 | 93.8 | ||
| K | AAA | 3 | 3.1 | |
| K or N | AAW | 1 | 1.0 | |
| D | GAT | 1 | 1.0 | |
| 223 | F | TTC | 18 | 18.8 |
| TTT | 78 | 81.3 | ||
| 226 | Y | TAT | 96 | 100.0 |
| 229 | E | GAA | 95 | 99.0 |
| GAG | 1 | 1.0 | ||
| 233 | Q | CAG | 37 | 38.5 |
| CAA | 59 | 61.5 | ||
| 234 | L | CTG | 73 | 76.0 |
| CTC | 14 | 14.6 | ||
| TTG | 7 | 7.3 | ||
| CTT | 1 | 1.0 | ||
| CTA | 1 | 1.0 |
Data are from 96 sequences of HIV-2/SIVsmm complete genomes in the 2014 HIV Sequence Compendium (Los Alamos National Laboratory [see http://www.hiv.lanl.gov/]). The amino acid and codon of GL-AN are shown at the top line for each amino acid position.
W is A or T.
vif transcript levels are highly divergent among natural variants found in the extended SA1prox.
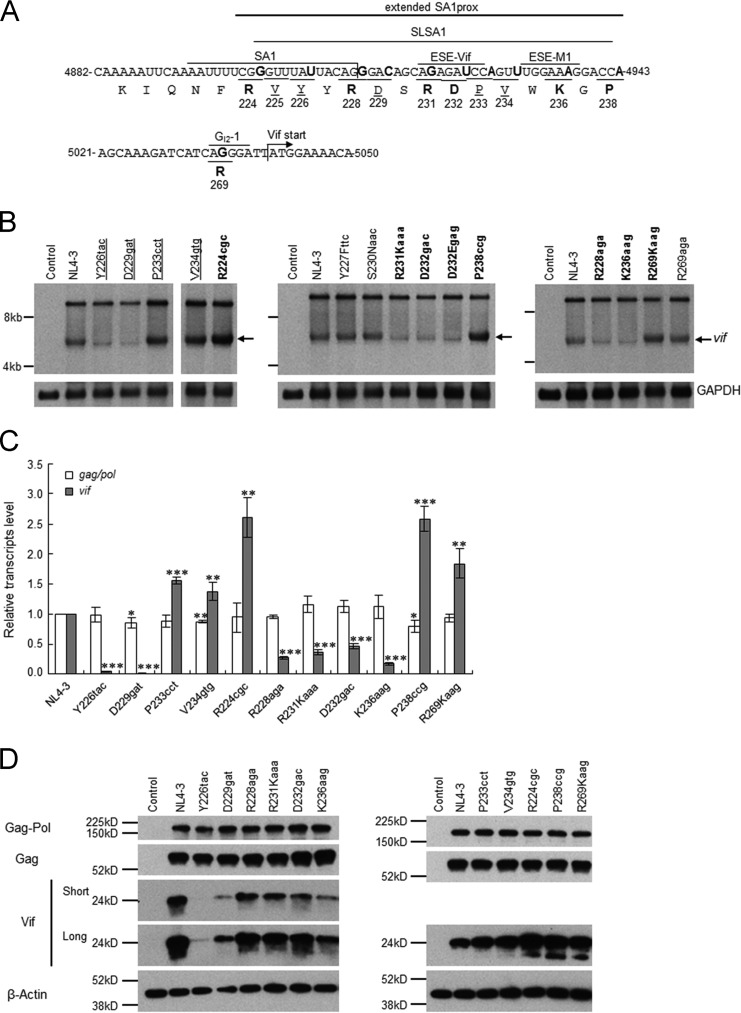
Natural variations (Y226tac, D229gat, P233cct, and V234gtg) that alter the Vif expression level and viral replication ability were scattered within the SLSA1 region (Fig. 2 and 4A). Furthermore, the introduction of artificial mutations into the known splicing regulatory elements close to SA1 (Fig. 4A) affected the expression pattern of vif mRNAs (13–15). Thus, we predicted that more natural variations around SA1prox (extended SA1prox in Fig. 4A) may alter Vif expression levels and viral replication potentials and searched for such mutations in the HIV-1/SIVcpz complete genomes in the 2014 HIV Sequence Compendium (199 strains). Of the 10 new sites examined (R224, Y227, R228, S230, R231, D232, W235, K236, G237, and P238) (Fig. 4A and Table 2), 8 (R224, Y227, R228, S230, R231, D232, K236, and P238) (Table 2) were found to have nucleotide variations. Based on higher codon frequencies at each site, seven variations (Y227Fttc, R228aga, S230Naac, R231Kaaa, D232gac, K236aag, and P238ccg) (Table 2) were selected for further analyses. The synonymous variation R224cgc was chosen for the R224 site, and D232Egag was chosen as a second variation for the D232 site (Table 2). In total, nine natural variations were selected from the extended SA1prox for functional studies. In addition, two single-nucleotide natural variations found in the GI2-1 element (55) (R269Kaag and R269aga) were selected for further study.
FIG 4.
Identification of new single-nucleotide variations in the extended SA1prox that affect vif mRNA and Vif expression levels. (A) Nucleotide and amino acid sequences in the regions around the SA1 and Vif start sites. NL4-3 sequences (nucleotides 4882 to 4943 and 5021 to 5050 and Pol-IN amino acids 219 to 238 and 269) are shown. The extended SA1prox (NL4-3 nucleotides 4899 to 4943), SA1 (7–10); SLSA1 (54); and three known splicing regulatory elements, ESE-Vif (13, 15), ESE-M1 (14), and GI2-1 (55), are indicated. Letters in boldface type in the sequences represent the sites at which natural variations that affect vif mRNA levels are located. The sites analyzed in Fig. 2 and previously (6) are indicated by underlined amino acids. (B) Identification of natural variations that affect vif mRNA levels. The proviral clones indicated were transfected into HEK 293T cells, and at 20 h posttransfection, poly(A)-selected total RNAs were prepared. After the DNase I treatment, samples were subjected to Northern blot analysis using a vif probe (Fig. 1). GAPDH was used as an internal control. RNA size markers are on the left. The position of the vif mRNA is indicated by arrows. Proviral clones that were analyzed for Vif protein expression levels in Fig. 2 are underlined. Letters in boldface type indicate newly identified natural variants expressing vif mRNA at a lower or higher level than that of NL4-3. Control, pUC19. The left panel was separated to remove an unnecessary lane. (C) Expression levels of gag/pol and vif transcripts. Total RNA was prepared from HEK 293T cells transfected with the proviral clones indicated and treated with DNase I. Samples were then subjected to qRT-PCR analysis using primer pairs specific for gag/pol (NL-9kb in Fig. 1) and vif (NL-vif in Fig. 1) transcripts. The expression levels of all HIV-1 transcripts (NL-U in Fig. 1) and GAPDH were analyzed by qRT-PCR in parallel for transfection and internal controls, respectively. The expression levels of gag/pol and vif transcripts in each sample were normalized by those of all HIV-1 transcripts and GAPDH. gag/pol and vif transcript levels relative to those of NL4-3 are presented. Mean values ± standard deviations from three independent experiments are shown. Significance relative to NL4-3 was determined by the Student t test (***, P < 0.01; **, P < 0.05; *, P < 0.1). (D) Expression levels of viral proteins. The proviral clones indicated were transfected into HEK 293T cells, and cell lysates were prepared on day 1 posttransfection for Western blot analysis using anti-p24, anti-Vif, and anti-β-actin antibodies. The migration positions of mass standards are on the left. Representative data from three independent experiments are shown. Control, pUC19; Short, short exposure; Long, long exposure.
TABLE 2.
Codon frequencies in the extended SA1prox of HIV-1/SIVcpza
| Amino acid position in NL4-3 Pol-IN | Amino acid | Codonb | No. of sequences | % of sequences |
|---|---|---|---|---|
| 224 | R | CGG | 195 | 98.0 |
| CGC | 1 | 0.5 | ||
| Q | CAG | 2 | 1.0 | |
| R or Q | CRG | 1 | 0.5 | |
| 227 | Y | TAC | 186 | 93.5 |
| TAT | 2 | 1.0 | ||
| F | TTC | 10 | 5.0 | |
| S | TCC | 1 | 0.5 | |
| 228 | R | AGG | 141 | 70.9 |
| AGA | 56 | 28.1 | ||
| AGR | 1 | 0.5 | ||
| Stop | TGA | 1 | 0.5 | |
| 230 | S | AGC | 190 | 95.5 |
| TCA | 1 | 0.5 | ||
| N | AAC | 7 | 3.5 | |
| A | GCC | 1 | 0.5 | |
| 231 | R | AGA | 197 | 99.0 |
| K | AAA | 2 | 1.0 | |
| 232 | D | GAT | 76 | 38.2 |
| GAC | 115 | 57.8 | ||
| GAY | 1 | 0.5 | ||
| E | GAA | 3 | 1.5 | |
| GAG | 1 | 0.5 | ||
| N | AAT | 1 | 0.5 | |
| AAC | 1 | 0.5 | ||
| G | GGC | 1 | 0.5 | |
| 235 | W | TGG | 199 | 100.0 |
| 236 | K | AAA | 197 | 99.0 |
| AAG | 2 | 1.0 | ||
| 237 | G | GGA | 199 | 100.0 |
| 238 | P | CCA | 190 | 95.5 |
| CCG | 9 | 4.5 |
Data are from 199 sequences of HIV-1/SIVcpz complete genomes in the 2014 HIV Sequence Compendium (Los Alamos National Laboratory ([see http://www.hiv.lanl.gov/]). The amino acid and codon of NL4-3 are shown at the top line for each amino acid position. Regarding codon frequencies for amino acids at positions 225, 226, 229, 233, and 234, see reference 6.
R is A or G, and Y is C or T.
In order to identify new natural variations that alter vif mRNA production in SA1prox and the GI2-1 element, we first carried out Northern blot analysis. Although this analysis is rather qualitative, it is suitable for the initial screening of variations that have a relatively large effect on vif mRNA production. Eleven proviral clones carrying each new variation described above were constructed and transfected into HEK 293T cells in parallel with parental pNL4-3 and the other natural variants (Y226tac, D229gat, P233cct, and V234gtg) (Fig. 2) (6). Poly(A)-selected total RNA was prepared at 20 h posttransfection and examined by Northern blotting using the vif probe (Fig. 1C). As shown in Fig. 4B, consistent with the expression of Vif (Fig. 2B), Y226tac/D229gat and P233cct/V234gtg generated decreased and increased vif mRNA levels, respectively, relative to NL4-3. Of the new natural variants, while Y227Fttc, S230Naac, and R269aga produced levels of vif mRNA similar to that of NL4-3, clones that expressed higher (R224cgc, P238ccg, and R269Kaag) or lower (R228aga, R231Kaaa, D232gac, D232Egag, and K236aag,) vif mRNA levels than that of NL4-3 were readily identified. Notably, R224cgc and P238ccg appeared to express a markedly higher level of vif mRNA than NL4-3. These results show that natural variations within the extended SA1prox and in the GI2-1 element frequently affect vif mRNA levels.
To confirm the results of Northern blot analysis, we quantitatively compared the effects of natural variations in the SA1prox and GI2-1 element on vif transcript expression. Since splicing efficiency at SA1 has been shown to influence gag/pol (9 kb) mRNA production (Fig. 1) (13, 15), its expression level was also quantitatively analyzed. NL4-3 and its variants (R224cgc, Y226tac, R228aga, D229gat, R231Kaaa, D232gac, P233cct, V234gtg, K236aag, P238ccg, and R269Kaag) were transfected into HEK 293T cells, and DNase I-treated total RNA preparations at 20 h posttransfection were subjected to qRT-PCR analysis. As shown in Fig. 4C, the gag/pol transcript level for most natural variants tested was similar to that for NL4-3. In contrast, the vif transcript level was always markedly different from that of NL4-3. Moreover, vif transcript levels remarkably varied among natural variants: with reduced levels for R228aga, R231Kaaa, D232gag, and K236aag; markedly reduced levels for Y226tac and D229gat; increased levels for R224cgc, P233cct, V234gtg, P238ccg, and R269Kaag; and markedly increased levels for R224cgc and P238ccg. Consistent with the vif and gag/pol transcript levels, these variants expressed their Vif proteins at different levels in transfected HEK 293T cells but exhibited similar expression levels of Gag and Gag-Pol proteins (Fig. 4D). On the basis of these results, natural variants were categorized into low-vif (Y226tac, R228aga, D229gat, R231Kaaa, D232gac, and K236aag), high-vif (P233cct, V234gtg, and R269Kaag), and excessive-vif (R224cgc and P238ccg) types. All of the natural variations tested here had more profound effects on vif transcript levels than on gag/pol transcript levels, and each variation had different impacts on vif transcript expression.
Naturally occurring combinations of some variations appear to exhibit an additive effect on vif transcript production.
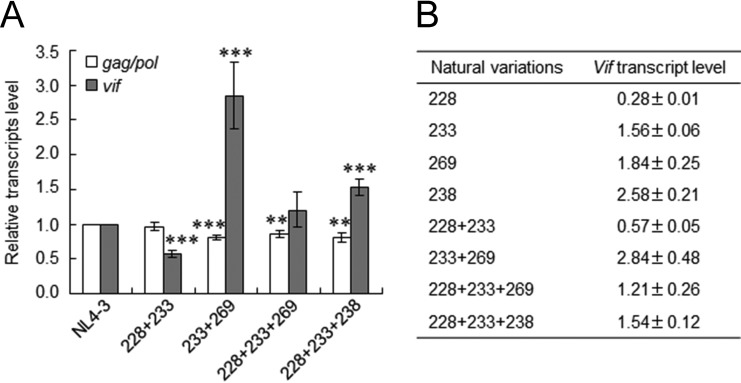
In the comparative investigation of HIV-1/SIVcpz sequences described above, we noted several HIV-1 strains carrying two or three variations (in combinations of R228aga, P233cct/ccc, R269Kaag, and/or P238ccg), which have different effects on vif transcript production (Fig. 4C and Table 3). We previously reported that P233cct and P233ccc exhibited similar growth phenotypes in permissive MT4/CCR5 cells (6). In order to examine how naturally occurring double or triple variations in the genome influence vif transcript levels, we constructed proviral clones carrying multiple variations (R228aga+P233cct, P233ccc+R269Kaag, R228aga+P233cct+R269Kaag, and R228aga+P233cct+P238ccg) found in the HIV-1/SIVcpz sequences. NL4-3 and proviral clones carrying natural combinations of the variations were transfected into HEK 293T cells, and DNase I-treated total RNA samples prepared at 20 h posttransfection were subjected to qRT-PCR analysis. As shown in Fig. 5A, the gag/pol transcript levels of NL4-3 and its natural combination variants were similar but slightly reduced for P233ccc+R269Kaag, R228aga+P233cct+R269Kaag, and R228aga+P233cct+P238ccg. While R228aga+P233cct+R269Kaag expressed a level of the vif transcript similar to that expressed by NL4-3, vif transcript levels for R228aga+P233cct and for P233ccc+R269Kaag/R228aga+P233cct+P238ccg were lower and higher, respectively, than that for NL4-3. As summarized in Fig. 5B, the results obtained by qRT-PCR analysis in Fig. 4C and 5A revealed that the effects of these natural variations on vif transcript levels appeared to be additive. A previous study suggested that several splicing regulatory elements around SA1 act competitively on the regulation of the vif mRNA expression level (13). Thus, the vif production level determined by combinations of variations may be complicatedly regulated by the relative strength of splicing regulatory sites around SA1.
TABLE 3.
Effects of natural single-nucleotide variations around SA1prox on vif mRNA levels and viral replication potentials
| Viral clone | Amino acid in IN | Parental NL4-3 |
Natural variant |
Relative vif transcript/mRNA levela | Growth kinetics in cellsb |
|||
|---|---|---|---|---|---|---|---|---|
| Amino acid | Codon | Amino acid | Codon | Low A3G | High A3G | |||
| NL4-3 | 1.00/++ | ++ | ++ | |||||
| N222aac | 222 | N | AAT | N | AAC | ++c | ||
| F223ttc | 223 | F | TTT | F | TTC | ++c | ||
| R224cgc | 224 | R | CGG | R | CGC | 2.61 | + | + |
| V225gtc | 225 | V | GTT | V | GTC | ++c | ||
| Y226tac | 226 | Y | TAT | Y | TAC | 0.04 | +++ | − |
| Y227Fttc | 227 | Y | TAC | F | TTC | ++ | ||
| R228aga | 228 | R | AGG | R | AGA | 0.28 | +++d | ++ |
| D229gat | 229 | D | GAC | D | GAT | 0.01 | +++ | + |
| S230Naac | 230 | S | AGC | N | AAC | ++ | ||
| R231Kaaa | 231 | R | AGA | K | AAA | 0.36 | +++d | ++ |
| D232gac | 232 | D | GAT | D | GAC | 0.47 | +++d | ++ |
| D232Egag | 232 | D | GAT | E | GAG | + | ||
| P233cct | 233 | P | CCA | P | CCT | 1.56 | + | ++ |
| P233ccc | 233 | P | CCA | P | CCC | +c | ||
| P233ccg | 233 | P | CCA | P | CCG | +c | ||
| V234gtg | 234 | V | GTT | V | GTG | 1.38 | + | ++ |
| K236aag | 236 | K | AAA | K | AAG | 0.16 | +++ | +e |
| P238ccg | 238 | P | CCA | P | CCG | 2.58 | + | + |
| R269aga | 269 | R | AGG | R | AGA | ++ | ||
| R269Kaag | 269 | R | AGG | K | AAG | 1.84 | + | ++ |
The relative vif mRNA levels are indicated by the average values obtained from the qRT-PCR analysis shown in Fig. 4. The relative levels of some variants (Y227Fttc, S230Naac, D232Egag, and R269aga) are shown semiquantitatively as ++ (NL4-3 level) and + (lower level than that of NL4-3) (see Fig. 4 for actual results). Blank spaces indicate that the experiments were not performed.
CEM-SS and/or MT4/CCR5 cells were used as low-A3G expressers. H9 and/or CEM cells were used as high-A3G expressers. +++, virus grew better than NL4-3; ++, virus grew similarly to NL4-3; +, virus grew more poorly than NL4-3; −, virus growth was undetectable during the observation period (15 or 21 days). Blank spaces indicate that the experiments were not performed.
The growth characteristics of these clones were previously described (6) but not tested in this study.
Data from multiple experiments showed that the variants appeared to grow slightly better than NL4-3.
Data from multiple experiments showed that the variant appeared to grow slightly more poorly than NL4-3.
FIG 5.
Effects of naturally occurring combinations of variations in SA1prox on vif transcript levels. (A) vif transcript levels. RNA samples were prepared from HEK 293T cells transfected with the proviral clones indicated, as described in the legend of Fig. 4, and subjected to qRT-PCR analysis using primer pairs specific for gag/pol (NL-9kb in Fig. 1) and vif (NL-vif in Fig. 1) transcripts. The expression levels of all HIV-1 transcripts (NL-U in Fig. 1) and GAPDH were analyzed by qRT-PCR in parallel for transfection and internal controls, respectively. The expression levels of gag/pol and vif transcripts in each sample were normalized by those of all HIV-1 transcripts and GAPDH. gag/pol and vif transcript levels relative to those of NL4-3 are presented. Mean values ± standard deviations from three independent experiments are shown. Significance relative to NL4-3 was determined by the Student t test (***, P < 0.01; **, P < 0.05). (B) Quantification of vif transcripts. vif transcript levels relative to that of NL4-3 for single (Fig. 4C) and combination (see above) variants determined by qRT-PCR are shown. 228, R228aga; 233, P233cct; 269, R269Kaag; 238, P238ccg; 228 + 233, R228aga+P233cct; 233 + 269, P233ccc+R269Kaag; 228 + 233 + 269, R228aga+P233cct+R269Kaag; 228 + 233 + 238, R228aga+P233cct+P238ccg.
There are some natural variations and their combinatorial variants that alter the vif transcript level in specific HIV-1 subtypes. For example, P238ccg, R228aga, P233ccc, P233cct, and/or their combinations are found mainly in some specific groups or subtypes. Since Vif proteins of different HIV-1 subtypes were previously shown to display different anti-A3G activities (56), it will be interesting to determine whether there is a relationship between Vif expression levels determined by the SA1prox nucleotide sequence and Vif function.
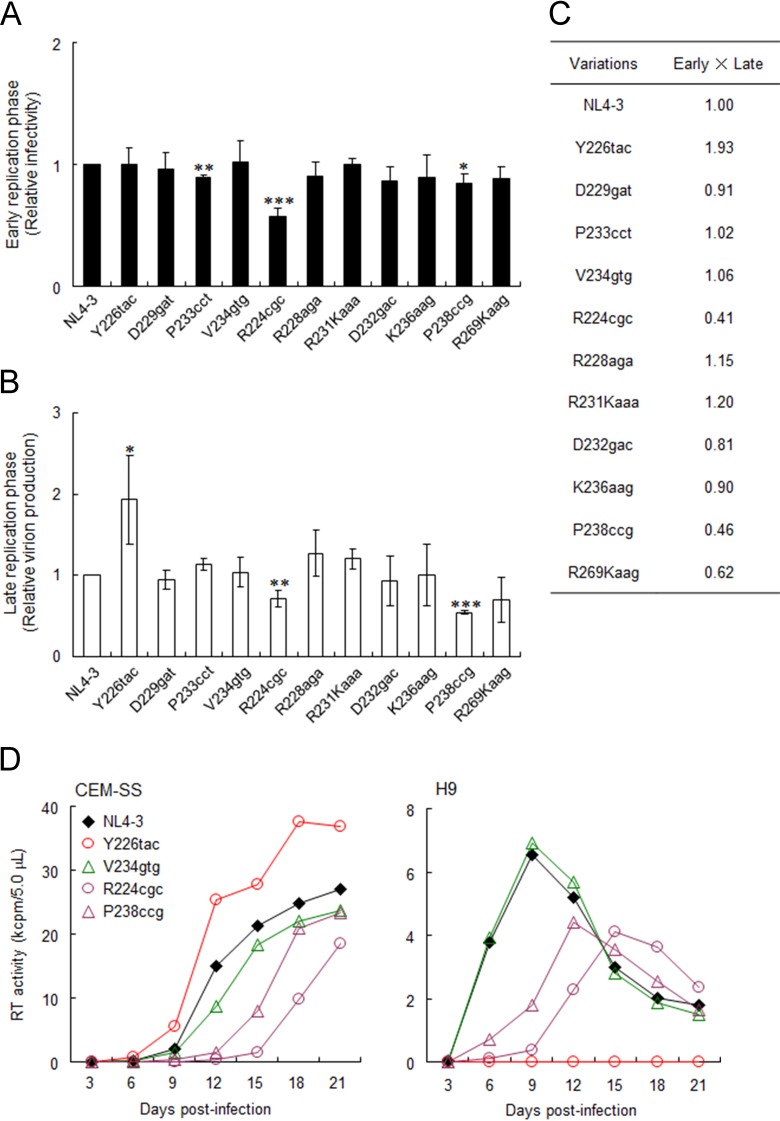
Excessive vif type variants (R224cgc and P238ccg) exhibit A3G-independent and decreased replication ability due to reduced virion infectivity/production.
Increases in vif mRNA/protein expression levels were previously shown to adversely affect viral infectivity and virion production (13, 57, 58). Therefore, we examined the effects of natural variations in the extended SA1prox on early and late viral replication phases. Viral infectivity at the early phase was monitored by using TZM-bl cells with undetectable A3G expression (Fig. 2C). TZM-bl is frequently and widely used as a standard reporter cell line to determine viral infectivity by measuring Tat-driven luciferase expression from an LTR reporter gene. Luciferase activity in TZM-bl cells has been shown to correlate well with infectious virion units (45, 46, 59–61). Viruses were prepared from HEK 293T cells transfected with NL4-3 or its natural variants (R224cgc, Y226tac, R228aga, D229gat, R231Kaaa, D232gac, P233cct, V234gtg, K236aag, P238ccg, and R269Kaag) and were then inoculated into TZM-bl cells. On day 1 postinfection, cell lysates were prepared for luciferase assays. The infectivity of most natural variants was similar to that of NL4-3, but P233cct (high vif) and R224cgc (excessive vif), especially R224cgc, exhibited significantly decreased viral infectivity (Fig. 6A). In order to monitor virion production, H9 cells were transfected with NL4-3 or its natural variants in the presence of the CXCR4 antagonist AMD3100, and on day 2 posttransfection, culture supernatants were prepared for Gag-p24 ELISAs. As shown in Fig. 6B, while the virion production levels of most variants were similar to that of NL4-3, Y226tac (low vif) and R224cgc/P238ccg (excessive vif) produced progeny virions at higher and lower levels, respectively. These results indicated that R224cgc impaired both early and late replication phases and that some variations (Y226tac and P238ccg) strongly influenced virion production. When replication ability indices were calculated (Fig. 6C), R224cgc, Y226tac, and P238ccg were clearly distinct from NL4-3.
FIG 6.
Effects of natural variations in SA1prox on viral replication. (A) Single-cycle infectivity. Viruses were prepared from HEK 293T cells transfected with the proviral clones indicated, and equal numbers of RT units were inoculated into TZM-bl cells. Cell lysates were prepared on day 1 postinfection for luciferase assays. Infectivity is presented as luciferase activity relative to that exhibited by NL4-3. Mean values ± standard deviations from three independent experiments are shown. Significance relative to NL4-3 was determined by the Student t test (***, P < 0.01; **, P < 0.05; *, P < 0.1). (B) Virion production. H9 cells were cotransfected with the proviral clones indicated and a luciferase reporter vector (pGL3) by using a Nucleofector kit. Virion production on day 2 posttransfection was measured by the amount of Gag-p24 in the culture supernatants. The CXCR4 antagonist AMD3100 (0.5 μM) was added to cultures throughout the experiment. The amount of p24 was normalized to the luciferase activity in cell lysates. The amount of p24 relative to that produced by NL4-3 is presented. Mean values ± standard deviations from three independent experiments are shown. Significance relative to NL4-3 was determined by the Student t test (***, P < 0.01; **, P < 0.05; *, P < 0.1). (C) Replication ability index. Indices for viral single-cycle infectivity were calculated by the multiplication of early (relative TZM-bl infectivity) and late (relative virion production) replication efficiencies. (D) Viral replication kinetics. Viruses were prepared from HEK 293T cells transfected with the proviral clones indicated, and equal numbers of RT units (103 units) were inoculated into 105 CEM-SS (low-A3G expresser) and H9 (high-A3G expresser) cells. Virus replication was monitored by the activity of RT released into the culture supernatants. Representative data from at least two independent experiments are shown. Red, green, and purple symbols/lines represent viral clones of low-, high-, and excessive-vif types, respectively.
In Fig. 2, we show that four natural variations in SA1prox altered viral growth kinetics in an A3G-dependent manner. In order to determine how this result is applicable to the excessive-vif-type variations, we compared the viral growth kinetics of R224cgc and P238ccg with those of the low (Y226tac)- and high (V234gtg)-vif-type variants in low-A3G CEM-SS and high-A3G H9 cells (Fig. 6D). In permissive CEM-SS cells, Y226tac grew better than NL4-3, but V234gtg grew slightly worse than NL4-3, as shown in Fig. 2. The growth kinetics of the excessive-vif variants R224cgc and P238ccg were significantly more retarded than those of NL4-3. In contrast to CEM-SS cells, Y226tac did not grow at all in nonpermissive H9 cells, whereas V234gtg grew similarly to NL4-3, as shown in Fig. 2. R224cgc and P238ccg again grew more poorly in H9 cells than did NL4-3. These results indicate that the viral replication potentials of variant clones are influenced by both Vif and A3G expression levels. The low (Y226tac)- and high (V234gtg)-vif types grew in a manner that was dependent on their anti-A3G activity under strong A3G pressure; however, in the absence of A3G restriction, their replication ability was reversed. Consistent with the replication ability indices (Fig. 6C), Y226tac grew the best among the viral clones tested in the absence of A3G. Although we did not observe any significant changes in single-cycle replication assays for V234gtg (Fig. 6A to C), multicycle infection experiments allowed us to detect the effect of high Vif levels on growth ability (Fig. 2 and 6D). Notably, regardless of the A3G levels in cells, an excessive-vif type (R224cgc and P238ccg) always grew more poorly than did NL4-3, which was consistent with the replication ability indices (Fig. 6C). However, the excessive-vif type that can counteract A3G grew considerably, but the markedly low-vif type Y226tac did not do so under high-A3G conditions. Collectively, the viral replication potentials of SA1prox natural variants are determined by their Vif expression levels and antagonizing ability against A3G.
Reduction in replication potential of excessive-vif-type variants occurs independently of early protein expression (Tat, Rev, or Nef).
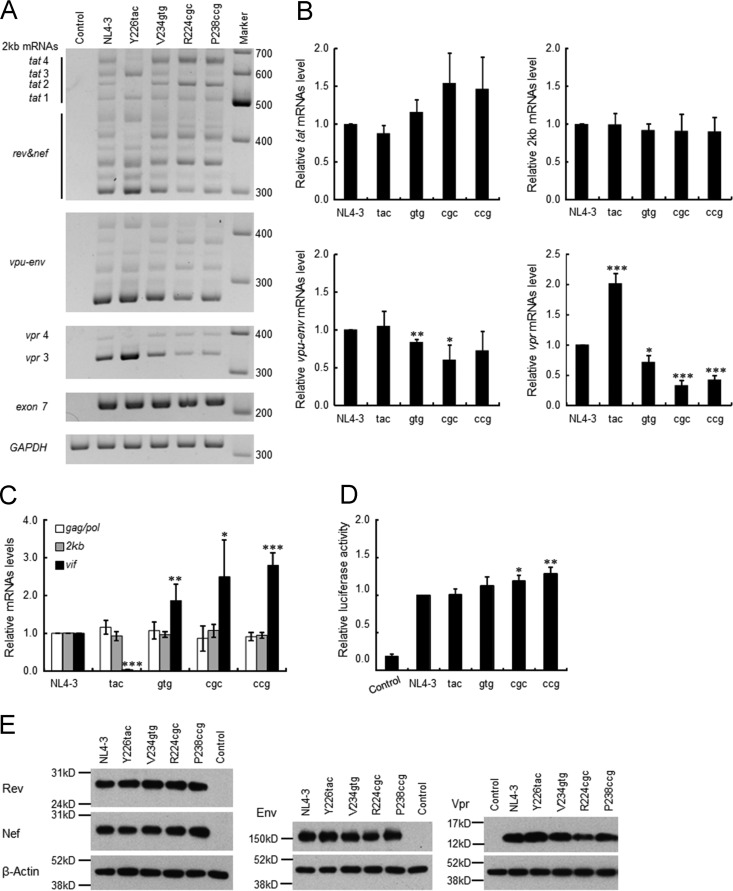
Unlike low- and high-vif-type variants, excessive-vif-type variants displayed an impeded replication ability relative to that of wild-type NL4-3 regardless of the A3G expression level (Fig. 6D). It has been shown that alternative splicing of HIV-1 RNA often occurs in a mutually exclusive manner and that the change in splicing efficiency at SA1, which produces vif mRNA, influences the splicing at the other site(s) (13, 15, 54, 55, 58, 62, 63). To investigate the possibility that the alteration of splicing efficiency at SA1 and the other sites in excessive-vif-type variants could have a negative effect on replication potential, we comparatively examined the expression patterns and levels of viral mRNA species for NL4-3, Y226tac (low-vif type), V234gtg (high-vif type), and R224cgc and P238ccg (excessive-vif types). Total RNA samples were prepared from HEK 293T cells transfected with proviral clones and subjected to semiquantitative RT-PCR analysis using the primer pairs for 2-kb (tat, rev, and nef), vpu-env, and vpr mRNA species (Fig. 1C). In the semiquantitative RT-PCR analysis, vif and vpr mRNA species of 2 kb were undetectable (data not shown). As shown in Fig. 7A, although the total HIV-1 mRNA levels of each sample (exon 7) were similar, the expression patterns and levels of 2-kb, vpu-env, and vpr mRNA species were different among NL4-3 and its variants. Y226tac variation increased tat 1 (exon content 1.4.7), tat 3 (1.3.4.7), and vpr 3 (1.3E) mRNA levels but decreased tat 2 (1.2.4.7), tat 4 (1.2.3.4.7), and vpr 4 (1.2.3E) mRNA levels relative to those of NL4-3 (Fig. 1B and C and 7A). In contrast, high- and excessive-vif-type variants (V234gtg, R224cgc, and P238ccg) exhibited increased levels in tat 2 (1.2.4.7) and tat 4 (1.2.3.4.7) mRNAs and decreased levels in tat 3 (1.3.4.7) and vpr 3 (1.3E) mRNAs relative to those of NL4-3. The results here imply an increase of exon 2 inclusion into viral mRNAs (e.g., tat 2 and tat 4) in high- and excessive-vif-type variants, suggesting increases in the activities of SD2 as well as of SA1 (Fig. 1 and 7A). This is consistent with previous reports that elevated production of vif mRNA promotes exon 2 inclusion into the other viral mRNAs (13, 58).
FIG 7.
Effect of natural variations in SA1prox on viral gene expression. (A) Semiquantitative RT-PCR analysis of spliced mRNAs. Total RNA samples were prepared from HEK 293T cells transfected with the indicated proviral clones and subjected to semiquantitative RT-PCR analysis using the primer pairs for 2-kb mRNA species, vpu-env mRNAs, and vpr mRNAs (Fig. 1C). The PCR products of all viral mRNAs (exon 7 in Fig. 1) and GAPDH were analyzed in parallel for transfection and internal controls, respectively. Representative data from three independent experiments are shown. vpr and tat mRNA species are indicated (see also Fig. 1). DNA size markers are on the right. Control, pUC19. (B) Quantitation of semiquantitative RT-PCR products. Signal intensities of semiquantitative RT-PCR products were quantitated. The intensities of tat, 2-kb (tat, rev, and nef), vpu-env, and vpr mRNAs in each sample were normalized to those of all viral mRNAs (exon 7) and GAPDH. The normalized mRNA intensities in each sample relative to those of NL4-3 are presented. (C) Expression levels of viral mRNAs. Samples used for semiquantitative RT-PCR analysis were subjected to qRT-PCR analysis using primer pairs specific for gag/pol (NL-9kb in Fig. 1), vif (NL-vif in Fig. 1), and 2-kb (NL-2kb in Fig. 1) mRNAs. The expression levels of HIV-1 mRNAs were calculated and are presented as described in the legend of Fig. 4C. (D) Tat trans-activation assay. The indicated proviral clones and 5RLTR-Luc (6) were cotransfected into HEK 293T cells, and on day 1 posttransfection, cells were lysed for luciferase assays. Luciferase activity relative to that exhibited by NL4-3 is presented. (E) Expression levels of viral proteins. The proviral clones indicated were transfected into HEK 293T cells, and cell lysates were prepared on day 1 posttransfection for Western blot analysis using anti-Rev, anti-Nef, anti-Env, anti-Vpr, and anti-β-actin antibodies. The migration positions of mass standards are on the left. Representative data from three independent experiments are shown. Control, pUC19. For experiments in panels B to D, mean values ± standard deviations from three independent experiments are shown. Significance relative to NL4-3 was determined by the Student t test (***, P < 0.01; **, P < 0.05; *, P < 0.1). Control, pUC19; tac, Y226tac; gtg, V234gtg; cgc, R224cgc; ccg, P238ccg.
Quantification of signal intensities of semiquantitative RT-PCR products revealed that the total expression levels of tat mRNAs (tat 1 to tat 4) and 2-kb mRNAs (tat, rev, and nef) were not significantly different among NL4-3 and its variants (Fig. 7B). In contrast, vpu-env and vpr mRNA levels varied significantly among various vif type variants (Fig. 7B). V234gtg and R224cgc showed a decreased tendency to express the vpu-env mRNA. Notably, the expression level of vpr mRNAs was higher for Y226tac but lower for V234gtg, R224cgc, and P238ccg than for NL4-3. In agreement with data from a previous report (63), this result suggests that vif and vpr mRNAs appear to be produced in a mutually exclusive manner. In order to ascertain the expression levels of 9-kb, vif, and 2-kb mRNAs in samples used for semiquantitative RT-PCR analysis, we performed qRT-PCR analysis using the primer pairs specific for these mRNAs (Fig. 1C). Consistent with the results described so far (Fig. 4C and 7B), expression levels of 2-kb and 9-kb mRNAs of various vif type variants (Y226tac, V234gtg, R224cgc, and P238ccg) were similar to those of NL4-3, whereas these variants expressed distinct levels of vif mRNAs (Fig. 7C).
Since HIV-1 mRNA expression was changed significantly by the variations tested here, we checked their effects on viral protein expression (Tat/Rev/Nef from 2-kb mRNAs and Env/Vpr from 4-kb mRNAs). Because an appropriate anti-Tat antibody is not available to us, trans-activation assays were carried out instead. Proviral clones and an LTR-driven luciferase reporter clone were cotransfected into HEK 293T cells, and cell lysates were subjected to luciferase assays. As shown in Fig. 7D, Tat activity of the variant clones was not lower than that of NL4-3. Western blot analysis of cell lysates from transfected HEK 293T cells showed that various vif type variants expressed Rev and Nef proteins at similar levels (Fig. 7E). On the other hand, the expression levels of Env and Vpr proteins were slightly different among the variants. R224cgc expressed Env and Vpr at a modestly low level (Fig. 7E). This feature of R224cgc may explain the decrease in its replication ability (Fig. 6). These results suggest that while the expression patterns and levels of viral mRNAs were distinctly altered by the variations, there were only small differences in the total expression levels of 2-kb/9-kb mRNAs and the encoded proteins among NL4-3 and the variants. Taken together, we concluded that the decline in viral replication potential in excessive-vif-type variants is not primarily due to inadequate expression of the regulatory protein Tat or Rev.
DISCUSSION
In the present study, we defined the extended SA1prox (Fig. 4 and Table 2) and clarified the biological significance of natural nucleotide variations there (NL4-3 nucleotides 4899 to 4943) for viral replication (Table 3). Most natural variations present within SA1prox of HIV-1 and HIV-2 were found to alter vif transcript expression levels (Fig. 3 and 4), suggesting the biological importance of SA1prox. Natural SA1prox variants of HIV-1 displayed different expression levels of Vif (low-, high-, and excessive-vif types) (Fig. 2 and 4). Viral replication potentials of the SA1prox variants were found to be determined by the expression levels of both Vif and A3G (Fig. 2 and 6). In cells with a low A3G expression level, Y226tac (low-vif type) replicated better than NL4-3, whereas V234gtg (high-vif type) exhibited a slightly slower growth phenotype than did NL4-3. Importantly, under various A3G-restrictive conditions, an adequate expression level of Vif was required to maintain a certain extent of viral replication (Fig. 2D and 6D). This implies that the outcome of antagonism between Vif and A3G has an essential effect on viral replication. We conclude that SA1prox is a key region that determines Vif expression levels through its nucleotide variations, thereby modulating the viral antagonistic ability against A3G and viral replication potential.
The excessive expression of Vif negatively acted on single-cycle infectivity and/or virion production for R224cgc and P238ccg, and thus, regardless of the A3G level, these variants were impeded in their replication ability (Fig. 4 and 6). This replication-restrictive phenotype was not associated with a lower expression level of Tat or Rev (Fig. 7). Regarding the effect of excessive vif expression on viral replication ability, there are currently four possible explanations. First, the impairment of the early replication phase for R224cgc may be caused by the high expression level of Vif, a detrimental effect of Vif, as previously reported (57). Second, the decrease in the virion production level (∼10-fold reduction) has been shown to result from the elevated expression of vif mRNA (∼20-fold increase) and concomitantly the reduced accumulation of gag/pol mRNA (13, 58). This scenario can explain the decrease in the replication ability of excessive-vif types in late and early phases; i.e., a lower level of production of gag/pol mRNA results in lower Gag/Gag-Pol expression levels and possibly lower levels of viral genome RNA packaging. Although we did not observe such a remarkable alteration in gag/pol transcript levels for our natural variants (Fig. 4 and 7), this may have been due to the smaller effects of our natural variations on vif expression levels than those of previously reported artificial mutations (13, 58). On one hand, we used different cell types for qRT-PCR and virion production analyses (HEK 293T and H9 cells, respectively). It is possible that the expression patterns of viral mRNAs may differ in both cell types. Third, the modest decrease in both env and vpr mRNA/protein levels in the excessive-vif-type variant could have a negative effect on viral replication (Fig. 7). Fourth, it can be speculated that an alteration(s) in overall viral mRNA expression of excessive-vif-type variants may perturb the balanced viral mRNA expression, e.g., the ratio of 4-kb mRNAs to 9-kb mRNAs, and consequently impair viral replication. Further studies are necessary to elucidate the relationship between the change in HIV-1 mRNA expression by SA1prox variations and the alteration in replication potential.
Several splicing regulatory elements associated with vif mRNA splicing are located around SA1 and the downstream region of SD2. HIV-1 vif mRNA production is complexly regulated by exonic splicing enhancer (ESE)-Vif, ESE-M1 and -M2, the GGGG silencer, and the GI2-1 element (13–15, 55). Here we demonstrated that natural variations were also present within these elements and that these variations altered Vif expression levels and viral replication ability (Fig. 4 and 6). Moreover, natural variations within HIV-2 SA1prox also exhibited similar effects (Table 1 and Fig. 3). These results emphasized the importance of nucleotide variations in SA1prox for the determination of Vif expression levels and viral replication ability. Table 3 summarizes the effects of natural nucleotide variations on vif expression levels and viral replication ability. Fluctuations in viral replication ability were clearly observed for natural variants of markedly low (Y226tac and D229gat)-, high (P233cct, V234gtg, and R269Kaag)-, and excessive (R224cgc and P238ccg)-vif types. However, the other low-vif types (R228aga, R231Kaaa, D232gac, and K236aag [vif levels relative to NL4-3 are 0.16 to 0.47]) (Table 3) exhibited mild phenotypic changes in growth properties in permissive and nonpermissive cells (Table 3). Furthermore, high- and excessive-vif types did not replicate better than NL4-3 in cells expressing a high level of A3G (Table 3). A certain range of vif expression levels may be required for efficient viral replication, being associated with A3G antagonism and the optimal ratio of vif and gag/pol mRNAs.
As discussed above, splicing to produce various HIV-1 mRNA species is tightly controlled, and splicing regulation is conducted by both nucleotide sequences and RNA structures (13–15, 54, 55, 58, 64–66). Since most meaningful natural variations identified in the present study were clustered within the SLSA1 structure (Fig. 4A), variations may affect splicing at the sequence and/or structure level. Thus, we examined whether stabilities of the SLSA1 structure are correlated with the vif mRNA levels (Table 4). The stem-loop RNA structures of SA1prox variants were predicted in silico, and their structural stabilities were evaluated based on their energies (ΔG). The results showed that the ΔG values of predicted structures were not always correlated with vif mRNA levels among NL4-3 and SA1prox variants, suggesting that stabilities of secondary RNA structure at the SLSA1 region may not be a major factor in the regulation of the levels of vif mRNA. Nevertheless, accumulating evidence suggested a possible role(s) of local RNA structure and long-range RNA structural interactions in splicing regulation (54, 64–66). Further studies are needed in order to elucidate the molecular basis for alterations in vif mRNA expression by variations in SA1prox.
TABLE 4.
Effect of SA1prox variations on stability of the SLSA1 structure
| Amino acid | Codon (NL4-3) | Codon (SA1prox variant) | vif mRNA level | SLSA1 |
||
|---|---|---|---|---|---|---|
| ΔG | ΔΔG | Stability | ||||
| NL4-3 | −5.7 | |||||
| R224 | CGG | CGC | Up | −5.2 | 0.5 | Down |
| Y226 | TAT | TAC | Down | −6.1 | −0.4 | Up |
| R228 | AGG | AGA | Down | −5.3 | 0.4 | Down |
| D229 | GAC | GAT | Down | −7.8 | −2.1 | Up |
| R231K | AGA | AAA | Down | −5.7 | 0 | |
| D232 | GAT | GAC | Down | −7.9 | −2.2 | Up |
| P233 | CCA | CCT | Up | −10.5 | −4.8 | Up |
| V234 | GTT | GTG | Up | −8.5 | −2.8 | Up |
| K236 | AAA | AAG | Down | −6.6 | −0.9 | Up |
| P238 | CCA | CCG | Up | −3.0 | 2.7 | Down |
In HIV-1-infected individuals, A3G mRNA levels were previously reported to be inversely correlated with plasma viral load/disease progression (30–34). A very recent study showed that the anti-A3G activity of HIV-1 Vif derived from elite controllers was attenuated (67). However, the molecular mechanism to explain this decrease in Vif activity has not yet been elucidated. These findings suggest that the power balance between Vif and A3G may affect viral replication properties and disease statuses in infected individuals. Here, we demonstrate for the first time that naturally occurring nucleotide variations within SA1prox in the HIV-1 genome can alter Vif expression levels, thereby modulating viral replication potential. Since variations that regulate Vif expression levels naturally exist in the HIV-1 population, one can speculate that nucleotide variations in SA1prox may be involved in viral replication/evolution in vivo. Indeed, we have found some variations in HIV-1 genome sequences derived from HIV-1-infected individuals that alter Vif expression levels (our unpublished observations). Among natural variations, excessive-vif types (R224cgc and P238ccg) exhibited weaker growth potentials than that of the parental NL4-3 clone regardless of the extent of A3G restriction. This property of these variants may not be disadvantageous to viral replication in vivo; virus replication is not completely blocked, and viruses can persist and survive. In contrast, the impeded replication phenotype of these variants may be advantageous for viral evolution. The G-to-A hypermutation that is induced by a sublethal level of A3G has been reported to contribute to HIV-1 diversification, immune escape, and drug resistance; however, this issue remains controversial (68–74). Regardless, it is conceivable that alterations in Vif expression levels by synonymous single-nucleotide mutations in various sites within SA1prox represent a rational evolution pathway, because this regulation at the nucleotide sequence level facilitates the adaptation of HIV-1 to various environments without any effects on Pol-IN or Vif functions.
In conclusion, we identified SA1prox as a regulatory region in the HIV-1 genome that modulated Vif expression levels. A new element (GI3-2) associated with vif mRNA production within the vpr coding region was recently reported (63), and thus, further studies to precisely disclose the regulatory elements for vif mRNA expression are needed. Defining the factors that affect the power balance and interactions between Vif and A3G is important for a better understanding of viral replication ability/evolution in vivo and for the development of a new target for antiviral therapy.
ACKNOWLEDGMENTS
We thank Kazuko Yoshida for her editorial assistance. We also thank Kei Miyakawa and Akihide Ryo of Yokohama City University School of Medicine, Japan, for the CEM and CEM-SS cells. We are indebted to the NIH AIDS Research and Reference Reagent Program for the antibodies. TZM-bl cells were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, from John C. Kappes, Xiaoyun Wu, and Tranzyme Inc. We appreciate the Support Center for Advanced Medical Sciences, Institute of Biomedical Sciences, Tokushima University Graduate School, for experimental facilities and technical assistance.
We declare that we have no competing interests.
REFERENCES
- 1.Blanco-Melo D, Venkatesh S, Bieniasz PD. 2012. Intrinsic cellular defenses against human immunodeficiency viruses. Immunity 37:399–411. doi: 10.1016/j.immuni.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris RS, Hultquist JF, Evans DT. 2012. The restriction factors of human immunodeficiency virus. J Biol Chem 287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirchhoff F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55–67. doi: 10.1016/j.chom.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Malim MH, Bieniasz PD. 2012. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nomaguchi M, Doi N, Fujiwara S, Saito A, Akari H, Nakayama EE, Shioda T, Yokoyama M, Sato H, Adachi A. 2013. Systemic biological analysis of the mutations in two distinct HIV-1mt genomes occurred during replication in macaque cells. Microbes Infect 15:319–328. doi: 10.1016/j.micinf.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Nomaguchi M, Miyake A, Doi N, Fujiwara S, Miyazaki Y, Tsunetsugu-Yokota Y, Yokoyama M, Sato H, Masuda T, Adachi A. 2014. Natural single-nucleotide polymorphisms in the 3′ region of the HIV-1 pol gene modulate viral replication ability. J Virol 88:4145–4160. doi: 10.1128/JVI.01859-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amendt BA, Si ZH, Stoltzfus CM. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol 15:4606–4615. doi: 10.1128/MCB.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell DFJ, Martin MA. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol 67:6365–6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz S, Felber BK, Benko DM, Fenyö EM, Pavlakis GN. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol 64:2519–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz S, Felber BK, Fenyö EM, Pavlakis GN. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J Virol 64:5448–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi M. 2011. The regulation of HIV-1 mRNA biogenesis, p 79–100. In Grabowski P. (ed), RNA processing. InTech, Rijeka, Croatia: http://www.intechopen.com/books/rna-processing/the-regulation-of-hiv-1-mrna-biogenesis. [Google Scholar]
- 12.Karn J, Stoltzfus CM. 2012. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2:a006916. doi: 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Exline CM, Feng Z, Stoltzfus CM. 2008. Negative and positive mRNA splicing elements act competitively to regulate human immunodeficiency virus type 1 vif gene expression. J Virol 82:3921–3931. doi: 10.1128/JVI.01558-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H. 2006. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology 3:89. doi: 10.1186/1742-4690-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandal D, Exline CM, Feng Z, Stoltzfus CM. 2009. Regulation of vif mRNA splicing by human immunodeficiency virus type 1 requires 5′ splice site D2 and an exonic splicing enhancer to counteract cellular restriction factor APOBEC3G. J Virol 83:6067–6078. doi: 10.1128/JVI.02231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes RK, Malim MH, Bishop KN. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci 32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. 2014. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol 426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 19.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 20.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 21.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog 4:e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res 35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XY, Guo F, Zhang L, Kleiman L, Cen S. 2007. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J Biol Chem 282:32065–32074. doi: 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- 24.Mbisa JL, Barr R, Thomas JA, Vandegraaff N, Dorweiler IJ, Svarovskaia ES, Brown WL, Mansky LM, Gorelick RJ, Harris RS, Engelman A, Pathak VK. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J Virol 81:7099–7110. doi: 10.1128/JVI.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jäger S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. 2011. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- 27.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2011. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 30.Biasin M, Piacentini L, Lo Caputo S, Kanari Y, Magri G, Trabattoni D, Naddeo V, Lopalco L, Clivio A, Cesana E, Fasano F, Bergamaschi C, Mazzotta F, Miyazawa M, Clerici M. 2007. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J Infect Dis 195:960–964. doi: 10.1086/511988. [DOI] [PubMed] [Google Scholar]
- 31.Eyzaguirre LM, Charurat M, Redfield RR, Blattner WA, Carr JK, Sajadi MM. 2013. Elevated hypermutation levels in HIV-1 natural viral suppressors. Virology 443:306–312. doi: 10.1016/j.virol.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin X, Brooks A, Chen H, Bennett R, Reichman R, Smith H. 2005. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol 79:11513–11516. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kourteva Y, De Pasquale M, Allos T, McMunn C, D'Aquila RT. 2012. APOBEC3G expression and hypermutation are inversely associated with human immunodeficiency virus type 1 (HIV-1) burden in vivo. Virology 430:1–9. doi: 10.1016/j.virol.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vázquez-Pérez JA, Ormsby CE, Hernández-Juan R, Torres KJ, Reyes-Terán G. 2009. APOBEC3G mRNA expression in exposed seronegative and early stage HIV infected individuals decreases with removal of exposure and with disease progression. Retrovirology 6:23. doi: 10.1186/1742-4690-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amoêdo ND, Afonso AO, Cunha SM, Oliveira RH, Machado ES, Soares MA. 2011. Expression of APOBEC3G/3F and G-to-A hypermutation levels in HIV-1-infected children with different profiles of disease progression. PLoS One 6:e24118. doi: 10.1371/journal.pone.0024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho SJ, Drechsler H, Burke RC, Arens MQ, Powderly W, Davidson NO. 2006. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol 80:2069–2072. doi: 10.1128/JVI.80.4.2069-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. 2008. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol 82:3125–3130. doi: 10.1128/JVI.01533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mous K, Jennes W, Camara M, Seydi M, Daneau G, Mboup S, Kestens L, Van Ostade X. 2012. Expression analysis of LEDGF/p75, APOBEC3G, TRIM5alpha, and tetherin in a Senegalese cohort of HIV-1-exposed seronegative individuals. PLoS One 7:e33934. doi: 10.1371/journal.pone.0033934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.An P, Bleiber G, Duggal P, Nelson G, May M, Mangeat B, Alobwede I, Trono D, Vlahov D, Donfield S, Goedert JJ, Phair J, Buchbinder S, O'Brien SJ, Telenti A, Winkler CA. 2004. APOBEC3G genetic variants and their influence on the progression to AIDS. J Virol 78:11070–11076. doi: 10.1128/JVI.78.20.11070-11076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng J, Ao Z, Matthews C, Wang X, Ramdahin S, Chen X, Li J, Chen L, He J, Ball B, Fowke K, Plummer F, Embree J, Yao X. 2013. A naturally occurring Vif mutant (I107T) attenuates anti-APOBEC3G activity and HIV-1 replication. J Mol Biol 425:2840–2852. doi: 10.1016/j.jmb.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog 1:e6. doi: 10.1371/journal.ppat.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh KK, Wang Y, Gray KP, Farhad M, Brummel S, Fenton T, Trout R, Spector SA. 2013. Genetic variants in the host restriction factor APOBEC3G are associated with HIV-1-related disease progression and central nervous system impairment in children. J Acquir Immune Defic Syndr 62:197–203. doi: 10.1097/QAI.0b013e31827ab612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawamura M, Sakai H, Adachi A. 1994. Human immunodeficiency virus Vpx is required for the early phase of replication in peripheral blood mononuclear cells. Microbiol Immunol 38:871–878. doi: 10.1111/j.1348-0421.1994.tb02140.x. [DOI] [PubMed] [Google Scholar]
- 45.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lebkowski JS, Clancy S, Calos MP. 1985. Simian virus 40 replication in adenovirus-transformed human cells antagonizes gene expression. Nature 317:169–171. doi: 10.1038/317169a0. [DOI] [PubMed] [Google Scholar]
- 48.Kamada K, Igarashi T, Martin MA, Khamsri B, Hatcho K, Yamashita T, Fujita M, Uchiyama T, Adachi A. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc Natl Acad Sci U S A 103:16959–16964. doi: 10.1073/pnas.0608289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nomaguchi M, Yokoyama M, Kono K, Nakayama EE, Shioda T, Doi N, Fujiwara S, Saito A, Akari H, Miyakawa K, Ryo A, Ode H, Iwatani Y, Miura T, Igarashi T, Sato H, Adachi A. 2013. Generation of rhesus macaque-tropic HIV-1 clones that are resistant to major anti-HIV-1 restriction factors. J Virol 87:11447–11461. doi: 10.1128/JVI.01549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol 62:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jablonski JA, Caputi M. 2009. Role of cellular RNA processing factors in human immunodeficiency virus type 1 mRNA metabolism, replication, and infectivity. J Virol 83:981–992. doi: 10.1128/JVI.01801-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piroozmand A, Yamamoto Y, Khamsri B, Fujita M, Uchiyama T, Adachi A. 2007. Generation and characterization of APOBEC3G-positive 293T cells for HIV-1 Vif study. J Med Invest 54:154–158. doi: 10.2152/jmi.54.154. [DOI] [PubMed] [Google Scholar]
- 54.Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, Weeks KM, Swanstrom R. 2013. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and nonconserved structural motifs. PLoS Pathog 9:e1003294. doi: 10.1371/journal.ppat.1003294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Widera M, Erkelenz S, Hillebrand F, Krikoni A, Widera D, Kaisers W, Deenen R, Gombert M, Dellen R, Pfeiffer T, Kaltschmidt B, Münk C, Bosch V, Köhrer K, Schaal H. 2013. An intronic G run within HIV-1 intron 2 is critical for splicing regulation of vif mRNA. J Virol 87:2707–2720. doi: 10.1128/JVI.02755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwabu Y, Kinomoto M, Tatsumi M, Fujita H, Shimura M, Tanaka Y, Ishizaka Y, Nolan D, Mallal S, Sata T, Tokunaga K. 2010. Differential anti-APOBEC3G activity of HIV-1 Vif proteins derived from different subtypes. J Biol Chem 285:35350–35358. doi: 10.1074/jbc.M110.173286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akari H, Fujita M, Kao S, Khan MA, Shehu-Xhilaga M, Adachi A, Strebel K. 2004. High level expression of human immunodeficiency virus type-1 Vif inhibits viral infectivity by modulating proteolytic processing of the Gag precursor at the p2/nucleocapsid processing site. J Biol Chem 279:12355–12362. doi: 10.1074/jbc.M312426200. [DOI] [PubMed] [Google Scholar]
- 58.Madsen JM, Stoltzfus CM. 2006. A suboptimal 5′ splice site downstream of HIV-1 splice site A1 is required for unspliced viral mRNA accumulation and efficient virus replication. Retrovirology 3:10. doi: 10.1186/1742-4690-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deymier MJ, Ende Z, Fenton-May AE, Dilernia DA, Kilembe W, Allen SA, Borrow P, Hunter E. 2015. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-α resistance. PLoS Pathog 11:e1005154. doi: 10.1371/journal.ppat.1005154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JH, Song H, Austin JL, Cheng W. 2013. Optimized infectivity of the cell-free single-cycle human immunodeficiency viruses type 1 (HIV-1) and its restriction by host cells. PLoS One 8:e67170. doi: 10.1371/journal.pone.0067170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Erkelenz S, Poschmann G, Theiss S, Stefanski A, Hillebrand F, Otte M, Stühler K, Schaal H. 2013. Tra2-mediated recognition of HIV-1 5′ splice site D3 as a key factor in the processing of vpr mRNA. J Virol 87:2721–2734. doi: 10.1128/JVI.02756-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Widera M, Hillebrand F, Erkelenz S, Vasudevan AA, Münk C, Schaal H. 2014. A functional conserved intronic G run in HIV-1 intron 3 is critical to counteract APOBEC3G-mediated host restriction. Retrovirology 11:72. doi: 10.1186/s12977-014-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbink TE, Berkhout B. 2008. RNA structure modulates splicing efficiency at the human immunodeficiency virus type 1 major splice donor. J Virol 82:3090–3098. doi: 10.1128/JVI.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jablonski JA, Buratti E, Stuani C, Caputi M. 2008. The secondary structure of the human immunodeficiency virus type 1 transcript modulates viral splicing and infectivity. J Virol 82:8038–8050. doi: 10.1128/JVI.00721-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacquenet S, Ropers D, Bilodeau PS, Damier L, Mougin A, Stoltzfus CM, Branlant C. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res 29:464–478. doi: 10.1093/nar/29.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kikuchi T, Iwabu Y, Tada T, Kawana-Tachikawa A, Koga M, Hosoya N, Nomura S, Brumme ZL, Jessen H, Pereyra F, Trocha A, Walker BD, Iwamoto A, Tokunaga K, Miura T. 2015. Anti-APOBEC3G activity of HIV-1 Vif protein is attenuated in elite controllers. J Virol 89:4992–5001. doi: 10.1128/JVI.03464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Armitage AE, Deforche K, Chang CH, Wee E, Kramer B, Welch JJ, Gerstoft J, Fugger L, McMichael A, Rambaut A, Iversen AK. 2012. APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete “all or nothing” phenomenon. PLoS Genet 8:e1002550. doi: 10.1371/journal.pgen.1002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ebrahimi D, Anwar F, Davenport MP. 2011. APOBEC3 has not left an evolutionary footprint on the HIV-1 genome. J Virol 85:9139–9146. doi: 10.1128/JVI.00658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jern P, Russell RA, Pathak VK, Coffin JM. 2009. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog 5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim EY, Lorenzo-Redondo R, Little SJ, Chung YS, Phalora PK, Maljkovic Berry I, Archer J, Penugonda S, Fischer W, Richman DD, Bhattacharya T, Malim MH, Wolinsky SM. 2014. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog 10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mulder LC, Harari A, Simon V. 2008. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A 105:5501–5506. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sadler HA, Stenglein MD, Harris RS, Mansky LM. 2010. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J Virol 84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, Gaschen B, Daniels M, Ferrari G, Haynes BF, McMichael A, Shaw GM, Hahn BH, Korber B, Seoighe C. 2009. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog 5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]