ABSTRACT
Herpes simplex virus 1 (HSV-1) establishes lifelong infection in the neurons of trigeminal ganglia (TG), cycling between productive infection and latency. Neuronal antiviral responses are driven by type I interferon (IFN) and are crucial to controlling HSV-1 virulence. Autophagy also plays a role in this neuronal antiviral response, but the mechanism remains obscure. In this study, HSV-1 infection of murine TG neurons triggered unusual clusters of autophagosomes, predominantly in neurons lacking detectable HSV-1 antigen. Treatment of neurons with IFN-β induced a similar response, and cluster formation by infection or IFN treatment was dependent upon an intact IFN-signaling pathway. The autophagic clusters were decorated with both ISG15, an essential effecter of the antiviral response, and p62, a selective autophagy receptor. The autophagic clusters were not induced by rapamycin or starvation, consistent with a process of selective autophagy. While clusters were triggered by other neurotropic herpesviruses, infection with unrelated viruses failed to induce this response. Following ocular infection in vivo, clusters formed exclusively in the infected ophthalmic branch of the TG. Taken together, our results show that infection with HSV and antiviral signaling in TG neurons produce an unorthodox autophagic response. This autophagic clustering is associated with antiviral signaling, the presence of viral genome, and the absence of HSV protein expression and may therefore represent an important neuronal response to HSV infection and the establishment of latency.
IMPORTANCE Herpes simplex virus type 1 (HSV-1) is a ubiquitous virus and a significant cause of morbidity and some mortality. It is the causative agent of benign cold sores, but it can also cause blindness and life-threatening encephalitis. The success of HSV-1 is largely due to its ability to establish lifelong latent infections in neurons and to occasionally reactivate. The exact mechanisms by which neurons defend against virus infection is poorly understood, but such defense is at least partially mediated by autophagy, an intracellular pathway by which pathogens and other unwanted cargoes are degraded. The study demonstrates and investigates a new autophagic structure that appears to be specific to the interaction between neurotropic herpesviruses and murine primary sensory neurons. This work may therefore have important implications for our understanding of latency and reactivation.
INTRODUCTION
Herpes simplex virus 1 (HSV-1) is a highly prevalent human pathogen that establishes lifelong infection in the neurons of sensory and autonomic ganglia (1, 2). Initial infection and lytic replication at mucosal sites are followed by infection of innervating axonal termini of sensory neurons. Virions then travel in a retrograde direction to the soma, where they may replicate or immediately establish latency, depending partly on the infected neuronal subtype (3). Virions resulting from periodic reactivation events travel in an anterograde direction along the axon, allowing reinfection of the oral epithelium, thereby facilitating viral shedding and host-to-host transmission (2).
Innate immune responses are critical in controlling virulence of HSV-1 and many other viruses through a variety of antiviral pathways (4–12), and viruses have coevolved to counter these host responses (13–20). Cells detect the presence of incoming virus through pattern recognition receptors (21, 22) that, in turn, lead to the activation of pivotal transcription factors such as IRF (5, 23) and NF-κB (24). These factors subsequently induce the production of type I interferon (IFN), which drives IFN-stimulated gene (ISG) expression (25) through a JAK-STAT-dependent pathway (26, 27). This further stimulates production of type I IFN and ISGs to establish an antiviral state that consists of transcriptional and translational arrest, cytokine production, and apoptosis (28, 29).
Defects in innate immunity are often associated with increased susceptibility to HSV infection in both mice and humans, with frequent neurological sequelae (8, 12, 30–32). That said, the IFN-driven responses of neurons themselves are muted and atypical (12, 33). Nondestructive clearance is especially critical for postmitotic adult neurons, a population of irreplaceable cells that is essential for both the success and survival of the host. Indeed, there is mounting evidence that neurons depend on autophagy rather than inflammatory or cell-destructive responses for the control of intracellular pathogens (34–37). Autophagy is a highly conserved response to starvation, during which a portion of the cytosol is engulfed in a double-walled membrane vesicle termed an autophagosome. The autophagosome then fuses with the lysosome, and its contents are degraded by lysosomal enzymes (38). Autophagosomes can also selectively target/engulf ubiquitinated cargo (selective autophagy [39]) such as aggresomes (aggrephagy [40, 41]), mitochondria (mitophagy [42–44]), or pathogens (xenophagy [45–47]).
Autophagy has several key functions in neurons. Constitutive autophagy is necessary for neural pathway formation during development, neuronal differentiation, and homeostasis (48), and it is essential for facilitating the clearance of misfolded proteins. Indeed, loss of neuronal autophagy leads to neuronal death and development of neurodegenerative disease (49, 50). There are also multiple lines of evidence for the involvement of neuronal autophagy in control of HSV infection. First, HSV-1 was observed within autophagosomes in sympathetic neurons with evidence of xenophagic degradation (34). Second, at least 2 HSV-1 genes (γ34.5 and US11) antagonize autophagy (34, 51), and viruses lacking γ34.5 accumulate within maturing xenophagosomes (34). Third, viruses lacking either the entire γ34.5 gene (Δ34.5) or the domain that binds the essential autophagy protein beclin 1 (Δbbd) exhibit significant neuroattenuation in vivo (13, 35, 52). Finally, Δbbd virus replicates poorly in neurons from the dorsal root ganglia (DRG), and this replication defect is restored in autophagy-deficient neurons (37). While these data support the idea that autophagy is a critical component of the neuronal antiviral response, several aspects of this process remain obscure. For example, it is not known how autophagy is induced in HSV-infected neurons and whether neuronal xenophagic clearance of the virus actually mediates the antiviral effect. Furthermore, the precise impact of neuronal autophagy on balancing the lytic-latent viral status in neurons remains unknown.
To elucidate these aspects of autophagy and neuronal defense, we cultured neurons from the trigeminal ganglia (TG) of adult mice (32), allowing us to examine neuron-intrinsic antiviral responses in mature neurons. We based our in vivo studies on the ocular infection model (53), to examine neuronal autophagy in a physiologically relevant setting. As expected, in both in vitro neuronal cultures and intact TG in vivo, many neurons contained conventional (0.5- to 1.5-μm) autophagosomes. Unexpectedly, however, a small subset of neurons presented with an atypical structure consisting of a >4-μm cluster of several autophagosomes. Cluster frequency and abundance were increased by infection with HSV-1, and clusters predominated in neurons that did not express detectable viral antigen. Accumulation of autophagic clusters was also induced by IFN-β and was dependent upon an intact IFN signaling pathway. Moreover, clusters colocalized with ISG15, an essential mediator of the antiviral response (28). Clusters strongly colocalized with the selective autophagy receptor p62 and were not induced by starvation or rapamycin treatment, suggesting a process of selective autophagy. In vivo, clusters were confined to infected areas of the TG. The mutual exclusivity of autophagosomal clustering and HSV protein expression suggests that these clusters are novel markers of the neuronal antiviral response or may accumulate in neurons that are nonpermissive for lytic infection. These studies are consistent with the hypothesis that clusters are either markers or mediators of neuronal responses to HSV that may tilt the lytic-latent balance.
MATERIALS AND METHODS
Neuron isolation and culture.
Neurons were isolated and cultured as previously reported (3, 33). Coverslips (12 mm) were coated with poly-d-lysine (BD Biosciences) at 20 μg/ml in Hanks balanced salt solution lacking calcium and magnesium (HBSS; Cellgro) for a minimum of 3 h. Coverslips were then washed three times with sterile distilled water and coated with natural mouse laminin (Invitrogen) at a concentration of 18 μg/ml in HBSS overnight. TG neurons were isolated as described previously, with a few modifications (3). Mice 6 to 10 weeks old were euthanized by CO2 and transcardially perfused with phosphate-buffered saline (PBS; HyClone). TG were harvested and enzymatically digested in a solution consisting of 40 U/ml of papain (Worthington) in HBSS with 2.75 mM l-cysteine (Sigma) and saturated NaHCO3 diluted 1:1,000 (Sigma) for 20 min at 37°C on a rotator. This was followed by a similar incubation in a solution of 5 mg/ml of collagenase type II (Invitrogen) and 5.5 mg/ml of neutral protease (Worthington) dissolved in HBSS. TG were then triturated in Neurobasal-A (NB-A) working medium (Neurobasal-A [Invitrogen], 2% B27 [StemCell], and 1% penicillin-streptomycin [pen-strep; HyClone]). The resulting homogenate was spun over a four-layer density gradient made with Optiprep and NB-A working medium. Optiprep (Sigma) was first diluted to 50.5% with 0.8% sodium chloride and then combined with NB-A working medium to obtain the following 1-ml-volume gradient layers (Optiprep/NB-A working medium ratios): 0.45:0.55, 0.35:0.65, 0.25:0.75, and 0.15:0.85. After a 20-min centrifugation at 800 × g, two bands of lower density were collected and washed three times. Neurons were counted and seeded at a density of 3,600 in a volume of 60 μl onto 12-mm coated coverslips. After 1 to 2 h, coverslips were transferred to 24-well plates and neurons were cultured in NB-A complete medium with the antimitotic 5′-fluoro-2′-deoxyuridine (FUDR; Sigma) for a minimum of 3 days prior to use. NB-A complete medium consisted of Neurobasal-A, 2% SM1, 1% GlutaMAX (Invitrogen), 1% pen-strep, 50 ng/ml of Neurturin (R&D Systems), 50 ng/ml of neuronal growth factor (NGF; Invitrogen), and 50 ng/ml of glial cell-derived neurotrophic factor (GDNF; R&D Systems). Neurons were initially identified in culture by β-III-tubulin or NeuN staining and later by morphology.
Viruses and mice.
HSV-1 strain 17 syn+ (54) (GenBank accession no. NC_001806), the Δbbd strain (35), the Δ34.5 strain (55), the Δ34.5 ΔUS11 strain (56), dlLAT1.8 (57), pseudorabies virus strain Becker (PrV) (58), and vesicular stomatitis virus (VSV) (33) were grown as previously described. Several investigators provided viruses as follows: Neal DeLuca, HSV-1 d92 (59); Todd Margolis, HSV-1 KOS-58 and KOS-62 (3); Lynda Morrison, HSV-2 strain 333 (60); Edward Usherwood, murine gammaherpesvirus 68 (MHV-68) and vaccinia virus (VACV) (61); Andrew Pachner, Theiler's murine encephalomyocarditis virus (TMEV) (62); and Charles Rice, yellow fever virus strain 17D (YFV) (63). Noboru Mizushima provided green fluorescent protein (GFP)-LC3 mice (64, 65), and Joan Durbin provided STAT1−/− mice (26). IFNR-αβγ−/− (66) and IRF3−/− (67) mice were described previously. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (68). The protocol was approved by the Dartmouth IACUC (5 June 2012, permit number leib.da.1). No surgery was performed, and all efforts were made to minimize suffering.
In vitro infection.
Neurons cultured on coverslips were infected in NB-A working medium in a volume of 250 μl for 1 h. They were then washed in NB-A working medium and refed with NB-A complete medium. Neurons were infected with HSV-1 strain 17 or HSV-1 mutants at a multiplicity of infection (MOI) of 25, 5, 1, or 0.2, as indicated below. For infection with other virus types, the MOI was set to result in ∼30% antigen-positive neurons, similar to that observed with HSV-1. The MOIs used were as follows: HSV-2, 5; PrV, 2; MHV-68, 2; VACV, 4; TMEV, 2; YFV, 10; and VSV, 2. For viral growth assays, culture supernatant was sampled at 24 or 48 h postinfection (hpi), frozen, and thawed, and then titers were determined via plaque assay on Vero cells as described previously (53).
Drug treatments.
When noted, cells were treated for 18 h with the following drugs: IFN-α (PBL Assay Science) at 100 U/ml, IFN-β (PBL Interferon Source) at 12.5 U/ml, IFN-γ (Miltenyi Biotec) at 100 ng/ml, IFN-λ (Peprotech) at 100 ng/ml, and rapamycin (Calbiochem) at 0.5 μM. Alternatively, cells were treated by nutrient deprivation comprising NB-A only. For Tat-beclin 1 peptide treatments, cells were treated for 3 h at 10 μM, washed with NB-A working medium, and then infected or incubated in NB-A complete medium. Bafilomycin 1 (Sigma) was used at 0.5 μM for 1 to 4 h. Nocodazole (Sigma) was used at 20 μg/ml for 1 to 4 h.
Immunocytochemistry.
Cells were fixed in 4% paraformaldehyde and then permeabilized by a 5-min incubation with 0.1% Triton X-100 (Sigma) in PBS containing 0.2% fish skin gelatin (FSG; Sigma). Alternatively, for LC3 immunostaining, after fixation cells were permeabilized with −20°C MeOH for 10 min and then blocked in PBS containing 0.2% FSG for 30 min at room temperature. Primary and secondary antibody incubations were carried out in 0.1% FSG for 1 h and 30 min, respectively, at room temperature. Primary antibodies used were rabbit anti-β-III-tubulin (Abcam; ab18207), mouse anti-NeuN (Millipore; MAB377), rabbit anti-HSV-1 (Dako; B0114), rabbit anti-MHV-68 (kindly provided by Edward Usherwood), mouse anti-vaccinia virus (Thermo Scientific; MA1-7484), mouse anti-TMEV (kindly provided by Andrew Pachner), mouse anti-YFV (Santa Cruz; 3576), mouse anti-VSV-G (Santa Cruz; P5D4), rabbit anti-β-galactosidase (anti-β-Gal; Rockland; 100-4136), mouse anti-ICP0 (Virusys; H1A027), mouse anti-VP16 (Santa Cruz; 7545), mouse anti-ICP5 (Abcam; ab6508), rabbit anti-LC3 (MBL [PD014] and Cell Signaling [12741]), rabbit anti-p62 (Novus; NBP1-48320), rat anti-LAMP1 (Santa Cruz; 1D4B), rabbit anti-PABP (Abcam; ab21060), goat anti-TIA (Santa Cruz; 1751), rabbit anti-HDAC6 (Assay Biotech; B0941), rabbit anti-γ-tubulin (Bioss; bs-1322R), and rabbit anti-ISG15 (Abgent; AP1150a). Secondary antibodies were goat or donkey anti-mouse, -rabbit, or -rat, conjugated to Alexa 405, 488, 555, or 647 (Invitrogen). Slides were mounted in medium containing 2% n-propyl-gallate and 75% glycerol. Images were acquired on an automated AxioVision microscope and analyzed using ZEN2012 or FIJI software. Virus staining was scored as positive or negative. LC3 patterns were scored as none, 0.5 to 1 μm, or >4 μm and later as cluster positive or cluster negative. Neurons were binned according to virus stain, and the number of cluster-positive neurons within each binning category was expressed as percentage of all neurons within that category. This was done for a minimum of 3 repeats on 200 to 1,500 cells per sample.
Live imaging.
Neurons were seeded on glass-bottomed dishes (MatTek Corporation), cultured in phenol red-free NB-A complete medium, and imaged using an Olympus IX83 TIRF-4 microscope equipped with a controlled environment stage. Images were acquired at 1- to 3-min intervals over the course of <2 h. Alternatively, images were acquired at 15-min intervals over the course of 5 h.
qPCR.
Neuron cultures were treated with 10 U/ml of IFN-β or infected with HSV-1 strain 17 at an MOI of 5. After 3 to 6 h (as indicated below), RNA was isolated by TRIzol extraction (Thermo Fisher) according to the manufacturer's instructions. RNA was treated with DNA-free kit (Ambion), and cDNA was synthesized using the SuperScriptIII reverse transcriptase kit (Invitrogen) with random primers (Promgea). For quantitative PCR (qPCR), SYBR Select master mix (Life Technologies) was used with primers for IFIT1 (forward [Fw], TGC TTT GCG AAG GCT CTG AAA GTG; reverse [Rv], TGG ATT TAA CCG GAC AGC CTT CCT), ISG15 (Fw, TGA GCA TCC TGG TGA GGA ACG AAA; Rv, AGC CAG AAC TGG TCT TCG TGA CTT), Atg5 (Fw, TTG CTT TTG CCA AGA GTC AGC; Rv, ATG CCA TTT CAG GGG TGT GC), p62 (Fw, GCC AGA GGA ACA GAT GGA GTC; Rv, AGC TTG GCC CTT CCG ATT C), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fw, CAT CTT CCA GGA GCG AGA TCC C; Rv, CAA ATG AGC CCC AGC CTT CTC C). IFIT1, ISG15, p62, and ATG5 values were calculated by the threshold cycle (2−ΔΔCT) method (69) and normalized to that for GAPDH, and values for infected or IFN-treated cells were normalized to that for mock-infected cells.
In vivo infections.
For corneal infection, 6- to 10-week-old male or female mice were anesthetized intraperitoneally with ketamine (87 mg/kg of body weight) and xylazine (13 mg/kg). Corneas were bilaterally scarified with a 25-gauge syringe needle, and virus was inoculated by adding 2 × 106 PFU in a 5-μl volume (54). Four days postinfection, mice were euthanized by CO2 and transcardially perfused first with PBS and then with 4% formaldehyde (Fisher Scientific). The TG were harvested, incubated in 30% sucrose overnight at 4°C, and embedded in Tissue-Tek OCT compound (Sakura). The tissue was then cut on the transverse plane into 5-μm sections, which were mounted directly onto charged glass slides (Thermo Scientific).
Immunohistochemistry.
Tissue sections were rehydrated, blocked, and permeabilized in PBS containing 5% normal goat serum (NGS; Vector Laboratories) and 0.1% Triton X-100 for 1 h at room temperature. All subsequent stains and washes were done in PBS containing 1% NGS. Primary antibodies (as described above) were incubated ON at 4°C. Secondary antibodies (as described above) were incubated for 1 h at room temperature. Samples were mounted in Vectashield (Hardset with 4′,6-diamidino-2-phenylindole [DAPI]; Vector Laboratories). Images were acquired and analyzed as described above. Each experiment was repeated in triplicate; one representative slice from each mouse was analyzed.
RESULTS
HSV-1 replication in TG neurons induces atypical autophagic patterns.
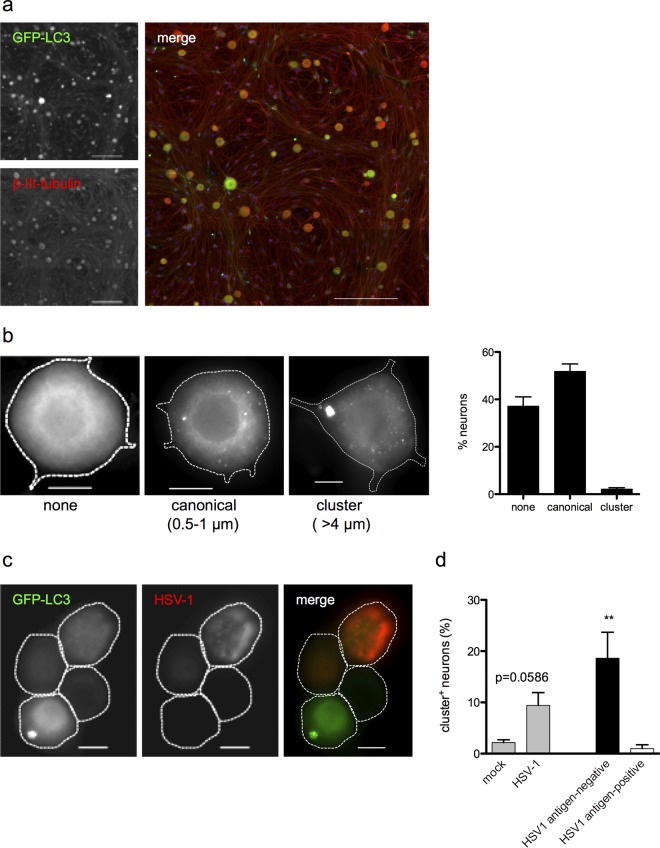
We initially examined the steady-state profile of autophagy in untreated cultured TG neurons derived from GFP-LC3 transgenic mice (64, 65). In these mice, soluble GFP-LC3 produces a nondescript cytoplasmic GFP haze (70), while autophagosome-bound GFP-LC3 manifests as distinct GFP puncta. These transgenic mice thus facilitate visualization of autophagic vesicles. TG neurons were cultured from adult GFP-LC3 mice and imaged after 4 days of growth in vitro (Fig. 1a). The majority of neurons (≥50%) had one or more GFP-LC3+ puncta (0.5 to 1 μm), while the remainder had none (Fig. 1b). These puncta partially colocalized with the lysosomal markers LAMP1 and LysoTracker, suggesting that they represent an ongoing autophagic process (71). A small number of neurons (<4%) contained a larger GFP-LC3+ structure of irregular shape, measuring >4 μm in area (Fig. 1b).
FIG 1.
TG neurons present atypical autophagic structures. (a) Overview image of TG neuron cultures isolated from GFP-LC3 mice and stained with β-III-tubulin (red). Bar = 200 μm. (b) Untreated TG neurons showing profiles of the three autophagic phenotypes observed (no discernible puncta, “none”; one or more puncta 0.5 to 1 μm in diameter, “canonical”; one or more GFP+ structures >4 μm in diameter, “cluster”). The graph shows the percentage of neurons presenting each phenotype, relative to all neurons counted (3 experiments; >200 neurons counted per experiment). Bar = 10 μm. (c) Representative image of TG neurons from GFP-LC3 mice infected with HSV-1 for 24 h. The left image shows GFP-LC3 (green), the middle image shows HSV-1 antigen (red), and the right image shows merged image. Bar = 10 μm. (d) Quantification of cultures described for panel c. The left bars show percentages of neurons with the cluster GFP-LC3 phenotype following mock or HSV-1 infection. Neurons in infected cultures were then categorized as either HSV antigen negative or HSV antigen positive, based on HSV staining. The right bars show percentages of neurons with the cluster GFP-LC3 phenotype in HSV antigen-negative or HSV antigen-positive categories. Four experiments were performed, with 200 to 800 neurons scored per experiment. Error bars represent standard errors of the means (SEM). Significance was evaluated by one-way analysis of variance (ANOVA) with Bonferroni posttests. **, P < 0.01.
To determine whether neuronal autophagy was changed following infection with HSV, we infected GFP-LC3 TG neurons with HSV-1 at an MOI of 25 for 24 h. We observed an increase in the percentage of neurons containing the larger (>4 μm) GFP-LC3+ structures (Fig. 1c and d). We hypothesized that HSV antigens were disrupting normal autophagy in these neurons. To test this, we performed immunofluorescence testing for the presence of HSV antigen (Fig. 1c). Unexpectedly, neurons containing the large LC3-positive structures did not express detectable viral antigen (P < 0.01) (Fig. 1d).
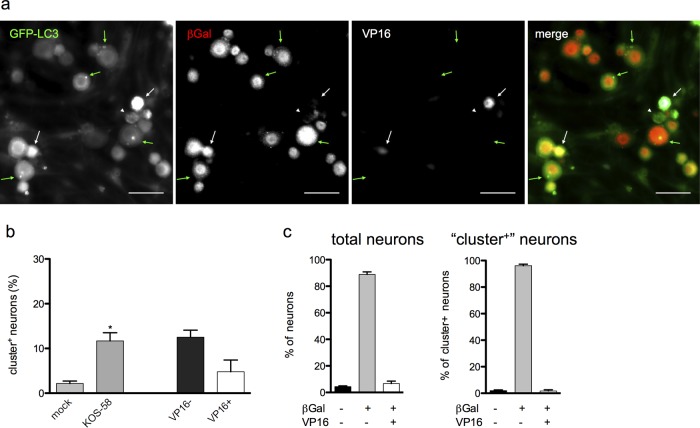
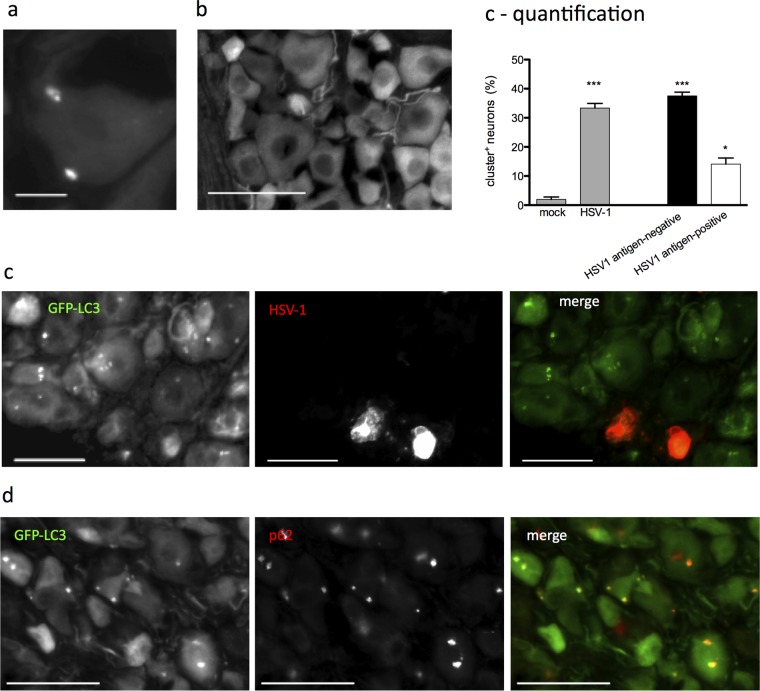
Previous characterization of this infection model shows that while >90% of neurons are infected, only ∼10% express detectable HSV antigens (3). It was therefore unclear whether the HSV antigen-negative neurons were HSV naive or harboring the HSV genome. To test this, we repeated the experiment using HSV-1 KOS-58 (Fig. 2a), which expresses β-galactosidase (β-Gal) specifically in neurons under the control of the neurofilament promoter in both lytically and latently infected cells (3). Infection with KOS-58 induced the formation of clusters in ∼11% of neurons, and clusters were found predominantly in HSV antigen-negative cells (Fig. 2a and b), thus recapitulating the effects of strain 17 infection. As previously reported (3), >90% of neurons in KOS-58-infected cultures express β-Gal (Fig. 2a and c). Moreover, most of the cluster-positive neurons were also β-Gal+ (Fig. 2a and c), indicating that these cells harbor HSV genome yet do not express detectable HSV antigen. Taken together, these findings demonstrate that TG neurons have atypical autophagic responses to HSV-1 infection and that large LC3-positive structures form predominantly in HSV genome-positive neurons that do not express HSV protein.
FIG 2.
Large GFP-LC3+ structures (clusters) form in neurons that harbor the HSV-1 genome. (a) GFP-LC3 neurons infected with KOS-58 and stained for β-Gal (red) and VP16 (white). Green arrows indicate neurons with clusters, β-Gal expression, and no VP16. White arrows indicate neurons with VP16 expression and no clusters. Bar = 50 μm. (b) Quantification of cultures described for panel a and analyzed as for Fig. 1d. (c) Quantification of β-Gal and VP16 prevalence among total neurons (left graph) or among cluster-positive neurons (right graph) in the cultures represented in panel a. A total of ≥3 experiments were performed, with >1,000 neurons scored per experiment. Error bars represent SEMs. Significance was evaluated by one-way analysis of variance (ANOVA) with Bonferroni posttests. *, P < 0.05.
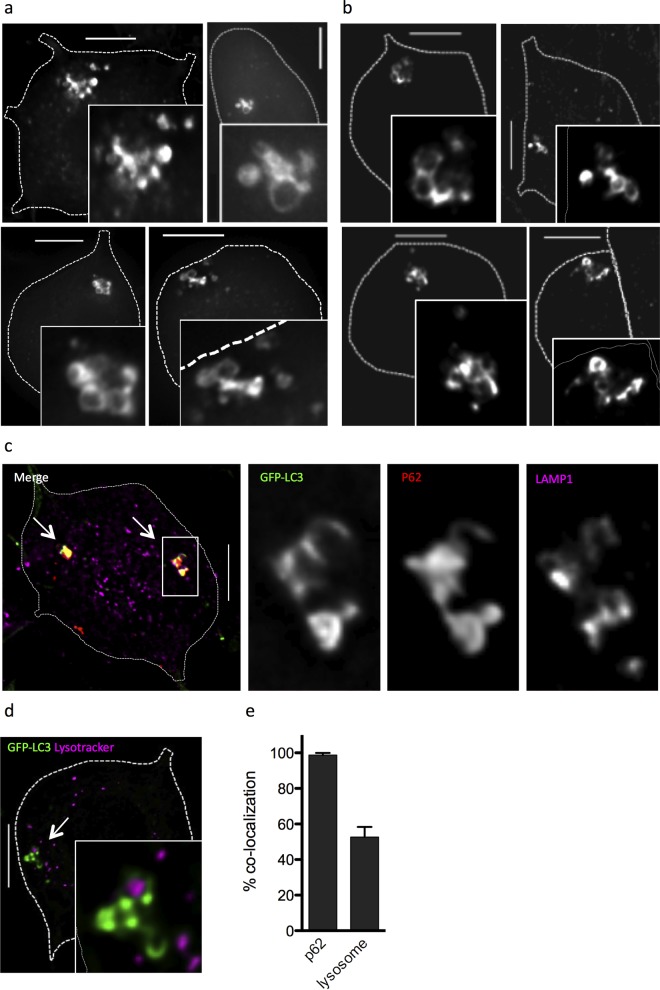
We next examined the GFP-LC3+ structures at higher resolution using deconvolution microscopy. We found that the large GFP-LC3+ structures consisted of clusters of smaller (0.5 to 1.5 μm) GFP-LC3+ puncta (Fig. 3a). Immunofluorescence staining for LC3 revealed comparable structures in TG neurons derived from C57BL/6J mice (Fig. 3b). This demonstrates that the LC3+ clusters were not artifacts arising from the GFP-LC3 fusion construct. To confirm that the GFP-LC3+ clusters represented an autophagic process, we stained the cultures with LysoTracker dye and performed immunocytochemistry for the lysosomal resident protein LAMP1 and the autophagosome cargo protein p62 (Fig. 3c). When colocalization was quantified, we found that >50% of the clusters colocalized with lysosomal compartments, and almost 100% colocalized with p62 (Fig. 3d), supporting the notion that these structures represent an autophagic process.
FIG 3.
Large GFP-LC3+ structures are clustered autophagosomes. (a) Deconvolution microscopy imaging of four representative neurons cultured from GFP-LC3 mice to show clustered autophagosomes, with enlarged insets to show detail. (b) Deconvolution microscopy imaging of four representative neurons cultured from WT C57BL/6J mice stained with an anti-LC3 antibody (Cell Signaling) to show clustered autophagosomes, with enlarged insets to show detail. (c) Merged fluorescence image of neuron from GFP-LC3 mice, with white arrows indicating clusters. Zoomed single-channel images of the boxed cluster show GFP-LC3 (green), p62 (red), and LAMP1 (purple). (d) Merged fluorescence image of neuron from GFP-LC3 mice stained with LysoTracker (purple), with a white arrow indicating a cluster. A zoomed image is shown as an inset. (e) Graph showing the percentage of GFP-LC3+ clusters that colocalized with p62 or lysosomal markers (LAMP1 or LysoTracker) expressed as a fraction of total clusters. The experiment was performed ≥3 times, and >200 clusters were scored per experiment. Error bars represent SEMs. Size bar = 10 μm.
Autophagosomal clustering is induced by IFN-β and requires antiviral signaling.
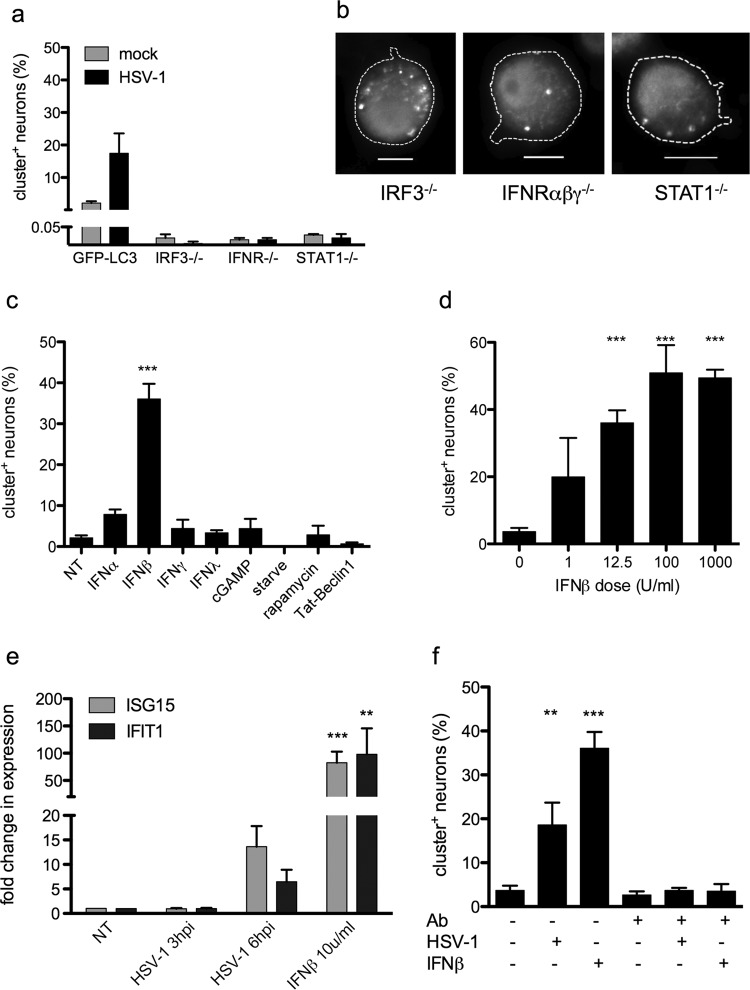
Based on our findings, we hypothesized that HSV-1 infection triggers antiviral signaling in TG neurons, which, in turn, induces autophagosomal clustering. Accordingly, TG neurons derived from mice deficient in IRF3, IFNR-αβγ, or STAT1 were unable to form the larger clusters in response to infection (Fig. 4a). In contrast, they did form typical 0.5- to 1.5-μm autophagosomes in response to starvation (Fig. 4b), ruling out a general deficit in their autophagic responses. Thus, antiviral signaling was necessary for cluster formation. Indeed, IFN-β efficiently induced cluster formation in the absence of viral infection (Fig. 4c). In contrast, IFN-α, IFN-γ, IFN-λ, or the STING agonist cGAMP did not trigger this response (Fig. 4c). This shows that IFN-β-mediated antiviral signaling is sufficient for cluster formation. The response to IFN-β was dose dependent, reaching significance at low dose (Fig. 4d), underscoring the sensitivity of neuronal autophagy to this signal. Importantly, canonical inducers of autophagy (starvation, rapamycin, and Tat-beclin peptide (70, 72) did not increase the prevalence of clusters (Fig. 4c).
FIG 4.
IFN-β induces autophagosomal clustering. (a) Neurons from IRF3−/−, IFNR-αβγ−/−, or STAT1−/− mice were infected with HSV-1 at an MOI of 25 or mock infected and then stained and analyzed as for Fig. 1d, presented relative to GFP-LC3. (b) Neurons from IRF3−/−, IFNR-αβγ−/−, or STAT1−/− mice were starved for 24 h, treated with bafilomycin for 2 h, and then stained for LC3. Bar = 10 μm. (c) GFP-LC3 neurons were treated with IFN-α, IFN-β, IFN-γ, IFN-λ, cGAMP, nutrient deprivation (“starve”), rapamycin, or Tat-beclin 1 peptide, and cluster formation was assessed. The graph indicates the percentage of neurons with GFP-LC3+ clusters relative to the total neurons observed. (d) GFP-LC3 neurons were treated with the indicated doses of IFN-β. The graph indicates the percentage of neurons with GFP-LC3+ clusters relative to the total neurons observed. (e) Neurons were infected with HSV-1 at an MOI of 5 or treated with 10 U/ml of IFN-β, and then expression of ISG15 and IFIT was assayed by qPCR. (f) GFP-LC3 neurons were untreated or pretreated with an IFN-blocking antibody and then either infected with HSV-1, treated with IFN-β, or left untreated. The graph indicates the percentage of neurons with GFP-LC3+ clusters relative to the total neurons observed or to the HSV-1 antigen-negative neurons. All experiments were performed at least 3 times, and 200 to 1,200 cells were scored per experiment. Error bars represent SEMs. Significance was evaluated by one-way ANOVA with Bonferroni posttests. **, P < 0.01; ***, P < 0.001.
Based on these findings, we hypothesized that TG cultures express IFN-β in response to HSV infection and that this was the cause of cluster formation after infection. qPCR analysis revealed that the ISGs IFIT1 and ISG15 (28) are upregulated in HSV-1-infected cultures (Fig. 4e). Consistent with this idea, an IFN-β receptor-blocking antibody inhibited cluster formation in HSV-infected neuronal cultures (Fig. 4f). Taken together, these data support the hypothesis that HSV-1 infection of TG neuronal cultures triggers antiviral signaling, which modulates the autophagic pathway to induce cluster formation.
Autophagic clusters are induced by other alphaherpesviruses.
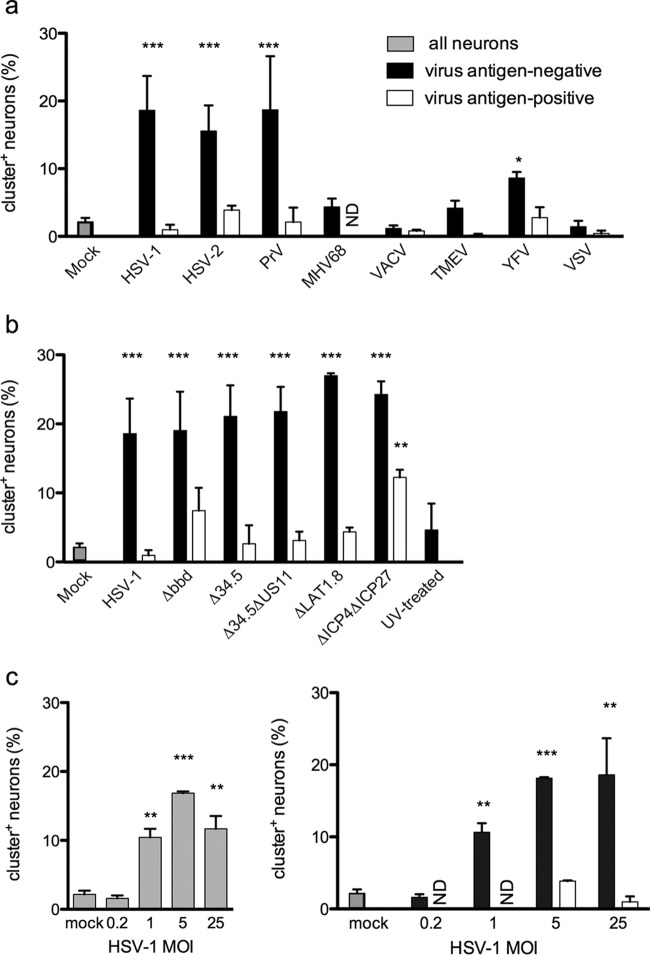
Since autophagosomal clustering was triggered by antiviral signaling and HSV infection, we reasoned that other viruses should also induce cluster formation. We therefore compared cluster formation in neuron cultures infected with a range of virus types: three related neurotropic alphaherpesviruses (HSV-1, HSV-2, and pseudorabies virus [PrV]), a lymphotropic gammaherpesvirus (murine gammaherpesvirus 68 [MHV-68]), an unrelated DNA virus (vaccinia virus [VACV]), a neurotropic RNA virus (Theiler's murine encephalomyelitis virus [TMEV]), and two unrelated RNA viruses (yellow fever virus [YFV] and vesicular stomatitis virus [VSV]) (Fig. 5a). Unexpectedly, only HSV-1, HSV-2, and PrV induced robust cluster formation, again predominantly in neurons that exhibited no viral antigen expression. YFV induced a modest increase in cluster prevalence, an interesting finding given its ability to establish persistent neuronal infections in immunocompromised mice (73). Overall, however, these data showed that autophagic clusters were most efficiently induced in response to alphaherpesviruses.
FIG 5.
Clusters are induced by alphaherpesvirus infection. (a) GFP-LC3 neurons were infected with the indicated viruses and then stained with anti-HSV-1 (HSV-1, HSV-2, and PrV), anti-MHV-68, anti-vaccinia virus, anti-YFV, or anti-VSV-G. Neurons were analyzed as for Fig. 1d. (b) GFP-LC3 neurons were infected with HSV-1 mutants and then stained for HSV antigen and analyzed as for Fig. 1d. (c) Neurons were infected with HSV-1 at the indicated MOIs and then stained and analyzed as for Fig. 1d. Each experiment was performed 3 or 4 times, and 200 to 1,200 neurons were scored per experiment. Error bars represent SEMs. Significance was evaluated by two-way ANOVA with Bonferroni posttests. *, P < 0.05; **, P < 0.01; ***, P < 0.001. ND, not detected.
Given that HSV-1 inhibits the antiviral response, it was possible that it might also inhibit the cluster response in antigen-positive cells. We reasoned that γ34.5 or US11 might be involved, since both interfere with antiviral and autophagy pathways (14, 33–35, 51, 74, 75). We therefore infected neuron cultures with HSV-1 lacking the beclin-binding region in the γ34.5 gene (Δbbd) (35), the entire γ34.5 gene (Δ34.5) (14), or the both γ34.5 and US11 genes (Δ34.5 ΔUS11) (56) and examined cluster prevalence and HSV antigen expression. Unexpectedly, there was no difference in the abundance of clusters induced by wild-type (WT) or mutant viruses, and there was no change in the mutual exclusivity of viral antigen expression and cluster formation (Fig. 5b). We therefore wondered whether cluster formation is influenced by the expression of latency-associated-transcripts (LAT) and early lytic genes within HSV-infected neurons. We explored this possibility using mutant viruses that lack either LAT expression (dlLAT1.8 [57]) or the key immediate early (IE) genes ICP4 and ICP27, such that lytic gene expression is very limited (d92 [59]). Both dlLAT1.8 and d92 were fully capable of inducing cluster formation (Fig. 5b). However, the near-complete mutual exclusivity of clusters and HSV antigen expression was broken in the d92-infected cultures. Treatment with UV-inactivated HSV-1 did not induce cluster formation (Fig. 5b). Moreover, cluster formation in response to wild-type HSV-1 was dose dependent and was significantly induced even at an MOI of 1 (Fig. 5c). Taken together, these findings demonstrate that limited expression of HSV lytic genes is sufficient to trigger the cluster autophagic response in TG neurons. They also suggest that cluster formation in antigen-positive cells is inhibited by early HSV gene expression.
Dynamics of cluster formation and clearance.
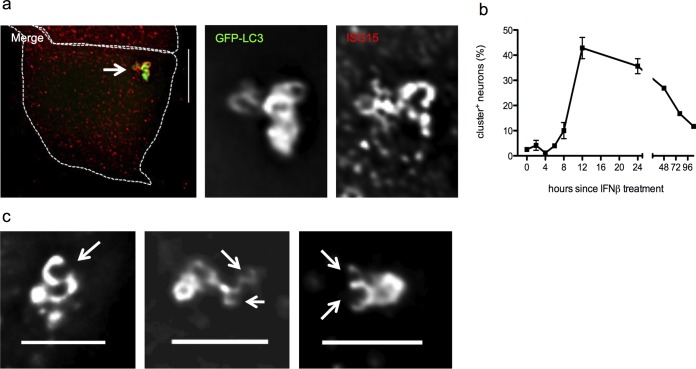
We next examined the process of cluster formation after IFN-β treatment. We used qPCR to detect transcripts of the autophagy genes Atg5 and p62 and found no change in their expression 3 to 6 h postinfection or 6 h after IFN-β treatment. This finding supports the notion that a generalized increase in autophagy is not necessary or sufficient to induce cluster formation (Fig. 4c). We therefore hypothesized that antiviral effectors directly modulate the autophagic machinery. In cancer cell lines, the ubiquitin-like molecule ISG15 (76) modifies beclin 1 (77) and can bind directly to p62 (78). We found that ISG15 colocalized with >70% of clusters (Fig. 6a). While colocalization is not proof of an interaction, these findings raise the possibility that neuronal antiviral signaling produces posttranslational modifications in the autophagy machinery.
FIG 6.
Cluster composition, kinetics, and form. (a) Merged fluorescence image of IFN-β-treated neurons from GFP-LC3 mice, with a white arrow indicating a cluster (size bar = 10 μm). Shown are zoomed single-channel images of the cluster show GFP-LC3 (green) and ISG15 (red). (b) GFP-LC3 neurons treated with IFN-β for the times indicated were scored for presence of clusters. Results are presented as percentages of neurons from the entire population scored. The experiment was performed ≥3 times, and >1000 neurons were scored per experiment. Error bars represent SEMs. (c) GFP-LC3 neurons treated with IFN-β. Images show representative clusters; white arrows indicate “open cups.” Bar = 5 μm.
We also sought to determine the kinetics of cluster formation. Cluster abundance increases approximately 8 h after IFN-β treatment, peaks at 12 h, and then steadily declines (Fig. 6b). We reasoned that individual autophagosomes might form at discrete locations in the cytosol and then be shuttled to a single site in a microtubule-dependent process, as described by Nunes et al. (79). Microtubule disruption with nocodazole, however, did not affect cluster formation (data not shown). When examined in live cells, it appeared that the clusters represent a stable, though not static, structure (see Movie S1 in the supplemental material). Moreover, deconvolution microscopy showed that some clusters appear to include open cups (Fig. 6c, white arrows, and Fig. 3c) that may represent elongating autophagosomes. Taken together, these data suggest that autophagic biogenesis may occur at the clusters.
Next, we sought to determine how clusters might be cleared. Live imaging revealed no evidence of autophagosome migration away from existing clusters (see Movie S1). Since clusters decline over the course of 72 h (Fig. 6b) and also colocalize with lysosomes (Fig. 3c and d), it seemed possible that lysosomal fusion might clear vesicles from the clusters. To test this idea, we treated neurons with IFN-β for 12 h to induce cluster formation and then treated them with bafilomycin 1 to inhibit the proton pump-dependent maturation. We expected that clusters would increase in size and abundance. However, live imaging revealed that autophagic trafficking around the cluster was dysregulated, and the stability of the clusters was compromised (see Movie S2). Thus, cluster stability likely depends on lysosomal acidification.
HSV-1 infection in vivo induces local cluster formation.
We utilized the ocular model of HSV-1 infection (53) in order to test whether our findings in vitro were a recapitulation of neuronal autophagosome dynamics in vivo. We infected GFP-LC3 mice via corneal scarification with HSV-1 strain 17 and 4 days postinfection (dpi) examined histological sections of TG. In three independent experiments, we observed that autophagic clusters had formed in the neurons of infected TG (Fig. 7a) but not in neurons of mock-infected animals (Fig. 7b). As observed in vitro, clusters were found predominantly in HSV-1 antigen-negative neurons (Fig. 7c), and most (∼80%) colocalized with p62 (Fig. 6d).
FIG 7.
Clusters are induced in the context of HSV-1 infection in vivo. GFP-LC3 mice were infected with HSV-1 (a, c, and d) or mock infected (b). TG were isolated and 10-μm sections were stained with anti-HSV-1 (c) or anti-p62 (d). Autophagic clusters were quantified for panel c as for Fig. 1d. Each experiment was performed 3 times; 1 representative slice was scored per mouse. Bars = 10 μm (a), 100 μm (b), and 50 μm (c and d). Error bars represent SEMs. Significance was evaluated by one-way ANOVA with Bonferroni posttests. *, P < 0.05; ***, P < 0.001.
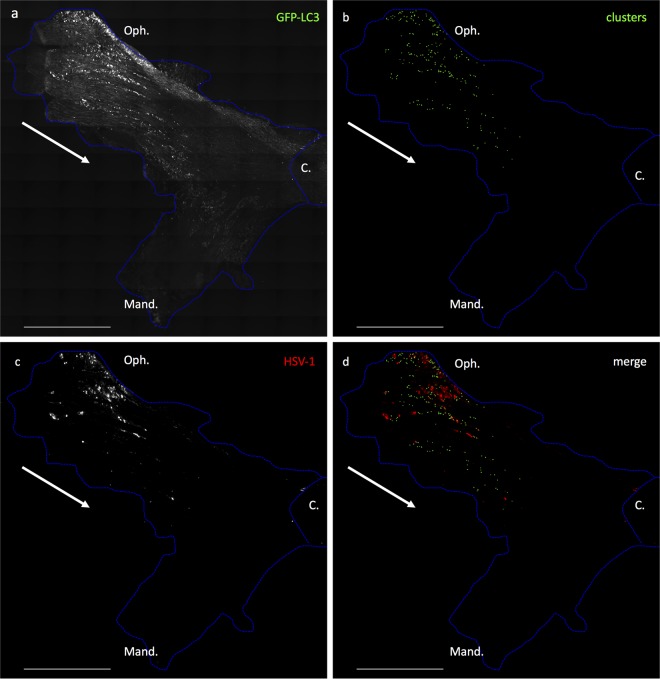
Since clusters form in response to HSV-1 infection and IFN-β stimulation, it was of interest to localize the cluster-containing neurons more precisely within the infected TG. We examined longitudinal sections comprising the entire TG to establish a map of the neurons that were positive for HSV antigen or GFP-LC3 clusters (Fig. 8). GFP-LC3 was detectable throughout the TG (Fig. 8a). However, autophagic clusters were restricted to neurons within the ophthalmic branch (Oph.), proximal to the infection site (Fig. 8b). Immunohistochemistry confirmed that this was the most HSV antigen-positive infected region of the TG (Fig. 8c). The merged image demonstrates that the autophagic clusters were in close proximity but not colocalized to the HSV-1 antigen-positive cells (Fig. 8d). Taken together, these data demonstrate that autophagosomal clustering occurs in vivo during acute infection in neurons that are proximal to those expressing HSV antigen.
FIG 8.
Clusters occur in close proximity to infected cells in TG in vivo. Shown are tiled images of a whole TG in a representative longitudinal section. The blue outline demarcates the boundaries of the tissue; the white arrow indicates the direction of viral spread. GFP-LC3 mice were infected with HSV via the corneal route. At 4 dpi the TG were stained for HSV-1 antigen. (a) GFP-LC3 in the entire TG. (b) After image acquisition, cluster locations were marked by addition of green pixels (no fluorescence is shown in this panel). (c) HSV-1 antigen. (d) Merged image of panels b and c. The experiment was performed 3 times. Bar = 1 mm. Oph., ophthalmic branch; Mand., mandibular branch; C., central nervous system.
DISCUSSION
We have studied the relationship between virus infection, the interferon response, and the induction of autophagy (35, 37, 55, 74) We recently showed that the neuronal antiviral response to HSV-1 is driven by IFN-β signaling and that HSV strains that cannot counter autophagy are more susceptible to this antiviral effect (12). While several studies have shown that IFN-β induces autophagy in cancer cells (80), to our knowledge a direct link between IFN-β signaling and autophagy induction has not been previously demonstrated in neurons. Our data support the notion that HSV infection and IFN-β signaling in primary murine TG neurons induce a distinct structure, consisting of clustered, and possibly selective, autophagosomes. Given that treatment with conventional pharmacological inducers of autophagy did not result in clusters, it appears that this process is dependent upon antiviral signaling pathways, which are both necessary and sufficient to induce cluster formation.
Three key observations in this study are suggestive of possible links between neuronal autophagosomal clustering and the HSV-1 life cycle. First, while >95% of neurons in infected cultures harbor HSV genomes, the clusters formed almost exclusively in viral antigen-negative neurons. The cluster-containing neurons are therefore largely HSV genome positive, and it is of interest to determine whether latency has been established in these cells. Second, TG neurons formed clusters efficiently and almost exclusively in response to infection with neurotropic alphaherpesviruses. Third, HSV gene expression was necessary to induce clusters, yet the limited immediate-early/early (IE/E) gene expression of d92 was sufficient in this regard. It is possible, therefore, that clusters form specifically in response to alphaherpesvirus IE gene expression. Accordingly, cluster formation in vivo was restricted to the branch of the TG that had been infected by HSV and showed antigen expression. Thus, TG neurons display both sensitivity and specificity to alphaherpesvirus infection, as judged by cluster formation. There is some precedent for the notion that TG neurons have adapted to respond specifically to alphaherpesviruses, since they express neuronal miR138, which mediates specific repression of HSV-1 ICP0 (32).
TG neurons are sensitive to IFN signaling, since clusters were induced by low levels of IFN. Neurons fail to produce detectable levels of IFN-β, and nonneuronal cells, such as glial cells or infiltrating immune cells, are the most likely source of IFN in vivo. Consistent with this idea, IFN production and cluster formation are modest in vitro, where the neurons are highly enriched relative to nonneuronal cells. Indeed, in vivo infection produced a higher percentage of clusters, comparable to the frequency induced by IFN treatment in vitro. These clusters were restricted to the acutely infected region of the TG, proximal to localized IFN-producing cells. Intriguingly, clusters were found predominantly in neurons which were HSV antigen negative. HSV is known to modulate IFN signaling in infected cells. It is of interest to determine both in vitro and in vivo whether the absence of clusters in antigen-positive neurons reflects either the absence of an antiviral response or immunomodulatory activity of HSV.
Autophagy is clearly a key player in the neuronal antiviral response (35, 37, 74), but the precise role for autophagic clusters in this critical pathway remains unclear. There are some clues, however, that implicate a role for selective autophagy that warrant further study. Virtually every cluster examined was p62 positive, consistent with a hypothesis that this overall process represents p62-mediated selective autophagy. p62 is known to recruit the autophagic machinery to large cargo (41, 81). We therefore stained IFN-β-treated neuron cultures for markers of aggresomes (γ-tubulin and HDAC6 [82, 83]) and stress granules (TIA and PABP [84, 85]). We also stained infected neuron cultures for ICP5, a component of the HSV capsid (55). While we were able to clearly observe these markers in their expected subcellular locations, they did not colocalize with clusters (our unpublished data). We are therefore investigating whether clusters facilitate selective autophagic degradation of smaller cargos, such as components of innate signaling pathways (86). The presence of a smaller cargo is, however, at odds with the presence of these closely associated clustered vesicles. A previous study with kidney cells showed that individual autophagic vesicles can migrate to a single location in a microtubule-driven fashion (79), but recent work has shown that nocodazole does not disrupt cluster formation (our unpublished data). Furthermore, our live-imaging movies show no evidence of vesicle migration into the clusters. Another line of investigation in our lab is driven by the fact that p62 self-oligomerizes, possibly promoting cluster formation (87). Alternatively, vesicle clustering may be promoted by innate mediator ISG15 (28, 88, 89) which decorates the clusters and is known to interact with beclin 1 and p62 (77, 78).
In summary, the autophagic clusters described here are novel reporters of heightened neuronal antiviral responses both in vitro and in vivo. Further characterization of these structures and their cargo may provide important insights into the relationship between HSV, sensory neurons, and the antiviral response and likely elucidate aspects of the establishment, maintenance, and reactivation of latency. Finally, the possible involvement of autophagy in the regulation of the antiviral state in neurons opens up new pathways for intervention and therapy for herpetic diseases.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants to D.A.L. (RO1 EY09083 and PO1 AI098681). The project was also supported by P30RR016437 from the National Center for Research Resources to Dartmouth. Support from Geisel School of Medicine Immunology Program Training Grant 2T32AI007363-21A1 to S.K. is gratefully acknowledged.
We also acknowledge Brian North for colony maintenance and genotyping, Audra Charron for critical reading of the manuscript, the Dartmouth Autophagy Work Group for valuable input, and Erika Holzbauer for providing the GFP-LC3 mice with permission from Noboru Mizushima.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02908-15.
REFERENCES
- 1.Lee S, Ives AM, Bertke AS. 2015. Herpes simplex virus 1 reactivates from autonomic ciliary ganglia independently from sensory trigeminal ganglia to cause recurrent ocular disease. J Virol 89:8383–8391. doi: 10.1128/JVI.00468-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith G. 2012. Herpesvirus transport to the nervous system and back again. Annu Rev Microbiol 66:153–176. doi: 10.1146/annurev-micro-092611-150051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertke AS, Swanson SM, Chen J, Imai Y, Kinchington PR, Margolis TP. 2011. A5-positive primary sensory neurons are nonpermissive for productive infection with herpes simplex virus 1 in vitro. J Virol 85:6669–6677. doi: 10.1128/JVI.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 5.Collins SE, Noyce RS, Mossman KL. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J Virol 78:1706–1717. doi: 10.1128/JVI.78.4.1706-1717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuchet D, Sykes A, Nicolas A, Orr A, Murray J, Sirma H, Heeren J, Bartelt A, Everett RD. 2011. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J Cell Sci 124:280–291. doi: 10.1242/jcs.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conrady CD, Zheng M, Fitzgerald KA, Liu C, Carr DJJ. 2012. Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal Immunol 5:173–183. doi: 10.1038/mi.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lafaille FG, Pessach IM, Zhang S-Y, Ciancanelli MJ, Herman M, Abhyankar A, Ying S-W, Keros S, Goldstein PA, Mostoslavsky G, Ordovas-Montanes J, Jouanguy E, Plancoulaine S, Tu E, Elkabetz Y, Al-Muhsen S, Tardieu M, Schlaeger TM, Daley GQ, Abel L, Casanova J-L, Studer L, Notarangelo LD. 2012. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature 491:769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM. 2015. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci U S A 112:E1773–E1781. doi: 10.1073/pnas.1424637112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagan JC. 2012. Defining the subcellular sites of innate immune signal transduction. Trends Immunol 33:442–448. doi: 10.1016/j.it.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carty M, Reinert L, Paludan SR, Bowie AG. 2014. Innate antiviral signalling in the central nervous system. Trends Immunol 35:79–87. doi: 10.1016/j.it.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Rosato PC, Leib DA. 2015. Neuronal interferon signaling is required for protection against herpes simplex virus replication and pathogenesis. PLoS Pathog 11:e1005028. doi: 10.1371/journal.ppat.1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou J, Kern ER, Whitley RJ, Roizman B. 1990. Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250:1262–1266. doi: 10.1126/science.2173860. [DOI] [PubMed] [Google Scholar]
- 14.Leib DA, Machalek MA, Williams BRG, Silverman RH, Virgin HW. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc Natl Acad Sci U S A 97:6097–6101. doi: 10.1073/pnas.100415697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Härle P, Sainz B, Carr DJJ, Halford WP. 2002. The immediate-early protein, ICP0, is essential for the resistance of herpes simplex virus to interferon-alpha/beta. Virology 293:295–304. doi: 10.1006/viro.2001.1280. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Zhang C, Chen X, Yu J, Wang Y, Yang Y, Du M, Jin H, Ma Y, He B, Cao Y. 2011. ICP34.5 protein of herpes simplex virus facilitates the initiation of protein translation by bridging eukaryotic initiation factor 2α (eIF2α) and protein phosphatase 1. J Biol Chem 286:24785–24792. doi: 10.1074/jbc.M111.232439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xing J, Ni L, Wang S, Wang K, Lin R, Zheng C. 2013. Herpes simplex virus 1-encoded tegument protein VP16 abrogates the production of beta interferon (IFN) by inhibiting NF-κB activation and blocking IFN regulatory factor 3 to recruit its coactivator CBP. J Virol 87:9788–9801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori I, Nishiyama Y. 2006. Accessory genes define the relationship between the herpes simplex virus and its host. Microbes Infect 8:2556–2562. doi: 10.1016/j.micinf.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Lanfranca MP, Mostafa HH, Davido DJ. 2014. HSV-1 ICP0: an E3 ubiquitin ligase that counteracts host intrinsic and innate immunity. Cells 3:438–454. doi: 10.3390/cells3020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medzhitov R, Preston-Hurlburt P, Janeway CA. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 22.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juang YT, Lowther W, Kellum M, Au WC, Lin R, Hiscott J, Pitha PM. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci U S A 95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 25.Der SD, Zhou A, Williams BR, Silverman RH. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A 95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durbin JE, Hackenmiller R, Simon MC, Levy DE. 1996. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84:443–450. doi: 10.1016/S0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 27.Meraz MA, White JM, Sheehan KCF, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. 1996. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84:431–442. doi: 10.1016/S0092-8674(00)81288-X. [DOI] [PubMed] [Google Scholar]
- 28.Sadler AJ, Williams BRG. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S-Y, Sanchez DJ, Cheng G. 2011. New developments in the induction and antiviral effectors of type I interferon. Curr Opin Immunol 23:57–64. doi: 10.1016/j.coi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furr SR, Chauhan VS, Moerdyk-Schauwecker MJ, Marriott I. 2011. A role for DNA-dependent activator of interferon regulatory factor in the recognition of herpes simplex virus type 1 by glial cells. J Neuroinflammation 8:99. doi: 10.1186/1742-2094-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinert LS, Harder L, Holm CK, Iversen MB, Horan KA, Dagnæs-Hansen F, Ulhøi BP, Holm TH, Mogensen TH, Owens T, Nyengaard JR, Thomsen AR, Paludan SR. 2012. TLR3 deficiency renders astrocytes permissive to herpes simplex virus infection and facilitates establishment of CNS infection in mice. J Clin Invest 122:1368–1376. doi: 10.1172/JCI60893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. 2014. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe 15:446–456. doi: 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosato PC, Leib DA. 2014. Intrinsic innate immunity fails to control herpes simplex virus and vesicular stomatitis virus replication in sensory neurons and fibroblasts. J Virol 88:9991–10001. doi: 10.1128/JVI.01462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tallóczy Z, Herbert Virgin I, Levine B. 2006. PKR-dependent xenophagic degradation of herpes simplex virus type 1. Autophagy 2:24–29. doi: 10.4161/auto.2176. [DOI] [PubMed] [Google Scholar]
- 35.Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the beclin 1 autophagy protein. Cell Host Microbe 1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Orvedahl A, MacPherson S, Sumpter R Jr, Tallóczy Z, Zou Z, Levine B. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. 2012. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe 12:334–345. doi: 10.1016/j.chom.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn WA., Jr 1994. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol 4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 39.Rogov V, Dötsch V, Johansen T, Kirkin V. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 40.Komatsu M, Waguri S, Koike M, Sou Y-S, Ueno T, Hara T, Mizushima N, Iwata J-I, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S-I, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 41.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun J-A, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 42.Geisler S, Holmström KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ, Springer W. 2010. PINK1/parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol 12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 43.Kawajiri S, Saiki S, Sato S, Sato F, Hatano T, Eguchi H, Hattori N. 2010. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett 584:1073–1079. doi: 10.1016/j.febslet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 44.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu W, Ko HS, Magrané J, Moore DJ, Dawson VL, Grailhe R, Dawson TM, Li C, Tieu K, Przedborski S. 2010. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. 2004. Autophagy defends cells against invading group A streptococcus. Science 306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 46.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 47.Orvedahl A, Sumpter R, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier R, Xie Y, Levine B. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature 480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto A, Yue Z. 2014. Autophagy and its normal and pathogenic states in the brain. Annu Rev Neurosci 37:55–78. doi: 10.1146/annurev-neuro-071013-014149. [DOI] [PubMed] [Google Scholar]
- 49.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 50.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 51.Lussignol M, Queval C, Bernet-Camard M-F, Cotte-Laffitte J, Beau I, Codogno P, Esclatine A. 2013. The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J Virol 87:859–871. doi: 10.1128/JVI.01158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolovan CA, Sawtell NM, Thompson RL. 1994. ICP34.5 mutants of herpes simplex virus type 1 strain 17syn+ are attenuated for neurovirulence in mice and for replication in confluent primary mouse embryo cell cultures. J Virol 68:48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rader KA, Ackland-Berglund CE, Miller JK, Pepose JS, Leib DA. 1993. In vivo characterization of site-directed mutations in the promoter of the herpes simplex virus type 1 latency-associated transcripts. J Gen Virol 74(Part 9):1859–1869. [DOI] [PubMed] [Google Scholar]
- 54.Smith KO. 1964. Relationship between the envelope and the infectivity of herpes simplex virus. Exp Biol Med 115:814–816. doi: 10.3181/00379727-115-29045. [DOI] [PubMed] [Google Scholar]
- 55.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. 2007. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol 81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol 77:10917–10928. doi: 10.1128/JVI.77.20.10917-10928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leib DA, Bogard CL, Kosz-Vnenchak M, Hicks KA, Coen DM, Knipe DM, Schaffer PA. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J Virol 63:2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Card JP, Dubin JR, Whealy ME, Enquist LW. 1995. Influence of infectious dose upon productive replication and transynaptic passage of pseudorabies virus in rat central nervous system. J Neurovirol 1:349–358. doi: 10.3109/13550289509111024. [DOI] [PubMed] [Google Scholar]
- 59.Samaniego LA, Webb AL, DeLuca NA. 1995. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J Virol 69:5705–5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duerst RJ, Morrison LA. 2004. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology 322:158–167. doi: 10.1016/j.virol.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 61.Fuse S, Zhang W, Usherwood EJ. 2008. Control of memory CD8+ T cell differentiation by CD80/CD86-CD28 costimulation and restoration by IL-2 during the recall response. J Immunol 180:1148–1157. doi: 10.4049/jimmunol.180.2.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pachner AR, Li L, Narayan K. 2007. Intrathecal antibody production in an animal model of multiple sclerosis. J Neuroimmunol 185:57–63. doi: 10.1016/j.jneuroim.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 63.Barba-Spaeth G, Longman RS, Albert ML, Rice CM. 2005. Live attenuated yellow fever 17D infects human DCs and allows for presentation of endogenous and recombinant T cell epitopes. J Exp Med 202:1179–1184. doi: 10.1084/jem.20051352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15:1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuma A, Mizushima N. 2008. Chromosomal mapping of the GFP-LC3 transgene in GFP-LC3 mice. Autophagy 4:61–62. doi: 10.4161/auto.4846. [DOI] [PubMed] [Google Scholar]
- 66.van den Broek MF, Müller U, Huang S, Aguet M, Zinkernagel RM. 1995. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J Virol 69:4792–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539–548. doi: 10.1016/S1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 68.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 69.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 70.Klionsky DJ, et al. 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Komatsu M, Ueno T, Waguri S, Uchiyama Y, Kominami E, Tanaka K. 2007. Constitutive autophagy: vital role in clearance of unfavorable proteins in neurons. Cell Death Differ 14:887–894. [DOI] [PubMed] [Google Scholar]
- 72.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B. 2013. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mateo RI, Xiao S-Y, Travassos da Rosa APA, Lei H, Guzman H, Lu L, Tesh RB. 2007. Yellow fever 17-D vaccine is neurotropic and produces encephalitis in immunosuppressed hamsters. Am J Trop Med Hyg 77:919–924. [PubMed] [Google Scholar]
- 74.Tallöczy Z, Jiang W, Virgin HW, Leib DA, Scheuner D, Kaufman RJ, Eskelinen E-L, Levine B. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc Natl Acad Sci U S A 99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gobeil PAM, Leib DA. 2012. Herpes simplex virus γ34.5 interferes with autophagosome maturation and antigen presentation in dendritic cells. mBio 3(5):e00267-12. doi: 10.1128/mBio.00267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loeb KR, Haas AL. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem 267:7806–7813. [PubMed] [Google Scholar]
- 77.Xu D, Zhang T, Xiao J, Zhu K, Wei R, Wu Z, Meng H, Li Y, Yuan J. 2015. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy 11:617–628. doi: 10.1080/15548627.2015.1023982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nakashima H, Nguyen T, Goins WF, Chiocca EA. 2015. Interferon-stimulated gene 15 (ISG15) and ISG15-linked proteins can associate with members of the selective autophagic process, histone deacetylase 6 (HDAC6) and SQSTM1/p62. J Biol Chem 290:1485–1495. doi: 10.1074/jbc.M114.593871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nunes P, Ernandez T, Roth I, Qiao X, Strebel D, Bouley R, Charollais A, Ramadori P, Foti M, Meda P, Féraille E, Brown D, Hasler U. 2013. Hypertonic stress promotes autophagy and microtubule-dependent autophagosomal clusters. Autophagy 9:550–567. doi: 10.4161/auto.23662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmeisser H, Bekisz J, Zoon KC. 2014. New function of type I IFN: induction of autophagy. J Interferon Cytokine Res 34:71–78. doi: 10.1089/jir.2013.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Komatsu M, Ichimura Y. 2010. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett 584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 82.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115:727–738. doi: 10.1016/S0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 83.García-Mata R, Bebök Z, Sorscher EJ, Sztul ES. 1999. Characterization and dynamics of aggresome formation by a cytosolic Gfp-chimera. J Cell Biol 146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins Tia-1 and Tiar link the phosphorylation of Eif-2α to the assembly of mammalian stress granules. J Cell Biol 147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, Deretic V. 2015. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol 210:973–989. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ciuffa R, Lamark T, Tarafder AK, Guesdon A, Rybina S, Hagen WJH, Johansen T, Sachse C. 2015. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep 11:748–758. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 88.Skaug B, Chen ZJ. 2010. Emerging role of ISG15 in antiviral immunity. Cell 143:187–190. doi: 10.1016/j.cell.2010.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao C, Collins M, Hsiang T-Y, Krug RM. 2013. Interferon-induced ISG15 pathway: an ongoing virus–host battle. Trends Microbiol 21:181–186. doi: 10.1016/j.tim.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.