Abstract
In natural and artificial aquatic environments, population structures and dynamics of photosynthetic microbes are heavily influenced by the grazing activity of protistan predators. Understanding the molecular factors that affect predation is critical for controlling toxic cyanobacterial blooms and maintaining cyanobacterial biomass production ponds for generating biofuels and other bioproducts. We previously demonstrated that impairment of the synthesis or transport of the O-antigen component of lipopolysaccharide (LPS) enables resistance to amoebal grazing in the model predator-prey system consisting of the heterolobosean amoeba HGG1 and the cyanobacterium Synechococcus elongatus PCC 7942 (R. S. Simkovsky et al., Proc Natl Acad Sci U S A 109:16678–16683, 2012, http://dx.doi.org/10.1073/pnas.1214904109). In this study, we used this model system to identify additional gene products involved in the synthesis of O antigen, the ligation of O antigen to the lipid A-core conjugated molecule (including a novel ligase gene), the generation of GDP-fucose, and the incorporation of sugars into the lipid A core oligosaccharide of S. elongatus. Knockout of any of these genes enables resistance to HGG1, and of these, only disruption of the genes involved in synthesis or incorporation of GDP-fucose into the lipid A-core molecule impairs growth. Because these LPS synthesis genes are well conserved across the diverse range of cyanobacteria, they enable a broader understanding of the structure and synthesis of cyanobacterial LPS and represent mutational targets for generating resistance to amoebal grazers in novel biomass production strains.
INTRODUCTION
Lipopolysaccharide (LPS) serves as a critical interface between a Gram-negative bacterium's internal physiology and its abiotic and biotic environments, protecting the cell from specific antimicrobials, evading the innate immune and complement systems, enabling symbiosis with eukaryotic hosts, acting as a recognition factor for phage infection, and protecting bacteria against digestion by predatory amoebae (1–5). Alterations to any of the three primary components of the LPS structure (the endotoxin lipid A, the core oligosaccharide, or the O-antigen polysaccharide), through changes in sugar content or sequence, acylation, phosphorylation, or other molecular modifications, can alter these interactions, modulate the toxicity of the endotoxin, or prevent cellular behaviors such as motility (6, 7).
While much is known about the structure and synthesis of LPS in enteric and pathogenic proteobacteria, little is known about either aspect of LPS in photosynthetic cyanobacteria and how the cyanobacterial LPS affects interactions with the environment (8–11). To date, only three cyanobacterial LPS structures have been reported: the partial structural determination of the filamentous freshwater cyanobacterium Oscillatoria planktothrix LPS (12) and the complete structural determination of the LPSs of the unicellular marine Synechococcus strains WH8102 and CC9311 (9). Beyond these structures, the remaining current body of knowledge concerning cyanobacterial LPS structure is derived from sugar and lipid composition studies. These structural and compositional studies highlight a few key differences between enteric and cyanobacterial LPSs: (i) many cyanobacterial LPSs completely lack or contain only trace amounts of 3-deoxy-d-manno-oct-2-ulosonic acid (KDO), which is ubiquitous in enteric Gram-negative bacteria; (ii) heptoses are almost universally absent from cyanobacterial LPS; and (iii) some cyanobacterial LPSs contain no phosphates, whereas others have variable amounts of phosphate (9, 10, 12–16). Due to the lack of acid-labile KDO in many cyanobacterial species (10), in only a few of the compositional studies was the LPS molecule able to be degraded and a purified fraction, such as the O antigen, characterized for composition. For example, the O antigen of Microcystis aeruginosa NIES-87 is composed solely of glucose (17), and the O antigen of Synechococcus elongatus PCC 6301 is reminiscent of the polymannose O antigen of Escherichia coli O8 and O9 (18).
Genes involved in cyanobacterial LPS synthesis have primarily been identified through homology with enteric genes, such as the prediction of the lpxA, lpxB, lpxC, and lpxD genes involved in the synthesis of lipid A (19, 20). We previously identified and characterized, through mutational studies, four genes in S. elongatus PCC 7942 involved in a Wzm/Wzt-like O-antigen synthesis and transport pathway homologous to that of E. coli serotypes O8 and O9 (21). Prior to that study, only a single publication on phage-resistant mutants in Anabaena sp. strain PCC 7120 had characterized genes involved in the synthesis of cyanobacterial LPS or its constituent polysaccharide, O antigen (22).
Cyanobacteria are currently being tested for large-scale outdoor cultivation for producing biofuels, feedstocks, nutraceuticals, and other high-value coproducts (23). To optimize the ecology of these production ponds for maximum yields and to develop crop protection strategies to prevent biomass losses due to infection or predation by contaminants such as viruses or protists, it is critical to understand the role cyanobacterial LPS has in modulating these interactions. Knowledge on how LPS regulates predation could further enable biotic mechanisms for controlling cyanobacterial harmful algal blooms, such as Microcystis blooms visible by satellite that contaminate drinking water supplies with the neurotoxin microcystin (24, 25). Elucidating the diversity of cyanobacterial LPS structures will enable the determination of its potential toxicity to humans that come in contact with production ponds or natural cyanobacterial blooms, an issue that remains highly debated in the literature (17, 26).
We recently demonstrated that removal of the O antigen from the LPS of S. elongatus PCC 7942 confers resistance to predation by a heterolobosean amoebal grazer, HGG1 (21). Strains that lack the O antigen display a visibly identifiable “rough” phenotype, which enabled the identification of new resistant mutants through random mutagenesis. Whereas the wild type (WT) remains planktonic, these rough strains autoflocculate and settle to the bottom of the culture, a phenotype beneficial for harvesting large-scale biomass. To extend our previous findings on the genes involved in LPS synthesis and to discover more gene targets for generating grazer-resistant cyanobacteria, in this study, we screened the unigene set (UGS) of insertional transposon mutants of S. elongatus (27, 28) for strains that display the rough phenotype and identified additional mutagenesis targets through bioinformatics analysis. In total, we identified six additional genes whose knockout mutations produce grazer resistance and impair the production of lipopolysaccharide. The elucidation and characterization of these LPS synthesis genes are critical to our understanding of how cyanobacteria interact with their environments, including predators, and how we can exploit these organisms for production purposes.
MATERIALS AND METHODS
Plasmids, strains, and culture conditions.
All vectors and strains used in this study are listed in Table 1. To generate novel constitutive expression vectors for complementation studies, open reading frames (ORFs) were PCR amplified from WT S. elongatus cells using Q5 DNA polymerase (NEB) and TOPO cloned into the pSyn_1/D-TOPO expression vector (Life Technologies) as previously reported (21). WT-like antibiotic-resistant strains (29), transposon-insertion knockout mutants (27, 28), and expression clones were derived from a WT strain of S. elongatus PCC 7942 (lab collection accession no. AMC06) through standard methods of transformation and homologous recombination that take advantage of its natural competence (30).
TABLE 1.
Strains and plasmids used in this study
| AMC no.a | Strain name | Genotype | Plasmid(s) to generate strainb | Antibiotic resistance(s)c | Source or reference |
|---|---|---|---|---|---|
| 6 | Synechococcus elongatus PCC 7942 (WT) | WT | Pasteur culture collection | ||
| 2180 | WT+1901cx | NS1::1901 | pAM4738 | Sp, Sm | This study |
| 2181 | WT+1902cx | NS1::1902 | pAM4739 | Sp, Sm | This study |
| 2017 | WT+1903cx | NS1::1903 | pAM4637 | Sp, Sm | 21 |
| 2182 | WT+1904cx | NS1::1904 | pAM4760 | Sp, Sm | This study |
| 1896 | WT+Kmr | NS2::nptI | pAM1579 | Km | 21 |
| 2177 | WT+Kmr+1901cx | NS2::nptI; NS1::1901 | pAM1579, pAM4738 | Km, Sp, Sm | This study |
| 2178 | WT+Kmr+1902cx | NS2::nptI; NS1::1902 | pAM1579, pAM4739 | Km, Sp, Sm | This study |
| 2019 | WT+Kmr+1903cx | NS2::nptI; NS1::1903 | pAM1579, pAM4637 | Km, Sp, Sm | 21 |
| 2179 | WT+Kmr+1904cx | NS2::nptI; NS1::1904 | pAM1579, pAM4760 | Km, Sp, Sm | This study |
| 2165 | 1901− | 1901::Tn5 | UGS 14C3 (8S6-L5) | Km | 27; this study |
| 2166 | 1901−+1901cx | 1901::Tn5; NS1::1901 | UGS 14C3 (8S6-L5), pAM4738 | Km, Sp, Sm | This study |
| 2167 | 1901−+1903cx | 1901::Tn5; NS1::1903 | UGS 14C3 (8S6-L5), pAM4637 | Km, Sp, Sm | This study |
| 2168 | 1902− | 1902::Tn5 | UGS 14C4 (8S6-D11) | Km | 27; this study |
| 2169 | 1902−+1902cx | 1902::Tn5; NS1::1902 | UGS 14C4 (8S6-D11), pAM4739 | Km, Sp, Sm | This study |
| 2170 | 1902−+1903cx | 1902::Tn5; NS1::1903 | UGS 14C4 (8S6-D11), pAM4637 | Km, Sp, Sm | This study |
| 1953 | 1903− | 1903::Tn5 | UGS 14C5 (8S6-F12) | Km | 21 |
| 2171 | 1903−+1901cx | 1903::Tn5; NS1::1901 | UGS 14C5 (8S6-F12), pAM4738 | Km, Sp, Sm | 27; this study |
| 2172 | 1903−+1902cx | 1903::Tn5; NS1::1902 | UGS 14C5 (8S6-F12), pAM4739 | Km, Sp, Sm | This study |
| 2021 | 1903−+1903cx | 1903::Tn5; NS1::1903 | UGS 14C5 (8S6-F12), pAM4637 | Km, Sp, Sm | 21 |
| 2173 | 1903−+1904cx | 1903::Tn5; NS1::1904 | UGS 14C5 (8S6-F12), pAM4760 | Km, Sp, Sm | This study |
| 2174 | 1904− | 1904::Tn5 | UGS 14C6 (8S6-C4) | Km | 27; this study |
| 2175 | 1904−+1903cx | 1904::Tn5; NS1::1903 | UGS 14C6 (8S6-C4), pAM4637 | Km, Sp, Sm | This study |
| 2176 | 1904−+1904cx | 1904::Tn5; NS1::1904 | UGS 14C6 (8S6-C4), pAM4760 | Km, Sp, Sm | This study |
| 1905− | 1905::Tn5 | UGS 14C7 (8S6-O7) | Km | 27; this study | |
| 2119 | WT + 2292cx | NS1::2292 | pAM4687 | Sp, Sm | This study |
| 2190 | WT + 2293cx | NS1::2293 | pAM4761 | Sp, Sm | This study |
| 1895 | WT+Cmr | NS2::cat | pAM1573 | Cm | 21 |
| 2189 | WT+Cmr+2293cx | NS2::cat; NS1::2293 | pAM1573, pAM4761 | Cm, Sp, Sm | This study |
| 2120 | WT+Kmr+2292cx | NS2::nptI; NS1::2292 | pAM1579, pAM4687 | Km, Sp, Sm | This study |
| 2074 | 2292− | 2292::Tn5 | UGS 10E8 (2D5-B-E8) | Km | 27; this study |
| 2122 | 2292−+2292cx | 2292::Tn5; NS1::2292 | UGS 10E8 (2D5-B-E8), pAM4687 | Km, Sp, Sm | This study |
| 2183 | 2292−+2293cx | 2292::Tn5; NS1::2293 | UGS 10E8 (2D5-B-E8), pAM4761 | Km, Sp, Sm | This study |
| 2184 | 2293− | 2293::Mu | UGS 3C6 (2F6-E1) | Cm | 27; this study |
| 2185 | 2293−+2292cx | 2293::Mu; NS1::2292 | UGS 3C6 (2F6-E1), pAM4687 | Cm, Sp, Sm | This study |
| 2186 | 2293−+2293cx | 2293::Mu; NS1::2293 | UGS 3C6 (2F6-E1), pAM4761 | Cm, Sp, Sm | This study |
| 1907 | 1126− | 1126::Mu | UGS 7D3 (1B3-L12) | Cm | 21 |
| 1977 | 1126−+1126cx | 1126::Mu; NS1::1126 | UGS 7D3 (1B3-L12), pAM4606 | Cm, Sp, Sm | 21 |
| 2123 | 1126−+2292cx | 1126::Mu; NS1::2292 | UGS 7D3 (1B3-L12), pAM4687 | Cm, Sp, Sm | This study |
| 2125 | 1903−+2292cx | 1903::Tn5; NS1::2292 | UGS 14C5 (8S6-F12), pAM4687 | Km, Sp, Sm | This study |
| 2187 | 2294− | 2294::Mu | UGS 3C7 (2F6-G1) | Cm | 27; this study |
| 2188 | 2295− | 2295::Mu | UGS 3C9 (1C2-L3) | Cm | 27; this study |
| 2236 | WT+1342cx | NS1::1342 | pAM4973 | Sp, Sm | This study |
| 2286 | WT+2027cx | NS1::2027 | pAM4994 | Sp, Sm | This study |
| 2237 | WT+Kmr+1342cx | NS2::nptI; NS1::1342 | pAM1579, pAM4973 | Sp, Sm | This study |
| 2287 | WT+Kmr+2027cx | NS2::nptI; NS1::2027 | pAM1579, pAM4994 | Sp, Sm | This study |
| 2240 | 1903−+1342cx | 1903::Tn5; NS1::1342 | UGS 14C5 (8S6-F12), pAM4973 | Km, Sp, Sm | This study |
| 2290 | 1903−+2027cx | 1903::Tn5; NS1::2027 | UGS 14C5 (8S6-F12), pAM4994 | Km, Sp, Sm | This study |
| 2228 | 1342− | 1342::Tn5 | UGS 25D5 (8S30-E10) | Km | 27; this study |
| 2238 | 1342−+1342cx | 1342::Tn5; NS1::1342 | UGS 25D5 (8S30-E10), pAM4973 | Km, Sp, Sm | This study |
| 2292 | 2027− | 2027::Tn5 | UGS 10B11 (3A9-B-F8) | Km | 27; this study |
| 2288 | 2027−+2027cx | 2027::Tn5; NS1::2027 | UGS 10B11 (3A9-B-F8), pAM4994 | Km, Sp, Sm | This study |
| 1126−+2292− | 1126::Mu; 2292::Tn5 | UGS 7D3 (1B3-L12), UGS 10E8 (2D5-B-E8) | Cm, Km | 27; this study | |
| 1903−+2293− | 1903::Tn5; 2293::Mu | UGS 14C5 (8S6-F12), UGS 3C6 (2F6-E1) | Cm, Km | 27; this study | |
| 1342−+2293− | 1342::Tn5; 2293::Mu | UGS 25D5 (8S30-E10), UGS 3C6 (2F6-E1) | Cm, Km | 27; this study | |
| 2196 | 2098− | 2098::Tn5 | UGS 15E1 (8S9-H9) | Km | 27; this study |
| 2198 | 2099− | 2099::Tn5 | UGS 15E2 (8S9-H1) | Km | 27; this study |
| 2193 | 2100− | 2100::Tn5 | UGS 15E3 (8S9-E8) | Km | 27; this study |
| 2204 | 2101− | 2101::Tn5 | UGS 15E4 (8S9-C7) | Km | 27; this study |
| 2192 | 0058− | 0058::Tn5 | UGS 18A3 (8S15-C11) | Km | 27; this study |
| 0579− | 0579::Tn5 | UGS 20E4 (8S19-J5) | Km | 27; this study | |
| 0948− | 0948::Tn5 | UGS 23B8 (8S25-I3) | Km | 27; this study | |
| 0949− | 0949::Tn5 | UGS 23B9 (8S25-L3) | Km | 27; this study | |
| 0950− | 0950::Tn5 | UGS 23B10 (8S25-J10) | Km | 27; this study | |
| 0466− | 0466::Tn5 | UGS 19G7 (8S17-F1) | Km | 27; this study | |
| 0973− | 0973::Tn5 | UGS 23D9 (8S26-M10) | Km | 27; this study | |
| 2151− | 2151::Mu | UGS 2F1 (1A8-V12) | Cm | 27; this study | |
| 1398− | 1398::Tn5 | UGS 25H6 (8S44-L4) | Km | 27; this study | |
| 0220− | 0220::Mu | UGS 5A3 (10B12-E5) | Cm | 27; this study | |
| 0133− | 0133::Tn5 | UGS 18E5 (8S37-A9) | Km | 27; this study | |
| 0471− | 0471::Mu | UGS 5H9 (2A10-D8) | Cm | 27; this study | |
| 1307− | 1307::Tn5 | UGS 25A7 (8S29-E5) | Km | 27; this study |
Catalog number of the strain in the lab's frozen cyanobacterial strain collection.
Plasmid numbers (e.g., pAM1573) refer to the catalog number of the strain in the lab's enterobacterial collection of plasmids. Vectors used to generate UGS mutants (e.g., 8S6-L5 for UGS mutant 14C3) were previously published in batch in the work of Chen et al. (27).
Antibiotic abbreviations are as follows: Cm, chloramphenicol; Km, kanamycin; Sm; streptomycin; Sp, spectinomycin.
As illustrated in Table 1, we herein abbreviate all S. elongatus genes by their four-digit Synpcc7942_ numbers (e.g., 1901 for Synpcc7942_1901). UGS-derived transposon-insertion knockout mutants are referenced as null for the gene product (e.g., 1901−). Expression strains that harbor an ORF expressed from the weak constitutive promoter Psc in neutral site 1 (NS1) are identified with a superscript “cx” after the ORF product (e.g., 1901cx), so that a complemented strain of the 1901 knockout is named 1901−+1901cx, while a 1901 knockout expressing a different ORF, such as 1902, is named 1901−+1902cx. The genotypes and complete segregation of strains were confirmed by standard colony or whole-cell PCR protocols using Taq, Phusion High-Fidelity, or Q5 DNA polymerases (NEB), as detailed in Fig. S1 to S3 in the supplemental material. Primers used for genotyping, segregation testing, cloning, and sequencing are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study for cloning, sequencing, and segregation analysis of genes of interest
| Name | Sequence (5′→3′)b | Description |
|---|---|---|
| LT 102Fa | AGTCGGCAAATAACCCTCGG | Forward sequencing primer for the pSyn_1/D-TOPO Life Technologies vector |
| LT 102Ra | CGTTTTATTTGATGCCTGGC | Reverse sequencing primer for the pSyn_1/D-TOPO Life Technologies vector |
| NS1-Fa | CGTCGAAGATGGAAAAGCTC | Forward primer for segregation analysis of the neutral site 1 locus |
| NS1-Ra | ATTGACCCGGTAGGGATTTC | Reverse primer for segregation analysis of the neutral site 1 locus |
| TOPO-1901F2 | caccATGCAGATTGGCATTATTGCTCCCAGCTC | Forward primer for cloning 1901 |
| TOPO-1901R2 | ctaTTATTTCAACAATTCTTTGAGGGCGATCGCGTAGTC | Reverse primer for cloning 1901 |
| TOPO-1902F | caccATGCGGATTGCGATCGCAACAG | Forward primer for cloning 1902 |
| TOPO-1902R | ttaCTACGCGAACGAAGAAGTTAAACGATTTACC | Reverse primer for cloning 1902 |
| TOPO-1904F | caccATGTCTATTTCATCGCCTACACCCAGTCC | Forward primer for cloning 1904 |
| TOPO-1904R | ttaCTAGCGAATTGTCTTGAGTGCATCACTACTACG | Reverse primer for cloning 1904 |
| TOPO-1905F2 | caccATGGTTGCTATTCCTTCGAACC | Forward primer for cloning 1905 and segregation analysis of the 1905 locus |
| TOPO-1905R2 | ctaTTAGTTCAAAGCATCTTGTTGACG | Reverse primer for cloning 1905 and segregation analysis of the 1905 locus |
| 1901genomeF | TTCAGCTGTGTAATTGTGGC | Forward primer for segregation analysis of the 1901 locus |
| 1901genomeR | CTCGCAAAGGTCTAAATTGC | Reverse primer for segregation analysis of the 1901 locus |
| 1902genomeF | GAGATAGGCTACGTTGATGC | Forward primer for segregation analysis of the 1902 locus |
| 1902genomeR | TAATGACACCCGCAAAGTTC | Reverse primer for segregation analysis of the 1902 locus |
| 1903genomeFa | TGATGCACTCAAGACAATTCG | Forward primer for segregation analysis of the 1903 locus |
| 1903genomeRa | ATGCAGTTCTGCACCTCCTC | Reverse primer for segregation analysis of the 1903 locus |
| 1904genomeF | ACGAGAATTCCTTTGCACTG | Forward primer for segregation analysis of the 1904 locus |
| 1904genomeR | TCAGTTAACGACTGTTGCTG | Reverse primer for segregation analysis of the 1904 locus |
| sp1901seq | TTTAGATAACGCCAAGCGAG | Sequencing primer for 1901 |
| sp1901seqR | GTGATACAGATGGCAGAGCC | Sequencing primer for 1901 |
| TOPO-2292F | caccATGATGCCATCTCCAGCCGGCTC | Forward primer for cloning 2292 |
| TOPO-2292R | tcattaTCAGGCGAGCGGAGAACTAGAGGTG | Reverse primer for cloning 2292 |
| TOPO-2293F | caccATGACTGTGCGATCGCGCTTTC | Forward primer for cloning 2293 |
| TOPO-2293R | ttaCTACTGTGCTGCTGGAGCTGACAG | Reverse primer for cloning 2293 |
| TOPO-2294F-2 | caccATGGCCGATCCCATTCG | Forward primer for segregation analysis of the 2294 locus |
| TOPO-2294R-2 | ttaTCAGACCTTGACCGGACG | Reverse primer for segregation analysis of the 2294 locus |
| TOPO-2295F-2 | caccATGCGGCGACTCATCGA | Forward primer for segregation analysis of the 2295 locus |
| TOPO-2295R-2 | ttaCTACAACCTTGGATTGCTAGCC | Reverse primer for segregation analysis of the 2295 locus |
| 2292genomeF | TTTAACTGTGCGGCAGTCCC | Forward primer for segregation analysis of the 2292 locus |
| 2292genomeR | ATCGGTTCTCGGTAGCGTTG | Reverse primer for segregation analysis of the 2292 locus |
| 2293genomeF | CTACCCACCTCTAGTTCTCC | Forward primer for segregation analysis of the 2293 locus |
| 2293genomeR | CTCCGGAACCAATGAAGATG | Reverse primer for segregation analysis of the 2293 locus |
| 2292seqF | TTGGTCTCGGACTACTCTGG | Sequencing primer for 2292 |
| TOPO-1342F | caccATGACCCGTAAGCGCGCC | Forward primer for cloning 1342 |
| TOPO-1342R | ttaTCACTCGAAGTAGTCGAAGGCC | Reverse primer for cloning 1342 |
| TOPO-2027F | caccGTGCCCAAGCTCTCTTTGATCATC | Forward primer for cloning 2027 |
| TOPO-2027R | ttaCTAAGCCAATCCTATGAGTTTACGGCG | Reverse primer for cloning 2027 |
| 1342genomeF | CCTGAAGGTTTGAGGAAGAG | Forward primer for segregation analysis of the 1342 locus |
| 1342genomeR | AGATTGGAAGTTGCTATCCAG | Reverse primer for segregation analysis of the 1342 locus |
| 2027genomeF | ACCTCTACTACATTGAGAACTGG | Forward primer for segregation analysis of the 2027 locus |
| 2027genomeR | AGATAGGGCAACAAACTGAG | Reverse primer for segregation analysis of the 2027 locus |
| TOPO-2098F | caccATGCAGATCCGCCACACCGC | Forward primer for segregation analysis of the 2098 locus |
| TOPO-2098R | ttaTCATATGAATACCTCCGCTTCCAAGAG | Reverse primer for segregation analysis of the 2098 locus |
| TOPO-2099F | caccATGAAGGTTTTACTGACAGGGGCTG | Forward primer for segregation analysis of the 2099 locus |
| TOPO-2099R | ttaTCATTGGTTCTTTGCAACTTGCGTC | Reverse primer for segregation analysis of the 2099 locus |
| TOPO-2100F | caccATGAAGATTTTGATTACGGGTGGTGCTG | Forward primer for segregation analysis of the 2100 locus |
| TOPO-2100R | ttaTCATACTGTGGTTTCCTGCTCCTGTG | Reverse primer for segregation analysis of the 2100 locus |
| TOPO-2101F | caccATGACCGAGGCACGGCGC | Forward primer for segregation analysis of the 2101 locus |
| TOPO-2101R | ttaTTAGAGAGGATTGTGCAAGAGGTCCAG | Reverse primer for segregation analysis of the 2101 locus |
| TOPO-0058F | caccATGCCAACTGAGTTACGAGCAACG | Forward primer for segregation analysis of the 0058 locus |
| TOPO-0058R | ttaTCATAATCCCGAGAGAAACGGTAAAGC | Reverse primer for segregation analysis of the 0058 locus |
| TOPO-0579F | caccATGCGCATCGCTCTCTTTACCG | Forward primer for segregation analysis of the 0579 locus |
| TOPO-0579R | ttaTCAGGCCGCTAAGGGTAAGC | Reverse primer for segregation analysis of the 0579 locus |
| TOPO-0948F | caccATGAAACCTCGATTCCGCTGG | Forward primer for segregation analysis of the 0948 locus |
| TOPO-0948R | ttaTTAGCCCTTCACTGCTCCGG | Reverse primer for segregation analysis of the 0948 locus |
| TOPO-0949F | caccATGACTCATCCGCCTCGTTGGC | Forward primer for segregation analysis of the 0949 locus |
| TOPO-0949R | ttaTCACAGTCCTCCCGACGGG | Reverse primer for segregation analysis of the 0949 locus |
| TOPO-0950F | caccATGCCGTTCTTGCGTTGTGG | Forward primer for segregation analysis of the 0950 locus |
| TOPO-0950R | ttaTCATGAGGCTGTTGCTCCTTGC | Reverse primer for segregation analysis of the 0950 locus |
| TOPO-0466F | caccATGACAAGGCCAATCAGCAGGC | Forward primer for segregation analysis of the 0466 locus |
| TOPO-0466R | ttaCTAGGAACGCTGCGGCACC | Reverse primer for segregation analysis of the 0466 locus |
| TOPO-0973F | caccTTGGCTGCTGGCGTCGC | Forward primer for segregation analysis of the 0973 locus |
| TOPO-0973R | ttaTTAGCGACCGATCCCGATGTAGC | Reverse primer for segregation analysis of the 0973 locus |
| TOPO-2151F | caccATGCAATTAAAACAACTGCGAAAACTTGCTCC | Forward primer for segregation analysis of the 2151 locus |
| TOPO-2151R | ttaTCAGCGGCTATTTGCCAAGGGATG | Reverse primer for segregation analysis of the 2151 locus |
| TOPO-1398F | caccATGCGTTTCCCCAACTTTCTACAGC | Forward primer for segregation analysis of the 1398 locus |
| TOPO-1398R | ttaTTAACCATTGTTGTAGCGCGTTAAAAGCG | Reverse primer for segregation analysis of the 1398 locus |
| TOPO-0220F | caccATGACTCAAATTGTTTCCGTACATTCGTTTCG | Forward primer for segregation analysis of the 0220 locus |
| TOPO-0220R | ttaCTAGTCACTGATCAAAGCATGCGCTAACT | Reverse primer for segregation analysis of the 0220 locus |
| TOPO-0133F | caccGTGCAGGAACTGCAAATGGCG | Forward primer for segregation analysis of the 0133 locus |
| TOPO-0133R | ttaCTATACGCAAGCGAGTACCTCACTCC | Reverse primer for segregation analysis of the 0133 locus |
| TOPO-0471F | caccGTGCGTCTCTCTGCTGGATTCCG | Forward primer for segregation analysis of the 0471 locus |
| TOPO-0471R | ttaTTATCCCTTCACACCACTGGCAGC | Reverse primer for segregation analysis of the 0471 locus |
| TOPO-1307F | caccATGAGTAGTCTCCTCGCTTCGACTG | Forward primer for segregation analysis of the 1307 locus |
| TOPO-1307R | ttaTTAGTCTTGCTGGCTGGCAAACTG | Reverse primer for segregation analysis of the 1307 locus |
| TOPO-0320F | caccGTGGCAGGGGCAACCATTCTG | Forward primer for segregation analysis of the 0320 locus |
| TOPO-0320R | ttaTCACGAGGGGCGATCGCA | Reverse primer for segregation analysis of the 0320 locus |
| TOPO-1608F | caccATGCTGGTTCCGGTCATCCTC | Forward primer for segregation analysis of the 1608 locus |
| TOPO-1608R | ttaTCAGCTACGACCGTAGTGGTCTTC | Reverse primer for segregation analysis of the 1608 locus |
| TOPO-2287F | caccATGAACCTGTCTCCGATCCGCC | Forward primer for segregation analysis of the 2287 locus |
| TOPO-2287R | ttaCTAAGGAGAGAACGACGGTTTTTTCCCG | Reverse primer for segregation analysis of the 2287 locus |
| TOPO-2290F | caccATGGTTCGAATTCTGGCAGTGATTCC | Forward primer for segregation analysis of the 2290 locus |
| TOPO-2290R | ctaTTAGCGTTGACTGGCCCATGCG | Reverse primer for segregation analysis of the 2290 locus |
| TOPO-0281F2 | caccATGACAGCCCCGGCTGCGCCTAC | Forward primer for segregation analysis of the 0281 locus |
| TOPO-0281R2 | tcattaTCAGGGAAGAGAACGGCGCGATCGCTG | Reverse primer for segregation analysis of the 0281 locus |
| TOPO-2025F | caccATGAGAGTTGCGATCGTTCACTATTGG | Forward primer for segregation analysis of the 2025 locus |
| TOPO-2025R | ttaCTAGAGCACCGACGTGAGGAAGC | Reverse primer for segregation analysis of the 2025 locus |
| TOPO-2028F | caccGTGGGCAATCTGTTAGTCAATTTGGCAATGG | Forward primer for segregation analysis of the 2028 locus |
| TOPO-2028R | tcattaCTAAAGAAATTTTTCGAGCACTTGGCAAGTTTCTTTACC | Reverse primer for segregation analysis of the 2028 locus |
| TOPO-0134F | caccTTGCGTATAGGGTCGATCCTGCG | Forward primer for segregation analysis of the 0134 locus |
| TOPO-0134R | ttaTCAGGCGCTTTGGGCCCG | Reverse primer for segregation analysis of the 0134 locus |
| TOPO-0357F | caccATGACTGTCTGGCAAACTCTGACTTTTGC | Forward primer for segregation analysis of the 0357 locus |
| TOPO-0357R | tcattaCTACATTTTTTCGTCTGAATGCTCGGCTTCTG | Reverse primer for segregation analysis of the 0357 locus |
| TOPO-0463F | caccATGACCGTCAAAGTACTGTTCGTCTGT | Forward primer for segregation analysis of the 0463 locus |
| TOPO-0463R | ttaTTAGCGGTGAGTTTTAATCAGTCCCTCTT | Reverse primer for segregation analysis of the 0463 locus |
| TOPO-2150F | caccATGACCAATACTCTCGGAATTGCTGCG | Forward primer for segregation analysis of the 2150 locus |
| TOPO-2150R | ttaTTATAGTCCAGCAAACTCGAATGGCTTGG | Reverse primer for segregation analysis of the 2150 locus |
| TOPO-1761F | caccGTGATCGGGAACAAGTGCAAATGTTG | Forward primer2 for segregation analysis of the 1761 locus |
| TOPO-1761R | ttaTCAGCCCCCCGCACGTG | Reverse primer for segregation analysis of the 1761 locus |
| TOPO-2148F | caccATGACGCTAGCCGTTCGTATTGAGC | Forward primer for segregation analysis of the 2148 locus |
| TOPO-2148R | ttaCTAACGTTGCATGGCGCGCTTTTTC | Reverse primer for segregation analysis of the 2148 locus |
| TOPO-2149F | caccATGACTACCACGCTACCGAAGTCTG | Forward primer for segregation analysis of the 2149 locus |
| TOPO-2149R | ttaCTAGCTTAGCGATCGCTTGAGGGC | Reverse primer for segregation analysis of the 2149 locus |
Originally published in the work of Simkovsky et al. (21).
Lowercase letters in the sequence indicate additional sequence that does not match the target genome or plasmid, such as the TOPO cloning tag (5′-cacc-3′) and additional stop codons.
Cyanobacterial cultures were grown in liquid or on solid BG-11 (31) or BG-11M medium (32) under 24-h constant light levels ranging from 200 to 350 μmol photons m−2 s−1 at 30°C. To prevent the appearance of secondary suppressor mutations in strains with growth defects, such as 1342− and 2027−, liquid cultures of these strains were grown under constant light at 70 μmol photons m−2 s−1. When appropriate, media were supplemented with chloramphenicol at 7.5 μg/ml, kanamycin at 5 μg/ml, spectinomycin at 2 μg/ml, or streptomycin at 2 μg/ml. Amoeba HGG1 was routinely maintained on lawns of WT S. elongatus by resuspending scrapings from the growing edge of a previously inoculated amoebal plaque plate in BG-11 medium and spotting ∼20,000 amoebae in 4 μl of liquid onto the center of a plate on which a lawn of WT S. elongatus had been grown at 30°C and 350 μmol photons m−2 s−1 for 7 days. After addition of amoebae, maintenance plates were incubated under ambient light at room temperature.
Plaque assays.
Unless otherwise noted, lawn plates were generated and plaque assays were performed as previously described (21). For experiments involving 1342− and 2027− strains, lawn plates were generated by using 1 ml (4×), 12.5 ml (50×), or 25 ml (100×) of late-log-phase cultures (approximate optical density [OD] at 750 nm of 0.5 to 0.7) concentrated to a final volume of 250 μl and spreading the concentrate onto BG-11 plates (1.5% [wt/vol] agar) containing appropriate antibiotics.
Purification and analysis of outer membranes.
Outer membrane purifications were performed by modification of the EDTA-sucrose method of Brahamsha (33) and Simkovsky et al. (21). Specifically, outer membrane preparations were made from 10 ml of 4- to 7-day-old cultures grown to an approximate OD at 750 nm of 0.5; samples were treated with 300 μl of stripping buffer, and the supernatants from stripped cells were not further purified by ultracentrifugation. Instead, samples were flash frozen with liquid nitrogen, concentrated in a CentriVap vacuum concentrator (Labconco) for 2 h at room temperature, resuspended vigorously in 1× SDS-PAGE sample buffer, boiled for 10 min, and digested with 30 μg of proteinase K (Sigma) for 1 h at 55°C. Samples were analyzed by SDS-PAGE with either a 10% (wt/vol) Tris-glycine SDS acrylamide gel with a 5% stacking layer or a 16% (wt/vol) Tris-Tricine SDS acrylamide gel with a 4% stacking layer. Gels were stained with the rapid, LPS-sensitive silver staining method of Fomsgaard et al. (34)
Nucleotide sugar analysis.
Nucleotides and nucleotide sugars were purified from cell pellets of 50 ml of 6-day-old late-log-phase cultures using the 10% trichloroacetic acid (TCA) and water-saturated diethyl ether methodology of Fairbanks et al. (35). Extracts were dissolved in Milli-Q water and analyzed by high-performance liquid chromatography (HPLC) and UV-visible (UV-Vis) absorbance on a Dionex DX600 HPLC (Sunnyvale, CA) equipped with a CarboPac PA1 column as described by Akizu et al. (36). For each sample, the peak that was eluted at the same retention time as a GDP-fucose standard was analyzed by liquid chromatography-mass spectrometry (LC-MS) using a LTQ-XL Orbitrap Discovery (Thermo Scientific) in negative ion electrospray ionization mode equipped with a porous graphitic carbon reversed-phase column.
Growth curve analysis.
The growth of flask cultures inoculated at an optical density of 0.02 at 750 nm was monitored using optical density and dry biomass measurements as previously described (21). Simultaneously, 1:5 serial dilutions of cultures were produced daily using BG-11 medium. Four microliters of each dilution was spotted onto BG-11 plates (1.5% [wt/vol] agar). Plates were grown under constant light levels ranging from 150 to 200 μmol photons m−2 s−1 at 30°C for 7 days prior to colony counting.
RESULTS
Screening of the UGS mutant library reveals novel rough mutants.
Our previous screen of the UGS collection revealed only a single grazer resistance gene, 1126, encoding an ABC transporter of O antigen. This single hit allowed us to bioinformatically identify three more O-antigen production genes whose inactivation confers grazer resistance (21). We reasoned that an alternative screen of the library and a broader bioinformatics analysis would reveal additional genes relevant for resistance. Because colonies of previous grazer-resistant mutants are identifiable via the visibly rough phenotype, we visually rescreened the library for mutants that display this rough appearance. This screen identified four new UGS mutants: 1901−, 1902−, 2292−, and 2293−. To confirm the rough phenotype and test for grazer resistance and impairment of O-antigen production, all mutants were regenerated from WT and appropriate constitutive expression strains were constructed to test for complementation. Because S. elongatus is oligoploid (37), complete segregation was confirmed to ensure homozygosity of the mutant alleles and expression constructs prior to phenotypic analysis (see Fig. S1 in the supplemental material).
1901 and 1902 are necessary for O-antigen synthesis and grazer susceptibility.
1901 and 1902 encode predicted glycosyltransferases. We hypothesized that these genes, which are downstream of the previously characterized wbdD O-antigen synthesis gene 1903 (Fig. 1A), are involved in the polymerization of O antigen and that their respective insertional knockout mutants, 1901− and 1902−, are resistant to amoebal grazing.
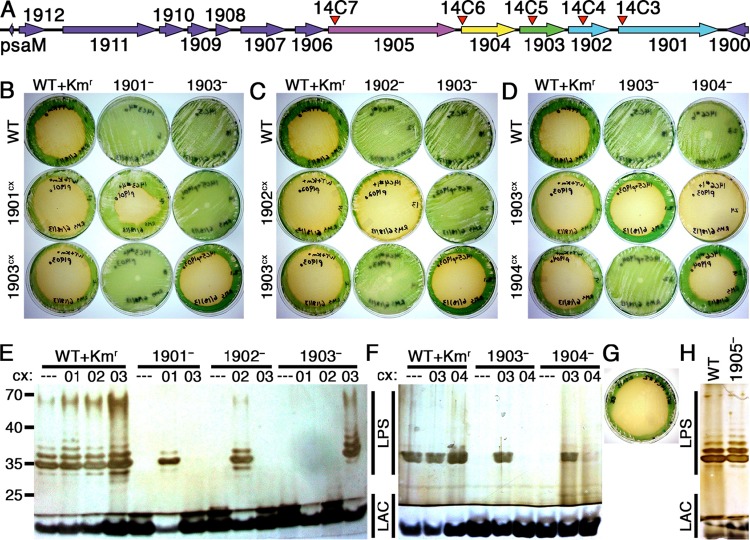
FIG 1.
Analysis of mutants defective for the loci from 1901 to 1905. (A) Diagram of the ORFs surrounding 1901 to 1905. The locations of transposon-insertion mutations 14C3 to 14C7 are indicated by red arrowheads. (B to D) Photographs of plaque plates taken 6 days following addition of amoebae to test the grazer resistance of 1901− (B), 1902− (C), 1904− (D), and related constitutive expression strains compared to WT+Kmr and grazer-resistant 1903− controls. The strain on each plate is identified by a combination of column and row labels. (E and F) Silver-stained Tris-glycine SDS gels of outer membrane preparations from 1901−, 1902−, 1903−, 1904−, and relevant control and expression strains. The sample loaded in each lane is identified by a combination of the parental strain on top and the constitutive expression (cx) construct below (---, WT NS1; 01, 1901cx; 02, 1902cx; 03, 1903cx; 04, 1904cx). (G) 1905− plaque plate. (H) Outer membrane analysis of 1905−. For all gels (E, F, and H), the regions representing banding patterns of LPS and lipid A-core (LAC) molecules are indicated and positions of molecular mass standards run on the gel (not shown) are noted on the left, in kilodaltons.
Amoeba plaque assays confirmed that 1901− and 1902− are resistant to grazing by HGG1 (Fig. 1B and C). Expression of the appropriate WT ORF from NS1 recovers amoeba susceptibility in the 1901−+1901cx and 1902−+1902cx strains, whereas constitutively expressing 1903 in strains 1901−+1903cx and 1902−+1903cx or expressing 1901 or 1902 in strains 1903−+1901cx and 1903−+1902cx does not recover amoebal susceptibility. SDS-PAGE analysis of outer membrane preparations demonstrated a lack of O antigen in 1901− and 1902− as previously observed in 1903− (Fig. 1E). O-antigen production was restored in mutants complemented with the appropriate WT ORF but not when an alternative ORF was expressed. Thus, each locus contributes individually to the resistance phenotype, and none of the resistance phenotypes results from a polar effect of mutation in an upstream locus.
Because 1901, 1902, and 1903 represent the end of a stretch of 12 ORFs on the same strand, with ORFs 1901 to 1907 being cotranscribed (38), we generated mutants defective for 1904 and 1905 to explore whether a larger operon beyond these three genes encodes O-antigen production or affects grazer resistance (Fig. 1A). 1904 encodes a predicted type I secretion adaptor protein between an ABC transporter and an outer membrane protein, and 1905 encodes a putative cyclic nucleotide-binding double-glycine protease and ABC transporter. The insertion in 1904 produced resistance, but susceptibility was restored by expression of either 1903 or 1904 from NS1 (Fig. 1D). Consistent with the amoebal grazing observations, 1904− lacks O antigen and recovers it upon expression of 1903 from NS1 in 1904−+1903cx (Fig. 1F). Interestingly, trans expression of 1904 only partially restores O-antigen production levels in 1904−+1904cx. Although these results suggest an unusual polar effect, we noted that heterologous expression of 1904 in the 1903−+1904cx strain did not complement the 1903− grazer resistance and O-antigen-defective phenotypes (Fig. 1D and F). We hypothesize that 1904− reduces 1903 protein activity levels through an uncharacterized transcriptional or posttranscriptional regulatory mechanism. Because 1905− did not result in grazer resistance (Fig. 1G) and did not alter O-antigen production (Fig. 1H), we did not pursue mutations in genes 1906 to 1912.
2292 and 2293 are necessary for O-antigen ligation and grazer susceptibility.
We previously proposed that 2292 encodes the WaaL O-antigen ligase (21). BLAST (39) analysis of the predicted 2293 protein sequence does not yield any similar known proteins; however, amino acid sequence analysis using CDD (40), PHOBIUS (41), and ScanProsite (42) predicts a single transmembrane helix from residues 20 to 38, a lipid attachment site at C31, and a domain of low similarity (E value, 1e−2) to pyruvate kinase at positions 54 to 88. Two genes exist downstream of these putative ligase genes: 2294, a homolog of the waaJ alpha-1,2-glucosyltransferase gene responsible for incorporating glucose into the outer core of lipid A, and 2295, which corresponds to a hypothetical secreted protein (Fig. 2A). To test all four genes for their potential roles in grazer resistance and LPS maturation, we generated insertional knockout mutants in each gene.
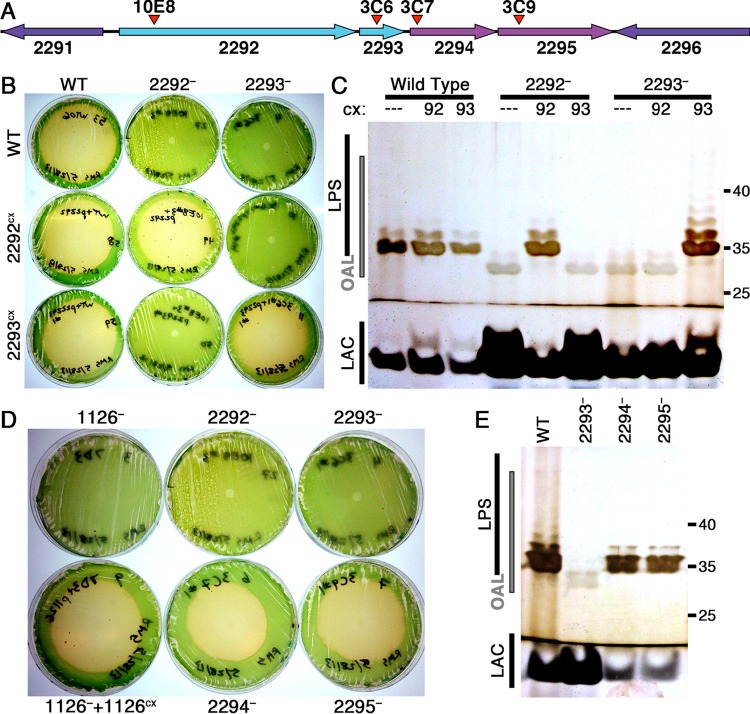
FIG 2.
Analysis of mutants defective for the loci from 2292 to 2295. (A) Diagram of the ORFs surrounding 2292 to 2295, with the locations of transposon-insertion mutations 10E8 and 3C6 to 3C9 indicated by red arrowheads. (B) Photographs of plaque plates taken 7 days following addition of amoebae on cultures of the WT, 2292−, 2293−, and related expression strains. The strain on each plate is identified by a combination of column and row labels as in Fig. 1. (C) Silver-stained Tris-glycine SDS gel of outer membrane preparations of the WT, 2292−, 2293−, and expression strains labeled as in Fig. 1, with the region containing bands representing O antigen attached to a lipid anchor (OAL) indicated on the left and molecular mass standards, in kilodaltons, on the right. ---, WT NS1; 92, 2292cx; 93, 2293cx. (D) Plaque plates of 3C7, 3C9, and controls photographed 7 days following amoeba addition. (E) Outer membrane analysis of 3C7 and 3C9, labeled as in panel C.
Mutations in 2292 and 2293 generated resistance to HGG1, whereas mutants defective for 2294 or 2295 are susceptible to amoebal grazing (Fig. 2B and D). Expression of each WT ORF from NS1 complemented the corresponding mutation (2292−+2292cx and 2293−+2293cx), but expression of 2292 did not complement 2293− and 2293 did not complement 2292−. This finding demonstrates that these genes encode functionally distinct proteins that are both necessary for maintaining grazer susceptibility.
The LPS ladder pattern of outer membrane preparations from 2292− and 2293− migrated faster on electrophoretic gels than did the WT (Fig. 2C), suggesting a significant mass decrease. Expression of 2292 from NS1 in 2292−+2292cx or 2293 in 2293−+2293cx restored the WT-like LPS band pattern, though expression of 2292 in 2293−+2292cx or 2293 in 2292−+2293cx did not. Rick et al. observed a similar ladder pattern shift in mutants of E. coli serotype O8 unable to ligate O antigen to LPS due to the inability to synthesize the core oligosaccharides that act as acceptors for the O-antigen ligase, and they interpreted the shifted band pattern as heterogeneous lengths of O-antigen molecules attached to the lipid carrier on which it was polymerized inside the cell (43).
To confirm this interpretation, Rick et al. subjected membrane preparations to mild treatment with either acid or alkali to separate sugar polymers from their lipid carriers (43). We performed these procedures for 2292− and 2293− to determine if the shifted bands break down into predictable components. Acid treatment did not alter the LPS ladder band of the WT, which is consistent with previous reports that, unlike with enteric LPS, mild acid hydrolysis does not release O antigen from cyanobacterial LPS derived from species closely related to S. elongatus PCC 7942 (9, 14). In addition, a slow-migrating band present in untreated samples runs at a similar position to the LPS-shifted band after either acid or alkali treatment and in the case of alkali treatment renders the results uninterpretable (see Fig. S4 in the supplemental material). Katz et al. reported observing contamination from glycogen-like material in gel filtration fractions of undegraded LPS from a strain nearly identical to S. elongatus PCC 7942 after exposure to strong acid hydrolysis (18). This explanation likely accounts for the interfering bands we observed.
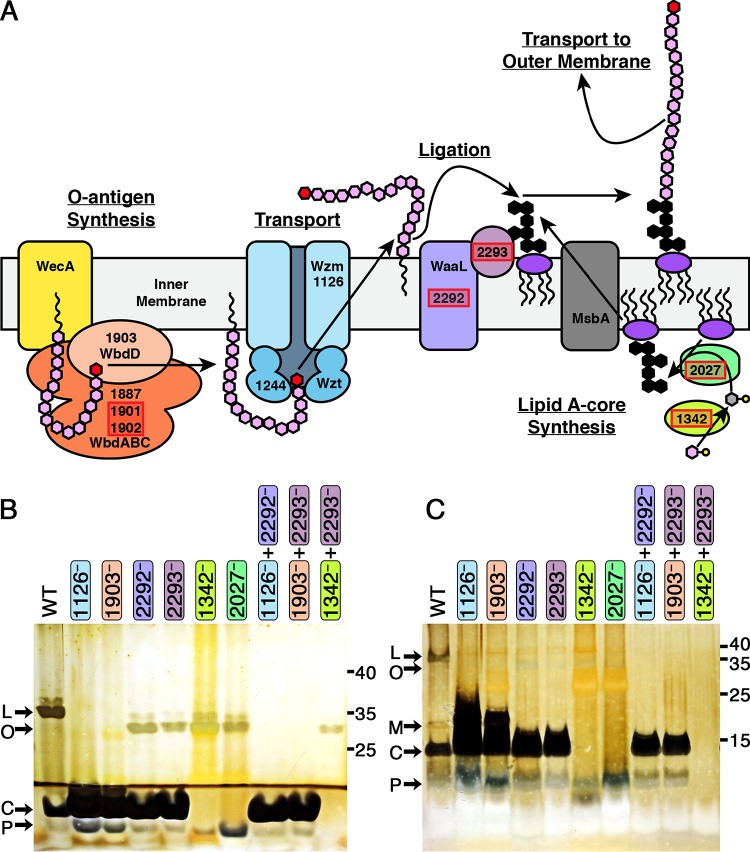
Although we could not identify the LPS-shifted bands biochemically, we observed that a second mutation in an O-antigen production gene in the double mutants 1126−+2292− and 1903−+2293− removed the shifted LPS ladder pattern without altering the lipid A-core conjugated molecule (lipid A-core) bands, compared to the results obtained with 2292− and 2293− (Fig. 3B and C). Because 1126− and 1903− are disrupted in the synthesis or transport of O antigen, these data support the interpretation of the shifted ladder pattern as O antigen attached to a lipid carrier; this interpretation supports the conclusions that 2292 encodes the WaaL O-antigen ligase and 2293 encodes a second protein product necessary for O-antigen ligation to lipid A-core that is functionally distinct from the 2292 ligase.
FIG 3.
Model of LPS synthesis in S. elongatus PCC 7942 and epistasis analysis of LPS mutants. (A) Diagram of the O-antigen transport and synthesis components previously discovered (21) along with the new components (highlighted in red boxes) described here for O-antigen synthesis, ligation, and lipid A-core synthesis. Sugars are represented by hexagons (pink, mannose; red, methyl-mannose; gray, fucose; black, lipid A-core, for which the precise structure and sugar composition are unknown). Nucleotide sugars are drawn as hexagons with an attached circle. The 2293 product is drawn to reflect its predicted location anchored to the inner membrane. (B and C) Outer membrane preparations were analyzed by silver staining Tris-glycine (B) or Tris-Tricine (C) SDS gels. The locations of primary bands representing lipopolysaccharide (L), O antigen attached to a lipid anchor (O), modified lipid A-core (M), lipid A-core (C), and lipid A suggested to be either with a partial core oligosaccharide or no core oligosaccharide (P) are indicated on the left, and molecular mass standards, in kilodaltons, are indicated on the right.
Mutations in 2294 and 2295 did not confer grazer resistance or detectably alter the LPS or lipid A-core bands (Fig. 2D and E). This result indicates that if 2294 incorporates sugars into the outer core of lipid A as predicted, then the absence of these sugars from the lipid A-core molecule in 2294− does not prevent O-antigen ligation or cause grazer resistance.
Bioinformatics identifies genes involved in sugar polymer production.
All data up to this point suggest that the synthesis and export of a complete LPS are necessary for amoebal predation of cyanobacteria. To test the hypothesis that alterations in the sugar composition of the O-antigen or the lipid A-core moiety would confer grazer resistance, we identified genes necessary for the generation and polymerization of these polysaccharides. Bioinformatics analysis using BLAST, the PFAM protein database (44), and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (45) identified S. elongatus homologs of genes encoding nucleotide sugar synthetases and glycosyltransferases (46, 47), including those responsible for activating and polymerizing glucose, rhamnose, mannose, fucose, or 3-deoxy-d-manno-oct-2-ulosonic acid (KDO) (see Fig. S5A in the supplemental material).
To test the role that these proteins play in LPS synthesis and grazer resistance, we attempted to generate transposon-insertion mutants impaired in the production, polymerization, or incorporation of these nucleotide sugars. Table S1 and Fig. S5A in the supplemental material summarize the mutants that we attempted to generate, whether fully segregated mutant clones were obtained, and the associated phenotypes of these mutants. Of particular note, we were unable to generate UGS insertional mutants predicted to impair the synthesis or activation of KDO, galactose, or mannose, suggesting that these nucleotide sugars or their downstream products are essential for cell viability. In contrast, we successfully generated mutants defective for the predicted nucleotide activation enzymes for glucose, rhamnose, fucose, or glucuronate. Successfully segregated insertional mutants were tested for resistance to grazing and for LPS alterations (Fig. S5B and C). Of these mutants, only 1342−, defective in the putative GDP-mannose 4,6-dehydratase, and 2027−, impaired in a putative glycosyltransferase, were resistant to HGG1 and displayed alterations in the LPS banding pattern.
1342− and 2027− are impaired in lipid A-core maturation, provide grazer resistance, and hinder growth.
Expression of the appropriate WT ORF from NS1 completely restored amoebal grazing for both 1342−+1342cx and 2027−+2027cx (Fig. 4C and D), in contrast to the amoeba resistance of 1342− and 2027− (see Fig. S5B and C). LPS analyses of these mutants revealed that both 1342− and 2027− produced O-antigen lipid bands similar to those observed in the ligation mutants, 2292− and 2293−, and lacked bands representative of lipid A-core (Fig. 4E and F). In the 1342−+1342cx and 2027−+2027cx strains, the LPS and lipid A-core defects were both complemented so that these bands appear identical to those observed in the WT. These results suggest that these mutants prevent the ligation of O antigen to the lipid A core oligosaccharide through defects in the synthesis of the lipid A-core molecule.
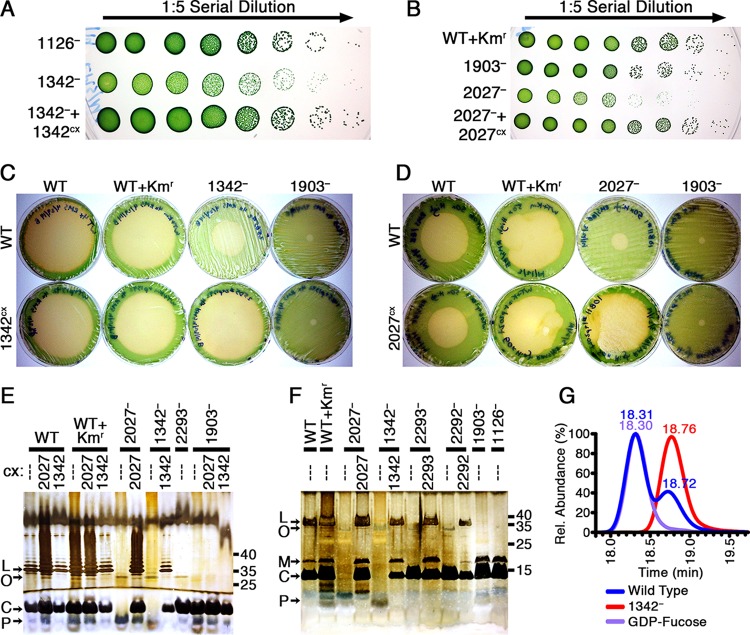
FIG 4.
Analysis of mutants in the 1342 and 2027 loci. (A and B) Serial dilution (1:5) spot plates of 8-day-old cultures of 1342− (A) and 2027− (B) strains, with increasing dilutions plated from left to right so that individual colonies are visible on the right. Plates were photographed after 7 days of growth. (C and D) Photographs of plaque plates taken 6 days following addition of amoebae to investigate the grazer resistance of 1342− (C), 2027− (D), and related expression strains compared to the WT, WT+Kmr, and 1903− controls. Plates in panel C were generated using 4× cell concentrates, whereas plates in panel D were made with 50× cell concentrates, as described in Materials and Methods. The strain on each plate is identified by a combination of column and row labels as in Fig. 1. (E and F) Outer membrane preparations were analyzed by silver staining Tris-glycine (E) and Tris-Tricine (F) SDS gels, labeled as in Fig. 3. The sample loaded in each lane is identified by a combination of the parental strain on top and the constitutive expression construct below it. (G) LC-MS chromatograms for samples of the WT and 1342− of HPLC fractions eluted at the same time as a GDP-fucose standard.
To test the prediction that 1342 encodes GDP-mannose 4,6-dehydratase, which produces GDP-fucose from GDP-mannose, we confirmed the presence of GDP-fucose in the WT and the absence of GDP-fucose in 1342− through UV-HPLC and LC-MS analysis of cell lysates (Fig. 4G). Because fucose is a known component of S. elongatus LPS (14, 18) and the lack of GDP-fucose in 1342− does not alter the O-antigen–lipid band pattern relative to that observed in 2292− and 2293−, we propose that the lipid A-core band defect of 1342− is due to the incomplete synthesis of the core oligosaccharide. Because the banding patterns observed with 2027− were nearly identical to those of 1342−, we propose that the product of 2027 is a glycosyltransferase that incorporates sugars, possibly fucose, into the lipid A core oligosaccharide.
The 1342− and 2027− mutants showed a striking growth defect, particularly at higher light intensities. We examined the growth kinetics of these strains, as well as strains expressing the respective WT ORFs from NS1 (1342−+1342cx and 2027−+2027cx), through serial dilution spot plates and growth curve analysis using optical densitometry, dry biomass, and counting CFU. The growth defect is most readily observable in the small size of 1342− and 2027− colonies (Fig. 4A and B) and in these strains' significantly reduced CFU counts (see Fig. S6 in the supplemental material) compared to the WT-like colony sizes and growth of the complemented strains 1342−+1342cx and 2027−+2027cx. We also observed the spontaneous appearance of WT-sized colonies from 1342− and 2027− cultures, which are presumed to result from spontaneous suppressor mutations (see Fig. S7C in the supplemental material).
Coincident with the appearance of WT-sized colonies, repetition of the amoeba resistance assay demonstrated that grazer resistance in the original mutants 1342− and 2027− was no longer complete, but rather the amoeba plaque grew to a reduced diameter compared to those of the WT and complemented strains (Fig. 4C and D). We have observed that the grazer resistance associated with previously published strains, such as 1126− and 1903−, can be reduced in a similar manner via a number of mechanisms: through mixing with small amounts of WT on the same plate, as a consequence of incomplete segregation of the mutant chromosome, or when lawn plates have low cell densities so that incomplete or patchy lawns are formed. We hypothesize that the impaired growth rate or spontaneous appearance of WT-like colonies in 1342− and 2027− cultures reduces the strength of their grazer resistance phenotype by reducing the cellular densities of the plate or mimicking the presence of WT cells, respectively. To test the plating density hypothesis, we generated plates with increasing concentrations of cyanobacterial cultures (see Fig. S7A and B). Plaques grew to reduced diameters with increasing cyanobacterial cell densities of 1342− and 2027−, well beyond that observed with the WT, indicating that the growth defect hinders but does not abolish the grazer resistance phenotype.
Epistasis confirms the pathway of LPS production.
To further confirm the hypotheses that 2292− and 2293− lack ligase activities while 1342− and 2027− are impaired in the synthesis of the lipid A core oligosaccharide, we generated a double mutant in lipid A core oligosaccharide synthesis and ligation (1342−+2293−) in addition to the 1126−+2292− and 1903−+2293− double mutants previously described. Although we attempted to generate strains defective in both O-antigen and lipid A-core syntheses (e.g., 1126−+1342−), we were never able to recover colonies from these transformations, suggesting that a lack of both O antigen and lipid A-core is synthetically lethal. In contrast to the lack of LPS-shifted bands in 1126−+2292− and 1903−+2293− outer membrane preparations, 1342−+2293− still retains the mass-shifted banding pattern but lacks all lipid A-core bands. This result supports the interpretation that this mass-shifted band is unligated O antigen and that O-antigen synthesis occurs prior to ligation.
Detailed analysis of the lipid A-related bands using a Tris-Tricine gel revealed that 1126− and 1903− produce a large quantity of a band with a higher apparent molecular weight than the lipid A-core band of the WT. This band is absent from the 2292− and 2293− single mutants and all of the double mutants (Fig. 3C). The identity of this band is unclear at this time, but it likely represents a modification of the lipid A-core molecule that depends upon the functionality of both ligation genes. Interestingly, 1342−+2293− lacks all bands in this region, while 1342−, 2027−, and all other strains retain a band that migrates faster than the lipid A-core band. We hypothesize that this apparently low-molecular-weight band represents either lipid A-core with an incomplete core oligosaccharide or lipid A without any core oligosaccharide.
DISCUSSION
Novel resistance genes enhance our model of LPS generation in S. elongatus.
Prior to this work, we predicted an initial model of the S. elongatus O-antigen synthesis pathway based on the four previously characterized amoeba-resistant mutants (21) and the known pathway in E. coli serotypes O8 and O9 (4). The discovery of six more mutants that disable O-antigen synthesis, nucleotide sugar production, lipid A-core synthesis, or ligation has refined the current working model for LPS synthesis in S. elongatus (Fig. 3A). For example, we previously predicted that two genes not discussed here, 0388 and 2028, encode the remaining Wbd O-antigen polymerases. However, the current work demonstrates that the protein products of two other genes, 1901 and 1902, function in the synthesis of the O antigen. In contrast, it is unlikely that 2028 contributes to the nonessential process of O-antigen polymerization because the 2028− mutant could not be obtained, suggesting that 2028 performs an essential function. In support of this conclusion, the entire operon containing 2028, 2027, and the genes for two other putative glycosyltransferases (2025 and 2026) has recently been identified as essential in S. elongatus via random barcode transposon site sequencing (RB-TnSeq) (48). Although we have demonstrated that 2027 is not essential, it is reasonable that growth-defective mutants such as 2027− would not survive the pooled, competitive growth required for RB-TnSeq. Based on the 2027− data presented here and the inability to generate 2028− and 2025−, we hypothesize that all four cotranscribed genes (38) contribute to the synthesis of the lipid A core oligosaccharide, with different gene mutations resulting in altered lipid A-core structures with various degrees of deleteriousness to cell viability.
2293 is a novel gene necessary for O-antigen ligation.
While our previous prediction that 2292 encodes an O-antigen ligase was confirmed, the discovery that 2293 encodes a necessary and functionally distinct second O-antigen ligase is unprecedented among all currently known LPS synthesis pathways. Although investigations on Salmonella enterica by Kaniuk et al. indicated that more components may be necessary for substrate recognition of the WaaL ligase (49), no other proteins essential to ligation were discovered. Given the uniqueness of this protein to S. elongatus, its precise function and evolutionary origins are unclear. Based on sequence analysis, we predict that it is anchored in the inner membrane by a single transmembrane helix and a covalently attached lipid. The low similarity to pyruvate kinase, a phosphotransferase, suggests that this enzyme is involved in the transfer of phosphates to or from the O-antigen-lipid carrier molecule as part of the ligation process. Alternatively, this enzyme may phosphorylate lipid A-core inside the cell, as occurs in the lipid A of other bacteria, including Pseudomonas aeruginosa, where phosphorylation of lipid A-core is required for LPS transport to the outer membrane (50). The presence of phosphorus has already been demonstrated in the LPS of Anacystis nidulans KM (18), a strain since considered S. elongatus.
Structure of LPS of S. elongatus.
The current model for LPS synthesis is consistent with the sugar composition of A. nidulans KM and S. elongatus PCC 6301, PCC 6908, PCC 6311, and PCC 6910, which comprise mannose with trace amounts of methyl-mannose and minor amounts of fucose, glucose, galactose, KDO, glucosamine, and mannosamine (14, 18). In analogy to the LPS synthesis pathway found in E. coli serotypes O8 and O9a (4), we propose that polymerization of the polymannose O antigen by 1887, 1901, and 1902 is terminated by methylation of the terminal mannose by 1903. The remaining sugars identified in the composition of the LPS are most likely components of the lipid A-core molecule. Our work indicates that fucose is a component of the lipid A core oligosaccharide required for O-antigen ligation. Although our failure to impair KDO synthesis and nucleotide activation indicates that this sugar may be an essential component of the lipid A-core structure, the inability to degrade S. elongatus LPS through mild acid hydrolysis suggests that it does not perform the same role as in enteric LPS (14, 18). These inferences are consistent with the observation that strong acid hydrolysis liberates fucose due to an acid-labile linkage and releases a single major fraction that lacks lipid A, fatty acids, fucose, and KDO but contains all of the other sugars, in ratios similar to those detected in S. elongatus LPS (18), suggesting that fucose may serve as a key link between O antigen and lipid A.
Knockouts in the pathway for rhamnose activation did not alter the LPS, which is consistent with rhamnose not being detected in the LPS of A. nidulans KM. This finding is in contrast to a number of LPS structures from marine Synechococcus strains, which incorporate rhamnose into their lipid A core oligosaccharides, do not utilize fucose or KDO, and have O antigens primarily composed of glucose (9).
Genes involved in LPS synthesis represent conserved targets for generating predator resistance.
Except for 2292 and 2293, whose translations showed poor similarity to known proteins via BLAST searches, the LPS synthesis genes that confer resistance to HGG1 are well conserved across the vast diversity of sequenced cyanobacteria (see Fig. S8 in the supplemental material). Notably, the marine Synechococcus and Prochlorococcus species do not have detectable homologs of 1126, encoding the Wzm ABC transporter of O antigen. Whether this difference is due to an evolutionary adaptation to marine salinity, a general lack of sequence conservation in the transmembrane domains of ABC transporters (51), or the different sugar composition of the LPS of marine species (9) is unclear. Overall, these conservation data support the hypothesis that the cyanobacteria evolved to use the Wzm-Wzt pathway for O-antigen synthesis and the Wzx-Wzy flippase mechanism for generating exopolysaccharides (52, 53). This strategy is in contrast to that of enteric bacteria, where the Wzx-Wzy mechanism is the dominant pathway for both O-antigen (2) and extracellular polysaccharide (54) syntheses.
The overall conservation makes these genes promising loci for generating grazer resistance through targeted markerless deletion (29) or random mutagenesis and screening in any potential cyanobacterial production strain for large-scale cultivation purposes. We previously generated nontransgenic grazer-resistant strains by UV mutagenesis that would not be considered novel under the regulations of the Environmental Protection Agency (21).
Tolerance limitations of lipid A-core production.
Although these amoebal predation resistance genes are useful mutational targets for crop protection, limitations exist on how much the LPS can be altered without affecting the viability and growth of the cell. At a minimum, impairment of lipid A-core resulted in impaired growth, which would be undesirable for industrial-scale biomass cultivation. Therefore, mutations in nucleotide sugar and lipid A-core synthesis homologs of 1342 and 2027 are not candidate loci for generating resistance to predation. The inability to generate a number of other mutations in lipid A-core or nucleotide sugar synthesis further demonstrates that alterations in these molecules can reduce cell viability. While this limitation decreases the number of target genes, these data help define the mutagenesis scheme for producing crop protection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nicholas Bearmar, Karen Tang, and Joshua Kenchel for assistance with strain preparation and plaque assays. We thank Life Technologies for donation of the pSyn_1/D-TOPO vector. Nucleotide sugar analysis was performed by Jeremy Van Vleet and Biswa Choudhury at the UCSD Glycotechnology Core.
This work was supported by grants from the U.S. Department of Energy (DOE grant DE-EE-0003373 to the Consortium for Algal Biofuels Commercialization), California Energy Commission (CEC Grant 500-10-039) to the San Diego Center for Algae Biotechnology, and Extremadura Government and European Regional Development Fund (ERDF grant GR10107 to research group CTS020).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00135-16.
REFERENCES
- 1.Seeger EM, Thuma M, Fernandez-Moreira E, Jacobs E, Schmitz M, Helbig JH. 2010. Lipopolysaccharide of Legionella pneumophila shed in a liquid culture as a nonvesicular fraction arrests phagosome maturation in amoeba and monocytic host cells. FEMS Microbiol Lett 307:113–119. doi: 10.1111/j.1574-6968.2010.01976.x. [DOI] [PubMed] [Google Scholar]
- 2.Raetz CR, Whitfield C. 2002. Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banemann A, Deppisch H, Gross R. 1998. The lipopolysaccharide of Bordetella bronchiseptica acts as a protective shield against antimicrobial peptides. Infect Immun 66:5607–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenfield LK, Whitfield C. 2012. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr Res 356:12–24. doi: 10.1016/j.carres.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 5.Lambris JD, Ricklin D, Geisbrecht BV. 2008. Complement evasion by human pathogens. Nat Rev Microbiol 6:132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgenstein RM, Clemmer KM, Rather PN. 2010. Loss of the WaaL O-antigen ligase prevents surface activation of the flagellar gene cascade in Proteus mirabilis. J Bacteriol 192:3213–3221. doi: 10.1128/JB.00196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoiczyk E, Hansel A. 2000. Cyanobacterial cell walls: news from an unusual prokaryotic envelope. J Bacteriol 182:1191–1199. doi: 10.1128/JB.182.5.1191-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder DS, Brahamsha B, Azadi P, Palenik B. 2009. Structure of compositionally simple lipopolysaccharide from marine Synechococcus. J Bacteriol 191:5499–5509. doi: 10.1128/JB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weckesser J, Drews G, Mayer H. 1979. Lipopolysaccharides of photosynthetic prokaryotes. Annu Rev Microbiol 33:215–239. doi: 10.1146/annurev.mi.33.100179.001243. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Qin S, Zhao F, Chi X, Zhang X. 2007. Comparison of envelope-related genes in unicellular and filamentous cyanobacteria. Comp Funct Genomics doi: 10.1155/2007/25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carillo S, Pieretti G, Bedini E, Parrilli M, Lanzetta R, Corsaro MM. 2014. Structural investigation of the antagonist LPS from the cyanobacterium Oscillatoria planktothrix FP1. Carbohydr Res 388:73–80. doi: 10.1016/j.carres.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt W, Drews G, Weckesser J, Mayer H. 1980. Lipopolysaccharides in four strains of the unicellular cyanobacterium Synechocystis. Arch Microbiol 127:217–222. doi: 10.1007/BF00427196. [DOI] [Google Scholar]
- 14.Schmidt W, Drews G, Weckesser J, Fromme I, Borowiak D. 1980. Characterization of the lipopolysaccharides from eight strains of the cyanobacterium Synechococcus. Arch Microbiol 127:209–215. doi: 10.1007/BF00427195. [DOI] [Google Scholar]
- 15.Weckesser J, Katz A, Drews G, Mayer H, Fromme I. 1974. Lipopolysaccharide containing l-acofriose in the filamentous blue-green alga Anabaena variabilis. J Bacteriol 120:672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keleti G, Sykora JL. 1982. Production and properties of cyanobacterial endotoxins. Appl Environ Microbiol 43:104–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii M, Sato Y, Ito H, Masago Y, Omura T. 2012. Monosaccharide composition of the outer membrane lipopolysaccharide and O-chain from the freshwater cyanobacterium Microcystis aeruginosa NIES-87. J Appl Microbiol 113:896–903. doi: 10.1111/j.1365-2672.2012.05405.x. [DOI] [PubMed] [Google Scholar]
- 18.Katz A, Weckesser J, Drews G, Mayer H. 1977. Chemical and biological studies on the lipopolysaccharide (O-antigen) of Anacystis nidulans. Arch Microbiol 113:247–256. doi: 10.1007/BF00492032. [DOI] [PubMed] [Google Scholar]
- 19.Opiyo SO, Pardy RL, Moriyama H, Moriyama EN. 2010. Evolution of the Kdo2-lipid A biosynthesis in bacteria. BMC Evol Biol 10:362. doi: 10.1186/1471-2148-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicolaisen K, Mariscal V, Bredemeier R, Pernil R, Moslavac S, Lopez-Igual R, Maldener I, Herrero A, Schleiff E, Flores E. 2009. The outer membrane of a heterocyst-forming cyanobacterium is a permeability barrier for uptake of metabolites that are exchanged between cells. Mol Microbiol 74:58–70. doi: 10.1111/j.1365-2958.2009.06850.x. [DOI] [PubMed] [Google Scholar]
- 21.Simkovsky R, Daniels EF, Tang K, Huynh SC, Golden SS, Brahamsha B. 2012. Impairment of O-antigen production confers resistance to grazing in a model amoeba-cyanobacterium predator-prey system. Proc Natl Acad Sci U S A 109:16678–16683. doi: 10.1073/pnas.1214904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X, Khudyakov I, Wolk CP. 1997. Lipopolysaccharide dependence of cyanophage sensitivity and aerobic nitrogen fixation in Anabaena sp. strain PCC 7120. J Bacteriol 179:2884–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones CS, Mayfield SP. 2012. Algae biofuels: versatility for the future of bioenergy. Curr Opin Biotechnol 23:346–351. doi: 10.1016/j.copbio.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Bláha L, Babica P, Marsalek B. 2009. Toxins produced in cyanobacterial water blooms—toxicity and risks. Interdiscip Toxicol 2:36–41. doi: 10.2478/v10102-009-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paerl HW, Otten TG. 2013. Harmful cyanobacterial blooms: causes, consequences, and controls. Microb Ecol 65:995–1010. doi: 10.1007/s00248-012-0159-y. [DOI] [PubMed] [Google Scholar]
- 26.Stewart I, Schluter PJ, Shaw GR. 2006. Cyanobacterial lipopolysaccharides and human health—a review. Environ Health 5:7. doi: 10.1186/1476-069X-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, Holtman CK, Taton A, Golden SS. 2012. Functional analysis of the Synechococcus elongatus PCC 7942 genome—functional genomics and evolution of photosynthetic systems. Adv Photosynth Respir 33:119–137. [Google Scholar]
- 28.Holtman CK, Chen Y, Sandoval P, Gonzales A, Nalty MS, Thomas TL, Youderian P, Golden SS. 2005. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res 12:103–115. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- 29.Andersson CR, Tsinoremas NF, Shelton J, Lebedeva NV, Yarrow J, Min H, Golden SS. 2000. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol 305:527–542. doi: 10.1016/S0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 30.Clerico EM, Ditty JL, Golden SS. 2007. Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol Biol 362:155–171. doi: 10.1007/978-1-59745-257-1_11. [DOI] [PubMed] [Google Scholar]
- 31.Allen MM. 1968. Simple conditions for growth of unicellular blue-green algae on plates. J Phycol 4:1–4. [DOI] [PubMed] [Google Scholar]
- 32.Bustos SA, Golden SS. 1991. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J Bacteriol 173:7525–7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brahamsha B. 1996. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc Natl Acad Sci U S A 93:6504–6509. doi: 10.1073/pnas.93.13.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fomsgaard A, Freudenberg MA, Galanos C. 1990. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J Clin Microbiol 28:2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fairbanks LD, Jacomelli G, Micheli V, Slade T, Simmonds HA. 2002. Severe pyridine nucleotide depletion in fibroblasts from Lesch-Nyhan patients. Biochem J 366:265–272. doi: 10.1042/bj20020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akizu N, Cantagrel V, Schroth J, Cai N, Vaux K, McCloskey D, Naviaux RK, Van Vleet J, Fenstermaker AG, Silhavy JL, Scheliga JS, Toyama K, Morisaki H, Sonmez FM, Celep F, Oraby A, Zaki MS, Al-Baradie R, Faqeih EA, Saleh MA, Spencer E, Rosti RO, Scott E, Nickerson E, Gabriel S, Morisaki T, Holmes EW, Gleeson JG. 2013. AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell 154:505–517. doi: 10.1016/j.cell.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griese M, Lange C, Soppa J. 2011. Ploidy in cyanobacteria. FEMS Microbiol Lett 323:124–131. doi: 10.1111/j.1574-6968.2011.02368.x. [DOI] [PubMed] [Google Scholar]
- 38.Vijayan V, Jain IH, O'Shea EK. 2011. A high resolution map of a cyanobacterial transcriptome. Genome Biol 12:R47. doi: 10.1186/gb-2011-12-5-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Käll L, Krogh A, Sonnhammer EL. 2007. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res 35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rick PD, Hubbard GL, Barr K. 1994. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J Bacteriol 176:2877–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehm BH. 2010. Bacterial polymers: biosynthesis, modifications and applications. Nat Rev Microbiol 8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 47.Samuel G, Reeves P. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr Res 338:2503–2519. doi: 10.1016/j.carres.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, Lowe LC, Curtin G, Arkin AP, Deutschbauer A, Golden SS. 2015. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci U S A 112:E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaniuk NA, Vinogradov E, Whitfield C. 2004. Investigation of the structural requirements in the lipopolysaccharide core acceptor for ligation of O antigens in the genus Salmonella: WaaL “ligase” is not the sole determinant of acceptor specificity. J Biol Chem 279:36470–36480. doi: 10.1074/jbc.M401366200. [DOI] [PubMed] [Google Scholar]
- 50.DeLucia AM, Six DA, Caughlan RE, Gee P, Hunt I, Lam JS, Dean CR. 2011. Lipopolysaccharide (LPS) inner-core phosphates are required for complete LPS synthesis and transport to the outer membrane in Pseudomonas aeruginosa PAO1. mBio 2(4):e00142-11. doi: 10.1128/mbio.00142-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilkens S. 2015. Structure and mechanism of ABC transporters. F1000Prime Rep 7:14. doi: 10.12703/P7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pereira S, Zille A, Micheletti E, Moradas-Ferreira P, De Philippis R, Tamagnini P. 2009. Complexity of cyanobacterial exopolysaccharides: composition, structures, inducing factors and putative genes involved in their biosynthesis and assembly. FEMS Microbiol Rev 33:917–941. doi: 10.1111/j.1574-6976.2009.00183.x. [DOI] [PubMed] [Google Scholar]
- 53.Pereira SB, Mota R, Vieira CP, Vieira J, Tamagnini P. 2015. Phylum-wide analysis of genes/proteins related to the last steps of assembly and export of extracellular polymeric substances (EPS) in cyanobacteria. Sci Rep 5:14835. doi: 10.1038/srep14835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. 2009. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev 73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.