Abstract
In Drosophila, molecular clocks control circadian rhythmic behavior through a network of ~150 pacemaker neurons. To explain how the network’s neuronal properties encode time, we performed brain-wide calcium imaging of groups of pacemaker neurons in vivo for 24 hours. Pacemakers exhibited daily rhythmic changes in intracellular Ca2+ that were entrained by environmental cues and timed by molecular clocks. However, these rhythms were not synchronous, as each group exhibited its own phase of activation. Ca2+ rhythms displayed by pacemaker groups that were associated with the morning or evening locomotor activities occurred ~4 hours before their respective behaviors. Loss of receptor for neuropeptide PDF promoted synchrony of Ca2+ waves. Thus neuropeptide modulation is required to sequentially time outputs from a network of synchronous molecular pacemakers.
Circadian clocks help animals adapt their physiology and behavior to local time. The clocks require a highly-conserved set of genes and proteins (1) operating through molecular feedback loops to generate robust rhythms that produce a 24 hour timing signal (2). These clocks are expressed by pacemaker neurons which themselves are assembled into an interactive network (3). Through network encoding and cellular interactions, pacemaker neurons in the suprachiasmatic nucleus (SCN) of the mammalian brain coordinate many circadian rhythmic outputs (4–7). To study how molecular clocks couple to network encoding, and how network encoding relates to specific behavioral outputs, we conducted an in vivo brain-wide analysis of the circadian pacemaker network in Drosophila across an entire 24 hour day.
This network contains ~150 synchronized pacemaker neurons (8,9) (Fig S1) yet it produces biphasic behavioral outputs – the morning and evening peaks of locomotor activity (Fig. 1A). The molecular clocks are entrained by environmental cues and by network interactions, for example by release of the neuropeptide pigment-dispersing factor (PDF) (10). Genetic mosaic studies indicate that morning and evening peaks of locomotor activity are controlled by distinct pacemaker groups (11–14) (Fig. 1B). We reasoned that: (i) synchronous signals from the pacemaker network might diverge in downstream circuits or (ii) the pacemaker network might itself generate different timing signals to downstream circuits. To explore this, we developed an in vivo imaging assay to monitor the intracellular Ca2+ concentration ([Ca2+]i), in pacemaker cell bodies over a ~24 hour period (Fig. 1C and methods). Intracellular Ca2+ dynamics directly reflect amounts of neuronal activity and Ca2+ imaging allows monitoring activity across neuronal ensembles (15).
Fig. 1. Ca2+ activity patterns in circadian pacemaker neurons in vivo.
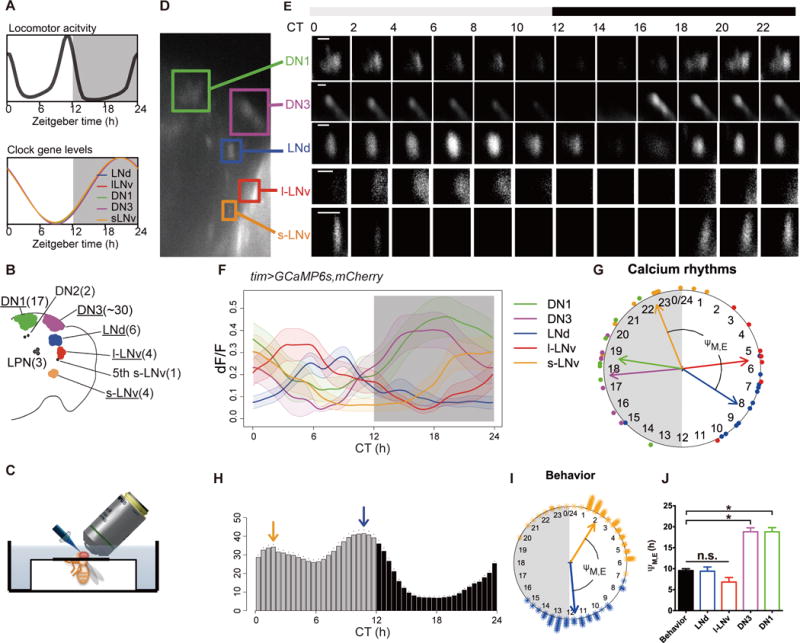
(A) Schematic representations of bimodal behavioral rhythms (top) that are driven by a pacemaker network that display synchronous, unimodal molecular clocks. (B) Map of the eight major clock pacemaker groups in the fly brain. Numbers in parentheses indicate the cell number per group. (C) Schematic to illustrate method for long-term in vivo imaging; the head is immersed in saline while the body remains in an air-filled enclosure (see Methods for details). (D) A representative image of tim>GCaMP6s signals showing the locations of five identifiable pacemaker groups. (E) Representative images showing 24-h Ca2+ activity patterns of five identifiable groups. (F) Average Ca2+ transients in the five pacemaker groups as a function of Circadian Time (n=13 flies). Gray aspect indicates the period of lights-off during the preceding six days of 12hour:12hour photoentrainment. (G) Phase distributions of 24 hour Ca2+ transients in different pacemaker groups (data from G). Each colored dot outside of the clockface represents the calculated peak phase of one group in one fly as described in Methods. Colored arrows are mean vectors for the different clock neuron groups. The arrow magnitude describes the phase coherence of Ca2+ transients in a specific pacemaker group among different flies (n=13, not all 5 groups were visible in each fly due to the size of the cranial windows – see Table S1). ΨM, E is the phase difference between M cells (s-LNv) and E cells (LNd). (H) The average activity histogram of tim>GCaMP6s,mCherry.NLS flies in the first day under constant darkness (DD1). Arrows indicate behavioral peak phases (orange: morning, blue: evening). Dots indicate SEM (n=47 flies). (I) Phase distributions of behavioral peaks indicated by arrows in (I) (asterisks: peak phases of individual flies; orange: morning, blue: evening). ΨM, E is the phase difference between morning and evening behavioral peaks. (J) Comparing phase differences between M cells (s-LNv) and other pacemaker groups (potential E cells): the difference between s-LNv and LNd (ΨM,LNd) best compared to the behavioral ΨM, E. n.s. = not significant; “*” denotes significantly different groups (P <0.05) by ANOVA followed by post hoc Tukey tests. ΨM,LNd matched behavioral ΨM, E (t-test, p=0.91; f-test, p=0.65). Error bars denote SEM.
We used objective-coupled planar illumination (OCPI) microscopy (16), which illuminates an entire focal plane simultaneously; this method accelerates volumetric imaging and reduces phototoxicity caused by repeated illumination. To permit imaging, we made cranial holes in the heads of living tim > GCaMP6s flies, which express the Ca2+ sensor GCaMP6s in all pacemaker neurons (15)(Fig. 1C), and monitored [Ca2+]i in five of the eight major pacemaker groups: small Lateral Neuron ventral (s-LNv), large Lateral Neuron ventral (l-LNv), Lateral Neuron dorsal (LNd), Dorsal Neuron 1 (DN1) and Dorsal Neuron 3 (DN3) (Fig. 1D). Each of the five groups displayed a prominent peak of [Ca2+]i during the 24 hour recording sessions and each peak had distinct timing (Fig. 1E). To test whether these Ca2+ dynamics reflected intrinsic circadian patterning, we began 24 hour recording sessions at different Zeitgeber times (ZT). In all such recordings, the peaks of Ca2+ activity reflected the pacemaker group identity, not the time at which recordings began (Fig. S2). Thus, Ca2+ varies in pacemaker neurons systematically as a function of the time of day based on biologically-defined rules of entrainment (Fig. 1F). Three Drosophila pacemaker groups (l-LNv, s-LNv and DN1p) show morning peaks of electrical activity when measured in acutely-dissected brains (17–19). Thus the phases of Ca2+ rhythms we observed are roughly coincident with, or slightly anticipate their peak electrical activity. Ca2+ rhythms produced by different pacemaker groups were similar in amplitude (Fig. 1F), yet different in waveform (Fig. S3) and phase (Fig. 1G). We confirmed our results using the FRET-based cameleon2.1 imaging method (20), for which the ratio of fluorescence from the Yellow Fluorescence Protein to that of the Cyan Fluorescent protein reflects [Ca2+]i, independent of the abundance of the sensor. [Ca2+]i estimated by this assay exhibited ~2 fold circadian variation with temporal patterns consistent with those obtained with GCaMP6s (Fig. S4). In addition, the [Ca2+]i rhythms did not result from experimental activation of CRYPTOCHROME (Fig. S5). These observations demonstrate that the Drosophila pacemaker network exhibits stereotyped and diverse spatiotemporal patterns of Ca2+ activity during the course of the 24 hour day.
We compared this diversity of Ca2+ activity patterns with the diversity of pacemaker functions. Pacemaker functions have been revealed by genetic mosaic experiments, as exemplified by the categorization of M (morning) and E (evening) cells (11–14). These autonomous oscillators primarily drive the morning and evening peaks of locomotor activity, respectively. The phase relationships (ΨM/E) between the peaks of Ca2+ rhythms in canonical M (s-LNv) and E (LNd) cells and the two daily peaks of locomotor activity were highly correlated (Fig 1H–J). In M cells, the Ca2+ rhythm peaked towards the end of the subjective night, whereas in E cells it peaked towards the end of the subjective day (Fig. 1F). The ~10 hour phase difference between Ca2+ rhythms in M and E pacemakers is similar to the ~10 hour phase difference between the morning and evening behavioral peaks (Fig. 1J). Thus, M and E pacemaker Ca2+ activations precede by ~4 hours the behavioral outputs they control. The distinct phases of Ca2+ rhythms in the other three pacemaker groups (l-LNv, DN1 and DN3) may also involve the morning and evening behavioral peaks, or may regulate other, distinct circadian-gated outputs.
The E category of pacemakers includes the LNd as well as the 5th s-LNv (11–14). However, the LNd is a heterogeneous group of neurons that exhibits diverse entrainment properties (21); likewise, the critical 5th s-LNv could not be unambiguously identified with tim-GAL4. To better understand the function of these subsets of E pacemakers, we used a PDF receptor (pdfr)(B) GAL4 driver (22); this driver restricts GCaMP6s expression to s-LNv, to three of six LNd and to the single 5th s-LNv (Fig. 2A). The three PDFR-expressing LNd and the 5th s-LNv displayed the same basic E cell pattern of Ca2+ activity – a peak in late subjective day, suggesting they both function as circadian pacemakers (Fig. 2B). Thus, the phase difference between Ca2+ rhythms in these PDFR-expressing M and E cell groups again matched that between the morning and evening behavioral activity peaks (Fig. 2, C through F).
Fig. 2. Spontaneous Ca2+ activity patterns are CRY-independent and reflect pacemaker functions.
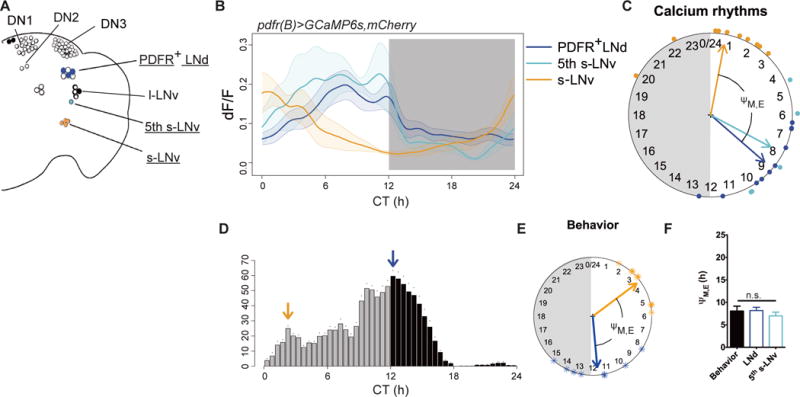
(A) Schematic of PDFR-expressing clock neurons: neuronal groups and sub-groups driven by pdfr(B)-gal4 are filled and color-coded; those imaged for GCaMP6s signals are underlined. (B–F) As Fig. 1G–K: (B) Ca2+ transients in three PDFR+ clock neuron groups and subgroups (n=10 flies): activities in the three PDFR+ LNd and in the single 5th s-LNv are similar (Pearson’s r=0.89). (C) Ca2+ rhythm phases from panel (B). (D) The DD1 locomotor activity of pdfr(B)>GCaMP6s,mCherry.NLS flies (n=8). (E) The phases of behavioral peaks from panel (D). (F) Phase differences from M cells (s-LNv) to both LNd and the 5th s-LNv matched behavioral ΨM, E (ANOVA, p=0.7239).
M and E cell categorization supports a classic model of seasonal adaptation (23) wherein a two-oscillator system responds differentially to light, and so can track dawn and dusk independently. For example, under long day conditions, light accelerates a “morning” clock and decelerates an “evening” clock. If these Ca2+ rhythms are critical output features of M and E cells, their properties may also reflect differences in photoperiodic entrainment. We entrained flies under either long day (16 hour light: 8 hour dark) or short day (8 hour light: 16 hour dark) conditions. In these flies, the phase difference between the morning and evening behavioral activity peaks tracked dawn and dusk (Fig S6). Likewise, the phases of pacemaker Ca2+ rhythms were also tracked dawn and dusk (Fig. 3, A and B, E and F, and Fig. S7). Regardless of the photoperiodic schedule, the s-LNv (M cells) always peaked around dawn, while the LNd (E cells) always peaked before dusk (Fig. 3, B through D and F through H). Thus, Ca2+ activity patterns within the pacemaker network correspond to the circadian temporal landmarks of dawn and dusk.
Fig. 3. Effects of environmental information and molecular clocks on the spatiotemporal patterns of Ca2+ activity in the pacemaker network.
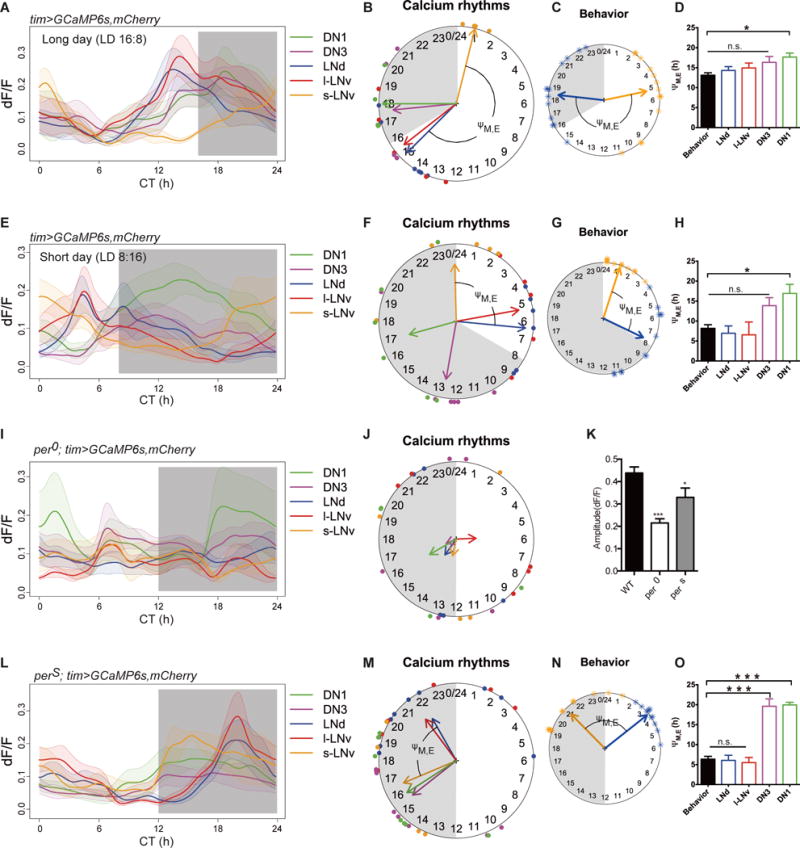
(A and E) Ca2+ transients: (A) under long (16:8 LD) photoperiod (n=6 flies) and (E) under short (8:16 LD) photoperiod (n=6 flies). (B and F) Ca2+ rhythm phases (B) under long photoperiod and (F) under short photoperiod. The shaded circular sectors indicate the 8 hours (B) and 16 hours (F) lights-out periods. Note that M cells (s-LNv, orange) peaked around lights-on and E cells (LNd, blue) peaked before lights-off regardless of photoperiod. (C and G) The phases of behavioral peaks in DD1 after 6 days of photoperiodic entrainment: (C) long photoperiod (n=13 flies) and (G) short photoperiod (n=12 flies). (See Fig S5 for more details). (D and H) ΨM,LNd matches behavioral ΨM, E under long photoperiod (t-test, p=0.32; f-test, p=0.88) and under short photoperiod (t-test, p=0.30; f-test, p=0.16). (I) Arrhythmic Ca2+ transients in per01 mutants (n=5 flies). (J) Phase coherence of Ca2+ transients was poor among per01 flies. (K) Amplitude of Ca2+ transients (maximum dF/F) was significantly smaller in per01 and in perS mutants (vs. control flies, Mann-Whitney test, *p<0.1, ***p<0.001). (L) Ca2+ transients in perS mutants (n=6 flies). (M) Ca2+ rhythm phases of perS mutants. (N) Phases of behavioral peaks corresponding to Ca2+ rhythm phases in panel (M) (n=16 flies). (O) ΨM,LNd matched behavioral ΨM, E (t-test, p=0.83; f-test, p=0.13).
We tested whether changes in the molecular oscillator would alter the patterns of [Ca2+]i. We used different alleles of the gene period, which encodes a state variable of the Drosophila circadian clock. In per01 (null) mutant flies, which lack inherent rhythmicity in their molecular oscillators and in free-running behavior (24,25) (Fig. S8), all clock neurons showed reduced rhythmicity in [Ca2+]i. The amplitudes of Ca2+ fluctuations were reduced by half (Fig. 3, I and K) and coherence was lost within groups (Rayleigh test, p>0.5; Fig. 3J and Table S1). In fast-running perS mutant flies, which have ~19 h free-running period (24,25) (Fig. S9), the Ca2+ rhythms were phase-shifted (Fig. 3, L and M, and Fig. S10) consonant with the direction of behavioral phase shifts (Fig. 3N and Fig. S9). The phase difference between Ca2+ rhythm peaks in perS M and E pacemakers still matched the phase difference between M and E behavioral peaks (Fig 3, N and O). Thus, molecular clocks determine the pace of Ca2+ rhythms in the pacemaker network.
To explore how synchronous molecular clocks can have staggered phases of Ca2+ activation by many hours, we tested whether PDF, which mediates interactions between pacemakers, was required. Flies bearing the severely hypomorphic han 5304 mutation of the PDF receptor show unimodal or arrhythmic behavior patterns under DD (26) (Fig. S11 and Table S2). In these flies, we found that the Ca2+ rhythms in M cells (s-LNv and DN1) were unaffected, but they were phase-shifted in LNd and DN3, such that these two groups now produced Ca2+ rhythms around dawn, roughly in synchrony with M cells (Fig. 4, A and B). The phase of l-LNv did not change, consistent with the absence of PDF sensitivity by this pacemaker group (27). The phase shifts in LNd and DN3 were fully restored by the expression of complete pdfr from a BAC transgene (Fig. 4, C through E “Rescue 1”, and Fig. S11). Thus PDF, which promotes synchronization of molecular clocks under constant conditions (10,28), is also needed to properly stagger their Ca2+ activity phases across the day. Whether the phases of the l-LNv and DN3 are set by other intercellular signals remains to be determined.
Fig. 4. Requirement of PDFR signaling for staggered waves of Ca2+ transients among the pacemaker groups.
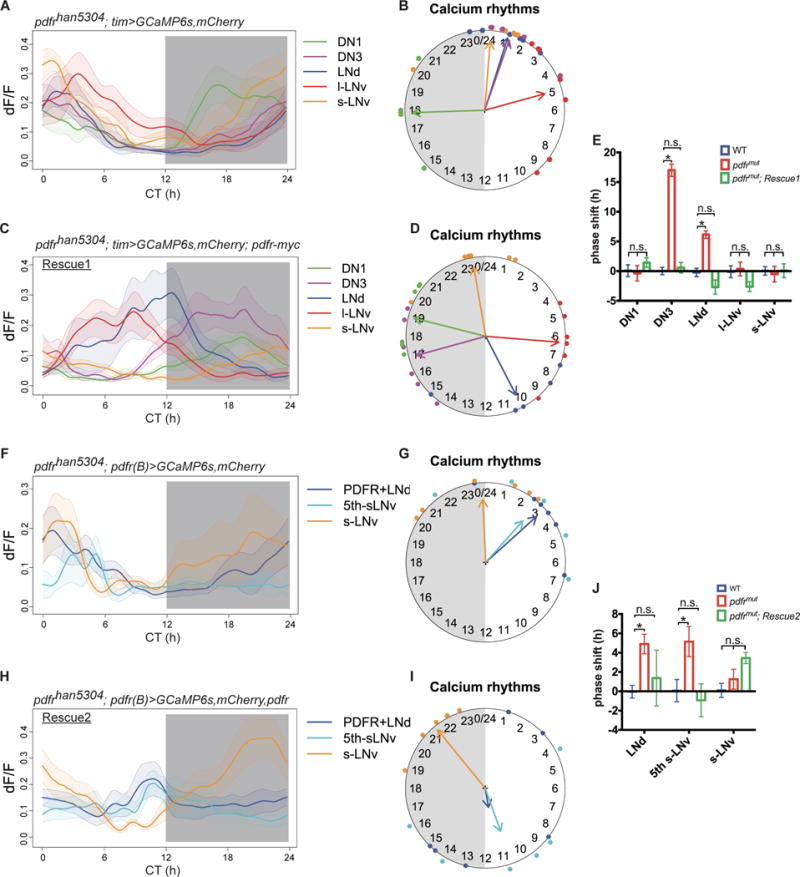
(A) Ca2+ transients in five pacemaker groups in pdfrhan5304 mutants (n=7 flies). (B) Ca2+ rhythm phases from panel (A): LNd and DN3 were phase-shifted towards s-LNv. (C) Ca2+ transients in pdfr mutant flies that are restored by a large BAC-recombineered pdfr-myc transgene (Rescue 1, n=6 flies). (D) Ca2+ rhythm phases from panel (C). (E) The phase shifts in mutants were fully rescued by restoring PDFR (two-way ANOVA followed by a Bonferroni post-hoc test, *p<0.001). Colors in this panel indicate genotype. (F) Ca2+ transients in three pacemaker groups targeted by pdfr(B)-gal4 in pdfrhan5304 mutants (n=6 flies). (G) Ca2+ rhythm phases from (F). (H) Ca2+ transients in pdfr mutant flies that are restored by pdfr(B)-gal4>pdfr (Rescue 2, n=6 flies). (I) Ca2+ rhythm phases from (H): the PDFR+ LNd and the single 5th s-LNv display restored phases, but lack strong phase coherence (Rayleigh test, p>0.1) (also see Fig. S12). (J) Phase shifts in mutant flies were partially restored by restoring pdfr in subsets of PDFR+ cells (two-way ANOVA followed by a Bonferroni post-hoc test, *p<0.001). Colors in this panel indicate genotype.
We further examined the pdfr mutant phenotype at higher cellular resolution (pdfr(B)> GCaMP6s; Fig. 2A). The PDFR-expressing E cell groups (the 3 PDFR-expressing LNd and the 5th s-LNv) displayed phase shifts similar to those of the entire LNd group (Fig. 4, F and G). When pdfr expression was restored just in these subsets of pacemaker neurons (with GAL4-UAS), both behavior and Ca2+ rhythms were partially restored (Fig. 4, H through J, “Rescue 2”, Fig. S11, and Table S2). The phase of the 5th s-LNv was fully restored, suggesting PDFR signaling is required for cell-autonomously setting of Ca2+ phase in this pacemaker group. However in “Rescue 2”, a single LNd typically remained active around dawn whereas two LNd were active around dusk (Fig. S12), which we interpret as a partial restoration or a non-autonomous phase-setting mechanism for LNd.
Our results show that molecular clocks drive circadian rhythms in the neural activity of pacemakers. Temporally patterned neural activity encodes different temporal landmarks of the day in a manner that reflects the different functions of the pacemaker groups. The homogeneous molecular clock produces sequential activity peaks by a mechanism dependent upon PDFR signaling. By generating diverse phases of neural activity in different pacemaker groups, the circadian clock greatly expands its functional output.
Supplementary Material
Acknowledgments
We thank W. Li and D. Oakley for technical assistance, D. Dolezel (CAS, Czech Republic) for technical advice, the Holy and Taghert laboratories for advice, E. Herzog (Washington Univ.) for comments on the manuscript, the Bloomington Stock Center, Janelia Research Center, J. Kim, and M. Affolter for sharing fly stocks, and M. Rosbash for anti-PER antibodies.
Funding: supported by the Washington University McDonnell Center for Cellular and Molecular Neurobiology and by grants from the National Institutes of Health (R01 NS068409 and R01 DP1 DA035081 to TEH) and (NIMH 2 R01 MH067122-11 to PHT).
Footnotes
Author contributions: XL, TEH, and PHT conceived the experiments; XL performed and analyzed all experiments; XL, TEH, and PHT wrote the manuscript.
Competing interests: TEH has a patent on OCPI microscopy.
Data and materials study: Materials are available upon request.
References and Notes
- 1.Lim C, Allada R. Emerging roles for post-transcriptional regulation in circadian clocks. Nature Neurosci. 2013;16:1544–1550. doi: 10.1038/nn.3543. [DOI] [PubMed] [Google Scholar]
- 2.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;2:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freeman GM, Jr, Krock RM, Aton SJ, Thaben P, Herzog ED. GABA networks destabilize genetic oscillations in the circadian pacemaker. Neuron. 2013;78:799–806. doi: 10.1016/j.neuron.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Nat’l Acad Sci U S A. 2007;104:7664–7649. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80:973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brancacci M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78:714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshii T, Vanin S, Costa R, Helfrich-Förster C. Synergic entrainment of Drosophila’s circadian clock by light and temperature. J Biol Rhythms. 2009;24:452–464. doi: 10.1177/0748730409348551. [DOI] [PubMed] [Google Scholar]
- 9.Roberts L, Leise TL, Noguchi T, Galschiodt AM, Houl JH, Welsh DK, Holmes TC. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr Biol. 2015;25:858–867. doi: 10.1016/j.cub.2015.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 12.Grima B, Chélot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- 13.Yoshii T, Funada, Ibuki-Ishibashi T, Matsumoto A, Tanimura T, Tomioka K. Drosophila cryb mutation reveals two circadian clocks that drive locomotor rhythm and have different responsiveness to light. J Insect Physiol. 2004;50:479–488. doi: 10.1016/j.jinsphys.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Liu Y, Bilodeau-Wentworth D, Hardin PE, Emery P. Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behavior. Curr Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holekamp TF, Turaga D, Holy TE. Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron. 2008;57:661–672. doi: 10.1016/j.neuron.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White WP, Dinner AR, Lear BC, Ren D, Diekman CO, Raman IM, Allada R. A conserved bicycle model for circadian clock control of membrane excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Nat’l Acad Sci U S A. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science (Wash DC) 2014;343:1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Im SH, Taghert PH. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. V. Pacemaker structure: A clock for all seasons. J Comp Physiol A. 1976;106:223–252. [Google Scholar]
- 24.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Nat’l Acad Sci U S A. 1971;68:2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardin PE, Hall JC, Rosbash M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature. 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 26.Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee L, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 27.Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshii T, Wülbeck C, Sehadova H, Veleri S, Bichler D, Stanewsky R, Helfrich-Förster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- 30.Caussinus E, Colombelli J, Affolter M. Tip-cell migration controls stalk-cell intercalation during Drosophila tracheal tube elongation. Curr Biol. 2008;18:1727–1734. doi: 10.1016/j.cub.2008.10.062. [DOI] [PubMed] [Google Scholar]
- 31.Fiala A, Spall T. In vivo calcium imaging of brain activity in Drosophila by transgenic cameleon expression. Sci STKE 2003. 2003;PL6 doi: 10.1126/stke.2003.174.pl6. [DOI] [PubMed] [Google Scholar]
- 32.Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Dolezelova E, Dolezel D, Hall JC. Rhythm defects caused by newly engineered null mutations in Drosophila’s cryptochrome gene. Genetics. 2007;177:329–345. doi: 10.1534/genetics.107.076513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stewart BA, Atwood HL, Renger JJ, Wang J, Wu CF. Improved stability of Drosophila larval neuromuscular preparations in haemolymph-like physiological solutions. J Comp Physiol A. 1994;175:179–191. doi: 10.1007/BF00215114. [DOI] [PubMed] [Google Scholar]
- 35.Turaga D, Holy TE. Organization of vomeronasal sensory coding revealed by fast volumetric calcium imaging. J Neurosci. 2012;32:1612–1621. doi: 10.1523/JNEUROSCI.5339-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thévenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- 38.Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enoki R, Kuroda S, Ono D, Hasan MT, Ueda T, Honma S, Honma K. Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc Nat’l Acad Sci U S A. 2012;109:21498–21503. doi: 10.1073/pnas.1214415110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci. 1997;17:676–696. doi: 10.1523/JNEUROSCI.17-02-00676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 42.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 43.Yoshii T, Todo T, Wülbeck C, Stanewsky R, Helfrich-Förster C. Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J Comp Neurol. 2008;508:952–966. doi: 10.1002/cne.21702. [DOI] [PubMed] [Google Scholar]
- 44.Fogle KJ, Parson KG, Dahm, Holmes TC. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science (Wash DC) 2011;331:1409–1413. doi: 10.1126/science.1199702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


