Abstract
The binding of MS2-GFP protein to arrays of MS2 sites in yeast mRNAs has been used extensively to visualize mRNA localization. We previously reported that arrays of MS2 sites bound by MS2 protein could inhibit Xrn1p and lead to the accumulation of 3′ mRNA decay fragments. We suggest that these decay fragments have the potential to complicate mRNA localization studies, as stated in an earlier study.
Keywords: MS2 RNA, MS2-MCP system, RNA decay, yeast, mRNA localization
In a rebuttal to our observations from Garcia and Parker (2015), Haimovich et al. (2016) use a variety of arguments to conclude “… that the MS2 system can accurately predict mRNA localization in living cells when used properly (e.g., for endogenously expressed mRNAs or upon careful analysis of the localization/stability of the overexpressed mRNA).” To address their findings and clarify their observations, we focus on two key issues raised by Haimovich et al. (2016). A more detailed discussion of their arguments can be found in the Supplemental Material.
Key issue #1: Do endogenously expressed MS2-tagged mRNAs lead to the accumulation of 3′ mRNA decay fragments?
Haimovich et al. (2016) performed RNA-seq data to identify 3′ decay fragments from immunopurified mRNAs tagged with 12 MS2 aptamers. They concluded that decay fragments are not readily detectable by RNA-seq for four of the 10 MS2-tagged mRNAs tested. For the other cases, they suggested that an enrichment of 3′ RNA-seq reads could come from the mechanism of library preparation; thereby implying the endogenously tagged mRNAs do not generally produce mRNA decay products.
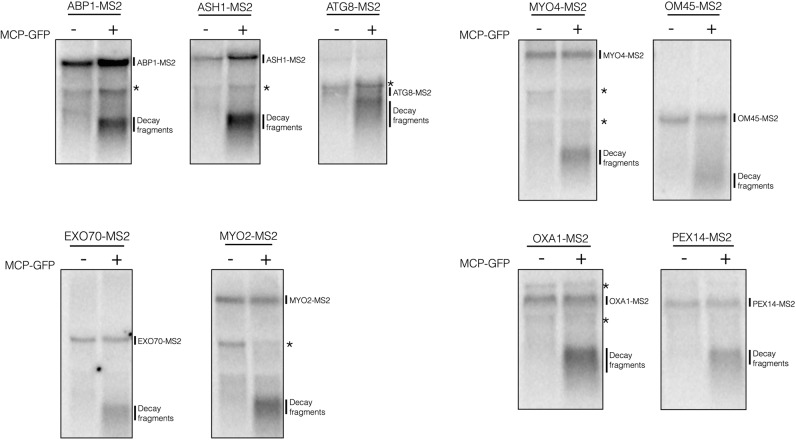
Haimovich et al. (2016) were kind enough to provide us with their strains and plasmids, so we could examine whether these endogenously tagged mRNAs produced decay fragments by Northern blots. Strikingly, we observed for all nine mRNAs examined clear 3′ mRNA decay fragments (Fig. 1, right lane of each panel), which were dependent on the coexpression of the MS2 protein (Fig. 1, left lane of each panel). We conclude from this experiment that yeast mRNAs with chromosomally integrated MS2 arrays ubiquitously accumulate 3′ mRNA decay fragments in the presence of MS2-binding protein.
FIGURE 1.
Decay fragments are detectable in mTAG mRNAs when the MCP-GFP plasmid is present. Total RNA was isolated from yeast strains harboring MS2-tagged mRNAs and visualized by Northern blotting using a 1.5% formaldehyde agarose gel. Right lane of each blot is total RNA isolated from strains transformed with pUG36-CP-GFP×3, while the left lane is obtained from untransformed strains. The mRNAs analyzed on the same blot are grouped together. Visualization of the MS2-tagged mRNAs and decay products was done using the 12×MS2 probe (5′-CTGCAGACATGGGTGATCCTCATGTTTTCT-3′). Strains were grown to OD600 0.6 to 0.8 in minimal media (−MCP) or –ura minimal media (+MCP) at 30°C. (*) Indicates background bands.
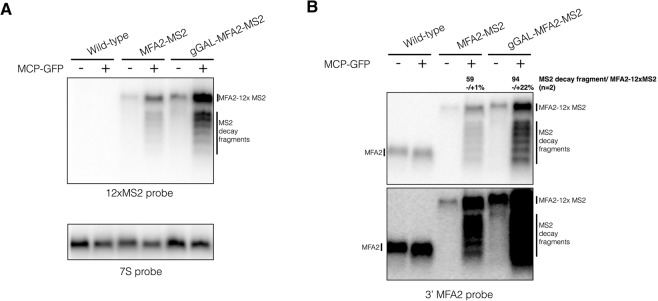
Haimovich et al. (2016) argue that overexpression of an MS2-tagged mRNA, specifically MFA2, can exacerbate the formation of decay fragments as assessed by RT-qPCR and lead to an observed colocalization of the MS2-tagged RNA with P-bodies (PB). By Northern blots of their strains, we confirm that a Gal-driven MS2-tagged MFA2 mRNA expressed chromosomally produces a higher percentage of mRNA decay fragments than a chromosomally tagged MFA2 mRNA when compared to the levels of the full-length MS2-tagged mRNA (Figure 2A,B). In contrast to Haimovich et al. (2016), we observed that MS2 protein expression was required for substantial accumulation of the mRNA decay product (Fig. 2A,B). Like in Figure 1, we do not observe any detectable mRNA decay product without MCP even in a darker exposure of the Northern blot (Fig. 2B). Yet, it remains possible that arrays of MS2 sites by themselves can also lead to some inhibition of Xrn1. Thus, both studies are in agreement that overexpression of mRNAs can exacerbate the accumulation of mRNA decay fragments.
FIGURE 2.
Decay fragments are detectable in MFA2-MS2 genomic constructs. Total RNA was isolated and visualized from strains expressing MFA2-12×MS2 from the chromosome. Grown as described in Haimovich et al. (2016). (A) Total RNA from wild-type, mfa2::MFA2-12×MS2 (MFA2-MS2) and mfa2::GALpromoter-MFA2-12×MS2 (gGAL-MFA2-MS2) plus and minus the pUG36-CP-GFP×3 plasmid was run on a 1.5% formaldehyde agarose gel, probed with 12×MS2 probe, and then stripped and reprobed with 7S probe (5′-GTCTAGCCGCGAGGAAGG-3′). (B) Same as above except visualized with a probe to the 3′ UTR of MFA2 (5′-GATGAGAGAATTGGAATAAATTAGTTTGCCAGC-3′) with a light (top panel) and a dark exposure (bottom panel) of the blot. Quantification of the percentage of decay fragment relative to full-length MS2-tagged mRNA was done using ImageJ to calculate the total intensity of the decay fragments over the total intensity of the full-length mRNA. Both values had the background values subtracted prior to calculation. This percentage represents the average of two biological replicates and its standard deviation.
Key issue #2: Can 3′ mRNA decay fragments containing MS2 stem–loops affect apparent mRNA localization?
Haimovich et al. (2016) suggest that previous literature has validated the MS2-MCP system and that decay fragments may not interfere with mRNA localization studies. While this may be true, it seems prudent to assume that such mRNA decay fragments will bind MS2-GFP fusion proteins and as such be detected as potentially localized mRNAs. Consistent with this possibility, mRNA decay fragments with a poly(G) block to Xrn1 mediated degradation have been localized to P-bodies (Sheth and Parker 2003). Haimovich et al. (2016) argue that a subset of foci is due to mRNA decay fragments, which can be manually removed from the population before assessing mRNA localization. Although possible, this assumes that all the mRNA fragments are in these foci, as well as making it impossible to identify mRNAs whose full-length form is actually in specific subcellular foci.
Bottom line: Can the MS2-MCP system be used to accurately visualize the localization mRNAs?
The possibility exists that the accumulation of mRNA decay fragments does not invalidate the use of the MS2-MCP system for localizing mRNAs. However, since fragments can exist, it suggests that one is prudent to (i) make sure any mRNA decay fragments for a given construct are not a large percentage of the observed mRNA and (ii) confirm any key localization results by other means (such as FISH, subcellular fraction followed by RT-qPCR, etc.). Ideally, refinements of the MS2-MCP system in manners that limit the production of mRNA decay products will remove this concern in future work.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.056325.116.
REFERENCES
- Garcia JF, Parker R. 2015. MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system. RNA 21: 1393–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Zabezhinsky D, Haas B, Slobodin B, Purushothaman P, Fan L, Levin JZ, Nusbaum C, Gerst JE. 2016. Use of the MS2 aptamer and coat protein for RNA localization in yeast: A response to “MS2 coat proteins bound to yeast mRNAs block 5′ to 3′ degradation and trap mRNA decay products: implications for the localization of mRNAs by MS2-MCP system.” RNA (this issue) 22: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.