 Novel glycoside amphiphile (NDT-C11) displays favorable behavior in maintaining both protein stability and conformational flexibility compared to DDM and MNG-3.
Novel glycoside amphiphile (NDT-C11) displays favorable behavior in maintaining both protein stability and conformational flexibility compared to DDM and MNG-3.
Abstract
Membrane proteins are amphipathic bio-macromolecules incompatible with the polar environments of aqueous media. Conventional detergents encapsulate the hydrophobic surfaces of membrane proteins allowing them to exist in aqueous solution. Membrane proteins stabilized by detergent micelles are used for structural and functional analysis. Despite the availability of a large number of detergents, only a few agents are sufficiently effective at maintaining the integrity of membrane proteins to allow successful crystallization. In the present study, we describe a novel class of synthetic amphiphiles with a branched tail group and a triglucoside head group. These head and tail groups were connected via an amide or ether linkage by using a tris(hydroxylmethyl)aminomethane (TRIS) or neopentyl glycol (NPG) linker to produce TRIS-derived triglucosides (TDTs) and NPG-derived triglucosides (NDTs), respectively. Members of this class conferred enhanced stability on target membrane proteins compared to conventional detergents. Because of straightforward synthesis of the novel agents and their favourable effects on a range of membrane proteins, these agents should be of wide applicability to membrane protein science.
Introduction
Integral membrane proteins (IMPs) account for ∼25% of the proteins encoded in genomes.1 They play a key role in cell physiology by mediating various cellular processes including metabolite transport, signal transduction, environmental response, and intercellular communication. Malfunction of IMPs is associated with a range of diseases including cancer, cystic fibrosis, Alzheimers, epilepsy, and hypertension.2 The importance of IMPs in disease is reflected by the fact that half of current drug molecules target these biomacromolecules.3 Thus, detailed information on the structure and function of these proteins is of major importance for biology4 and human health.5 However, in spite of their immense biological and pharmaceutical significance, understanding of the precise mechanism of action of many of these proteins, particularly those from eukaryotes, remains limited. A comparatively low number of high resolution structures of membrane proteins are available; they comprise approximately 1% of all proteins with known structure.6 The major difficulty arises from the amphipathic character associated with membrane protein architecture. Lipid bilayers, called membranes, provide the requisite environment for the retention of structure and function of these proteins, but are not compatible with membrane protein analysis. The proteins must be extracted from the bilayers for structural characterization. However, extraction of the membrane protein into a non-native environment leads to rapid protein denaturation and aggregation because of the incompatibility between the large hydrophobic surface of protein and the polarity of aqueous media.7
Detergents are amphipathic agents which can mimic lipid bilayers and are thus widely used to maintain the structural and functional integrity of target proteins in the course of membrane protein solubilisation, purification and crystallization.8 Currently over 120 conventional detergents are available which can be classified into three main categories depending upon the nature of the head group: ionic, zwitterionic and non-ionic. Each class of detergents has advantages and disadvantages, but nonionic detergents are most widely used for structure determination of membrane protein. Notably, the five most popular detergents, OG (n-octyl-β-d-glucopyranoside), NG (n-nonyl-β-d-glucopyranoside), DM (n-decyl-β-d-maltoside), DDM (n-dodecyl-β-d-maltoside), and LDAO (lauryldimethylamine-N-oxide), have facilitated ∼70% of α-helical membrane proteins with known structure.9 However, many membrane proteins solubilized even in these popular detergents are prone to structural degradation.10 Conventional detergents typically have a simple architecture, possessing a flexible alkyl tail connected to a hydrophilic head group. The limited utility of conventional detergents in membrane protein study is likely to originate from the small variability in detergent architecture. In contrast, membrane proteins are highly variable in terms of their propensity to aggregate and denature, related to the large variability in their 3D structures. Therefore, a major research effort is focused on development of novel amphiphiles with varying architectures that have high efficacy for membrane protein solubilisation and stabilization.11
Over the last two decades a number of novel amphiphiles with unique structures have been developed. These non-conventional amphiphiles can be classified into four main categories: variants of conventional detergents (e.g., Chae's glyco-tritons (CGTs)12a and deoxycholate-based glycosides (DCGs)),12b peptide-based amphiphiles (e.g., lipopeptide detergents (LPDs)13a and β-peptides (BPs)),13b membrane-mimetic systems with an amphipathic polymer (e.g., amphipols (Apols),14a,b nanodiscs (NDs)14c and nanolipodisq particles14d), and rigid hydrophobic group-bearing agents (e.g., glyco-diosgenin (GDN)15a and tripod amphiphiles (TPAs)15b–d). Despite the large diversity in detergent architecture, only a small number of classes have shown to be successful for membrane protein crystallization, exemplified by calixarene-based detergents,16a facial amphiphiles (FAs)16b,c and neopentyl glycol (NG) class amphiphiles.16d–g NG class agents include glucose-neopentyl glycol amphiphiles (GNGs)16c,d and maltose-neopentyl glycol amphiphiles (MNGs).16e,f GNG-3 and MNG-3 have contributed to the determination of 20 new membrane protein structures including the β2 adrenergic,17a–e acetylcholine17f,g and opioid G-protein coupled receptors17h,i in the last four years. These results highlight the potential that novel amphiphiles have with respect to structural elucidation of membrane proteins of both biological and pharmaceutical significance. In this study, we designed and prepared two sets of novel agents, designated tris(hydroxymethyl)aminomethane (TRIS)-derived triglucosides (TDTs) and neopentyl glycol (NPG)-derived triglucosides (NDTs). When evaluated with three membrane proteins, some of these glucoside agents were both effective at solubilisation and conferred greater stability than one of most popular conventional detergents, DDM.18
Results and discussion
Detergent structures and physical characterizations
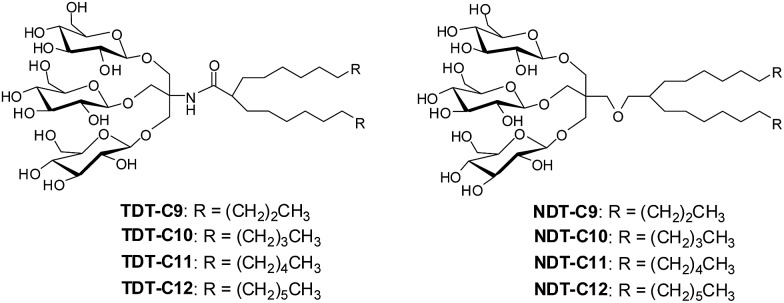
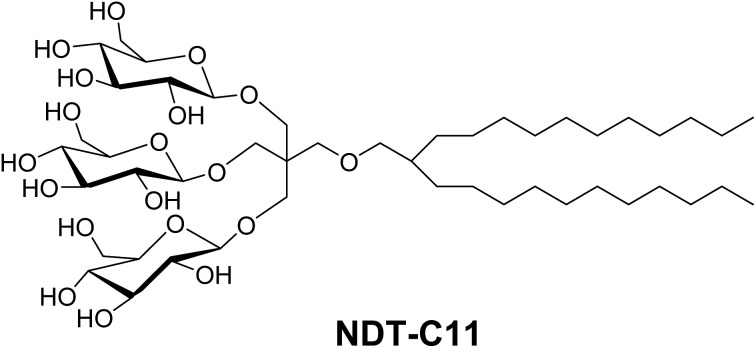
The design of TDTs and NDTs features two alkyl chains and a triglucoside head group, connected by an amide linkage in the case of TDTs and by an ether linkage in the case of NDTs (Fig. 1). TRIS and NPG were used as linkers for the preparation of TDTs and NDTs, respectively. Each set (TDTs or NDTs) has variation in the carbon chain length ranging from C9 to C12, which was used for detergent designation. Both TDTs and NDTs were synthesized via straightforward synthetic schemes. The syntheses of TDTs were completed in five steps, comprising dialkylation of dimethylmalonate, Krapcho's decarboxylation, amide coupling with TRIS, glycosylation and deprotection (see ESI† for details). In the case of syntheses of NDTs, NPG coupling was used instead of TRIS coupling (see ESI† for details). The ease of synthesis along with high synthetic efficiency makes it possible to synthesize the designed amphiphiles in multi-gram quantities.
Fig. 1. Chemical structures of newly prepared TRIS-derived triglucosides (TDTs) and neopentyl glycol-derived triglucosides (NDTs).
All TDTs and NDTs except NDT-C12 and TDT-C12 are water soluble up to 10%; NDT-C12 and TDT-C12 are water-soluble up to 5%. Interestingly, these two agents tend to form hydrogels, particularly at a low temperature. The critical micelle concentration for each new agent was determined by the aid of a fluorescent dye, diphenylhexatriene (DPH).19 The sizes of micelles formed by the new agents were measured as hydrodynamic radii (Rh) through dynamic light scattering (DLS) experiments. The summarized data are presented in Table 1. The CMC values of TDTs/NDTs are much smaller than that of DDM. For example, TDT-12 and NDT-12 with the longest alkyl chain (C12) have CMCs >100 times smaller than DDM. The small CMCs reflect the greater propensity of these agents to form micellar structures. Note that the CMC values of TDTs are higher than those of NDTs when comparing amphiphiles with the same chain length. Both sets of new amphiphiles (TDTs and NDTs) displayed an inverse relationship between the alkyl chain length and their CMC values, which can be explained by detergent hydrophobicity increasing with the long alkyl chain. In terms of micelle size, NDTs lie within a narrow window, ranging from 3.1 to 3.8 nm, and are thus comparable to DDM (3.4 nm). In contrast, a large variation was observed for TDTs, ranging from 3.4 to 53 nm. Thus, the small difference in the chemical structure (i.e., amide or ether linkage) appeared to significantly affect the morphology of the self-aggregates in an aqueous medium. Micelles formed by TDTs and NDTs increase in size with increasing alkyl chain length as the shape of the detergent molecules becomes closer to a cylinder. All amphiphiles displayed a single set of populations in the number-averaged size distribution of micelles (Fig. S1†).
Table 1. Molecular weights (MWs) and critical micelle concentrations (CMCs; n = 2) of new glucosides (TDTs and NDTs) and a conventional detergent (DDM), and the hydrodynamic radii (Rh; n = 4) of their micelles.
| Detergent | MW a | CMC (μM) | CMC (wt%) | R h b (nm) |
| TDT-C9 | 902.1 | 47 ± 1.5 | 0.0042 ± 0.0001 | 3.4 ± 0.4 |
| TDT-C10 | 930.1 | 14 ± 1.0 | 0.0013 ± 0.0001 | 4.5 ± 0.2 |
| TDT-C11 | 958.2 | 11 ± 1.5 | 0.0011 ± 0.0001 | 37 ± 8.0 |
| TDT-C12 | 986.2 | 6.0 ± 0.1 | 0.0006 ± 0.0000 | 53 ± 1.2 |
| NDT-C9 | 903.1 | 26 ± 4.0 | 0.0023 ± 0.0004 | 3.1 ± 0.1 |
| NDT-C10 | 931.2 | 12 ± 0.5 | 0.0011 ± 0.0000 | 3.2 ± 0.1 |
| NDT-C11 | 959.2 | 6.1 ± 1.8 | 0.0005 ± 0.0002 | 3.5 ± 0.0 |
| NDT-C12 | 987.3 | 2.4 ± 0.9 | 0.0002 ± 0.0001 | 3.8 ± 0.4 |
| DDM | 510.1 | 170 | 0.0087 | 3.4 ± 0.0 |
aMolecular weight of detergents.
bHydrodynamic radius of detergents measured at 1.0 wt% by dynamic light scattering.
Detergent evaluation with membrane proteins
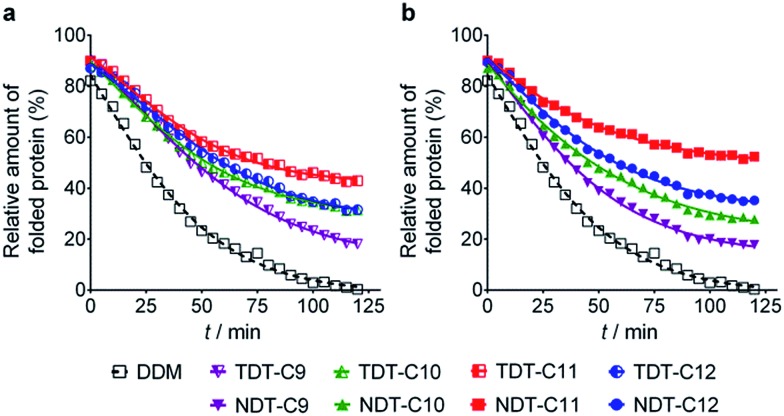
The new agents (TDTs and NDTs) were first evaluated with a membrane protein transporter, UapA. This protein is a uric acid–xanthine/H+ symporter from Aspergillus nidulans.20 Protein stability was assessed using fluorescence spectroscopy with the assistance of a sulfhydryl-specific fluorophore, N-[4-(7-diethylamino-4-methyl-3-coumarinyl) phenyl]maleimide (CPM).21 The free sulfhydryl groups of cysteine residues are normally buried within the core of the protein but become solvent-accessible upon protein unfolding. The CPM reacts with the free thiols and becomes fluorescent, thereby serving as an unfolding sensor. For the thermal stability assay, UapA protein was solubilized and purified in DDM and the DDM-purified protein was diluted into buffer solutions including individual amphiphiles at CMC + 0.04 wt%. Immediately following addition of CPM the samples were incubated at 40 °C for 120 min. The fluorescence of the individual samples was measured at regular intervals. Since the UapA was the least stable in DDM, the amounts of folded proteins in the other agents (TDTs and NDTs) were normalized with respect to DDM. TDT agents bearing an amide linkage were all better than DDM at maintaining the folded state of the protein (Fig. 2). Of the TDTs, TDT-C9 resulted in the least stable protein while TDT-C11 was best. TDT-C10 and TDT-C12 were comparable to each other. The NDTs also resulted in improved stability of the UapA compared to DDM, with NDT-C11 being the best performing agent. When detergent concentration was increased to CMC + 0.2 wt%, the same overall trend in detergent efficacy was observed (Fig. S2†). However, differences in stabilities conferred by DDM and the novel agents were more prominent. TDTs are markedly better than DDM, and the NDTs were even superior to TDTs, again with NDT-C11 the best performing agent. The increased differences in detergent efficacy observed here could be due, at least in part, to the harsh nature of DDM at this high detergent concentration; excess detergent micelles are known to be harmful for protein stability. In contrast, TDTs and NDTs are effective at stabilizing UapA even at the high detergent concentration indicating that the general architecture of the new agents is favorable for membrane protein stability. NDT-C11 showed a slightly enhanced efficacy relative to MNG-3, the best MNG, in maintaining the folded state of the protein, when these agents were tested at CMC + 0.2 wt% (Fig. S3†).
Fig. 2. Thermal denaturation profile of UapA protein purified in DDM and then exchanged into novel TDTs (a) and NDTs (b) at detergent concentrations of CMC + 0.04 wt%. Thermal stability of the protein was monitored by CPM assay performed at 40 °C for 120 min. The relative amounts of folded protein were normalized relative to the most destabilizing condition in this experiment, that is, protein denaturation in DDM after 2 h incubation. Mean standard deviations (n = 2) for DDM, TDT-C9, TDT-C10, TDT-C11, TDT-C12, NDT-C9, NDT-C10, NDT-C11 and NDT-C12 are 4.9, 9.3, 2.3, 5.8, 6.1, 2.7, 9.4, 10.2, 9.5, respectively.
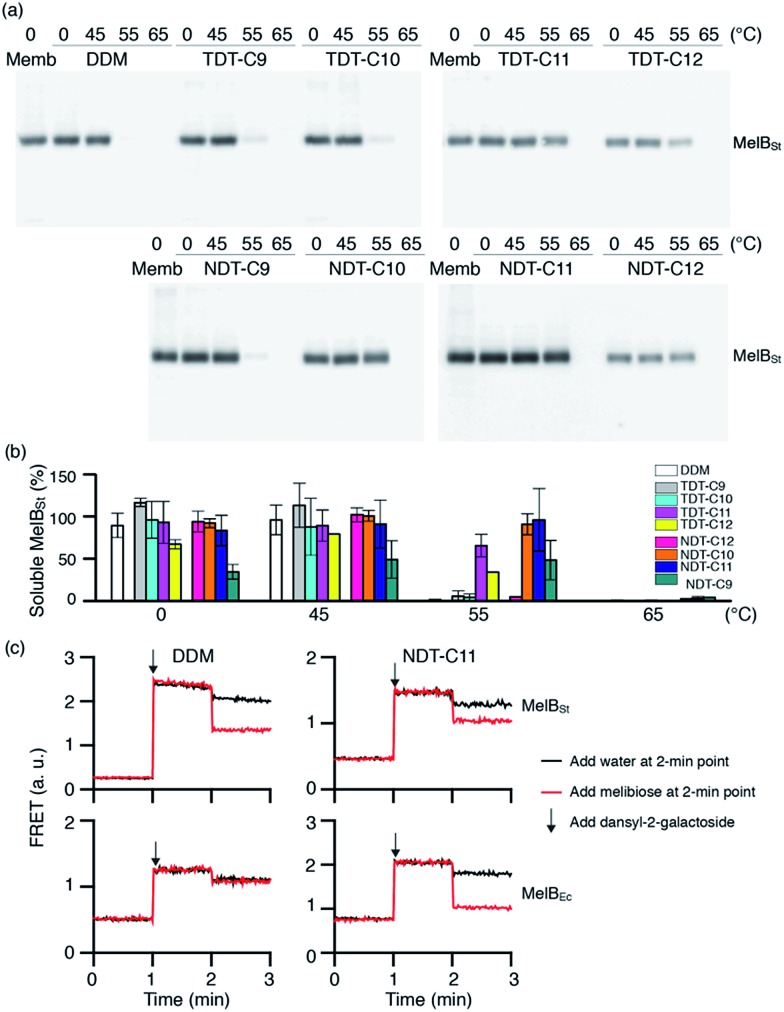
As a second target, the melibiose permease of Salmonella typhimurium (MelBSt) was used for assessing solubilisation efficiency of the new amphiphiles.22 MelBSt is the major facilitator superfamily permease catalysing cotransport of galactosides with either a proton, sodium, or lithium ion. To test the TDT and NDT amphiphiles, membrane fractions of E. coli cells overexpressing MelBSt were treated with 1.5% TDTs, NDTs or DDM for 90 min, and subjected to ultracentrifugation to remove the insoluble fraction. The amount of soluble MelBSt was assessed by SDS-PAGE and Western immunoblotting. All tested detergents efficiently extracted MelBSt from the membranes at 0 °C (Fig. S4†), except for TDT-C12 and NDT-C12; these agents with the C12 alkyl chain produced soluble MelBSt with ∼70% and ∼44% yields, respectively. The poor solubilisation efficiency of these agents is likely a result of their tendency to form hydrogels, particularly at a low temperature. In order to further explore the protein stabilization efficacy, the thermostability of MelBSt was estimated by performing a similar assay at elevated temperatures (45, 55 and 65 °C). Only the soluble fraction after ultracentrifugation was analyzed and quantitatively expressed as a percentage of total MelBSt protein of the membrane control (Fig. 3a). Following 90 min incubation at 45 °C, the amounts of MelBSt solubilized by TDTs and NDTs with C9, C10 or C11 alkyl chains were comparable to that solubilized by DDM. TDT-C12 and NDT-C12 showed increased solubilisation efficiency at this elevated temperature; the solubilisation efficiency rises from 44% at 0 °C to 68% at 45 °C for NDT-C12 (Fig. 3b). It is likely that the increase in solubilized MelBSt is a consequence of enhanced solubility of these agents at the elevated temperature as the tendency to form hydrogels decreases with increasing temperature. When the incubation temperature was increased further to 55 °C, no soluble MelBSt was detected in DDM while small amounts of soluble protein were detectable for most of TDTs. Notably, TDT-C11 yielded a substantial amount of soluble protein (∼65%). In contrast to the TDTs, most of NDTs were superior to DDM and TDTs with the exception of NDT-C9. The best performance was achieved by NDT-C11, followed by NDT-C10 and NDT-C12. None of the novel detergents could effectively protect MelBSt from aggregation at 65 °C. Overall, this result indicates that some of the new amphiphiles, particularly NDT-C10 and NDT-C11, are not only favourable for membrane protein solubilisation, but also remarkably effective at maintaining MelBSt in a soluble state in an aqueous medium. In order to explore the functional state of the detergent-solubilized MelB protein, we utilized Förster resonance energy transfer (FRET) from tryptophans (Trp) to the fluorescent ligand, 2′-(N-dansyl)aminoalkyl-1-thio-β-d-galactopyranoside (D2G).22a,23 MelB protein bound to D2G is fluorescent due to the close proximity of this FRET pair. Upon melibiose addition, however, fluorescence intensity decreases if detergent solubilisation produces an active protein because melibiose replaces the bound D2G molecule. MelBSt solubilized in DDM or NDT-C11 was subjected to melibiose reversal of Trp → D2G FRET. As can be seen in Fig. 3c, DDM and NDT-C11 produced functional MelBSt proteins, as observed for MNG-3 in a previous study.24 In order to differentiate detergent efficacy, MelBEc, less stable than MelBSt, was used for comparison.24 When extracted by DDM, MelBEc underwent complete loss of melibiose binding. In contrast, MelBEc solubilized with NDT-C11 retained functionality (Fig. 3c). These results indicate that NDT-C11 is capable of maintaining the melibiose binding activity of both MelBSt and MelBEc while DDM is only effective for the more stable MelBSt.
Fig. 3. Thermosolubility and functional profiles of detergent-solubilised MelBSt. The solubility test at elevated temperatures was carried out as described in the ESI.† (a) Solubilised materials after ultracentrifugation of detergent-treated membranes were analysed by SDS-15%PAGE and Western blot. The total amount of MelBSt protein used in each assay is shown by the untreated membrane sample (Memb). (b) Histogram of band density. The solubilisation efficiency of MelBSt is expressed as a percentage of band density relative to the untreated membrane sample. The density was measured by ImageQuant software. Error bars, SEM, n = 2–4. (c) Galactoside binding. Right-side-out (RSO) membrane vesicles containing MelBSt or MelBEc were solubilised with DDM or NDT-C11 as described in the ESI.† After ultracentrifugation, the supernatant was used to test melibiose reversal of Trp to dansyl-2-galacotside (D2G) FRET. Note the difference in FRET response of the D2G bound MelB to melibiose or water addition at the 2 min point.
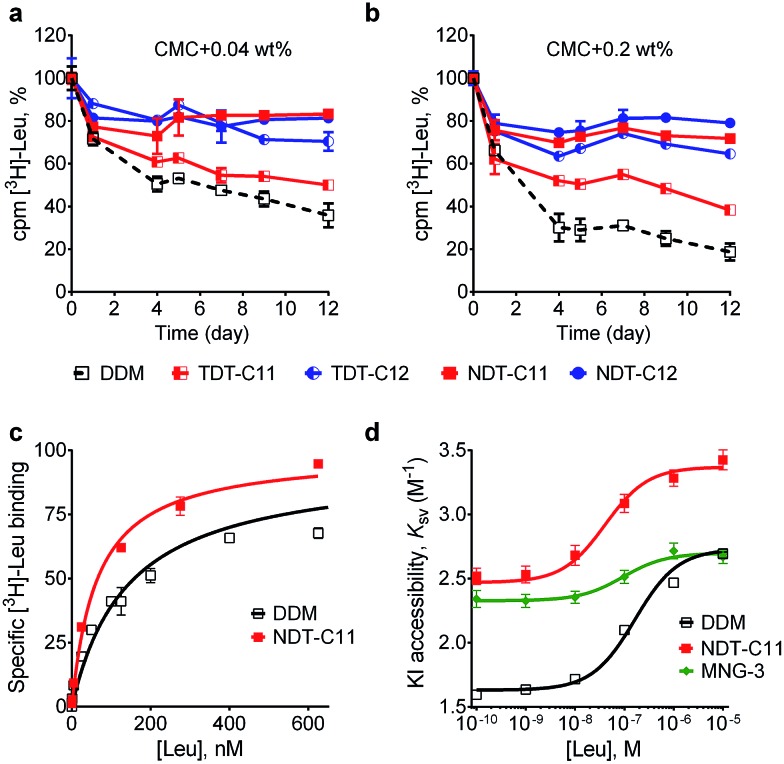
The intriguing results obtained for the TDTs and NDTs prompted us to evaluate these agents with another membrane protein, the bacterial leucine transporter (LeuT). LeuT from Aquifex aeolicus is a prokaryotic homologue of the mammalian neurotransmitter/sodium symporters (NSSs family).25 Based on the results with UapA and MelBSt, we selected some of the most promising TDTs (TDT-C11 and TDT-C12) and NDTs (NDT-C10, NDT-C11, NDT-C12) for evaluation with LeuT. To begin with, LeuT was solubilized and purified in DDM. DDM-purified LeuT was diluted into buffer solutions containing individual NDT and TDT agents to reach a final detergent concentration of CMC + 0.04 wt% or CMC + 0.2 wt%. LeuT activity was monitored as a function of time by incubating protein samples for 12 days at room temperature. The binding affinity of the transporter for a radio-labeled ligand ([3H]leucine) was measured by scintillation proximity assay (SPA).26 As can be seen in Fig. 4a and b, both TDTs (TDT-C11 and TDT-C12) were superior to DDM at both detergent concentrations tested, with TDT-C12 producing more stable protein than TDT-C11. All tested NDT agents (NDT-C10, NDT-C11 and NDT-C12) were superior to both DDM and the TDTs (Fig. S5†). The best detergents showed dependency on detergent concentration. Specifically, NDT-C11 was best at a detergent concentration of CMC + 0.04 wt% while NDT-C10/NDT-C12 was best at CMC + 0.2 wt%. Overall, all tested NDTs were excellent at preserving the transporter activity under the assay conditions. In addition to LeuT stability, we also wished to assess the ability of NDT-C11 to preserve conformational dynamics of the transporter. Accordingly, a cysteine residue was inserted at position 192 of LeuT (E192C) and coupled to the thiol-reactive fluorophore (tetramethylrhodamine-5-maleimide; TMR). This TMR-conjugated LeuT, LeuT E192CTMR, is a highly sensitive system to monitor conformational transition as a response to ligand binding.27 Upon binding of leucine, LeuT undergoes a conformational change in a detergent solution.28 This conformational transition renders TMR more accessible to the aqueous environment and therefore more accessible for the water-soluble quencher, iodide (I–). TMR quenching intensity as a function of leucine binding can then be plotted in a Stern–Volmer plot for direct measurement of conformational flexibility in the protein. Ligand binding of TMR-labelled transporter was measured with increasing concentrations of [3H]leucine using SPA (Fig. 4c). NDT-C11-solubilized transporter had slightly lower Kd value than DDM-solubilized protein (64 vs. 142 nM, Table S1†). Fluorescence quenching of LeuT E192CTMR was measured with increasing concentration of iodide along with various leucine concentrations (Fig. S6†). From this data, we plotted the Stern–Volmer constant (KSV) as a function of the added leucine concentration (Fig. 4d). From the plot of KSVvs. [Leu], a saturation response was observed with an EC50 value of 163 nM, 41 nM and 94 nM for DDM, NDT-C11 and MNG-3-solubilized transporters, respectively (Table S1†). The observed EC50 values correspond to the [3H]leucine affinity measured by SPA. The change in relative TMR accessibility by I– (ΔKSV) is an indication of conformational constraint imposed by the detergent micelles. NDT-C11 displayed a ΔKSV value comparable to that of DDM (0.9 M–1 and 1.1 M–1, respectively) while MNG-3 gave a much smaller ΔKSV (0.4 M–1vs. 1.1 M–1) (Table S1†). These data suggests that, in contrast to MNG-3, NDT-C11 allows conformational rearrangement in LeuT to an equal extent as DDM. Interestingly, the KSV in the absence of leucine is markedly increased in NDT-C11 relative to DDM (from 1.6 in DDM to 2.5 in NDT-C11). This suggests that initial TMR accessibility is more pronounced in NDT-C11 possibly because of less shielding by detergent molecules. A decreased tendency of detergent molecules to occupy intracellular loop regions could indicate a higher propensity to form crystal contacts. Taken together, NDT-C11 is superior to DDM and comparable to MNG-3 in maintaining LeuT stability, but retains LeuT conformational flexibility as observed in DDM.
Fig. 4. Long-term activity of wild type leucine transporter (LeuT), ligand binding affinity and KI accessibility of the LeuT E192CTMR. Long-term stability was measured by using the transporter solubilized in novel amphiphiles (TDT-C11, TDT-C12, NDT-C11 and NDT-C12) and a conventional detergent (DDM). The detergents were used at CMC + 0.04 wt% (a) and CMC + 0.2 wt% (b). Protein activity for LeuT was measured by scintillation proximity assay (SPA). Results are expressed as % activity relative to activity at day 0 (mean ± s.e.m., n = 2). (c) Saturation binding of [3H]leucine assessed by SPA for mutant protein, LeuT E192CTMR, in either CMC + 0.04 wt% DDM or NDT-C11. Data are fitted to a single site model. Data points are means ± s.e.m. with n = 3–4. (d) KSV values were plotted as a function of leucine concentration at CMC + 0.04 wt% detergent concentration. A conventional detergent (DDM), newly prepared NDT-11, and previously reported MNG-3 were used for comparison. Data points are means ± s.e.m. with n = 3–4.
Detergent efficacy is often substantially affected by a small change in detergent structure. In the current study, TDTs and NDTs have the same overall architecture with two flexible alkyl chains connected to a triglucoside head group via a rigid linker (TRIS and NPG, respectively). The only structural difference between these two sets of detergents is the functional group in the linker region. TDTs have an amide linkage while NDTs have an ether linkage. Despite such a small variation in the chemical structure, detergent efficacy between these two sets is different for all tested membrane proteins (UapA, MelBSt and LeuT) with the NDTs (e.g., NDT-C11) markedly better than TDTs. A large difference was also observed for the micelle sizes formed by these two sets of amphiphiles. The micelles formed by TDTs were larger than those formed by NDTs when compared with each other with the same chain length. The precise reason for these interesting findings is unclear. We suggest that the difference in bond rigidity of the amide and ether linkages is responsible for both detergent micelle size and detergent stabilization efficacy for membrane proteins. Specifically, because of higher flexibility, the alkyl chains connected by the ether linkage can pack more effectively in the interior of detergent micelles than those attached by the amide linkage. This tight packing, reflected by the small CMC values of NDTs relative to those of TDTs, could reduce the size of self-aggregates. The flexibility of the ether bond would also affect packing density of detergent alkyl chains when associated with membrane proteins, thereby playing a key role in enhancing the stability of a target protein. The effect of the functional groups in the linker region on detergent micelle size and membrane protein stability has not yet been reported and discussed. Such detergent structure-property-efficacy relationships will play an important role in the future design of novel amphiphiles.
A detergent with a small head group (e.g., glucoside) tends to form small protein–detergent complexes (PDCs). A small PDC size is known to be favorable for membrane protein crystallization by providing a large hydrophilic protein surface area. Crystal lattice formation is facilitated by interactions between the hydrophilic parts of membrane proteins. This advantage of a small detergent head group is consistent with the general notion that conventional glucoside detergents (OG and NG) are widely used for membrane protein crystallization, although are generally less favourable than maltoside detergents (DM and DDM) for membrane protein stabilization.11a A similar trend can be found for novel amphiphiles. For example, GNG-3 has facilitated crystal structure determination of a few membrane proteins in the last three years17j–l and FA-5 showed promising behaviour in the crystallization of a couple of target proteins;16a both agents have glucoside head groups. Despite such favourable properties, the glucoside head group has not been popularly utilized in novel amphiphile design. This is mainly due to the general perception that glucoside amphiphiles are less stabilizing than maltoside agents, as can be seen in the comparison of OG vs. DDM or GNG vs. MNG. To date, there are no glucoside detergents that confer consistently greater stability to a range of membrane proteins than DDM. Remarkably, this seems to be the case for the TDTs and NDTs. Furthermore, NDT-C11 was superior to MNG-3, one of most promising novel amphiphiles, in providing conformational flexibility essential for protein function, in addition to high efficacy for membrane protein stabilization. This result indicates that NDT-C11 could be an optimal novel agent for biophysical studies requiring both stable and functional proteins.
Conclusions
In summary, the novel triglucoside amphiphiles with a TRIS or NPG linker were prepared and evaluated with a few membrane proteins. In this evaluation, the novel agents were consistently better than DDM in stabilizing the native structures of the target membrane proteins. Interestingly, NPG-derived triglucoside agents (NDTs) were markedly superior to TRIS-based analogs (TDTs) for all tested target membrane proteins, indicating the important role of the functional group in the linker region in determining detergent efficacy. Of NDTs, NDT-C11 conferred the most enhanced stability on the target membrane proteins, presumably originating from its optimal hydrophile–lipophile balance (HLB). The protein stabilizing efficacy of NDT-C11 and its ability to retain protein conformational flexibility together with the presence of the glucoside head group and the straightforward synthesis protocol, strongly indicate that these agents hold significant potential for membrane protein structural and functional study.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number 2008-0061891 and 2013R1A2A2A03067623 to P. S. C. and A. S.). The work was also supported by Biotechnology and Biological Sciences Research Council (grant BB/K017292/1 to B. B.), by the National Science Foundation (grant MCB-1158085 to L. G.) and by the National Institutes of Health (grant R01 GM095538 to L. G.), by the Danish Council for Independent Research Sapere Aude program 0602-02100B (to C. J. L.) and the Lundbeck Foundation R108-A10755 (to C. J. L.).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sc02900g
References
- (a) Wallin E., von Heijne G. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fagerberg L., Jonasson K., von Heijne G., Uhlen M., Berglund L. Proteomics. 2010;10:1141–1149. doi: 10.1002/pmic.200900258. [DOI] [PubMed] [Google Scholar]
- (a) Drake M. T., Shenoy S. K., Lefkowitz R. J. Circ. Res. 2006;99:570–582. doi: 10.1161/01.RES.0000242563.47507.ce. [DOI] [PubMed] [Google Scholar]; (b) Lappano R., Maggiolini M. Nat. Rev. Drug Discovery. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- (a) Sanders C. R., Myers J. K. Annu. Rev. Biophys. Biomol. Struct. 2004;33:25–51. doi: 10.1146/annurev.biophys.33.110502.140348. [DOI] [PubMed] [Google Scholar]; (b) Overington J. P., Al-Lazikani B., Hopkins A. L. Nat. Rev. Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- (a) Rosenbaum D. M., Rasmussen S. G. F., Kobilka B. K. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) McCusker E. C., Bagneris C., Naylor C. E., Cole A. R., D'Avanzo N., Nichols C. G., Wallace B. A. Nat. Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ghosh E., Kumari P., Jiaman D., Shukla A. K. Nat. Rev. Mol. Cell Biol. 2015;16:69–81. doi: 10.1038/nrm3933. [DOI] [PubMed] [Google Scholar]
- (a) Nam H. J., Jouhyun J., Sanguk K. BMB Rep. 2009;42:697–704. doi: 10.5483/bmbrep.2009.42.11.697. [DOI] [PubMed] [Google Scholar]; (b) Deschamps J. R. AAPS J. 2005;7:813–819. doi: 10.1208/aapsj070478. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Murray C. W., Blundell T. L. Curr. Opin. Struct. Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html .
- (a) White S. H., Wimley W. C. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; (b) Bowie J. U. Curr. Opin. Struct. Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; (c) Lacapere J. J., Pebay-Peyroula E., Neumann J. M., Etchebest C. Trends Biochem. Sci. 2007;32:259–327. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]; (d) Philips R., Ursell T., Wiggins P., Sens P. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Seddon A. M., Curnow P., Booth P. J. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]; (b) Yang Z., Wang C., Zhou Q., An J., Hildebrandt E., Aleksandrov L. A., Kappes J. C., DeLucas L. J., Riordan J. R., Urbatsch I. L., Hunt J. F., Brouillette C. G. Protein Sci. 2014;23:769–789. doi: 10.1002/pro.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J. L., Newstead S. Protein Sci. 2012;21:1358–1365. doi: 10.1002/pro.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Serrano-Vega M. J., Magnani F., Shibata Y., Tate C. G. Proc. Natl. Acad. Sci. U. S. A. 2008;105:877–882. doi: 10.1073/pnas.0711253105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Newstead S., Ferrandon S., Iwata S. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) He Y., Wang K., Yan N. Protein Cell. 2014;5:658–672. doi: 10.1007/s13238-014-0086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Privé G. G. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; (b) Chae P. S., Laible P. D., Gellman S. H. Mol. BioSyst. 2010;6:89–94. doi: 10.1039/b915162c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Zhang Q., Tao H., Hong W.-X. Methods. 2011;55:318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Chae P. S., Wander M. J., Cho K. H., Laible P. D., Gellman S. H. Mol. BioSyst. 2013;9:626–629. doi: 10.1039/c3mb25584k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bae H. E., Gotfryd K., Thomas J., Hussain H., Ehsan M., Go J., Loland C. J., Byrne B., Chae P. S. ChemBioChem. 2015;16:1454–1459. doi: 10.1002/cbic.201500151. [DOI] [PubMed] [Google Scholar]
- (a) McGregor C.-L., Chen L., Pomroy N. C., Hwang P., Go S., Chakrabartty A., Privé G. G. Nat. Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]; (b) Tao H., Lee S. C., Moeller A., Roy R. S., Siu F. Y., Zimmermann J., Stevens R. C., Potter C. S., Carragher B., Zhang Q. Nat. Methods. 2013;10:759–761. doi: 10.1038/nmeth.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Tribet C., Audebert R., Popot J.-L. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Popot J. L., Althoff T., Bagnard D., Banères J. L., Bazzacco P., Billon-Denis E., Catoire L. J., Champeil P., Charvolin D., Cocco M. J., Crémel G., Dahmane T., de la Maza L. M., Ebel C., Gabel F., Giusti F., Gohon Y., Goormaghtigh E., Guittet E., Kleinschmidt J. H., Kühlbrandt W., Le Bon C., Martinez K. L., Picard M., Pucci B., Sachs J. N., Tribet C., van Heijenoort C., Wien F., Zito F., Zoonens M. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]; (c) Nath A., Atkins W. M., Sligar S. G. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]; (d) Orwick-Rydmark M., Lovett J. E., Graziadei A., Lindholm L., Hicks M. R., Watts A. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- (a) Chae P. S., Rasmussen S. G. F., Rana R. R., Gotfryd K., Kruse A. C., Nurva S., Gether U., Guan L., Loland C. J., Byrne B., Kobilka B. K., Gellman S. H. Chem.–Eur. J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) McQuade D. T., Quinn M. A., Yu S. M., Polans A. S., Krebs M. P., Gellman S. H. Angew. Chem., Int. Ed. 2000;39:758–761. [PubMed] [Google Scholar]; (c) Chae P. S., Wander M. J., Bowling A. P., Laible P. D., Gellman S. H. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Chae P. S., Cho K. H., Wander M. J., Bae H. E., Gellman S. H., Labile P. D. Biochim. Biophys. Acta. 2014;1838:278–286. doi: 10.1016/j.bbamem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Matar-Merheb R., Rhimi M., Leydier A., Huche F., Galian C., Desuzinges-Mandon E., Ficheux D., Flot D., Aghajari N., Kahn R., Di Pietro A., Jault J. M., Coleman A. W., Falson P. PLoS One. 2011;6:e18036. doi: 10.1371/journal.pone.0018036. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lee S. C., Bennett B. C., Hong W.-X., Fu Y., Baker K. A., Marcoux J., Robinson C. V., Ward A. B., Halpert J. R., Stevens R. C., Stout C. D., Yeager M. J., Zhang Q. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1203–1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chae P. S., Gotfryd K., Pacyna J., Miercke L. J. W., Rasmussen S. G. F., Robbins R. A., Rana R. R., Loland C. J., Kobilka B., Stroud R., Byrne B., Gether U., Gellman S. H. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Chae P. S., Rana R. R., Gotfryd K., Rasmussen S. G. F., Kruse A. C., Cho K. H., Capaldi S., Carlsson E., Kobilka B. K., Loland C. J., Gether U., Banerjee S., Byrne B., Lee J. K., Gellman S. H. Chem. Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Cho K. H., Bae H. E., Das M., Gellman S. H., Chae P. S. Chem. Asian J. 2014;9:632–638. doi: 10.1002/asia.201301303. [DOI] [PubMed] [Google Scholar]; (f) Chae P. S., Rasmussen S. G. F., Rana R. R., Gotfryd K., Chandra R., Goren M. A., Kruse A. C., Nurva S., Loland C. J., Pierre Y., Drew D., Popot J. L., Picot D., Fox B. G., Guan L., Gether U., Byrne B., Kobilka B., Gellman S. H. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Cho K. H., Byrne B., Chae P. S. ChemBioChem. 2013;14:452–455. doi: 10.1002/cbic.201200759. [DOI] [PubMed] [Google Scholar]
- (a) Rasmussen S. G. F., Choi H. J., Fung J. J., Pardon E., Casarosa P., Chae P. S., DeVree B. T., Rosenbaum D. M., Thian F. S., Kobilka T. S., Schnapp A., Konetzki I., Sunahara R. K., Gellman S. H., Pautsch A., Steyaert J., Weis W. I., Kobilka B. K. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Rosenbaum D. M., Zhang C., Lyons J., Holl R., Aragao D., Arlow D. H., Rasmussen S. G. F., Choi H.-J., DeVree B. T., Sunahara R. K., Chae P. S., Gellman S. H., Dror R. O., Shaw D. E., Weis W. I., Caffrey M., Gmeiner P., Kobilka B. K. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rasmussen S. G. F., DeVree B. T., Zou Y., Kruse A. C., Chung K. Y., Kobilka T. S., Thian F. S., Chae P. S., Pardon E., Calinski D., Mathiesen J. M., Shah S. T. A., Lyons J. A., Caffrey M., Gellman S. H., Steyaert J., Skiniotis G., Weis W. I., Sunahara R. K., Kobilka B. K. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Ring A. M., Manglik A., Kruse A. C., Enos M. D., Weis W. I., Garcia K. C., Kobilka B. K. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Shukla A. K., Westfield G. H., Xiao K., Reis R. I., Huang L.-Y., Shukla P. T., Qian J., Li S., Blanc A., Oleskie A. N., Dosey A. M., Su M., Liang C.-R., Gu L. L., Shan J. M., Chen X., Hanna R., Choi M., Yao X. J., Klink B. U., Kahsai A. W., Sidhu S. S., Koide S., Penczek P. A., Kossiakoff A. A., Woods Jr V. L., Kobilka B. K., Skiniotis G., Lefkowitz R. J. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Kruse A. C., Hu J., Pan A. C., Arlow D. H., Rosenbaum D. M., Rosemond E., Green H. F., Liu T., Chae P. S., Dror R. O., Shaw D. E., Weis W. I., Wess J., Kobilka B. K. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Haga K., Kruse A. C., Asada H., Kobayashi T. Y., Shiroishi M., Zhang C., Weis W. I., Okada T., Kobilka B. K., Haga T., Kobayashi T. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Manglik A., Kruse A. C., Kobilka T. S., Thian F. S., Mathiesen J. M., Sunahara R. K., Pardo L., Weis W. I., Kobilka B. K., Granier S. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Granier S., Manglik A., Kruse A. C., Kobilka T. S., Thian F. S., Weis W. I., Kobilka B. K. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Kellosalo J., Kajander T., Kogan K., Pokharel K., Goldman A. Science. 2012;337:473–476. doi: 10.1126/science.1222505. [DOI] [PubMed] [Google Scholar]; (k) Quigley A., Dong Y. Y., Pike A. C. W., Dong L., Shrestha L., Berridge G., Stansfeld P. J., Sansom M. S. P., Edwards A. N., Bountra C., Von Delft F., Bullock A. N., Burgess-Brown N. A., Carpenter E. P. Science. 2013;339:1604–1607. doi: 10.1126/science.1231513. [DOI] [PubMed] [Google Scholar]; (l) Frick A., Eriksson U. K., de Mattia F., Oberg F., Hedfalk K., Neutze R., Grip W. J., Deen P. M. T., Tornroth-Horsefield S. Proc. Natl. Acad. Sci. U. S. A. 2014;111:6305–6310. doi: 10.1073/pnas.1321406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Sonoda Y., Newstead S., Hu N. J., Alguel Y., Nji E., Beis K., Yashiro S., Lee C., Leung J., Cameron A. D., Byrne B., Iwata S., Drew D. Structure. 2011;19:17–25. doi: 10.1016/j.str.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Caffrey M., Li D., Dukkipati A. Biochemistry. 2012;51:6266–6288. doi: 10.1021/bi300010w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay A., London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- Takano J., Noguchi K., Yasumori M., Kobayashi M., Gajdos Z., Miwa K., Hayashi H., Yoneyama T., Fujiwara T. Nature. 2002;420:337–340. doi: 10.1038/nature01139. [DOI] [PubMed] [Google Scholar]
- Alexandrov A., Mileni M., Chien E. Y., Hanson M. A., Stevens R. C. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- (a) Guan L., Nurva S., Ankeshwarapu S. P. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ethayathulla A. S., Yousef M. S., Amin A., Leblanc G., Kaback H. R., Guan L. Nat. Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Amin A., Ethayathulla A. S., Guan L. J Bacterol. 2014;196:3134–3139. doi: 10.1128/JB.01868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehrel C., Cordat E., Mus-Veteau I., Leblanc G. J. Biol. Chem. 1998;273:33192–33197. doi: 10.1074/jbc.273.50.33192. [DOI] [PubMed] [Google Scholar]
- Amin A., Hariharan P., Chae P. S., Guan L. Biochemistry. 2015;54:5849–5855. doi: 10.1021/acs.biochem.5b00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert G., Warren P. V., Gaasterland T., Young W. G., Lenox A. L., Graham D. E., Overbeek R., Snead M. A., Keller M., Aujay M., Huber R., Feldman R. A., Short J. M., Olsen G. J., Swanson R. V. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- (a) Hart H. E., Greenwald E. B. Mol. Immunol. 1979;16:265–267. doi: 10.1016/0161-5890(79)90065-8. [DOI] [PubMed] [Google Scholar]; (b) Quick M., Javitch J. A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billesbølle C. B., Kruger M. B., Shi L., Quick M., Li Z., Stolzenberg S., Kniazeff J., Gotfryd K., Mortensen J. S., Javitch J. A., Weinstein H., Loland C. J., Gether U. J. Biol. Chem. 2015;290:26725–26738. doi: 10.1074/jbc.M115.677658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Terry D., Shi L., Weinstein H., Blanchard S. C., Javitch J. A. Nature. 2010;465:188–193. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.