Abstract
DNA-protein recognition has shown us the importance of DNA shapes in the recognition process. Specific high-affinity targeting of DNA shapes by small molecules is desirable for many biological applications that involve regulation of DNA based processes. Here, the effect of linker length and rigidity on the affinity of a conjugated neomycin dimer for a specific DNA shape (B* form) AT-rich DNA was explored. Binding constants approximating 108 M−1 for optimal linker lengths of 18–19 atoms are reported herein.
Keywords: Nucleic Acid, Aminoglycoside Derivatives, Major Groove Binders, Fluorescent Intercalator Displacement, Isothermal Calorimetry Titration
Graphical Abstract
1. Introduction
DNA secondary structures enable many key biological processes including storage of genetic information, replication, transcription, and translation, which makes them important molecular targets of therapeutic interest.1–3 However, DNA recognition using small molecule ligands with selectivity, high efficacy, and limited toxicity has remained an elusive goal in contemporary medicinal chemistry. Small molecule based nucleic acid recognition is advantageous because DNA offers a comparatively simpler target in comparison to protein targets when probing the molecular basis of protein-nucleic acid interactions. Small molecules of both natural and synthetic origin that interact with DNA via non-covalent interactions fall within the category of either intercalators or groove binders.2, 3 In recent years, considerable interest has developed in structure based design of small molecules that target specific sequences of DNA.2 Inspired by the DNA recognition ability of naturally occurring distamycin, Dervan and coworkers have engineered small molecules called polyamides that can specifically target DNA sequences of interest.4–6 Among others, carbohydrate based small molecules have attracted considerable attention in recent years for their ability to target nucleic acids.7 Our group has demonstrated that aminosugars, a class of carbohydrates, bind to a variety of nucleic acid geometries, including both RNA and DNA triple helices,8 hybrid triple helices,9 poly(rA),10 and even high order nucleic acid structures including DNA quadruplexes.11, 12 Furthermore, we explored the binding of aminosugars with B-form DNA13–15 and DNA/RNA hybrid, and we have demonstrated the expansion of aminosugars’ ability to recognize a variety of diverse nucleic acid structures by conjugating them with small molecule intercalators,16, 17 and minor groove binders.15
Recently, we have shown that a conjugated neomycin dimer (DPA 79) can specifically bind to a ~10–12 base pair stretch of B*-form DNA with a binding constant in the range of Ka ~108 M−1.13 This neomycin dimer was one of the first “all carbohydrate” ligands to bind duplex DNA with such high affinity, without any planar or intercalative moieties, and showed clear preference in binding to B*DNA containing A tracts over DNA with alternating AT or GC rich sequences. The ITC derived binding profile suggested two different sets of binding sites, in which the first binding event was entropically driven, whereas the second binding event was enthalpically driven. The entropic contribution for the first binding event was suggested to arise from the destabilization of the spine of hydration which results in liberation of water molecules from the major groove into bulk solvent during the binding interaction. The first binding event was highly specific, and demonstrated an affinity for the DNA target which was ~100–500- fold higher than the affinity shown for the secondary binding event.13, 18–20
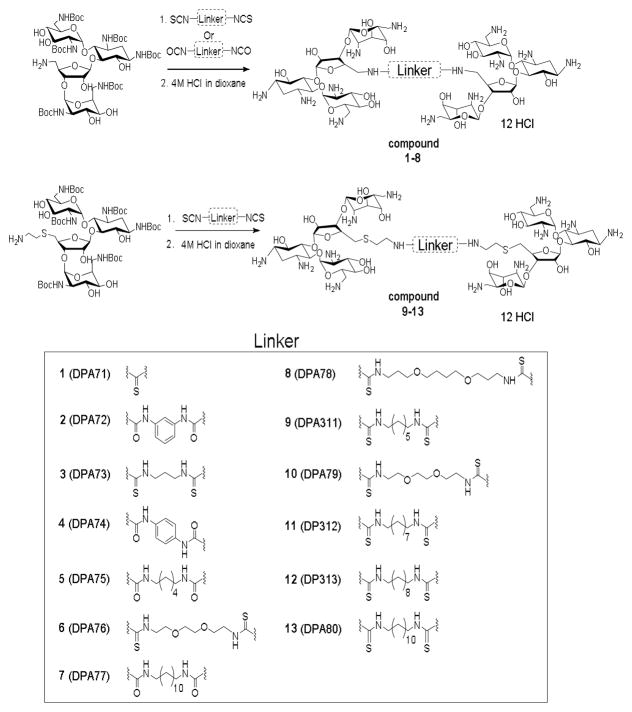
Recognition of subtle differences in double-stranded DNA architectures by groove-binding small molecules, such as the ones we describe, is an essential step in a comprehensive approach to DNA targeting. Previously, our work showed that a neomycin dimer had a binding preference that changed with DNA shape and sequence, and favoured B* form DNA.13 This DNA sub-structure is exemplified by a narrower minor groove and a comparatively more rigid structure, overall, with respect to its standard B form counterpart, and is commonly adopted by continuous AT tracts. Herein, we report the synthesis of a library of neomycin dimers with varying linker lengths (Figure 1), designed to achieve optimal binding affinity with AT rich DNA. The range of the linker length varies from L = 1 to 22, where L = number of carbon and/or oxygen atoms between two neomycin units starting after 5″-amine. The binding was characterized using two independent techniques; isothermal titration calorimetry (ITC) and fluorescent intercalator displacement (FID) assay. The results obtained from both techniques agree well and corroboration of these data allowed for observation of trends in binding affinities for the dimers and their contiguous AT rich DNA targets.
Figure 1.
2. Materials
2.1 Chemicals
All chemicals were purchased from commercial suppliers and used without further purification. Neomycin B tri-sulfate was purchased from MP Biomedicals (Solon, OH), di-tert-butyl dicarbonate (Boc anhydride) was purchased from Advanced ChemTech (Louisville, KY), SC (Sodium cacodylate), EDTA (Ethylenediamine tetraacetic acid), KCl, and sodium phosphate (mono- and di-) salts were purchased from Fisher Scientific (Hampton, NH). DMAP (4-dimethyl-aminopyridine) was purchased from Acros organics (Fair Lawn, NJ). TCDP (1,1′-thiocarbonyldi-2(1H)-pyridone), TPS-Cl (2,4,6-triisopropylbenzenesulfonyl chloride), and 4M HCl in dioxane were purchased from Sigma Aldrich (St. Louis, MO). All the diamines and diisocyanates were purchased from Sigma Aldrich (St. Louis, MO). Silica plates (w/UV254, aluminum backed) and silica gel (particle size = 40–63 micron, 230 × 400 mesh) for flash column chromatography were purchased from Sorbent Technologies (Atlanta, GA). All solvents were purchased from VWR. Reaction solvents were distilled over calcium hydride [pyridine, DCM (dichloromethane)]. EtOH (Ethanol) was first distilled with sodium metal and then redistilled over magnesium turnings. Reactions were conducted under N2 using a dry solvent, unless otherwise noted.
2.2 Nucleic Acids
The 12-mer duplex d[5′-A12-x-T12-3′] (x= hexaethylene glycol) oligonucleotide was synthesized on an Applied Biosystem 8890 using standard phosphoramidite chemistry and purified by HPLC on a Gen-Pak FAX (4.6 x 100 mm) ion exchange column, eluting with buffer A (25 mM Tris HCl, 1 mM EDTA, 10 % CH3CN, pH 8.0) from 98% to 50% and buffer B (25 mM Tris HCl, 1 mM EDTA, 1 M NaClO4, 10 % CH3CN, pH 8.0) from 2% to 50% in a 15 minute process. All other oligonucleotides were purchased from IDT DNA (Coraville, IA), the concentrations of which were determined by UV using the following extinction coefficients [L/ (mole·cm)]:ε260 = 157,900 for d[5′-G2A6T6C2-3′], ε260 = 59,844 for d[5′-A12-x-T12-3′], ε260 = 605,700 for d[5′-A30T30-3′].
2.3 Instrumentation
1H NMR spectra were collected on a Bruker 500 MHz FT-NMR spectrometer, and the MS (MALDI-TOF) spectra were collected using a Bruker Omniflex MALDI-TOF mass spectrometer. Isothermal titration calorimetric measurements were performed on a MicroCal VP-ITC (MicroCal, Inc. Northampton, MA). FID assays were performed in 96-well plates (Black w/flat bottom, Greiner Bio-one, Monroe, NC) on a Genios Multi-Detection Microplate Reader, TECAN (McLean, VA) with Magellan software.
3. Methods
3.1 Isothermal Titration Calorimetry (ITC)
In a typical experiment, an aliquot (9 μL) of neomycin dimer (125 μM in 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8) was injected at 25 °C into an isothermal sample chamber containing an oligonucleotide duplex solution (1.42 mL, 4 μM/duplex in 100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8) via a rotary syringe (300 rpm, vol. 296 μL). The interval between each injection was 300 seconds, the duration of each injection was 10 s, and the initial delay prior to the first injection was 60 s. Injection of neomycin dimer at identical concentrations into a buffer solution at 25 °C was used as a blank. Each injection generated a heat burst curve of microcalories per second. The area under each heat burst curve was determined by integration using the Origin (version 5.0, MicroCal, Inc. Northampton, MA) software to obtain a measure of the heat associated with that injection. The heat associated with each drug-buffer injection was subtracted from the corresponding heat associated with each drug-DNA injection to yield the heat of drug binding for that injection. The final corrected injection heats were plotted as a function of molar ratio ([drug]/[DNA]) and either fitted with a single binding site model or with two independent binding site models. In the case of two binding site model, the heat curves were fit by allowing free floating of N1 and N2 (binding stoichiometric ratios, drug/DNA). The fits were optimized until no more change in the chi-squared value was observed.
3.2 Fluorescence Intercalator Displacement Assay (FID)
FID assay for oligomeric DNA duplexes were conducted in a 96 well plate. For FID single point assay, a thiazole orange (TO) solution (0.5 equivalent/base pair, 200 μL) in sodium cacodylate buffer (100 mM KCl, 10 mM SC, 0.5 mM EDTA, pH 6.8) was prepared and added to an oligomeric duplex solution with final concentration of 1 μM/strand (for d[5′-A12-x-T12-3′]) or 1 μM/duplex (for d[5′-G2A6T6C2-3′]). The fluorescence of the solution in each well was measured and normalized to a 100% relative fluorescence. An aliquot of the stock solution of ligand (5 μM to 200 μM) was added into the pre-incubated solution of DNA+TO and the fluorescence was measured after 5 min at 20 °C. The FID assay were conducted in triplicate. For FID titrations, the addition of ligand was continued until no more change in the fluorescence was detected. For FID assay, the final concentrations were corrected for dilution (less than 5% of the total volume). Fluorescence readings were reported as a percentage of fluorescence relative to the control wells. The reference fluorescence was defined as [TO+DNA] providing 100% fluorescence and [TO] alone provides 0% fluorescence. The reported error for FID assay was standard error which is calculated from three independent experiments.
4. Results and discussion
4.1. Synthesis of Neomycin dimers
The library of neomycin dimers was synthesized using thiourea- and urea-based coupling chemistry. Hexa-N-Boc deoxy-neomycin-5″-amine with short and long (Figure 1) linkers were synthesized from commercially available neomycin sulphate using previously published methods.13, 18, 19, 21, 22 The diisothiocyanate linkers used in the synthesis of neomycin dimers were prepared by reacting commercially available diamines with TCDP (1,1′-thiocarbonyldi-2(1H)-pyridone) in almost quantitative yields (see supplementary information Table S1). The Hexa-N-Boc deoxy-neomycin-5″-amines were then reacted with diisothiocyanates and diisocyanates in the presence of pyridine (dry) and DMAP (catalytic amounts) for 12 hours at room temperature to produce the Boc protected neomycin dimers with various linker lengths (DPA71–DPA80, see supporting information Scheme S1). The acid labile Boc protected neomycin dimers were then reacted with 4M HCl/dioxane for 30 minutes to obtain the neomycin dimers as their hydrochloride salts in good yields (60%–71% yield for the two steps) (Figure 1).
4.2. FID assay reveals longer linker neomycin dimers are stronger binder than short linker dimers
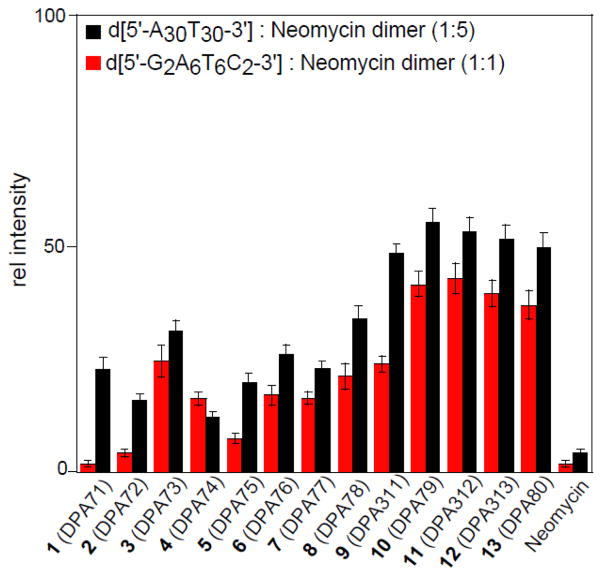
A single point FID assay23 was performed with the neomycin dimers and DNA duplexes ([5′-G2A6T6C2-3′] and [5′-A30T30-3′]). DNA duplexes were incubated with thiazole orange, an exogenous intercalator, at a stoichiometric ratio of 1: 0.5 (DNA base pair: thiazole orange) followed by the addition of neomycin dimers. The binding site size for neomycin dimer is ~12 base pairs/molecule, therefore we used molar ratios of 1:1 and 1:5 for [5′-G2A6T6C2-3′] and [5′-A30T30-3′] respectively. The decrease in fluorescence intensity can be directly correlated with the affinity of neomycin dimers (Figure 2). In all the studies carried out, including ITC and FID, neomycin a weak binder of AT-rich DNA served as a control. There are few important trends that emerged from the FID study: (1) Neomycin dimers, in general, exhibit stronger binding to AT-rich DNA than neomycin (2) Neomycin dimers with long linkers, specifically L= 16 to 22, exhibit 30–50 fold higher affinity to AT rich DNA sequences than neomycin. (3) Neomycin dimers (L=17–22) displayed higher affinity than short/rigid linker based neomycin dimers (L= 1 to 12). In the case of d[5′-G2A6T6C2-3′], DPA312 (L=19) attenuated the fluorescence intensity by a factor of ~25 than DPA71 (L=1). (4) The activity trend observed for neomycin dimers is similar towards both the AT-rich DNA target sequences, however there is a slight difference in % fluorescence change which might be attributed to the difference in the DNA sequences. It appears from the FID assay that neomycin dimers, DPA79 (L=18) and DPA312 (L=19) exhibit the greatest activity for the AT rich DNA duplexes. The significant decrease in the activity for neomycin dimers with short linkers (DPA71–DPA76) is presumably because of the decreased conformational freedom that the short linkers allow in comparison to the longer linkers. This would result in a decrease in the necessary flexibility required for optimal binding.
Figure 2.
Bar graph represents the FID-derived change in % fluorescence of AT-rich DNA-Thiazole Orange complex in the presence of indicated ligands.
4.2. The ITC-derived binding affinities of neomycin dimers corroborate with FID based binding activities
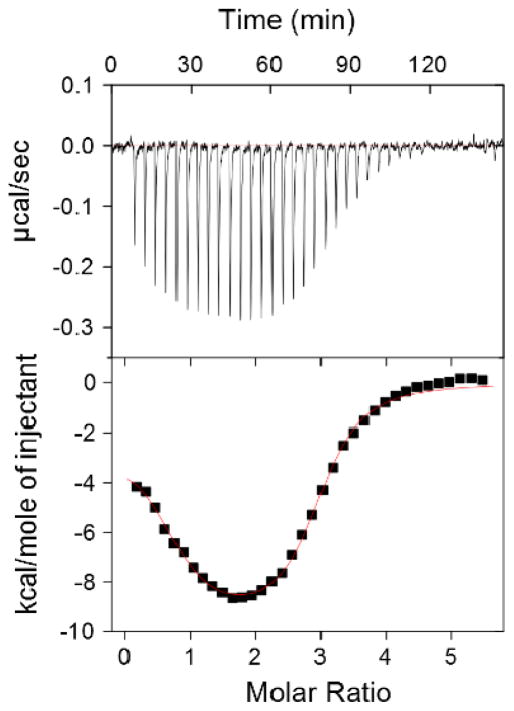
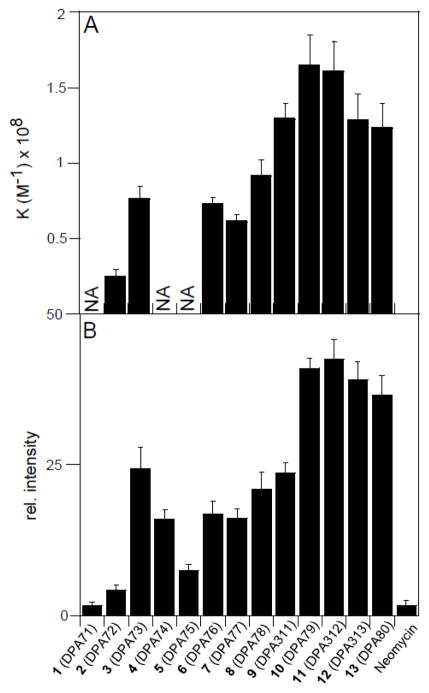
ITC was employed to gain insight into the thermodynamics of binding between d[5′-G2A6T6C2-3′] and neomycin dimers (DPA71–DPA80) (Figure 3, 4, and supporting information Table S3). The ITC studies were performed at pH = 5.5 to ensure that the dimers were substantially protonated (lowest pKa for an amine group of neomycin = 5.91).19 The following observations were noted from the thermodynamic characterization. (1) Similar to our earlier reported work, neomycin dimers with linker length L=12 to 22 exhibit two sets of binding sites, in which the first is predominantly entropically driven, with a binding stoichiometry of 1:1 (neomycin dimer:DNA) while the second is enthalpically driven with a binding stoichiometry of ~3:1 (neomycin dimer:DNA). The binding stoichiometry for the first binding event was ~10–12 base pairs per neomycin dimer, which is also validated by CD and FID titrations in our earlier published work.13 (2) Under otherwise matched conditions, the ITC profile for neomycin dimers with linker length below L=12 is either not saturated or exhibits multiple sets of binding sites and precluded us from extracting any meaningful thermodynamic data (Fig. 4A). We conjecture this is due to weak binding interactions, likely caused by electrostatic interaction between the multi-cationic neomycin dimers and the negatively charged DNA backbone. (3) DPA79 (L=18) with a binding constant ~108 M−1 exhibits ~4–5 fold higher affinity than DPA72 (L=7). (4) ITC-derived binding constants for neomycin dimers with d[5′-G2A6T6C2-3′] agreed well with the FID assay based binding activity. Additionally, the trend for ITC derived binding constant was in fair agreement with FID based AC50 values (aminoglycoside concentration required for 50% fluorescence change) calculated for neomycin dimer with an AT rich DNA duplex ([5′-A12–x-T12-3′]) (see supporting information Figure S69). It is intriguing that the binding interaction between AT-rich DNA and neomycin dimers is saturated at linker length of 18 and did not change significantly afterwards (upto L=22). The lack of structural information prohibited us in offering an explanation.
Figure 3.
ITC based binding profile of DPA312 with d[5′-G2A6T6C2-3′]. The upper panel represents the heat burst curves, each of which is the result of a 9 μL injection of 125 μM of DPA312 serially added to the DNA (4 μM/duplex) duplex.
Figure 4.
Comparison of binding activity of indicated ligands with d[5′-G2A6T6C2-3′] using ITC (A) and FID assay (B). *NA-Not able to extract thermodynamic data.
5. Conclusion
The targeting of DNA by small molecules is an extremely desirable goal in our quest for specific gene regulation. Despite the therapeutic potential of compounds, the discovery of DNA-binding ligands that are both DNA shape-specific and demonstrate high affinity for their target has proven to be difficult. In this work, the effect of linker length and flexibility is probed for a series of neomycin conjugates that show promise as high affinity AT rich DNA duplex-binding ligands. The preferential binding and recognition of DNA sub-structures such as B* DNA is an important first step towards the development of DNA-targeted shape and sequence recognition. In summary, a series of neomycin dimers targeting B* DNA was synthesized, and a change in linker length from L=12 to L=18 resulted in a significant difference in the binding constant. ITC and FID assays suggest that the highest binding affinity was exhibited by neomycin dimers with linker lengths of L=18 to L=19. In contrast, the ITC profiles for short linker based neomycin dimers suggested weak or nonspecific binding to B* form AT rich DNA duplexes. These studies underscore the importance of linker length and flexibility optimization in the design of novel DNA-binding small molecule conjugates.
Supplementary Material
Acknowledgments
The authors thank the National Institutes of Health for support of this work (R15CA125724 and R41AI114114 to D.P.A.).
Footnotes
Supplementary material that may be helpful in the review process should be prepared and provided as a separate electronic file. That file can then be transformed into PDF format and submitted along with the manuscript and graphic files to the appropriate editorial office.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Hecht SM. J Am Chem Soc. 2009;131:3791. doi: 10.1021/ja9012485. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB. Bioorg Med Chem. 2001;9:2215. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 3.Neidle S. Nat Prod Rep. 2001;18:291. doi: 10.1039/a705982e. [DOI] [PubMed] [Google Scholar]
- 4.Mrksich M, Dervan PB. J Am Chem Soc. 1995;117:3325. [Google Scholar]
- 5.White S, Baird EE, Dervan PB. Biochemistry. 1996;35:12532. doi: 10.1021/bi960744i. [DOI] [PubMed] [Google Scholar]
- 6.Dervan PB, Bürli RW. Curr Opin Chem Biol. 1999;3:688. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 7.Willis B, Arya DP. Curr Org Chem. 2006;10:663. [Google Scholar]
- 8.Arya DP, Coffee RL, Jr, Willis B, Abramovitch AI. J Am Chem Soc. 2001;123:5385. doi: 10.1021/ja003052x. [DOI] [PubMed] [Google Scholar]
- 9.Arya DP, Coffee RL, Jr, Charles I. J Am Chem Soc. 2001;123:11093. doi: 10.1021/ja016481j. [DOI] [PubMed] [Google Scholar]
- 10.Xi H, Gray D, Kumar S, Arya DP. FEBS Lett. 2009;583:2269. doi: 10.1016/j.febslet.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Ranjan N, Andreasen KF, Kumar S, Hyde-Volpe D, Arya DP. Biochemistry. 2010;49:9891. doi: 10.1021/bi101517e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arya DP. Acc Chem Res. 2011;44:134. doi: 10.1021/ar100113q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S, Xue L, Arya DP. J Am Chem Soc. 2011;133:7361. doi: 10.1021/ja108118v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willis B, Arya DP. Biochemistry. 2006;45:10217. doi: 10.1021/bi0609265. [DOI] [PubMed] [Google Scholar]
- 15.Arya DP, Willis B. J Am Chem Soc. 2003;125:12398. doi: 10.1021/ja036742k. [DOI] [PubMed] [Google Scholar]
- 16.Arya DP, Xue L, Tennant P. J Am Chem Soc. 2003;125:8070. doi: 10.1021/ja034241t. [DOI] [PubMed] [Google Scholar]
- 17.Shaw NN, Xi H, Arya DP. Bioorg Med Chem Lett. 2008;18:4142. doi: 10.1016/j.bmcl.2008.05.090. [DOI] [PubMed] [Google Scholar]
- 18.Charles I, Xue L, Arya DP. Bioorg Med Chem Lett. 2002;12:1259. doi: 10.1016/s0960-894x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Spano MN, Arya DP. Biopolymers. 2014;101:720. doi: 10.1002/bip.22448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamilton P, Arya DP. Nat Prod Rep. 2012;29:134. doi: 10.1039/c1np00054c. [DOI] [PubMed] [Google Scholar]
- 21.Kirk SR, Luedtke NW, Tor Y. J Am Chem Soc. 2000;122:980. [Google Scholar]
- 22.Xue L, Xi H, Kumar S, Gray D, Davis E, Hamilton P, Skriba M, Arya DP. Biochemistry. 2010;49:5540. doi: 10.1021/bi100071j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. J Am Chem Soc. 2001;123:5878. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.