Abstract
Higher order chromatin structure establishes domains that organize the genome and coordinate gene expression. However, the molecular mechanisms controlling transcription of individual loci within a topological domain (TAD) are not fully understood. The cystic fibrosis transmembrane conductance regulator (CFTR) gene provides a paradigm for investigating these mechanisms. CFTR occupies a TAD bordered by CTCF/cohesin binding sites within which are cell-type-selective cis-regulatory elements for the locus. We showed previously that intronic and extragenic enhancers, when occupied by specific transcription factors, are recruited to the CFTR promoter by a looping mechanism to drive gene expression. Here we use a combination of CRISPR/Cas9 editing of cis-regulatory elements and siRNA-mediated depletion of architectural proteins to determine the relative contribution of structural elements and enhancers to the higher order structure and expression of the CFTR locus. We found the boundaries of the CFTR TAD are conserved among diverse cell types and are dependent on CTCF and cohesin complex. Removal of an upstream CTCF-binding insulator alters the interaction profile, but has little effect on CFTR expression. Within the TAD, intronic enhancers recruit cell-type selective transcription factors and deletion of a pivotal enhancer element dramatically decreases CFTR expression, but has minor effect on its 3D structure.
INTRODUCTION
The role of the architectural proteins CCCTC-binding factor (CTCF) and cohesin complex in organizing the higher order structure of the genome and facilitating coordinated gene expression is now well-established (1–6). Moreover, genome-wide studies using a variety of novel approaches showed the integrated functions of cell-selective transcription factors within the topological domains established by the architectural proteins. Though the global view of chromatin organization and its inter-relationship with gene expression is now becoming clearer, the details, which can be provided by in-depth investigation of gene clusters or single gene loci, require further analysis. Much has been learnt from analysis of cis-regulatory elements and cell-specific 3D structures within the globin and homeobox (HOX) gene clusters (7–15). In contrast there are few single gene loci that are as well studied as these gene clusters, however one exception is the cystic fibrosis transmembrane conductance regulator (CFTR) gene (reviewed in (16,17)). CFTR is a large (∼190 kb) gene lying within a single transcriptional unit flanked by CTCF- and cohesin complex-binding elements. The gene is expressed primarily in specialized epithelial cells, though the relative abundance of the transcript shows substantial variation in different tissues. Mutations in CFTR cause the devastating inherited disorder cystic fibrosis (CF). The CFTR locus adopts a looped structure which is limited by architectural proteins, but within which intronic and extragenic cis-regulatory elements are brought into close association with the gene promoter by additional chromatin tethers (18–21). The CFTR promoter appears to lack tissue-specific regulatory elements and cell-type control is provided by the recruitment of different sets of cis-elements in airway and intestinal epithelial cells. To date the best-characterized CFTR cis-elements are enhancers, which bind diverse transcription factors in the individual differentiated cell types. In intestinal epithelial cells enhancers in introns 1 and 11 of the gene are critical in driving gene expression. A transcriptional network including forkhead box A1/A2 (FOXA1/A2), hepatocyte nuclear factor 1 (HNF1) and caudal-type homeobox 2 (CDX2) is recruited to these elements (22,23). In contrast, in some airway epithelial cells open chromatin regions at −44 kb and −35 kb 5′ to the CFTR translational start site encompass elements that respond to antioxidants and interferon regulatory factors, respectively (24–26). Another cell type that expresses abundant CFTR mRNA is the male genital duct epithelium, where primary cells show a combination of the cis-regulatory elements of both airway and intestinal epithelium.
Here we take a global view of the CFTR locus and the role of individual classes of cis-regulatory elements in coordinating its higher order chromatin structure and gene expression. To achieve this we used CRISPR/Cas9-mediated deletion of a CTCF-binding insulator element 5′ to the locus and a pivotal intronic enhancer, followed by an unbiased chromosome conformation capture technique, 4C-seq. Our data reveal a mechanism coordinating regulatory elements across the locus, which senses structural perturbations and apparently responds to maintain normal gene expression levels. However, the impact of loss of a key intronic enhancer on CFTR transcription cannot be rescued by structural changes in the locus.
MATERIALS AND METHODS
Cell culture
Caco2 (27) and Calu3 (28) cell lines were obtained from ATCC and grown in DMEM (Dulbecco's Modified Eagle's medium) with 10% FBS (fetal bovine serum). For all experiments with Caco2 cells they were harvested 48 h post-confluence, a time at which CFTR expression is close to maximum levels (29). Human bronchial epithelial (HBE) cells were donated by Dr Scott Randell (UNC) and cultured in Bronchial Epithelial Cell Growth Medium, Lonza. Adult human epididymis cells (caput) were described previously (30). Skin fibroblasts (GM08333) from Coriell Institute were grown in Ham's F-10 media supplement with 15% FBS.
Transient siRNA depletion experiments
40 nM Stealth™ CTCF and RAD21 siRNAs (Life Technologies, LT) (6) along with non-targeting medium GC negative control siRNAs were forward transfected with RNAiMAX™ (LT) into Caco2 cells, 48 h after plating. Cells were harvested 72 h after transfection at which time they were 48 h post-confluent.
Reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted with TRIzol and cDNA prepared with the TaqMan reverse transcription kit (LT). CFTR mRNA levels were assayed using a primer/probe set spanning exons 5 and 6 (29). The data were normalized to 18s rRNA as an endogenous control. To verify that the deletion of the intron 11 enhancer had not caused aberrant splicing of the CFTR transcript RT-PCR was performed with primers B1R and B1L (31) and products visualized on a 1% agarose gel.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described (18,22). Antibodies were specific for FOXA2 (Santa-Cruz Biotechnology sc-6554x) and goat IgG (sc-2028), CTCF (Millipore 07–729), RNAP II (sc-9001x) and rabbit IgG (Millipore12–370). Primer sequences used for qPCR can be found in Supplementary Table S1.
DNase-seq
DNase-seq libraries were prepared from Caco2, Calu3, adult human epididymis and HBE cells as previously described (32,33). The first 20 bases of sequencing reads were used for mapping to the hg19 genome using Bowtie (34), allowing a maximum of 2 mismatches. The aligned reads were then processed with Homer software (35) to generate tag density files and peak files. DNAse-seq data are available at GEO (GSE74709).
4C-seq
4C-seq libraries were generated from cultured cells as described previously (36). NlaIII and DpnII or Csp6I were used as the primary or secondary restriction enzymes, respectively. Enzyme pairs and primer sequences used to generate 4C-seq libraries for each viewpoint are shown in Supplementary Table S2. Some of the primers contain a 2 nucleotide barcode at the 3′ end of P5 linker to enable multiplexing of libraries generated from same viewpoint in the same Hi-Seq flow cell. Samples were sequenced on Illumina Hi-Seq machines using standard protocols. The sequencing data were processed using the 4Cseqpipe protocol (37). All 4C-seq images were generated using default parameters of the pipeline.
Fluorescence in situ hybridization (FISH) and data analysis
FISH experiments were performed as described previously (21). Bacterial artificial chromosomes (BACs) (LT) were used as probes for the −80.1 kb (CTD-2100H12) and +48.9 kb (CTD-3014G19) sites, and a downstream control region (CTD-2329K1). All images were acquired on a Nikon A1R+ line scanning confocal microscope equipped with photo-multiplier tubes. Imaging was done with a Plan Apo VC 60x Oil, 1.4 NA, objective as a multi-dimensional z-stack with a consistent step size of 0.25 μm using the Nikon Elements software. Nuclei, as defined by DAPI staining, were segmented using the method of Otsu (38) which reduces a grey level image to a binary image based on the assumption that the histogram of grey levels present in the image will be bi-modal and describes two classes of pixels; foreground and background. This method was performed on the bi-modal histogram generated by the collection of intensity values from every z-plane in the complete image stack. Nuclei that were close together and thus not well resolved by this method were further split by visual inspection and manual segmentation. Individual FISH signals were identified through manual intensity based segmentation with a focus on minimizing the total number of pixels in each signal. The FISH signal/genomic location was then calculated as the intensity weighted centroid of the thresholded signal. Given that Caco2 have three copies of Chr7, k-means clustering was performed for three clusters with the centroids of all the FISH signals within a single nucleus. The resultant clusters describe loci on the same chromosome and were used to measure the Euclidian distance in three dimensions between loci of interest.
CRISPR guide design, CRISPR/Cas9 transfection and screening
Two pairs of gRNAs flanking the −20.9 kb CTCF site and the intron 11 enhancer core were identified using the CRISPR Design Program (http://crispr.mit.edu) (Supplementary Table S3). gBlocks from Integrated DNA Technologies (Iowa), were amplified with the gRNA primers and Phusion polymerase and cloned into pSCB (Agilent). Caco2 cells were seeded onto 6-well plates and transfected after 48 h with 0.11 pmol each of pMJ290 (wild-type Cas9 plasmid tagged with GFP) (Addgene, plasmid #42234), 5′ gRNA, and the 3′ gRNA using Lipofectamine 2000 (LT). 48 h after transfection, cells were trypsinized and single cell suspensions were generated for fluorescence-activated cell sorting of GFP positive cells. Single cells were plated into individual wells of a 96-well plate and clones expanded for screening. Effective deletion of each cis-regulatory element was confirmed by PCR of genomic DNA using primers flanking the gRNA PAM sites and sequencing.
RESULTS
The 3D structure of the CFTR locus shows a cell-type-specific conformation
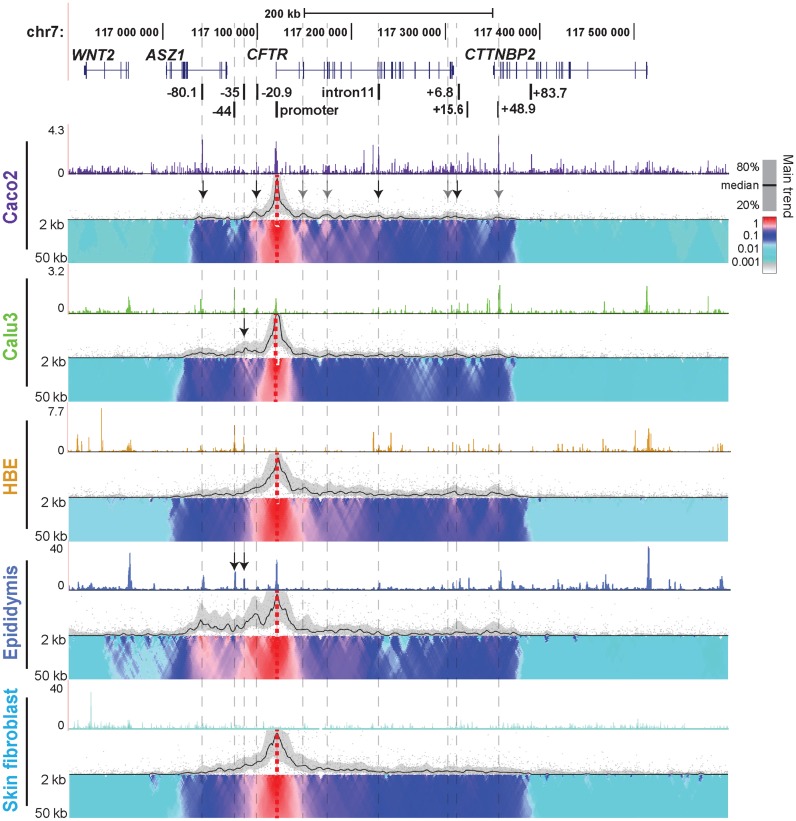
In order to characterize the 3D structure of the active CFTR locus in epithelial cells from different tissues involved in CF pathology, we used 4C-seq (Figures 1 and 2). Epithelial cell lines from intestine (Caco2 adenocarcinoma) and lung (Calu3 adenocarcinoma), primary human bronchial epithelial cells (HBE) and primary human epididymis (caput) epithelial cells were investigated. Skin fibroblasts, which do not express CFTR provided a negative control representing the inactive gene. 4C-seq libraries were generated at least twice for each cell line and from two independent donor cultures for primary cells. CFTR expression levels in each cell type were established previously (18,24) and verified in primary cells by RT-qPCR (not shown). Open chromatin for each cell type was mapped by DNase-seq and is shown above each panel in Figures 1 and 2. To identify potentially novel cis-regulatory elements we first interrogated interactions genome-wide with an NlaIII restriction fragment viewpoint at the CFTR promoter (Figure 1, dotted red vertical line). Each panel in Figure 1 shows a different cell type and black lines mark the main trend of relative interactions across the locus, while domainograms (39) underneath show interaction frequency with multiple scales. Relative interactions are always strongest close to the viewpoint due to the physical proximity of the restriction fragments (20,37). Specific long-range interactions are indicated by peaks of the main trend, together with a local increase in intensity of the domainogram. In Caco2 cells, consistent with previous 3C results (18,20,21), strong interactions are evident between the CFTR promoter and −80.1 kb, −20.9 kb, introns 10/11, and several elements at the 3′ end of the gene (indicated by black arrows). In addition to these sites, the 4C-seq data reveal novel regions interacting with the promoter including within introns 2, 4, the 3′ end of the gene, and +48.9 kb downstream of the last exon (marked by gray arrows). In contrast, in Calu3 cells, no interactions were seen between the CFTR promoter and any intronic elements, though those with the −80.1 kb and +48.9 kb sites, and the 3′ end of the gene, are evident. In addition, a strong interaction is seen between the promoter and an element at −35 kb 5′ to the gene (black arrow) in Calu3 cells. This element encompasses an airway-selective enhancer for the CFTR promoter that responds to interferon regulatory factors (IRF1/2) (25). In HBE cells, the promoter interacts with the same region of intron 2 that is seen to interact in Caco2 cells and also with multiple sites between intron 4 and the middle of intron 10. Elements immediately 3′ to the gene and the +48.9 kb element are also brought into close association with the promoter in HBE cells, though the intron 11 intestinal enhancer is not. Specific interactions upstream of the promoter are less evident in these cells. The CFTR 4C profile in primary epididymis cells is similar to that of HBE cells within the locus and 3′ to it. However, unique to the epididymis are the extensive strong interactions of upstream sequences with the promoter, particularly elements at −80.1, −44, −35 and −20.9 kb to the gene. Weak interactions are also detected far upstream of the −80.1 kb CTCF binding site, within the Wingless-Type MMTV Integration Site Family Member 2 (WNT2) locus, which are not seen in other cell types. The high interaction frequency seen at −44 and −35 kb corresponds to two DHS that we observed previously using DNase-chip in fetal primary epididymis cells (18) and DNase-seq analysis of adult epididymis caput cells (indicated by arrows). However, these elements may not be occupied by the same factors in epididymis and airway cells. Though the DHS associated with the strong intestinal enhancer in intron 11 is also evident in fetal and adult epididymis, only weak interactions were seen between the CFTR promoter and this site in the epididymis 4C-seq profile. In comparison to the epithelial cells with active CFTR loci, the locus is inactive in skin fibroblasts, consistent with the absence of significant interactions across the region, though the topological domain encompassing the gene is evident.
Figure 1.
Cell-type-specific chromatin structure at the CFTR locus: 4C-seq profiles with a CFTR promoter viewpoint. A schematic at the top shows the genomic location of CFTR and adjacent genes on chromosome 7 and below are known cis-regulatory elements for the CFTR locus. The nomenclature represents distance in kb (−) 5′ to the translational start site and (+) 3′ to the last coding base. Open chromatin mapped by DNase-seq and 4C-seq data are shown for Caco2, Calu3, HBE, epididymis and skin fibroblast cells. Representative data from one of the replicates are shown here. DNase-seq data are presented as histograms from screen shots of data sets uploaded to the UCSC genome browser. 4C-seq data are presented in alignment with the DNase-seq data and have two subpanels. The upper panel indicates the main trend of contact profile using a 5-kb window size. Relative interactions are normalized to the strongest point (which is set to 1) within each panel. The lower panel is a domainogram (39), which uses color coded intensity values to show relative interactions with window sizes varying from 2 to 50 kb. Here, red denotes the strongest interactions and dark blue, through turquoise, to gray represent gradually decreasing frequencies. Arrows denote important data features described in the results.
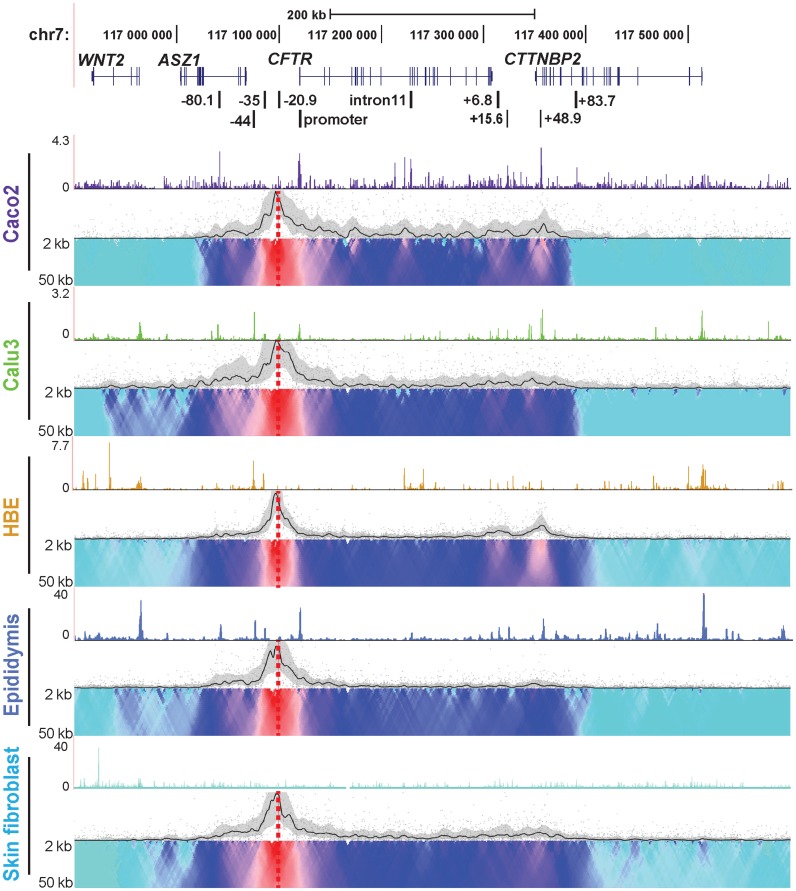
Figure 2.
Cell-type-specific chromatin structure at the CFTR locus: 4C-seq profiles with a −20.9 kb CTCF-binding insulator viewpoint. Cell types, DNase-seq data and 4C-seq data presentation as described in Figure 1.
Next, 4C-seq was done in the same cell types using a viewpoint at the −20.9 kb enhancer blocking insulator site, which binds CTCF (4,40) (Figure 2, red dotted line). All the cells showed some level of association between this viewpoint and elements at the 3′ end of the gene and the +48.9 kb CTCF binding sites. These interactions were strongest in Caco2 and HBE, though also present in Calu3 and weakly in primary epididymis cells, suggesting a higher order chromatin structure that was independent of cell type. Also of note is that interactions decreased dramatically upstream of −80.1 kb and downstream of +48.9 kb (shown by the domainogram) in all cell types, with both promoter and −20.9 kb viewpoint. This is indicative of a potential topological domain or sub-domain at the CFTR locus. In cell types with promoter: enhancer interactions at −35 and −44 kb (Calu3 and epididymis), a low level of interaction is also seen upstream of the −80.1 kb site. Caco2 cells show unique interacting regions with the −20.9 kb viewpoint in intron 1, intron 4 and intron 10 which may reflect cis-regulatory elements specific to intestinal cells. In all cases, there was a strong correlation between 4C interactions and regions of open chromatin shown by DNase-seq. Some interactions are seen between the −20.9 kb bait and the WNT2 gene in Calu3 airway cells and primary human epididymis cells. This peak of interaction corresponds to a cell type selective CTCF binding site (ENCODE data), though it is much weaker than interactions with other CTCF sites across the locus and in CTTNBP2 on the 3′ side.
The CFTR locus is demarcated by a topological domain in which cell-type-specific chromosome looping occurs
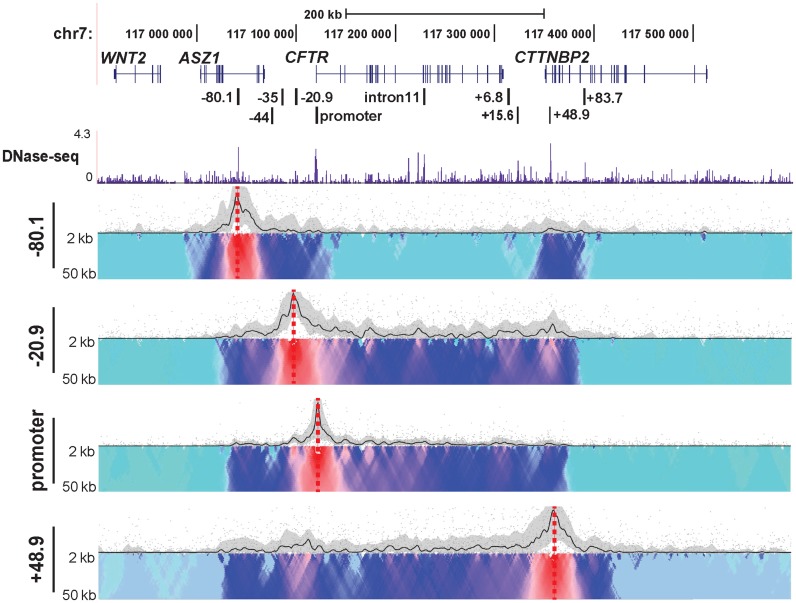
Topological domains (TADs) are usually conserved across cell types and their borders are enriched for CTCF sites (41). To further characterize role of CTCF in the chromatin structure at the CFTR locus, we designed additional 4C-seq viewpoints at the −80.1 and +48.9 kb CTCF binding sites flanking the locus (21). 4C-seq data from each viewpoint in Caco2 cells are shown in Figure 3. A lack of interactions with all viewpoints beyond the −80.1 and +48.9 kb sites, suggested that these two CTCF sites define the boundary of a TAD or sub-TAD at the CFTR locus. To verify this interpretation, we used published human fibroblast Hi-C data (http://www.3dgenome.org) to define the TADs surrounding CFTR and observed an identical domain to that illustrated by our 4C-seq data (Supplementary Figure S1). The 4C data also showed that the two boundary CTCF binding sites at −80.1 and +48.9 kb, interact strongly with each other, but show relatively weaker associations with the gene body in the middle of the TAD. In contrast, the −20.9 kb CTCF binding site viewpoint interacts strongly with both the boundary CTCF sites and with intronic elements, which also associate with the promoter. These data reveal two levels of chromosome looping involving CTCF at the CFTR locus: the first maintains the overall 3D structure of the topological domain, while the second facilitates recruitment of cis-regulatory elements to the gene promoter. To verify the interaction between the boundary CTCF sites, we performed FISH using BAC probes (CTD-2100H12 covers the −80.1 kb site and CTD-3014G19 encompasses the +48.9 element). A BAC (CTD-2329K1) mapping downstream of the +48.9 kb element, at an equal genomic distance as the −80.1 to +48.9 kb interval, was chosen as a control probe (Supplementary Figure S2A). Quantification of the FISH signals (Supplementary Figure S2B) in 94 nuclei shows that the −80.1 and +48.9 kb sites are significantly closer together than are the +48.9 kb site and the control probe (Supplementary Figure S2C).
Figure 3.
Chromatin structure at the CFTR locus in Caco2 cells analyzed from multiple viewpoints. 4C-seq data and DNase-seq data presentation in Caco2 cells as described in Figure 1. Viewpoints are CTCF-binding sites at −80.1, −20.9 and +48.9 kb and at the promoter.
CTCF/cohesin complex are required for maintaining the 3D chromatin structure and proper regulation of the CFTR locus
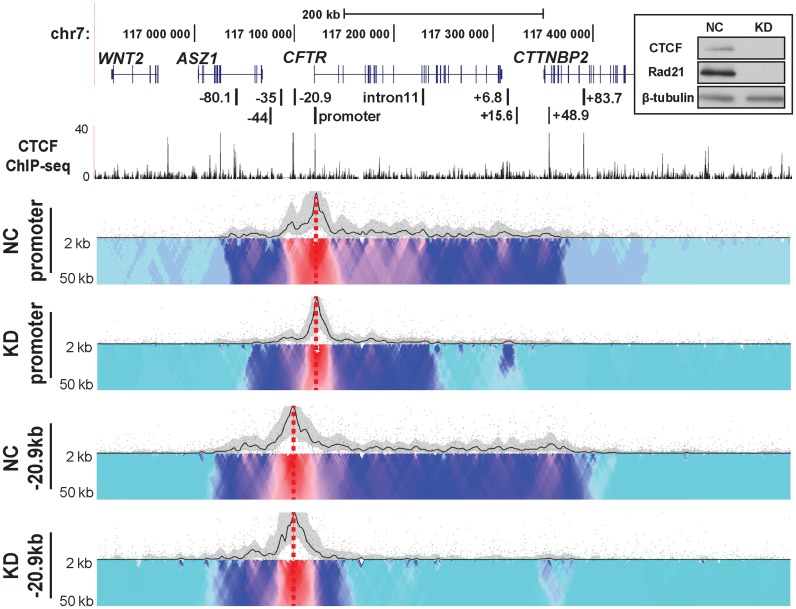
We previously showed that CTCF and cohesin complex had distinct roles in modulating the higher order structure of the CFTR gene (21). While both were involved in maintaining the 3D structure of the locus, cohesin had an additional role in stabilizing the interactions between the gene promoter and specific cis-regulatory elements. Moreover, loss of both CTCF and Rad21 (a subunit of the cohesin complex) increased CFTR expression levels (21). Having established by 4C-seq that the interaction of CTCF binding sites governed the structural organization of the active CFTR locus, we next investigated the direct contribution of these architectural proteins. CTCF and Rad21 were simultaneously depleted in Caco2 cells by siRNA (Figure 4 inset) and the impact on the CFTR locus interaction profile was measured by 4C-seq (Figure 4). Using viewpoints at either the promoter or −20.9 kb in comparison to negative control siRNA, the CTCF/Rad21 siRNAs caused a dramatic and global decrease of interactions across the whole topological domain. The interactions between the promoter viewpoint and CTCF sites at −80.1, −20.9 and 48.9 kb were either greatly reduced or lost completely, as were associations with intronic regions 3′ to the middle of the locus. Intronic regions 5′ to and including intron 11 still showed some interactions with the promoter but at a greatly reduced frequency. The downstream boundary of the topological domain was barely evident using either the promoter or the −20.9 kb viewpoint, though overall the loss of associations with the latter was greater. The interaction profile upstream of the −20.9 kb viewpoint was less affected by CTCF/cohesin depletion, implicating other factors in maintenance of these structures. We showed previously that CTCF/RAD21 depletion in Caco2 cells does not alter expression of the genes immediately adjacent to CFTR on the 5′ and 3′ side (21).
Figure 4.
Depletion of CTCF and cohesin complex has a dramatic impact on chromatin structure at the CFTR locus. 4C interaction profiles of the CFTR locus in Caco2 cells after CTCF/Rad21 knockdown (KD). Top right inset. Efficacy of CTCF/Rad21 KD: western blots of cell lysates from Caco2 cells co-transfected with CTCF, RAD21-specific or negative control (NC) siRNAs, probed with antibodies to CTCF, RAD21 and β-tubulin. 4C-seq viewpoints are at the CFTR promoter and −20.9 kb in negative control or CTCF/Rad21 double knockdown (KD) Caco2 cells. CTCF ChIP-seq data combined from multiple cell types are from ENCODE (62).
Loss of a CTCF-binding insulator element at −20.9 kb enhances CTCF recruitment at adjacent sites, maintaining locus architecture and CFTR expression
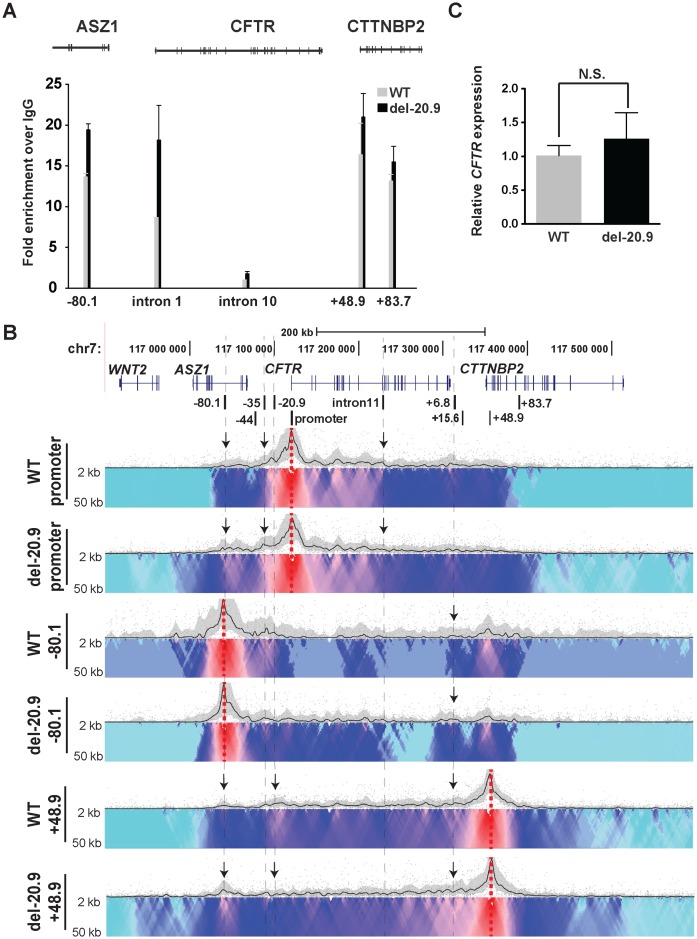
To determine the contribution of a single CTCF binding site to the higher order structure and expression of CFTR, the −20.9 kb insulator (40) was removed from the endogenous locus in Caco2 cells. CRISPR/Cas9 technology (42) with two flanking guides was used to remove an ∼1.2 kb fragment encompassing the −20.9 kb element from all three CFTR alleles in these cells (Caco2 is trisomic for chromosome 7) (Supplementary Figure S3A, B). Two independent deletion clones were evaluated together with non-targeted WT clones arising from the same experiment to exclude artifacts of clonal variation. First, we examined CTCF occupancy at known binding sites across the locus by ChIP. CTCF sites were described previously (21) and additional sites within the locus were also assayed (Figure 5A). Loss of the −20.9 kb element caused a statistically significant increase in CTCF occupancy at the −80.1 kb site, when compared to wild-type, and also increases at sites in intron 1 and 3′ to the locus at +48.9 kb. These data suggested that loss of a critical CTCF site caused a re-distribution of the architectural proteins to maintain the 3D structure of the active locus. Consistent with the CTCF ChIP data are 4C-seq results (Figure 5B) showing changes in looping interactions across the locus after removal of the −20.9 kb site. Though the overall TAD remained intact, as would be predicted since this element is not at the TAD boundary, interactions between CTCF binding sites at −80.1 and +48.9 kb and with the gene promoter were altered. With the +48.9 kb viewpoint, a reduction in interactions with −20.9 kb region was accompanied by increased interactions with −80.1 kb and 3′ end of CFTR in deletion clones (indicated by arrows). With the −80.1 kb viewpoint, interactions were increased across the locus and with the +6.8 kb CTCF site, in the del-20.9 clones (indicated by arrows). The promoter viewpoint showed that deletion of −20.9 kb increased interactions with the −80.1 kb site and −35 kb sites but reduced interactions with specific, but not all enhancer elements in the 5′ half of the locus (indicated by arrows). Next we performed RT-qPCR to measure CFTR expression levels in the −20.9 kb deletion clones in comparison to WT clones and showed that the minor increase (1.25-fold) was not significant (Figure 5C). This suggests that the −20.9 kb site primarily plays a structural role at the locus and in its absence other CTCF sites can at least partially compensate for its function.
Figure 5.
Deletion of the −20.9 kb CTCF site from the CFTR locus disrupts its chromatin structure but has little effect on gene expression. −20.9 kb CTCF site removed from endogenous locus in Caco2 cells by CRISPR/Cas9 (Supplementary Figure S3A, B). (A) ChIP for CTCF in wildtype (WT, gray) and del-20.9 kb (black) Caco2 clones. CTCF occupancy measured by qPCR at CTCF sites (−80.9 kb, intron 1, +48.8 kb and +83.7 kb) and at the intron 11 enhancer. Results are pooled from three independent experiments from two WT and two deletion clones. Error bars represent S.E.M. (B) 4C-seq interaction profiles of WT and del-20.9 kb Caco2 clones using viewpoints at the promoter, −80.1 kb, and +48.9 kb. (C) CFTR expression measured by RT-qPCR relative to 18s RNA in WT and del-20.9 kb Caco2 clones, showing no significant change. Results are pooled from three independent experiments from two WT and two deletion clones. Error bars represent S.E.M.
To confirm that the alteration in CTCF occupancy after removal of the −20.9 kb element was specific to the CFTR locus we examined CTCF binding at the gel-forming mucin gene cluster on chromosome 11. We previously reported the role of CTCF in the organization of this multi-gene cluster (43). No difference in CTCF occupancy was observed by ChIP at two critical CTCF binding sites in the region (sites IV and IX) in WT and −20.9 kb deletion clones (Supplementary Figure S5).
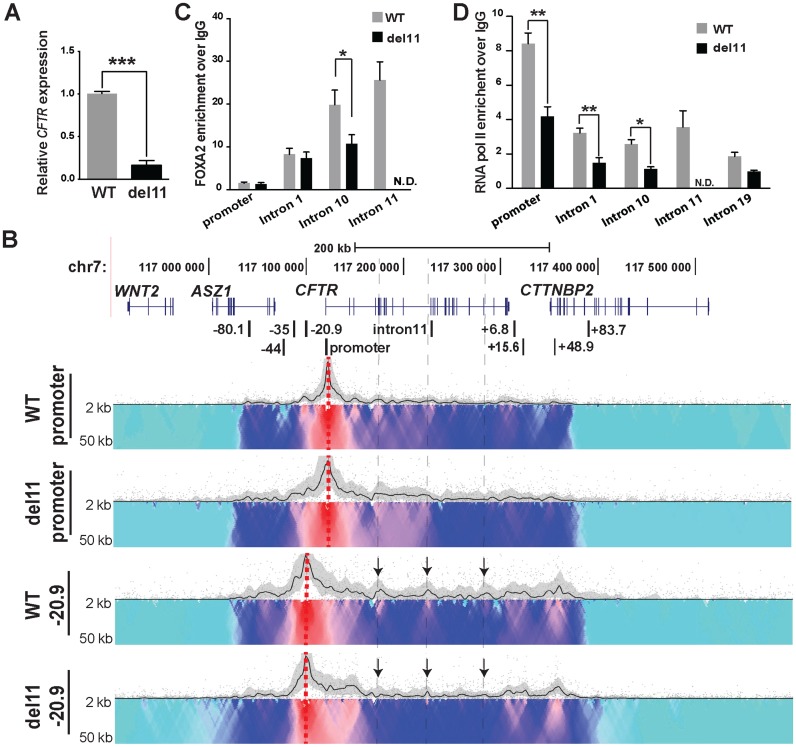
Deletion of a strong enhancer in intron 11 has little impact on the 3D architecture, but represses CFTR gene expression
Previously we characterized a strong intestinal selective enhancer in intron 11 of CFTR (DHS11) (18,22). To determine how cis-regulatory elements such as this enhancer contribute to higher order chromatin structure and gene expression we again used CRISPR/Cas9 technology to remove it from Caco2 cells. Approximately 1.2 kb encompassing the binding sites for critical transcription factors driving the intron 11 enhancer (FOXA1/2, HNF1 and CDX2 (18,22,23)) were removed as shown in Supplementary Figure S3C. Deletion of DHS11 (del11) from all three CFTR alleles in targeted Caco2 cell clones was verified by PCR of genomic DNA (Supplementary Figure S3C and D). Three wild-type (non-targeted) and three del11 clones were used for further analysis. CFTR expression in these clones was measured by RT-qPCR and deletion of the enhancer reduced transcript levels by more than 80% (Figure 6A). To verify that the deletion of DHS11 did not interfere with the normal splicing of CFTR mRNA, we performed RT-PCR with primers spanning from exon 6 through exon 13 of the transcript (44). The results show no differences in splicing between WT and del11 clones (Supplementary Figure S4). 4C–seq was next performed on both WT and del11 clones and though no substantial changes in the 3D structure of the locus were evident with a promoter viewpoint, interactions with −20.9 kb became weaker in some intronic regions marked by arrows (Figure 6B). Del11 clones also showed minor changes in the interaction profile between both viewpoints and the intron 4 to intron 10 regions. Since chromatin structure of the locus did not show major changes after loss of DHS11, we examined whether del11 altered recruitment of critical transcription factors to other elements in the locus. FOXA2, a pioneer transcription factor (45), regulates CFTR (22,23) by recruitment to several intronic sites. We recently confirmed by ChIP-seq in Caco2 cells that FOXA2 is enriched at the CFTR promoter and at regulatory elements within introns 1, 10 and 11 of the locus among other sites (46). ChIP for FOXA2 in WT and del11 clones, followed qPCR for key binding sites across the locus showed a significant loss of FOXA2 binding at a site in intron 10 (Figure 6C), though no significant change in occupancy at the promoter or an enhancer in intron 1. To investigate further the mechanism of del11 induced loss of CFTR expression, we measured enrichment of RNA polymerase II (RNAP II) across the locus by ChIP (Figure 6D). In addition to occupancy at the CFTR promoter in WT Caco2 cells, RNAP II is enriched at the active cis-regulatory elements in intron 1, 10 and 11. A regulatory site in intron 19 that is not utilized in Caco2 cells is also bound by RNAP II, illustrating the active transcriptional state of the locus. The del11 clones show a significant reduction in RNAP II occupancy at the promoter, intron 1, 10 and 11, suggesting a global loss of the polymerase across the CFTR locus. These data suggest that the intron 11 enhancer participates in recruitment of RNAP II to the CFTR promoter as well as to intronic regions.
Figure 6.
Deletion of the intron 11 enhancer from the CFTR locus has little impact on chromatin structure but substantially reduces gene expression. (A) RT-qPCR analysis showing CFTR expression relative to 18s RNA in WT (gray) and del11 (black) Caco2 clones. Values are mean ± S.E.M., n = 3, ***P < 0.001. (B) 4C-seq interaction profiles of WT and del11 Caco2 clones using viewpoints at the promoter and −20.9 kb. (C) FOXA2 ChIP in WT (gray) and del11 (black) Caco2 clones, followed by qPCR with primers for the promoter and intronic cis-regulatory elements in CFTR. (D) RNA pol II ChIP in WT (gray) and del11 (black) Caco2 clones, followed by qPCR. ChIP results are pooled from three independent experiments from two WT and two deletion clones. Data present the mean ± S.E.M., *P < 0.05, **P < 0.01.
DISCUSSION
Our high-resolution analysis of CFTR 3D chromatin architecture reveals novel features of the behavior of a single locus in different cell lineages. We detect a conserved topological domain or sub-domain spanning ∼320 kb with boundaries at −80.1 kb and +48.9 kb flanking the CFTR gene, irrespective of its expression levels. Consistent with the original description of the domain structure of the mammalian genome by Hi-C and 5C techniques (41,47), we show that the boundaries of the TAD at the CFTR locus are cell type independent and enriched for CTCF and cohesin complex (21). Our data also suggest that there are two discrete but interrelated types of chromatin looping at the locus: the first establishes and maintains the TAD, while the second brings enhancers and other cis-regulatory elements into close association with the gene promoter and the CTCF structural network. The latter loops are likely formed hierarchically within the topological domain and are cell-type dependent. Consistent with this model is the general lack of intra-domain interactions within the CFTR TAD in the fibroblast cells, which do not express the gene and the unique loop footprints seen in intestinal, airway and epididymis epithelial cells, all of which express abundant CFTR. Further evidence is provided by the observation that disruption of the looping interactions by depletion of CTCF Rad21 alters CFTR expression.
CTCF and cohesin complex occupy many of the same sites genome-wide and both mediate long-range interactions (3,4,6,48–52). Our data provide direct evidence for the maintenance of higher order chromatin structures being dependent on CTCF and cohesin complex, and are in agreement with observations at the β-globin locus (14), at a complex gene cluster (Hox) through development (53) and genome-wide (5,14,54). The high-resolution studies on a single locus (CFTR) that are described here contribute a more detailed level of understanding of the underlying mechanisms. We show that depletion of CTCF/Rad21 not only destroys the TAD structure, but also disrupts the intra-domain enhancer-promoter interactions. Looking in more detail at the two levels of chromatin looping, for example in Caco2 cells, the CTCF/cohesin sites at either end of the TAD ((21), site I, −80.9 kb and site IV, +48.9 kb), seem to isolate the CFTR locus from regulatory signals from outside the domain. Moreover, CTCF/RAD21 depletion has little or no impact on interactions between elements within the TAD and adjacent regions outside it. This suggests that either minimal occupancy of these proteins is required to maintain the boundaries, other proteins may compensate for their loss or that the TAD has inherent structural features based on DNA sequence alone. It is of interest that structural disruption of TADs directly impacts promoter enhancer interactions and is associated with disease phenotypes (55).
In contrast, though the CTCF/cohesin site at −20.9 kb has enhancer blocking insulator activity (40) and interacts with the CTCF sites at the TAD boundaries (−80.1 kb and +48.9 kb), it also has a pivotal role in looping within the TAD, which recruits cis-regulatory elements to the gene promoter. siRNA-mediated depletion of CTCF/Rad21 at this site may be largely responsible for the global loss of interactions of intronic sites with the gene promoter. This suggestion is supported by the observation that deletion of the −20.9 kb element decreases interactions between the CFTR promoter and specific intronic regions of the gene. It is of interest that despite this loss of intragenic looping CFTR gene expression is not significantly affected by the deletion. There are many potential explanations for this observation. These include an inbuilt redundancy of CTCF/cohesin binding sites, so that increased CTCF occupancy and interactions with other nearby sites (for example immediately adjacent to −20.9 kb, or within intron 1 of CFTR) could compensate for the loss of the −20.9 kb site. Alternatively, removal of the −20.9 kb site facilitates interaction between the CFTR promoter and 5′ enhancers at −35 and −44 kb that are not normally active in Caco2 cells (25,26). The 4C -seq data would support this suggestion (Figure 5), however the critical factors that drive these 5′ enhancers in airway cells may not be present in intestinal cells. It is also possible that removal of the −20.9 kb CTCF site destabilizes the CFTR locus so facilitating interactions outside the domain, though these appear generally to be weak. Alternatively, recruitment of critical transcription factors to the intronic enhancers may transmit signals to the gene promoter by looping-independent mechanisms.
The high-resolution structure/function data on CFTR presented here greatly advance our understanding of the cell-type specific regulatory mechanisms for this complex gene. Previously, a strong intestinal enhancer element was identified in intron 11 of CFTR gene in Caco2 cells, which was shown to interact with the gene promoter by 3C assays (18,22). The unbiased 4C-seq data presented here not only confirmed the 3C data, but also identified multiple additional interacting regions that could contribute to the gene regulation. Some of these regions may contain novel enhancers, as suggested by histone modifications (56), while others may be bystanders in the looping mechanism. Consistent with this suggestion are the results of deleting the intron 11 enhancer in Caco2 cells. Removal of this site represses CFTR expression by more than 80%, but does not abolish global interactions between the 5′ half of the locus and the gene promoter, implicating additional sites in driving the looping. The deletion also results in increased interactions with other sites within and outside the gene. Our observation that removal of the intron 11 enhancer reduces RNAP II occupancy at the gene promoter by only ∼50% likewise predicts the interaction of multiple intronic enhancers with the CFTR promoter. Indeed, we reported the involvement of several critical intronic enhancers in Caco2 cells in previous work (18). It is also of note that RNAP II occupancy is reduced to the same extent (∼50%) at several other sites across the locus suggesting that deletion of the intron 11 enhancer does not impact CFTR transcript elongation.
In Calu3 airway cells, intronic interactions are much less evident than in Caco2 cells, but a relatively strong interaction is seen with the −35 kb region. This is of interest since we previously identified an airway-selective CFTR enhancer at −35 kb that responds to interferon regulatory factors (24,25). Its recruitment to the CFTR promoter by looping events in airway cells might be predicted. Primary human epididymis (caput) cells, from more than one biological replica, show a distinct 4C-seq profile with very strong interactions between the gene promoter and upstream elements. Epididymis cells express very high levels of the CFTR transcript (18,30,57) and these data suggest that CFTR expression may be driven by additional 5′ cis-regulatory elements in these cells.
Finally, we integrated the cell type dependent chromatin structures we observed at the CFTR locus with our open chromatin data, ChIP-seq data for both transcription factors (TFs) and histone modifications (56). In Caco2 cells, interacting regions identified by 4C show a high degree of overlap with open chromatin regions revealed by DNase-seq data (Figures 1 and 2). Similarly, these regions coincide with peaks of occupancy by TFs, which participate in the network activating CFTR expression in intestinal epithelial cells. Important TFs include FOXA2, CDX2, HNF1 and HNF4α (22,23,46,58), (Yang et al. submitted). These observations suggest a strong relationship between the regulatory network and 3D chromatin structure. This suggestion is supported by our previous results showing that depletion of FOXA2 concurrently represses CFTR expression and interactions between the gene promoter and intronic regulatory elements (23). It also coincides with data from pluripotent stem cells, showing that loss of important transcription factors disrupted pluripotency-specific long range interactions (59–61). The correlation between cell-specific genomic interactions and open chromatin is also seen in Calu3 (56) and human primary epididymis cells (Figure 1). In conclusion, the CFTR gene provides a paradigm for understanding detailed regulatory mechanisms at a single locus, and can inform studies on other loci. These mechanisms involve the interplay of architectural proteins, transcription factors binding to cis-regulatory elements and chromatin modifications, to establish and facilitate dynamic responses of the 3D chromatin structure.
Supplementary Material
Acknowledgments
We thank Dr Scott Randell and colleagues for HBE cells; also Dr Wouter de Laat and Dr Omer Schwartzman for help with 4C experiments and data analysis; and Dr P Faber and staff at the University of Chicago Genomics Core.
Footnotes
Present address: Jenny L. Kerschner, UNC Lineberger Comprehensive Cancer Center and Department of Biochemistry and Biophysics, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01HD068901 to A.H.); NIGMS New Innovator Award [DP2 OD008717 to S.T.K.]; Cystic Fibrosis Foundation (Harris 11G0 and 14P0). Funding for open access charge: National Institutes of Health and others.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 2.Giles K.E., Gowher H., Ghirlando R., Jin C., Felsenfeld G. Chromatin boundaries, insulators, and long-range interactions in the nucleus. Cold Spring Harb. Symp. Quant. Biol. 2010;75:79–85. doi: 10.1101/sqb.2010.75.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hadjur S., Williams L.M., Ryan N.K., Cobb B.S., Sexton T., Fraser P., Fisher A.G., Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nativio R., Wendt K.S., Ito Y., Huddleston J.E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J.M., Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuin J., Dixon J.R., van der Reijden M.I., Ye Z., Kolovos P., Brouwer R.W., van de Corput M.P., van de Werken H.J., Knoch T.A., van I.W.F., et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U.S.A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 7.Bau D., Sanyal A., Lajoie B.R., Capriotti E., Byron M., Lawrence J.B., Dekker J., Marti-Renom M.A. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat. Struct. Mol. Biol. 2011;18:107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berlivet S., Paquette D., Dumouchel A., Langlais D., Dostie J., Kmita M. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 2013;9:e1004018. doi: 10.1371/journal.pgen.1004018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forrester W.C., Epner E., Driscoll M.C., Enver T., Brice M., Papayannopoulou T., Groudine M. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 10.Gavrilov A.A., Razin S.V. Spatial configuration of the chicken alpha-globin gene domain: immature and active chromatin hubs. Nucleic Acids Res. 2008;36:4629–4640. doi: 10.1093/nar/gkn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgs D.R., Vernimmen D., Wood B. Long-range regulation of alpha-globin gene expression. Adv. Genet. 2008;61:143–173. doi: 10.1016/S0065-2660(07)00005-3. [DOI] [PubMed] [Google Scholar]
- 12.Hughes J.R., Lower K.M., Dunham I., Taylor S., De Gobbi M., Sloane-Stanley J.A., McGowan S., Ragoussis J., Vernimmen D., Gibbons R.J., et al. High-resolution analysis of cis-acting regulatory networks at the alpha-globin locus. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 2013;368:20120361. doi: 10.1098/rstb.2012.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim A., Dean A. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 2003;23:8099–8109. doi: 10.1128/MCB.23.22.8099-8109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vernimmen D., Marques-Kranc F., Sharpe J.A., Sloane-Stanley J.A., Wood W.G., Wallace H.A., Smith A.J., Higgs D.R. Chromosome looping at the human alpha-globin locus is mediated via the major upstream regulatory element (HS -40) Blood. 2009;114:4253–4260. doi: 10.1182/blood-2009-03-213439. [DOI] [PubMed] [Google Scholar]
- 16.Gillen A.E., Harris A. Transcriptional regulation of CFTR gene expression. Front. Biosci. 2012;4:587–592. doi: 10.2741/401. [DOI] [PubMed] [Google Scholar]
- 17.Gosalia N., Harris A. Chromatin dynamics in the regulation of CFTR expression. Genes. 2015;6:543–558. doi: 10.3390/genes6030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ott C.J., Blackledge N.P., Kerschner J.L., Leir S.H., Crawford G.E., Cotton C.U., Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19934–19939. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott C.J., Suszko M., Blackledge N.P., Wright J.E., Crawford G.E., Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J. Cell. Mol. Med. 2009;13:680–692. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gheldof N., Smith E.M., Tabuchi T.M., Koch C.M., Dunham I., Stamatoyannopoulos J.A., Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–4336. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosalia N., Neems D., Kerschner J.L., Kosak S.T., Harris A. Architectural proteins CTCF and cohesin have distinct roles in modulating the higher order structure and expression of the CFTR locus. Nucleic Acids Res. 2014;42:9612–9622. doi: 10.1093/nar/gku648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerschner J.L., Harris A. Transcriptional networks driving enhancer function in the CFTR gene. Biochem. J. 2012;446:203–212. doi: 10.1042/BJ20120693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerschner J.L., Gosalia N., Leir S.H., Harris A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics. 2014;9:557–565. doi: 10.4161/epi.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Ott C.J., Lewandowska M.A., Leir S.H., Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J. Cell. Mol. Med. 2012;16:1321–1330. doi: 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z., Leir S.H., Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol. Cell. Biol. 2013;33:2843–2853. doi: 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z., Leir S.H., Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am. J. Respir. Cell Mol. Biol. 2015;52:387–396. doi: 10.1165/rcmb.2014-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogh J., Wright W.C., Loveless J.D. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J. Natl. Cancer Inst. 1977;58:209–214. doi: 10.1093/jnci/58.2.209. [DOI] [PubMed] [Google Scholar]
- 28.Shen B.Q., Finkbeiner W.E., Wine J.J., Mrsny R.J., Widdicombe J.H. Calu-3: a human airway epithelial cell line that shows cAMP-dependent Cl- secretion. Am. J. Physiol. 1994;266:L493–L501. doi: 10.1152/ajplung.1994.266.5.L493. [DOI] [PubMed] [Google Scholar]
- 29.Mouchel N., Henstra S.A., McCarthy V.A., Williams S.H., Phylactides M., Harris A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem. J. 2004;378:909–918. doi: 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leir S.H., Browne J.A., Eggener S.E., Harris A. Characterization of primary cultures of adult human epididymis epithelial cells. Fertil. Steril. 2015;103:647–654. doi: 10.1016/j.fertnstert.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chalkley G., Harris A. Lymphocyte mRNA as a resource for detection of mutations and polymorphisms in the CF gene. J. Med. Genet. 1991;28:777–780. doi: 10.1136/jmg.28.11.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bischof J.M., Ott C.J., Leir S.H., Gosalia N., Song L., London D., Furey T.S., Cotton C.U., Crawford G.E., Harris A. A genome-wide analysis of open chromatin in human tracheal epithelial cells reveals novel candidate regulatory elements for lung function. Thorax. 2012;67:385–391. doi: 10.1136/thoraxjnl-2011-200880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Splinter E., de Wit E., van de Werken H.J., Klous P., de Laat W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods. 2012;58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 37.van de Werken H.J., Landan G., Holwerda S.J., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Oz Y., Bouwman B.A., et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods. 2012;9:969–972. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 38.Otsu N. IEEE Trans. Syst. Man. Cybern. Vol. 9. IEEE; 1979. pp. 62–66. [Google Scholar]
- 39.de Wit E., Braunschweig U., Greil F., Bussemaker H.J., van Steensel B. Global chromatin domain organization of the Drosophila genome. PLoS Genet. 2008;4:e1000045. doi: 10.1371/journal.pgen.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blackledge N.P., Carter E.J., Evans J.R., Lawson V., Rowntree R.K., Harris A. CTCF mediates insulator function at the CFTR locus. Biochem. J. 2007;408:267–275. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gosalia N., Leir S.H., Harris A. Coordinate regulation of the gel-forming mucin genes at chromosome 11p15.5. J. Biol. Chem. 2013;288:6717–6725. doi: 10.1074/jbc.M112.437400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hull J., Shackleton S., Harris A. Analysis of mutations and alternative splicing patterns in the CFTR gene using mRNA derived from nasal epithelial cells. Hum. Mol. Genet. 1994;3:1141–1146. doi: 10.1093/hmg/3.7.1141. [DOI] [PubMed] [Google Scholar]
- 45.Sekiya T., Muthurajan U.M., Luger K., Tulin A.V., Zaret K.S. Nucleosome-binding affinity as a primary determinant of the nuclear mobility of the pioneer transcription factor FoxA. Genes Dev. 2009;23:804–809. doi: 10.1101/gad.1775509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gosalia N., Yang R., Kerschner J.L., Harris A. FOXA2 regulates a network of genes involved in critical functions of human intestinal epithelia cells. Physiol. Genomics. 2015;47:290–297. doi: 10.1152/physiolgenomics.00024.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hou C., Dale R., Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 51.Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., Lieberman P.M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D. Transcription. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou C., Zhao H., Tanimoto K., Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R., et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fossum S.L., Mutolo M.J., Yang R., Dang H., O'Neal W.K., Knowles M.R., Leir S.H., Harris A. Ets homologous factor regulates pathways controlling response to injury in airway epithelial cells. Nucleic Acids Res. 2014;42:13588–13598. doi: 10.1093/nar/gku1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris A., Coleman L. Ductal epithelial cells cultured from human foetal epididymis and vas deferens: relevance to sterility in cystic fibrosis. J. Cell Sci. 1989;92:687–690. doi: 10.1242/jcs.92.4.687. [DOI] [PubMed] [Google Scholar]
- 58.Verzi M.P., Shin H., He H.H., Sulahian R., Meyer C.A., Montgomery R.K., Fleet J.C., Brown M., Liu X.S., Shivdasani R.A. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell. 2010;19:713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Apostolou E., Ferrari F., Walsh R.M., Bar-Nur O., Stadtfeld M., Cheloufi S., Stuart H.T., Polo J.M., Ohsumi T.K., Borowsky M.L., et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712. doi: 10.1016/j.stem.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Wit E., Bouwman B.A., Zhu Y., Klous P., Splinter E., Verstegen M.J., Krijger P.H., Festuccia N., Nora E.P., Welling M., et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231. doi: 10.1038/nature12420. [DOI] [PubMed] [Google Scholar]
- 61.Wei Z., Gao F., Kim S., Yang H., Lyu J., An W., Wang K., Lu W. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47. doi: 10.1016/j.stem.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 62.Rosenbloom K.R., Sloan C.A., Malladi V.S., Dreszer T.R., Learned K., Kirkup V.M., Wong M.C., Maddren M., Fang R., Heitner S.G., et al. ENCODE data in the UCSC genome browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.