Abstract
Chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) is a key technique in chromatin research. Although heavily applied, existing ChIP-seq protocols are often highly fine-tuned workflows, optimized for specific experimental requirements. Especially the initial steps of ChIP-seq, particularly chromatin shearing, are deemed to be exceedingly cell-type-specific, thus impeding any protocol standardization efforts. Here we demonstrate that harmonization of ChIP-seq workflows across cell types and conditions is possible when obtaining chromatin from properly isolated nuclei. We established an ultrasound-based nuclei extraction method (NEXSON: Nuclei EXtraction by SONication) that is highly effective across various organisms, cell types and cell numbers. The described method has the potential to replace complex cell-type-specific, but largely ineffective, nuclei isolation protocols. By including NEXSON in ChIP-seq workflows, we completely eliminate the need for extensive optimization and sample-dependent adjustments. Apart from this significant simplification, our approach also provides the basis for a fully standardized ChIP-seq and yields highly reproducible transcription factor and histone modifications maps for a wide range of different cell types. Even small cell numbers (∼10 000 cells per ChIP) can be easily processed without application of modified chromatin or library preparation protocols.
INTRODUCTION
Chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) is a powerful technique for the genome-wide mapping of DNA-binding proteins and histone modifications (1,2). In recent years ChIP-seq has been applied systematically to a large variety of samples obtained from many cell types and analyzed by different research groups. While this has resulted in comprehensive resources, such as from ENCODE (3) and the NIH Roadmap (4), the lack of standardization in the very first steps of the protocol still represents a formidable challenge for comparative studies, not only across large consortia, but also among individual labs.
A typical ChIP-seq workflow includes cell fixation to covalently bind proteins to the DNA, chromatin extraction, immunoprecipitation with the antibody of interest, library preparation and deep sequencing. Many steps of the described workflow are extensively reviewed in literature, including antibody choice, library preparation, deep sequencing technologies and data analysis (5–9). In contrast, the initial steps of ChIP-seq procedures (including nuclei isolation, nuclei lysis and chromatin sonication) vary greatly across protocols and cell types (e.g., as shown in (7,10–13)). Despite a great deal of efforts aimed to improve ChIP-seq, given this diversity of cell types and experimental conditions, it has been nearly impossible to define common guidelines appropriate for all situations (6).
Especially the chromatin sonication step is notoriously difficult to optimize and standardize between different laboratories, cell types and samples. Large chromatin fragments (exceeding 800 bp) can compromise chromatin quality and lead to the failure of ChIP-seq (14). Aiming for DNA fragment sizes from 100 bp to 800 bp, trial-and-error approaches are typically used to optimize formaldehyde fixation time, buffer composition and sonication settings suitable for the respective experiment. Such extensive and labor-intensive protocol re-adjustments make ChIP-seq assays extremely material consuming, difficult to reproduce and expensive. Highly specialized workflows can also affect the comparability of results (15) and the need for iterative testing greatly limits ChIP-seq applicability to scarce samples (e.g., patient-derived specimen, sorted cells). Until now, the problems with small amounts of input material had been tackled using refined library preparation methods (16–19) to reduce artifacts from PCR amplification, but this does not address limitations in the very first steps of the chromatin extraction.
In this study we find that the above problems derive from the insufficient extraction of nuclei from formaldehyde-fixed cells, and we have developed a novel method to solve them. We show, for the first time, that ChIP-seq workflows are completely independent of the cell type if chromatin is extracted from properly isolated nuclei. Therefore we developed a new, sonication-assisted nuclei extraction procedure called NEXSON (Nuclei EXtraction by SONication). While existing nuclei extraction methods are largely ineffective on fixed cells, NEXSON allows efficient nuclei isolation using a simple and reproducible procedure. By including NEXSON in ChIP-seq protocols, we generate high-quality genome-wide chromatin maps across many different cell types. Furthermore, without any additional protocol modifications (20) or adjusted library preparation, we are able to significantly scale down the number of cells per assay (about 10 000 cells/histone modification ChIP and 100 000 cells/transcription factor ChIP). Our method can replace cell-type-specific protocols and will significantly improve the comparability of chromatin maps from different research groups and consortia.
MATERIALS AND METHODS
Cell culture and isolation
Cell lines were cultured as follows: IMR-90 (ATCC# CCL-186) and HepG2 (ATCC# HB-8065) were grown to the passage 20 in Eagle's Minimal Essential Medium (EMEM, ATCC, 30–2003) supplemented with 10% FBS, 2 mM L-Glutamine and Penicillin-Streptomycin mixture (100 units/ml). Mouse ES were cultured in 2i Medium. Human monocytes from two healthy male blood donors and human hepatocytes from one healthy female donor were obtained after written informed consent and anonymized. Blood cells were collected by leukapheresis in a Spectra cell separator (Gambro BCT, CO, USA) followed by counterflow elutriation (21). Cells were elutriated in the following order: platelets, lymphocytes, monocytes and then granulocytes. Aliquots of the different cell fractions were analyzed for cell purity on a BD FACSCanto flow cytometer (Becton Dickinson). Macrophages were differentiated in vitro seeding 1 × 106 elutriated monocytes per ml in macrophage serum-free medium (Invitrogen, Germany) supplemented with 50 ng/ml recombinant human monocyte-colony stimulating factor (rhMCSF; R&D Systems, USA), followed by 5 days incubation at 37°C with 5% CO2. Hepatocytes were isolated immediately after tissue resection, via liver perfusion (22). Buttoned cannulae were inserted into the largest vessels at the resection site of the liver piece and glued. Liver was perfused using minimum 300 ml of perfusion solution I (100 mM Hepes, 142 mM NaCl, 6.7 mM KCl, 5 mM, N-acetyl-L-cysteine 5 mM, 2.4 M EGTA) to remove residual blood. Tissues have been digested recirculating warm collagenase P in perfusion solution II (1.3 l of solution A containing 100 mM Hepes, 67 mM NaCl, 6.7 mM KCl, 0.5% Albumin were mixed with 150 ml of solution B, containing 4.8 mM CaCl2; pH was adjusted to 7.5 prior to use). Digestion was stopped after 10–20 min by pouring stop solution (20% Fetal Calf Serum in PBS) on the sample. Hepatocytes have been released by cutting up the liver piece into two halves and by gentle tweezing. Cell suspension was filtered to eliminate tissue debris, centrifuged for 5 min at 100 g, and resuspended in cell culture media. Viable cells were then isolated using density gradient centrifugation; pellets were collected and PBS-washed. Adipocytes samples were isolated by collagenase treatment (Type II, 0.5% in D-MEM containing 1.5% BSA) for 5 min at 37°C under shaking followed by 5 min of collagenase inactivation with 10 mM EDTA and 5 mM EGTA; after centrifugation the fat layer was collected.
Cell fixation
All cell types were fixed in 1% methanol-free formaldehyde (Thermo Scientific, 28906) in D-MEM for 5 min (unless otherwise indicated) at room temperature, followed by 5 min blocking in 125 mM glycine. Cells were rinsed two times in ice-cold PBS and pelleted (250 g, 10 min, 4°C). Adherent cultured cells were fixed in the plate and harvested by scraping after PBS washes. Prior fixation, whole Drosophila embryos were collected after the stage five and dechorionated with 50% bleach followed by permeabilization with 0.1% Triton X-100. All fixed cell pellets were flash-frozen in liquid nitrogen prior chromatin preparation.
Nuclei isolation (conventional)
Samples were resuspended in 1 ml of Farnham Lab (FL) buffer (5 mM PIPES pH 8; 85 mM KCl; 0.5% Igepal CA-630) (23) and incubated for 15 min at 4°C in an Eppendorf Thermomixer at 750 rpm mixing speed. To enhance nuclei extraction, incubation times were prolonged up to 60 min. Furthermore, other nuclei extraction buffer with different detergent composition were tested: FL with 1% Igepal and a commercial buffer (Covaris cell lysis buffer, cat no. 010128). After chemical treatment, nuclei were mechanically extracted by applying several (30 to 100) strokes of Dounce homogenizer (type B ‘tight’ pestle). Samples were centrifuged (1000 g, 5 min, 4°C) and washed once in equal volumes of the nuclei extraction buffer.
Nuclei isolation by NEXSON
For all cell types, 10 000 to 12 million cells were resuspended in 1 ml of Farnham lab (FL) buffer (5 mM PIPES pH 8; 85 mM KCl; 0.5% Igepal CA-630) supplemented with Complete Protease Inhibitor Cocktail, EDTA-free (Roche, 11873580001). Cell suspensions were sonicated in 12 × 12 AFA tubes (Covaris, 520081) using a Covaris S220 focused ultrasonicator, at peak power 75 W, duty factor 2% and 200 Cycles/burst at 4°C. For Drosophila embryos, duty factor was increased to 10% to prevent embryo sinking. The nuclei extraction progress was inspected every 30 s, using a benchtop phase-contrast microscope (Primo Vert, Zeiss) and stopped when more than 70% of nuclei were isolated (compare with Figure 3A). The specific treatment time for NEXSON varies across cell types or fixation time. For NEXSON, blood cells were treated for 60–90 s, except CD4+ cells requiring longer sonication time (5 min), mouse and human hepatocytes were treated for 4–5 min, HepG2 and IMR-90 for 2–3 min, mouse ES cell line for 30–60 s, adipocytes for 2 min, Drosophila embryos for 30–40 s and Paramecium samples for 2–3 min. Nuclei were collected by centrifugation (1000 g, 5 min, 4°C) and washed once in 1 ml of FL buffer.
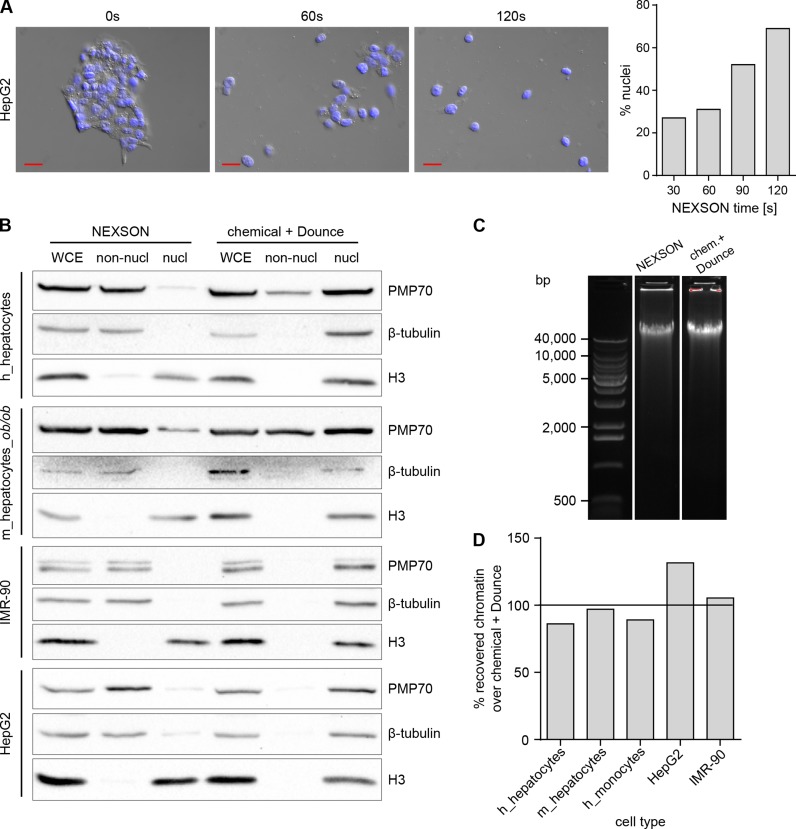
Figure 3.
NEXSON quality controls. (A) NEXSON time-course on HepG2. Fixed cell pellet was resuspended in FL buffer and treated by NEXSON using increasing amount of time. A small aliquot of cells was collected at 0, 30, 60, 90 and 120 s of ultrasound treatment and inspected with a microscope. Microscope images show the merge between DAPI and differential interference contrast (DIC) channels at the indicated time points. Red scale bar: 20 μm. For nuclei counting, 150 cells/nuclei were counted; bar chart shows the percentage of isolated nuclei over the total at the indicated time point. (B) Western blot analysis of non-nuclear (non-nucl) and nuclear (nucl) fractions obtained after NEXSON or chemical + Dounce treatment were conducted to check the effectiveness of the respective nuclei extraction protocol. Whole cell extract (WCE) was collected prior nuclei extraction and served as control. Cytoplasmic (PMP70 and β-tubulin) and nuclear (histone H3) markers were used to inspect the nucleus-cytoplasm fractionation. Abbreviations: h: human, m: mouse, m_hepatocytes_ob/ob: hepatocytes extracted from leptin-deficient obese mice. (C) Quality check of DNA integrity after NEXSON or chemical + Dounce treatment. Fixed cells were treated with the indicated nuclei extraction procedure; afterwards, an aliquot of chromatin was de-crosslinked and DNA was purified. Equal DNA amounts were loaded on a 0.7% agarose gel to inspect DNA integrity. Main base pairs (bp) of the molecular weight marker are indicated. (D) Comparison of chromatin recovery after NEXSON or chemical + Dounce treatment for nuclei extraction. Fixed cells were resuspended in FL buffer, splitted in two aliquots and treated with either NEXSON or the Chemical + Dounce nuclei isolation protocol. Nuclear preparations were resuspended in shearing buffer, chromatin was sheared and DNA purified and quantified. Bar chart shows the percentage of chromatin recovered after NEXSON over the chromatin obtained after chemical and Dounce treatment.
For NEXSON using either the Bioruptor Plus or the Bioruptor Pico sonicator (Diagenode), HepG2 were resuspended in 0.5 ml of FL buffer and treated with the following parameters: Bioruptor Plus at low power with three cycles (15 s on and 30 s off); Bioruptor Pico with six cycles (10 s on and 30 s off).
Microscope analysis
For picture acquisition and to check the nuclei quality, nuclei were DAPI-stained (24). All pictures were taken in the DAPI and differential interference contrast (DIC) channels using the Zeiss Axioimager Apotome microscope equipped with Zeiss AxioVision software.
Western blot
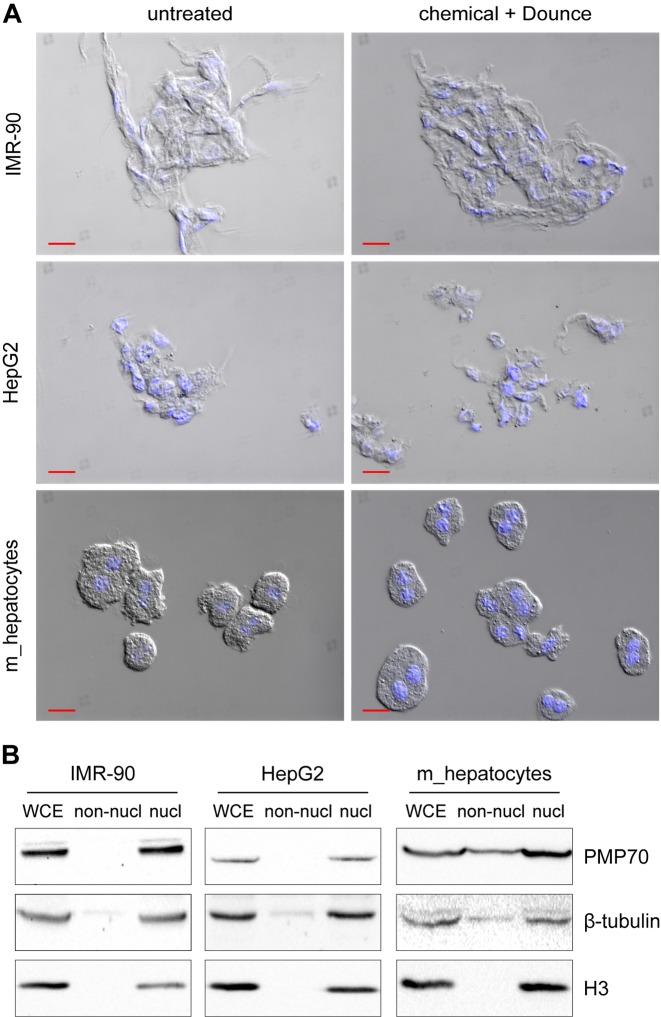
To inspect the effectiveness of nuclei isolation, equal volumes (one twentieth of the sample) of supernatant (non-nuclear fraction) and nuclear fraction were collected during the treatment. Equal amounts of unfractionated sample (WCE: Whole Cell Extract) were also harvested prior to nuclei extraction as a control. All fractions were denatured in Laemmli buffer, supplemented with β-mercaptoethanol (3% final), and heated. Equal volumes of the fractions were loaded on a 10–12% SDS-PAGE gel and transferred on a nitrocellulose membrane. H3 (∼17 kDa) and PMP70 (∼70 kDa) were separated by cutting up the membrane and incubated overnight with the primary antibodies (anti-PMP70: Millipore, ABT12, 1:10 000; anti-H3: Diagenode, C15310135, 1:10 000) in TBS-T 5% Milk. β-tubulin (∼50 kDa) has been detected afterwards through re-incubation of the same membrane with anti-β-tubulin (Santa Cruz, sc9104, 1:500). All antibodies were detected with the secondary antibody goat anti-Rabbit IgG, HRP conjugated (Millipore, AP132P, 1:20 000) and visualized by chemiluminescence (Supersignal West Pico, Thermo scientific, 34087) using the ChemiDoc XRS+ system (Bio-Rad).
Chromatin preparation and ChIP
All NEXSON-isolated nuclei were resuspended in 1 ml of shearing buffer (10 mM Tris-HCl pH 8; 0.1% SDS; 1 mM EDTA) supplemented with Complete Protease Inhibitor Cocktail EDTA-free (Roche, 11873580001) and sheared for 15–20 min to a fragment size distribution of 100–800 bp (Covaris S220 focused ultrasonicator, Peak Power: 140 W; Duty factor: 5%; Cycles/burst: 200, water temperature 4°C). For the protocol comparison experiment, the same batch of fixed cells was used to test the following commonly used protocols: ‘BLUEPRINT’ (13), ‘Young’ (12), ‘ENCODE’ (7) and NEXSON (this study). All samples were sonicated with the Covaris instrument to reach a fragment size distribution of 100–800 bp (Supplementary Figure S8). Automatic ChIP was performed using the SX-8G Compact IP-Star liquid handler from Diagenode in combination with Diagenode Auto Histone Kits (Diagenode, C01010022). Using the pre-programmed method ‘indirect ChIP’, ChIP reactions were carried out in a final volume of 200 μl for 10 h followed by 3 h beads incubation and 5 min washes (at 4°C). After ChIP, eluates were recovered, RNase A-treated, decrosslinked and deproteinized for 30 min at 37°C and 4 h at 65°C, and DNA was purified using MinElute columns (Qiagen, 28006). The following antibodies were used for ChIP-seq (1–2 μg per ChIP reaction): anti-H3K4me3 (C15410003) and anti-H3K27me3 (C15410195) from Diagenode, and anti-CTCF (5 μg per ChIP) from Santa Cruz (sc-15914).
Chromatin quality controls
To inspect DNA concentration and fragment size distribution after chromatin preparation, DNA was purified (Qiagen PCR purification kit, 28106) from aliquots of decrosslinked and proteinase K-treated chromatin. DNA was quantified using the Qubit dsDNA High Sensitivity assay (Invitrogen, Q32851). Fragment size distribution was analyzed by capillary electrophoresis (Agilent 2100 Bioanalyzer) using the High Sensitivity DNA ChIP kit (Agilent, 5067–4626).
Library preparation and deep sequencing
Sequencing libraries were prepared using the NEBNext Ultra DNA Library Prep kit for Illumina (E7370S, NEB). Starting amount of fragmented DNA varied between sub-nanograms and 2–5 ng. Following adaptor ligation (1.5 μM), size selection was omitted and adaptor-ligated DNA was directly purified using AMPure XP beads (Beckman Coulter). Subsequently adaptor-ligated DNA was enriched using 10 PCR cycles. Final libraries were quantified with Qubit dsDNA HS assay (Invitrogen, Q32851) and size distribution was monitored by capillary electrophoresis (Agilent 2100 Bioanalyzer, High Sensitivity DNA Chips (Agilent, 5067–4626)). Libraries were sequenced paired-end with a read length of 50 bp on a HiSeq 2500 instrument (Illumina).
ChIP-seq data deposition
Human sequencing data have been deposited at the European Genome-Phenome Archive (EGA) under accession numbers EGAS00001000719 (human monocytes) and EGAS00001000972 (human hepatocytes). Cell line sequencing data have been deposited at the European Nucleotide Archive under accession numbers PRJEB7356 (HepG2 presented in Figure 5 and Supplementary Figure S7) and PRJEB7177 (IMR-90, HepG2 presented in Figures 7 and 8).
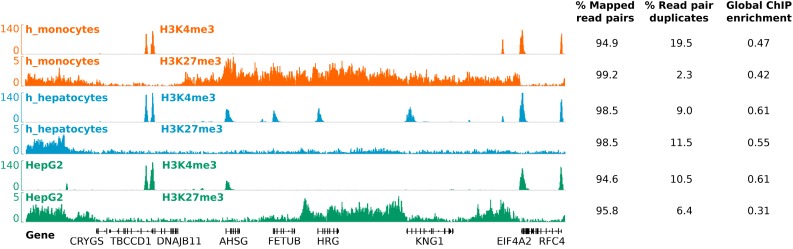
Figure 5.
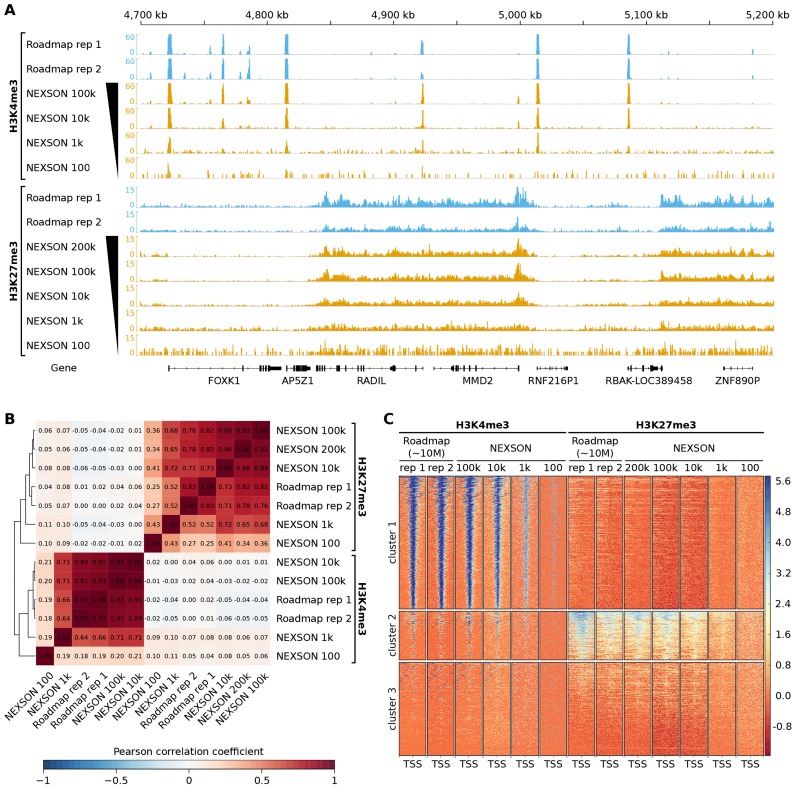
ChIP-seq of NEXSON-treated cells yields high-quality signals. Read coverage profiles of histone modification ChIP-seq for human monocytes (orange), hepatocytes (blue) and HepG2 cell line (green) prepared by NEXSON. Read mapping statistics and global ChIP enrichment (FRiP, Fraction of mapped Reads in Peaks) are given next to each sample. Per ChIP reaction, between 300 000 and 850 000 cells were used. Please note cell-type-specific epigenetic marking of transcript start sites (TSS) and gene bodies, e.g. of fetuin-A (AHSG), which is produced exclusively by the liver. ChIP-seq tracks show a 320 kb region of human chromosome 3; the signal of each sample is normalized with respect to sequencing depth (26).
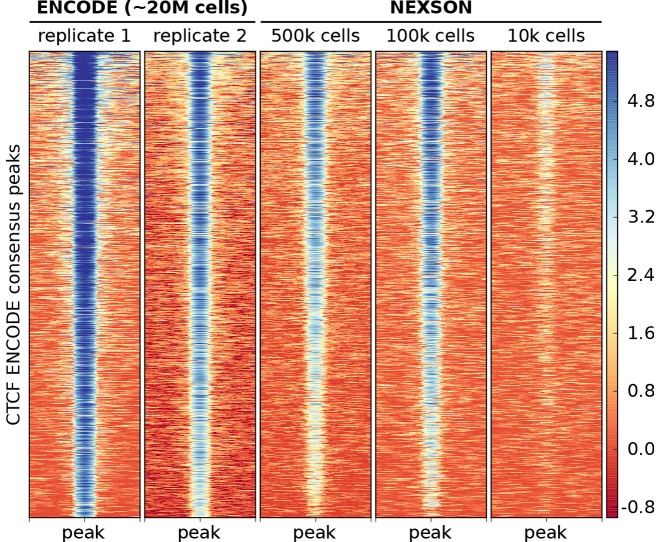
Figure 7.
CTCF transcription factor ChIP-seq works reliably with NEXSON down to 100 000 cells. CTCF ChIP-seq enrichment in HepG2 over a consensus set of CTCF peaks. The heatmap shows all CTCF peaks ± 1 kb with each row representing a distinct peak. The signal intensity is measured in log2 read coverage normalized by sequencing depth. NEXSON-treated samples used medium (500 000) to low (100 000 or 10 000) cell numbers per ChIP reaction as indicated. External ENCODE samples used a high cell number (∼20 million cells/ChIP).
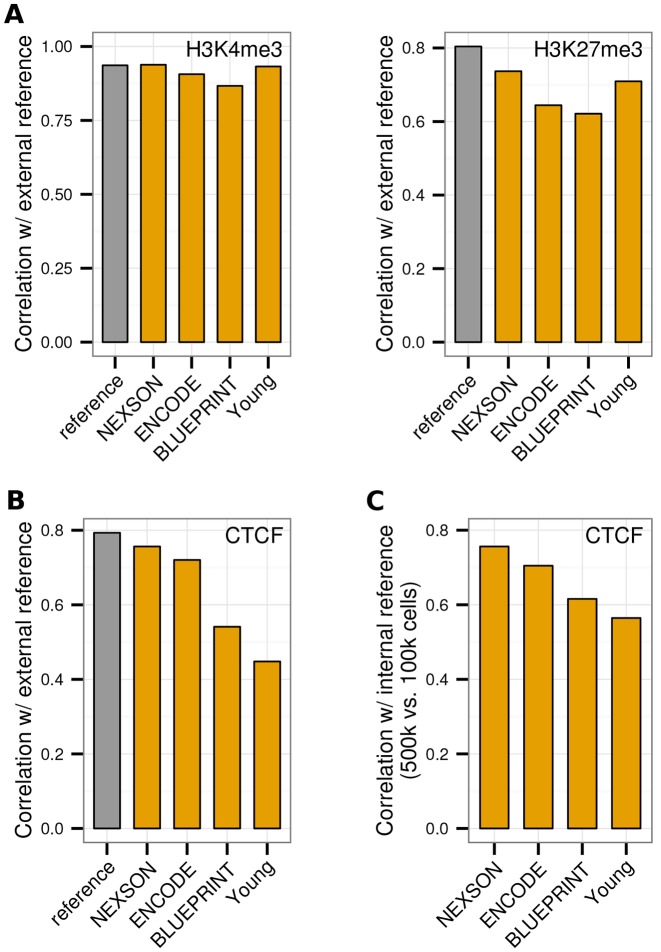
Figure 8.
NEXSON ensures higher ChIP-seq reproducibility than other methods when using low cell numbers. Genome-wide correlation of ChIP-seq data from HepG2 cells treated with NEXSON compared to three existing protocols (ENCODE, BLUEPRINT, Young) and external ENCODE data (3). Pairwise correlation of (A) H3K4me3 and H3K27me3 histone modification ChIP-seq data (10 000 cells/ChIP), and (B) CTCF transcription factor ChIP-seq data (100 000 cells/ChIP), all produced in this study, with external ENCODE reference data (orange). External ENCODE data sets from two different labs are additionally compared with each other (grey). (C) Intra-protocol comparability of CTCF ChIP-seq, quantified by pairwise correlation of samples obtained from medium (500 000) versus low (100 000) cell numbers per ChIP reaction. All plots show the Pearson correlation coefficient on 1 kb bins excluding signal artifact regions.
External reference ChIP-seq data
IMR-90 ChIP-seq data produced by the NIH Roadmap Epigenomics Mapping Consortium (4) were obtained from Sequence Read Archive. FASTQ files were extracted with the fastq-dump tool. Samples included H3K27me3 ChIP (GEO accessions GSM469968 and GSM521889 for biological replicate 1 and 2, respectively), H3K4me3 ChIP (GSM469970 and GSM521901) and the corresponding input chromatin (GSM521926/7 and GSM521929). HepG2 ChIP-seq data were obtained as FASTQ files with raw sequencing reads and BED files with optimal IDR thresholded peaks from the ENCODE Consortium (3) portal. Samples included H3K27me3 ChIP (ENCODE accessions ENCFF000BFU and ENCFF001FLT for biological replicate 1 and 2, respectively), H3K4me3 ChIP (ENCFF000BGE and ENCFF001FMH) and CTCF ChIP (ENCFF000PHE and ENCFF001HNT).
Analysis of ChIP-seq data
Monocytes, hepatocytes, HepG2 and IMR-90 ChIP-seq raw sequencing reads were aligned to human genome assembly hs37d5 with bowtie2 (25) (parameter “-X 1000” for paired-end reads, default parameters otherwise). Paired-end reads were used for monocytes, hepatocytes and HepG2 data (presented in Figure 5, Supplementary Figure S7); single-end reads were used for IMR-90 and HepG2 sequencing data produced within this study (presented in Figures 6–8), and by the Roadmap and ENCODE consortia. Fragment coverage bigWig files were computed at 25 bp resolution, 200 bp average fragment size and normalization to sequencing depth using deepTools (26). Log2 ratios of ChIP over input signal were computed with deepTools using normalization based on read counts; reads not aligned in pairs were extended to 200 bp fragment size. Peak regions were called with MACS2 (27) (parameters “-g 2.9e9 --keep-dup all”, default parameters otherwise) providing input chromatin data as control; calling of broad peaks was enabled for H3K27me3. For monocytes, hepatocytes and HepG2 paired-end data, the median fragment size was determined with Picard tools and provided as input parameter to MACS2. For IMR-90 single-end data, MACS2 estimated the fragment size by strand cross-correlation. Read coverage profiles were visualized with IGV (28).
Figure 6.
ChIP-seq with NEXSON gives high-quality and reproducible enrichment signals even for limited cell numbers. Comparison of H3K4me3 and H3K27me3 IMR-90 ChIP-seq samples prepared by NEXSON with external data from the NIH Roadmap Consortium (4). NEXSON-treated samples used either 200 000, 100 000, 10 000, 1 000 or 100 cells per ChIP reaction as indicated. Roadmap samples used a high cell number (∼10 million cells/ChIP). (A) Read coverage profiles of NEXSON (orange) and Roadmap (blue) samples. The read coverage of all samples is sequencing-depth normalized. Shown is a 500 kb region of human chromosome 7. (B) Genome-wide correlation of read coverage. The heatmap shows the pairwise Pearson correlation coefficient based on read coverage of 1 kb bins excluding signal artifact regions. (C) ChIP-seq enrichment over all TSS annotated in RefSeq genes. The heatmap shows regions of 5 kb up- and downstream of the TSS with each row representing a distinct TSS. The signal intensity is measured in log2 read coverage normalized by sequencing depth. Regions were clustered by the k-means algorithm.
Comparison of ChIP-seq samples
Before comparison of ChIP-seq samples, duplicated reads were identified with Picard tools and removed. Correlations between samples were computed with the UCSC tool wigCorrelate (29) based on genomic read coverage of 1 kb bins excluding regions with artificially high read coverage as compiled in the ENCODE DAC Blacklist (3). Global ChIP enrichment strength was evaluated by FRiP (30) (Fraction of mapped Reads in Peak regions) as the number of reads overlapping peak regions, divided by the total number of mapped reads. For IMR-90 samples, FRiP values were computed for the union of significant peak regions identified in the Roadmap biological replicates. Heatmaps for visualizing and clustering of the ChIP-seq signal intensity around all distinct RefSeq transcription start sites (TSS) and ENCODE consensus CTCF peaks were generated with deepTools (26). RefSeq gene annotation for human genome assembly hg19 was obtained from UCSC Genome Browser (31) at 22 May 2014. A consensus CTCF peak set for HepG2 was computed by the intersection of the optimal IDR thresholded peak regions provided by ENCODE.
RESULTS
Traditional methods fail to isolate nuclei from fixed cells
Traditional nuclei extraction methods generally include a chemical treatment with hypotonic buffers that contain non-ionic detergents such as Igepal CA-630 (23). The composition of the lysis buffer in use varies between protocols and no standard method is available to select the appropriate buffer composition. Furthermore, a mechanical treatment, using a Dounce homogenizer (32), can be applied to enforce nuclei release. We discovered that state-of-the-art nuclei extraction methods, typically included in many ChIP-seq workflows, are largely ineffective when applied to formaldehyde-fixed cells. DIC microscopy and DAPI staining revealed a large fraction of nuclei surrounded by cytoplasm when nuclei were extracted from fixed cells using state-of-the-art nuclei extraction methods (Figure 1A). Western blot analysis of the fractions collected prior and after nuclei extraction, to detect protein markers located in the cytoplasm (β-tubulin and the 70-kDa peroxisomal membrane protein PMP70) and in the nucleus (histone H3), confirmed that proper fractionation does not occur when cells were formaldehyde-fixed prior to nuclei extraction (Figure 1B). Even harsher treatments like more stringent detergent conditions, additional Dounce homogenizer strokes and increased incubation time, resulted in no or very poor nuclei extraction (Supplementary Figure S1). Notice that the described nuclei extraction procedures can be effectively applied to unfixed cells (Supplementary Figure S2).
Figure 1.
Traditional methods fail to isolate nuclei from formaldehyde-fixed cells. (A) Microscope pictures of fixed samples resuspended in phosphate-buffered saline, prior (untreated) or after (chemical + Dounce) nuclei extraction. Pictures show the merge between DAPI (blue: nuclei) and differential interference contrast (DIC) channels. Red scale bar: 20 μm. Different cell types were fixed for 5 min using formaldehyde; subsequently, cells pellets were freeze-thawed and nuclei were extracted using detergent-containing hypotonic buffers in conjunction with mechanical homogenization (30 Dounce homogenizer strokes with a tight pestle). (B) Western blot analysis of fractions collected prior (WCE: whole cell extract) or after nuclei extraction treatment (non-nucl: non-nuclear equals supernatant after treatment; nucl: nuclear extract equals pellet after treatment). PMP70, β-tubulin (cytoplasmic markers) and histone H3 (nuclear marker) were visualized with the respective antibodies on the same experiment to confirm successful nuclei extraction. Abbreviation: m: mouse.
Development of an efficient nuclei extraction method for fixed cells
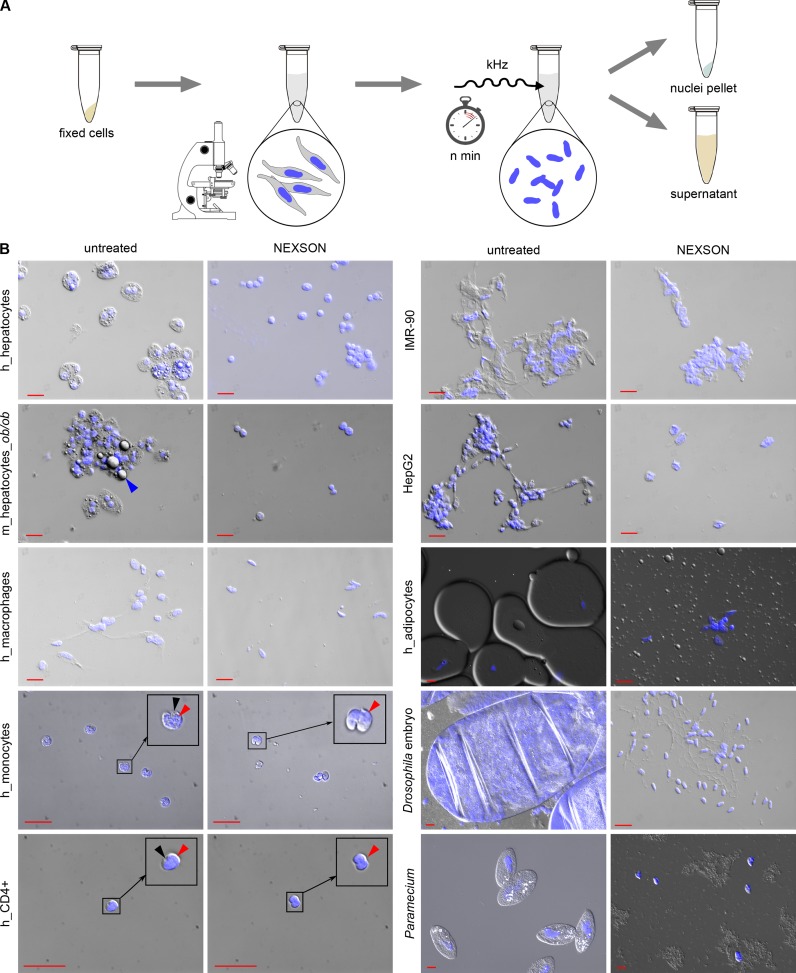
Since traditional methods fail to properly extract nuclei from fixed cells, we established a novel method that does not rely on chemical treatments and mechanical homogenization to extract nuclei from fixed cells, but uses instead ultrasound to lyse the cell membrane (NEXSON). As outlined in Figure 2A, NEXSON involves two simple steps: resuspension of the cell pellet in a buffer compatible with nuclei extraction followed by moderate sonication to isolate nuclei from fixed cells. The utilized buffer must maintain nuclei intact; therefore it should not contain ionic detergents that lyse the nuclear membrane. The progression of nuclei extraction is controlled by visual inspection of a small amount of sample using a benchtop phase-contrast microscope. Nuclei are counted at definite time points of treatment and NEXSON is stopped when a sufficient number of nuclei has been extracted (over 70%, Figure 3A). The adjustments in the sonication time, which vary between cell types (30 sec–5 min) and fixation conditions, are controlled in real-time by microscope inspection on the sample of interest. Note that the sonication power used for NEXSON is much lower compared to chromatin sonication (see Materials and Methods). While we are mostly using a Covaris S220 ultrasonicator, NEXSON works equally well with other sonicators (Supplementary Figure S3), highlighting the independence of this protocol from specific devices.
Figure 2.
Nuclei extraction by sonication (NEXSON) from formaldehyde-fixed cells. (A) Schematic representation of NEXSON. Formaldehyde-fixed cells are suspended in a buffer compatible with nuclei extraction and subsequent ChIP, and treated with ultrasound. Progression of nuclei extraction is controlled in real-time using a benchtop phase-contrast microscope. Based on the cell type in use, the sonication time needs marginal adjustments. Nuclear and non-nuclear fractions are separated by centrifugation when most of the nuclei were isolated. The whole procedure can be completed in less than 20 min. (B) Various fixed cell types or whole organisms (Drosphila embryos in cellularization stage five, Paramecium) were treated by NEXSON. Formaldehyde-fixed cells were resuspended in nuclei extraction buffer and treated by sonication to induce nuclei extraction. Pictures in DAPI (blue, nuclei) and differential interference contrast (DIC) channels were taken before (untreated) and after (NEXSON) ultrasound treatment. The analyzed cell types included blood cells (in vitro-derived macrophages, monocytes and CD4+ cells), high fat-containing cells (adipocytes and hepatocytes), as well as cell lines (HepG2, IMR-90). Monocyte and CD4+ pictures were enlarged for better visualization of nucleus and cytoplasm (encircled): red arrows indicate the nuclei, black arrows the cytoplasm. Fat droplets are highlighted in the m_hepatocytes_ob/ob sample (blue arrow). Red scale bar: 20 μm. Abbreviations: h: human, m: mouse, m_hepatocytes_ob/ob: hepatocytes extracted from leptin-deficient obese mice.
We demonstrate effectiveness of NEXSON on various cell types differing greatly in size, fat and cytoskeletal content. Figure 2B shows microscopy quality controls performed on NEXSON-treated fixed hepatocytes, high fat-containing hepatocytes (ob/ob hepatocytes from leptin-deficient mice) and cell lines like IMR-90 myofibroblasts and HepG2. After the treatment, the microscope analysis revealed that all NEXSON-treated samples are depleted from cytoplasm and isolated nuclei are present as intended. We also applied NEXSON to more challenging samples, notoriously difficult to process, such as fixed blood cells (monocytes, in vitro-derived macrophages, CD4+ cells) and human adipocytes, organisms from different kingdoms (Paramecium), and even whole fixed Drosophila embryos. As shown in the microscopy images in Figure 2B, intact nuclei have been isolated from those samples, including macronuclei of Paramecium. Western blot analysis of the fractions collected after NEXSON revealed an enrichment of cytoplasmic markers only in the non-nuclear fractions and their reduction in the nuclear fractions (Figure 3B), confirming the effective nucleus-cytoplasm fractionation. Importantly, NEXSON does neither compromise the DNA integrity in the nuclei (Figure 3C) nor the chromatin recovery after chromatin preparation (Figure 3D).
NEXSON maintains its reproducibility when applied to samples fixed for longer times (up to 20 min, which is generally considered a prolonged fixation; Supplementary Figure S4A) and it performs equally well even in buffers not intended for nuclei extraction purposes (phosphate-buffered saline, Supplementary Figure S4B), highlighting the independence of NEXSON from nuclei extraction-specific buffers.
Reproducible and optimal chromatin shearing across cell types
A prerequisite of successful ChIP-seq is proper fragmentation of the chromatin to sizes between 100 and 800 bp, marking the trade-off between epitope binding and integrity as well as the size range required for high-throughput sequencing. In ChIP-seq workflows the desired size distribution is typically achieved by iterative variation of shearing buffer composition (e.g., increasing the amount of detergents), and tuning fixation and sonication time.
By including NEXSON in the chromatin preparation step, we achieve reproducible and optimal shearing profiles across samples and cell types without varying shearing buffer composition or sonication power. Notice that of the 15 different cell types tested only for CD4+ cells the sonication time was prolonged from 15 to 20 min to achieve the desired size distribution (Supplementary Figure S5).
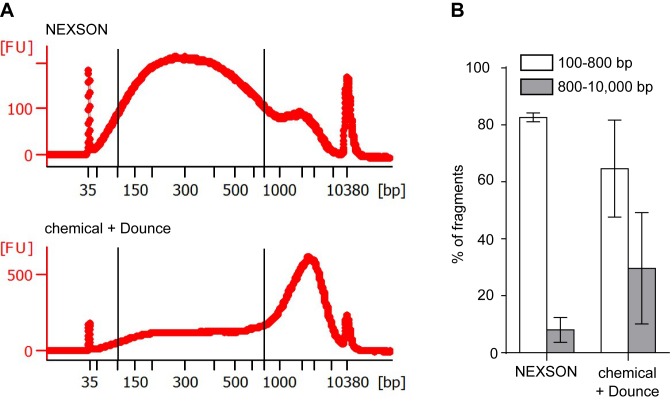
In contrast, when chromatin is prepared using chemical nuclei isolation enforced with Dounce homogenization, shearing profiles vary largely across different samples, as exemplified in Figure 4. Figure 4A shows significant differences in the chromatin fragment size distribution after ultrasound-assisted (NEXSON) or chemical nuclei extraction. Considering a fragment size distribution of 100–800 bp as ideal for ChIP-seq assays, a significantly lower percentage of fragments resides in the desired size range when nuclei are not properly isolated prior to chromatin shearing. Furthermore, variability in shearing profiles can be reduced upon successful nuclei extraction (Figure 4B). Notice that in some cases failure of conventional nuclei extraction can lead to partially shearable chromatin and influence chromatin quality (Supplementary Figure S6).
Figure 4.
NEXSON enhances the reproducibility of chromatin shearing. (A) Size distribution after chromatin shearing when extracting nuclei with either NEXSON or chemical and Dounce treatment from a single, formaldehyde-fixed cell batch (human monocytes). Chromatin was sheared with identical sonicator settings and DNA size distribution was analyzed using capillary electrophoresis. Note that differences in ultrasound exposure between the two workflows (NEXSON or chemical + Dounce) were normalized prior chromatin shearing. Electropherograms, generated with Agilent expert 2100 software, show the size distribution of the respective samples. Optimal chromatin size distribution for ChIP-seq is located between the two bars (100–800 bp). x axis: base pairs (bp), y axis: fluorescence units (FU). (B) Quantitative analysis of the chromatin size distribution of three different samples after nuclei extraction with NEXSON or chemical + Dounce homogenizer treatment. Bars show the percentage of DNA fragments in the optimal (between 100 and 800 bp, white bars) or inadequate (800–10 000 bp, gray bars) size range for ChIP-seq. Percentages of fragments in the respective region are calculated with Agilent Bioanalyzer software and averaged (error bars indicate s.d.; n = 3: fixed monocytes, IMR-90 and hepatocytes samples).
Overall, these observations indicate that the inclusion of NEXSON in chromatin preparation for ChIP-seq is beneficial to achieve proper and reproducible chromatin size distribution without additional shearing buffer or sonication power optimizations.
High-quality histone modification maps from NEXSON-based ChIP-seq
To illustrate the applicability of NEXSON for ChIP-seq, we applied ChIP-seq to chromatin prepared by NEXSON from various fixed cells, including human cells derived from biopsies and cell lines alike. After NEXSON treatment, samples were subjected to chromatin shearing. The shearing was followed by immunoprecipitation using antibodies against histone modifications marking transcriptionally active (H3K4me3) and inactive (H3K27me3) chromatin. Genome-wide ChIP-seq profiles were generated for human monocytes and hepatocytes extracted from whole tissues, and for the human cell lines HepG2 and IMR-90.
For all samples, we observe a strong ChIP enrichment signal and low background noise, as can be seen for selected loci and also on a genome-wide scale (Figures 5 and 6A). Moreover, samples prepared by NEXSON show a very high mapping rate (≥95%) and low duplication rates (≤20%). Genome-wide profiles from several other cell types presented in Figure 2B and Supplementary Figure S5 are of the same quality (data not shown). As expected, we can observe cell-type-specific differences in the epigenetic profiles (Figure 5). Unsupervised clustering of the ChIP-seq signal around all annotated TSS confirms cell-type-specific promoter activity on a genome-wide level (Supplementary Figure S7).
For the IMR-90 cell line, we compared our results to publicly available reference epigenomes from NIH Roadmap (4) and observe excellent agreement as quantified by genome-wide correlation of the read coverage (Figure 6B). Distribution and intensity of the H3K4me3 and H3K27me3 signal at TSS is in very good agreement between NEXSON and Roadmap samples (Figure 6C), again highlighting the concordance and quality of NEXSON results for both localized and broadly distributed histone modifications.
NEXSON works robustly for low cell numbers
When generating histone modification maps in an epigenomic context, typically a comprehensive set of different histone marks is studied by individual ChIP-seq experiments. Therefore we investigated how many ChIP reactions could be conducted from a single sample without changing chromatin extraction, ChIP and sequencing library preparation protocols. In particular, we did not apply more refined procedures (such as proposed in (16,18,33)) and only used standard protocols. To assess the ChIP and library preparation sensitivity limit, NEXSON-prepared chromatin from 1 million formaldehyde-fixed IMR-90 cells was serially diluted down to 100 cells. Visual inspection shows that the ChIP signal is still detectable at 100 cells/ChIP, although the signal-to-noise ratio and the library complexity deteriorate markedly below 1000 cells/ChIP (Figure 6; Supplementary Table S1). On the other hand, histone profiles are highly reproducible down to 10 000 cells per ChIP reaction (Figure 6), which is three orders of a magnitude below the cell number used for the Roadmap samples. These results demonstrate the robustness of sample preparation by NEXSON even for very small cell numbers, without compromising the overall quality of the data (Supplementary Table S1). The results further suggest that a population of 1 million cells is sufficient for up to 100 individual histone ChIPs in high-throughput assays.
NEXSON works reliably for transcription factor ChIP-seq using small amounts of cells
To illustrate that NEXSON also helps the more challenging analysis of transcription factors (TFs), we performed ChIP-seq against CTCF in the HepG2 cell line using different amounts of starting material. Our results were compared to reference data that was generated by two different labs from the ENCODE consortium (3). We see a high genome-wide concordance of NEXSON-derived signal down to 100 000 cells per ChIP reaction (Figure 7). Our CTCF samples show a clear enrichment in the regions that were identified as consistently enriched across ENCODE replicates. These results demonstrate the applicability of NEXSON to TF ChIP-seq when the amount of starting material is limited. Please note that, for very low cell numbers (10 000 and below), we still detect enrichment in the annotated CTCF binding sites, but the signal-to-noise ratio is compromised and we do not suggest to use such a small cell number for de novo detection of CTCF sites.
NEXSON-based ChIP-seq has higher reproducibility than other widely used ChIP-seq protocols
We assessed the quality and robustness of ChIP-seq on NEXSON-prepared chromatin by a side-by-side comparison with three established protocols for chromatin preparation (7,12,13), starting from the same batch of formaldehyde-fixed HepG2 cells. Compared to the other protocols, shearing of NEXSON-treated chromatin required shorter time and no further adjustments to achieve the desired fragment size range (Supplementary Figure S8). Using the chromatin obtained by each method, we then conducted ChIP-seq against CTCF (500 000 and 100 000 cells starting material) and against the histone modifications H3K4me3 and H3K27me3 (10 000 cells starting material). The results were compared to publicly available reference data from two ENCODE labs working with high cell numbers (∼20 million cells/ChIP). ChIP-seq signals obtained by NEXSON have the highest genome-wide agreement with the external references, both for the studied histone modifications (Figure 8A) as well as for the TF (Figure 8B). Furthermore, we would like to stress that NEXSON is the only of the four tested protocols that works equally well for studying both transcription factor binding and histone modifications. Another important issue is the intra-protocol comparability of ChIP-seq signals, especially when the amount of starting material varies due to limited sample availability. When comparing the CTCF profiles obtained from medium versus low cell numbers by the same protocol, we find that NEXSON ensures the best reproducibility of the genome-wide ChIP-seq signal of all compared protocols (Figure 8C). This observation is completely in line with the robustness already seen for histone marks (IMR-90, Figure 6).
Overall, our results demonstrate the reliability of NEXSON to generate high-quality genome-wide ChIP-seq signals for both transcription factors and histone modifications, and suggest its application to low cell number ChIP-seq experiments without the need for any protocol modifications.
DISCUSSION
Chromatin preparation was deemed to be too difficult to be standardized across various cell types and hence it had not been investigated systematically. We identified nuclear extraction to be a key determinant of a reproducible ChIP-seq. Indeed, we observed that common chemical treatments and Dounce homogenization frequently fail to isolate nuclei from fixed cells. Failures at this early stage can undermine all subsequent standardization efforts. For example, they typically result in highly variable chromatin shearing profiles and size distributions that are unsuitable for further processing. We speculate that these problems might be due to covalent bonds induced by the crosslinking step. Such bonds could anchor the nuclei to a robust mesh of cytoplasmic components and detergents will be unable to break such bonds.
In our efforts to standardize ChIP-seq, we developed a simple and universal method designed to yield high quality and quantity of nuclei from fixed cells. The main idea is to resolve the cell membrane and extract nuclei by low-energy sonication, which is able to break covalent bonds. This basic approach is straightforward to control and real-time visual inspection can be used to determine the only free parameter in the protocol: the sonication time. As soon as a sufficient number of nuclei (e.g., >70%) are released, one can proceed with chromatin shearing. No iterative and material-consuming optimization is required, which makes NEXSON particularly suitable for cases where only limited material is available, such as for many clinical samples.
Starting from a large fraction of isolated nuclei removes many of the cell-type-specific problems that have plagued chromatin research, and provides a purer substrate for subsequent steps, notably chromatin shearing. This major improvement of NEXSON can clearly be seen and quantified by the shearing profiles which are highly reproducible in our approach. We tested our approach extensively for a large variety of fixation conditions, heterogeneous cell types and even whole embryos. In all cases, NEXSON yields chromatin that can easily be sheared to the desired size distribution. There is no need for fine-tuned conditions or extensive shearing that often accompany ‘difficult’ cell types.
Overall our chromatin extraction is much gentler compared to traditional approaches. In particular, nuclei isolated by NEXSON are more easily sonicated in mild shearing buffers and with short shearing time to obtain appropriate size distribution of chromatin. This might help to preserve epitope integrity and binding to the DNA. Supporting this hypothesis, we are able to pull down sufficient amount of DNA also from very scarce starting material. Even though we are relying only on standard protocols for immunoprecipitation and library preparation, we are able to generate high-quality histone maps from as few as 10 000 cells/ChIP, and maps of transcription factors from 100 000 cells/ChIP and below. This can be quantified by very high mapping rates, low duplication rates, high signal-to-noise ratios and excellent genome-wide reproducibility at the level of current reference standards and better. Furthermore, NEXSON-based ChIP-seq has higher reproducibility than other widely used ChIP-seq protocols and performs equally well independent of the analyzed target.
The sensitivity of the NEXSON approach and its independence of detergents also bodes well for other applications that require nuclear extraction and intact DNA, such as Chromosome Conformation Capture and derived techniques (34), chromatin accessibility assays (35) and also proteomic analysis (36), although in this latter case more refined analyses might be needed to investigate potential co-purification with other cellular compartments. Even for unfixed cells, NEXSON might be beneficial in situations where nuclei extraction is challenging or requires softer treatment, e.g. DNase I chromatin accessibility (37).
In conclusion, NEXSON is an extremely simple and reproducible method for isolating nuclei from a wide range of fixed cells, which eliminates sample-to-sample variability of sonication and results in many improvements for downstream applications. This makes the method optimal for standardized chromatin research, large-scale collaborations and clinical studies. Given the proven quality of NEXSON-treated chromatin even for small cell numbers, our method can also be used in combination with ongoing efforts to increase the sensitivity of ChIP-seq. Therefore we propose to use NEXSON as a first common step for all ChIP-seq protocols and whenever a standardized nuclei extraction protocol is required.
Supplementary Material
Acknowledgments
We would like to thank C. Cadenas (IfADo, Dortmund) for providing hepatocytes, S. Wallner (University Hospital Regensburg) for monocytes and human adipocytes, J. Polansky (DRFZ, Berlin) for CD4+ cells, H. Pahl and C. Köllerer (Medical Center University of Freiburg) for granulocytes, N. Iovino and E. Löser for Drosophila embryos (MPI-IE, Freiburg), K. Dalgaard and J. Longinotto for mouse adipocytes (MPI-IE, Freiburg), M. Simon and M. Cheaib for Paramecium samples (Saarland University, Saarbrücken), K. Hoffmeyer for mouse embryonal stem cells (MPI-IE, Freiburg) and N. and G. Gasparoni for providing the HepG2 cell line and HepaRG fixed pellets (Saarland University, Saarbrücken).
We further would like to thank T. Jenuwein for support and critical comments, A. Pospisilik and F. Dündar for critical reading of the manuscript and suggestions, F. Ramírez for help with project-specific adjustments to deepTools, and Bärbel Felder and Gideon Zipprich for support with sequence data submission. We thank the imaging facility of the MPI-IE for providing support and instrumentation for picture acquisition.
Author contributions: L.A. conceived and executed the study, U.B., E.B. and K.B. performed sequencing and library preparation, A.S.R. performed the bioinformatic and comparative analysis, S.D. implemented the pipeline for primary sequence analysis and quality control, U.B. and T.M. provided advice and supervised the project, L.A., A.S.R., U.B. and T.M. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Federal Ministry of Education and Research (German Epigenome Programme ‘DEEP’’) [01KU1216G]; German Research Foundation collaborative research centre ‘Medical Epigenetics’ [CRC992]. Funding for open access charge: own funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 3.ENCODE Project Consortium. Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Kheradpour P., Zhang Z., Heravi-Moussavi A., Liu Y., Amin V., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egelhofer T.A., Minoda A., Klugman S., Lee K., Kolasinska-Zwierz P., Alekseyenko A.A., Cheung M.-S, Day D.S., Gadel S., Gorchakov A.A., et al. An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furey T.S. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat. Rev. Genet. 2012;13:840–852. doi: 10.1038/nrg3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasper W.C., Marinov G.K., Pauli-Behn F., Scott M.T., Newberry K., DeSalvo G., Ou S., Myers R.M., Vielmetter J., Wold B.J. Fully automated high-throughput chromatin immunoprecipitation for ChIP-seq: identifying ChIP-quality p300 monoclonal antibodies. Sci. Rep. 2014;4:5152. doi: 10.1038/srep05152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Browne J.A., Harris A., Leir S.-H. An optimized protocol for isolating primary epithelial cell chromatin for ChIP. PLoS One. 2014;9:e100099. doi: 10.1371/journal.pone.0100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen R., Mandrup S. Genome-wide profiling of transcription factor binding and epigenetic marks in adipocytes by ChIP-seq. Methods Enzymol. 2014;537:261–279. doi: 10.1016/B978-0-12-411619-1.00014-8. [DOI] [PubMed] [Google Scholar]
- 12.Lee T.I., Johnstone S.E., Young R.A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saeed S., Quintin J., Kerstens H.H.D., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.-C., Ratter J., Berentsen K., van der Ent M.A., et al. Epigenetic programming during monocyte to macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Geen H., Echipare L., Farnham P.J. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol. Biol. 2011;791:265–286. doi: 10.1007/978-1-61779-316-5_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidder B.L., Hu G., Zhao K. ChIP-Seq: technical considerations for obtaining high-quality data. Nat. Immunol. 2011;12:918–922. doi: 10.1038/ni.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shankaranarayanan P., Mendoza-Parra M.-A., Walia M., Wang L., Li N., Trindade L.M., Gronemeyer H. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat. Methods. 2011;8:565–567. doi: 10.1038/nmeth.1626. [DOI] [PubMed] [Google Scholar]
- 17.Adli M., Bernstein B.E. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 2011;6:1656–1668. doi: 10.1038/nprot.2011.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidl C., Rendeiro A.F., Sheffield N.C., Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng J.-H., Kumar V., Muratani M., Kraus P., Yeo J.-C., Yaw L.-P., Xue K., Lufkin T., Prabhakar S., Ng H.-H. In vivo epigenomic profiling of germ cells reveals germ cell molecular signatures. Dev. Cell. 2013;24:324–333. doi: 10.1016/j.devcel.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 20.Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S., et al. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallner S., Grandl M., Konovalova T., Sigrüner A., Kopf T., Peer M., Orsó E., Liebisch G., Schmitz G. Monocyte to macrophage differentiation goes along with modulation of the plasmalogen pattern through transcriptional regulation. PLoS One. 2014;9:e94102. doi: 10.1371/journal.pone.0094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godoy P., Hewitt N.J., Albrecht U., Andersen M.E., Ansari N., Bhattacharya S., Bode J.G., Bolleyn J., Borner C., Böttger J., et al. Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME. Arch. Toxicol. 2013;87:1315–1530. doi: 10.1007/s00204-013-1078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd K.E., Farnham P.J. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Louwers M., Splinter E., van Driel R., de Laat W., Stam M. Studying physical chromatin interactions in plants using Chromosome Conformation Capture (3C) Nat. Protoc. 2009;4:1216–1229. doi: 10.1038/nprot.2009.113. [DOI] [PubMed] [Google Scholar]
- 25.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H., Jiang H., Ma W., Johnson D.S., Myers R.M., Wong W.H. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karolchik D., Barber G.P., Casper J., Clawson H., Cline M.S., Diekhans M., Dreszer T.R., Fujita P.A., Guruvadoo L., Haeussler M., et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–D770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dounce A.L., Witter R.F., Monty K.J., Pate S., Cottone M.A. A method for isolating intact mitochondria and nuclei from the same homogenate, and the influence of mitochondrial destruction on the properties of cell nuclei. J. Biophys. Biochem. Cytol. 1955;1:139–153. doi: 10.1083/jcb.1.2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adli M., Zhu J., Bernstein B.E. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat. Methods. 2010;7:615–618. doi: 10.1038/nmeth.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon J.M., Giresi P.G., Davis I.J., Lieb J.D. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat. Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu H., Wang Y. Exploring DNA-binding proteins with in vivo chemical cross-linking and mass spectrometry. J. Proteome Res. 2009;8:1983–1991. doi: 10.1021/pr8009319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.