Abstract
FANCM is a highly conserved DNA remodeling enzyme that promotes the activation of the Fanconi anemia DNA repair pathway and facilitates replication traverse of DNA interstrand crosslinks. However, how FANCM interacts with the replication machinery to promote traverse remains unclear. Here, we show that FANCM and its archaeal homolog Hef from Thermoplasma acidophilum interact with proliferating cell nuclear antigen (PCNA), an essential co-factor for DNA polymerases in both replication and repair. The interaction is mediated through a conserved PIP-box; and in human FANCM, it is strongly stimulated by replication stress. A FANCM variant carrying a mutation in the PIP-box is defective in promoting replication traverse of interstrand crosslinks and is also inefficient in promoting FANCD2 monoubiquitination, a key step of the Fanconi anemia pathway. Our data reveal a conserved interaction mode between FANCM and PCNA during replication stress, and suggest that this interaction is essential for FANCM to aid replication machines to traverse DNA interstrand crosslinks prior to post-replication repair.
INTRODUCTION
Interstrand DNA crosslinks (ICLs) are covalent and irreversible linkages between nucleotides of opposite DNA strands within the double-helix. They prevent strand separation during replication and transcription (1), and therefore, constitute a serious threat to genomic stability and cell viability (2). How ICLs are repaired or bypassed during replication remains poorly understood. In vertebrate cells, the majority of ICLs (about 60%) encountered by replication forks are processed through a replication-traverse pathway, in which the ICLs are left unrepaired, but are traversed by the replication machinery to allow DNA synthesis to resume on the other side. This enables cells to complete replication, which is essential for viability, at the expense of leaving the damage behind (3). The unrepaired ICLs are subsequently removed during a post-replication repair process and the gaps are filled. Conversely, a minority of ICLs (about 20–30%) block progression of replication forks, in forms of either single-fork collision or dual-fork collision (in which two conversing forks collide with the same ICL).
It was shown that the dual fork-collision with ICLs can activate the Fanconi anemia (FA) pathway (4). FA patients are characterized by hypersensitivity towards DNA crosslinking agents, increased chromosomal instability, congenital abnormalities, bone-marrow failure, cancer predisposition and infertility (5). So far, 19 FA proteins and several additional associated factors have been identified (6). They constitute the FA pathway that connects to several DNA repair systems - nucleotide excision repair (NER), homologous recombination (HR) and translesion synthesis (TLS) - to remove ICLs in a replication-dependent process (7,8). The DNA translocase FANCM in complex with its DNA binding partners FAAP24 and MHF1/2 recognizes the stalled replication fork to activate the FA pathway (9–12). FANCM then recruits the core complex (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG (XRCC9), FANCL and the associated proteins FAAP100 and FAAP20) to the lesion (10–13). The main function of the core complex is to monoubiquitinate two downstream FA proteins, FANCI and FANCD2 (ID-complex), by the integrated E3 ubiquitin ligase FANCL and the E2 ubiquitin conjugating enzyme FANCT (UBE2T) (14–17). The ubiquitinated ID-complex then interacts with FANCS (BRCA1), FANCD1 (BRCA2), FANCJ (BRIP1) and FANCN (PALB2), triggering downstream repair reactions that involve FANCO (Rad51C), FANCR (RAD51), FANCP (SLX4) and FANCQ (XPF) (18–22). However, the exact steps toward removal of the ICL damage still remain elusive.
The replication-traverse pathway depends on a conserved DNA remodeling complex consisting of FANCM and MHF1/2 (3). FANCM possesses specific binding and translocase activity for branched DNA, such as replication forks and Holiday junctions (9,23), is conserved from archaea to human (24,25), and its translocase activity is required to promote replication traverse (3). In higher eukaryotes, FANCM-MHF1/2 is also part of the FA core complex and its DNA binding activity is important to recruit the FA core complex to damaged DNA to monoubiquitinate the FANCD2-FANCI complex, a key step of the FA pathway (23). The traverse pathway and the FA pathway are independent of each other, as the former but not the latter requires the FANCM translocase activity; whereas the latter but not the former requires the FA core complex (3). The traverse pathway seems to play a lesser role than the FA pathway in cellular resistance to ICLs, because FANCM-knockout cells, which lack the first pathway but have a partially active second pathway, display weaker sensitivity to ICLs compared to cells inactivated of the FA core complex (25,26).
How FANCM interacts with the replication machinery to promote the traverse pathway remains unclear. To date, none of the FANCM-interacting partners (MHF, BLM complex, FA core complex and FAAP24) are replication factors. Interestingly, a recent study shows that an archaeal homolog of FANCM, Hef from Thermococcus kodakarensis (tkHef), interacts with proliferating cell nuclear antigen (PCNA) (27), the structurally highly-conserved DNA sliding clamp known to enhance the processivity of replicative polymerases (28–30). Moreover, several studies in mammalian cells have also linked RAD18-mediated PCNA ubiquitination to regulation of the FA pathway and ICL repair (31,32). PCNA is located at replication forks and serves as a landing platform for multiple proteins that are involved in DNA replication and repair (33). It often interacts with its partners through a PIP-box (PCNA-interacting peptide) with the consensus sequence Qxxhxxaa (Q = glutamine, x = undefined, h = hydrophobic, a = aromatic) (28,34). However, in the case of tkHef no obvious PIP-box has been found and its PCNA interaction is mediated by a non-conserved region (27). Thus, it is unclear whether the observed PCNA interaction is limited to tkHef alone or a general mechanism for other Hef proteins and FANCM exists.
Contrary to FANCM that lacks an active nuclease motif, most archaeal Hef proteins comprise an active nuclease entity at the C-terminus (25,35). Interestingly, Hef from Thermoplasma acidophilum (taHef) differs from other archaeal Hef proteins and exclusively comprises the N-terminal helicase entity that is characteristic for FANCM. This raised the possibility that taHef and vertebrate FANCM may share common mechanisms, including identical interacting partners. Here, we utilized taHef as a model for its eukaryotic FANCM homologs and identified a direct interaction between taHef and PCNA that is mediated by a PIP-box located at the C-terminus of taHef. Importantly, we identified a similar PIP-box in vertebrate FANCM proteins that mediates their direct interaction with PCNA. In vivo studies revealed that this interaction plays important roles in both replication traverse and FA pathways in vertebrate cells. These data thus unraveled a highly conserved interaction module that plays a critical role in replication-dependent DNA repair.
MATERIALS AND METHODS
Cloning and mutagenesis
The genes encoding Hef (NCBI: CAC12619) and PCNA (NCBI: Q9HJQ0) from Thermoplasma acidophilum DSM 1728 (taHef, taPCNA) were cloned from genomic DNA (ATCC 25905) into the bacterial expression vectors pBADM-11 and pETM-11 (EMBL), respectively. Human PCNA (NCBI: CAG38740, hsPCNA) was cloned from the MegaMan Human Transcriptome Library (Agilent) into the vector pBADM-11. After PCR amplification (primers for taHef: 5′TTTCAGGGCGCCATGGTTATTCAGCCCAGGGAATATCAATTAAACG3′ and 5′GTGCGGCCGCAAGCTTGCAGAGATAGGCAGCAGGACAGATTAAAAGTCG3′ / primers for taPCNA: 5′CTTTATTTTCAGGGCGCCATGATCAGACTGAACCTTTCGGTTAAGAATC3′ and 5′GTGCGGCCGCAAGCTTTACTGCTCCATTCTCGGAGCCAG3′ / primers for hsPCNA: 5′CTTTATTTTCAGGGCGCCATGTTCGAGGCGCGCCTGGTCCAGGGCTCC3′ and 5′GTGCGGCCGCAAGCTTGTCCTAAGATCCTTCTTCATCCTCGATCTTGGG3′) the genes were inserted into NcoI and HindIII digested vectors using the In-Fusion® (Clontech) cloning method. Site-directed mutagenesis for taHef ΔC7 and I505R/F508S, K31A was performed using the QuikChange™ protocol (Stratagene) and reactions were carried out as suggested by the manufacturer's instructions (primers for taHef ΔC7: 5′GAAGAGCGATGTTTAGAAAACAATATTCGAC3′ and 5′GTCGAATATTGTTTTCTAAACATCGCTCTTC3′ / primers for taHef I505R/F508S: 5′CAAGATTCGACTCTTAATCTGTCC3′ and 5′GATTAAGAGTCGAATCTTGTTTTC3′). All constructs were verified by sequencing.
Protein purification
taHef wild-type (WT), ΔC7, I505R/F508S, K31A and taPCNA as well as hsPCNA were recombinantly expressed as N-terminally His6-tagged proteins in Escherichia coli BL21-CodonPlus (DE3)-RIL cells (Stratagene). Cells were grown at 37°C in LB medium supplemented with the required antibiotics until an OD of ∼0.6 was reached. taHef (WT & mutants), hsPCNA (pBADM-11) and taPCNA (pETM-11) expression was induced with 0.02% arabinose and 0.5 mM isopropyl-β-thiogalactoside (IPTG), respectively. After induction cell growth was continued at 15°C for 18 h. All proteins were purified to homogeneity by metal affinity chromatography (NiMAC; Novagen) followed by size-exclusion chromatography (HiLoad 26/60 Superdex 200 prep grade, GE Healthcare) in 20 mM Tris/HCl, 200 mM NaCl, 5 mM MgCl2, pH 7.5 at 4°C. The oligomeric state and correct folding of the purified proteins were confirmed by dynamic light scattering (DLS) and circular dichroism (CD) measurements (Supplementary Figure S1) (36,37). Protein concentrations refer to the monomeric specimen. Protein samples were pooled, concentrated to 20–50 mg/ml, flash frozen in liquid nitrogen and stored at −80°C.
Analytical size exclusion chromatography
Analytical size exclusion chromatography was performed using a Superdex™ 200 10/300 GL column at an Äkta Purifier FPLC system (GE Healthcare) at 4°C. The protein with an initial concentration of 50 μM in a volume of 500 μl was incubated for 15 min at room temperature and then eluted isocratically with 20 mM Tris/HCl, 200 mM NaCl, 5 mM MgCl2, pH 7.5. Elution fractions were collected and subsequently analyzed by SDS-PAGE (38).
Isothermal titration calorimetry
Protein samples were extensively dialyzed against either 20 mM Tris/HCl, 200 mM NaCl, 5 mM MgCl2, pH 7.5 (taHef WT, ΔC7 or I505R/F508S and taPCNA) or 20 mM Tris/HCl, 50 mM KCl, 5 mM MgCl2, pH 7.5 (hsFANCM peptide and hsPCNA) for 18 h at 4°C and subsequently centrifuged for 20 min at 25 000 x g to remove aggregates. The buffer used for dialysis was also the reference buffer for the ITC measurements. All experiments were performed using a MicroCal ITC 200 instrument (GE Healthcare) at 25°C. A total of 40 μl of a solution containing 380 μM of recombinant taHef WT, ΔC7 or I505R/F508S in the syringe were titrated into the 200 μl sample cell containing 35 μM of recombinant taPCNA. A total of 20 injections were carried out with an injection volume of 2 μl each with the exception of the first injection at a volume of 0.8 μl, resulting in a final molar ratio of 2.2:1 (taHef:taPCNA). Measurements with 1.23 mM hsFANCM peptides (purchased from GenScript) as the titrant and recombinant 120 μM hsPCNA in the cell were conducted in 20 mM Tris/HCl, 50 mM KCl, 5 mM MgCl2, pH 7.5 under the same conditions as mentioned before. The signal of the first injection was discarded. The data were fitted to a single binding site model using the MicroCal ITC data evaluation software with no fixed parameters.
Small-angle X-ray solution scattering
Protein samples were prepared by an additional size exclusion chromatography purification step using a Superdex™ 200 10/300 GL column in 20 mM Tris/HCl, 200 mM NaCl, 5 mM MgCl2, pH 7.5 at 4°C and subsequently concentrated to ∼10 mg/ml. Polydispersity below 10% was validated by dynamic light scattering (37). To prevent radiation damage of the samples, immediately prior to the measurements 5 mM DTT were added and a dilution row from ∼10 mg/ml to ∼0.5 mg/ml of the protein was monitored. Small-angle X-ray scattering experiments were conducted at the ESRF BioSAXS beamline ID14-eh3 with additional measurements at the upgraded endstation BM29 (39,40). Standard experimental protocols were used with sample handling and scripting via the robotic sample changer and automated data collection available at the beamline. Ten individual frames were collected for every exposure, each 2 s in duration using the Pilatus 1M detector (Dectris). Individual frames were processed automatically and independently within the EDNA framework, yielding individual radially averaged curves of normalized intensity versus scattering angle s = 4πSINθ/λ. Additional data reduction within EDNA utilizes the automatic data processing tools of the EMBL-Hamburg ATSAS package, to combine timeframes, excluding any data points affected by aggregation induced by radiation damage, yielding the average scattering curve for each exposure series (41). Matched buffer measurements taken before and after every sample were averaged and used for background subtraction. Merging of separate concentrations and further analysis steps were performed manually using the tools of the ATSAS package. The forward scattering I(0) and the radius of gyration (Rg) were calculated from the Guinier approximation, the hydrated particle volume was computed using the Porod invariant and the maximum particle size (Dmax) was determined from the pair distribution function computed by GNOM using PRIMUS (42–44). Forty ab initio models were calculated for the PCNA construct using DAMMIF, and then averaged, aligned and compared using DAMAVER (45,46). The most representative model for each construct was selected by DAMAVER. The theoretical scattering of the known structures was calculated with CRYSOL and OLIGOMER was used for fitting of a combination of multiple structures simultaneously (47). Rigid body modeling using the high resolution structures of the known domains was performed with CORAL (48).
Vertebrate cell lines, plasmids and antibodies
HEK293 cells, HeLa cells and FANCM−/− DT40 cells were cultured as previously described (12,49). Human FANCM antibody and FANCM expression vectors were described in (24). The expression vectors for six overlapping fragments of FANCM were kindly provided by Dr. A. Deans (50). The HA-tagged FANCM-Helicase domain expression vector was described in (23). The chicken FANCM and FANCD2 antibodies were described in (12). The chicken FANCM expression vector (pcDNA2.1-Zeocin) was described in (12). The FANCM-PIP-box mutant plasmid was made by introducing L8S/W12R double point mutations into the pcDNA3.1 (zeocin)-FANCM vector using standard molecular biology techniques. The Flag-PCNA expression vector (pIRESneo3 Flag-hsPCNA) was generated using the pBADM-11 hsPCNA construct as a template. In a first step the linker between the upstream His6-tag and the TEV-site was substituted by the Flag-tag (primers: ACAAAGACGATGACGACAAGATCCCCACTACTGAGAATCTTTATTTTCAGG and TCGTCATCGTCTTTGTAGTCGCTCATGGGGTGATGGTGATGGTGATGTTTC). Then, in a second step the entire expression cassette was extracted from pBADM-11 and inserted into the pIRESneo3 vector using sequence and ligation-independent cloning (SLIC) (51). The PCNA antibody was obtained from Abcam (PC10/ab29). The Digoxigenin antibody was obtained from Invitrogen.
Cell transfection and immunoprecipitation
For immunoprecipitation experiments, 20 ml of suspension HEK293 cells (1 × 106 cells/ml) were transiently transfected with 20 μg of human Flag-FANCM expression vectors with polyethylenimine (PEI) as described (49). After 52 h, one aliquot of cells was treated with MMC at a final concentration of 70 ng/ml. At 54 h, another aliquot of cells were treated with 1 mM HU. All cells were harvested at 72 h. Preparation of cell lysates and subsequent immunoprecipitation using the Flag antibody were performed as described (49). The immunoprecipitate eluted from the anti-Flag antibody beads (Sigma) was analyzed by standard immunoblotting protocols.
FANCM knockout DT40 cells (1 × 106 cells/ml) were transfected with the pcDNA 3.1.zeocin expression vector carrying either FANCM-wildtype or the PIP-box mutant (1–5 μg) using the Amaxa Nucleofector kit in Solution T. The clones stably expressing either wild-type or mutant FANCM were selected by Western blotting as described (12). Cells were untreated or treated with 1 mM HU before harvest. Whole cell lysate was prepared as described (12). Western blotting using FANCM and FANCD2 antibodies were described (12).
Replication traverse assay
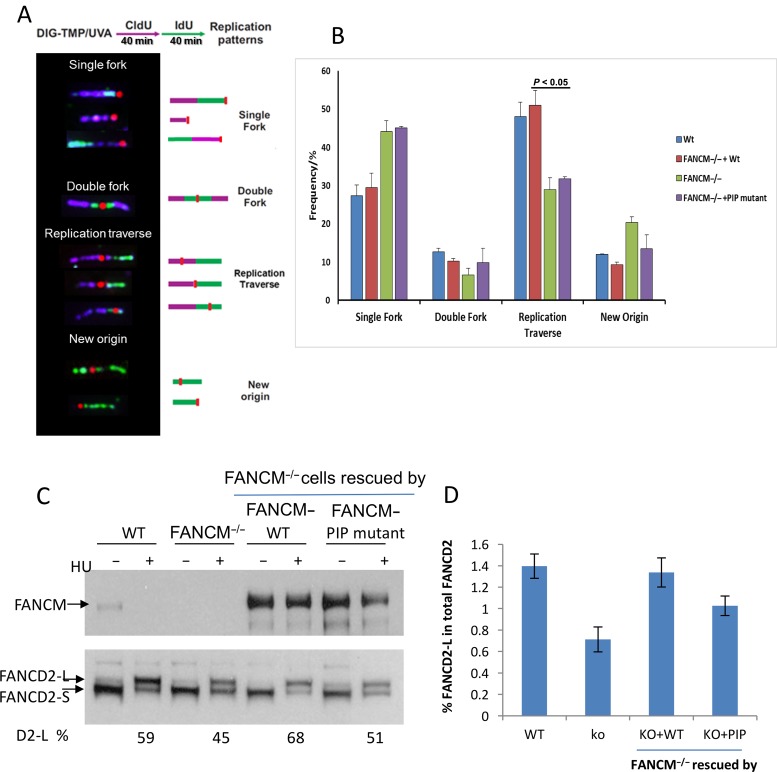
Following 1 h treatment with 5 μM Digoxigenin tagged trimethylpsoralen (Dig-TMP) (Supplementary Figure S3A) cells were exposed to UVA irradiation in a Rayonet chamber at 3 J/cm. They were then incubated with 20 μM CldU for 20 min and then for 20 min with 100 μM IdU. The cells were trypsinized and mixed with 0.5% SDS in 200 mM Tris/HCl, 50 mM EDTA, pH 7.5 on a silanated glass slide (Newcomer Supply). After tilting, the slides were air dried and fixed in 3:1 methanol/acetic acid, incubated in 2.5 M HCl for 60 min, neutralized in 0.4 M Tris/HCl, pH 7.5 for 5 min, washed in phosphate buffered saline (PBS), and immuno-stained with antibodies against Digoxigenin, CldU and IdU. Fluorescent secondary antibodies were used to display the CldU and the IdU, while a Qdot 655 secondary was used to illuminate the Dig tag (3). Imaging was performed on a Zeiss Axiovert 200 M microscope. The quantum dot signal was imaged with a Qdot 655 filter. The sequence of the differentially colored CldU and IdU tracks defines the direction of replication forks. Replication tracts terminating in a Dig signal reflect single forks stalled at a psoralen ICL, while a Dig dot in the midst of the second color track bounded on each side by tracts of the first color represent double fork collisions. Sequential CldU and IdU tracts with a Dig dot in the midst of either are indicative of a traverse event in which replication continues on the distal side of an ICL (Figure 5). Three independent experiments were performed.
Figure 5.
The FANCM PIP-box mutant is defective in promoting replication traverse, and is not fully active in promoting FANCD2 monoubiquitination. (A) Patterns of replication tracts in the vicinity of Dig-TMP ICLs (marked by the red dots) on DNA fibers. The purple and green tracks indicate DNA fibers labeled by CIdU and IdU, as illustrated by the protocol on the top. The forks are moving from magenta to green. (B) Frequency of patterns in wildtype DT40 cells, FANCM deficient DT40 cells and FANCM deficient cells complemented with either the wildtype FANCM gene or the PIP-box mutant (L8S/W12R). Three independent experiments were performed. The patterns of replication forks that encounter Dig-TMP were analyzed. The number of encounters analyzed in individual experiments: 96, 83, 75 in the wildtype cells; 84, 80, 76 in the FANCM−/− cells supplemented with wildtype FANCM; 66, 61, 89 in FANCM−/- cells; 86, 90, 74 in the FANCM−/− cells supplemented with the FANCM PIP mutant. (C) Representative immunoblotting images show that the FANCM PIP-box mutant is partially defective in promoting FANCD2 monoubiquitination in HU-treated DT40 cells. Monoubiquitinated and non-ubiquitinated FANCD2 are indicated by FANCD2-L (long form) and FANCD2-S (short form), respectively. The percentage of FANCD2-L in total FANCD2 (FANCD2-L + FANCD2-S) is indicated below the lanes. Cells are either untreated or treated with HU, as indicated on top. (D) Graphical representation showing the mean percentage of FANCD2-L in total FANCD2 after quantification of the immunoblotting images of FANCD2 monoubiquitination using 4 pairs of FANCM-knockout cells that were rescued by either FANCM WT or the FANCM PIP-box mutant protein (Supplementary Figure S2B). Error bars indicate standard deviations.
Proximity ligation assay
Cells were seeded in 35 mm glass bottom dishes (MatTek) and treated with or without 1 mM HU overnight. In experiments with Flag-tag FANCM, the cells were transfected with Flag-tagged wildtype FANCM or the PIP-box mutant FANCM plasmid with the PolyJet™ DNA Transfection Reagent (SignaGen Laboratories). Twenty-four hours after transfection, the cells were reseeded in 35 mm glass bottom dishes and treated with or without 1 mM HU overnight. The cells were washed with cold PBS and then 2 × 5 min in CSK buffer (10 mM PIPES, pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.5% Triton X-100). The cells were washed with cold PBS and fixed with 2% formaldehyde for 10 min at room temperature. After a PBS wash, they were incubated with −20°C methanol for 20 min. After washing with PBS they were treated with RNAse (100 μg/ml) for 30 min at 37°C. Following another PBS wash the cells were incubated with the blocking buffer in the Duolink kit (Sigma) and the manufacturer's instructions were followed through the amplification step. The primary antibodies were mouse anti PCNA (Abcam 1/200) and rabbit anti FANCM (Abcam 1/200) or rabbit anti-Flag (Sigma, 1/200). To allow visualization of the individual target proteins, after amplification, the cells were incubated with goat anti rabbit Alexa Fluor 488 conjugate (Invitrogen, 1/1000) and goat anti mouse Alexa Fluor 568 conjugate (Invitrogen, 1/1000) for 45 min. After washing with PBS the mounting medium containing DAPI (ProlongGold, Invitrogen) was added and the cells placed under a coverslip. Imaging was performed on a Zeiss Axiovert 200 M microscope. The number of spots in individual nuclei were determined and reported in dot plots. Two independent experiments were performed.
RESULTS
Archaeal Hef and vertebrate FANCM contain a conserved PCNA binding motif
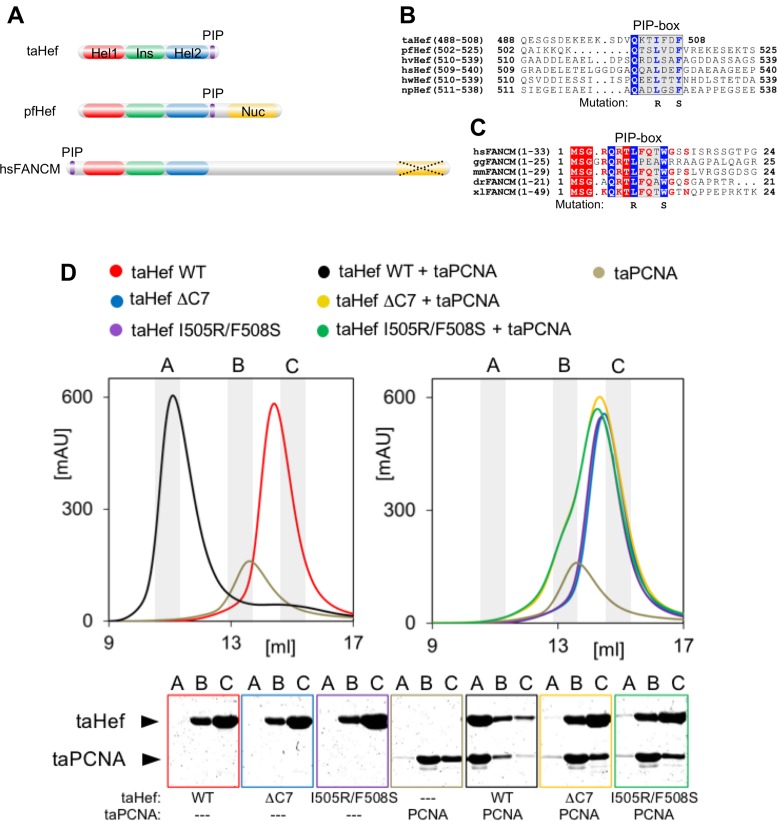
A common feature of the archaeal Hef proteins from taHef and Pyrococcus furiosus (pfHef) as well as vertebrate FANCM proteins is the presence of the two RecA-like motor domains (Hel1 and Hel2) characteristic for SF2 helicases and the thumb-like insertion domain (Ins) decisive for substrate specificity (Figure 1A) (52). In addition, pfHef and the vertebrate FANCM proteins contain an ERCC4-like nuclease domain which is absent in taHef. We searched for additional motifs conserved in this family of proteins and identified a potential PIP-box (purple) at the C-terminus of the helicase domain in taHef, pfHef and other archaeal Hef proteins (Figure 1A and B). This result prompted further analysis of the eukaryotic FANCM proteins and we identified a conserved PIP-box at their very N-terminus which is most likely accessible for protein–protein interactions (Figure 1A and C). Interestingly, the PIP-box from Hef and vertebrate FANCM contain only one conserved aromatic residue, whereas the known PIP-box consensus sequence has two. However, this deviation has been reported in multiple other PCNA interaction partners (53). We were unable to identify any potential PIP-box in tkHef, which has recently been shown to interact with PCNA (27). Possibly, the PCNA-interacting motif in tkHef has diverged from the PIP-box in sequence, so that it cannot be recognized in a sequence alignment.
Figure 1.
Hef interacts with proliferating cell nuclear antigen (PCNA) through a conserved PIP-box. (A) Domain organization of archaeal Hef and its human homolog FANCM. A common feature is the helicase core that comprises the RecA-like motor domains Hel1 (red) and Hel2 (blue) containing the seven SF2 helicase motifs and the enclosed thumb-like insertion domain Ins (green). Thermoplasma acidophilum (taHef) contains only the helicase core, while pfHef is extended by the additional C-terminal ERCC4-like nuclease domain (yellow) which is present in hsFANCM but enzymatically inactive (illustrated by the dotted cross). (B) Sequence alignment of the C-terminus of the helicase portion and the beginning of the adjacent linker toward the nuclease domain of Hef from different archaeal organisms (ta = Thermoplasma acidophilum; pf = Pyrococcus furiosus; hv = Haloferax volcanii; hs = Halobacterium salinarum; hw = Haloquadratum walsbyi; np = Natronomonas pharaonis). taHef is the only member that does not comprise the C-terminal nuclease domain. The conserved canonical PIP-box within an undefined and unstructured region is highlighted by the grey box. The double point mutation generated in taHef is shown below the alignment. The alignment was generated with ClustalW2 (66). (C) Alignment of the N-terminal regions of FANCM proteins from various higher eukaryotes. Conserved residues are shown by red letters, and strictly conserved residues in white with a red background. The N-terminally conserved putative canonical PIP-box motif is highlighted by a grey background. The double point mutation generated in the hsFANCM peptide is shown below the alignment. The alignment was generated with ClustalW2. The species are: hs: human; gg: chicken; mm: mouse; dr: zebrafish; xl: frog. (D) Analytical size exclusion chromatography (SEC) of taHef WT (red), taPCNA (grey), and a mixture of both (black; ϵ taPCNA / ϵ taHef WT = 0.35). The peak at a lower elution volume (∼11 ml) representing a higher molecular weight indicates stable complex formation. SEC profiles of taHef ΔC7 (blue) and taHef I505R/F508S (purple) show a similar elution behavior as the wildtype protein (red). Combinations of taHef ΔC7 and taPCNA (yellow) as well as taHef I505R/F508S and taPCNA (green) do not show any complex formation. Peak fractions A, B and C (labels above the gels) were analyzed via SDS-PAGE. Corresponding elution chromatograms are indicated by the respective frame colors and the particular content is additionally specified below the gels.
The predicted PIP-box mediates interactions between taHef and PCNA in vitro
The presence of the highly conserved PIP-box in archaeal Hef proteins (taHef residues 502–508) predicts a possible interaction interface for PCNA binding. Analytical size exclusion chromatography (SEC) of a 3:1 mixture of the taHef monomer and the taPCNA homotrimer showed that the two proteins fractionate together in a single peak distinct from those by each protein alone, suggesting that they form a stable complex (Figure 1D). Specifically, the taHef protein (red) elutes from the calibrated SEC column with a single peak at 14.3 ml (calculated MW: 59 kDa), and taPCNA (grey) at 13.6 ml (calculated MW: 86 kDa) (Figure 1D). These values correspond to a taHef monomer (actual MW: 61 kDa) and a taPCNA homotrimer (actual MW: 93 kDa; monomer: 31 kDa). The single peak including both proteins (black) is shifted to 11.1 ml (∼280 kDa), which corresponds to the combined molecular weight of 3 taHef monomers and 1 taPCNA homotrimer. The results thus imply that one taHef monomer interacts with one monomeric subunit of PCNA.
To verify the predicted PIP-box (Figure 1B) as the PCNA interaction site of taHef, we deleted this patch (taHef ΔC7, grey box in Figure 1D and Supplementary Figure S1), and analyzed its SEC profile either alone, or as a 3:1 mixture with taPCNA. We found that the profile of the mixture was nearly identical to that of the taHef ΔC7 protein alone (about 13.6 ml; 61 kDa), but distinct from that of the wildtype taHef-PCNA complex (11.1 ml; 280 kDa) (Figure 1D), indicating that the predicted PIP-box is critical for the formation of the taHef-PCNA complex.
It was previously shown that PIP-boxes specifically bind to a hydrophobic pocket in PCNA by constituting a 310-helix. Hydrophobic and aromatic residues within the PIP-box consensus sequence are crucial for this particular 310-helix formation (54). To investigate whether the taHef PIP-box employs the same mechanism of interaction, we generated a taHef double-mutant carrying a substitution of the conserved hydrophobic and aromatic residues, I505R/F508S. We found that the SEC profile of this double mutant in the presence of PCNA (magenta in Figure 1D) was indistinguishable from that of the protein in the absence of PCNA. This feature is similar to the ΔC7 deletion mutant, indicating that the PIP-box of taHef employs the same mode of interactions with PCNA as do the consensus PIP-boxes, possibly through a 310-helix formation.
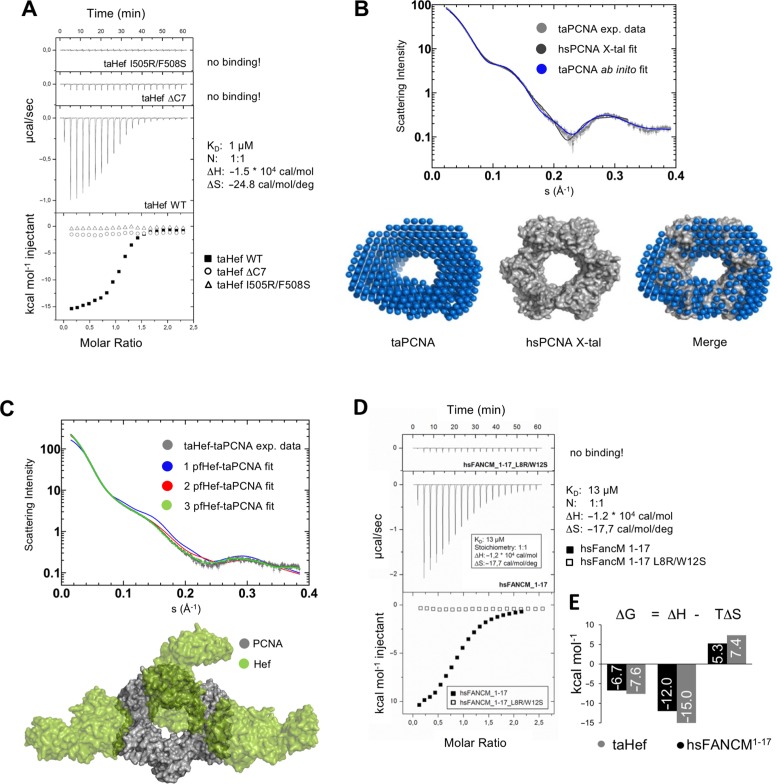
To further validate the interaction between taHef and taPCNA and also to gain insight into the binding kinetics of the complex, isothermal titration calorimetry (ITC) was pursued. The results show that taHef WT bound taPCNA with a dissociation constant (KD) of 1 μM and in a 1:1 ratio of monomer to monomer stoichiometry (Figure 2A). Since taPCNA forms a homotrimer, the complex may comprise three taHef monomers resulting in a heterohexamer. The Gibb's free energy (ΔG) of this complex formation constitutes −7.6 kcal mol−1 and shows a spontaneous reaction. The free enthalpy (ΔH) contribution of −15 kcal mol−1 indicates an exothermic reaction and the reduction of entropy (ΔS) of −24.8 cal mol−1 K−1 can be partially explained by the transition of the PIP-box from an unstructured state to a well-defined 310-helix (55). The entropy term (TΔS) accounts for −7.4 kcal mol−1 and counteracts the overall enthalpy driven reaction. In accordance to our analytical size exclusion chromatography studies, both PIP-box variants (taHef ΔC7 and taHef I505R/F508S) were deficient in taPCNA binding in the ITC experiments (Figure 2A). These results further confirm the C-terminal PIP-box of taHef (residues 502–508) as the major interaction interface for taPCNA binding.
Figure 2.
Biochemical analysis of Hef, FANCM and PCNA interactions. (A) Isothermal titration calorimetry measurements of taHef (WT & mutants) and taPCNA. taHef WT-taPCNA in vitro binding kinetics feature a dissociation constant (KD) of 1 μM and a 1:1 stoichiometry (considering both proteins as monomers). Thermodynamic parameters of the binding reaction: ΔH = −15 kcal mol−1, ΔS = −24.8 cal mol−1 K−1 and calculated ΔG = −7.6 kcal mol−1. For taHef ΔC7 and taHef I505R/F508S no heat release and thus no binding can be detected. (B) 1D SAXS data of taPCNA (black) overlaid with the theoretical scattering of the human PCNA structure (grey) yielding a chi value of 4.0 and the taPCNA ab initio model (blue) with a corresponding chi value of 3.7 indicating that the shape of the structure is compatible with the solution data but suggesting some flexibility. Overlay of the ab initio model with the human PCNA structure (PDB entry 1VYM) indicating that the PCNA ring is likely to bend slightly and expand and contract leading to a more oval average structure. (C) Experimental SAXS data for the taHef-taPCNA complex (grey) with indicated error (light grey) and CORAL rigid body fit for one (blue; chi = 361.93), two (red; chi = 13.62), and three (green; chi = 1.47) Hef proteins bound to the taPCNA trimer. SAXS rigid body model of the complex containing three Hef monomers (green) bound to the PCNA trimer (grey). For PCNA modeling the human crystal structure (PDB entry 1VYM) and for taHef the Pyrococcus furiosus (pfHef) structure (PDB entry 1WP9) was used. (D) Isothermal titration calorimetry measurements of peptides representing the N-terminal 17 amino acids of hsFANCM in its wildtype or mutated state (see also Figure 1C) and hsPCNA. hsFANCM1–17-hsPCNA in vitro binding kinetics feature a KD of 13 μM and a 3:1 stoichiometry, i.e. three peptides binds to one PCNA homotrimer. The thermodynamic parameters of the binding reaction are: ΔH = −12 kcal mol−1, ΔS = −17.7 cal mol−1 K−1 and calculated ΔG = −6.7 kcal mol−1. For hsFANCM1–17 L8R/W12S no heat release and thus no binding can be detected. (E) Comparison of the different parameters of binding reactions between the human FANCM PIP-box and PCNA, and those between the taHef PIP-box and PCNA.
In solution model of the taHef-taPCNA complex
To gain structural insight into the taHef-taPCNA interaction we utilized SAXS. As control experiments, we also analyzed apo taPCNA (Figure 2B) via SAXS to facilitate modeling and interpretation of the taHef-taPCNA complex (Figure 2C). The first step was to verify that the taPCNA ring can be approximated as a single rigid body. Since there is no crystal structure of taPCNA available, we utilized the structure of human PCNA (hsPCNA; PDB entry 1VYM) which matched the experimental data reasonably well (chi 4.9), though systematic deviations consistent with a small amount of flexibility were observed (Figure 2B) (56). This flexibility was visualized in the superposition of the ab initio model with hsPCNA, which appeared slightly bent and more elliptical, indicating the possibility of twisting, expansion and contraction of the ring (Figure 2B). Although taPCNA displays some flexibility, the data show that the hsPCNA structure is a reasonable approximation of the average position in solution. Moreover, any effects caused by the flexibility of taPCNA will be minor compared to the possible movement of taHef relative to taPCNA due to a linker region comprising 26 amino acids N-terminal to the PIP motif in taHef. Thus the hsPCNA structure was also used for the modeling of the taHef-taPCNA complex. For modeling of taHef within the complex we utilized the crystal structure of the pfHef helicase entity (PDB entry 1WP9) (57).
We analyzed the possibility of one, two or three taHef monomers bound to taPCNA using CORAL, with no symmetry applied. The best fit was obtained with 3 Hef molecules bound, followed closely by 2 Hef molecules, whereas 1 Hef provided a very poor fit (chi 1.47, 13.62 and 361.93, respectively) (Figure 2C). Thus the SAXS data show that the 3 taHef bound model was the preferred stoichiometry in solution which is in agreement with our ITC and analytical size exclusion experiments.
The PIP-box motif of FANCM interacts with PCNA in vitro
To investigate whether the N-terminal PIP-box motif of FANCM also interacts with PCNA we performed ITC experiments using a synthetic peptide (FANCM1–17) containing the PIP-box which showed that the wildtype FANCM1–17 peptide bound to hsPCNA, with a KD of 13 μM and in a 3:1 stoichiometry, i.e. 3 peptides bind to 1 PCNA homotrimer (Figure 2D).
The observed interactions between the N-terminal FANCM PIP-box (residues 5–12) and PCNA confirmed our prediction based on the taHef data, and indicates that the PIP-box mediated PCNA interaction is conserved in the Hef/FANCM protein family. To further verify the interaction via the PIP box we utilized a peptide harboring the L8R/W12S mutation for additional ITC experiments. The mutated PIP-box (L8R/W12S) was generated in analogy to the taHef I505R/F508S double point mutant, which substituted two conserved residues critical for PCNA interaction. The mutated FANCM peptide revealed no detectable binding to PCNA (Figure 2D) indicating a similar interaction mechanism for both FANCM and taHef with PCNA. Indeed, various parameters of the binding reaction (ΔG, ΔH and ΔS) between human FANCM1-17 and PCNA are comparable to those between taHef and PCNA (Figure 2E). Specifically, the ΔG of the binding reaction between hsFANCM1-17 WT and PCNA has a value of −6.7 kcal mol-1, ΔH comprises −12 kcal mol-1 and ΔS is −17.7 cal mol-1 K-1. The data indicate a transition from a disordered toward a defined conformation which is comparable to the taHef-taPCNA interaction (Figure 2E). Likewise, the TΔS value of −5.3 kcal mol-1 counteracts the overall enthalpy driven reaction. It is possible that the PIP-box of human FANCM forms a 310-helix to interact with PCNA similarly as other PIP-boxes, and the L8R/W12S double mutation prevents formation of the helix and thus disrupts the PCNA interaction.
FANCM and PCNA interact in cells under replication stress
After establishing the in vitro interaction of a FANCM peptide with PCNA we investigated whether full length FANCM interacts with PCNA in vivo. Previous work found that FANCM-associated polypeptides immunoprecipitated by a FANCM antibody from HeLa cells (12) contain no peptides derived from PCNA, although they include known interacting partners of FANCM, such as MHF, FAAP24, components of the FA core complex and the BLM helicase complex. This implied that if PCNA interacts with FANCM, the interaction might be transient and/or weak, and thus not strong enough to survive the extensive washing of the immunoprecipitation procedure which is also supported by the high KD value determined in the ITC experiments.
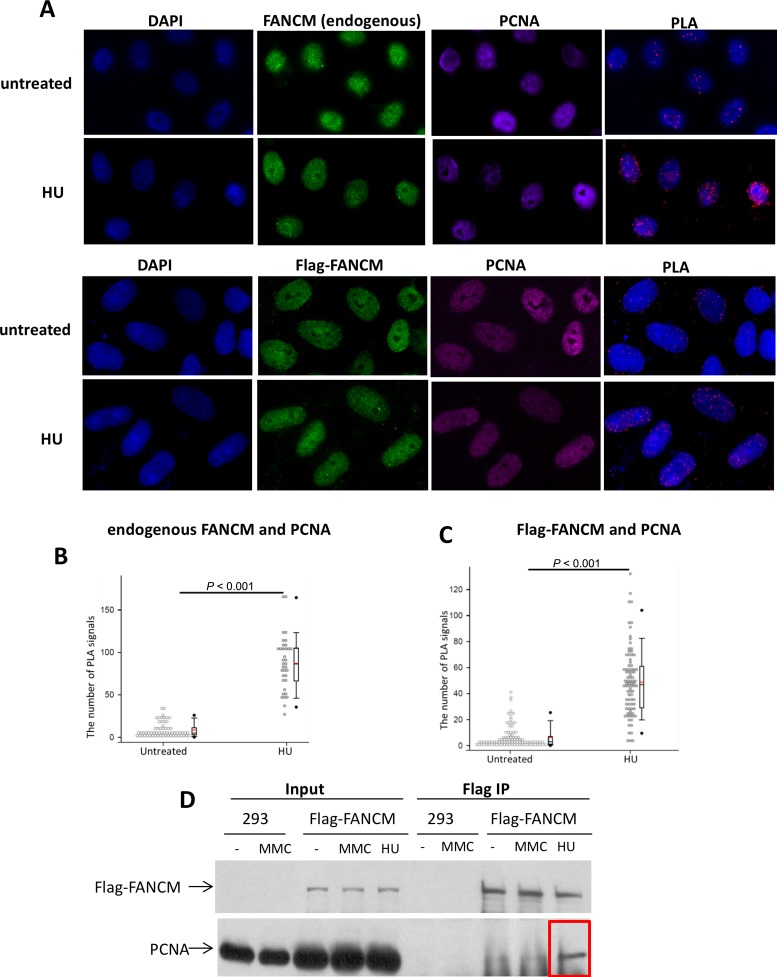
Thus, we investigated whether FANCM and PCNA interact in cells using the in situ proximity ligation assay (PLA), a method widely used to detect endogenous protein interactions in cells with high specificity and sensitivity (58). Moreover, it allows the detection of regulated protein complex formation in response to DNA damage and replication stress. Briefly, we used secondary antibodies conjugated with specific oligonucleotides to recognize primary antibodies (rabbit, mouse) bound to FANCM and PCNA in HeLa cells. When the two proteins form a complex, the oligonucleotides on the two secondary antibodies will be brought to close proximity to hybridize to connector oligonucleotides, which, following ligation, can then be detected by rolling circle amplification. The amplification products are visualized by fluorescence microscopy as discrete dots. We found that in HeLa cells untreated with DNA damaging agents, the PLA signal that reflects the FANCM-PCNA interaction was low (Figure 3A and B). However, when cells were treated with hydroxyurea (HU), which induces replication stress by inhibiting the production of deoxyribonucleotides, the PLA signal (reported as the number of spots per nucleus) was strongly increased (at least 10-fold). We also introduced Flag-tagged FANCM into HeLa cells, and performed PLA using antibodies against Flag and PCNA; and obtained similar results (Figure 3A and C). These data indicate that FANCM and PCNA can form a complex in vivo, and formation of this complex is considerably enhanced when cells are under replication stress.
Figure 3.
FANCM and PCNA interact in cells under replication stress. (A) Immunofluorescence images, and (B,C) quantitation of the PLA assay showing the FANCM and PCNA interaction in HeLa cells under replication stress. Cells untreated or treated with HU are indicated. FANCM was stained with either an antibody against endogenous FANCM (top 2 rows), or a Flag antibody that reacts with Flag-FANCM ectopically expressed in HeLa cells. DNA is co-stained with DAPI. In the quantitation plots each spot represents the number of PLA signals in an individual nucleus. Two independent experiments were performed for each analysis. In the experiments with exogenous FANCM a total of 144 nuclei from the untreated cells and 162 nuclei from the HU treated cells were examined. In the experiment with the Flag-FANCM a total of 157 nuclei from non-treated cells and 156 nuclei from HU treated cells were analyzed. (D) Immunoblotting shows that Flag-FANCM transfected in HEK293 cells co-immunoprecipitates with PCNA in cells treated with HU, but not in untreated cells, or in cells treated with MMC.
To further analyze the FANCM-PCNA complex, we introduced Flag-FANCM into HEK293 cells and performed immunoprecipitation with the Flag antibody coupled by Western blotting. We detected the presence of PCNA in the FANCM immunoprecipitate only in cells treated with HU, but not in untreated cells (Figure 3D). These findings are consistent with the PLA data, indicating that FANCM and PCNA form a complex in response to HU-induced replication stress.
We failed to detect the presence of PCNA in the Flag-FANCM immunoprecipitate from cells treated with MMC (Figure 3D), a DNA interstrand crosslinking drug that can block progression of replication forks. One possible reason may be that HU treatment can lead to a collapse of all replication forks, whereas MMC can only block a subset of forks that encounter the DNA crosslinks. Thus, HU may trigger a stronger DNA damage response than MMC, which is reflected by a higher level of FANCM hyperphosphorylation and FANCD2 monoubiquitination (24), leading to a stronger induction of the FANCM-PCNA complex.
The predicted PIP-box mediates the interaction between FANCM and PCNA
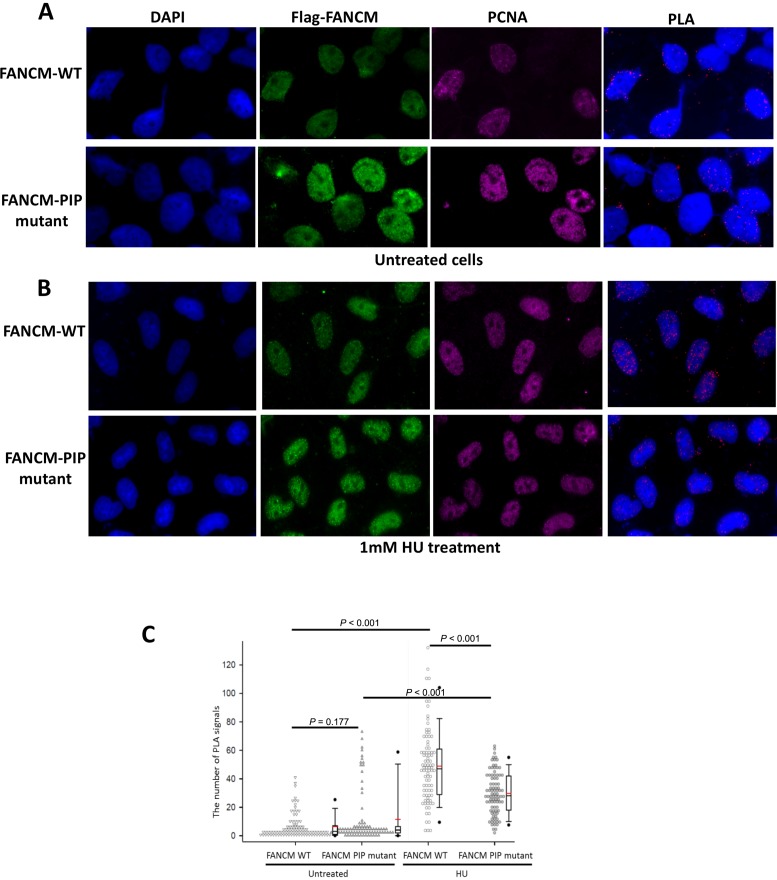
To confirm that the predicted PIP-box at the N-terminus of FANCM mediates PCNA interaction, we introduced the L8R/W12S PIP-box mutation that has been shown to disrupt FANCM-PCNA interaction in vitro into full length Flag-FANCM, and investigated whether the mutation disrupts FANCM-PCNA association in vivo using the PLA assay described above. We found that in cells untreated with DNA damaging reagents, the PLA signals were low for both the wildtype and the PIP-box mutant; and their mean values are not statistically different from each other (Figure 4A–C), suggesting a low frequency of interaction between FANCM and PCNA without DNA damage. In cells treated with HU, the PLA signals of both the wildtype and mutant proteins were substantially increased (about 10-fold and 3-fold, respectively) (Figure 4B and C), consistent with the notion that the FANCM-PCNA interaction is induced by replication stress. Notably, the PLA signal for the FANCM-PIP box mutant was about 50% of that of the FANCM wildtype protein, suggesting that the L8R/W12S PIP-box mutation partially disrupts the FANCM-PCNA association under replication stress.
Figure 4.
The replication stress-induced FANCM-PCNA interaction is reduced by the FANCM PIP-box mutation. (A,B) Immunoflurescence images and (C) quantitation of the PLA assay showing that the HU-induced interaction between Flag-FANCM and PCNA is reduced by the FANCM PIP-box mutation. HeLa cells were transfected with either Flag-FANCM wildtype or the PIP-box mutant (L8S/W12R); and are either untreated (A) or treated with HU (B). The cells were co-stained with antibodies against Flag and PCNA. DNA was co-stained with DAPI. Two independent experiments were performed. In the quantitation plots each spot represents the number of PLA signals in an individual nucleus. In the experiment with untreated cells 157 nuclei expressing wildtype FANCM and 149 nuclei expressing the PIP mutant were analyzed. In the experiment with cells treated with HU 156 nuclei expressing wild type FANCM and 153 nuclei expressing the PIP mutant were analyzed.
We also introduced the L8R/W12S PIP-box mutation into an HA-tagged N-terminal FANCM fragment 1–754, and examined whether this fragment displays a defective association with Flag-tagged PCNA in a co-IP assay after their co-transfection in HEK293 cells. This FANCM fragment was chosen because it contains the intact helicase domain, as well as intact binding and ATP-dependent remodeling activities for branched DNA structures, such as Holliday junctions and forks (23). We found that whereas the HA-tagged FANCM fragment carrying the wildtype PIP-box co-immunoprecipitated with Flag-PCNA after they were co-transfected into HEK293 cells, the FANCM fragment carrying the PIP-box mutant obtained a lower level of PCNA that is indistinguishable from that of a mock immunoprecipitation from cells transfected with Flag-PCNA alone (Supplementary Figure S2A, compare lanes 8 and 6), indicating that the predicted N-terminal PIP-box is required for normal FANCM-PCNA interaction. Combined with the results of the PLA assays where the interaction was reduced, but not eliminated by the L8R/W12S mutation in the full length FANCM, we infer that in addition to the predicted N-terminal PIP-box, other regions in FANCM might also contribute to the association with PCNA.
The FANCM PIP-box mutant is defective in promoting replication traverse
FANCM and its interacting partner MHF have been shown to be required for replication forks to traverse ICLs (3). Because PCNA plays a key role in replication, we used the same assay to study whether the FANCM-PCNA interaction is required for replication forks to traverse ICLs induced by UV-activated DIG-TMP (Supplementary Figure S3A). Psoralen preferentially intercalates at 5′TA sites in DNA and, following photoactivation by UVA, forms cyclobutane adducts with the thymines on opposite strands (Supplementary Figure S3B and C). Cells were treated with Dig-TMP/UVA and then sequentially pulsed with CldU and IdU to label replication tracts (see Methods). Consistent with previous findings, chicken DT40 cells inactivated of FANCM have a lower level of replication traverse compared to the wildtype cells (about 30% versus 50%) (Figure 5A and B). Re-expression of wildtype FANCM restored the traverse level to that of the wildtype cells (about 50%), but re-expression of the FANCM-PIP-box mutant (FANCM L8R/W12S) did not (remained at about 30%). The results show that the FANCM-PIP-box mutant is deficient in promoting replication traverse, suggesting that the PIP-box mediated FANCM-PCNA interaction is important for the replication machinery to traverse the crosslinks. We noted that the level of the FANCM-PIP mutant protein in the transfected DT40 cells was comparable to that of the FANCM-wildtype protein (Figure 5C), suggesting that the effect of the PIP-box mutation is not due to destabilization of FANCM through disruption of protein folding.
FANCM and its various partners (MHF and FAAP24) have been shown to promote FANCD2 monoubiquitination in response to DNA damage. We thus studied whether the FANCM-PIP box mutant is defective in promoting FANCD2 monoubiquitination in cells treated with HU. In agreement with earlier findings (25), FANCM-knockout DT40 cells exhibited a lower level of monoubiquitinated FANCD2 compared to the wildtype DT40 cells, as reflected by the reduced percentage of the FANCD2-long form (ubiquitinated form) in total FANCD2 (Figure 5C and D). When FANCM−/− cells were stably transfected with wildtype FANCM protein, the monoubiquitinated FANCD2 level was largely restored to that of the wildtype cells. However, when FANCM−/− cells were stably transfected with the FANCM-PIP box mutant (L8R/W12S), the level of monoubiquitinated FANCD2 was slightly lower than that of the cells stably expressing the FANCM wildtype protein, but higher than that of the FANCM−/− cells (Figure 5C and D, and Supplementary Figure S2B). This feature differs from the previously reported FANCM mutants deleted of its helicase domain, which are completely defective in promoting FANCD2 monoubiquitination (23,59). The data thus suggest that the FANCM-PIP box mutant is a hypomorphic mutant that is not fully active in promoting FANCD2 monoubiquitination compared to the wildtype FANCM protein, but is partially active compared to the FANCM-helicase deletion mutants; the PIP-box-mediated PCNA interaction is thus not as important as the helicase domain-mediated FANCM-DNA interaction in promoting FANCD2 monoubiquitination.
DISCUSSION
The FANCM-Hef protein family interacts with PCNA through a conserved PIP-box
An earlier study has shown that an archaeal Hef (tkHef) forms a stable complex with PCNA (27). However, tkHef has no recognizable PIP-box and its PCNA interaction is mediated through a non-conserved region. Here, we report the presence of a PIP-box motif in not only Hef proteins from many archaeal species, but also in the vertebrate homolog of Hef - FANCM. Importantly, we demonstrate that the predicted PIP-box mediates PCNA interaction in both archaeal Hef (taHef) and vertebrate FANCM. Thus, our data suggest that PCNA interaction is a general activity conserved in the Hef-FANCM protein family. Our ITC experiments show that taHef binds to PCNA with a KD of 1 μM, whereas the FANCM-PIP box has a KD of 13 μM. These values are intermediate compared to known PCNA binding proteins (human p21: KD = 80 nM; FEN-1: KD = 60 μM) (60). On the basis of these values, we classify taHef as a moderate PCNA binding partner, and FANCM may act as a weak one based on our peptide interaction data.
The FANCM-PCNA complex is induced by replication stress
In vertebrate cells, the majority of ICLs are bypassed by the replication traverse pathway, which depends on FANCM and its DNA translocase activity (3). However, how FANCM interacts with the replication machinery to allow the traverse to occur remains unclear. Our studies reveal that FANCM interacts with PCNA, a cofactor for replicative DNA polymerases. PCNA differs from all known FANCM-interacting partners, because its association with FANCM is transient and can only be appreciably detected in cells under replication stress. In contrast, other FANCM partners (MHF, FAAP24, BLM complex and FA core complex) can form highly stable complexes with FANCM, which are resistant to at least 0.5 M salt wash and can be isolated from cells in the absence of DNA damage (10,61). In fact, the interaction between the N-terminal FANCM PIP-box and PCNA is quite weak (KD = 13 μM) compared to that between FANCM and forked DNA, which is much stronger (KD is in the nM range or less) (9). The data imply that FANCM may bind stalled replication forks first before interacting with PCNA at the same forks. In cells that are in the non-replication phase or in normal replication, no stalled forks are present so that FANCM does not stably interact with PCNA. Only in cells under replication stress FANCM recognizes the stalled forks through its strong DNA binding activity and then interacts with PCNA at the same forks through its PIP-box.
FANCM works with PCNA to promote replication traverse of ICLs
Interstrand crosslinks have long been considered absolute blocks to replication. Current scenarios propose that following collision with an ICL, replication can be completed either by lesion repair prior to the resumption of DNA synthesis (single fork model), or by completion of synthesis by a replication fork from the opposite direction (double fork model) (62). The recently discovered replication traverse model (3) is consistent with much earlier work describing replication restart following leading strand encounters with UV photoproducts (63–65). In those pathways the replicative helicase can unwind DNA past the photoproduct, after which repriming occurs and synthesis resumes, leaving the adduct in a gap to be repaired at a later time. The ICL presents a special challenge as, unlike many single strand adducts, it is an obstacle to the replicative helicase, which encircles a single strand. However, FANCM is a translocase and can move along duplex DNA without unwinding (24). The interaction with PCNA might provide the opportunity for FANCM to translocate the replication machinery from one side of the ICL to the other. Additionally, the interaction might be important for the restart of DNA synthesis on the distal side of the ICL. These possibilities are not exclusive of one another. Additional experiments will be required to elucidate the contribution of the FANCM-PCNA interaction to replication traverse of the ICLs.
Supplementary Material
Acknowledgments
We thank the ESRF for beamtime and the staff of the BioSAXS beamline (formally ID14eh3 and now located at BM29) for technical support.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (DFG) [Forschungszentrum FZ-82 to C.K.]; Intramural Research Program of the National Institute on Aging [AG000688–07 in part]; National Institute of Health (NIH). Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG) [Forschungszentrum FZ-82 to C.K.]; Intramural Research Program of the National Institute on Aging [AG000688–07 in part]; National Institute of Health (NIH).
Conflict of interest statement. None declared.
REFERENCES
- 1.Deans A.J., West S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schärer O.D. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 3.Huang J., Liu S., Bellani M.A., Thazhathveetil A.K., Ling C., de Winter J.P., Wang Y., Wang W., Seidman M.M. The DNA translocase FANCM/MHF promotes replication traverse of DNA interstrand crosslinks. Mol. Cell. 2013;52:434–446. doi: 10.1016/j.molcel.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang J., Dewar J.M., Budzowska M., Motnenko A., Cohn M.A., Walter J.C. DNA interstrand cross-link repair requires replication-fork convergence. Nat. Struct. Mol. Biol. 2015;22:242–247. doi: 10.1038/nsmb.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auerbach A.D. Fanconi anemia and its diagnosis. Mutat. Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kottemann M.C., Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niedzwiedz W., Mosedale G., Johnson M., Ong C.Y., Pace P., Patel K.J. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol. Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Knipscheer P., Raschle M., Smogorzewska A., Enoiu M., Ho T.V., Schärer O.D., Elledge S.J., Walter J.C. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–1701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gari K., Decaillet C., Stasiak A.Z., Stasiak A., Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 10.Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., Meetei A.R., Laghmani el H., Joenje H., McDonald N., de Winter J.P., et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Singh T.R., Saro D., Ali A.M., Zheng X.F., Du C.H., Killen M.W., Sachpatzidis A., Wahengbam K., Pierce A.J., Xiong Y., et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan Z., Delannoy M., Ling C., Daee D., Osman F., Muniandy P.A., Shen X., Oostra A.B., Du H., Steltenpool J., et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Z., Guo R., Paramasivam M., Shen W., Ling C., Fox D. 3rd, Wang Y., Oostra A.B., Kuehl J., Lee D.Y., et al. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol. Cell. 2012;47:61–75. doi: 10.1016/j.molcel.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida Y.J., Machida Y., Chen Y., Gurtan A.M., Kupfer G.M., D'Andrea A.D., Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Meetei A.R., de Winter J.P., Medhurst A.L., Wallisch M., Waisfisz Q., van de Vrugt H.J., Oostra A.B., Yan Z., Ling C., Bishop C.E., et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 16.Virts E.L., Jankowska A., MacKay C., Glaas M.F., Wiek C., Kelich S.L., Lottmann N., Kennedy F.M., Marchal C., Lehren E., et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum. Mol. Genet. 2015;24:5093–5108. doi: 10.1093/hmg/ddv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hira A., Yoshida K., Sato K., Okuno Y., Shiraishi Y., Chiba K., Tanaka H., Miyano S., Shimamoto A., Tahara H., et al. Mutations in the gene encoding the E2 conjugating enzyme UBE2T cause Fanconi anemia. Am. J. Hum. Genet. 2015;96:1001–1007. doi: 10.1016/j.ajhg.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawyer S.L., Tian L., Kahkonen M., Schwartzentruber J., Kircher M., Majewski J., Dyment D.A., Innes A.M., University of Washington Centre for Mendelian Genomics and FORGE Canada Consortium et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Somyajit K., Subramanya S., Nagaraju G. Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: implications for Fanconi anemia and breast cancer susceptibility. J. Biol. Chem. 2012;287:3366–3380. doi: 10.1074/jbc.M111.311241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y., Lach F.P., Desetty R., Hanenberg H., Auerbach A.D., Smogorzewska A. Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 2011;43:142–146. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., Raams A., Trujillo J.P., Minguillon J., Ramirez M.J., Pujol R., et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am. J. Hum. Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K., et al. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xue Y., Li Y., Guo R., Ling C., Wang W. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum. Mol. Genet. 2008;17:1641–1652. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meetei A.R., Medhurst A.L., Ling C., Xue Y., Singh T.R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C.G., et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J.B., Johnson M., Langevin F., Pace P., Patel K.J. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., Leung J.W., Jiang Y., Lowery M.G., Do H., Vasquez K.M., Chen J., Wang W., Li L. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol. Cell. 2013;49:997–1009. doi: 10.1016/j.molcel.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishino S., Yamagami T., Kitamura M., Kodera N., Mori T., Sugiyama S., Ando T., Goda N., Tenno T., Hiroaki H., et al. Multiple interactions of the intrinsically disordered region between the helicase and nuclease domains of the archaeal hef protein. J. Biol. Chem. 2014;289:21627–21639. doi: 10.1074/jbc.M114.554998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moldovan G.L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 29.McNally R., Bowman G.D., Goedken E.R., O'Donnell M., Kuriyan J. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct. Biol. 2010;10:1–14. doi: 10.1186/1472-6807-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garg P., Burgers P.M. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 31.Geng L., Huntoon C.J., Karnitz L.M. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J. Cell Biol. 2010;191:249–257. doi: 10.1083/jcb.201005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song I.Y., Palle K., Gurkar A., Tateishi S., Kupfer G.M., Vaziri C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J. Biol. Chem. 2010;285:31525–31536. doi: 10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haynes B., Saadat N., Myung B., Shekhar M.P. Crosstalk between translesion synthesis, Fanconi anemia network, and homologous recombination repair pathways in interstrand DNA crosslink repair and development of chemoresistance. Mutat. Res. Rev. Mutat. Res. 2015;763:258–266. doi: 10.1016/j.mrrev.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warbrick E. PCNA binding through a conserved motif. BioEssays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 35.Komori K., Hidaka M., Horiuchi T., Fujikane R., Shinagawa H., Ishino Y. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 2004;279:53175–53185. doi: 10.1074/jbc.M409243200. [DOI] [PubMed] [Google Scholar]
- 36.Kelly S.M., Jess T.J., Price N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Berne B.J., Pecora R. Dynamic light scattering: with applications to chemistry, biology, and physics. 3rd edn. NY: Dover Publications; 2000. [Google Scholar]
- 38.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 39.Pernot P., Theveneau P., Giraud T., Nogueira Fernandez R., Nurizzo D., Spruce D., Surr J., McSweeney S., Round A., Felisaz F., et al. New beamline dedicated to solution scattering from biological macromolecules at the ESRF. J. Phys. 2010;247:012009. [Google Scholar]
- 40.Pernot P., Round A., Barrett R., De Maria Antolinos A., Gobbo A., Gordon E., Huet J., Kieffer J., Lentini M., Mattenet M., et al. Upgraded ESRF BM29 beamline for SAXS on macromolecules in solution. J. Synchrotron Radiat. 2013;20:660–664. doi: 10.1107/S0909049513010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petoukhov M.V., Konarev P.V., Kikhney A.G., Svergun D.I. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 2007;40:s223–s228. [Google Scholar]
- 42.Guinier A. The diffusion of X-rays under the extremely weak angles applied to the study of fine particles and colloidal suspension. C. R. Acad. Sci. Hebd. Acad. Sci. D. 1938;206:1374–1376. [Google Scholar]
- 43.Porod G. In: General theory. small-angle X-ray scattering. Glatter O, Kratkv O, editors. London: Academic Press; 1982. pp. 17–51. [Google Scholar]
- 44.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 45.Franke D., Svergun D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkov V.V., Svergun D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svergun D., Barberato C., Koch M.H.J. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 48.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M., Gorba C., Mertens H.D.T., Konarev P.V., Svergun D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Yang J., et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013;16:1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deans A.J., West S.C. FANCM connects the genome instability disorders Bloom's Syndrome and Fanconi anemia. Mol. Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Li M.Z., Elledge S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 52.Hall M.C., Matson S.W. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 53.Matsumiya S., Ishino S., Ishino Y., Morikawa K. Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus. Genes Cells. 2002;7:911–922. doi: 10.1046/j.1365-2443.2002.00572.x. [DOI] [PubMed] [Google Scholar]
- 54.Chapados B.R., Hosfield D.J., Han S., Qiu J., Yelent B., Shen B., Tainer J.A. Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell. 2004;116:39–50. doi: 10.1016/s0092-8674(03)01036-5. [DOI] [PubMed] [Google Scholar]
- 55.Sakurai S., Kitano K., Yamaguchi H., Hamada K., Okada K., Fukuda K., Uchida M., Ohtsuka E., Morioka H., Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kontopidis G., Wu S.Y., Zheleva D.I., Taylor P., McInnes C., Lane D.P., Fischer P.M., Walkinshaw M.D. Structural and biochemical studies of human proliferating cell nuclear antigen complexes provide a rationale for cyclin association and inhibitor design. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1871–1876. doi: 10.1073/pnas.0406540102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishino T., Komori K., Tsuchiya D., Ishino Y., Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 58.Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 59.Rosado I.V., Niedzwiedz W., Alpi A.F., Patel K.J. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37:4360–4370. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruning J.B., Shamoo Y. Structural and thermodynamic analysis of human PCNA with peptides derived from DNA polymerase-delta p66 subunit and flap endonuclease-1. Structure. 2004;12:2209–2219. doi: 10.1016/j.str.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 61.Meetei A.R., Sechi S., Wallisch M., Yang D., Young M.K., Joenje H., Hoatlin M.E., Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J., Walter J.C. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst) 2014;19:135–142. doi: 10.1016/j.dnarep.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rupp W.D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 64.Yeeles J.T., Marians K.J. Dynamics of leading-strand lesion skipping by the replisome. Mol. Cell. 2013;52:855–865. doi: 10.1016/j.molcel.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lehmann A.R., Fuchs R.P. Gaps and forks in DNA replication: rediscovering old models. DNA Rep. 2006;5:1495–1498. doi: 10.1016/j.dnarep.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 66.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.