SUMMARY
Congenital heart defects (CHDs) develop through a complex interplay between genetic variants, epigenetic modifications and maternal environmental exposures. Genetic studies of CHDs have commonly tested single genetic variants for association with CHDs. Less attention has been given to complex gene-by-gene and gene-by-environment interactions. In this study, we applied a recently developed likelihood-ratio Mann-Whitney (LRMW) method to detect joint actions among maternal variants, fetal variants and maternal environmental exposures, allowing for high-order statistical interactions. All subjects are participants from the National Birth Defect Prevention Study, including 623 mother-offspring pairs with CHD-affected pregnancies and 875 mother-offspring pairs with unaffected pregnancies. Each individual has 872 single nucleotide polymorphisms encoding for critical enzymes in the homocysteine, folate, and transsulfuration pathways. By using the LRMW method, three variants (fetal rs625879, maternal rs2169650 and maternal rs8177441) were identified with a joint association to CHD risk (nominal P-value=1.13e-07). These 3 variants are located within gene BHMT2, GSTP1 and GPX3, respectively. Further examination indicated that maternal SNP rs2169650 may interact with both fetal SNP rs625879 and maternal SNP rs8177441. Our findings suggest that the risk of CHD may be influenced by both the intra-generational interaction within the maternal genome and the inter-generational interaction between maternal and fetal genomes.
Keywords: gene-by-gene interaction, high-order interactions, congenital heart defects, maternal variants, fetal variants, U-Statistics, likelihood ratio
INTRODUCTION
Congenital heart defects (CHDs) are the most prevalent and severe type of birth defects, occurring in approximately 1 of every 100 live births (Moller et al., 1993, Botto et al., 2001, Hoffman & Kaplan, 2002, Hoffman et al., 2004). A substantial portion of infants born with serious CHDs die in infancy, and many of those who survive may require repeated surgeries and lengthy hospitalizations (Cleves et al., 2003, Gilboa et al., 2010, Nembhard et al., 2001). CHDs are a major concern for pediatric morbidity and mortality, and there are currently few strategies for reducing the public health impact of these conditions (Jenkins et al., 2007).
The genetic susceptibility of CHDs has been recognized for decades (Hobbs et al., 2002). As demonstrated by twin studies, the concordance rates of CHD phenotypes were significantly higher among monozygotic twins (10.0%) than dizygotic twins (2.5%) (Berg et al., 1989). Animal studies have identified several biological pathways controlling the development of the fetal heart. For example, two transcription factors cytoplasma 1 (the nuclear factor of activated T-cells; NF-ATc) and Smad6 were suggested to be involved in the formation of cardiac valves (Galvin et al., 2000). Mice lacking NF-ATc exhibit fatal defects in valve formation and disruption of Smad6 leads to abnormally thickened valves (High & Epstein, 2008). Meanwhile, maternal environmental exposures may have an effect modification on key developmental genes that shape cardiac development (Ashworth & Antipatis, 2001, Keen et al., 2003, Doolin et al., 2002). It is believed that more than 85% of CHDs result from complex interactions between genetic variants, epigenetic modifications and maternal environmental exposures (Botto & Correa, 2003).
To identify genetic and environmental factors that foster the development of CHDs, extensive candidate-gene-based and genome-wide association studies have been conducted (Hobbs et al., 2006, Goldmuntz et al., 2008, Hobbs et al., 2011, Wessels & Willems, 2010, Pediatric Cardiac Genomics et al., 2013, Zaidi et al., 2013). Multiple genes and genetic variants are associated with CHDs. However, most of these studies, including our own, have adopted a single-locus analysis strategy, by testing each single genetic variant individually. As discussed above and elsewhere, CHDs are usually caused by the interplay between multiple genetic variants and environmental factors. If a gene operates primarily through a complex mechanism involving multiple other genes, the effect may often be missed if one examines it in isolation, without allowing for its potential interactions with other genes (Cordell, 2009). Particularly for maternal and perinatal research, two types of gene-gene interactions are possible during pregnancy: intra-generational interaction within either maternal or fetal genes, and inter-generational interaction between maternal and fetal genes (Sinsheimer et al., 2010). Both maternal and fetal genes may interact with other genes within each individual genome, while the maternal and fetal genes may also interact across genomes, leading to either conflicting or beneficial environment for fetal growth, which may influence the phenotypes of both mothers and babies (Haig, 2004). To our knowledge, only one intra-generational interaction between variants from maternal genes MTHFR and CBS has been reported for association with CHDs (Lupo et al., 2010). We and others have proposed to detect inter-generational interactions with a penalized logistic regression model, and applied it to the studies of CHDs (Li et al., 2010, Li et al., 2014b, Li et al., 2014a). However, those studies are limited to interactions between genes from the same genomic region. Further, all existing interaction studies have adopted a two-way interaction strategy, by testing the interaction effect between two genetic variants or interaction effect between one genetic variant and one environmental factor (Lupo et al., 2010, Li et al., 2014a, Li et al., 2014b, Tang et al., 2014, Tang et al., 2015). Although convenient and easy to interpret, such a strategy cannot consider higher order interactions (e.g., three-way interactions), or the complex gene-by-gene (GXG) and gene-by-environment (GXE) interactions between maternal variants, fetal variants and environmental exposures.
In reality, higher-order interactions (e.g., three-way interactions) could exist (Moore, 2003, Ritchie et al., 2001). By not limiting the search to two-way interactions, we may have a better chance of identifying potentially important GXG/GXE interactions, which may elucidate how maternal and fetal genetic variants interact with one another and interplay with environmental factors to cause CHDs. However, detecting high-order GXG/GXE interactions underlying complex human diseases have remained to be a major challenge in genetic studies (Cordell, 2009, Thomas, 2010). A main difficulty for the conventional regression-based methods, such as logistic regression model, is the specification of underlying disease models. By specifying a disease model, certain inheritance of disease risks is implicitly assumed. When such assumption is violated, the effect estimate could be biased and the type I errors be inflated. Further, the number of parameters to be specified in the model increases exponentially with the order of interaction being analyzed. It may become impractical to specify such a model for high dimensional data (Carlborg & Haley, 2004, Peduzzi et al., 1996). To address these difficulties, non-parametric dimension-reduction methods have gained popularity for GXG/GXE research in the past few years, especially for detecting high-order GXG/GXE interactions (Ritchie et al., 2001, Lou et al., 2007, Lu et al., 2012, Li et al., 2011). These proposed methods have been widely applied to population-based studies of complex human diseases, such as breast cancer and drug addiction. It would be of great interest to extend these methods for detecting GXG/GXE interactions in maternal and prenatal research.
Our current study is motivated by this gap in the existing literature, aiming to detect joint actions among a large number of maternal variants, fetal variants and maternal environmental exposures, while allowing for high-order statistical interactions. The current study included 872 maternal SNPs, 872 fetal SNPs selected from 62 candidate genes, and 19 maternal environmental exposures. We applied a recently developed likelihood-ratio Mann-Whitney (LRMW) method to search for GXG/GXE combinations among a large number of genetic and environmental factors. Finally, we explore possible biological mechanisms with respect to the GXG/GXE combinations that jointly alter CHD risk.
MATERIALS AND METHODS
Ethics statement
The study was approved by University of Arkansas for Medical Sciences’ Institutional Review Board and the National Birth Defects Prevention Study (NBDPS), with protocol oversight by the Centers for Disease Control and Prevention (CDC) and National Center on Birth Defects and Developmental Disabilities. All study subjects gave informed consent. For minors, informed consent was obtained from their legal guardian or themselves, if emancipated.
Study Population
All subjects were participants of the National Birth Defects Prevention Study, the largest population-based case control study conducted in the US of nonsyndromic birth defects, covering an annual birth population of 482,000, or 10 % of U.S. births. CHD cases were ascertained from birth defect registries in ten participating states that had similar inclusion criteria: Arkansas, California, Georgia, Iowa, Massachusetts, New Jersey, New York, North Carolina, Texas, and Utah. NBDPS began in 1996, and a detailed description of the study can be found elsewhere (Yoon et al., 2001, Rasmussen et al., 2002, Gallagher et al., 2011). In the current study, we included all available mother-offspring pairs with estimated dates of delivery between 1997 and 2010, which is based on the due date reported by the mother during interview. Case pairs were defined as those in which the child had at least one type of conotruncal heart defect or obstructive heart defect. Control pairs were defined as those in which the child had no structural birth defect. Control families were randomly selected from birth certificate and/or birth hospital records and thus represent a random sample from the general population (Yoon et al., 2001). For the current analyses, the study population comprised 1,498 mother-offspring pairs, including 623 case pairs and 875 control pairs. The maternal characteristics are summarized in Table 1. The cases and control families were balanced with respect to maternal age, race and ethnicity, maternal education, household income, folic acid intake, alcohol consumption, cigarette smoking and maternal obesity (all p-values>0.05).
Table 1.
Maternal characteristics for 623 case families and 875 control families
| Control (N=875) | Case (N=623) | P-value | |
|---|---|---|---|
|
|
|||
| Age at delivery, mean (SD) | 27.7 (5.9) | 28.2 (5.9) | 0.10 |
| Mother’s race | 0.65 | ||
| African American | 88 (10%) | 53 (8.5%) | |
| Caucasian | 620 (71%) | 456 (73%) | |
| Hispanic | 124 (14%) | 81 (13%) | |
| Others | 42 (4.8%) | 32 (5.1%) | |
| Missing information | 1 | 1 | |
| Mother’s education, N (%) | 0.65 | ||
| <12 years | 117 (13%) | 72 (12%) | |
| High school degree | 209 (24%) | 162 (26%) | |
| 1–3 years of college | 244 (28%) | 174 (28%) | |
| At least 4 years of college or Bachelor degree | 305 (35%) | 215 (35%) | |
| Missing information | 0 | 0 | |
| Household income, N (%) | 0.97 | ||
| Less than 10 Thousand | 112 (14%) | 84 (14%) | |
| 10 to 30 Thousand | 236 (29%) | 167 (28%) | |
| 30 to 50 Thousand | 190 (23%) | 142 (24%) | |
| More than 50 Thousand | 285 (35%) | 203 (34%) | |
| Missing information | 52 | 27 | |
| Folic acid supplementation, N (%) | 0.15 | ||
| Unexposed | 372 (43%) | 288 (46%) | |
| Exposed | 503 (57%) | 335 (54%) | |
| Missing information | 0 | 0 | |
| Alcohol consumption, N (%) | 0.62 | ||
| Unexposed | 681 (78%) | 479 (77%) | |
| Exposed | 191 (22%) | 143 (23%) | |
| Missing information | 3 | 1 | |
| Cigarette smoking, N (%) | 0.91 | ||
| Unexposed | 720 (82%) | 511 (82%) | |
| Exposed | 154 (18%) | 111 (18%) | |
| Missing information | 1 | 1 | |
| Maternal Obesity, N (%) | 0.23 | ||
| Non-Obese (BMI<30) | 681(81%) | 467 (77%) | |
| Obese (BMI>=30) | 158 (19%) | 138 (23%) | |
| Missing information | 26 | 18 | |
Genotyping and Quality Control
Our research team commissioned a custom 1,536 SNP panel covering 62 genes in the homocysteine, folate, and transsulfuration pathways potentially related to the development of CHDs, using the Illumina® GoldenGate custom genotyping platform, as detailed elsewhere (Chowdhury et al., 2012). Laboratories that use NBDPS archival DNA samples must demonstrate their proficiency in genotyping techniques by passing an External Quality Assessment (EQA). The EQA for High-throughput genotyping platforms, such as the Illumina Golden Gate assay employed in this project, consists of genotyping of a single SNP in one externally supplied blood-buccal trio. Results from the laboratory must be concordant (99%) for paired blood-derived and buccal-derived DNA and between genomic and whole genome amplified DNA. In addition, inter-lab results must be concordant as well as concordance between pre-characterized DNA and third-party results. Negative controls must not yield results and genotypes of trios must be consistent with Mendelian inheritance. Arkansas Center for Birth Defects Research and Prevention has consistently scored 100% on NBDPS EQA.
GenomeStudio, the software toolkit developed by Illumina, was used for the initial genotype calling. We found that the quality of genotype clustering varied substantially among SNPs, which we attribute to the in silico design of the custom SNP panel without the subsequent quality checks that would be applied to a standard commercial array. To ensure the data quality, we updated the genotype clustering by using SNPMClust, a bivariate Gaussian model-based genotype clustering and calling algorithm, which is available as an R package on the Comprehensive R Archive Network (CRAN).
After running SNPMClust, clustering and classification plots for all SNPs were visually inspected, leading to dropping a SNP from analysis or running SNPMClust under non-default settings in some cases. To ensure high-quality genotypes, we applied stringent quality control measures and excluded SNPs with obviously poor clustering behavior, no-call rates > 10%, Mendelian error rates > 5%, minor allele frequencies < 5%, or significant deviation from Hardy-Weinberg Equilibrium in at least one racial group (P-value < 10−4). After the quality assessment, the analytical data had 872 variants in total.
Statistical Methods
Recently, we and others have proposed a likelihood ratio Mann-Whitney (LRMW) method for detecting genetic variants and environmental factors that are jointly associated with disease phenotypes (Lu et al., 2012). The LRMW method searches for joint action among a large number of genetic variants, such as genome-wide data, and environmental factors. It extends the traditional univariate Mann-Whitney test to assess the joint association of multiple genetic variants and environmental factors simultaneously, while allowing for high-order statistical interactions. We have demonstrated through simulations that the LRMW method has improved power over the conventional Mann-Whitney method when higher order interaction exists among loci. The application of the LRMW method also identified a four-locus interaction among SNPs for type II diabetes, which was replicated in an independent dataset (Lu et al., 2012). The method has been implemented in a C++ software package, referred to as genome-wide gene-gene interaction analyses (GWGGI) (Wei & Lu, 2014). In this article, we extend the LRWM method in the context of mother-offspring pair data, aiming to detect the joint action among maternal variants, fetal variants and maternal environmental exposures. Following the same notation, we briefly describe our method below. The theoretical details can be found elsewhere (Lu et al., 2012).
Assuming that multiple SNPs and environmental factors may be jointly associated with the disease outcome, we expect all mother-offspring pairs can be partitioned into various groups based on their gene-by-gene and gene-by-environment (GXG/GXE) combinations, among which the likelihood of disease may not be all equal. Determining GXG/GXE groups on high-dimensional data could be statistically complicated and computationally intense, since a large number of GXG/GXE groups can be formed. To search for GXG/GXE groups that are most significantly associated with disease phenotype, we adopt a forward selection algorithm, which has shown promising performance in previous studies (Li et al., 2011, Lu et al., 2012, Lu & Elston, 2008). The algorithm starts with a null model by including all mother-offspring pairs as one group, and then uses forward selection to introduce SNPs or environmental factors into the model to partition all mother-offspring pairs into various GXG/GXE groups. In the first step, all SNPs and environmental factors were examined one at a time, and the one factor with the maximum LRMW statistic described below was selected. For example, if a bi-allelic SNP with alleles denoted as A and a, is selected in the first step, all pairs can be partitioned into two groups in two possible ways, assuming either dominant or recessive inheritance, respectively:
G1={AA or Aa} and G2={aa} (carrying one copy of risk allele will increase disease risk)
G1={AA} and G2={Aa or aa} (carrying two copies of risk alleles will increase disease risk)
After that, an environmental factor with two levels (e.g. smoker vs non-smoker) can further partition all pairs into 4 groups (i.e. G1/smoker, G1/non-smoker, G2/smoker, and G2/non-smoker). Or, a second SNP with alleles denoted as B and b can further partition all individuals into 4 groups in two possible ways:
G1/BB or Bb, G1/bb, G2/BB or Bb, G2/bb (Assuming a dominant inheritance of allele B)
G1/BB, G1/Bb or bb, G2/BB, G2/Bb or bb (Assuming a recessive inheritance of allele B)
Furthermore, because the first SNP and the second SNP may be a maternal SNP and a fetal SNP (or vice versa), respectively, the selected GXG/GXE groups may represent joint gene actions within maternal or fetal genomes or the joint gene actions between maternal and fetal genomes.
In practice, the SNPs and environmental factors are selected forwardly, forming GXG/GXE groups adaptively to maximize a LRMW statistic defined below. Suppose the selected SNPs and environmental factors can form R GXG/GXE groups, G1, G2,……GR, we first define a likelihood ratio (LR) for each risk group:
where D represents cases and D̄ represents controls. The LRMW statistic can be defined as:
where and are the number of case-mother pairs carrying genotype Gi and the number of control-mother pairs carrying genotype Gj, respectively; and φ[.] is a kernel function to compare the LR risk score between two groups. A commonly used kernel function is
which leads to a Mann-Whitney statistic comparing the difference in LR risk scores between cases and controls (Mann & Whitney, 1947, Wilcoxon, 1945). In our study, we model each mother-offspring pair as a unit, and build a LRWM statistic for case-mother and control-mother pairs.
In order to avoid over-fitting of data, we conduct a 10-fold cross-validation, i.e., randomly dividing the study population into 10 subsets, to determine the most parsimonious statistical model based on GXG/GXE groups. The joint association is also evaluated by permutation testing, which randomly shuffles the phenotype and applies the same procedure, including forward selection of GXG/GXE groups and ten-fold cross-validation. An empirical p-value can be obtained by repeating the permutation a large number of times (e.g. 5,000 times).
RESULTS
We applied the LRMW method to our dataset, aiming to search for GXG/GXE combinations among 872 maternal SNPs, 872 fetal SNPs, and 4 maternal environmental exposures, including folic acid supplementation, alcohol drinking, smoking and obesity. These maternal lifestyle factors have been shown to be associated with CHD development (Hobbs et al., 2011). For example, maternal intake of folic acid containing supplements may reduce the risk of CHDs (Shaw et al., 1995, van Beynum et al., 2010), while smoking, drinking and obesity may increase the risk (Jenkins et al., 2007, Malik et al., 2008, Stothard et al., 2009). After tenfold cross validation, LRMW identified a final model with 3 SNPs, which gave the maximum LRMW statistic, indicating a joint association among 3 SNPs. None of the environmental factors was selected into the final model. The 3 SNPs are fetal rs625879, maternal rs2169650 and maternal rs8177441, located within gene BHMT2, GSTP1 and GPX3, respectively.
Eight GXG groups were formed by the combination of three SNPs. The three SNPs and corresponding grouping strategies are summarized in Table 2. Three SNPs were identified through forward selection, and the corresponding GXG groups had an increasing area under the curve (AUC), consistent with an increasing ability to predict cardiac defects (i.e. from 0.54, 0.58 to 0.60).
Table 2.
Three SNPs identified by using the LRMW method, indicating a possible joint association with CHDs.
| SNP, Maternal/Fetal | Gene | Ch. | Position | Allele1 | MAF2 | Call Rate | Genotype Group | AUC |
|---|---|---|---|---|---|---|---|---|
| rs625879, Fetal | BHMT2 | 5 | 79085866 | A/C | 42.6% | 99.0% | {CC} Vs {AA or AC} | 0.54 |
| rs2169650, Maternal | GSTP1 | 11 | 67596446 | A/G | 5.12% | 92.5% | {GG} Vs {AA or AG} | 0.58 |
| rs8177441, Maternal | GPX3 | 5 | 151026433 | C/G | 21.6% | 94.3% | {GG} Vs {CC or CG} | 0.60 |
minor allele is bolded.
minor allele frequency
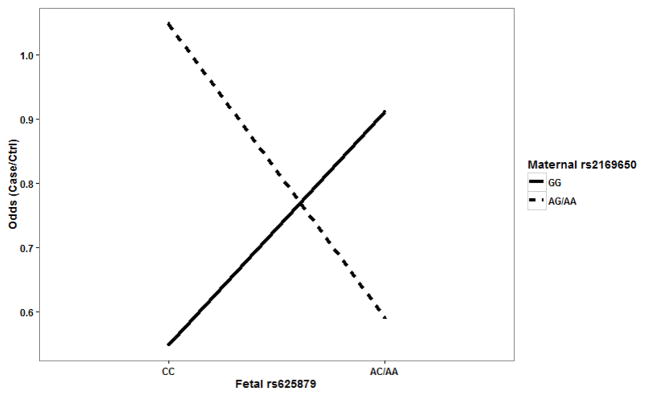
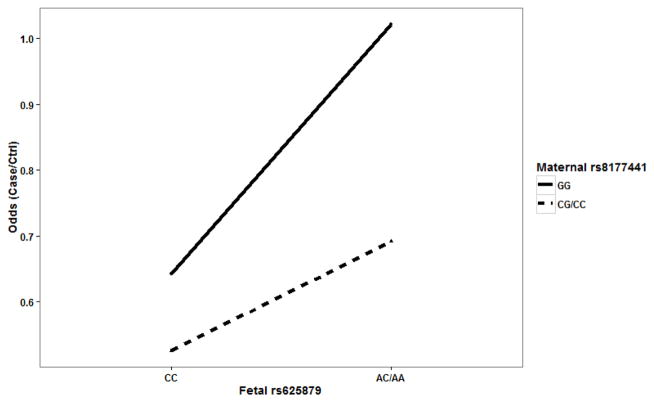
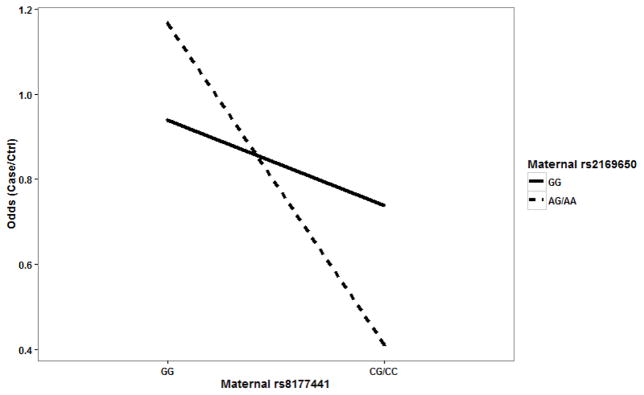
It is worthwhile to note that LRMW method is entirely non-parametric. It allows complex interactions without specifying any interaction model explicitly. Therefore, it is possible for the identified SNPs to have a joint action through either additive or interaction effects. In order to examine the underlying genetic mechanism, we looked into each SNP pair to identify patterns of risk for CHDs among the corresponding GXG groups. For each SNP pair, all mother-offspring pairs can be partitioned into 4 groups based on their GXG combinations. For example, fetal SNP rs625879 and maternal SNP rs2169650 together form four two-SNP genotypes: G1={fetal rs625879 = CC and maternal rs2169650 = GG}, G2={fetal rs625879 = CC and maternal rs2169650 = AA or AG}, G3={fetal rs625879 = AA or AC and maternal rs2169650 = GG}, and G4={fetal rs625879 = AA or AC and maternal rs2169650 = AA or AG}. We found that the effect of fetal SNP rs625879 was modified differently by the genotypes of maternal SNP rs2169650 (see. Figure 1), indicating a potential interaction effect between the two SNPs. In particular, it should be noted that this interaction is “essential” and not completely removable by a monotonic transformation of the data (Wu et al., 2009). The examination of the other two SNP pairs also indicated an additive effect between fetal SNP rs625879 and maternal SNP rs8177441 (see Figure 2), and an interaction effect between maternal SNP rs2169650 and maternal SNP rs8177441 (see Figure 3).
Figure 1.
Fetal SNP rs625879 and maternal SNP rs2169650 showed a possible interaction effect.
Figure 2.
Fetal SNP rs625879 and maternal SNP rs8177441 showed a possible additive effect.
Figure 3.
Maternal SNP rs2169650 and maternal SNP rs8177441 showed a possible interaction effect.
The proposed LRMW method is non-parametric, and does not assume any mode of inherence of disease. However, for the same reason, it does not provide any estimation of effect sizes. In order to estimate the effect sizes among identified genotype groups, we fit a logistic regression model based on the observed patterns of disease risk. The final model included main effects of three SNPs, and two interaction effects: 1) between fetal SNP rs625879 and maternal SNP rs2169650; 2) between maternal SNP rs2169650 and maternal SNP rs8177441. The results are summarized in Table 3. The results showed that a mother-offspring dyad had the highest risk of disease (OR=2.47; 95% C.I. [1.20, 5.08]), when genotype CC, AG or AA, and GG were observed for fetal rs625879, maternal rs2169650 and maternal rs8177441, respectively. The logistic regression attained a significant joint association for three SNPs (P-value=1.13e-07). It should also be noted that this is a nominal p-value without accounting for the selection of SNPs in LRMW method. Permutation test of LRMW with 5,000 replicates obtained an empirical p-value of 0.11, which did not reach the conventional 0.05 threshold. However, we think it may still suggest a potential significant association, considering the fact we had limited sample size (i.e. 1,498 samples) but a very high-dimensional search space (i.e. 1,748 genetic and environmental factors together and their high-order combinations).
Table 3.
Risk of disease by genotype combination formed by 3 SNPs (estimated through a logistic regression model)
| Genotype Combinations | # of Case Pairs | # of Ctrl Pairs | OR [95% C.I.] | P-values | Overall P-Value | ||
|---|---|---|---|---|---|---|---|
| Fetal rs625879 | Maternal rs2169650 | Maternal rs8177441 | |||||
| CC | GG | GG | 97 | 146 | Ref | Ref | 1.13e-07 |
|
| |||||||
| CC | GG | CG or CC | 51 | 100 | 0.78 (0.61, 0.99) | 0.04 | |
|
| |||||||
| CC | AG or AA | GG | 14 | 12 | 2.47 (1.20, 5.08) | 0.01 | |
|
| |||||||
| CC | AG or AA | CG or CC | 8 | 8 | 0.88 (0.39, 1.96) | 0.75 | |
|
| |||||||
| AC or AA | GG | GG | 249 | 227 | 1.66 (1.30, 2.13) | 5.8e-05 | |
|
| |||||||
| AC or AA | GG | CG or CC | 136 | 158 | 1.30 (0.92, 1.83) | 0.14 | |
|
| |||||||
| AC or AA | AG or AA | GG | 28 | 24 | 1.50 (0.86, 2.61) | 0.15 | |
|
| |||||||
| AC or AA | AG or AA | CG or CC | 8 | 31 | 0.53 (0.27, 1.04) | 0.07 | |
DISCUSSION
Our study builds on previous hypotheses and studies demonstrating that the etiology of nonsyndromic CHDs is a process involving multiple genetic variants, both maternal and fetal, maternal environmental exposures as well as complex GXG and GXE interactions. Existing studies typically have examined each single genetic variant for its association with CHDs. Relatively few studies have been conducted to examine potential CHD-related GXG and GXE interactions. Our understanding of the complex GXG/GXE interaction underlying CHDs is still in its infancy, especially for high-order interactions. Further, detecting those GXG/GXE interaction is especially challenging in maternal and prenatal research, since both intra-generational interaction and inter-generational interactions may occur, and both maternal and fetal genes may further interact with maternal environmental exposures (Sinsheimer et al., 2010). Our current study is motivated by the limitations and challenge in existing studies, aiming to detect complex GXG/GXE interactions among maternal variants, fetal variants and maternal environmental exposures, with the consideration of possible high-order interactions. Although, none of the environmental factors was identified to be significant in this study, to our knowledge, our finding is the first three-way interaction among maternal and fetal variants, involving two types of interactions: an intra-generational interaction within maternal genome (i.e. maternal GSTP1 and GPX3) and an inter-generational interaction between maternal and fetal genome (i.e. maternal GPX3 and fetal BHMT2). Findings from previous studies indicated that the interactions between maternal genes in the folate metabolic pathway may influence the risk of CHDs (Lupo et al., 2010), and interactions between maternal and fetal genes may also influence birth defects, such as neural tube defects (Lupo et al., 2014). The findings in our study are in line with previous findings, further suggesting that the interaction effects among maternal and fetal genes could be complex, and genes from homocysteine and transsulfuration may interact with one another to influence CHD development jointly. The current study also extends our previous investigation of maternal-fetal genotype interactions to variants from different genomic regions (Li et al., 2014b, Li et al., 2014a).
For comparison purpose, we also applied a conventional logistic regression model to the same dataset. Similar to our LRMW method, a forward selection strategy was used for logistic regression by minimizing Akaike Information Criteria (AIC). By using a logistic regression model, a total number of 358 SNPs were selected, forming a highly complex final model. It will require a large number of parameters to further investigate their interactions. In addition, it is less straightforward for interpretation.
In our study, we identified a joint action among three variants, fetal SNP rs625879, maternal SNPs rs216950 and rs8177441. It has been suggested that SNP rs625879 was associated with various complex diseases and birth defects, such as coeliac disease (Hozyasz et al., 2012), endometriosis-associated infertility (Szczepanska et al., 2011) and orofacial clefts (Mostowska et al., 2010). Further, the three identified SNPs are located in gene BHMT2, GSTP1 and GPX3, respectively. Gene BHMT2, or betaine--homocysteine S-methyltransferase 2, encodes one of two methyl transferases that can catalyze the transfer of the methyl group from betaine to homocysteine, which plays a crucial role in methylation reactions. Anomalies in homocysteine metabolism have been implicated in disorders ranging from vascular disease to birth defects, such as spina bifida and CHDs (Giusti et al., 2010, McGeachie et al., 2009, Weisberg et al., 2003, Shaw et al., 2009). Gene GSTP1, or glutathione S-transferase pi 1, belongs to a family of enzymes that play an important role in detoxification by catalyzing the conjugation of many hydrophobic and electrophilic compounds with reduced glutathione. GSTP1 proteins were suggested to function in xenobiotic metabolism and play a role in susceptibility to various diseases, such as esophageal squamous cell carcinoma and gastric cardia cancer (Zendehdel et al., 2009). Gene GPX3, or glutathione peroxidase 3 (plasma), belongs to the glutathione peroxidase family, which functions in the detoxification of hydrogen peroxide. GPX3 protects cells and enzymes from oxidative damage, by catalyzing the reduction of hydrogen peroxide, lipid peroxides and organic hydroperoxide. It was also suggested that decreased GPX3 activity may lead to inadequate nitric oxide (NO) levels, which disrupts platelet inhibitory mechanisms and increases arterial thrombosis (Kenet et al., 1999). GPX3 was also found to be associated with ischaemic stroke among children and young adults (Voetsch et al., 2007). While it is biologically plausible that those three identified genes may jointly alter the risk of CHDs, it is necessary for further studies to validate and replicate this finding.
A few limitations should also be noted. First, the permutation test of LRMW method attained an empirical P-value of 0.11, which did not reach the commonly used 0.05 threshold. We think this can be largely due to our limited sample size (i.e.1,498 samples) and high-dimensional search space (i.e. 1,748 genetic and environmental factors together with consideration of their high order combinations). However, NBDPS is the largest study ever conducted in the U.S., and we have included all the available samples collected to date. We expect power to increase as NBDPS moves further with more samples. Second, no environmental factors were identified in our study. One possible reason is that our LRMW method utilized a forward selection strategy, which search for GXG/GXE combinations forwardly. A major advantage is that the forward search is computational efficient and feasible for high dimensional data, such as genome-wide data. However, to detect an interaction effect between two factors, our method requires at least one of them to have a main effect. In our framework, a genetic variant or environmental factor without a main effect may only be introduced into the final model if it interacts with other factors having significant main effects (Lu et al., 2012). An interaction effect without any main effect of all factors will require exhaustive search among all factors, which may substantially increase the computational intensity and possibly reduce power (Li et al., 2011). We also expect “pure interaction” to be a rare scenario in disease etiology. Third, we have combined conotruncal heart defects (332 case-mother pairs) and obstructive heart defects (291 case-mother pairs) as cases in our study. It is possible that disease heterogeneity may exist between two subtypes of CHDs. However, previous studies have demonstrated that various types of pediatric heart disease may share same causal genes. For example, cardiac transcription factor, NKX2.5, has been associated with a wide variety of cardiac phenotypes such as, atrial septal defect, atrioventricular block, double outlet right ventricle, tetralogy of fallot, and ventricular septal defect (Benson, 2010). Further, stratified analysis with each particular subtype will significantly reduce our sample size, limiting our power to detect interactions, especially for high-order interactions.
Acknowledgments
We want to thank numerous families for their generous participation in the National Birth Defects Prevention Study that made this research possible. We also thank the Centers for Birth Defects Research and Prevention in Arkansas, California, Georgia, Iowa, Massachusetts, New Jersey, New York, North Carolina, Texas, and Utah for their contribution of data and manuscript review. We also want to thank Ashley S. Block for assistance in the preparation of this manuscript.
This work is supported in part by the National Institute of Child Health and Human Development under award number 5R01HD039054, the National Center on Birth Defects and Developmental Disabilities under award number 5U01DD000491, the Translational Research Institute through the NIH National Center for Research Resources and the National Center for Advancing Translational Sciences under Award Number UL1TR000039 and KL2TR000063, the University of Arkansas for Medical Sciences College of Medicine Children’s University Medical Group Fund Grant Program, the Arkansas Children’s Hospital Research Institute and the Arkansas Bioscience Institute.
Footnotes
The authors declare no conflict of interest.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH and CDC.
References
- Ashworth CJ, Antipatis C. Micronutrient programming of development throughout gestation. Reproduction. 2001;122:527–35. [PubMed] [Google Scholar]
- Benson DW. Genetic origins of pediatric heart disease. Pediatr Cardiol. 2010;31:422–9. doi: 10.1007/s00246-009-9607-y. [DOI] [PubMed] [Google Scholar]
- Berg KA, Astemborski JA, Boughman JA, Ferencz C. Congenital cardiovascular malformations in twins and triplets from a population-based study. American journal of diseases of children. 1989;143:1461–3. doi: 10.1001/archpedi.1989.02150240083023. [DOI] [PubMed] [Google Scholar]
- Botto LD, Correa A. Decreasing the Burden of Congenital Heart Anomalies: An Epidemiologic Evaluation of Risk Factors and Survival. Progress in Pediatric Cardiology. 2003;18:111–121. [Google Scholar]
- Botto LD, Correa A, Erickson JD. Racial and temporal variations in the prevalence of heart defects. Pediatrics. 2001;107:E32. doi: 10.1542/peds.107.3.e32. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nature reviews. Genetics. 2004;5:618–25. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Hobbs CA, Macleod SL, Cleves MA, Melnyk S, James SJ, Hu P, Erickson SW. Associations between maternal genotypes and metabolites implicated in congenital heart defects. Molecular genetics and metabolism. 2012;107:596–604. doi: 10.1016/j.ymgme.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleves MA, Ghaffar S, Zhao W, Mosley BS, Hobbs CA. First-year survival of infants born with congenital heart defects in Arkansas (1993–1998): a survival analysis using registry data. Birth defects research. Part A, Clinical and molecular teratology. 2003;67:662–8. doi: 10.1002/bdra.10119. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nature reviews. Genetics. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolin MT, Barbaux S, Mcdonnell M, Hoess K, Whitehead AS, Mitchell LE. Maternal genetic effects, exerted by genes involved in homocysteine remethylation, influence the risk of spina bifida. American journal of human genetics. 2002;71:1222–6. doi: 10.1086/344209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher ML, Sturchio C, Smith A, Koontz D, Jenkins MM, Honein MA, Rasmussen SA. Evaluation of mailed pediatric buccal cytobrushes for use in a case-control study of birth defects. Birth defects research. Part A, Clinical and molecular teratology. 2011;91:642–8. doi: 10.1002/bdra.20829. [DOI] [PubMed] [Google Scholar]
- Galvin KM, Donovan MJ, Lynch CA, Meyer RI, Paul RJ, Lorenz JN, Fairchild-Huntress V, Dixon KL, Dunmore JH, Gimbrone MA, Jr, Falb D, Huszar D. A role for smad6 in development and homeostasis of the cardiovascular system. Nature genetics. 2000;24:171–4. doi: 10.1038/72835. [DOI] [PubMed] [Google Scholar]
- Gilboa SM, Salemi JL, Nembhard WN, Fixler DE, Correa A. Mortality resulting from congenital heart disease among children and adults in the United States, 1999 to 2006. Circulation. 2010;122:2254–63. doi: 10.1161/CIRCULATIONAHA.110.947002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti B, Saracini C, Bolli P, Magi A, Martinelli I, Peyvandi F, Rasura M, Volpe M, Lotta LA, Rubattu S, Mannucci PM, Abbate R. Early-onset ischaemic stroke: analysis of 58 polymorphisms in 17 genes involved in methionine metabolism. Thrombosis and haemostasis. 2010;104:231–42. doi: 10.1160/TH09-11-0748. [DOI] [PubMed] [Google Scholar]
- Goldmuntz E, Woyciechowski S, Renstrom D, Lupo PJ, Mitchell LE. Variants of folate metabolism genes and the risk of conotruncal cardiac defects. Circulation. Cardiovascular genetics. 2008;1:126–32. doi: 10.1161/CIRCGENETICS.108.796342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Evolutionary conflicts in pregnancy and calcium metabolism--a review. Placenta. 2004;25(Suppl A):S10–5. doi: 10.1016/j.placenta.2004.01.006. [DOI] [PubMed] [Google Scholar]
- High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nature reviews. Genetics. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- Hobbs CA, Cleves MA, Simmons CJ. Genetic epidemiology and congenital malformations: from the chromosome to the crib. Archives of pediatrics & adolescent medicine. 2002;156:315–20. doi: 10.1001/archpedi.156.4.315. [DOI] [PubMed] [Google Scholar]
- Hobbs CA, James SJ, Jernigan S, Melnyk S, Lu Y, Malik S, Cleves MA. Congenital heart defects, maternal homocysteine, smoking, and the 677 C>T polymorphism in the methylenetetrahydrofolate reductase gene: evaluating gene-environment interactions. American journal of obstetrics and gynecology. 2006;194:218–24. doi: 10.1016/j.ajog.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Hobbs CA, Macleod SL, Jill James S, Cleves MA. Congenital heart defects and maternal genetic, metabolic, and lifestyle factors. Birth defects research. Part A, Clinical and molecular teratology. 2011;91:195–203. doi: 10.1002/bdra.20784. [DOI] [PubMed] [Google Scholar]
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. Journal of the American College of Cardiology. 2002;39:1890–900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- Hoffman JI, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. American heart journal. 2004;147:425–39. doi: 10.1016/j.ahj.2003.05.003. [DOI] [PubMed] [Google Scholar]
- Hozyasz KK, Mostowska A, Szaflarska-Poplawska A, Lianeri M, Jagodzinski PP. Polymorphic variants of genes involved in homocysteine metabolism in celiac disease. Molecular biology reports. 2012;39:3123–30. doi: 10.1007/s11033-011-1077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins KJ, Correa A, Feinstein JA, Botto L, Britt AE, Daniels SR, Elixson M, Warnes CA, Webb CL American Heart Association Council on Cardiovascular Disease in The, Y. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:2995–3014. doi: 10.1161/CIRCULATIONAHA.106.183216. [DOI] [PubMed] [Google Scholar]
- Keen CL, Clegg MS, Hanna LA, Lanoue L, Rogers JM, Daston GP, Oteiza P, Uriu-Adams JY. The plausibility of micronutrient deficiencies being a significant contributing factor to the occurrence of pregnancy complications. The Journal of nutrition. 2003;133:1597S–1605S. doi: 10.1093/jn/133.5.1597S. [DOI] [PubMed] [Google Scholar]
- Kenet G, Freedman J, Shenkman B, Regina E, Brok-Simoni F, Holzman F, Vavva F, Brand N, Michelson A, Trolliet M, Loscalzo J, Inbal A. Plasma glutathione peroxidase deficiency and platelet insensitivity to nitric oxide in children with familial stroke. Arteriosclerosis, thrombosis, and vascular biology. 1999;19:2017–23. doi: 10.1161/01.atv.19.8.2017. [DOI] [PubMed] [Google Scholar]
- Li M, Cleves MA, Mallick H, Erickson SW, Tang X, Nick TG, Macleod SL, Hobbs CA National Birth Defect Prevention, S. A genetic association study detects haplotypes associated with obstructive heart defects. Human genetics. 2014a;133:1127–38. doi: 10.1007/s00439-014-1453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Erickson SW, Hobbs CA, Li J, Tang X, Nick TG, Macleod SL, Cleves MA National Birth Defect Prevention, S. Detecting maternal-fetal genotype interactions associated with conotruncal heart defects: a haplotype-based analysis with penalized logistic regression. Genetic epidemiology. 2014b;38:198–208. doi: 10.1002/gepi.21793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Romero R, Fu WJ, Cui Y. Mapping haplotype-haplotype interactions with adaptive LASSO. BMC genetics. 2010;11:79. doi: 10.1186/1471-2156-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Ye C, Fu W, Elston RC, Lu Q. Detecting genetic interactions for quantitative traits with U-statistics. Genetic epidemiology. 2011;35:457–68. doi: 10.1002/gepi.20594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou XY, Chen GB, Yan L, Ma JZ, Zhu J, Elston RC, Li MD. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application to nicotine dependence. American journal of human genetics. 2007;80:1125–37. doi: 10.1086/518312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Elston RC. Using the optimal receiver operating characteristic curve to design a predictive genetic test, exemplified with type 2 diabetes. American journal of human genetics. 2008;82:641–51. doi: 10.1016/j.ajhg.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Wei C, Ye C, Li M, Elston RC. A likelihood ratio-based Mann-Whitney approach finds novel replicable joint gene action for type 2 diabetes. Genetic epidemiology. 2012;36:583–93. doi: 10.1002/gepi.21651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo PJ, Goldmuntz E, Mitchell LE. Gene-gene interactions in the folate metabolic pathway and the risk of conotruncal heart defects. Journal of biomedicine & biotechnology. 2010;2010:630940. doi: 10.1155/2010/630940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo PJ, Mitchell LE, Canfield MA, Shaw GM, Olshan AF, Finnell RH, Zhu H National Birth Defects Prevention, S. Maternal-fetal metabolic gene-gene interactions and risk of neural tube defects. Molecular genetics and metabolism. 2014;111:46–51. doi: 10.1016/j.ymgme.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Cleves MA, Honein MA, Romitti PA, Botto LD, Yang S, Hobbs CA National Birth Defects Prevention, S. Maternal smoking and congenital heart defects. Pediatrics. 2008;121:e810–6. doi: 10.1542/peds.2007-1519. [DOI] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. On a test of whether one of 2 random variables is stochastically larger than the other. Ann Math Stat. 1947;1:50–60. [Google Scholar]
- Mcgeachie M, Ramoni RL, Mychaleckyj JC, Furie KL, Dreyfuss JM, Liu Y, Herrington D, Guo X, Lima JA, Post W, Rotter JI, Rich S, Sale M, Ramoni MF. Integrative predictive model of coronary artery calcification in atherosclerosis. Circulation. 2009;120:2448–54. doi: 10.1161/CIRCULATIONAHA.109.865501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller JH, Allen HD, Clark EB, Dajani AS, Golden A, Hayman LL, Lauer RM, Marmer EL, Mcanulty JH, Oparil S, et al. Report of the task force on children and youth. American Heart Association. Circulation. 1993;88:2479–86. doi: 10.1161/01.cir.88.5.2479. [DOI] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Mostowska A, Hozyasz KK, Biedziak B, Misiak J, Jagodzinski PP. Polymorphisms located in the region containing BHMT and BHMT2 genes as maternal protective factors for orofacial clefts. European journal of oral sciences. 2010;118:325–32. doi: 10.1111/j.1600-0722.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- Nembhard WN, Waller DK, Sever LE, Canfield MA. Patterns of first-year survival among infants with selected congenital anomalies in Texas, 1995–1997. Teratology. 2001;64:267–75. doi: 10.1002/tera.1073. [DOI] [PubMed] [Google Scholar]
- Pediatric Cardiac Genomics, C. Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, Kaski JP, Kim R, Kline J, Mercer-Rosa L, Porter G, Roberts A, Rosenberg E, Seiden H, Seidman C, Sleeper L, Tennstedt S, Kaltman J, Schramm C, Burns K, Pearson G, Rosenberg E. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circulation research. 2013;112:698–706. doi: 10.1161/CIRCRESAHA.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peduzzi P, Concato J, Kemper E, Holford TR, Feinstein AR. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol. 1996;49:1373–9. doi: 10.1016/s0895-4356(96)00236-3. [DOI] [PubMed] [Google Scholar]
- Rasmussen SA, Lammer EJ, Shaw GM, Finnell RH, Mcgehee RE, Jr, Gallagher M, Romitti PA, Murray JC National Birth Defects Preventionm, S. Integration of DNA sample collection into a multi-site birth defects case-control study. Teratology. 2002;66:177–84. doi: 10.1002/tera.10086. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. American journal of human genetics. 2001;69:138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, Lu W, Zhu H, Yang W, Briggs FB, Carmichael SL, Barcellos LF, Lammer EJ, Finnell RH. 118 SNPs of folate-related genes and risks of spina bifida and conotruncal heart defects. BMC medical genetics. 2009;10:49. doi: 10.1186/1471-2350-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, O’malley CD, Wasserman CR, Tolarova MM, Lammer EJ. Maternal periconceptional use of multivitamins and reduced risk for conotruncal heart defects and limb deficiencies among offspring. Am J Med Genet. 1995;59:536–45. doi: 10.1002/ajmg.1320590428. [DOI] [PubMed] [Google Scholar]
- Sinsheimer JS, Elston RC, Fu WJ. Gene-gene interaction in maternal and perinatal research. Journal of biomedicine & biotechnology. 2010;2010 doi: 10.1155/2010/853612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stothard KJ, Tennant PW, Bell R, Rankin J. Maternal overweight and obesity and the risk of congenital anomalies: a systematic review and meta-analysis. JAMA. 2009;301:636–50. doi: 10.1001/jama.2009.113. [DOI] [PubMed] [Google Scholar]
- Szczepanska M, Mostowska A, Wirstlein P, Lianeri M, Marianowski P, Skrzypczak J, Jagodzinski PP. Polymorphic variants of folate and choline metabolism genes and the risk of endometriosis-associated infertility. European journal of obstetrics, gynecology, and reproductive biology. 2011;157:67–72. doi: 10.1016/j.ejogrb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Tang X, Cleves MA, Nick TG, Li M, Macleod SL, Erickson SW, Li J, Shaw G, Hobbs CA. Risk of obstructive heart defects associated with genetic variants and maternal behaviors. Am J Med Genet. 2015 doi: 10.1002/ajmg.a.36867. To Appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Nick TG, Cleves MA, Erickson SW, Li M, Li J, Macleod SL, Hobbs CA. Maternal obesity and tobacco use modify the impact of genetic variants on the occurrence of conotruncal heart defects. PloS one. 2014;9:e108903. doi: 10.1371/journal.pone.0108903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. Gene--environment-wide association studies: emerging approaches. Nature reviews. Genetics. 2010;11:259–72. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beynum IM, Kapusta L, Bakker MK, Den Heijer M, Blom HJ, De Walle HE. Protective effect of periconceptional folic acid supplements on the risk of congenital heart defects: a registry-based case-control study in the northern Netherlands. Eur Heart J. 2010;31:464–71. doi: 10.1093/eurheartj/ehp479. [DOI] [PubMed] [Google Scholar]
- Voetsch B, Jin RC, Bierl C, Benke KS, Kenet G, Simioni P, Ottaviano F, Damasceno BP, Annichino-Bizacchi JM, Handy DE, Loscalzo J. Promoter polymorphisms in the plasma glutathione peroxidase (GPx-3) gene: a novel risk factor for arterial ischemic stroke among young adults and children. Stroke; a journal of cerebral circulation. 2007;38:41–9. doi: 10.1161/01.STR.0000252027.53766.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Lu Q. GWGGI: software for genome-wide gene-gene interaction analysis. BMC genetics. 2014;15:101. doi: 10.1186/s12863-014-0101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg IS, Park E, Ballman KV, Berger P, Nunn M, Suh DS, Breksa AP, 3rd, Garrow TA, Rozen R. Investigations of a common genetic variant in betaine-homocysteine methyltransferase (BHMT) in coronary artery disease. Atherosclerosis. 2003;167:205–14. doi: 10.1016/s0021-9150(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Wessels MW, Willems PJ. Genetic factors in non-syndromic congenital heart malformations. Clinical genetics. 2010;78:103–23. doi: 10.1111/j.1399-0004.2010.01435.x. [DOI] [PubMed] [Google Scholar]
- Wilcoxon F. Individual comparisons by ranking methods. Biometr Bull. 1945;6:80–83. [Google Scholar]
- Wu C, Zhang H, Liu X, Dewan A, Dubrow R, Ying Z, Yang Y, Hoh J. Detecting essential and removable interactions in genome-wide association studies. Statistics and its interface. 2009;2:161–170. doi: 10.4310/sii.2009.v2.n2.a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon PW, Rasmussen SA, Lynberg MC, Moore CA, Anderka M, Carmichael SL, Costa P, Druschel C, Hobbs CA, Romitti PA, Langlois PH, Edmonds LD. The National Birth Defects Prevention Study. Public health reports. 2001;116(Suppl 1):32–40. doi: 10.1093/phr/116.S1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, Depalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe’er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H, Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–3. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zendehdel K, Bahmanyar S, Mccarthy S, Nyren O, Andersson B, Ye W. Genetic polymorphisms of glutathione S-transferase genes GSTP1, GSTM1, and GSTT1 and risk of esophageal and gastric cardia cancers. Cancer causes & control : CCC. 2009;20:2031–8. doi: 10.1007/s10552-009-9399-7. [DOI] [PubMed] [Google Scholar]