Abstract
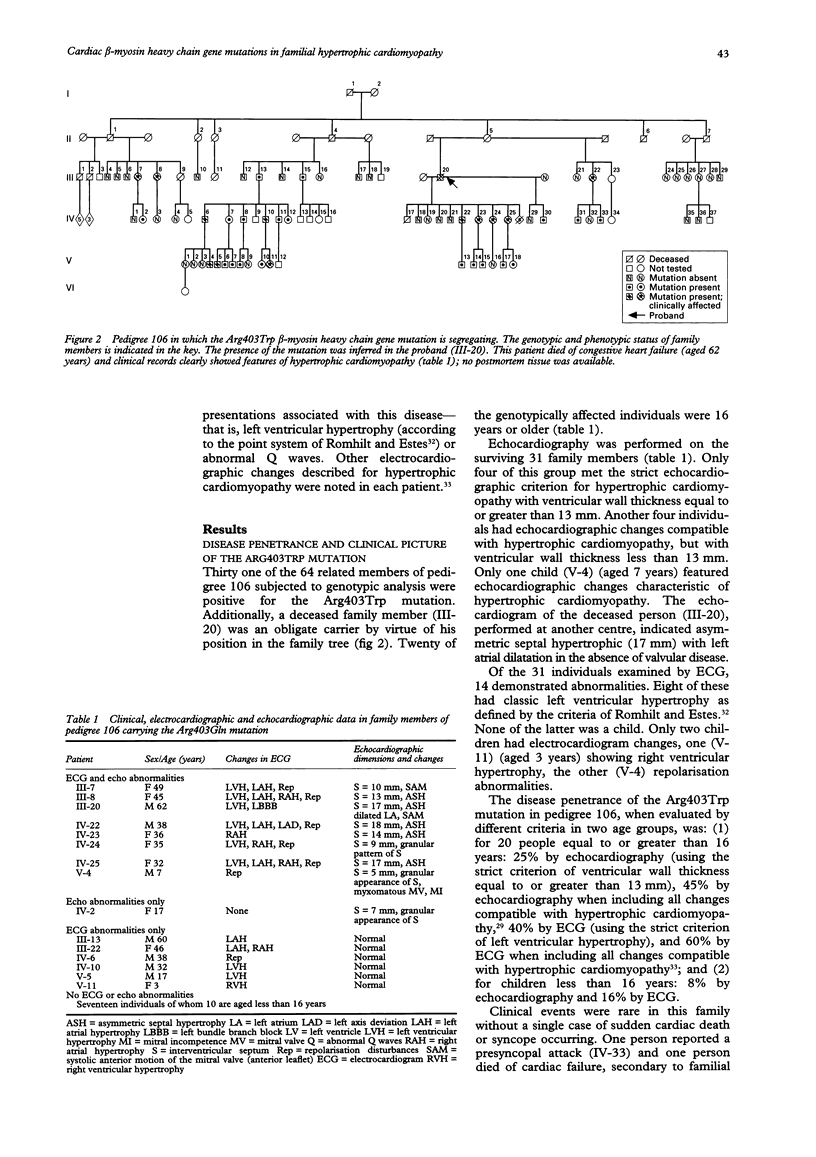
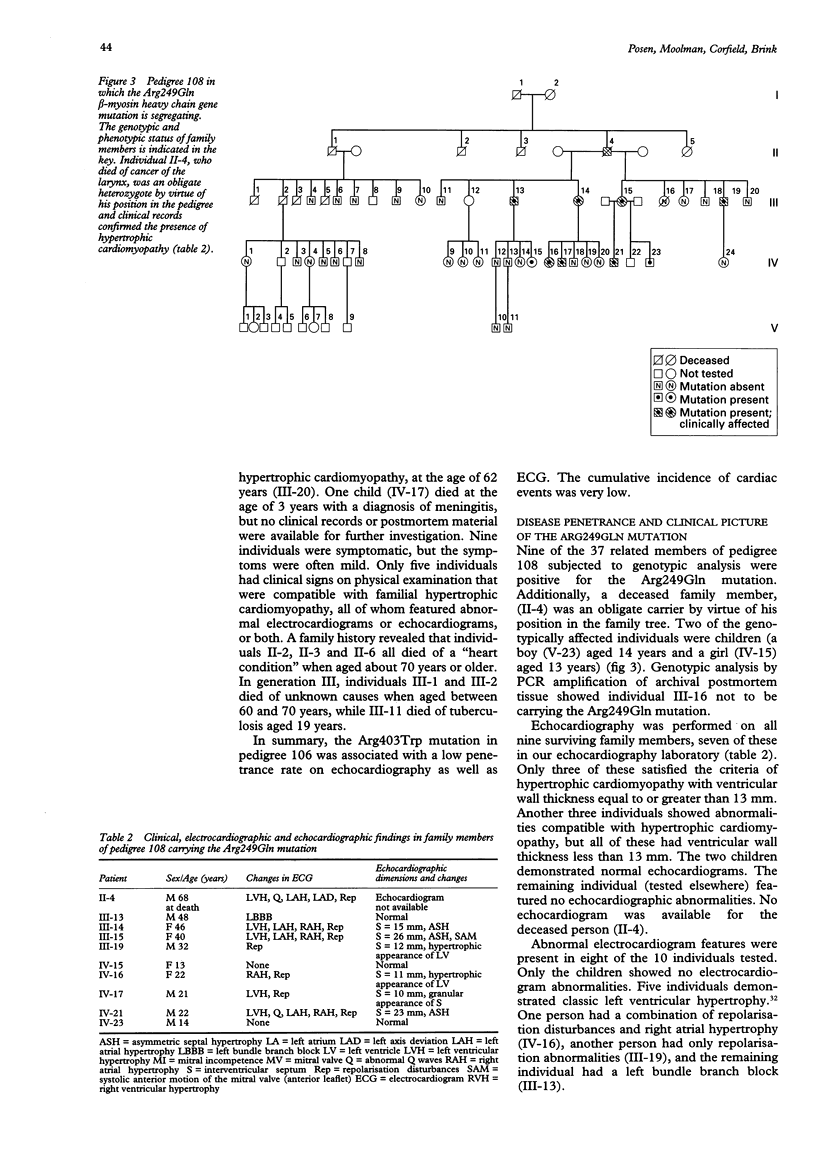
BACKGROUND--Familial hypertrophic cardiomyopathy is the most common inherited cardiac disorder, with sudden cardiac death at a young age the most frequent cause of death in affected individuals. Some cases of familial hypertrophic cardiomyopathy are caused by missense mutations of the beta myosin heavy chain (beta MHC) gene on chromosome 14 and at least 17 such mutations have been described. Recent reports suggest that a correlation exists between a specific beta MHC gene mutation and prognosis in familial hypertrophic cardiomyopathy. This premise is currently being used as a basis to provide counselling for affected families. This mutation/prognosis association, however, has not been widely assessed as yet. The clinical and prognostic features of two South African families of mixed racial descent, in which different beta MHC gene mutations were segregating, were studied to evaluate this correlation. The results were compared with those of previously published reports of European families carrying the same mutations. METHODS--The beta MHC gene missense mutations in two affected families were identified by single strand conformation polymorphism analysis and sequencing (pedigree 106: Arg403Trp; pedigree 108: Arg249Gln). All family members were subjected to genotypic analysis using polymerase chain reaction amplification and restriction enzyme based mutation detection techniques. Clinical, electrocardiographic, and echocardiographic studies were performed on genotypically affected individuals in these two kindreds. RESULTS--The number of individuals identified in pedigree 106 with the Arg403Trp mutation was 32.10 individuals bore the Arg249Gln mutation in pedigree 108. The penetrance rate in adults (equal to or greater than 16 years), using the strict echocardiographic criterion of maximum left ventricular wall thickness > or = 13 mm, was 25% for pedigree 106 and 33% for pedigree 108. Familial hypertrophic cardiomyopathy compatible electrocardiographic and echocardiographic abnormalities were seen in 60% of genotypically positive individuals aged > or = 16 years in pedigree 106 and 80% in pedigree 108. The prognosis was uniformly benign in the two families. For pedigree 106 this corresponded to a report of no early sudden cardiac deaths in a French family with the Arg403Trp mutation. For pedigree 108 the absence of such deaths was in apparent contrast to the four cases reported in 24 genotypically affected individuals in a study of a kindred of European ancestry bearing the Arg249Gln mutation. CONCLUSION--This study of a large South African kindred confirmed the benign nature of the Arg403Trp mutation suggested in a previous report. The number and the relatively young age of affected individuals in a second South African family must be considered when comparing the absence of familial hypertrophic cardiomyopathy associated deaths with the intermediate survival reported for the Arg249Gln mutation in a European family. This investigation lends support to current evidence relating specific beta MHC gene mutations to prognosis, which may be used as a basis to provide counselling for affected families.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carrier L., Hengstenberg C., Beckmann J. S., Guicheney P., Dufour C., Bercovici J., Dausse E., Berebbi-Bertrand I., Wisnewsky C., Pulvenis D. Mapping of a novel gene for familial hypertrophic cardiomyopathy to chromosome 11. Nat Genet. 1993 Jul;4(3):311–313. doi: 10.1038/ng0793-311. [DOI] [PubMed] [Google Scholar]
- Corfield V. A., Moolman J. C., Martell R., Brink P. A. Polymerase chain reaction-based detection of MN blood group-specific sequences in the human genome. Transfusion. 1993 Feb;33(2):119–124. doi: 10.1046/j.1537-2995.1993.33293158042.x. [DOI] [PubMed] [Google Scholar]
- Dausse E., Komajda M., Fetler L., Dubourg O., Dufour C., Carrier L., Wisnewsky C., Bercovici J., Hengstenberg C., al-Mahdawi S. Familial hypertrophic cardiomyopathy. Microsatellite haplotyping and identification of a hot spot for mutations in the beta-myosin heavy chain gene. J Clin Invest. 1993 Dec;92(6):2807–2813. doi: 10.1172/JCI116900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein N. D., Cohn G. M., Cyran F., Fananapazir L. Differences in clinical expression of hypertrophic cardiomyopathy associated with two distinct mutations in the beta-myosin heavy chain gene. A 908Leu----Val mutation and a 403Arg----Gln mutation. Circulation. 1992 Aug;86(2):345–352. doi: 10.1161/01.cir.86.2.345. [DOI] [PubMed] [Google Scholar]
- Epstein N. D., Fananapazir L., Lin H. J., Mulvihill J., White R., Lalouel J. M., Lifton R. P., Nienhuis A. W., Leppert M. Evidence of genetic heterogeneity in five kindreds with familial hypertrophic cardiomyopathy. Circulation. 1992 Feb;85(2):635–647. doi: 10.1161/01.cir.85.2.635. [DOI] [PubMed] [Google Scholar]
- Fananapazir L., Epstein N. D. Genotype-phenotype correlations in hypertrophic cardiomyopathy. Insights provided by comparisons of kindreds with distinct and identical beta-myosin heavy chain gene mutations. Circulation. 1994 Jan;89(1):22–32. doi: 10.1161/01.cir.89.1.22. [DOI] [PubMed] [Google Scholar]
- Geisterfer-Lowrance A. A., Kass S., Tanigawa G., Vosberg H. P., McKenna W., Seidman C. E., Seidman J. G. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990 Sep 7;62(5):999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- Hejtmancik J. F., Brink P. A., Towbin J., Hill R., Brink L., Tapscott T., Trakhtenbroit A., Roberts R. Localization of gene for familial hypertrophic cardiomyopathy to chromosome 14q1 in a diverse US population. Circulation. 1991 May;83(5):1592–1597. doi: 10.1161/01.cir.83.5.1592. [DOI] [PubMed] [Google Scholar]
- Hengstenberg C., Schwartz K. Molecular genetics of familial hypertrophic cardiomyopathy. J Mol Cell Cardiol. 1994 Jan;26(1):3–10. doi: 10.1006/jmcc.1994.1002. [DOI] [PubMed] [Google Scholar]
- Jaenicke T., Diederich K. W., Haas W., Schleich J., Lichter P., Pfordt M., Bach A., Vosberg H. P. The complete sequence of the human beta-myosin heavy chain gene and a comparative analysis of its product. Genomics. 1990 Oct;8(2):194–206. doi: 10.1016/0888-7543(90)90272-v. [DOI] [PubMed] [Google Scholar]
- Jarcho J. A., McKenna W., Pare J. A., Solomon S. D., Holcombe R. F., Dickie S., Levi T., Donis-Keller H., Seidman J. G., Seidman C. E. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989 Nov 16;321(20):1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- Lawson J. W. Hypertrophic cardiomyopathy: current views on etiology, pathophysiology, and management. Am J Med Sci. 1987 Sep;294(3):191–210. doi: 10.1097/00000441-198709000-00011. [DOI] [PubMed] [Google Scholar]
- Maron B. J., Bonow R. O., Cannon R. O., 3rd, Leon M. B., Epstein S. E. Hypertrophic cardiomyopathy. Interrelations of clinical manifestations, pathophysiology, and therapy (1). N Engl J Med. 1987 Mar 26;316(13):780–789. doi: 10.1056/NEJM198703263161305. [DOI] [PubMed] [Google Scholar]
- Maron B. J., Bonow R. O., Cannon R. O., 3rd, Leon M. B., Epstein S. E. Hypertrophic cardiomyopathy. Interrelations of clinical manifestations, pathophysiology, and therapy (2). N Engl J Med. 1987 Apr 2;316(14):844–852. doi: 10.1056/NEJM198704023161405. [DOI] [PubMed] [Google Scholar]
- Maron B. J., Gottdiener J. S., Epstein S. E. Patterns and significance of distribution of left ventricular hypertrophy in hypertrophic cardiomyopathy. A wide angle, two dimensional echocardiographic study of 125 patients. Am J Cardiol. 1981 Sep;48(3):418–428. doi: 10.1016/0002-9149(81)90068-0. [DOI] [PubMed] [Google Scholar]
- Maron B. J., Lipson L. C., Roberts W. C., Savage D. D., Epstein S. E. "Malignant" hypertrophic cardiomyopathy: identification of a subgroup of families with unusually frequent premature death. Am J Cardiol. 1978 Jun;41(7):1133–1140. doi: 10.1016/0002-9149(78)90870-6. [DOI] [PubMed] [Google Scholar]
- Maron B. J., Roberts W. C., McAllister H. A., Rosing D. R., Epstein S. E. Sudden death in young athletes. Circulation. 1980 Aug;62(2):218–229. doi: 10.1161/01.cir.62.2.218. [DOI] [PubMed] [Google Scholar]
- Maron B. J., Savage D. D., Wolfson J. K., Epstein S. E. Prognostic significance of 24 hour ambulatory electrocardiographic monitoring in patients with hypertrophic cardiomyopathy: a prospective study. Am J Cardiol. 1981 Aug;48(2):252–257. doi: 10.1016/0002-9149(81)90604-4. [DOI] [PubMed] [Google Scholar]
- McKenna W. J., Camm A. J. Sudden death in hypertrophic cardiomyopathy. Assessment of patients at high risk. Circulation. 1989 Nov;80(5):1489–1492. doi: 10.1161/01.cir.80.5.1489. [DOI] [PubMed] [Google Scholar]
- McKenna W. J., England D., Doi Y. L., Deanfield J. E., Oakley C., Goodwin J. F. Arrhythmia in hypertrophic cardiomyopathy. I: Influence on prognosis. Br Heart J. 1981 Aug;46(2):168–172. doi: 10.1136/hrt.46.2.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolman J. C., Brink P. A., Corfield V. A. Identification of a new missense mutation at Arg403, a CpG mutation hotspot, in exon 13 of the beta-myosin heavy chain gene in hypertrophic cardiomyopathy. Hum Mol Genet. 1993 Oct;2(10):1731–1732. doi: 10.1093/hmg/2.10.1731. [DOI] [PubMed] [Google Scholar]
- Panza J. A., Maron B. J. Relation of electrocardiographic abnormalities to evolving left ventricular hypertrophy in hypertrophic cardiomyopathy during childhood. Am J Cardiol. 1989 May 15;63(17):1258–1265. doi: 10.1016/0002-9149(89)90187-2. [DOI] [PubMed] [Google Scholar]
- Romhilt D. W., Estes E. H., Jr A point-score system for the ECG diagnosis of left ventricular hypertrophy. Am Heart J. 1968 Jun;75(6):752–758. doi: 10.1016/0002-8703(68)90035-5. [DOI] [PubMed] [Google Scholar]
- Savage D. D., Seides S. F., Clark C. E., Henry W. L., Maron B. J., Robinson F. C., Epstein S. E. Electrocardiographic findings in patients with obstructive and nonobstructive hypertrophic cardiomyopathy. Circulation. 1978 Sep;58(3 Pt 1):402–408. doi: 10.1161/01.cir.58.3.402. [DOI] [PubMed] [Google Scholar]
- Shibata D. K., Arnheim N., Martin W. J. Detection of human papilloma virus in paraffin-embedded tissue using the polymerase chain reaction. J Exp Med. 1988 Jan 1;167(1):225–230. doi: 10.1084/jem.167.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierfelder L., MacRae C., Watkins H., Tomfohrde J., Williams M., McKenna W., Bohm K., Noeske G., Schlepper M., Bowcock A. A familial hypertrophic cardiomyopathy locus maps to chromosome 15q2. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6270–6274. doi: 10.1073/pnas.90.13.6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins H., MacRae C., Thierfelder L., Chou Y. H., Frenneaux M., McKenna W., Seidman J. G., Seidman C. E. A disease locus for familial hypertrophic cardiomyopathy maps to chromosome 1q3. Nat Genet. 1993 Apr;3(4):333–337. doi: 10.1038/ng0493-333. [DOI] [PubMed] [Google Scholar]
- Watkins H., Rosenzweig A., Hwang D. S., Levi T., McKenna W., Seidman C. E., Seidman J. G. Characteristics and prognostic implications of myosin missense mutations in familial hypertrophic cardiomyopathy. N Engl J Med. 1992 Apr 23;326(17):1108–1114. doi: 10.1056/NEJM199204233261703. [DOI] [PubMed] [Google Scholar]



