Abstract
Three signaling systems play the fundamental roles in modulating cell activities: chemical, electrical, and mechanical. While the former two are well studied, the mechanical signaling system is still elusive because of the lack of methods to measure structural forces in real time at cellular and subcellular levels. Indeed, almost all biological processes are responsive to modulation by mechanical forces that trigger dispersive downstream electrical and biochemical pathways. Communication among the three systems is essential to make cells and tissues receptive to environmental changes. Cells have evolved many sophisticated mechanisms for the generation, perception and transduction of mechanical forces, including motor proteins and mechanosensors. In this review, we introduce some background information about mechanical dynamics in live cells, including the ubiquitous mechanical activity, various types of mechanical stimuli exerted on cells and the different mechanosensors. We also summarize recent results obtained using genetically encoded FRET (fluorescence resonance energy transfer)-based force/tension sensors; a new technique used to measure mechanical forces in structural proteins. The sensors have been incorporated into many specific structural proteins and have measured the force gradients in real time within live cells, tissues, and animals.
Keywords: Structural force, Mechanical dynamics, Motor proteins, Mechanosensors, Fluorescence-based force sensors
1. Ubiquitous mechanical activity in non-muscle cells
Biomechanical activity was first recognized and studied in muscle cells. However, all cells, from osteocytes and endothelial cells to neurons and immune cells, are mechanically sensitive and can generate force. All biological processes, including differentiation, mitosis, meiosis, motility, apoptosis and homeostasis, involve the modulation and response to mechanical force [1]. Mechanical force obviously varies in different tissues and organs; whether it is periodical preload or postload in ventricles, fluid shear force on the vascular endothelium, strain in skin or compression, and bending of bones and cartilage. Along with chemical free energy and electrical free energy, mechanical force completes the sources of free energy available to cells, and all the free energy sources are coupled to downstream pathways that regulate physiological activity. For instance, stem cell phenotypy is affected by mechanical factors including substrate stiffness and surface topography. When mesenchymal stem cells are cultured on soft substrates that mimic the elasticity of brain tissue, the cells differentiate into neuronal precursors; matrices with intermediate stiffness that mimic muscle and induce myogenic commitment; while rigid matrices mimicking collagenous bone directly differentiate into osteogenic lineages [2]. The mechanical forces’ connection to cell biology has been understudied, but is clearly an essential component of physiology and pathology.
2. Mechanical dynamics in live cells
Living cells generate and sense force; this interaction is used for homeostasis [3]. The cytoskeleton plays a fundamental role in conducting information and sensing mechanical force. The cytoskeleton is attached to the membrane through adhesion complexes involving integrins to facilitate mechanical communication between the cell and its surroundings. These differentiated couplers can translate mechanical force into electrical or biochemical signals through conformational changes [4,5]. The cytoskeleton also serves as tracks for motor proteins, including dynein, kinesin and myosin, which is the driving force behind most active transport of proteins and vesicles in the cytoplasm, as well as cell movement. For example, myosin is responsible for sliding actin filaments and cellular contractility; and the dynein motor is responsible for the wave like motion of the cilia, or the beat of the flagella. Furthermore, force generated by the movement of motor proteins is passed on along the cytoskeleton to mechanosensors, which can perceive and translate the force, activating various downstream signaling pathways that play important roles in the regulation of cell morphogenesis and polarity. The generation and transduction of cellular mechanical force all depend on the cytoskeleton, therefore, it is classified as a structural force.
Many factors, including extracellular mechanical stimuli, osmotic pressure, movement of motor proteins and mechanosensors, have real time effects on structural force. The system forms a network for the generation, perception, and transmission, as well as the translation of mechanical forces to maintain homeostasis in a mechanically changing environment. We are calling this the mechanical dynamics of living cells.
3. Mechanical stimuli and effects
Based on the origin of the force in and on cells, it can be classified into two groups: exogenous and endogenous. The former includes gravity, compression, stretch, strain, fluid shear, etc. Endogenous forces include osmotic pressure and the movement of motor proteins. As mentioned above, these mechanical factors play vital roles in the life of cells.
3.1. Exogenous mechanical stimuli
Investigations aimed at studying the effects of exogenous mechanical stimuli on cells in situ is difficult. Studying cell mechanics in vitro requires implementing a method to mimic the force that cells undergo in their physiological environment. There are a variety of such experimental methods and some are summarized in Table 1.
Table 1.
Exogenous mechanical stimuli.
| Types of mechanical force |
Experimental techniques |
Principles/mechanisms | Parameters | Comments |
|---|---|---|---|---|
| Stretch/strain | Substrate stretch [6] | Stretching of the substrate in two or three dimesions strains the cells or tissues. This will affect cell growth and differentiation. Engineered tissues that are mechanically stretched may have improved organization and functionality |
Spatial extent, waveform | Provides spatial homogeneity of strain and multiple parameters to modify force. |
| Magnetic fields [7] | Magnetic microspheres are bound to specific cellular receptors and exert linear stress or torque on the cell cortex when exposed to magnetic fields. |
Amplitude and the direction of the magnetic flux |
Can provide constant or variable stimuli and precise quantification. |
|
| Optical stretcher [8] | Laser traps exert force on beads attached to cells. | Percent of reflected laser beam, the total laser power |
No physical contact with the sample, able to apply force to sub pN range. Possible photo damage effect on biological sample at high laser power, the limited maximum force. Maximal forces are in the range of 10s of pN. |
|
| Micropipette aspiration [9] | Micropipette is used to aspirate a single cell, stretching the cortex monitored by changes in cell dimensions and the elongation of the region sucked into the pipette. Data is recorded by video microscopy. |
Diameter of the micropipette, suction pressure and waveform |
Able to measure the cortex elasticity and viscosity. Able to determine the lipid and protein distribution under stress with fluorescence techniques. Stress concentration at the pipette edge and pipette friction may interfere with the mechanical response of the cell during aspiration. |
|
| Stiffness of substrates [10,11] |
Stiff substrates | A stiff matrix resists cellular force more than a soft one. This leads the cell to be more rigid and extend about its periphery. Most cell types, including endothelial cells, myocytes and fibroblasts spread further, adhere better, and appear to survive better on stiffer matrices, and some cannot grow on soft surfaces. |
The substrates’ elastic modulus | Substrate stiffness modifies cell motility, adhesion, differentiation, and other cytoskeleton dependent activities. Studying the relationship between phenotypes and physical environment is important; not only for designing implantable devices, but also for understanding pathological sequelae that |
| Soft substrates | A soft substrate is more compliable, exerting less resistance to cells and leading to diffuse and dynamic adhesions. Some cell types, such as neurons, extend processes more avidly and appear to survive better on soft substrates. |
follow insult or disease progression. | ||
| Shear force [12] | Cone-and-plate flow chamber |
Rotation about a cone’s axis that is oriented perpendicular to a flat plate immersed in fluid generates spatially homogeneous fluid shear stress to cells on the plate. |
Angular, velocity of cone rotation | The conic taper and angular velocity provide a wide range of shear stress. |
| Parallel plate flow chamber |
A pressure differential is created at the ends of two parallel plates, creating uniform laminar shear force. |
Pressure differential between the inlet and outlet manifolds |
Homogeneity of the stress, simplicity of the equipment, ease of medium sampling/exchange, and small volumes. |
|
| Indentation | Atomic force microscopy [13–15] |
Controllable indentation on the top of cells. | Topography; stiffness; force | Imaging of cell surface morphology and membrane topography; probing of various properties and interactions at single molecule and single-cell levels; ligand–receptor-interaction, molecular recognition; and single cell manipulation. |
| Optical trapping/laser tweezer [16] |
Focus a laser to a diffraction-limited spot with a high numerical aperture microscope objective. |
Energy gradient at focus; laser power; polarization size and composition of the bead |
Study particles ranging from 20 nm to several micrometers and even whole cells and lipid vesicles. |
|
| Hydrostatic pressure [17] | Cells in the culture chamber are compressed by hydrostatic pressure. |
Magnitude, frequency | Simplicity of the equipment, spatial homogeneity of the stimulus, ease of configuring multiple loading replicates. Not well suited for long term experiments due to the changes of pH, pO2 and pCO2 [18]. |
|
| Platen abutment pressurization [19] |
Tissue explant or cells seeded in a matrix or carrier are directly compressed by a platen. Compression of a three dimensional matrix containing cells. |
Closely approximates the physiologic situation of minimal transverse strain at an articular surface and provides a wide range of cell deformations. Non-homogeneity of mechanical stimuli about the cells. |
3.2. Endogenous mechanical stimuli
3.2.1. Movement of motor proteins
Motor proteins are a class of molecular motors, consisting of dynein, myosin and kinesin, that are able to move along the cytoskeleton. They play a major role in bidirectional transport in cytoplasm, which is essential for cell physiology, plasticity, morphogenesis, and survival [20]. They also link chemical catalysis to the production of directed force along protein filaments [21].
Dynein superfamily proteins are mechanoenzymes that move along microtubules, and they are comprised of two major groups: cytoplasmic dyneins and axonemal dyneins (also called ciliary or flagellar dyneins) [22]. Dyneins operate as complexes built around force-generating sub-units called heavy chains, which contain the motor domain. The tail specifies oligomerization properties and serves as a platform for the binding of several types of associated subunits, which in turn mediate interactions with cargo either via direct binding or through the recruitment of adaptor proteins. Dynein also has an important associated protein complex called dynactin, which regulates dynein activity and the binding capacity of dynein for its cargos [23]. Cytoplasmic dynein performs a variety of cellular functions including: (1) Cytoplasmic dynein powers the transport of membrane bound vesicles and tubules, together with their resident molecules toward microtubule minus ends [24]. (2) Dyneins tethered to the cell cortex can apply a pulling force on the microtubule network by either walking toward the minus end of a microtubule or coupling to a disassembling plus end. This force is essential to cell division [25–27]. (3) At the outer nuclear envelope, dynein has been reported to contribute to nuclear rotation [28] and positioning [29], centrosome separation [30], and the breakdown of the nuclear envelope for open mitosis [31]. (4) At cell division, cytoplasmic dynein assists in assembling microtubules into the chromosome-segregating device known as the spindle [32,33]. (5) Cytoplasmic dynein localizes to the kinetochore; this dynein has an important role in the molecular surveillance mechanism that aids faithful chromosome segregation [34]. Dysfunctions of cytoplasmic dynein and dynactin contribute to many neurodegenerative and neurodevelopmental diseases, including short-rib polydactyly syndrome [35,36], motor neuron disease, ALS [37–39], lissencephaly [40,41], Alzheimer’s disease [42], etc.
The kinesin superfamily proteins (KIFs) comprise three major groups based on the position of the motor domain: N-terminal motor domain KIFs (N-KIFs), middle motor domain KIFs (M-KIFs), and C-terminal motor domain KIFs (C-KIFs) [43]. N-KIFs and C-KIFs are composed of a motor domain, a stalk domain, and a tail region. The motor domain consists of ATP- and microtubule-binding sites which enable it to bind to microtubules and to move them along by hydrolyzing ATP. In general, the tail regions, and less frequently the stalk regions, recognize and bind to the cargo(s) [20,43,44]. Kinesins play a major role in intracellular transport and they can be classified into many groups based on the cargos transported and the location of the transport activity [43]: (1) Anterograde axonal transport, such as synaptic vesicle precursor and mitochondrial transport along the axon. (2) Dendritic transport in neurons, like the transport of NMDA and AMPA receptors and mRNA. (3) Conventional transport, including transport between the endoplasmic reticulum and Golgi apparatus, lysosomal transport, transport from the trans-Golgi network to the plasma membrane, and endosomal recycling. KIFs are also closely involved in various diseases, such as kinesin-1 in spastic paraplagia [45,46], amyotrophic lateral sclerosis (ALS) [47,48] and Alzheimer disease [49–51]; kinesin-3 in Char-cot–Marie–Tooth disease [52,53] and multiple sclerosis [54,55]; kinesin-4 in congenital fibrosis of the extraocular muscles type 1 (CFEOM1) [56], etc.
Myosins are the only known actin-based motor proteins[57] and are classified into eighteen classes [58]. Most myosins form a dimer, consisting of a conserved catalytic motor domain (head) with actin-and ATP-binding sites, a neck region and a tail region that binds to cargos [57,59]. Myosins have various functions: (1) They are mechanoenzymes and their conformational changes associated with nucleotide binding, hydrolysis, and product release are crucial for the productive motility of myosin enzymes [60]. (2) Myosins, particularly the myosinII, have long coiled-coil domains that allow multimerization to take place. Thus, interactions between charged residues within these dimers mediate the formation of bipolar thick filaments that are responsible for contraction of muscle and cytokinesis [61,62]. (3) Myosins participate in many trafficking and anchoring events, such as directed migration of pigment containing vesicles involved with myosinVa in melanocytes [63] and correct targeting of the megalin receptors involved with myosinVI [64]. (4) Myosins play a role in actin-based projections including stereocilia and microvilli, which are essential for the normal function of hair cells in the inner ear and epithelial cells in the intestine and kidneys [65]. As for pathology, abnormalities of myosins are key factors in the development of diverse diseases, including autosomal dominant deafness (Myosin I) [66,67], May-Hegglin anomaly, Fechtner syndrome and Sebastian syndrome (Non-muscle Myosin II) [68,69], autosomal recessive deafness (Myosin III) [70], Griscelli syndrome and microvillus inclusion disease (Myosin V) [71,72], Usher syndrome type IB and unsyndromic deafness (Myosin VII) [73,74], loss of heterozygosity in lung cancer (Myosin XVIII) [75], etc.
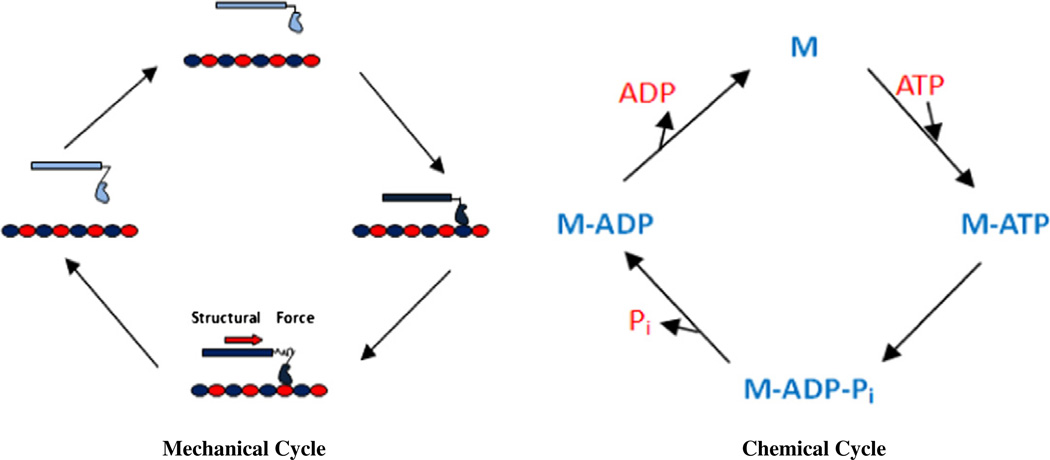
The mechanism of force production by motor proteins is thought to involve structural changes in a deformable element of the motor that undergoes changes in structure under load, which creates strain followed by a strain-relieving structural change that causes the element to recoil back into its original conformation, producing force. In this process, motor proteins couple a chemical cycle of ATP hydrolysis to a mechanical cycle of motor interactions with its filament by the motor (Fig. 1) [21,76]. Take dynein for example; ATP induces dissociation of the motor–microtubule complex [77]. After detaching from the microtubule, the motor rearranges, becoming primed for a subsequent structural change (termed the powerstroke) that is thought to generate the force. Following a diffusive search, rebinding of the motor to a new site on the microtubule stimulates the release of ATP hydrolysis products, thus triggering the powerstroke [78]. Mechanical force produced by this mechanism is not only responsible for the cytoplasmic transport, but also essential for the generation of mechanical effects on the cytoskeleton; which can change its structure and eventually the shape of cells, making them most adaptable to the mechanically changing environment.
Fig. 1.
Motor mechanochemical cycles. M: motor protein M-ATP: motor protein binding with ATP M-ADP: motor protein binding with ADP Mechanical (left) and chemical (right) cycles of a simple ATP-fueled molecular motor protein; coupling of the mechanical and chemical cycles produces the motor force-generating cycle.
3.2.2. Osmotic pressure
Concentration differences of diffusible solutes between cytoplasmic and extracellular environments produce hydrostatic pressure that exerts mechanical stimulation on constraints such as the bilayer and the cytoskeletal gel. That leads to deformation of cell morphology and changes in lipid bilayer tension bymodulating the shrinkage or swelling of cells. In this process, both ion channels and aquaporins (AQPs) play a major role, by gating the influx and outflow of ions and water, leading to shape change of cells, which eventually exerts mechanical force to cells [79,80]. Among ion channels, TRP channels and 2P domain K+ channels are the most important, which can be activated by hypertonicity [81], hypotonicity [82] or membrane stretching [83]. In addition, as mechanosensitive ion channels, they can also function as mechanosensors. AQPs play a central role in the flow of water across a cell membrane; cell volume regulation (CVR) is a necessary mechanistic component, which comprises regulatory volume decrease (RVD) in response to hypotonicity induced cell swelling and regulatory volume increase (RVI) that responds to hypertonicity induced cell shrinkage [84]. In the traditional model of RVD, the activation of K+ channels allows efflux of K+ from the cells and subsequent water loss by osmosis either through AQPs or directly through the lipid bilayer. However, we must note that an electrogenic ion flux cannot change the osmotic pressure because the membrane potential will limit ion flux to a negligible proportion of the resident ions. Cell volume can only be changed by electroneutral fluxes such as a combined flux of K+ and Cl−. In RVI, the activation of Na+–H+ exchangers and Na+–K+–2Cl− co-transporters (NKCCs) cause cellular influx of Na+ and subsequent volume increase by osmotic movement of water [80,85,86]. An important omission from this model is the elastic stresses in a cross linked cytoskeleton as water fills or empties. This process is known as poroelasticity. To add water, you must stretch the cytoskeleton to make room, and that work raises the hydrostatic pressure of water within the pores of the cytoskeleton. Most of the energy produced by exposing cells to osmotic pressure is distributed in three dimensions within the cytoskeleton and not concentrated at the cortex as in the traditional model [87–89].
4. Mechanosensors
Mechanosensory proteins have two characteristics: physiological levels of mechanical force can lead to their conformational changes; the conformational changes can activate downstream signaling pathways. This translation of mechanical stimuli into chemical or electrical signals modulates the biochemical and electrophysiological activity. Mechanotransduction is essential for many sensory functions, such as hearing, sensing, touch, balance, proprioception, and visceroception. Mechanosensors can be classified into the following four groups based on their distribution and structures.
4.1. Mechanosensitive ion channels
Mechanosensitive ion channels (MSCs) form a special group of mechanosensors that can serve as both sensors and effectors as they modify the electrical potential of the cell and mediate a flux of ions across the plasma membrane. They detect and transduce external mechanical force into electrical and/or chemical signals. MSCs appear to be present in all types of animal cells, including red blood cells [90]. Here, we focus mainly on MSCs in eukaryotes, including the cation selective group of the TRP (transient receptor potential) superfamily and Piezo channels, the Na+ selective DEG/ENaC superfamily, and the K+ selective 2P–domain-channels [91,92].
4.1.1. TRP channels
TRP channels are composed of four subunits and each contains six putative transmembrane segments (S1 to S6) with a pore-forming re-entrant loop between S5 and S6. TRP channels are present in all cellular membranes with the exception of the nuclear envelope and mitochondria. Most TRP channels are localized in the plasma membrane, where they have an essential role in the influx and/or transcellular machinery that transports Ca2+, Mg2+ and trace metal ions, modulating the membrane potential [93].
Several mechanisms that lead to TRP channel activation in response to mechanical stimuli have been described [94]: (1) Shear stress by fluid flow physically that bends a primary cilium leads to activation of TRP channels. Subsequent Ca2+ influx affects vascular smooth muscle cell (VSMC) cytoskeletal dynamics by inducing the release of vasodilators, such as NO and endothelium-derived hyperpolarizing factor (EDHF). (2) Shear stress and force applied directly to the cell–matrix or cell– cell adhesions locally activate TRP channels by interacting with adhesion molecules, such as integrins. TRP channel-mediated Ca2+ influx affects cell–matrix and cell–cell adhesion turnover by regulating cytoskeletal contractility; modulating the activity of FAK (cell–matrix) and activating the Ca2+ dependent proteases like calpain that cleaves adhesion plaque molecules. (3) Mechanically activated G-protein coupled receptors (GPCRs), such as the angiotensin II type 1 receptor, can trigger TRP channel opening through activation of PLC signaling. Subsequent Ca2+ influx activates the calcineurin/NFAT pathway, leading to changes in transcription of genes involved in cytoskeletal dynamics.
4.1.2.2P domain K+ channels
The 2P domain K+ channels each comprise four transmembrane (TM) segments and two pore domains in tandem with an extended extracellular loop between M1 and P1. They function as dimers in which both N- and C-termini face the cytosol. The four pore domains of a dimer form the aqueous pore that allows passage of ions through these channels [90]. The 2P domain K+ channels can be divided into six main functional classes; such as, TREK-1, TREK-2 and TRAAK, which are mechanogated. The transmembrane segments (primarily the fourth one) play an essential role in the mechanosensitivity, while some COOH-terminal tail fragments are also necessary for proper function [95–97].
The relationship between channel activity and membrane tension is described by a sigmoidal Boltzmann relationship [98,99]. Mechanical force may be transmitted directly to the channel via tension the lipid bilayer that may not require the integrity of the cytoskeleton, or possibly by direct mechanical links to the cytoskeleton. The cytoskeleton also acts to modulate the tension in the bilayer, allowing a dynamic tuning of the stretch sensitivity [95,100]. Opening of these channels by membrane stretch hyperpolarizes, and thus decreases neuronal excitability [101], and this emulates, perhaps specifically, the mechanism of the “knockout punch”. These channels display the common property of MSCs to be sensitive to amphipaths that can alter the local stress about the channels. This has been well examined by Honore’s group who showed that channel activation by general anesthetics occurs at concentrations that match those used for anesthesia [102].
4.1.3. DEG/ENaC
The DEG/ENaC proteins have two transmembrane domains with intracellular amino and carboxyl termini, and a large extracellular loop including a conserved cysteine-rich region [103]. The channels are generally selective for Na+ and can be blocked by the diuretic amiloride.
The DEG channel is formed of the MEC-4, MEC-6 and MEC-10 membrane proteins, and the integral membrane protein MEC-2. MEC-2 connects the MS channel to microtubules. The MEC-1 and MEC-5 proteins comprise the structural mantle which can be pushed by shearing or bending and results in the opening of the channels and depolarization and potentially action potentials. MEC-9 is an extracellular protein, which is proposed to form the gating spring between the MS channel and the mantle. MEC-2 and MEC-9 are thought to exert pressure on the channel, conferring gating stress when the channel is stretched between these two contact points [104–106]. However, the coupling of the mechanical stimulus to channel gating remains an open problem.
4.1.4. Piezo channels
Piezo1 (Fam38A) and related Piezo2 (Fam38B) are vertebrate multipass membrane proteins, containing about 2100 to 4700 amino acids, with an estimated 24 to 39 TM segments. Piezo proteins form tetramers, and these appear to form functional channels. Piezo proteins are expressed in many tissues including bladder, colon and lung [107–109]. Piezo2 is enriched in dorsal root ganglion (DRG) neurons [91]. The conformational changes of Piezos when activated by stretch is currently unknown, although the dimensional changes between closed and open have been measured to be in the range of 20 nm2 [98]. The research on Piezo channels is quite young and more detailed measurements are needed to establish the mechanism by which the Piezo channels are activated by mechanical force [110].
It is clear that MSCs play a vital role in the modulation of physiological activity and cells’ adaptation to a mechanically changing environment. Meanwhile, abnormalities of MSC, most likely produced by modulation of membrane stresses by the cytoskeleton as well as channel mutations, contribute to dysfunctions and diseases, including neuronal and muscular degeneration, muscular dystrophy [111], cardiac arrhythmias, hypertension and polycystic kidney disease [97]. For example, the TRPV4 gene is prone to many mutations that cause malfunctions of the encoded Ca2+ permeable cation channels; which in turn mediate skeletal dysplasia and motor and sensory neuropathies [112], such as brachyolmia [113], spondylometaphyseal dysplasia Kozlowski (SMDK), spondylo-epimetaphyseal dysplasia maroteaux pseudomorquio type 2 (SEDM-PM2) [114], congenital distal spinal muscle atrophy, scapuloperoneal spinal muscle atrophy [115,116] and Charcot-Marie-Tooth disease type 2C (CMT2C) [117].
4.2. Membrane mechanosensors
We will use the term “mechanosensors” to refer to sensors bound to cell membrane with transmembrane domains. The mechanosensors may link the extracellular matrix (ECM) with intracellular structures like the cytoskeleton or function as bilayer bound sensors like ion channels.
4.2.1. Endothelial glycocalyx
The endothelial glycocalyx (GCX) is a network of membrane-bound proteoglycans and glycoproteins, covering the endothelium lumina. Both endothelium- and plasma-derived soluble molecules integrate into this complex. Bound to the endothelial membrane are proteoglycans (PG), with long unbranched glycosaminoglycan side-chains (GAG-chain); and glycoproteins, with short branched carbohydrate sidechains. GAG includes heparin sulfate (HS), chondroitin sulfate (CS) and hyaluronic acid (HA). In addition, heparin sulfate proteoglycans (HSPG) including syndecan family and glypican family, can connect to HS and CS and play an essential role in transduction of shear force [118].
The extended GAGs are drag sensors that transmit shear force from the flow to the core proteins. The fluid shear force dissipated in the outer region of the glycocalyx imposes a torque on the relatively stiff core proteins that are transmitted to the actin cortical cytoskeleton via transmembrane domains [119]. There are two mechanisms of the downstream effects: the decentralized and centralized [120].
In the centralized mechanism, the former mechanotransduction activates PKCα regulated by Syndecans, which have an established association with the cytoskeleton. PKCα can mediate activation of eNOS by phosphorylation at residue Ser1179 and increase NO production [121]. Also, glypicans reside in caveolae along with inactive eNOS, G-protein-coupled receptors, G-proteins, mitogen-activated protein kinases, PKC isoforms, the L-arginine recycling enzymes and its transporter, and a number of other eNOS interacting proteins having transient associations with caveolin-1; such as heat shock protein and protein kinase A, both of which are linked to the shear-NO response [122,123].
In the decentralized mechanism, syndecans can decentralize the signals through cytoskeleton by spreading them to multiple sites within the cell, such as nucleus, organelles, focal adhesions, and junctions. For example, at the cell–cell border between platelets-endothelial cells, the adhesion molecule (PECAM) associates with the cytoskeleton through catenins and has been linked to shear-induced eNOS activation [124].
As for the pathology, GCX deterioration is associated with the onset of endothelial dysfunction, a phenomenon that is involved in many disorders, including atherosclerosis [125], diabetes [126,127] and ischemia/reperfusion [128].
4.2.2. Integrins
Integrins are expressed on almost all cell types in a widely varying pattern. The integrin family represents a major class of heterodimeric, transmembrane cell surface receptors; these comprise twenty-four subtypes formed out of eighteen different α and eight different β subunits. The two subunits are associated noncovalently and consist both of an extracellular, a transmembrane, and a short cytoplasmic domain [129].
Integrins enable adhesion, proliferation, and migration of cells by recognizing binding motifs in extracellular matrix proteins. Of all the mechanosensors, integrin not only can sense the mechanical stimuli, but also it is the only one that presents specificity for ligands. Structural and molecular studies have indicated that integrin adhesions associate with the actin cytoskeleton. As transmembrane linkers between the cytoskeleton and the ECM, they are able to recruit a huge variety of proteins and bidirectionally influence signaling pathways (Fig. 2), affecting regulation of gene expression and cell survival [4,130,131].
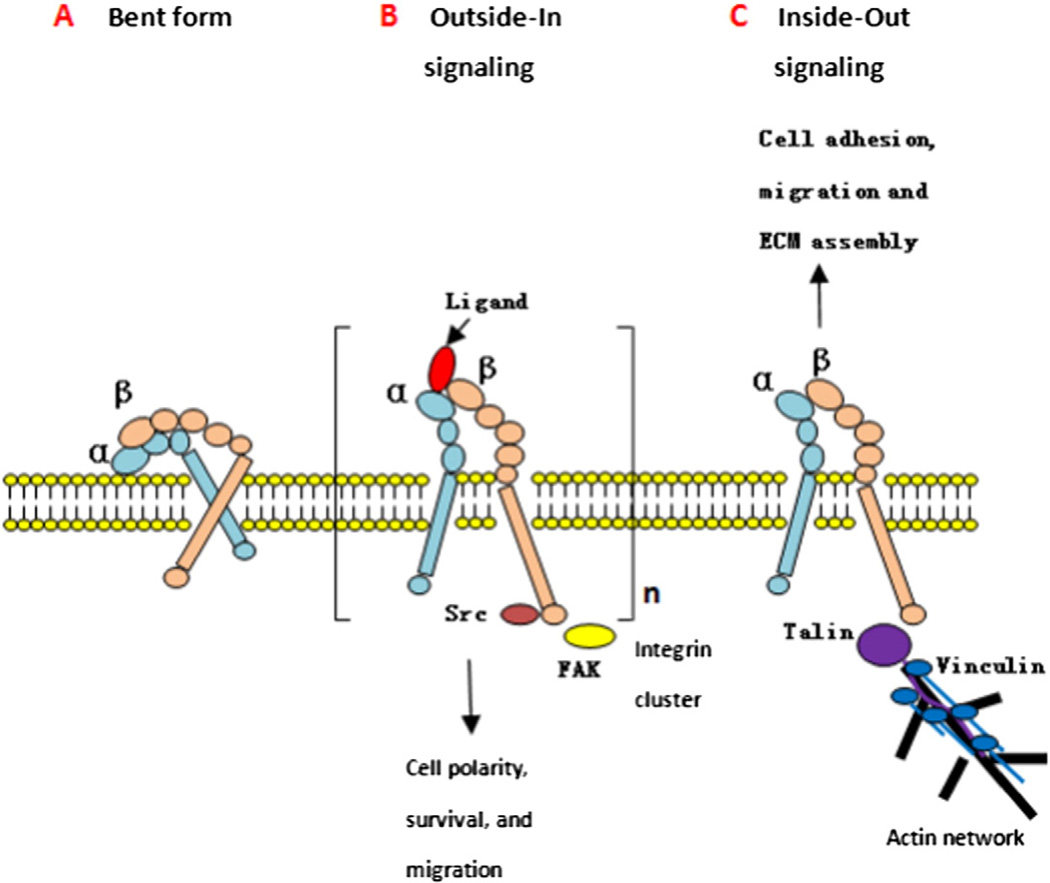
Fig. 2.
Bidirectional activation of integrin. A. Inactive state: the integrin head group points toward the cell surface and has a low affinity for ligands. B. Outside-in signaling: a ligand binds to the integrin and can induce, because of multivalency and integrin clustering. Activation of a signal cascade leads to intracellular signals, which regulate cell polarity, survival, migration, changes in cytoskeleton and gene expression. C. Inside-out signaling: an intracellular activator binds to the β-subunit, induces a conformational change leading to increased affinity for extracellular ligands. This process is known to regulate cell adhesion, migration, and invasion.
In the inactive state (Fig. 2A), the integrin headpiece is in a bent conformation with TM domains of α and β subunits associated together, pointing toward the membrane, thus possessing low affinity for ligands [132].
During outside-in signaling (Fig. 2B), ligand binding leads to dissociation of the transmembrane units and induces integrin clustering, forming so-called focal adhesions as a consequence of linkage of the intracellular domain to the actin cytoskeleton via talin and vinculin [133]. FAK and Src are recruited to β integrin cytoplasmic tail and activated, which then phosphorylate scaffold protein paxillin and p130Cas. Phos-phorylated p130Cas recruits guanine exchange factors DOCK, leading to Rac activation. ILK is also recruited to β integrin cytoplasmic domain and promotes Akt activity. Src also activates Akt and can promote cell proliferation through the Ras–Erk pathway [134]. Activation of these signal cascades play key roles in regulating cell polarity, survival, migration, changes in cytoskeleton, and gene expression.
During inside-out signaling (Fig. 2C), binding of proteins to the intracellular domain of the β-subunit can induce a conformational switch in the extracellular domain, accompanied by increasing affinity for ligands and binding (activation). This process is known to regulate cell adhesion, migration, and invasion. The specific binding to the cytoplasmic tail of integrin’s β subunit by the intracellular protein talin is the key step of inside-out signaling, and talin is an important intracellular tension sensor.
Integrins are involved in many pathological processes including periodontal diseases like periodontitis, hematological disorders such as Glanzmann’s thrombasthenia and LAD-III, muscular dystrophies, cardiovascular diseases like atherosclerosis, skin blistering disorders, and cancer [135].
4.2.3. Primary cilium
The primary cilium is observed in many cell types including renal tubular epithelia. It functions both as a mechanosensor and a chemosensor in renal tubular epithelia, and is involved in the determination of left-right sidedness during development. It is also a key factor in the development of polycystic kidney disease as well as a number of other abnormalities. The primary cilium has a so-called 9 + 0 axoneme, which refers to its nine peripherally located microtubule pairs and the absence of the central microtubule pair seen in 9 + 2 cilia [136].
In renal tubular epithelia, primary cilia act as flow sensors. Fluid shear force or direct bending of the cilium causes a calcium influx, which can spread from the stimulated cells to its neighbors by diffusion of a second messenger through gap junctions, leading to the downstream physiological effects [137]. Flow sensing is shown to reside in the cilium itself and to involve the proteins polycystin 1 and 2, defects that are associated with the majority of cases regarding human polycystic kidney disease. Some studies also suggested that the primary cilium plays a role in flow-dependent potassium secretion through the collecting tubule as well as sensing the chemical components of the luminal fluid. In the chondrocyte, primary cilium plays a role of detection and transmission of mechanical stimulation, in which the process mechanical loading bends the primary cilium and activates calcium influx. Additionally, primary cilium serves as a compartment for many signaling pathways that are involved in mechanotransduction in the chondrocyte, including integrins, Ihh, ion channels, connexins and cAMP second messenger signaling [138,139].
Mutations disrupt ciliary function and lead to distinctive developmental and/or degenerative phenotypes in the retina, kidney, nervous system and so on, such as osteoarthritic (OA) cartilage, primary ciliary dyskinesia (PCD), Leber congenital amaurosis (LCA) and retinitis pigmentosa (RP) [140–142].
4.3. GPCR AT1R
It has been proposed that G-protein-coupled receptors (GPCR) may play a role in mechanotransduction, besides neurohormonal signaling. The Gq/11-coupled angiotensin II AT1 receptor (AT1R) was the first GPCR claimed to be a mechanosensor in cardiac cells, smooth muscle cells of the small renal, and cerebral resistance arteries [143,144]. The structure of the AT1R is characterized by seven-transmembrane spanning α-helices with an extracellular N-terminus and a cytoplasmic C-terminus [145].
Mechanical stress, such as a rise of intravascular pressure, exerts a stretch force on the membrane which can induce a dislocation and an anticlockwise rotation of transmembrane domain 7 toward the agonist-binding pocket. Then, transmembrane domain 6 dislocates and rotates out of the binding pocket, thus increasing the distance between trans-membrane domains 3 and 6 [146,147]. This membrane-stretch-induced conformational change leads to activation of Gq/11-coupled receptors, PLC/PLA2 and subsequently, TRPC channels, resulting in depolarization of the membrane potential. Finally, depolarization causes an increase in the open probability of L-type voltage-gated calcium channels, which results in calcium entry and an elevation of the free intracellular calcium concentration leading to force development, cell shortening and vasoconstriction (Fig. 3) [144,148]. With regard to how the AT1 receptor senses mechanical stress and undergoes a conformational switch, there are two reasonable mechanisms: (1) Membrane tension causes thinning of the lipid bilayer but not more than ∼ 1% [149], which may trigger tilting of TM7 of AT1 receptor to avoid hydrophobic mismatch and to rectify lateral pressure profile [150]. (2) Mechanical stretch may activate specific mechanosensors, such as integrin-linked kinase [151], melusin [152], and stretch-sensitive ion channels [150,153], which can secondarily activate the AT1 receptor.
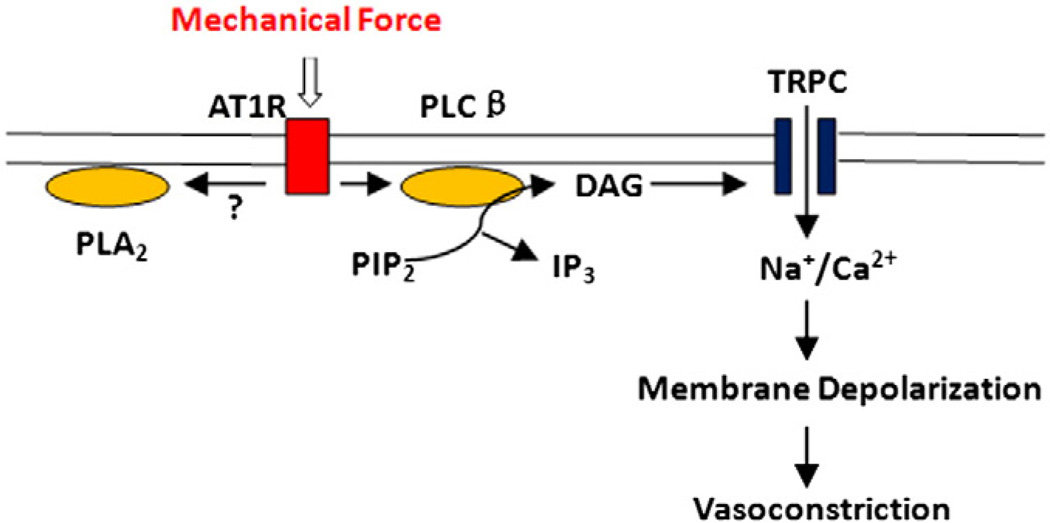
Fig. 3.
Mechanical force induced AT1R activation and downstream effects. TRPC channels are activated by mechanical stimulation of AT1R via a PLC-dependent signaling pathway, resulting in membrane depolarization, opening of L type voltage-gated calcium channels (not shown), and elevation of the free intracellular calcium concentration; which leads to force development, cell shortening and vasoconstriction.
Constitutively active mutants (CAMs) of AT1R are responsible for hypertension, cardiac hypertrophy and hyperaldosteronism, which causes vasoconstriction and aldosterone secretion [154].
4.4. Intracellular mechanosensitive proteins
4.4.1. p130Cas
p130Cas (Cas) acts as a primary force-sensor and is involved in various cellular events such as migration, survival, transformation, and invasion. The domains of p130Cas include Src homology 3 (SH3) domain, the proline-rich region (PR), the substrate domain (SD), the serine-rich region (SR), the Src-binding domain (SB), and the C-terminal region. The SD is located in the center of Cas and is flanked by the SH3 and the SB, which anchor Cas molecules to the cytoskeletal complex in order that the SD could be extended upon cytoskeleton stretching. The SD has fifteen characteristic repeats of a tyrosine-containing motif (YxxP) for Src family kinases (SFKs). The intramolecular interactions within the SD constrain its conformation in the absence of traction force, which is required to expose YxxP sites to kinases [155–158].
The mechanical stretch experienced by the cell, such as shear stress from blood flow, or cytoskeletal stretch in spreading cells, extends p130Cas and increases exposure of YxxP repeats within the p130Cas substrate binding domain, structurally priming these sites for tyrosine phos-phorylation [158]. Crk adaptor proteins bind phospho-tyrosine residues within the YxxP motifs and link p130Cas to downstream effectors, such as the guanine nucleotide exchange factors DOCK180–ELMO (Engulfment and cell motility) and C3G, which catalyze the activity of the small GTPases Rac and Rap respectively. This process of signal transduction brings about changes in the actin cytoskeleton to facilitate cell motility for migration and invasion [159–161]. Protein tyrosine phosphatase-PEST (PTP-PEST) associates with the SH3 domain of p130Cas and dephos-phorylates phospho-tyrosine residues, resulting in decreased p130Cas– Crk association and decreased pro-migratory signaling [162]. In addition, p130Cas functions as a mechanosensor via the interaction with the integrin. The generally short non-catalytic cytoplasmic domains of integrins associate with adaptor proteins, including p130Cas, to transduce outside-in signaling from the ECM [163]. Also, integrins containing sub-units α7, β1, and β3 have been shown to induce p130Cas phosphorylation, allowing signaling regulation in response to ECM composition, which is required for normal cell spreading and migration on various ECM substrates [164,165].
Due to the varied physiological significance of p130Cas, its dysfunction can lead to many diseases such as myocyte hypertrophy, hypertension, pulmonary hypertension and vasculoproliferative pathologies, like atherosclerosis in the cardiovascular system, cystic kidney disease, and familial juvenile nephronophthisis in the urinary system. It can also increase the invasiveness of cancer cells [166].
4.4.2. Filamins
Filamins (FLN) are conserved, modular, multidomain cytoplasmic proteins that are well positioned to serve as mechanosensors. In nonmuscle cells, filamins colocalize with F-actin along stress fibers at the cell cortex, and along the leading edge. Filamins are also enriched at the end of actin stress fibers and at the leading edge, the trailing ends of mature focal adhesions concentrating in adhesion sites after application of force to cells [166–168].
The N-terminal actin-binding domain (ABD) containing two calponin-homology domains (CH1 and CH2), allows filamins to bind to filamentous actin and induce potent actin filament gelationis. ABD is followed by 24 immunoglobulin-like domains (IgFLN), repeats of ∼96 amino acids each. The repeats are interrupted by two hinge regions (Hinge 1 and Hinge 2) and fold into antiparallel β-sheets. The C-terminal twenty-fourth repeat is in the dimerization domain. Repeats 1–15 make up rod domain 1, and repeats 16–24 make up rod domain 2 [168–170].
By interacting with transmembrane receptors, filamins provide a mechanical link between the ECM, the plasma membrane, and the actin cytoskeleton. Filamins cross-link and anchor actin filaments, regulate actin cytoskeleton remodeling, stabilize the plasma membrane, and contribute to the mechanical stability of the cell cortex [170,171]. In addition, filamins are important for tuning cellular responses to ECM stiffness [172] and other mechanical forces. IgFLN domains undergo conformational changes and unfolding, allowing filamin to extend its length, and in turn, protect the linkage between the membrane and the cytoskeleton [173,174]. The interdomain interactions among IgFLNa16–21 can mask ligand binding to the CD-face of IgFLNa19 and IgFLNa21. Pulling forces transmitted through filamin are locally targeted to the masking β-strand, causing it to open in a zipper-like fashion and facilitate binding of other ligand proteins, which, in turn, can lead to downstream signaling pathways [175,176]. For example, FLNa associates with a wide range of signaling molecules, including the small GTPases Rac, Rho, CdC42 and RalA, and factors upstream and downstream of the GTPases [177–179].
There has been a lot of information about diseases caused by filamin mutations. There is an X-chromosome-linked brain malformation known as periventricular heterotopia, which results from FLNa loss-of-function mutations [180]. In addition, specific missense mutations of FLNa can lead to four other X-linked human disorders, including otopalatodigital syndrome types 1 (OPD1) and 2 (OPD2), frontometaphyseal dysplasia (FMD) and Melnik-Needles syndrome (MNS) [181]. Moreover, mutations in FLNb disrupt bone morphogenesis, such as Spondylocarpotarsal syndrome (SCT), Larsen syndrome and Atelosteogenesis I and III [182].
4.4.3. Talin/vinculin
Talin is a highly abundant cytosolic protein important for cytoskeleton organization and cell–ECM adhesion [183]. Talin is made of a head domain containing a F0 domain followed by a FERM (four-point-one, ezrin, radixin, moesin) domain divided into three subdomains (F1, F2 and F3). F2 and F3 form an actin filament binding domain (ABD), while F3 interacts with the cytoplasmic tail of the subunit of integrins. The head domain is followed by a rod domain divided into thirteen helix bundles. The rod contains eleven vinculin binding sites (VBSs), a central ABD, and a C-terminal ABD (THATCH) [184,185]. Structural mapping experiments also have shown that an interdomain complex between talin-F3 and the C-terminal rod fragment (talin-RS) represents the principal structural unit for talin autoinhibition [186]. Vinculin is a ubiquitously expressed actin binding protein (ABP), made of a head domain (Vh) and a tail domain (Vt) separated by a flexible linker. In the inactive state of vinculin, the intramolecular interaction between Vh and Vt prevents Vt from interacting with actin filaments. The activation of vinculin requires a concomitant binding of talin to Vh and actin filaments to Vt [187–189].
Talin has long been recognized as a mechanical linker between the ECM and actin cytoskeleton to regulate cell adhesion and morphology. Furthermore, the binding of talin-FERM to integrin β cytoplasmic tails can promote the conversion of integrins from a low-affinity into a high-affinity ligand-binding states; a dynamic process termed integrin inside-out signaling or integrin activation (Fig. 4) [190–192].
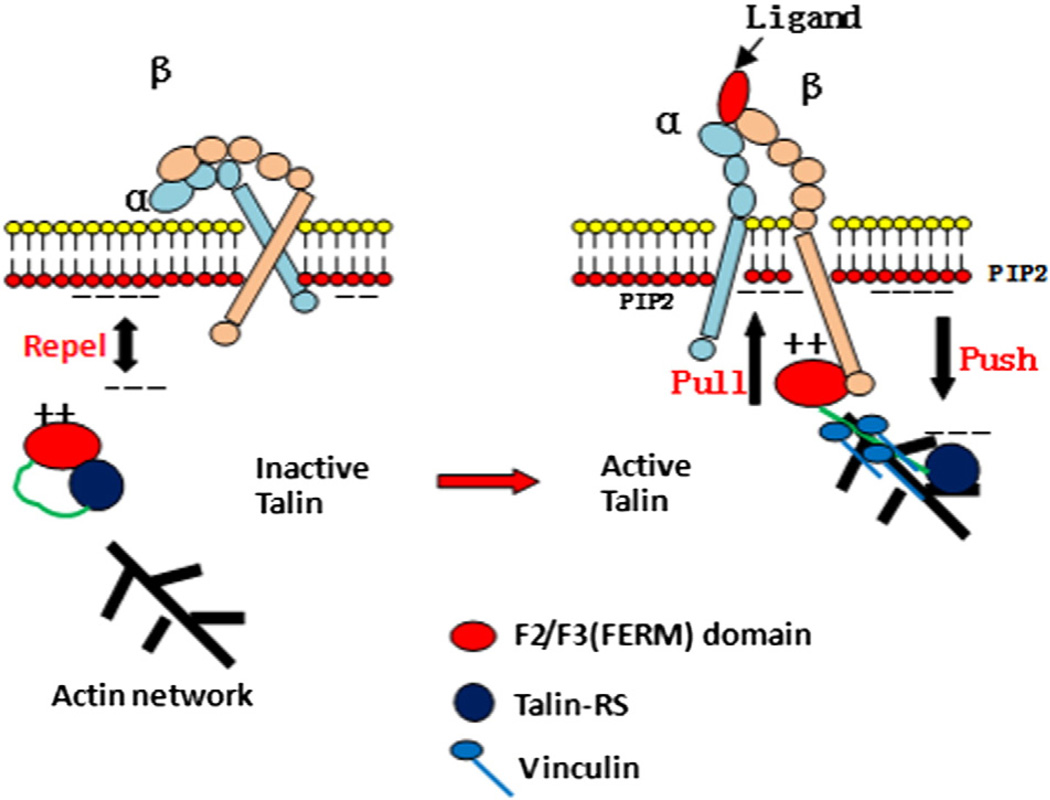
Fig. 4.
The role of talin in integrin inside-out signaling. In unstimulated cells, talin is autoinhibited and hindered from accessing the membrane. Local enrichment of PIP2 activates talin via an electrostatic pull–push mechanism. Activated talin is able to bind to integrin and vinculin, forming a link between FAs and actin.
In an inactivated state, the α and β cytoplasmic tails are in proximity, which constrains the integrin to a low-affinity conformation. The talin-RS domain binds to the talin-F3 domain, preventing the F3 domain from accessing the β-tail of integrin. At the same time, the negatively charged surface of talin-RS repels the talin-F2F3/talin-RS unit away from membrane [186,190].
This autoinhibition can be disrupted by talin activator, PIP2. When the membrane is locally enriched with PIP2, the binding affinity of PIP2 to positively charged talin-F2F3 is so strong that the repelling force from negatively charged talin-RS on the same face can no longer prevent the talin-F2F3 from docking on the membrane to access integrin β-tail. Instead, the talin-F2F3/talin-RS interface may cause mismatch by the repelling force from the membrane on talin-RS. This pull– push mechanism enables the β-tail-binding region of F3 to be available for interaction with integrins, causing dissociation of the two tails from each other and integrin activation [190–192]. In addition, the force applied on a fragment of the talin rod domain breaks the structure to expose VBSs for the binding of vinculin head (Vh) and induces the release of the actin binding tail of vinculin (Vt) [193,194]. Vinculin tails interact with both the side and the barbed end of an actin filament, reinforcing the coupling between focal adhesions (FAs), and the retrograde flow of actin or resist the actomyosin force. Furthermore, vinculin may provide a secondary path to transmit force to talin and thus stretch its rod domain, leading to a positive feedback loop [188,195].
Talin-mediated integrin activation plays a definitive role in integrin-mediated signaling and induction of downstream survival pathways, leading to protection from anoikis and consequently resulting in cancer progression to metastasis [196]. A single gene mutation in talin is associated with cardiomyopathy, such as ischemic heart disease (atherosclerosis), hypertension, and heart failure [197].
4.4.4. Zyxin
Zyxin is a focal adhesion protein that has been implicated in the modulation of cell adhesion and motility, and is hypothesized to be a mechanosensor in integrin-mediated responses to mechanical force. Zyxin is a member of LIM domain protein family that includes lipoma-preferred partner (LPP) and thyroid receptor-interacting proteins (TRIP6). LIM domain proteins possess two distinct motifs: (1) A proline-rich N-terminal region containing an unclear export signaling sequence. (2) A C-terminal region consisting of three LIM domains (termed by the initials of Lin-11, Isl-1, and Mec-3) [198,199]. Through the LIM domain and the N-terminal proline-rich domain-mediated protein–protein interactions, zyxin forms complexes with molecules such as α-actinin [200] and Mena/VASP [201].
Zyxin mainly localizes to FAs, but it may translocate to actin stress fibers with certain stimuli and cause a release of cytoskeletal stress allowing zyxin to dissociate from focal adhesions into the cytosol. Zyxin plays a role in modulating cytoskeleton and signaling behavior in mechanotransduction [202–204]. The N-terminal proline-rich domain of zyxin has been shown to form complexes with Mena/VASP family proteins in facilitating actin-polymerization at focal adhesions and stress fibers in fibroblast, which is critically involved in the localized cellular contractile response to FN-mediated mechanical force [201]. Also, zyxin possesses an N-terminal nuclear transport signal and has been postulated to shuttle between focal adhesion sites and the cell nucleus as a way of mediating protein expression changes that accompany cellular mechanotransduction [205,206].
Zyxin defects are related to many diseases. Zyxin interacts with the E6 oncoprotein of human papillomavirus type 6 via its LIM domain region, which is commonly associated with genital warts [207]. The mutation of LPP gene can lead to benign human lipomas and pulmonary chondroid hamartomas [208].
4.4.5. Titin
Titin is the giant elastic ruler protein of striated muscle sarcomeres, which span from Z-disk (N-terminus) to the M-band (C-terminus), and is responsible for passive stiffness of the sarcomere [209]. The canonical titin sequence has nine immunoglobulin-like (Ig) domains in the Z-disk segment, interspersed with Z-repeats and unique sequences. The elastic I-band portion contains three types of extensible segments: tandem Ig segments, the spring-like N2B element and the spring-like PEVK element. Titin also contains a single catalytic kinase domain (Titin kinase, TK) near its C-terminal M-band-associated end, which is a complex, autoinhibited conformation. The C-terminal autoinhibitory tail (AI) is formed from three secondary structure elements, αR1, αR2 and βR1, wrapping around the catalytic domain and tightly occluding the ATP binding site. In mammals, the catalytic base titin is blocked by Y170 [210–212].
TK could indeed function as a force sensor by switching between a closed and open conformations. Force acting at low velocities can lead to the sequential unfolding of the autoinhibitory tail with the main autoinhibitory αR2 being pulled out of the ATP binding site and exposing Y170, thus opening the active ATP binding site while preserving the catalytic core. This open conformation binds ATP and promotes further steps in TK activation, by exposing the autoinhibitory Y170 for auto- or trans-phosphorylation through protein kinases. Phosphorylation significantly alters the stiffness of the PEVK and N2B spring elements of titin, allowing for rapid adjustment of titin stiffness and rapid adaptations of striated muscle to match a mechanical changing environment, such as hemodynamic loads [213,214].
The dysfunction of titin is mainly associated with diseases in cardiovascular and musculoskeletal systems. The mutations of TTN, which encodes titin, have been found in patients with dilated cardiomyopathy (DCM), hypertrophic cardiomyopathy (HCM), restrictive cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, and various types of skeletal myopathies. Also, post-translational modifications can also lead to many diseases. For example, reduced basal levels of phosphorylation of the PKA/PKG sites, predicted to increase passive tension, is related to heart failure with preserved ejection fraction (HfpEF) [215–217].
5. FRET-based force sensors to estimate force in live cells
Traditionally, the dominant techniques used to study cell mechanics include microfabricated posts [218,219], traction force microscopy [220], atomic force microscopy (AFM) [221,222], laser tweezers [16, 223], magnetic tweezers [224], and DNA calibrated force sensors [225]. However, none of these techniques can be used to measure intracellular or intramolecular forces in real time, nor can they be easily applicable in situ measures in live cells and animals. For example, microfabricated posts and traction force microscopy was designed to measure the mechanical interactions between whole cells and the substrate, but that won’t tell us the forces between neighboring cells in three dimensions. Laser tweezers can apply small calibrated forces to micron sized beads bound to cells, and these measurements are mostly confined to the extracellular surface of cells where the beads can be readily applied. AFM can apply force through submicron microfabricated probes and this method can be used for measurements of single molecules from the subpN range [87] to whole cell measurement in the > nN range. Like the laser tweezers, the AFM can only be applied to the extracellular surface of cells [226]. As for the DNA based force sensor composed of single-stranded DNA oligomer flanked by two dyes, the sensor cannot be encoded into specific proteins, so, these sensors are limited to in vitro open systems.
To estimate the forces and their distribution in time and space in live cells, genetically coded FRET based probes that make up for the deficiency of traditional techniques have been developed. These sensors have emerged as a suitable technique and can be integrated into individual proteins, turning structural forces into optical signals to estimate the intracellular force in specific proteins, including the structural proteins (e.g. actinin, vinculin, spectrin, actin, etc.) in live cells, tissues, organs and animals [227–229].
5.1. Principles of FRET
Forster (or fluorescence) resonance energy transfer (FRET) refers to the dipole–dipole interactions of two fluorescent molecules with overlapping excitation and emission wavelengths [230,231]. Upon excitation, the electrons of the donor fluorophore jump from their ground state to a higher energy level. As they return to the ground state, a photon is emitted [232]. FRET efficiency depends on the inverse sixth power of the distance between donor and acceptor and the angles between the dipole moments. FRET can function typically over a distance < 10 nm. FRET was introduced by Forster in 1948 using two fluorescent dyes, one as the energy donor, the other one as the acceptor. There are many fluorescence dyes and fluorescence proteins (FPs) that are suitable for FRET pairs [233,234]. However, we will only discuss the probes that are genetically encoded.
5.2. FRET-based force sensors
The FRET efficiency depends on several factors: a proper spectral overlap between the donor and the acceptor, the distance between the two fluorophores, and the relative orientation of donor and acceptor [233–236]. Generally, a typical FRET-based force sensor consists of a pair of fluorescence proteins and an elastic linker. Protein domains, such as a α-helix or β sheet, are biological analogs to mechanical springs and can serve as linkers [226]. With tension, the biological spring is stretched, resulting in a decrease in FRET efficiency and providing a linear change in efficiency with strain [227]. Furthermore, in a circularly permuted stretch sensitive FRET (cpstFRET) sensor, tension changes the relative orientation between the donor and acceptor, which also decreases FRET efficiency [235]. Changes in the distance or orientation between the donor and acceptor can be readily detected and analyzed quantitatively.
Several types of genetically encoded FRET-based force sensors have been created to observe mechanical effects in structural proteins. To date these probes include stretch-sensitive FRET (stFRET), spectrin stFRET (sstFRET), tension sensor module (TSMod) and cpstFRET. Their characteristics are listed below (Table 2):
Table 2.
Types and characteristics of FRET-based force sensors.
| Types | FRET pair | Linker | Properties of the linker | Characteristics of the sensor |
|---|---|---|---|---|
| stFRET [227] | GFP variants: Cerulean and Venus |
A stable α-helix | 5 nm long at rest, R0 of the pair |
The first genetically encoded FRET-based force sensor. |
| sstFRET [228] | GFP variants: Cerulean and Venus |
Spectrin repeat | Composed of three folded α-helices |
Matching the mechanical compliance of common hosts. |
| TSMod [237] | GFP variants: Venus and mTFP1 |
(GPGGA)8 | Elastic linker | The first FRET-based force sensor calibrated with a laser trap and suitable for measuring pN forces. |
| cpstFRET [235] | GFP variants: cpCerulean and cpVenus |
Five glycine peptides | Closely link the pair | Taking angular orientation as the dominant variable; having a much wider dynamic range than the previous ones and smaller and less likely to interfere with host function. Calibrated using DNA springs. |
When probes are incorporated into specific proteins, to minimize possible interference with their physiological function, the sensors are best located near the middle of the fiber and away from known functional domains. Based on these principles, the mechanical dynamics of several structural proteins have been measured and analyzed using FRET-based force sensors:
-
(1)
α-actinin: Transfection with actinin-stFRET in 3T3 cells revealed that during migration, tension in actinin is lower in domains close to the lagging edge where adhesion to the substrate is released, and higher at the leading edge where adhesions pull the cell forward [227]. When α-actinin-sstFRET was expressed in Madin-Darby Canine Kidney (MDCK) cells, fluid shear stress produced a remarkable decrease in cytoskeletal stress, and the cytoskeleton required time to adapt to norm stress levels [238]. In addition, inserting sstFRET into α-actinin in human embryonic kidney cells (HEKs) and bovine aortic endothelial cells (BAECs) showed constitutive stress and force modulation during cell contraction, extension and migration [239].
-
(2)
β-actin: To better understand the mechanical stress in stem cells, cpstFRET was inserted into β-actin [240]. The authors then reprogrammed HEK-293 and MDCK cells into stem cells by culturing them on soft substrate (PDMS) without using transcription factors. The stem cells showed a close association of stemness to high actin stress, implicating that cell mechanics can control stem cell reprogramming and differentiation. Differentiating the cells back onto glass removed most actin stress. The association of pheotypy with substrate mechanics is well known [241–243].
-
(3)
vinculin: Integrating the TSMod into vinculin between the head and tail domains after amino acid 883, the authors concluded that FA stabilization under force requires both vinculin recruitment and force transmission, and these processes can be controlled independently [237].
-
(4)
spectrin: cpstFRET was incorporated into the linker region of spectrin between repeat domains ten and eleven of the α-subunit, and the data showed that, in general, spectrin, like all other investigated structural proteins, is under prestress (tension in a resting cell) and can undergo time-dependent modulation during cell migration [235].
-
(5)
collagen-19: Transgenic C. elegans labeled in collagen-19 produced a behaviorally and anatomically normal animal with constitutive stress in the cuticle in the living animals and infixed animals. When the worms were stretched with micromanipulators, the labeled COL-19 showed a decrease in FRET efficiency stretch, and had positive and negative responses in flexed regions [228]. The creation of transgenic animals (including Drosophila) showed that the probes were generally nontoxic and most likely could be extended to function in transgenic mice.
-
(6)
E-cadherin: TSMod was inserted into the cytoplasmic domain of E-cadherin, where it could sense force transmitted between the transmembrane domain and the β-catenin-binding domain. Data revealed that E-cadherin played a fundamental role in transducing mechanical force between the actomyosin cytoskeleton and the plasma membrane at cell–cell junctions and throughout cell surface [244].
5.3. Quantitative analysis
From above, it is clear that FRET-based force sensors can be used to show the existence, location, and changes of force in living cells; however, the precise magnitude of the force is still unknown. The existing relationship of FRET efficiency and force is constrained by the fact that we know the FRET value with no force applied. We also know the FRET value with ∼10 pN applied with a DNA spring [245,246], and if we approximate the relationship as a straight line between those two points we can convert the FRET ratio to tension in pN. More precise experiments are underway to refine the precision of this relationship.
There are four general strategies for measuring FRET that have proven useful: acceptor photobleaching, sensitized emission including the use of spectral imaging, fluorescence lifetime imaging microscopy (FLIM) and polarization anisotropy imaging [233,234].
5.3.1. Distance-dependent analysis
stFRET, sstFRET, TSMod take distance as their dominant variable. Hooke’s law states:
| (1) |
where F represents the force applied, x is the displacement of the spring and k is the spring constant. GFPs can withstand approximately 100 pN force before unfolding, and physiological force are typically in the range of 0–20 pN; therefore, the GFP based fluorophores will remain intact during measurements in live cells, and the FRET changes can faithfully represent the distance changes caused by stretching the linker [6]. In the host protein, stress creates strain in the probe linker and that changes the distance between the donor and the acceptor, leading to the change in FRET efficiency (E) with a nearly linear relationship (Fig. 5) [234].
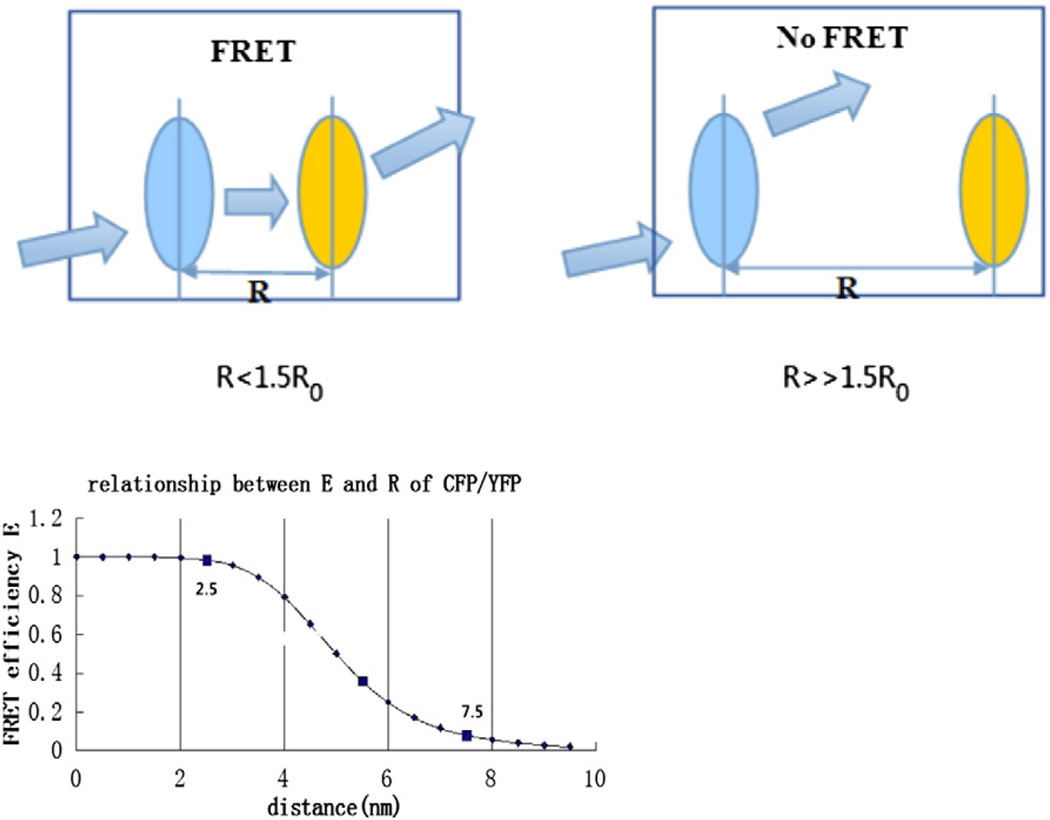
Fig. 5.
Distance-dependent analysis. Under force, if the distance between the donor and the acceptor is < ∼1.5R0, FRET will occur. R0 for CFP/YFP is approximately 5.0 nm and there is a nearly linear relationship between E and R when the distance ranges from 0.5R0 to 1.5R0.
E is given as:
| (2) |
R refers to the distance separating the donor and acceptor, and R0 is called the characteristic distance of a specific FRET pair, at which distance efficiency of energy transfer between donor and acceptor is 50%.
| (3) |
In this equation, QD is the fluorescence quantum yield of the donor in the absence of the acceptor, εA is the maximal acceptor extinction coefficient (mol−1 cm−1), and J(λ) is the spectral overlap integral between the normalized donor fluorescence, fD(λ), and the acceptor excitation spectra, εA(λ).
| (4) |
κ2 is usually assumed to be 2/3, which is the average value integrated over all possible angles and rarely achieved for bound probes. This value is obtained when both fluorochromes have the maximal degrees of freedom, i.e., are freely rotating and can be considered to be isotropically oriented during the excited state lifetime [234]. The main drawback of this analysis is that the dynamic range of these sensors is limited by the nearly linear relationship between FRET efficiency and strain and that provides a small dynamic range.
5.3.2. Orientation-dependent analysis
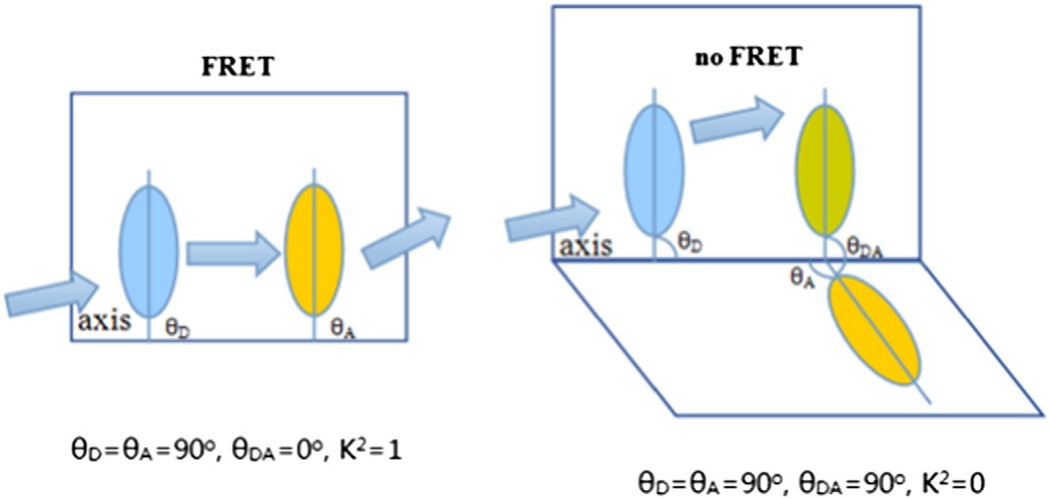
As discussed above, cpstFRET uses the angular dependence of FRET as the dominant variable. Force modulates the angle between the adjacent donor and acceptor much more than the distance between them, endowing this sensor with a greater dynamic range [235]. The angular dependence of the dipole interaction is described by the orientation factor, k2, where k2 is defined as [234]:
| (5) |
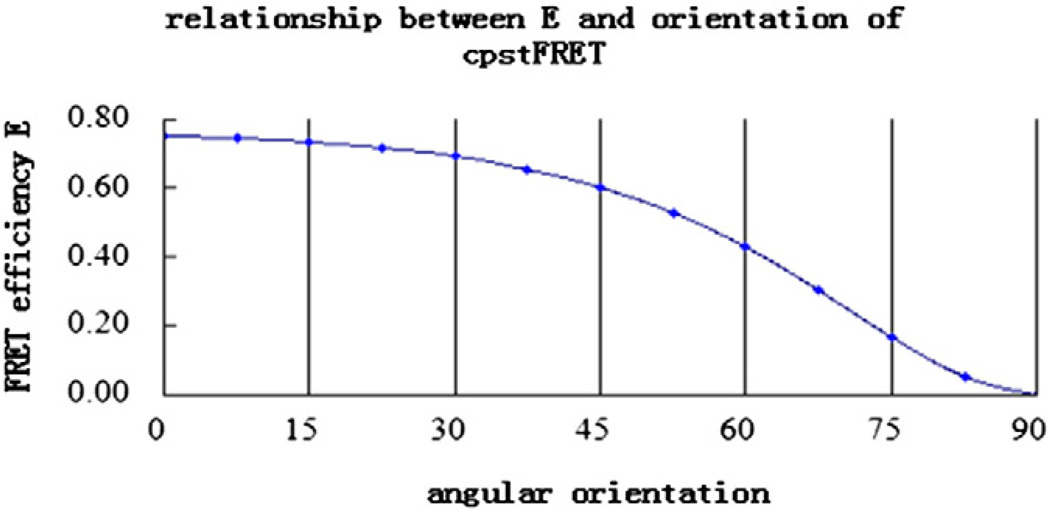
θA is the angle between acceptor and the linker dipole axis, θD is the angle between donor and linker axis, and θDA is the angle between donor (D) and acceptor (A) axis. The value for k2 can range from 0 to 1 depending on the relative orientation of the donor and acceptor. At zero stress, k2 = 1, which yields the highest FRET at a fixed distance with an efficiency E = ∼75%. Torsion will twist the dipoles toward perpendicular where k2 = 0 (Fig. 6) [234,235]. Since the cpstFRET probe is nearly parallel and side by side at rest, applied forces will decrease FRET efficiency.
Fig. 6.
At zero stress, θA = θD = 90° and θDA = 0°, thus k2 =1, which yields the highest FRET, with an efficiency E = 75%. When at a perpendicular position under force, θDA = 90° and k2 = 0, with an efficiency E = 0.
As mentioned above, force changes orientation much more than the distance between the donor and the acceptor, thus assuming that R stays permanent during FRET is reasonable. Qd, εA and J(λ) are constant when the two fluorochromes are determined. Based on the similarities of the two fluorophores in cpstFRET, the donor and the acceptor dipoles are nearly parallel at rest and approximately perpendicular to the linker; thus, θA and θD are both 90°. However, θDA changes with force. Combining Eqs. (2) to (5), a new equation describes the relationship between E and the relative orientation θDA can be achieved (Fig. 7):
| (6) |
Fig. 7.
FRET efficiency is predicted to decrease with the increase with force, changing the relative orientation between the donor and the acceptor from near 0° to 90°.
6. Perspectives of mechanical dynamics in live cells and FRET-based force sensing
Structural protein forces exist and function in almost all cells and tissues. The system of force generation, perception, translation and effect or processing is termed mechanical dynamics, and this is influenced by many factors. The drivers include extracellular mechanical stimuli, motor proteins, osmotic pressure and the effectors are the mechanosensors such as channels or force sensitive proteins [247]. The realtime observation and analysis of structural forces that affected and regulated by these factors is at the heart of cellular biomechanics. Inspired by this, the FRET-based force sensors have emerged as the tool of choice. The sensors can be genetically encoded and inserted in the middle of structural proteins to measure the force in real time. By turning force into optical signals, the magnitude, distribution, duration, and direction of structural forces can be visualized and analyzed. However, like all probes, all of the FRET-based force sensors mentioned above have some shortcomings. There is the potential for interference in the normal functions of host proteins, although this has been shown to not be significant. The future improvements include: (1) Probes containing only one fluorescent protein. (2) Adding new pairs with a spectral separation so that the dynamics of multiple proteins can be observed in synchrony. (3) Transgenic mice where special promoters can regulate the expression of mechanosensors in specific cells and tissues. (4) And as with all fluorophores, less bleaching.
At long last we can ask the questions about how biomechanics function at the level of single molecules in situ. The probes promise to uncover more mysteries than they answer, but that is why science is fun.
Acknowledgments
The work was supported by grants from the National Natural Science Foundation of China (No. 81170714) and an NIH grant to F.S.
Footnotes
Conflict of interest
We have no conflict of interest to declare. We confirm that our manuscript has not been, or will not be submitted elsewhere for publishing, and all listed authors in the report have concurred with the submission. The final manuscript is also approved by all authors including the responsible authorities in the laboratories where the work was carried out. If accepted, the article shall not be published elsewhere in the same form, in either the same or another language, without the consent of the Editors and Publisher of BBA-Mol Cell Res.
References
- 1.Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137(9):1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nava MM, Raimondi MT, Pietrabissa R. Controlling self-renewal and differentiation of stem cells via mechanical cues. J. Biomed. Biotechnol. 2012;2012:797410. doi: 10.1155/2012/797410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yunfei Cai, Sheetz Michael P. Force propagation across cells: mechanical coherence of dynamic cytoskeletons. Curr. Opin. Cell Biol. 2009;21:47–50. doi: 10.1016/j.ceb.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Zhu YH, Chi C, Wu HW, Guo J. Role of cytoskeleton in axonal regeneration after neurodegenerative diseases and CNS injury. Rev. Neurosci. 2014;25(4):527–542. doi: 10.1515/revneuro-2013-0062. [DOI] [PubMed] [Google Scholar]
- 6.Riehl BD1, Park JH, Kwon IK, Lim JY. Mechanical stretching for tissue engineering: two-dimensional and three-dimensional constructs. Tissue Eng. B Rev. 2012;18(4):288–300. doi: 10.1089/ten.teb.2011.0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desmaële D, Boukallel M, Régnier S. Actuation means for the mechanical stimulation of living cells via microelectromechanical systems: a critical review. J. Biomech. 2011;44(8):1433–1446. doi: 10.1016/j.jbiomech.2011.02.085. [DOI] [PubMed] [Google Scholar]
- 8.Guck J, Ananthakrishnan R, Mahmood H, Moon TJ, Cunningham CC, Käs J. The optical stretcher: a novel laser tool to micromanipulate cells. Biophys. J. 2001;81(2):767–784. doi: 10.1016/S0006-3495(01)75740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hochmuth RM. Micropipette aspiration of living cells. J. Biomech. 2000;33(1):15–22. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- 10.Georges PC, Janmey PA. Cell type-specific response to growth on soft materials. J. Appl. Physiol. (1985) 2005;98(4):1547–1553. doi: 10.1152/japplphysiol.01121.2004. [DOI] [PubMed] [Google Scholar]
- 11.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310(5751):1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 12.Brown TD. Techniques for mechanical stimulation of cells in vitro: a review. J. Biomech. 2000;33(1):3–14. doi: 10.1016/s0021-9290(99)00177-3. [DOI] [PubMed] [Google Scholar]
- 13.Shi X, Zhang X, Xia T, Fang X. Living cell study at the single-molecule and single-cell levels by atomic force microscopy. Nanomedicine (London) 2012;7(10):1625–1637. doi: 10.2217/nnm.12.130. [DOI] [PubMed] [Google Scholar]
- 14.Guo XE, Takai E, Jiang X, Xu Q, Whitesides GM, Yardley JT, Hung CT, Chow EM, Hantschel T, Costa KD. Intracellular calcium waves in bone cell networks under single cell nanoindentation. Mol. Cell. Biomech. 2006;3(3):95–107. [PubMed] [Google Scholar]
- 15.Langer MG, Koitschev A, Haase H, Rexhausen U, Hörber JK, Ruppersberg JP. Mechanical stimulation of individual stereocilia of living cochlear hair cells by atomic force microscopy. Ultramicroscopy. 2000;82(1–4):269–278. doi: 10.1016/s0304-3991(99)00136-9. [DOI] [PubMed] [Google Scholar]
- 16.Ashkin A, Dziedzic JM. Optical trapping and manipulation of viruses and bacteria. Science. 1987;235(4795):1517–1520. doi: 10.1126/science.3547653. [DOI] [PubMed] [Google Scholar]
- 17.Tanck E, van Driel WD, Hagen JW, Burger EH, Blankevoort L, Huiskes R. Why does intermittent hydrostatic pressure enhance the mineralization process in fetal cartilage? J. Biomech. 1999;32(2):153–161. doi: 10.1016/s0021-9290(98)00165-1. [DOI] [PubMed] [Google Scholar]
- 18.Ozawa H, Imamura K, Abe E, Takahashi N, Hiraide T, Shibasaki Y, Fukuhara T, Suda T. Effect of a continuously applied compressive pressure on mouse osteoblast-like cells (MC3T3-E1) in vitro. J. Cell. Physiol. 1990;142(1):177–185. doi: 10.1002/jcp.1041420122. [DOI] [PubMed] [Google Scholar]
- 19.Torzilli PA, Grigiene R, Huang C, Friedman SM, Doty SB, Boskey AL, Lust G. Characterization of cartilage metabolic response to static and dynamic stress using a mechanical explant test system. J. Biomech. 1997;30(1):1–9. doi: 10.1016/s0021-9290(96)00117-0. [DOI] [PubMed] [Google Scholar]
- 20.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms roles in brain function, development, and disease. Neuron. 2010;68(4):610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 21.Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 22.Pfister KK, Shah PR, Hummerich H, Russ A, Cotton J, Annuar AA, King SM, Fisher EM. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karki S, Holzbaur EL. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr. Opin. Cell Biol. 1999;11(1):45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- 24.Sakakibara H, Oiwa K. Molecular organization and force-generating mechanism of dynein. FEBS J. 2011;278(17):2964–2979. doi: 10.1111/j.1742-4658.2011.08253.x. [DOI] [PubMed] [Google Scholar]
- 25.Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, López MP, Vale RD, Jülicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling force that position microtubule asters. Cell. 2012;148(3):502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendricks AG, Lazarus JE, Perlson E, Gardner MK, Odde DJ, Goldman YE, Holzbaur EL. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr. Biol. 2012;22(7):632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNally FJ. Mechanisms of spindle positioning. J. Cell Biol. 2013;200(2):131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy JR, Holzbaur EL. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 2008;121(Pt 19):3187–3195. doi: 10.1242/jcs.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007;10(8):970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 30.Raaijmakers JA, van Heesbeen RG, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31(21):4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108(1):97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 32.Merdes A, Ramyar K, Vechio JD, Cleveland DW. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87(3):447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 33.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382(6590):420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 34.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013;14(1):25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 2005;287(2):378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 36.Merrill AE, Merriman B, Farrington-Rock C, Camacho N, Sebald ET, Funari VA, Schibler MJ, Firestein MH, Cohn ZA, Priore MA, Thompson AK, Rimoin DL, Nelson SF, Cohn DH, Krakow D. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am. J. Hum. Genet. 2009;84(4):542–549. doi: 10.1016/j.ajhg.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Jr, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nat. Genet. 2003;33(4):455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 38.Lai C, Lin X, Chandran J, Shim H, Yang WJ, Cai H. The G59S mutation in p150(glued) causes dysfunction of dynactin in mice. J. Neurosci. 2007;27(51):13982–13990. doi: 10.1523/JNEUROSCI.4226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy JR, Sumner CJ, Caviston JP, Tokito MK, Ranganathan S, Ligon LA, Wallace KE, LaMonte BH, Harmison GG, Puls I, Fischbeck KH, Holzbaur EL. A motor neuron disease-associated mutation in p150Glued perturbs dynactin function and induces protein aggregation. J. Cell Biol. 2006;172(5):733–745. doi: 10.1083/jcb.200511068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O’Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000;2(11):784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 41.Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, Yano Y, Mimori-Kiyosue Y, Nakamura T, Itoh K, Fushiki S, Setou M, Wynshaw-Boris A, Torisawa T, Toyoshima YY, Hirotsune S. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27(19):2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimura N, Imamura O, Ono F, Terao K. Aging attenuates dynactin–dynein interaction: down-regulation of dynein causes accumulation of endogenous tau and amyloid precursor protein in human neuroblastoma cells. J. Neurosci. Res. 2007;85:2909–2916v. doi: 10.1002/jnr.21408. [DOI] [PubMed] [Google Scholar]
- 43.Hirokawa N, Noda Y. Intracellular transport kinesin superfamily proteins, KIFs: structure function, and dynamics. Physiol. Rev. 2008;88(3):1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- 44.Alexander Marx Andreas Hoenger Eckhard Mandelkow. Structures of kinesin motor proteins. Cell Motil. Cytoskeleton. 2009;66(11):958–966. doi: 10.1002/cm.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, Graham FL, Gaskell PC, Dearlove A, Pericak-Vance MA, Rubinsztein DC, Marchuk DA. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am. J. Hum. Genet. 2002;71(5):1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xia CH, Roberts EA, Her LS, Liu X, Williams DS, Cleveland DW, Goldstein LS. Abnormal neurofilament transport caused by targeted disruption of neuronal kinesin heavy chain KIF5A. J. Cell Biol. 2003;161(1):55–66. doi: 10.1083/jcb.200301026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devon RS1, Orban PC, Gerrow K, Barbieri MA, Schwab C, Cao LP, Helm JR, Bissada N, Cruz-Aguado R, Davidson TL, Witmer J, Metzler M, Lam CK, Tetzlaff W, Simpson EM, McCaffery JM, El-Husseini AE, Leavitt BR, Hayden MR. Als2-deficient mice exhibit disturbances in endosome trafficking associated with motor behavioral abnormalities. Proc. Natl. Acad. Sci. U. S. A. 2006;103(25):9595–9600. doi: 10.1073/pnas.0510197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hadano S, Benn SC, Kakuta S, Otomo A, Sudo K, Kunita R, Suzuki-Utsunomiya K, Mizumura H, Shefner JM, Cox GA, Iwakura Y, Brown RH, Jr, Ikeda JE. Mice deficient in the Rab5 guanine nucleotide exchange factor ALS2/alsin exhibit age-dependent neurological deficits and altered endosome trafficking. Hum. Mol. Genet. 2006;15(2):233–250. doi: 10.1093/hmg/ddi440. [DOI] [PubMed] [Google Scholar]
- 49.Bowman AB, Kamal A, Ritchings BW, Philp AV, McGrail M, Gindhart JG, Goldstein LS. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell. 2000;103(4):583–594. doi: 10.1016/s0092-8674(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 50.Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32(3):389–401. doi: 10.1016/s0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- 51.Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS. Kinesin-mediated axonal transport of a membrane compartment containing b-secretase and presenilin-1 requires APP. Nature. 2001;414(6864):643–648. doi: 10.1038/414643a. [DOI] [PubMed] [Google Scholar]
- 52.Zhao C, Takita J, Tanaka Y, Setou M, Nakagawa T, Takeda S, Yang HW, Terada S, Nakata T, Takei Y, Saito M, Tsuji S, Hayashi Y, Hirokawa N. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105(5):587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- 53.Niwa S, Tanaka Y, Hirokawa N. KIF1Bbeta- and KIF1A–mediated axonal transport of presynaptic regulator Rab3 occurs in a GTP-dependent manner through DENN/ MADD. Nat. Cell Biol. 2008;10(11):1269–1279. doi: 10.1038/ncb1785. [DOI] [PubMed] [Google Scholar]
- 54.Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, Link J, Lundström W, Greiner E, Dessa Sadovnick A, Goossens D, Van Broeckhoven C, Del-Favero J, Ebers GC, Oostra BA, van Duijn CM, Hintzen RQ. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat. Genet. 2008;40(12):1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 55.Lyons DA, Naylor SG, Scholze A, Talbot WS. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat. Genet. 2009;41(7):854–858. doi: 10.1038/ng.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada K, Andrews C, Chan WM, McKeown CA, Magli A, de Berardinis T, Loewenstein A, Lazar M, O’Keefe M, Letson R, London A, Ruttum M, Matsumoto N, Saito N, Morris L, Del Monte M, Johnson RH, et al. Heterozygous mutations of the kinesin KIF21A in congenital fibrosis of the extraocular muscles type 1 (CFEOM1) Nat. Genet. 2003;35(4):318–321. doi: 10.1038/ng1261. [DOI] [PubMed] [Google Scholar]
- 57.Hartman MA, Finan D, Sivaramakrishnan S, Spudich JA. Principles of unconventional myosin function and targeting. Annu. Rev. Cell Dev. Biol. 2011;27:133–155. doi: 10.1146/annurev-cellbio-100809-151502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc. Natl. Acad. Sci. U. S. A. 2006;103(10):3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arnold DB, Gallo G. Structure meets function: actin filaments and myosin motors in the axon. J. Neurochem. 2014;129(2):213–220. doi: 10.1111/jnc.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin su-perfamily. Curr. Opin. Cell Biol. 2004;16(1):61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 61.Eddinger TJ, Meer DP. Myosin II isoforms in smooth muscle: heterogeneity and function. Am. J. Physiol. Cell Physiol. 2007;293(2):C493–C508. doi: 10.1152/ajpcell.00131.2007. [DOI] [PubMed] [Google Scholar]
- 62.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10(11):778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Gele M, Dynoodt P, Lambert J. Griscelli syndrome: a model system to study vesicular trafficking. Pigment Cell Melanoma Res. 2009;22(3):268–282. doi: 10.1111/j.1755-148X.2009.00558.x. [DOI] [PubMed] [Google Scholar]
- 64.Naccache SN, Hasson T, Horowitz A. Binding of internalized receptors to the PDZ domain of GIPC/synectin recruits myosin VI to endocytic vesicles. Proc. Natl. Acad. Sci. U. S. A. 2006;103(34):12735–12740. doi: 10.1073/pnas.0605317103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hartman MA, Spudich JA. The myosin superfamily at a glance. J. Cell Sci. 2012;125(Pt 7):1627–1632. doi: 10.1242/jcs.094300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, Ferrara A, Lanzara C, Ficarella R, Declau F, Pusch CM, Nürnberg P, Melchionda S, Zelante L, Ballana E, Estivill X, Van Camp G, Gasparini P, Savoia A. Nonmuscle myosin heavy-chain gene MYH14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (DFNA4) Am. J. Hum. Genet. 2004;74(4):770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laakso JM, Lewis JH, Shuman H, Ostap EM. Myosin I can act as a molecular force sensor. Science. 2008;321(5885):133–136. doi: 10.1126/science.1159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seri M1, Cusano R, Gangarossa S, Caridi G, Bordo D, Lo Nigro C, Ghiggeri GM, Ravazzolo R, Savino M, Del Vecchio M, d’Apolito M, Iolascon A, et al. Mutations in MYH9 result in the May-Hegglin anomaly and Fechtner and Sebastian syndromes. Nat. Genet. 2000;26(1):103–105. doi: 10.1038/79063. [DOI] [PubMed] [Google Scholar]
- 69.Wilson CA, Tsuchida MA, Allen GM, Barnhart EL, Applegate KT, Yam PT, Ji L, Keren K, Danuser G, Theriot JA. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 2010;465(7296):373–377. doi: 10.1038/nature08994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies’ eyes to our ears: mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Natl. Acad. Sci. U. S. A. 2002;99(11):7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349(6311):709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 72.Wang Z, Edwards JG, Riley N, Provance DW, Jr, Karcher R, Li XD, Davison IG, Ikebe M, Mercer JA, Kauer JA, Ehlers MD. Myosin Vb mobilizes recycling endosomes and AMPA receptors for postsynaptic plasticity. Cell. 2008;135(3):535–548. doi: 10.1016/j.cell.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374(6517):62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- 74.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374(6517):60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 75.Nishioka M, Kohno T, Tani M, Yanaihara N, Tomizawa Y, Otsuka A, Sasaki S, Kobayashi K, Niki T, Maeshima A, Sekido Y, Minna JD, Sone S, Yokota J. MYO18B, a candidate tumor suppressor gene at chromosome 22q12.1, deleted mutated and methylated in human lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2002;99(19):12269–12274. doi: 10.1073/pnas.192445899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kull FJ, Endow SA. Force generation by kinesin and myosin cytoskeletal motor proteins. J. Cell Sci. 2013;126(Pt 1):9–19. doi: 10.1242/jcs.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc. Natl. Acad. Sci. U. S. A. 2007;104(41):16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mogami T, Kon T, Ito K, Sutoh K. Kinetic characterization of tail swing steps in the ATPase cycle of Dictyostelium cytoplasmic dynein. J. Biol. Chem. 2007;282(30):21639–21644. doi: 10.1074/jbc.M701914200. [DOI] [PubMed] [Google Scholar]
- 79.Noda M, Sakuta H. Central regulation of body-fluid homeostasis. Trends Neurosci. 2013;36(11):661–673. doi: 10.1016/j.tins.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 80.Borgnia M, Nielsen S, Engel A, Agre P. Cellular and molecular biology of the aquaporin water channels. Annu. Rev. Biochem. 1999;68:425–458. doi: 10.1146/annurev.biochem.68.1.425. [DOI] [PubMed] [Google Scholar]
- 81.Sharif Naeini R, Witty MF, Séguéla P, Bourque CW. An N-terminal variant of Trpv1 channel is required for osmosensory transduction. Nat. Neurosci. 2006;9(1):93–98. doi: 10.1038/nn1614. [DOI] [PubMed] [Google Scholar]
- 82.Liedtke W, Choe Y, Martí-Renom MA, Bell AM, Denis CS, Sali A, Hudspeth AJ, Friedman JM, Heller S. Vanilloid receptor-related osmotically activated channel (VR-OAC) a candidate vertebrate osmoreceptor. Cell. 2000;103(3):525–535. doi: 10.1016/s0092-8674(00)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patel AJ, Honoré E, Maingret F, Lesage F, Fink M, Duprat F, Lazdunski M. A mammalian two pore domain mechano-gated S-like K+ channel. EMBO J. 1998;17(15):4283–4290. doi: 10.1093/emboj/17.15.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez Di Giorgio J, Soto G, Alleva K, Jozefkowicz C, Amodeo G, Muschietti JP, Ayub ND. Prediction of aquaporin function by integrating evolutionary and functional analyses. J. Membr. Biol. 2014;247(2):107–125. doi: 10.1007/s00232-013-9618-8. [DOI] [PubMed] [Google Scholar]
- 85.Day RE, Kitchen P, Owen DS, Bland C, Marshall L, Conner AC, Bill RM, Conner MT. Human aquaporins: regulators of transcellular water flow. Biochim. Biophys. Acta. 2014;1840(5):1492–1506. doi: 10.1016/j.bbagen.2013.09.033. [DOI] [PubMed] [Google Scholar]
- 86.Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol. Rev. 2009;89(1):193–277. doi: 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- 87.Beyder A, Sachs F. Electromechanical coupling in the membranes of Shaker-transfected HEK cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106(16):6626–6631. doi: 10.1073/pnas.0808045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Charras GT, Coughlin M, Mitchison TJ, Mahadevan L. Life and times of a cellular bleb. Biophys. J. 2008;94(5):1836–1853. doi: 10.1529/biophysj.107.113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mitchison TJ, Charras GT, Mahadevan L. Implications of a poroelastic cytoplasm for the dynamics of animal cell shape. Semin. Cell Dev. Biol. 2008;19(3):215–223. doi: 10.1016/j.semcdb.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vandorpe DH, Xu C, Shmukler BE, Otterbein LE, Trudel M, Sachs F, Gottlieb PA, Brugnara C, Alper SL. Hypoxia activates a Ca2 + -permeable cation conductance sensitive to carbon monoxide and to GsMTx-4 in human and mouse sickle erythrocytes. PLoS One. 2010;5(1):e8732. doi: 10.1371/journal.pone.0008732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xiao R, Xu XZ. Mechanosensitive channels: in touch with Piezo. Curr. Biol. 2010;20(21):936–938. doi: 10.1016/j.cub.2010.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martinac B. Mechanosensitive ion channels: molecules of mechanotransduction. J. Cell Sci. 2004;117(12):2449–2460. doi: 10.1242/jcs.01232. [DOI] [PubMed] [Google Scholar]
- 93.Nilius B, Owsianik G. The transient receptor potential family of ion channels. Genome Biol. 2011;12(3):218–228. doi: 10.1186/gb-2011-12-3-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuipers AJ, Middelbeek J, van Leeuwen FN. Mechanoregulation of cytoskeletal dynamics by TRP channels. Eur. J. Cell Biol. 2012;91(11–12):834–846. doi: 10.1016/j.ejcb.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 95.Patel AJ, Lazdunski M, Honoré E. Lipid and mechano-gated 2P domain K+ channels. Curr. Opin. Cell Biol. 2001;13(4):422–428. doi: 10.1016/s0955-0674(00)00231-3. [DOI] [PubMed] [Google Scholar]
- 96.Mirkovic K, Palmersheim J, Lesage F. Behavioral characterization of mice lacking Trek channels. Front. Behav. Neurosci. 2012;6:60. doi: 10.3389/fnbeh.2012.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Enyedi P, Czirják G. Molecular background of leak K currents: two-pore domain potassium channels. Physiol. Rev. 2010;90(2):559–605. doi: 10.1152/physrev.00029.2009. [DOI] [PubMed] [Google Scholar]
- 98.Bae C, Gnanasambandam R, Nicolai C, Sachs F, Gottlieb PA. Xerocytosis is caused by mutations that alter the kinetics of the mechanosensitive channel PIEZO1. Proc. Natl. Acad. Sci. U. S. A. 2013;110(12):E1162–E1168. doi: 10.1073/pnas.1219777110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Markin VS, Sachs F. Thermodynamics of mechanosensitivity. Phys. Biol. 2004;(1–2):110–124. doi: 10.1088/1478-3967/1/2/007. [DOI] [PubMed] [Google Scholar]
- 100.Patel AJ1, Honoré E, Maingret F. A mammalian two pore domain mechano-gated S-like K+ channel. EMBO J. 1998;17(15):4283–4290. doi: 10.1093/emboj/17.15.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Honoré E. The neuronal background K2P channels: focus on TREK1. Nat. Rev. Neurosci. 2007;8(4):251–261. doi: 10.1038/nrn2117. [DOI] [PubMed] [Google Scholar]
- 102.Patel AJ, Honoré E, Lesage F, Fink M, Romey G, Lazdunski M. Inhalational anesthetics activate twopore-domain background K+ channels. Nat. Neurosci. 1999;2(5):422–426. doi: 10.1038/8084. [DOI] [PubMed] [Google Scholar]
- 103.Mano I, Driscoll M. DEG/ENaC channels:a touchy superfamily that watches its salt. Bioessays. 1999;21(7):568–578. doi: 10.1002/(SICI)1521-1878(199907)21:7<568::AID-BIES5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 104.Eastwood Amy L, Goodman Miriam B. Insight into DEG/ENaC channel gating from genetics and structure. Physiology (Bethesda) 2012;27(5):282–290. doi: 10.1152/physiol.00006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bianchi L, Driscoll M. Protons at the gate: DEG/ENaC ion channels help us feel and remember. Neuron. 2002;34(3):337–340. doi: 10.1016/s0896-6273(02)00687-6. [DOI] [PubMed] [Google Scholar]
- 106.Tavernarakis N, Driscoll M. Degenerins: at the core of the metazoan mechanotransducer? Ann. N. Y. Acad. Sci. 2001;940:28–41. [PubMed] [Google Scholar]
- 107.Coste B, Mathur J, Schmidt M, Earley TJ. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330(6000):55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nilius B, Honoré E. Sensing pressure with ion channels. Trends Neurosci. 2012;35(8):477–486. doi: 10.1016/j.tins.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 109.McHugh BJ, Buttery R, Lad Y. Integrin activation by Fam38A uses a novelmechanism of R-Ras targeting to the endoplasmic reticulum. J. Cell Sci. 2010;123(1):51–61. doi: 10.1242/jcs.056424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Volkers L, Mechioukhi Y, Coste B. Piezo channels: from structure to function. Pflugers Arch. 2015;467(1):95–99. doi: 10.1007/s00424-014-1578-z. [DOI] [PubMed] [Google Scholar]
- 111.Suchyna T, Sachs F. Mechanical and electrical properties of membranes from dystrophic and normal mouse muscle. J. Physiol. Lond. 2007;581:369–387. doi: 10.1113/jphysiol.2006.125021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nilius B, Voets T. The puzzle of TRPV4 channelopathies. EMBO Rep. 2013;14(2):152–163. doi: 10.1038/embor.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rock MJ, et al. Gain-of-function mutations in TRPV4 cause autosomal dominant brachyolmia. Nat. Genet. 2008;40(8):999–1003. doi: 10.1038/ng.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nishimura G, et al. Spondylo-epiphyseal dysplasia, Maroteaux type (pseudo-Morquio syndrome type 2), and parastremmatic dysplasia are caused by TRPV4 mutations. Am. J. Med. Genet. A. 2010;152A(6):1443–1449. doi: 10.1002/ajmg.a.33414. [DOI] [PubMed] [Google Scholar]
- 115.Main M, Kairon H, Mercuri E, Muntoni F. The Hammersmith functional motor scale for children with spinal muscular atrophy: a scale to test ability and monitor progress in children with limited ambulation. Eur. J. Paediatr. Neurol. 2003;7(4):155–159. doi: 10.1016/s1090-3798(03)00060-6. [DOI] [PubMed] [Google Scholar]
- 116.Rossor AM, Kalmar B, Greensmith L, Reilly MM. The distal hereditary motor neuropathies. J. Neurol. Neurosurg. Psychiatry. 2012;83(1):6–14. doi: 10.1136/jnnp-2011-300952. [DOI] [PubMed] [Google Scholar]
- 117.Pareyson D, Saveri P, Piscosquito G. Charcot–Marie–Tooth disease and related hereditary neuropathies: from gene function to associated phenotypes. Curr. Mol. Med. 2014 Oct 10; doi: 10.2174/1566524014666141010154205. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 118.Reitsma S, Slaaf DW, Vink H, van Zandvoort MA, oude Egbrink MG. The endothelial glycocalyx: composition functions and visualization. Pflugers Arch. 2007;454(3):345–359. doi: 10.1007/s00424-007-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Weinbaum S1, Zhang X, Han Y, Vink H, Cowin SC. Mechanotransduction and flow across the endothelial glycocalyx. Proc. Natl. Acad. Sci. U. S. A. 2003;100(13):7988–7995. doi: 10.1073/pnas.1332808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Davies PF. Flow-mediated endothelial mechanotransduction. Physiol. Rev. 1995;75(3):519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Partovian C, Zhuang Z, Moodie K, Lin M, Ouchi N, Sessa WC, Walsh K, Simons M. PKCalpha activates eNOS and increases arterial blood flow in vivo. Circ. Res. 2005;97(5):482–487. doi: 10.1161/01.RES.0000179775.04114.45. [DOI] [PubMed] [Google Scholar]
- 122.Solomonson LP, Flam BR, Pendleton LC, Goodwin BL, Eichler DC. The caveolar nitric oxide synthase/arginine regeneration system for NO production in endothelial cells. J. Exp. Biol. 2003;206(Pt 12):2083–2087. doi: 10.1242/jeb.00361. [DOI] [PubMed] [Google Scholar]
- 123.Gratton JP, Bernatchez P, Sessa WC. Caveolae and caveolins in the cardiovascular system. Circ. Res. 2004;94(11):1408–1417. doi: 10.1161/01.RES.0000129178.56294.17. [DOI] [PubMed] [Google Scholar]
- 124.Dusserre N, L’Heureux N, Bell KS, Stevens HY, Yeh J, Otte LA, Loufrani L, Frangos JA. PECAM-1 interacts with nitric oxide synthase in human endothelial cells: implication for flow-induced nitric oxide synthase activation. Arterioscler. Thromb. Vasc. Biol. 2004;24(10):1796–1802. doi: 10.1161/01.ATV.0000141133.32496.41. [DOI] [PubMed] [Google Scholar]
- 125.Nieuwdorp M, Meuwese MC, Vink H, Hoekstra JB, Kastelein JJ, Stroes ES. The endothelial glycocalyx: a potential barrier between health and vascular disease. Curr. Opin. Lipidol. 2005;16(5):507–511. doi: 10.1097/01.mol.0000181325.08926.9c. [DOI] [PubMed] [Google Scholar]
- 126.Nieuwdorp M, Mooij HL, Kroon J, Atasever B, Spaan JA, Ince C, Holleman F, Diamant M, Heine RJ, Hoekstra JB, Kastelein JJ, Stroes ES, Vink H. Endothelial glycocalyx damage coincides with microalbuminuria in type 1 diabetes. Diabetes. 2006;55(4):1127–1132. doi: 10.2337/diabetes.55.04.06.db05-1619. [DOI] [PubMed] [Google Scholar]
- 127.Nieuwdorp M, van Haeften TW, Gouverneur MC, Mooij HL, van Lieshout MH, Levi M, Meijers JC, Holleman F, Hoekstra JB, Vink H, Kastelein JJ, Stroes ES. Loss of endothelial glycocalyx during acute hyperglycemia coincides with endothelial dysfunction and coagulation activation in vivo. Diabetes. 2006;55(2):480–486. doi: 10.2337/diabetes.55.02.06.db05-1103. [DOI] [PubMed] [Google Scholar]
- 128.Mulivor AW, Lipowsky HH. Inflammation- and ischemia-induced shedding of venular glycocalyx. Am. J. Physiol. Heart Circ. Physiol. 2004;286(5):H1672–H1680. doi: 10.1152/ajpheart.00832.2003. [DOI] [PubMed] [Google Scholar]
- 129.Humphries MJ. Integrin structure. Biochem. Soc. Trans. 2000;28(4):311–339. [PubMed] [Google Scholar]
- 130.Schiller HB, Fässler R. Mechanosensitivity and compositional dynamics of cell- matrix adhesions. EMBO Rep. 2013;14(6):509–519. doi: 10.1038/embor.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Geiger B, Yamada KM. Molecular architecture and function of matrix adhesions. Cold Spring Harb. Perspect. Biol. 2011;3(5) doi: 10.1101/cshperspect.a005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xiao T, Takagi J, Coller BS, Wang JH, Springer TA. Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature. 2004;432(7013):59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nat. Cell Biol. 2002;4(4):E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- 134.Hu P, Luo BH. Integrin bi-directional signaling across the plasma membrane. J. Cell. Physiol. 2013;228(2):306–312. doi: 10.1002/jcp.24154. [DOI] [PubMed] [Google Scholar]
- 135.Winograd-Katz SE, Fässler R, Geiger B, Legate KR. The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 2014;15(4):273–288. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 136.Poole CA, Jensen CG, Snyder JA, Gray CG, Hermanutz VL, Wheatley DN. Confocal analysis of primary cilia structure and colocalization with the Golgi apparatus in chondrocytes and aortic smooth muscle cells. Cell Biol. Int. 1997;21(8):483–494. doi: 10.1006/cbir.1997.0177. [DOI] [PubMed] [Google Scholar]
- 137.Praetorius HA, Spring KR. The renal cell primary cilium functions as a flow sensor. Curr. Opin. Nephrol. Hypertens. 2003;12(5):517–520. doi: 10.1097/00041552-200309000-00006. [DOI] [PubMed] [Google Scholar]
- 138.Nguyen AM, Jacobs CR. Emerging role of primary cilia as mechanosensors in osteocytes. BOne. 2013;54(2):196–204. doi: 10.1016/j.bone.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Muhammad H, Rais Y, Miosge N, Ornan EM. The primary cilium as a dual sensor of mechanochemical signals in chondrocytes. Cell. Mol. Life Sci. 2012;69(13):2101–2107. doi: 10.1007/s00018-011-0911-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mahjoub MR. The importance of a single primary cilium. Organogenesis. 2013;9(2):61–69. doi: 10.4161/org.25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST. Primary cilia signaling pathways in mammalian development health and disease. Nephron Physiol. 2009;111(3):39–53. doi: 10.1159/000208212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Christensen ST, Pedersen LB, Schneider L, Satir P. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 2007;8(2):97–109. doi: 10.1111/j.1600-0854.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- 143.Zou Y, Akazawa H, Qin Y, Sano M, Takano H, Minamino T, Makita N, Iwanaga K, Zhu W, et al. Mechanical stress activates angiotensin II type 1 receptor without the involvement of angiotensin II. Nat. Cell Biol. 2004;6(6):499–506. doi: 10.1038/ncb1137. [DOI] [PubMed] [Google Scholar]
- 144.Mederos y Schnitzler M, Storch U, Meibers S, Nurwakagari P, Breit A, Essin K, Gollasch M, Gudermann T. Gq-coupled receptors as mechanosensors mediating myogenic vasoconstriction. EMBO J. 2008;27(23):3092–3103. doi: 10.1038/emboj.2008.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gether U. Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 2000;21(1):90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 146.Yasuda N, Miura S, Akazawa H, Tanaka T, Qin Y, Kiya Y, Imaizumi S, Fujino M, Ito K, Zou Y, et al. Conformational switch of angiotensin II type 1 receptor underlying mechanical stress-induced activation. EMBO Rep. 2008;9(2):179–186. doi: 10.1038/sj.embor.7401157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karnik SS, Gogonea C, Patil S, Saad Y, Takezako T. Activation of G-protein-coupled receptors: a common molecular mechanism. Trends Endocrinol. Metab. 2003;14(9):431–437. doi: 10.1016/j.tem.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 148.Voets T, Nilius B. TRPCs, GPCRs and the Bayliss effect. EMBO J. 2009;28(1):4–5. doi: 10.1038/emboj.2008.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Akinlaja J, Sachs F. The breakdown of cell membranes by electrical and mechanical stress. Biophys. J. 1998;75(1):247–254. doi: 10.1016/S0006-3495(98)77511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Orr AW, Helmke BP, Blackman BR, Schwartz MA. Mechanisms of mechanotransduction. Dev. Cell. 2006;10(1):11–20. doi: 10.1016/j.devcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 151.Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, Just S, Trano N, Katus HA, Fishman MC, Rottbauer W. Integrin-linked kinase a novel component of the cardiac mechanical stretch sensor controls contractility in the zebrafish heart. Genes Dev. 2006;20(17):2361–2372. doi: 10.1101/gad.1448306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Brancaccio M, Fratta L, Notte A, Hirsch E, Poulet R, Guazzone S, De Acetis M, Vecchione C, Marino G, Altruda F, Silengo L, Tarone G, Lembo G. Melusin, a muscle-specific integrin beta1-interacting protein is required to prevent cardiac failure in response to chronic pressure overload. Nat. Med. 2003;9(1):68–75. doi: 10.1038/nm805. [DOI] [PubMed] [Google Scholar]
- 153.Kung C. A possible unifying principle for mechanosensation. Nature. 2005;436(7051):647–654. doi: 10.1038/nature03896. [DOI] [PubMed] [Google Scholar]
- 154.Petrel C, Clauser E. Angiotensin II AT1 receptor constitutive activation: from molecular mechanisms to pathophysiology. Mol. Cell. Endocrinol. 2009;302(2):176–184. doi: 10.1016/j.mce.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 155.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. A novel signaling molecule p130, forms stable complexes in vivo with v-Crk and v-Src in a tyrosine phosphorylation-dependent manner. EMBO J. 1994;13(16):3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Barrett A, Pellet-Many C, Zachary IC, Evans IM, Frankel P. p130Cas: a key signalling node in health and disease. Cell. Signal. 2013;25(4):766–777. doi: 10.1016/j.cellsig.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 157.Nakamoto T, Sakai R, Honda H, Ogawa S, Ueno H, Suzuki T, Aizawa S, Yazaki Y, Hirai H. Requirements for localization of p130cas to focal adhesions. Mol. Cell. Biol. 1997;17(7):3884–3897. doi: 10.1128/mcb.17.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sawada Y, Tamada M, Dubin-Thaler BJ, Cherniavskaya O, Sakai R, Tanaka S, Sheetz MP. Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell. 2006;127(5):1015–1026. doi: 10.1016/j.cell.2006.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Radha V, Mitra A, Dayma K, Sasikumar K. Signalling to actin: role of C3G a multitasking guanine-nucleotide-exchange factor. Biosci. Rep. 2011;31(4):231–244. doi: 10.1042/BSR20100094. [DOI] [PubMed] [Google Scholar]
- 160.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4(8):574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 161.Gustavsson A, Yuan M, Fällman M. Temporal dissection of beta1-integrin signaling indicates a role for p130Cas-Crk in filopodia formation. J. Biol. Chem. 2004;279(22):22893–22901. doi: 10.1074/jbc.M309693200. [DOI] [PubMed] [Google Scholar]
- 162.Garton AJ, Burnham MR, Bouton AH, Tonks NK. Association of PTP-PEST with the SH3 domain of p130cas; a novel mechanism of protein tyrosine phosphatase substrate recognition. Oncogene. 1997;15(8):877–885. doi: 10.1038/sj.onc.1201279. [DOI] [PubMed] [Google Scholar]
- 163.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285(5430):1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 164.Bouton AH, Riggins RB, Bruce-Staskal PJ. Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene. 2001;20(44):6448–6458. doi: 10.1038/sj.onc.1204785. [DOI] [PubMed] [Google Scholar]
- 165.Cary LA, Han DC, Polte TR, Hanks SK, Guan JL. Identification of p130Cas as a mediator of focal adhesion kinase-promoted cell migration. J. Cell Biol. 1998;140(1):211–221. doi: 10.1083/jcb.140.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J. Biol. Chem. 1998;273(3):1689–1698. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- 167.Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adhes. Migr. 2011;5(2):160–169. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2(2):138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 169.van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim. Biophys. Acta. 2001;1538(2–3):99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 170.Razinia Z, Mäkelä T, Ylänne J, Calderwood DA. Filamins in mechanosensing and signaling. Annu. Rev. Biophys. 2012;41:227–246. doi: 10.1146/annurev-biophys-050511-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J. Cell Biol. 2007;179(5):1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Gehler S, Baldassarre M, Lad Y, Leight JL, Wozniak MA, Riching KM, Eliceiri KW, Weaver VM, Calderwood DA, Keely PJ. Filamin A-beta1 integrin complex tunes epithelial cell response to matrix tension. Mol. Biol. Cell. 2009;20(14):3224–3238. doi: 10.1091/mbc.E08-12-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Furuike S, Ito T, Yamazaki M. Mechanical unfolding of single filamin A (ABP-280) molecules detected by atomic force microscopy. FEBS Lett. 2001;498(1):72–75. doi: 10.1016/s0014-5793(01)02497-8. [DOI] [PubMed] [Google Scholar]
- 174.Yamazaki M, Furuike S, Ito T. Mechanical response of single filamin A (ABP-280) molecules and its role in the actin cytoskeleton. J. Muscle Res. Cell Motil. 2002;23(5–6):525–534. doi: 10.1023/a:1023418725001. [DOI] [PubMed] [Google Scholar]
- 175.Lad Y, Kiema T, Jiang P, Pentikäinen OT, Coles CH, Campbell ID, Calderwood DA, Ylänne J. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 2007;26(17):3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Chen HS, Kolahi KS, Mofrad MR. Phosphorylation facilitates the integrin binding of filamin under force. Biophys. J. 2009;97(12):3095–3104. doi: 10.1016/j.bpj.2009.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat. Cell Biol. 2006;8(8):803–814. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 178.Ohta Y, Suzuki N, Nakamura S, Hartwig JH, Stossel TP. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. U. S. A. 1999;96(5):2122–2128. doi: 10.1073/pnas.96.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ueda K, Ohta Y, Hosoya H. The carboxy-terminal pleckstrin homology domain of ROCK interacts with filamin-A. Biochem. Biophys. Res. Commun. 2003;301(4):886–890. doi: 10.1016/s0006-291x(03)00048-2. [DOI] [PubMed] [Google Scholar]
- 180.Ekşioğlu YZ, Scheffer IE, Cardenas P, Knoll J, DiMario F, Ramsby G, Berg M, Kamuro K, Berkovic SF, Duyk GM, Parisi J, Huttenlocher PR, Walsh CA. Periventricular heterotopia: an X-linked dominant epilepsy locus causing aberrant cerebral cortical development. Neuron. 1996;16(1):77–87. doi: 10.1016/s0896-6273(00)80025-2. [DOI] [PubMed] [Google Scholar]
- 181.Robertson SP, Twigg SR, Sutherland-Smith AJ, Biancalana V, Gorlin RJ, Horn D, Kenwrick SJ, Kim CA, Morava E, Newbury-Ecob R, Orstavik KH, Quarrell OW, Schwartz CE, Shears DJ, Suri M, Kendrick-Jones J, Wilkie AO. OPD-spectrum Disorders Clinical Collaborative Group Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat. Genet. 2003;33(4):487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- 182.Krakow D, Robertson SP, King LM, Morgan T, Sebald ET, Bertolotto C, Wachsmann-Hogiu S, Acuna D, Shapiro SS, Takafuta T, Aftimos S, Kim CA, Firth H, Steiner CE, Cormier-Daire V, Superti-Furga A, Bonafe L, Graham JM, Jr, Grix A, Bacino CA, Allanson J, Bialer MG, Lachman RS, Rimoin DL, Cohn DH. Mutations in the gene encoding filamin B disrupt vertebral segmentation. joint formation and skeletogenesis, Nat. Genet. 2004;36(4):405–410. doi: 10.1038/ng1319. [DOI] [PubMed] [Google Scholar]
- 183.Burridge K, Connell L. A new protein of adhesion plaques and ruffling membranes. J. Cell Biol. 1983;97(2):359–367. doi: 10.1083/jcb.97.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Critchley DR. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 2009;38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 185.Moser M, Legate KR, Zent R, Fässler R. The tail of integrins talin and kindlins. Science. 2009;324(5929):895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- 186.Goult BT, Xu XP, Gingras AR, Swift M, Patel B, Bate N, Kopp PM, Barsukov IL, Critchley DR, Volkmann N, Hanein D. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: implications for talin activation. J. Struct. Biol. 2013;184(1):21–32. doi: 10.1016/j.jsb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Thoss F, Dietrich F, Punkt K, Illenberger S, Rottner K, Himmel M, Ziegler WH. Metavinculin: new insights into functional properties of a muscle adhesion protein. Biochem. Biophys. Res. Commun. 2013;430(1):7–13. doi: 10.1016/j.bbrc.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 188.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16(9):453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 189.Chen H, Choudhury DM, Craig SW. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 2006;281(52):40389–40398. doi: 10.1074/jbc.M607324200. [DOI] [PubMed] [Google Scholar]
- 190.Wang JH. Pull and push: talin activation for integrin signaling. Cell Res. 2012;22(11):1512–1514. doi: 10.1038/cr.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Song X, Yang J, Hirbawi J, Ye S, Perera HD, Goksoy E, Dwivedi P, Plow EF, Zhang R, Qin J. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell Res. 2012;22(11):1533–1545. doi: 10.1038/cr.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, Campbell ID. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 2009;28(22):3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hytönen VP, Vogel V. How force might activate talin’s vinculin binding sites: SMD reveals a structural mechanism. PLoS Comput. Biol. 2008;4(2):e24. doi: 10.1371/journal.pcbi.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Margadant F, Chew LL, Hu X, Yu H, Bate N, Zhang X, Sheetz M. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 2011;9(12):e1001223. doi: 10.1371/journal.pbio.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ciobanasu C, Faivre B, Le Clainche C. Integrating actin dynamics mechanotransduction and integrin activation: the multiple functions of actin binding proteins in focal adhesions. Eur. J. Cell Biol. 2013;92(10–11):339–348. doi: 10.1016/j.ejcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 196.Desiniotis A, Kyprianou N. Significance of talin in cancer progression and metastasis. Int. Rev. Cell Mol. Biol. 2011;289:117–147. doi: 10.1016/B978-0-12-386039-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Zemljic-Harpf A, Manso AM, Ross RS. Vinculin and talin: focus on the myocardium. J. Investig. Med. 2009;57(8):849–855. doi: 10.231/JIM.0b013e3181c5e074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Beckerle MC. Zyxin: zinc fingers at sites of cell adhesion. Bioessays. 1997;19(11):949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- 199.Smith MA, Hoffman LM, Beckerle MC. LIM proteins in actin cytoskeleton mechanoresponse. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.04.009. http://dx.doi.org/10.1016/j.tcb.2014.04.009. [DOI] [PMC free article] [PubMed]
- 200.Crawford AW, Michelsen JW, Beckerle MC. An interaction between zyxin and alpha-actinin. J. Cell Biol. 1992;116(6):1381–1393. doi: 10.1083/jcb.116.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hoffman LM, Jensen CC, Kloeker S, Wang CL, Yoshigi M, Beckerle MC. Genetic ablation of zyxin causes Mena/VASP mislocalization increased motility and deficits in actin remodeling. J. Cell Biol. 2006;172(5):771–782. doi: 10.1083/jcb.200512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ngu H, Feng Y, Lu L, Oswald SJ, Longmore GD, Yin FC, Effect of focal adhesion proteins on endothelial cell adhesion. motility and orientation response to cyclic strain. Ann. Biomed. Eng. 2010;38(1):208–222. doi: 10.1007/s10439-009-9826-7. [DOI] [PubMed] [Google Scholar]
- 203.Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J. Cell Biol. 2005;171(2):209–215. doi: 10.1083/jcb.200505018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hirata H, Tatsumi H, Sokabe M. Mechanical force facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J. Cell Sci. 2008;121(Pt 17):2795–2804. doi: 10.1242/jcs.030320. [DOI] [PubMed] [Google Scholar]
- 205.Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 1997;138(5):1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr. Opin. Cell Biol. 2006;18(5):524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 207.Degenhardt YY, Silverstein S. Interaction of zyxin a focal adhesion protein with the e6 protein from human papillomavirus type 6 results in its nuclear translocation. J. Virol. 2001;75(23):11791–11802. doi: 10.1128/JVI.75.23.11791-11802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Wang Y, Gilmore TD. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta. 2003;1593(2–3):115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 209.Granzier HL, Irving TC. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys. J. 1995;68(3):1027–1044. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mayans O, van der Ven PF, Wilm M, Mues A, Young P, Fürst DO, Wilmanns M, Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395(6705):863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- 211.Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270(5234):293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- 212.Labeit S, Gautel M, Lakey A, Trinick J. Towards a molecular understanding of titin. EMBO J. 1992;11(5):1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gräter F, Shen J, Jiang H, Gautel M, Grubmüller H. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 2005;88(2):790–804. doi: 10.1529/biophysj.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schäfer LV, Brandmeier B, Gräter F, Grubmüller H, Gaub HE, Gautel M. Mechanoenzymatics of titin kinase. Proc. Natl. Acad. Sci. U. S. A. 2008;105(36):13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.LeWinter MM, Wu Y, Labeit S, Granzier H. Cardiac titin: structure, functions and role in disease. Clin. Chim. Acta. 2007;375(1–2):1–9. doi: 10.1016/j.cca.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 216.Granzier H, Labeit S. Structure-function relations of the giant elastic protein titin in striated and smooth muscle cells. Muscle Nerve. 2007;36(6):740–755. doi: 10.1002/mus.20886. [DOI] [PubMed] [Google Scholar]
- 217.LeWinter MM, Granzier HL. Titin is a major human disease gene. Circulation. 2013;127(8):938–944. doi: 10.1161/CIRCULATIONAHA.112.139717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Moraes C, Sun Y, Simmons CA. Microfabricated platforms for mechanically dynamic cell culture. J. Vis. Exp. 2010;26(46):2224. doi: 10.3791/2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tan JL1, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles:An approach to isolate mechanical force. Proc. Natl. Acad. Sci. U. S. A. 2003;100(4):1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Munevar S, Wang Y, Dembo M. Traction force microscopy of migrating normal and h-ras transformed 3t3 fibroblasts. 2001;80(4):1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Kirmizis D, Logothetidis S. Atomic force microscopy probing in the measurement of cell mechanics. Int. J. Nanomedicine. 2010;5:137–145. doi: 10.2147/ijn.s5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Besch S, Snyder KV, Zhang RC, Sachs F. Adapting the Quesant (c) Nomad (TM) atomic force microscope for biology and patch-clamp atomic force microscopy. Cell Biochem. Biophys. 2003;39(3):195–210. doi: 10.1385/cbb:39:3:195. [DOI] [PubMed] [Google Scholar]
- 223.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibro-nectin and the cytoskeleton depends on talin. Nature. 2003;424(6946):334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 224.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA moleculesby using magnetic beads. Science. 1992;258(5085):1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 225.Shroff H, Reinhard BM, Siu M, Agarwal H, Spakowitz A, Liphardt J. Biocompatible force sensor with optical readout and dimensions of 6 nm3. Nano Lett. 2005;5(7):1509–1514. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
- 226.Lee CK, Wang YM, Huang LS, Lin S. Atomic force microscopy: determination of unbinding force off rate and energy barrier for protein-ligand interaction. Micron. 2007;38(5):446–461. doi: 10.1016/j.micron.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 227.Meng F, Suchyna TM, Sachs F. A FRET based mechanical stress sensor for specific proteins in situ. FEBS J. 2008;275(12):3072–3087. doi: 10.1111/j.1742-4658.2008.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Meng F, Suchyna TM, Lazakovitch E. Real time FRET based detection of mechanical stress in cytoskeletal and extracellular matrix proteins. Cell. Mol. Bioeng. 2011;4(2):148–159. doi: 10.1007/s12195-010-0140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Guo J, Sachs F, Meng F. Fluorescence-based force/tension sensors: a novel tool to visualize mechanical force in structural proteins in live cells. Antioxid. Redox Signal. 2014;20(6):986–999. doi: 10.1089/ars.2013.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Forster T. Experimentelle und theoretische Untersuchungdes zwischenmolekularen Ubergangs von Elektronenanregungsenergie. Z. Naturforsch. A. 1949;4:321–327. [Google Scholar]
- 231.Forster VT. Zwischenmolekulare energiewanderung undfluoreszenz. Ann. Phys. 1948;6:54–75. [Google Scholar]
- 232.Kemp-O’Brien K, Parsons M. Using FRET to analyse signals controlling cell adhesion and migration. J. Microsc. 2013;251(3):270–278. doi: 10.1111/j.1365-2818.2012.03686.x. [DOI] [PubMed] [Google Scholar]
- 233.Day Richard N, Davidson Michael W. Fluorescent proteins for FRET microscopy: monitoring protein interactions in living cells. Bioessays. 2012;34(5):341–350. doi: 10.1002/bies.201100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ishikawa-Ankerhold HC, Ankerhold R, Drummen GP. Advanced fluorescence microscopy techniques—FRAP FLIP, FLAP FRET and FLIM. Molecules. 2012;17(4):4047–4132. doi: 10.3390/molecules17044047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Meng F, Sachs F. Orientation-based FRET sensor for real-time imaging of cellular force. J. Cell Sci. 2012;125(3):743–750. doi: 10.1242/jcs.093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Loura LM, de Almeida RF, Silva LC, Prieto M. FRET analysis of domain formation and properties in complex membrane systems. Biochim. Biophys. Acta. 2009;1788(1):209–224. doi: 10.1016/j.bbamem.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 237.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466(7303):263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rahimzadeh J, Meng F, Sachs F, Wang J, Verma D, Hua SZ. Real-time observation of flow-induced cytoskeletal stress in living cells. Am. J. Physiol. Cell Physiol. 2011;301(3):646–652. doi: 10.1152/ajpcell.00099.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Meng F, Sachs F. Visualizing dynamic cytoplasmic force with acompliance-matched FRET sensor. J. Cell Sci. 2011;124(2):261–269. doi: 10.1242/jcs.071928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Guo J, Wang Y, Sachs F, Meng F. Actin stress in cell reprogramming. Proc. Natl. Acad. Sci. U. S. A. 2014;111(49):E5252–E5261. doi: 10.1073/pnas.1411683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 242.Chowdhury F, Na S, Li D, Poh YC, Tanaka TS, Wang F, Wang N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2010;9(1):82–88. doi: 10.1038/nmat2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Majkut S, Dingal PC, Discher DE. Stress sensitivity and mechanotransduction during heart development. Curr. Biol. 2014;24(10):R495–R450. doi: 10.1016/j.cub.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Borghi N, Sorokina M, Shcherbakova OG. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc. Natl. Acad. Sci. U. S. A. 2012;109(31):12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tseng CY, Wang A, Zocchi G, Rolih B, Levine AJ. Elastic energy of protein–DNA chimeras. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2009;80(6 Pt 1):061912. doi: 10.1103/PhysRevE.80.061912. [DOI] [PubMed] [Google Scholar]
- 246.Sanchez DS, Qu H, Bulla D, Zocchi G. The elastic energy of sharply bent nicked DNA. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2013;87(2):022710. doi: 10.1103/PhysRevE.87.022710. [DOI] [PubMed] [Google Scholar]
- 247.Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc. Natl. Acad. Sci. U. S. A. 2008;105(18):6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]