Abstract
Metazoan photochemistry involves cis-trans isomerization of a retinylidene chromophore bound to G protein coupled receptors. Successful production of chromophore is critical for photoreceptor production and survival. For chromophore production, animals have to choose from more than 600 naturally occurring carotenoids and process them by oxidative cleavage and geometric isomerization of double bonds. Vertebrates employ three carotenoid cleavage oxygenases to tailor the carotenoid precursor in the synthesis of 11-cis-retinal (vitamin A1). Lepidoptera (butterfly and moth) possess only one such enzyme, NinaB, which faces the challenge to catalyze these reactions in unison to produce 11-cis-3-hydroxy-retinal (vitamin A3). We here showed that key to this multitasking is a bipartite substrate recognition site that conveys regio- and stereo-selectivity for double bond processing. One side performed the specific C11, C12 cis-isomerization and preferentially binds 3-OH-β-ionone rings sites. The other side maintained a trans configuration in the resulting product and preferentially binds non-canonical ionone ring sites. Concurrent binding of carotenoids containing two cyclohexyl rings to both domains is required for specific oxidative cleavage at position C15, C15’ of the substrate. The unique reaction sequence follows a dioxygenase mechanism with a carbocation/radical intermediate. This ingenious quality control system guarantees 11-cis-3-hydroxy-retinal production, the essential retinoid for insect (vitamin A3) vision.
Photoreception is an elegant example of nature’s engineering exploits to evolve complex biological systems from a limited number of genes. Though eyes have evolved several times during evolution, arthropods and vertebrates share common transcription factors for early development. Moreover, the photochemistry of all metazoan eyes employs visual G-protein coupled receptors (opsins) and cis-trans isomerization of the Schiff’ base bound retinylidene chromophore. Four different chromophores have been discovered in animals. Vertebrates use retinal (vitamin A1) and 3-dehydroretinal (vitamin A2). Arthropods use retinal and 3-hydroxy-retinal (vitamin A3) and in some crustaceans 4-hydroxy-retinal (vitamin A4) is found.1
Successful production of chromophore is critical for photoreceptor function and survival. Binding of chromophore to opsin promotes visual pigment maturation and determines their spectral sensitivity.2 For instance, 3-dehydoretinal absorbs at longer wavelengths than retinal, and so do the respective visual pigments.3 On the contrary, binding of 3-hydroxy-retinal to opsin shifts the spectral sensitivity of the visual pigments to shorter wavelengths when compared to the same molecule with bound retinal.4 Thus, the overwhelming majority of species use a single chromophore to avoid ambiguity in spectral sensitivity of different types of photoreceptors.
More than 600 naturally occurring carotenoids have been identified in nature and therefore mechanisms must have evolved, in animals, to differentiate between them for chromophore synthesis. Individual carotenoids differ in length of their polyene chromophore, shifts of their conjugated double bonds, their terminal ring sites, and their functional groups. The enormous diversity of these pigments poses a challenge to vitamin A biology. Enzymes involved in chromophore production must display stereo- and regio- selectivity for ring sites and specific double bonds of their carotenoid precursors. Production of non-canonical byproducts may impair visual pigment maturation, shift their spectral sensitivities, and eventually lead to photoreceptor cell death and blindness.
Key to chromophore synthesis is a class of structurally related carotenoid cleavage oxygenases (CCOs). These enzymes share a characteristic fold with a unique iron-coordination system. Hydrophobic tunnels and hydrophobic patches at the tunnel entrance of these structures are critical for the interaction with their lipophilic substrates. Vertebrates employ three different family members in chromophore production. β,β-carotene-15,15′-dioxygenase (BCO1) cleaves carotenoids with at least one β-ionone ring (provitamin A) symmetrically to produce all-trans-retinal.5–8 The primary cleavage product is reduced and ultimately converted to all-trans-retinyl esters by lecithin: retinol acyltransferase.7, 9, 10 Then, isomerization of all-trans-retinyl esters into 11-cis-retinol is carried out by the other CCO family member, the retinal pigment epithelium-specific 65 kDa protein (RPE65).11–13 Loss-of-function mutations in RPE65 cause devastating blinding diseases such as Leber congenital amaurosis and retinitis pigmentosa.14, 15 A third vertebrate CCO, carotenoid-9’, 10’-dioxygenase (BCO2), localizes to the mitochondria where it cleaves carotenoids asymmetrically at the C9, C10 double bond.16 Mounting evidence suggests that BCO2 tailors asymmetric provitamin A carotenoids by removing the non-canonical ring site and in doing so, control the homeostasis of non-provitamin A carotenoids. 17–20
Insect genomes encode a single CCO family member encoded by the gene, neither inactivation nor after potential B (ninaB). 2, 21–23 Similar to RPE65 in humans, mutations in the NinaB gene in Drosophila, render these insects blind.24 Using NinaB from the great wax moth (Galleria mellonella), we previously showed that the reaction is carried out in a single step, wherein a carotenoid substrate is converted into virtually equimolar amounts of 11-cis and all-trans retinoid isomers.23 Here we characterized the biochemical foundation of substrate specificity, regio-selectivity of oxidative cleavage and geometric isomerization, as well as catalytic mechanism. NinaB was found to contain two domains within its structure. One domain performs the specific C11, C12 cis-isomerization and preferentially binds 3-OH-β-ionone rings sites. The other domain maintains an all-trans configuration in the resulting retinoid product. Concurrent binding of carotenoids containing two cyclohexyl rings to both domains is required for specific isomerization and oxidative cleavage. The unique reaction sequence follows a dioxygenase mechanism with a carbocation/radical intermediate to produce the 11-cis-3-hydroxy-retinal, the unique chromophore of insects.
RESULTS
NinaB polypeptide Catalyzes Oxidative Cleavage and Geometric Isomerization
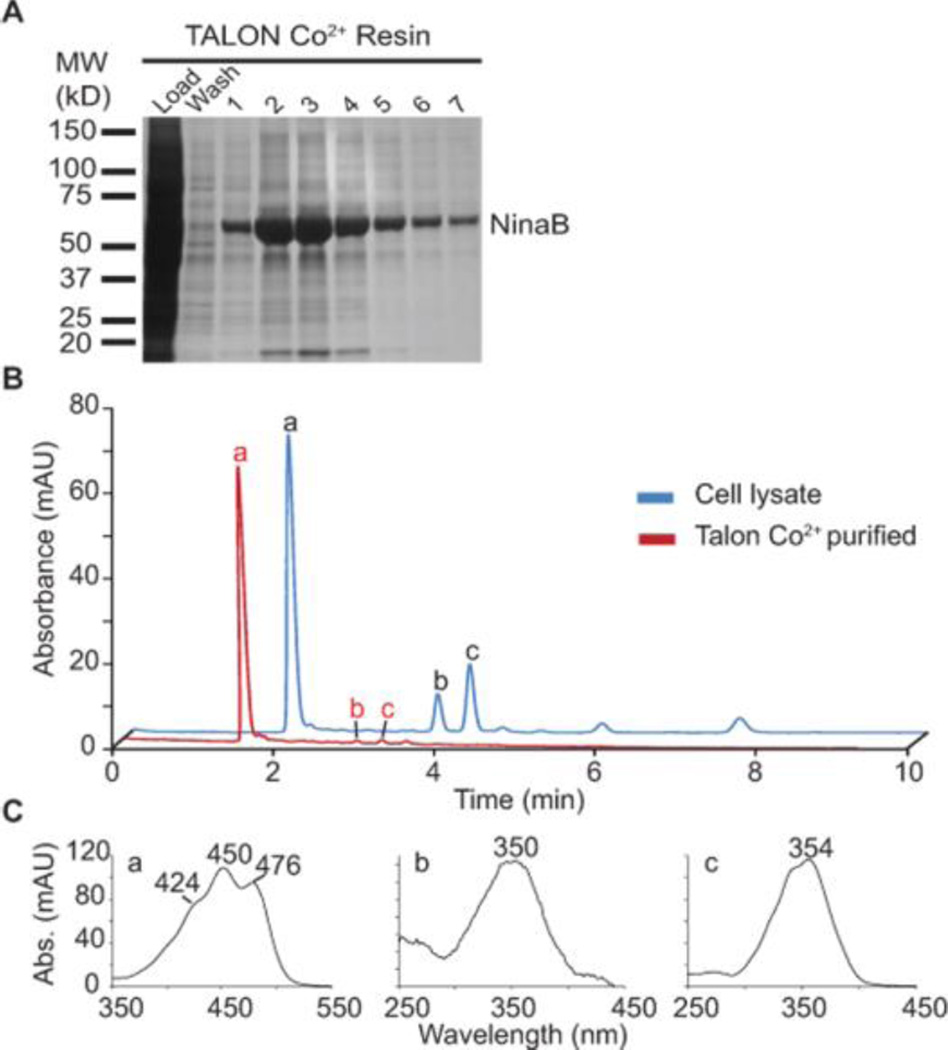
Expression of active, recombinant NinaB (Galleria mellonella) containing an N-terminal His-tag was achieved with the baculovirus expression system and Sf9 insect cells. Talon Co2+ purification yielded apparently pure enzyme (Figure 1A). Enzyme assays testing the isomerooxygenase activity of the recombinant purified enzyme with β-carotene showed enzymatic activity (Figure 2B and Figure 1B, blue traces). Notably, β-carotene conversion by NinaB yielded 11-cis-retinal and all-trans-retinal in nearly equal amounts. Other retinal stereoisomers were largely absent. Thus, we concluded that the NinaB polypeptide can catalyze oxidative cleavage at C15, C15’ and geometric isomerization of the C11, C12 double bonds in the absence of any added cofactor.
Figure 1.
Purification and tests for enzymatic activity of NinaB on β,β-carotene. (A) Coomasie stained SDS-PAGE gel showing purity of recombinant NinaB after purification with Talon Co2+ resin. (B) HPLC profiles at 360 nm of lipid extracts from in vitro tests for enzymatic activity from NinaB cell lysate (blue trace) and Talon purified NinaB enzyme (red trace). (C) UV/Vis spectra of corresponding peaks in (B) peak. a, β,β-carotene; b, 11-cis-retinal; c, all-trans-retinal.
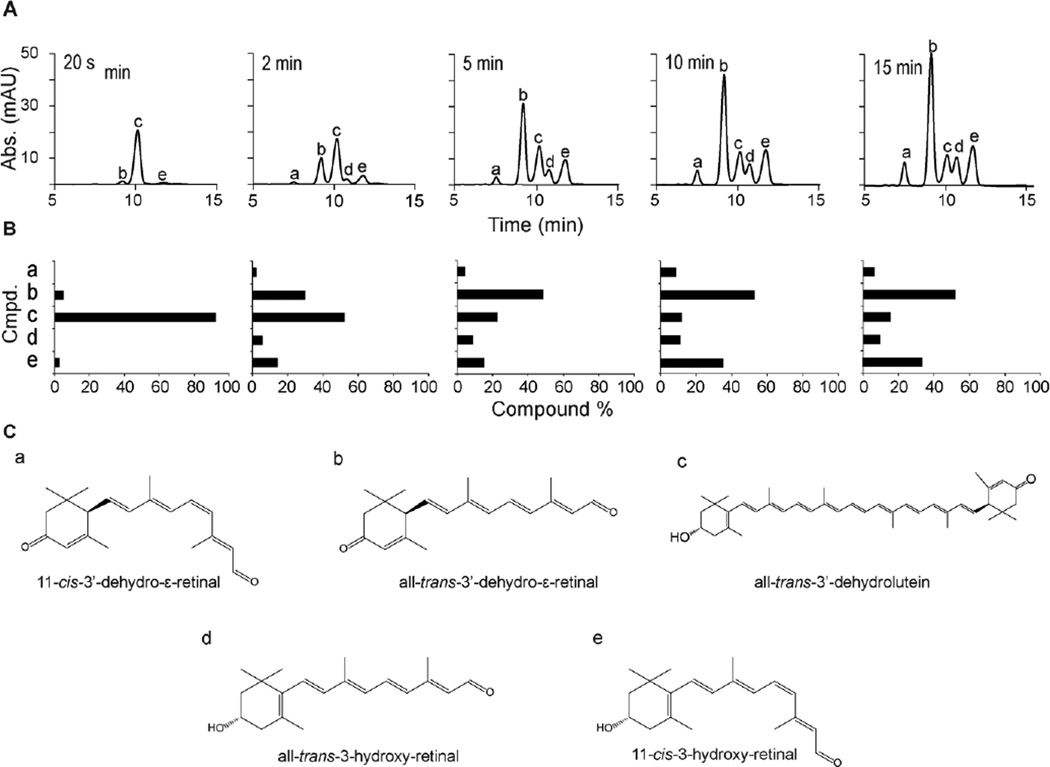
Figure 2.
Analysis of NinaB’s enzymatic action on all-trans-3’-dehydrolutein. (A) HPLC profiles at 420 nm of lipid extracts from timed in vitro tests for enzymatic activity from NinaB cell lysate. (B) Graphical representation of compound percentage from data shown in (A). (C) Chemical structures of the resulting products detected from in vitro enzymatic activity of NinaB on all-trans-3’-dehydrolutein.a, 11-cis-3’-dehydro-ε-retinal; b, all-trans-3’-dehydro-ε-retinal; c, all-trans-3’-dehydrolutein; d, all-trans-3-hydroxy-retinal; e, 11-cis-3-hydroxy-retinal.
NinaB’s Bipartite Substrate Recognition Site
We previously detected 3-hydroxy-β, β-carotene (β-cryptoxanthin), 3, 3’-dihydroxy-β, β-carotene (zeaxanthin) and 3, 3’-dihydroxy-β,ε-carotene (lutein) as major carotenoids of the wax moth.23 Additionally, we showed that NinaB preferentially isomerizes the 3-hydroxy-β-ionone ring site of these carotenoid precursors to produce 11-cis-3-hydroxy-retinal, the unique chromophore of insects. To clarify the biochemical foundation of this action we employed various synthetic and natural carotenoids in tests with NinaB protein extracts. First, we carried out enzymatic assays with the asymmetrically, engineered substrate, all-trans-3’-dehydrolutein (Figure 2 and Figure 3A and B). Incubation for 20 s produced only two of four possible products, all-trans-3’-dehydro-ε-retinal and 11-cis-3-hydroxy-retinal (b and e respectively, Figure 2C). Longer incubations of 2 to 15 min did produce the alternative products, i.e. 11-cis-3’-dehydro-ε-retinal and all-trans-3-hydroxy-retinal in approximately equimolar amounts (a and d respectively, Figure 2), but a consistent preferred production of all-trans-3’-dehydro-ε-retinal and 11-cis-3-hydroxy-retinal was observed. With this asymmetrical carotenoid, we demonstrated NinaB’s catalytic preference for isomerization on 3-OH-β-ionone ring sides as opposed to 3’-dehydro-ε-ring sides.
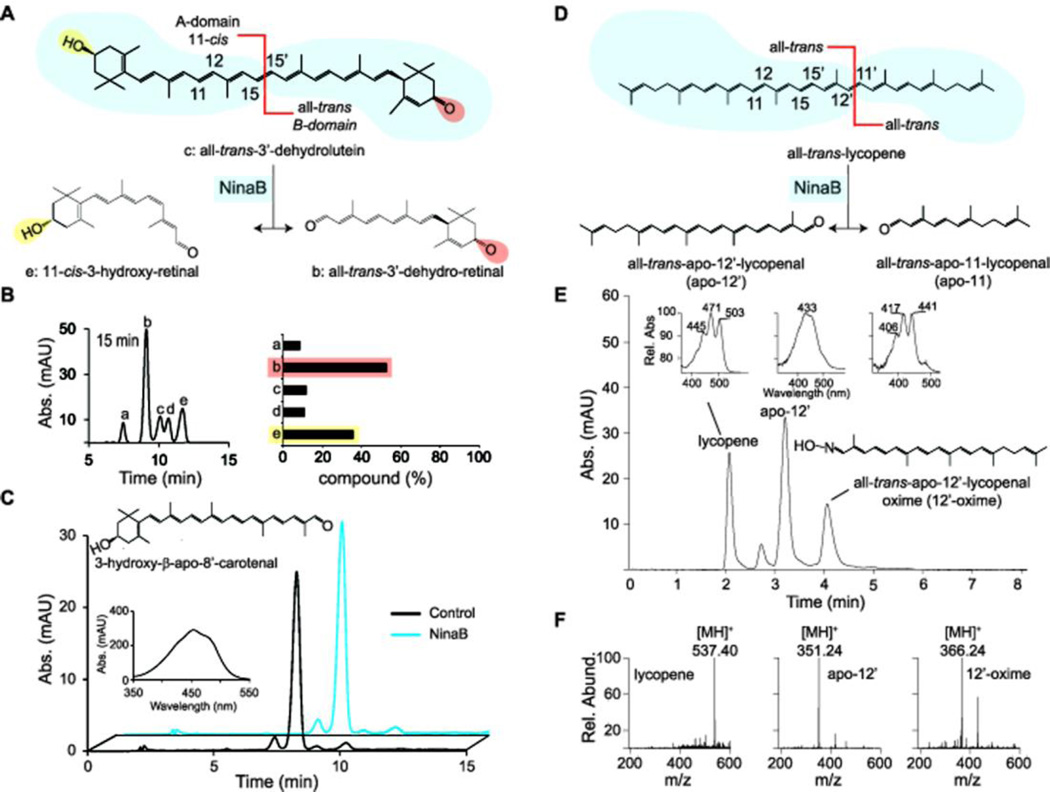
Figure 3.
Regio-selectivity of oxidative cleavage and geometric isomerization (A) Schematic representing the preferred stereoselectivity of NinaB on the asymmetric carotenoid, all-trans-3’-dehydrolutein. (B) HPLC profile at 420 nm of lipid extracts from in vitro enzymatic assays with all-trans-3’-dehydrolutein after 15 min incubations at 28°C. The graph gives the composition of different cleavage products (a, 11-cis-3’-dehydro-ε-retinal; b, all-trans-3’-dehydro-ε-retinal; c, all-trans-3’-dehydrolutein; d, all-trans-3-hydroxy-retinal, e, 11-cis-3-hydroxy-retinal) as percent of total products. (C) HPLC profiles at 420 nm of lipid extracts from in vitro assays of NinaB cell lysate (blue trace) and control (black trace) with the apocarotenoid, 3-hydroxy-β-apo-8’-carotenal. (D) Schematic of all-trans-lycopene’s asymmetrical oxidative cleavage by NinaB. (E) HPLC chromatogram of lipid extracts at 360 nm of lipid extracts from in vitro enzymatic assays of NinaB and lycopene. Insets: UV spectra of the substrates and products. (F) Mass spectral analysis of the lipid extracts from (E). apo-12, all-trans-apo-12’-lycopenal; 12’-oxime, all-trans-apo-12’-lycopenal oxime.
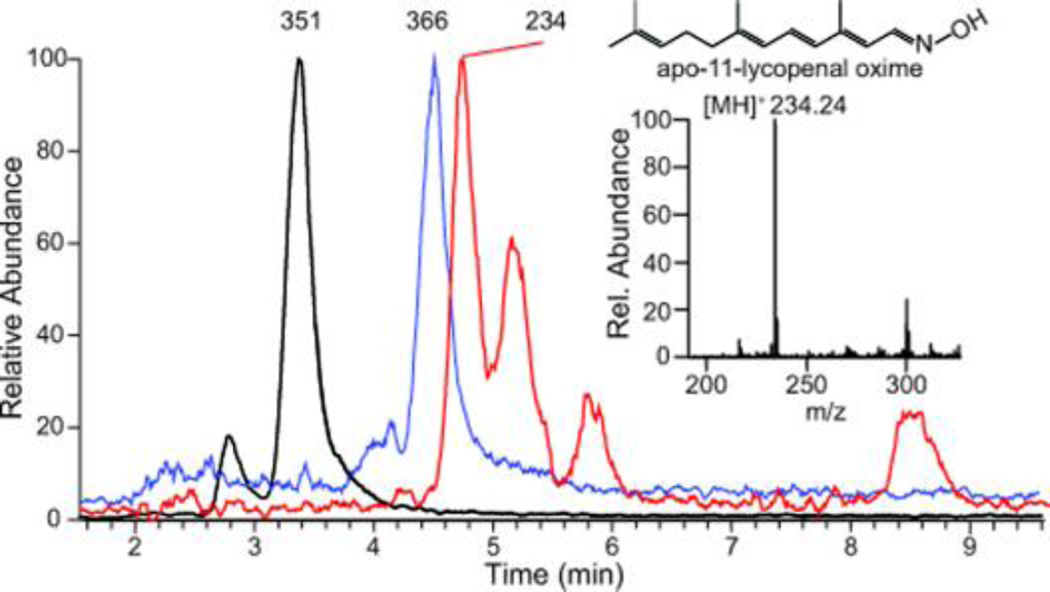
We next tested NinaB’s activity on apocarotenoids with one β-ionone ring site. NinaB cell lysate enzymatic assays with 3-hydroxy-β-apo-8’-carotenal (Figure 3C) and β-apo-8’-carotenal did not produce detectable products under two trialed methodologies. Inhibition studies were also carried out, but apocarotenoids were unable to inhibit β-carotene cleavage by NinaB. This indicates that the binding affinities of these apocarotenoids are magnitudes lower than those of carotenoids with two cyclohexyl rings. In contrast, vertebrate BCO1 and BCO2 readily catalyzed conversion of respective apocarotenoids, thus excluding that the enzyme had no access to the substrates under the applied test conditions.17, 20 To further analyze the role of ionone ring sites for proper carotenoid processing by NinaB, we subjected the acyclic all-trans-lycopene to NinaB enzymatic activity assays. We then subjected lipids extracted in the presence of hydroxylamine to UV/Vis-LC and LC-MS analysis. This analysis revealed the production of apo-12’-lycopenal and apo-12’-lycopenal oxime, (Figure 3D–F) with confirmed m/z’s of 351.24 [MH]+ and 366.24 [MH]+, respectively (Figure 3E). Due to the volatile nature of the second product, apo-11-lycopenal, UV/Vis detection was not possible, but the extracted ion MS chromatogram with m/z=234.24 [MH]+, the oxime derivative, demonstrated its enzymatic production (Figure 4, red plot).
Figure 4.
Extracted ion chromatograms of lipid extracts from NinaB cell lysate in vitro enzymatic assays with lycopene. Inset, mass spectrum analysis identifying apo-11-lycopenal as a product of lycopene oxidative cleavage by NinaB.
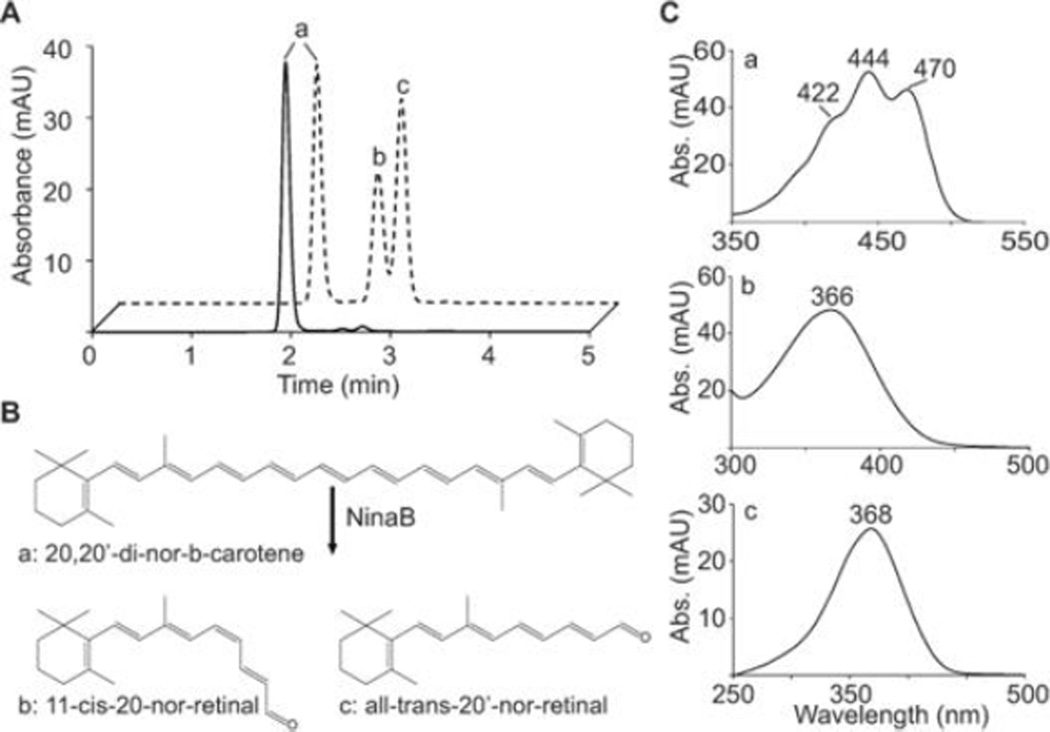
We also assessed the contribution of the methyl groups in the carbon backbone of the carotenoid substrate using 20, 20’-di-nor-β-carotene for enzymatic assays (Figure 5). HPLC analyses revealed the production of 11-cis-20-nor-retinal and all-trans-20-nor-retinal. The dual lack of methyl groups at carbons 20 and 20’ did not affect NinaB’s enzymatic activity, further suggesting that the cyclohexyl rings contain the major points for substrate recognition. Together, these analyses revealed that a minimal molecular diameter of the substrate is required for enzymatic processing by NinaB. The finding that all-trans-lycopene is cleaved asymmetrically at C11, C12 led us to infer that cyclohexyl rings orient the substrate to determine regio-selectivity of oxidative cleavage at the C15, C15’ double bond. Finally, we observed that the 3-OH-β-ionone ring side is preferentially, geometrically isomerized at C11, C12 in an asymmetric bicyclic carotenoid substrate and that the C20 methyl group in the polyene backbone plays a minor role in substrate enzyme interaction.
Figure 5.
Analysis of NinaB’s enzymatic action on 20, 20’-di-nor-β-carotene. (A) HPLC profiles at 360 nm of lipid extracts from in vitro tests for enzymatic activity from NinaB cell lysate (dashed trace) and control (solid trace). (B) Schematic depicting of the catalytic action on 20, 20’-di-nor-β-carotene by NinaB. (C) UV/Vis spectra of the corresponding products and the substrate in (A).
NinaB Displays a Hydrophobic Substrate Binding Cleft
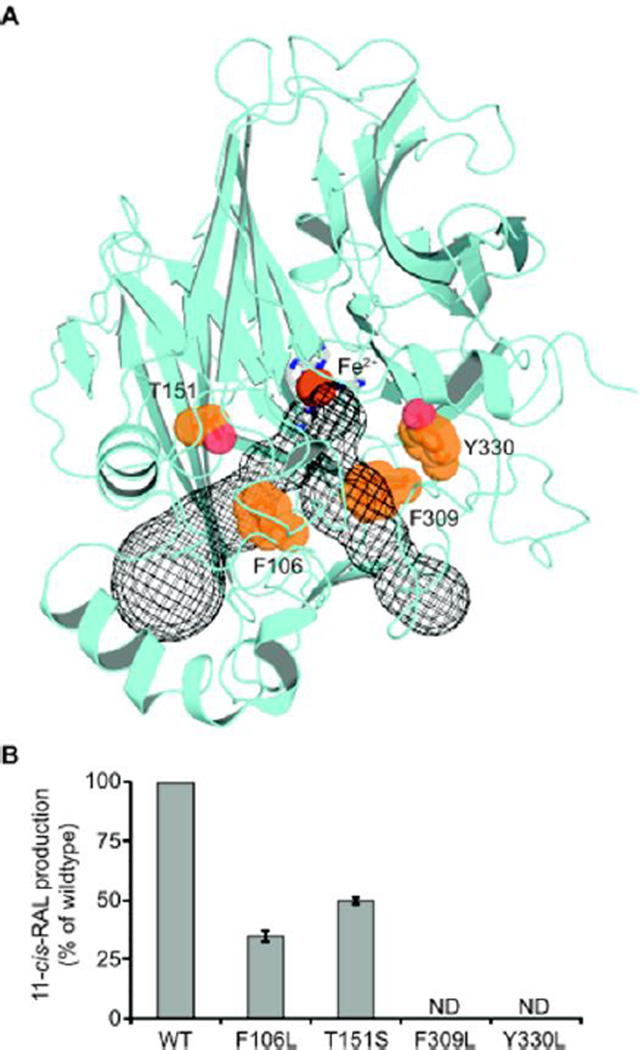
Recently, native bovine RPE65 has been crystalized in complex with the retinoid-mimetic drug ACU-4429 (emixustat).25 This study revealed that the active site, retinoid-binding cavity is located near the membrane-interacting surface as well as a Fe-bound palmitate ligand positioned in an adjacent pocket. The architecture of the arrangement of the enzyme/substrate complex (Michaelis complex) of a canonical double bond- cleaving CCO has not yet been solved. From analysis with various symmetric and asymmetric substrates, we infer that NinaB possesses a similar bipartite substrate binding. To provide evidence that NinaB adopts such a fold we generated a 3D homology structures, using the crystal structures of RPE65 (PDB 4F2Z) and ACO (PDB 2BIW) as templates (Figure 6A). This model predicted a basic structural fold consisting of a rigid seven-bladed β-propeller similar to other CCOs. A structural space in the active site domain near the four, absolutely conserved histidines, ligated to the iron co-factor was present. Also present was the presumed dual purposed, substrate entry and product exit membrane tunnel solved in RPE65; however the ACO-based NinaB model, predicts a secondary opening that is connected to the substrate binding site and the membrane tunnel (Figure 6A, blue mesh).
Figure 6.
NinaB structural model highlighting, in orange, residues within the substrate binding tunnel (blue mesh). (A) Effects site-directed mutagenesis on residues shown in (B) on NinaB catalytic activity as measured by the percent production of 11-cis-retinoids when compared to wild-type NinaB. Values given are the mean ± S.D. (error bars) of three independent measurements, ND, not detectable.
Previously, it was reported that specific residues of RPE65 stabilize the carbocation intermediate and facilitate the production of different cis-isomers, specifically 11- and 13-cis.26 Some of these residues were conserved in the NinaB substrate tunnel (Figure 6). Mutating two of these conserved residues, F106L and T151S in NinaB, diminished the enzymatic activity as measured by the production of 11-cis-retinal to 40 and 50% of control levels, respectively (Figure 6B). Mutation of F309 and Y330, two aromatic residues present within the predicted binding cleft of NinaB into leucine, abolished the enzyme’s activity entirely (Figure 6B). Thus, molecular modeling and mutagenesis studies provided evidence that a bipartite substrate tunnel is well conserved in NinaB and that aromatic amino acid residues play an important role in the substrate enzyme interaction.
Utilization of Dioxygen by NinaB
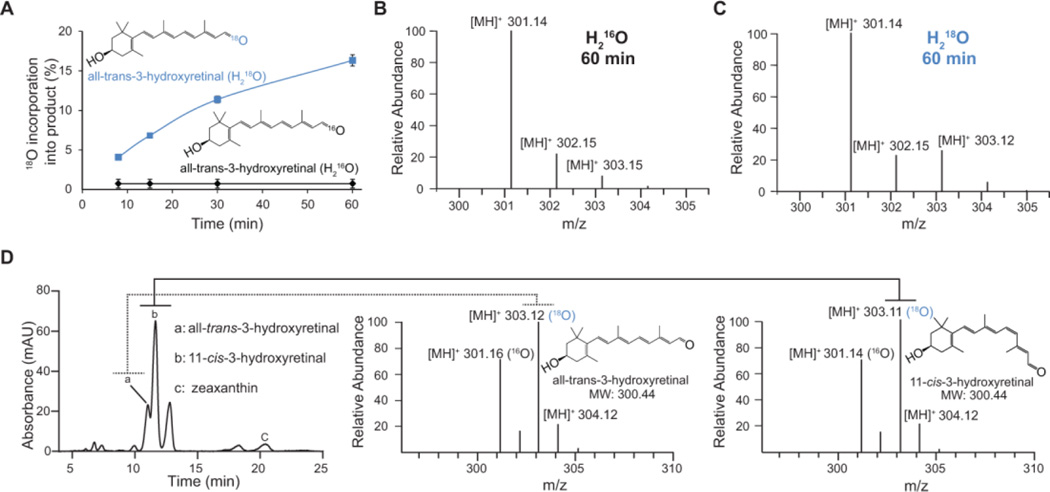
Recent studies proposed a reaction mechanism for retinoid isomerization by RPE65 25–27. In this sequence, Fe (II) catalyzes an alkyl bond cleavage followed by a trans-cis isomerization of the generated carbocation. Finally, the carbocation is quenched by the solvent water. This indicates a pronounced difference between the reaction catalyzed by RPE65 and canonical double bond cleaving CCOs. In fact we showed that canonical CCOs are dioxygenases whereas RPE65 catalyzes retinoid isomerization oxygen-independently.28 NinaB combines oxidative cleavage and geometric isomerization and is an intriguing hybrid type enzyme. To clarify how this isomerooxygenase reaction proceeds, we conducted heavy isotope labeling experiments with both H2 18O and 18O in combination with 16O and H2 16O, respectively. Heavy isotope incorporation into the all-trans-3-hydroxyretinal product formed under H2 16O/16O (control) and H2 18O/16O was tracked by LC-MS (Figure 7A–C) and found to be time-dependent. For the longest incubation period tested, 60 min, a 15% of 18O incorporation from H2 18O into all-trans-3-hydroxyretinal was recorded whereas only 4% was observed for our standard 8 min testing condition (Figure 7A). This indicated that the 18O incorporated into all-trans-3-hydroxyretinal is due to enzyme-independent oxygen exchange of the aldehyde with heavy water as opposed to enzymatic incorporation.
Figure 7.
The isomerooxygenase reaction proceeds via a dioxygenases reaction mechanism. (A) Analysis of oxygen exchange, from heavy isotope water (H218O) (blue trace), into the all-trans-retinoid product over time compared to naturally abundant, H216O water (black trace). (B) Mass spectra of resulting retinoid isotopologues of water exchange after a 60 min incubation period with H216O. (C) Mass spectra of resulting retinoid isotopologues of water exchange after a 60 min incubation period with H218O. (D) LC-MS analysis of 18O2 incorporation into the all-trans and 11-cis-retinoid products of zeaxanthin cleavage and isomerization by NinaB.
Monooxygenase-type mechanisms require 50:50 incorporation of molecular oxygen and oxygen from bulk water into the product. These results clearly indicated that the oxidative cleavage does not follow a monooxygenase mechanism; therefore we proceeded to test if the catalysis is performed via a dioxygenase mechanism. Mass spectral analysis of the cleavage reaction performed for 8 min under an 18O atmosphere showed an equal ratio of 18O incorporation into both products. The percent of incorporation, as determined by the ratio of the integrals of the extracted ion chromatograms of 18O-isotopologue to total isotopologues, was 58.4% for 11-cis-3-hydroxyretinal and 58.3% for all-trans-3-hydroxyretinal (Figure 7D). Theoretically, incorporation of molecular 18O should reach 100%, but incomplete labeling can be attributed to partial degassing of the reaction buffer. Together, these experiments provided evidence that oxidative cleavage at C15, C15’ follows a dioxygenase reaction mechanism as has been shown for other CCOs. 5, 29
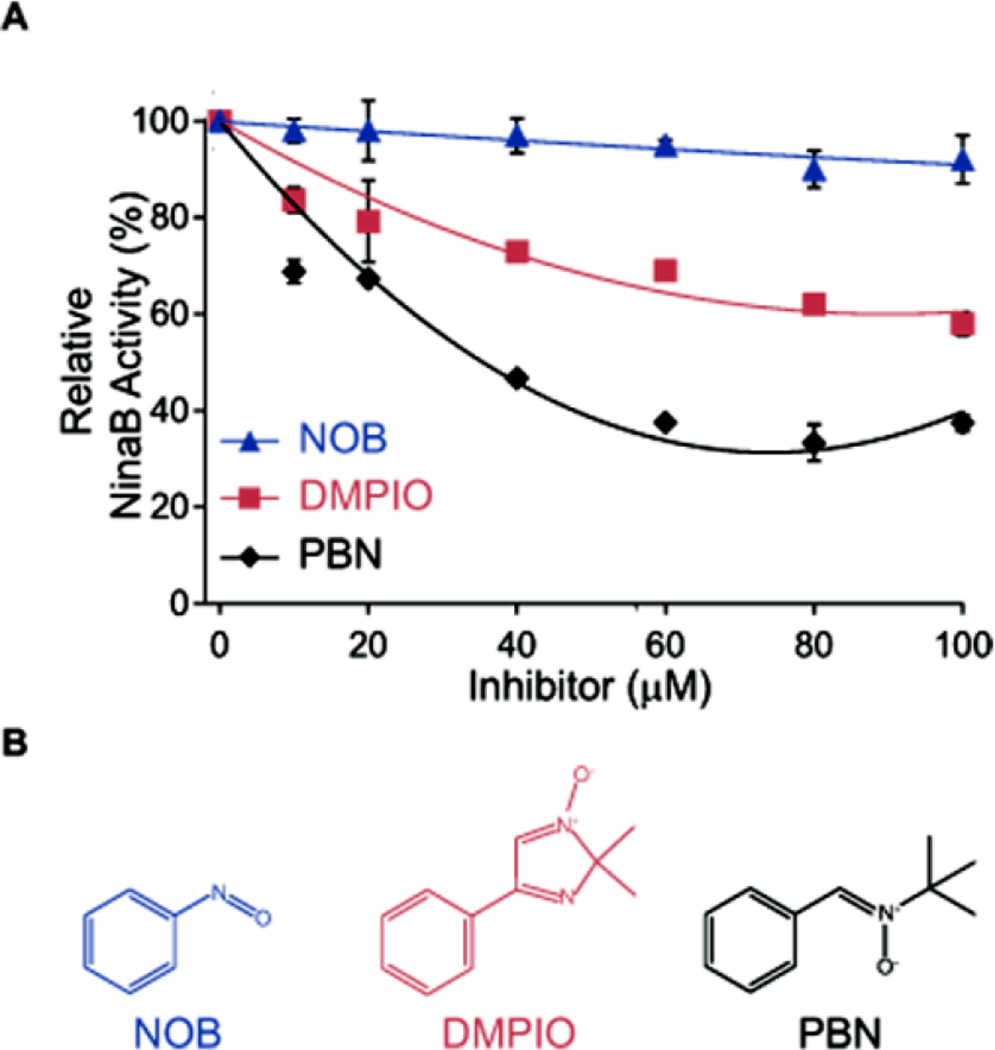
Redmond and coworkers have provided evidence that the isomerization reaction catalyzed by RPE65 involves a radical cation intermediate stabilized around the 11, 12-carbon double bond by specific residues 26, 27. These residues are conserved in NinaB (F106 and T151) and, as shown above, play a role in enzyme catalysis (Figure 6). To investigate the possibility that isomerization at C11, C12 may also occur via a radical intermediate, the NinaB reaction was probed in the presence of aromatic spin traps (Figure 8). All spin traps tested inhibited NinaB’s activity but to varying degrees. The most potent inhibitor was N-tertiary-butyl nitrone (PBN) followed by 2, 2-dimethyl-1-oxido-4-phenylimidazol-1-ium (DMPIO) and least potent was nitrosobenzene (NOB) (Figure 8). At equimolar levels of substrate, in this instance 20 µM of β-carotene, PBN reduced the enzyme’s activity by 30% whereas at 100 µM, the activity was reduced by 60%. Enzyme activity was measured as the production of all-trans and 11-cis-retinal. Thus, heavy isotope labeling provided evidence that NinaB is a dioxygenase and inhibition by spin traps suggests that the reaction proceeds via a radical intermediate.
Figure 8.
NinaB reaction is inhibited by various spin traps. (A) Enzymatic activity of NinaB in the presence of incremental concentrations of spin trap compounds. The activity is given in percent normalized to assays lacking spin traps. Each point value represents the mean ± S.D. (error bars) of three independent measurements. (B) Chemical structures of the spin traps used in (A). NOB, nitrosobenzene; DMPIO, 2,2-dimethyl-1-oxido-4-phenylimidazol-1-ium; PBN, N-tert-Butyl-a-phenylnitrone.
DISCUSSION
Our previous studies in Drosophila revealed that chromophore production takes place in the eyes since an eyeless mutant accumulates carotenoids but completely lacks retinoids.24 Additionally, carotenoids are mandatory for this process and its conversion to chromophore occurs light-independently and requires no additional enzyme.2, 30, 31 Drosophila similar to the moth Galleria, contain symmetric and asymmetric carotenoids, namely zeaxanthin, cryptoxanthin, and lutein. Previously, we showed that all these carotenoids are substrates for NinaB.23 Thus, without a directed interaction of NinaB with these substrates non-canonical retinoids would be synthesized. However, only vitamin A3 is found in the eyes of these insects.32 We have evidence to propose that NinaB contains a bipartite substrate recognition site that provides this specificity. One side is responsible for geometric isomerization across the 11, 12-carbon double bond and the other for the maintenance of all-trans geometry of the non-canonical ring site. Analyses with asymmetric substrates revealed that one side displays a greater affinity for 3-OH-β-ionone rings across which isomerization preferentially takes place. Proper binding of carotenoids with two cyclohexyl rings, symmetrical or asymmetrical, into both recognition sites provides the necessary alignment for oxidative cleavage across the 15, 15’-carbon double bond. When presented with lycopene, an acyclic carotenoid, cleavage occurs asymmetrically across the 11, 12-double bond. Also, we found that NinaB cannot oxidatively cleave apocarotenoids, further establishing the enzyme’s need for two rings to produce chromophore. Thus, we conclude that the asymmetry of NinaB’s bipartite substrate binding and catalytic domains ensure production of chromophore with a correct geometry as it is required for maturation of insect visual pigments. 33 In vertebrates, this task is distributed among three distinct CCO family members: BCO1 cleaves at C15, C15’, BCO2 removes non-canonical ring sites to ensure quality control of vitamin A1 production, and RPE65 isomerizes the C11, C12 double bond.34 In some species, an eye-specific cytochrome P450 family member, Cyp27c1, mediates the conversion of vitamin A1 to vitamin A2.35 In contrast to NinaB, all three vertebrate CCOs readily convert apocarotenoids with only one cyclohexyl-ring site. We speculate that this property of the vertebrate enzymes evolved to enable continuous re-isomerization of the chromophore after bleaching through the visual cycle.
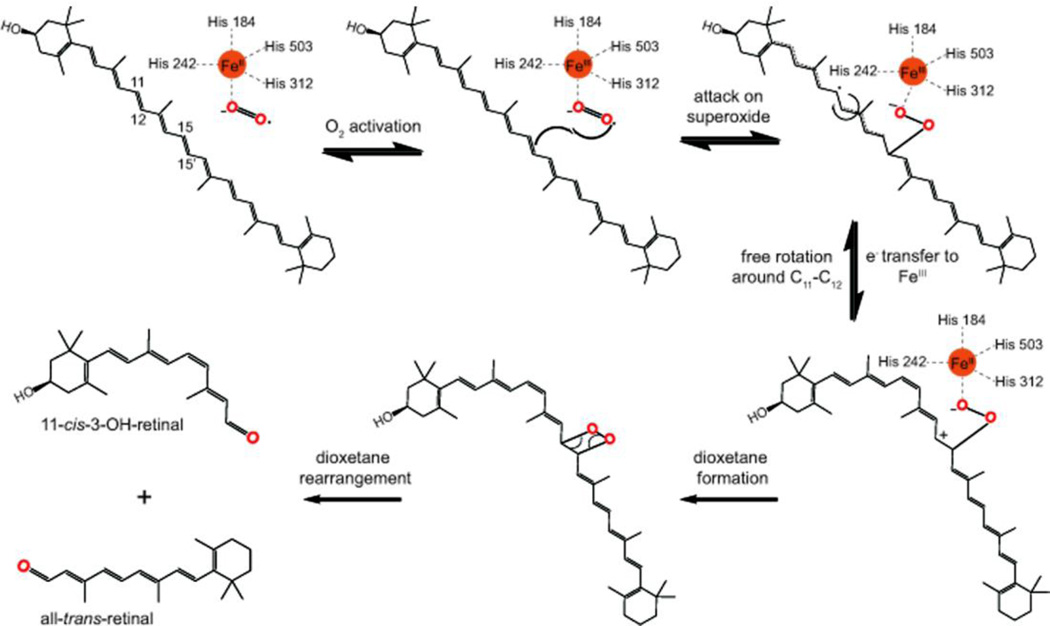
Aside from proper ring site selection, utilization of one CCO in insects instead of three in vertebrates poses an additional challenge to chromophore production. NinaB must catalyze the oxidative cleavage and geometric isomerization reactions in unison. Our present study, together with previous biochemical, structural and computational analyses allow us to propose a mechanism for this isomerooxygenase reaction. 26, 27, 29, 36, 37 The sequence comprises an initial geometric isomerization at C11, C12 followed by oxidative cleavage at C15, C15’ (Figure 9). A natural substrate such as β-cryptoxanthin would be acquired from membranes and discriminately oriented across the structurally defined tunnel with the 3-hydroxy-β-ionone ring site aligned into the isomerization domain. As with other non-heme iron oxygen-activating enzymes, the 2+ oxidation state of iron allows binding of molecular O2 and subsequent activation by a single electron transfer to produce a superoxide anion.38 At this stage, the inherent nature of carotenoids as antioxidants causes the C15, C15’ double bond to attack the superoxide and quench the free radical.37, 39 This forms a C15-O bond and a stable radical across the extensive π-electron backbone that allows the formation of a cis-configuration, a process that has precedence in non-heme iron dioxygenases.40 Rotation of the C11-C12 bond from a trans to cis-configuration occurs due to the radical being partially stabilized across C11 from complimentary interactions with F106 and T151, as evidenced by structural data from RPE65 and site directed mutagenesis of NinaB (Figure 6).25 The occurrence of this intermediate is further corroborated by inhibition of NinaB by spin traps (Figure 8). Then, an electron transfer reconstitutes iron (II) in the active center and produces a stabilized carbocation on substrate C15, priming an attack by oxygen to form a dioxetane intermediate. Quantum chemical analyses of ACO’s reaction mechanism, favors a dioxetane over an epoxide intermediate, despite the proposal that both lead to a dioxygenase labeling pattern.37 Accordingly, the heavy isotope labeling data provided here clearly show that NinaB is a dioxygenase. The final step in this reaction constitutes dioxetane rearrangement which effectively produces 3-OH-11-cis and all-trans-retinal. In the insect eyes, the chromophore then readily binds to the opsin moiety to establish visual pigments.
Figure 9.
Schematic representation of the proposed catalytic mechanism for NinaB.
CONCLUSION
Vision begins in photoreceptors, where light is absorbed and signaled to the nervous system. Throughout the animal kingdom, this process depends on a protein bound chromophore. This C20 retinoid metabolically derives from a C40 carotenoid precursor that is processed by successive oxidative cleavage and geometric isomerization reactions of double bonds. Vertebrates devote three structurally related enzymes to this task. Insects possess only one such enzyme that faces the challenge to catalyze these reactions in unison. We here show that this multitasking is achieved by a bipartite substrate binding site of NinaB that conveys regio- and stereo-selectivity for double bond processing of many naturally occurring carotenoids. The unique reaction sequence follows a dioxygenase mechanism with a carbocation/radical intermediate. Our study provides the biochemical basis of 11-cis-3-hydroxy-retinal production, the essential chromophore for insect (vitamin A3) vision.
METHODS
Synthesis of Compounds
All-trans-3’-dehydrolutein, 3-hydroxy-β-apo-8’-carotenal and 20, 20-di-nor-β-carotene were generous gifts from DSM Nutritional Products and were synthesized respectively according to 41–43.
Expression of NinaB and Cell Lysis
NinaB (Galleria mellonella) was expressed in one of two ways. First, 800 mL of Sf9 cells at a concentration of 1.6 × 106 cells / mL were infected with NinaB baculovirus and incubated at 28°C while shaking at 100 rpm for two days. Cells then were centrifuged at 4500 × g for 15 mins and cell pellets were stored at −80°C. Frozen Sf9 cell pellets (~4 g) infected with NinaB baculovirus were re-suspended in 10 mL of 20 mM Tricine (pH 7.5) sample buffer containing 200 mM NaCl, 1 mM dithiothreitol (DTT, Roche, Mannheim, Germany), one protease inhibitor cOmplete ULTRA Mini EDTA-free Tablet (Roche, Mannheim, Germany), 1 mM TCEP and decyl maltose neopentyl glycol (DMN). Cells were lysed in a glass homogenizer with 20 strokes. The lysate was centrifuged at 100,000 g for 1 h at 4°C (Beckman Coulter Optima L-90K Ultracentrifuge). The supernatant (10 mL) was collected and kept on ice until purification. Second, XL1-Blue E.coli cells were transformed with the expression vector for NinaB. Bacteria were grown at room temperature with constant shaking at 200 rpm until an OD600 of 0.6 was achieved. Protein expression was induced with L-arabinose at a final concentration of 0.02%. At the point of induction, FeSO4 and ascorbic acid were added to a final concentration of 5 µM and 10 mM, respectively and the temperature was decreased to 25°C. Protein expression proceeded for six hours. Then cells were collected by centrifugation at 4500 g for 20 min and cell pellets were stored at −80°C. Cell pellets were thawed on ice and their net weight was determined. Lysing reagent was prepared by combining 50 mL of B-PER Bacterial Protein Extraction Reagent (Life Technologies, Grand Island, NY) with one protease inhibitor cOmplete ULTRA Mini EDTA-free Tablet (Roche, Mannheim, Germany), final concentration of 2 mM ascorbic acid solution (Sigma Life Science), 1mM TCEP (Thermo Scientific, Rockford, IL), 20 U of recombinant, RNase-free DNase I. The manufacturer’s recommendation of 4 mL of lysing reagent per 1 gram of cell pellet was used to lyse and re-suspend cells. Re-suspended cell pellets were gently vortexed and allowed to incubate at room temperature for 10 min. The cell lysate was then cooled on ice and subjected to centrifugation at 100,000 g at 4°C for 30 min.
NinaB Purification
The supernatant containing NinaB, was loaded onto a 50 mL column containing 1.5 mL TALON Co2+-resin suspension (Clontech) pre-equilibrated with 5 column volumes of ice cold sample buffer. After flow-through collection, the TALON column was washed with 1 column volume of ice cold sample buffer and then with 1 column volume of sample buffer containing 1 mM imidazole. NinaB was eluted with cold sample buffer containing 50 mM imidazole (elution buffer). Aliquots of eluted NinaB fractions were collected for gel analysis. All eluted NinaB fractions were pooled and concentrated in a 30K Amicon Ultra Centrifugal Filter (Millipore) before being loaded onto a Superdex 200 10/300 GL size exclusion column (GE Healthcare Life Sciences). NinaB fractions were eluted from the column in 0.5 mL fractions at a flow rate of 0.4 mL/min. with sample buffer. Aliquots of these fractions were also collected for gel analysis. Fractions containing purified enzyme were then pooled, concentrated within a 30K Amicon Ultra Centrifugal Filter. To determine the monomeric state of purified NinaB, a mix of proteins (Bio-Rad Gel filtration Standard) with known molecular weights were run simultaneously on the size exclusion column.
Three Dimensional Structural Modeling
Homology models of Galleria NinaB, based on the structure of RPE65 (PDB accession code 4F2Z) and apocarotenoid oxygenase (PDB accession code 2BIW) were generated using the Phyre2 webserver (The Phyre2 web portal for protein modeling, prediction and analysis. Active site residue locations were consistent between these models. The ACO-based model was used for analysis owing to its preserved dual access to the active site, which is present in the alkene-cleaving CCO structures described to date. The structural model was visualized using PyMOL (Schrodinger).
Enzymatic Assays
Enzymatic assays were carried out as previously described. 30, 44 DMN micelles loaded with substrate were prepared as follows: 33 µL 3% DMN detergent solution was mixed with 10 µM (final concentration) of substrate, dissolved in acetone, in a 2-mL Eppendorf tube. This mixture was then dried in a Speedvac (Eppendorf Vacufuge Plus). To substrate, 100 µL of cell lysate was added and vortexed vigorously for 20 s and then placed on an Eppendorf thermo-shaker set at 28°C for 8 min at 300 rpm. Control assays were performed with un-induced E.coli cell pellet lysates. Reactions were stopped by adding 100 µL of water and 400 µL of acetone. Lipids were extracted by adding 400 µL of diethyl ether and 100 µL petroleum ether, then vortexed for 3 × 10 s periods, centrifuged at 15,000 × g for 1 min and finally the resulting organic phase was collected. The extraction was performed twice and the collected organic phase was dried in a Speedvac. The dried supernatant then was re-dissolved in mobile phase, 70:30 hexane:ethyl acetate and subjected to either HPLC or LC-MS analysis.
Isotope Labeling Experiments
For labeling experiments with H2 18O, NinaB cell lysate was mixed with H2 18O in a 20:80 (v:v) ratio, respectively. Assays were carried out as described above. Labeling experiments with 18O2 were performed in 10-mL screw-capped glass vials with a gastight Teflon septum. All volumes in the preparation of substrate micelles were increased 5-fold and dried accordingly in the glass vial and kept on ice. NinaB cell lysate (500 µL) was added to the glass vial and the mixture was saturated with 18O2 by aerating the solution on ice for 1 min with a release syringe placed into the septum. Then, after removal of the release syringe, the solution was saturated with18O2 once more for 2 min. The sample was vortexed and incubated at 28°C for 8 min while magnetically stirred. Reactions were stopped by adding 200 µL of water and 800 µL of acetone. This mixture was separated equally into two 2 mL Eppendorf tubes for lipid extraction. Lipids were extracted as described above with 400 µL of diethyl ether and 500 µL petroleum ether and subjected to LC-MS analyses.
HPLC and LC-MS Systems
HPLC analyses were carried out with an Agilent 1260 Infinity Quaternary HPLC system (Santa Clara, CA, USA) equipped with a pump (G1312C) with an integrated degasser (G1322A), a thermostatted column compartment (G1316A), an autosampler (G1329B), a diode-array detector (G1315D), and online analysis software (Chemstation). Analyses were performed at 25 °C using a normal-phase Zorbax Sil (5 µm, 4.6 × 150 mm) column (Agilent Technologies, Santa Clara, CA) protected with a guard column. Carotenoid and retinal separation was achieved using an isocratic composition of 70:30 (v:v) of hexane: ethyl acetate. The flow rate for all systems was 1.4 ml/min. Detection of carotenoids and apocarotenoids was performed at 420 nm wavelength. For LC-MS analyses, the eluate was directed into a LXQ linear ion trap mass spectrometer (Thermo Scientific) through atmospheric pressure chemical ionization (APCI) source working in the positive mode. To ensure optimal sensitivity, the instrument was tuned with Z as well as with retinal standards.
Site-directed Mutagenesis
Mutagenesis of NinaB was performed using the QuikChange XL site-directed mutagenesis kit (Agilent). Mutants were verified by sequence analysis of DNA maxipreps and validated plasmids purified by QIAquick purification kits (Qiagen). The primers used were F106L: 5'-GTTGTCACAGAGTTAGGTACGCGGGCCGTG-3' and 5'-CACGGCCCGCGTACCTAACTCTGTGACAAC-3'; T151S: 5'-GGAGATGAAATTTATGCTTTCTCTGAAGGTCCTG-3' and 5'-CAGGACCTTCAGAGAAAGCATAAATTTCATCTCC-3'; F309L: 5'-CGGAGCCGCTCTTGTACTTGCACATC-3' and 5'-GATGTGCAAGTACAAGAGCGGCTCCG-3'; Y330L: 5'-GACCTCTGCGCTTTAAAGGACGCCAAGGC-3' and 5'-GCCTTGGCGTCCTTTAAAGCGCAGAGGTC-3'.
Acknowledgments
This work was supported by grants from National Eye Institute (EY020551, EY023948, EY11373, EY007157). K.P. is John Hord Professor of Pharmacology.
Footnotes
AUTHOR CONTRIBUTIONS
JVL and DB designed the study and wrote the paper. DB performed all enzymatic assays and mass spectrometry analysis. MG performed mass spectrometry analysis. PDK provided 3D structure model. A.W. provided materials for experiments. All authors analyzed the data and approved the final version of the manuscript.
CONFLICT OF INTEREST
The authors declare no competing financial interest.
REFERENCES
- 1.Goldsmith TH. Evolutionary tinkering with visual photoreception. Vis Neurosci. 2013;30:21–37. doi: 10.1017/S095252381200003X. [DOI] [PubMed] [Google Scholar]
- 2.Voolstra O, Oberhauser V, Sumser E, Meyer NE, Maguire ME, Huber A, von Lintig J. NinaB is essential for Drosophila vision but induces retinal degeneration in opsin-deficient photoreceptors. J Biol Chem. 2010;285:2130–2139. doi: 10.1074/jbc.M109.056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enright JM, Toomey MB, Sato SY, Temple SE, Allen JR, Fujiwara R, Kramlinger VM, Nagy LD, Johnson KM, Xiao Y, How MJ, Johnson SL, Roberts NW, Kefalov VJ, Guengerich FP, Corbo JC. Cyp27c1 Red-Shifts the Spectral Sensitivity of Photoreceptors by Converting Vitamin A1 into A2. Curr Biol. 2015;25:3048–3057. doi: 10.1016/j.cub.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gartner W, Ullrich D, Vogt K. Quantum yield of CHAPSO-solubilized rhodopsin and 3-hydroxy retinal containing bovine opsin. Photochem Photobiol. 1991;54:1047–1055. doi: 10.1111/j.1751-1097.1991.tb02128.x. [DOI] [PubMed] [Google Scholar]
- 5.Dela Sena C, Riedl KM, Narayanasamy S, Curley RW, Jr, Schwartz SJ, Harrison EH. The human enzyme that converts dietary provitamin a carotenoids to vitamin a is a dioxygenase. J Biol Chem. 2014;289:13661–13666. doi: 10.1074/jbc.M114.557710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leuenberger MG, Engeloch-Jarret C, Woggon W-D. The Reaction Mechanism of the Enzyme-Catalyzed Central Cleavage of β-Carotene to Retinal. Angew. Chem. Int. Ed. 2001;40:2613–2617. doi: 10.1002/1521-3773(20010716)40:14<2613::AID-ANIE2613>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 7.Wyss A, Wirtz G, Woggon W, Brugger R, Wyss M, Friedlein A, Bachmann H, Hunziker W. Cloning and expression of beta,beta-carotene 15,15'-dioxygenase. Biochem Biophys Res Commun. 2000;271:334–336. doi: 10.1006/bbrc.2000.2619. [DOI] [PubMed] [Google Scholar]
- 8.Redmond TM, Gentleman S, Duncan T, Yu S, Wiggert B, Gantt E, Cunningham FX., Jr Identification, expression, and substrate specificity of a mammalian beta-carotene 15,15’-dioxygenase. J Biol Chem. 2001;276:6560–6565. doi: 10.1074/jbc.M009030200. [DOI] [PubMed] [Google Scholar]
- 9.Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Gudas LJ. Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J Biol Chem. 2005;280:40226–40234. doi: 10.1074/jbc.M509643200. [DOI] [PubMed] [Google Scholar]
- 11.Jin M, Li S, Moghrabi WN, Sun H, Travis GH. Rpe65 is the retinoid isomerase in bovine retinal pigment epithelium. Cell. 2005;122:449–459. doi: 10.1016/j.cell.2005.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moiseyev G, Chen Y, Takahashi Y, Wu BX, Ma JX. RPE65 is the isomerohydrolase in the retinoid visual cycle. Proc Natl Acad Sci U S A. 2005;102:12413–12418. doi: 10.1073/pnas.0503460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redmond TM, Poliakov E, Yu S, Tsai JY, Lu Z, Gentleman S. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. Proc Natl Acad Sci U S A. 2005;102:13658–13663. doi: 10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marlhens F, Bareil C, Griffoin JM, Zrenner E, Amalric P, Eliaou C, Liu SY, Harris E, Redmond TM, Arnaud B, Claustres M, Hamel CP. Mutations in RPE65 cause Leber's congenital amaurosis. Nat Genet. 1997;17:139–141. doi: 10.1038/ng1097-139. [DOI] [PubMed] [Google Scholar]
- 15.Bowne SJ, Humphries MM, Sullivan LS, Kenna PF, Tam LC, Kiang AS, Campbell M, Weinstock GM, Koboldt DC, Ding L, Fulton RS, Sodergren EJ, Allman D, Millington-Ward S, Palfi A, McKee A, Blanton SH, Slifer S, Konidari I, Farrar GJ, Daiger SP, Humphries P. A dominant mutation in RPE65 identified by whole-exome sequencing causes retinitis pigmentosa with choroidal involvement. Eur J Hum Genet. 2011;19:1074–1081. doi: 10.1038/ejhg.2011.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiefer C, Hessel S, Lampert JM, Vogt K, Lederer MO, Breithaupt DE, von Lintig J. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J Biol Chem. 2001;276:14110–14116. doi: 10.1074/jbc.M011510200. [DOI] [PubMed] [Google Scholar]
- 17.Amengual J, Lobo GP, Golczak M, Li HN, Klimova T, Hoppel CL, Wyss A, Palczewski K, von Lintig J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011;25:948–959. doi: 10.1096/fj.10-173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babino D, Palczewski G, Widjaja-Adhi MA, Kiser PD, Golczak M, von Lintig J. Characterization of the Role of beta-Carotene 9,10-Dioxygenase in Macular Pigment Metabolism. J Biol Chem. 2015;290:24844–24857. doi: 10.1074/jbc.M115.668822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palczewski G, Amengual J, Hoppel CL, von Lintig J. Evidence for compartmentalization of mammalian carotenoid metabolism. FASEB J. 2014;28:4457–4469. doi: 10.1096/fj.14-252411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amengual J, Widjaja-Adhi MA, Rodriguez-Santiago S, Hessel S, Golczak M, Palczewski K, von Lintig J. Two carotenoid oxygenases contribute to mammalian provitamin A metabolism. J Biol Chem. 2013;288:34081–34096. doi: 10.1074/jbc.M113.501049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 22.Seki T, Fujishita S, Ito M, Matsuoka N, Kobayashi C, Tsukida K. A fly, Drosophila melanogaster, forms 11-cis 3-hydroxyretinal in the dark. Vision Res. 1986;26:255–258. doi: 10.1016/0042-6989(86)90020-9. [DOI] [PubMed] [Google Scholar]
- 23.Oberhauser V, Voolstra O, Bangert A, von Lintig J, Vogt K. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc Natl Acad Sci U S A. 2008;105:19000–19005. doi: 10.1073/pnas.0807805105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Lintig J, Dreher A, Kiefer C, Wernet MF, Vogt K. Analysis of the blind Drosophila mutant ninaB identifies the gene encoding the key enzyme for vitamin A formation in vivo. Proc Natl Acad Sci U S A. 2001;98:1130–1135. doi: 10.1073/pnas.031576398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiser PD, Zhang J, Badiee M, Li Q, Shi W, Sui X, Golczak M, Tochtrop GP, Palczewski K. Catalytic mechanism of a retinoid isomerase essential for vertebrate vision. Nature chemical biology. 2015;11:409–415. doi: 10.1038/nchembio.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redmond TM, Poliakov E, Kuo S, Chander P, Gentleman S. RPE65, visual cycle retinol isomerase, is not inherently 11-cis-specific: support for a carbocation mechanism of retinol isomerization. J Biol Chem. 2010;285:1919–1927. doi: 10.1074/jbc.M109.027458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poliakov E, Parikh T, Ayele M, Kuo S, Chander P, Gentleman S, Redmond TM. Aromatic lipophilic spin traps effectively inhibit RPE65 isomerohydrolase activity. Biochemistry. 2011;50:6739–6741. doi: 10.1021/bi200532m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sui X, Golczak M, Zhang J, Kleinberg KA, von Lintig J, Palczewski K, Kiser PD. Utilization of Dioxygen by Carotenoid Cleavage Oxygenases. J Biol Chem. 2015;290:30212–30223. doi: 10.1074/jbc.M115.696799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt H, Kurtzer R, Eisenreich W, Schwab W. The carotenase AtCCD1 from Arabidopsis thaliana is a dioxygenase. J Biol Chem. 2006;281:9845–9851. doi: 10.1074/jbc.M511668200. [DOI] [PubMed] [Google Scholar]
- 30.Voolstra O, Kiefer C, Hoehne M, Welsch R, Vogt K, von Lintig J. The Drosophila class B scavenger receptor NinaD-I is a cell surface receptor mediating carotenoid transport for visual chromophore synthesis. Biochemistry. 2006;45:13429–13437. doi: 10.1021/bi060701u. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Wang T, Jiao Y, von Lintig J, Montell C. Requirement for an enzymatic visual cycle in Drosophila. Curr Biol. 2010;20:93–102. doi: 10.1016/j.cub.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seki T, Vogt K. Evolutionary Aspects of the Diversity of Visual Pigment Chromophores in the Class Insecta. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 1998;119:53–64. [Google Scholar]
- 33.Ozaki K, Nagatani H, Ozaki M, Tokunaga F. Maturation of major Drosophila rhodopsin, ninaE, requires chromophore 3-hydroxyretinal. Neuron. 1993;10:1113–1119. doi: 10.1016/0896-6273(93)90059-z. [DOI] [PubMed] [Google Scholar]
- 34.Lobo GP, Amengual J, Palczewski G, Babino D, von Lintig J. Mammalian carotenoid-oxygenases: key players for carotenoid function and homeostasis. Biochim Biophys Acta. 2012;1821:78–87. doi: 10.1016/j.bbalip.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enright JM, Toomey MB, Sato SY, Temple SE, Allen JR, Fujiwara R, Kramlinger VM, Nagy LD, Johnson KM, Xiao Y, How MJ, Johnson SL, Roberts NW, Kefalov VJ, Guengerich FP, Corbo JC. Cyp27c1 Red-Shifts the Spectral Sensitivity of Photoreceptors by Converting Vitamin A into A. Curr Biol. 2015 doi: 10.1016/j.cub.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiser PD, Farquhar ER, Shi W, Sui X, Chance MR, Palczewski K. Structure of RPE65 isomerase in a lipidic matrix reveals roles for phospholipids and iron in catalysis. Proc Natl Acad Sci U S A. 2012;109:E2747–E2756. doi: 10.1073/pnas.1212025109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borowski T, Blomberg MR, Siegbahn PE. Reaction mechanism of apocarotenoid oxygenase (ACO): a DFT study. Chemistry. 2008;14:2264–2276. doi: 10.1002/chem.200701344. [DOI] [PubMed] [Google Scholar]
- 38.Costas M, Mehn MP, Jensen MP, Que L., Jr Dioxygen activation at mononuclear nonheme iron active sites: enzymes, models, and intermediates. Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 39.Palozza P, Krinsky NI. beta-Carotene and alpha-tocopherol are synergistic antioxidants. Arch Biochem Biophys. 1992;297:184–187. doi: 10.1016/0003-9861(92)90658-j. [DOI] [PubMed] [Google Scholar]
- 40.Spence EL, Langley GJ, Bugg TDH. Cis−Trans Isomerization of a Cyclopropyl Radical Trap Catalyzed by Extradiol Catechol Dioxygenases: Evidence for a Semiquinone Intermediate. JACS. 1996;118:8336–8343. [Google Scholar]
- 41.Matsuno T, Katsuyama M, Maoka T, Hirono T, Komori T. Reductive metabolic pathways of carotenoids in fish (3S,3′S)-astaxanthin to tunaxanthin a, b and c. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 1985;80:779–789. [Google Scholar]
- 42.Pfander H, Lachenmeier A, Hadorn M. Citrus-Carotinoide. 2. Mitteilung. Synthese von (3R)-β-Citraurin, (3R)-β-Citraurol und (3R)-β-Citraurinin; Aufklärung der Konfiguration von Citrus-Carotinoiden. Helv. Chim. Acta. 1980;63:1377–1382. [Google Scholar]
- 43.Inhoffen HH, Bohlmann F, Rummert G. Synthesen in der Carotinoid-Reihe, X. 7,7′-Bis-desmethyl-β-carotin. Justus Liebigs Ann. Chem. 1950;569:226–237. [Google Scholar]
- 44.Lindqvist A, Andersson S. Biochemical properties of purified recombinant human beta-carotene 15,15’-monooxygenase. J Biol Chem. 2002;277:23942–23948. doi: 10.1074/jbc.M202756200. [DOI] [PubMed] [Google Scholar]